- 1Centre National de Recherche Scientifique, Institut de Génomique Fonctionnelle de Lyon, Lyon, France
- 2Institut de Génomique Fonctionnelle de Lyon, Ecole Normale Supérieure de Lyon, Lyon, France
- 3Department of Genetics and Evolution, Faculty of Science, Institute of Genetics and Genomics in Geneva, University of Geneva, Geneva, Switzerland
Hox genes are major regulators of embryonic development. One of their most conserved functions is to coordinate the formation of specific body structures along the anterior-posterior (AP) axis in Bilateria. This architectural role was at the basis of several morphological innovations across bilaterian evolution. In this review, we traced the origin of the Hox patterning system by considering the partnership with PBC and Meis proteins. PBC and Meis belong to the TALE-class of homeodomain-containing transcription factors and act as generic cofactors of Hox proteins for AP axis patterning in Bilateria. Recent data indicate that Hox proteins acquired the ability to interact with their TALE partners in the last common ancestor of Bilateria and Cnidaria. These interactions relied initially on a short peptide motif called hexapeptide (HX), which is present in Hox and non-Hox protein families. Remarkably, Hox proteins can also recruit the TALE cofactors by using specific PBC Interaction Motifs (SPIMs). We describe how a functional Hox/TALE patterning system emerged in eumetazoans through the acquisition of SPIMs. We anticipate that interaction flexibility could be found in other patterning systems, being at the heart of the astonishing morphological diversity observed in the animal kingdom.
Introduction
The phenotypic diversity observed in the animal kingdom arose from genetic innovations that modulate developmental processes, a step in evolution that often precedes speciation events (Gould, 1992; Arthur, 2002). A major challenge in biology is to characterize these genetic innovations and to understand how they impact developmental processes. Remarkably, the specification of body plans and body parts in species as different as humans or flies is controlled by a relatively small and highly conserved genetic repertoire called the “genetic toolkit” (True and Carroll, 2002; Erwin, 2009). This genetic toolkit, which acts at restricted stages of embryonic development, encodes for molecules involved in cell-cell communication, and gene regulation (Mann and Carroll, 2002). Components of the genetic toolkit are described in several bilaterian species to form character identification networks (Wagner, 2007), or kernels (Davidson and Erwin, 2006), which are part of large developmental networks that underlie body plan development (Davidson and Erwin, 2006). Several members of the genetic toolkit are also expressed in choanoflagellates, indicating that they originated prior to the emergence of the first metazoans (King et al., 2003; King, 2004; Wenger and Galliot, 2013).
The large majority of contemporary animals belong to Bilateria, which are characterized by three embryonic germ layers (ectoderm, mesoderm, endoderm) and a bilateral symmetry that results from the orthogonal intersection of two longitudinal axes, the anterior-posterior (AP) axis (also referred to as the primary axis), and the dorso-ventral (DV) axis (also referred to as the secondary axis). Bilaterians radiated during the Cambrian period some 500–550 million years ago. Other extant non-bilaterian species belong to Porifera (sponges), Ctenophora, Placozoa (Trichoplax), and Cnidaria, whose ancestors predate the Cambrian explosion, thus often named early-branched phyla (Figure 1). With the exception of Placozoa, species from these early-branched phyla display different types of symmetry, either radial (as seen in sponge larvae, some adult sponges, and in most cnidarians), or biradial (as seen in ctenophores), or partly bilateral (as seen in sea anemone species that belong to the anthozoan class of cnidarians). These various symmetries are especially evident during embryogenesis and larval stages and depend on the formation of a primary body axis (Ryan and Baxevanis, 2007).
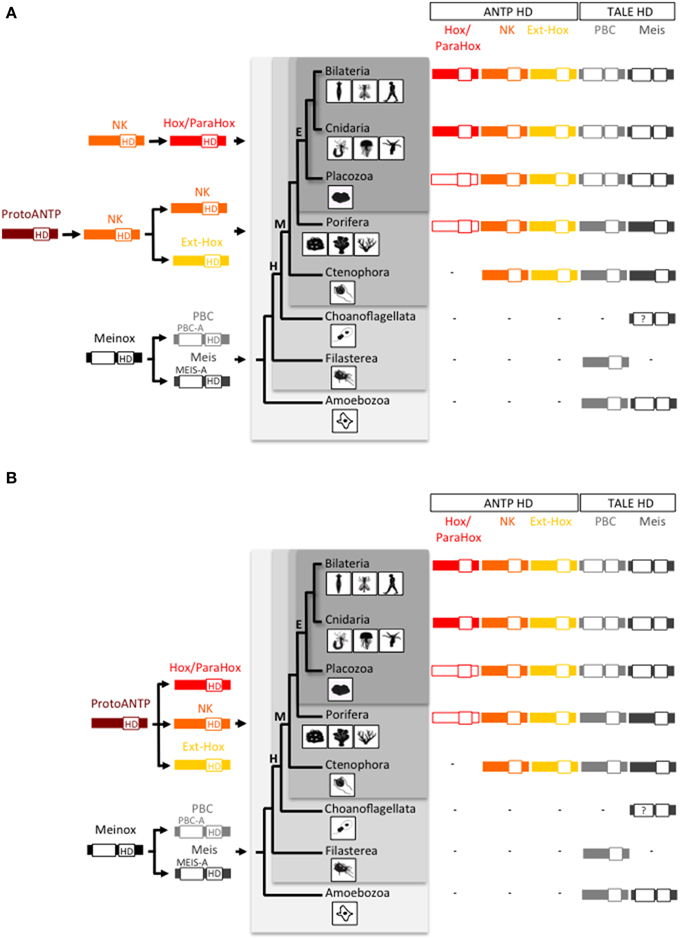
Figure 1. Origin and early evolution of ANTP- and TALE-class gene families. (A) First evolutionary scenario whereby the Hox/ParaHox family would have derived from a NK member in the Eumetazoan ancestor. (B) Second evolutionary scenario, also named the “ghost loci hypothesis,” whereby the main homeobox gene families (Hox/ParaHox, NK, and Ext-Hox) would have derived from a ProtoANTP cluster of homeobox genes already present in the Last Common Ancestor (LCA) of metazoans. The recent finding of a ParaHox-like gene in Porifera (Fortunato et al., 2014) actually supports the second scenario. Note that only one ParaHox member (symbolized by the absence of red filling) is found in Placozoa [annotated as a Gsx-like: (Schierwater et al., 2008a)] and Porifera [annotated as a Cdx-like: (Fortunato et al., 2014)] and that no Hox or ParaHox gene has been annotated in Ctenophora so far. In comparison, the PBC and Meis families originated earlier in the life tree, with representatives already present in unicellular phyla (Amoebozoa and Filasterea). Graded gray backgrounds highlight Eumetazoa (E), Metazoa (M), Holozoa (H), and Unikonta (U) super phyla. The homeodomain (HD) is indicated in each protein. PBC-A and Meis-A are domains required for the PBC/Meis partnership. Question mark in Choanoflagellata is for incomplete protein sequence of Meis. Animal drawings were taken from Ryan and Baxevanis (2007).
Cnidaria, a sister group to Bilateria, share with them typical features of eumetazoans, i.e., an ectodermal layer that differentiates as an epidermis, an endodermal layer that differentiates as a gut, and a nervous system, which, at the oral pole/extremity, allows an active feeding behavior. Also, Cnidaria includes a large variety of taxa with a wide spectrum of morphological diversity. All together, these characteristics place Cnidaria at a key phylogenetic position for tracing the emergence of molecular innovations that underlie developmental changes and diversification in animal evolution (Steele et al., 2011). Representative(s) of the main gene families involved in the specification of eumetazoan features are also found in Cnidaria (Martindale, 2005). Their study is however more challenging, due to the lack of advanced genetic tools that could allow establishing transgenic animals for stable gene expression or extinction in a tissue- and/or stage-specific manner.
Among the different conserved developmental gene families are the Hox genes, which are considered as the “Rosetta Stone” of the genetic toolkit. Hox genes were initially discovered in Drosophila, then rapidly investigated in vertebrate species, showing striking conserved features throughout bilaterian lineages (Lewis, 1978; McGinnis and Krumlauf, 1992; Kmita and Duboule, 2003). These conserved properties have been discussed in several reviews and relate to their clustered genomic organization that constrains embryonic expression (Duboule, 2007), but also to the presence of several typical protein signatures (Ogishima and Tanaka, 2007; Merabet et al., 2009). Modifications in Hox gene expression or in Hox protein function have been linked to several morphological innovations during the evolution of bilaterians (Pearson et al., 2005; Heffer et al., 2013). The presence of Hox genes in Cnidaria therefore raised the question of their role in the emergence of innovations shared by cnidarians and bilaterians, as well as in the emergence of innovations responsible for the morphological diversity observed among cnidarian species.
The most spectacular observation came from the embryo of the cnidarian sea anemone Nematostella vectensis, where several Hox-related genes show a staggered-like expression pattern along the oral-aboral (OA) axis. This expression profile led to the proposition that the cnidarian OA axis could be homologous to the bilaterian AP axis (Finnerty et al., 2004; Matus et al., 2006). The OA expression profile of Nematostella Hox genes is however neither conserved in other cnidarian lineages nor strictly following the collinear rules normally observed in Bilateria (Gauchat et al., 2000; Finnerty et al., 2004; Kamm et al., 2006; Ryan et al., 2007; Chiori et al., 2009). These additional observations led to the opposite conclusion that a Hox patterning system is likely not existing in Cnidaria (Kamm et al., 2006).
Surprisingly, the question of the evolution of Hox patterning mechanisms is rarely approached at the protein level, in particular by considering members of the PBC and Meis families. PBC and Meis are crucial patterning cofactors of Hox proteins along the AP axis, a partnership that is evolutionarily-conserved throughout Bilateria (Moens and Selleri, 2006; Mann et al., 2009). PBC and Meis belong to the TALE (Three Amino acids Loop Extension) class of homeodomain (HD)-containing transcription factors (Bürglin, 1997), are widely conserved across metazoans and can therefore be used as a molecular hallmark of the Hox patterning system. In this review, we report how the intricate interaction properties between Hox and TALE proteins were progressively acquired in pre-bilaterian animal evolution to eventually constitute a major patterning system.
Origin and Early Evolution of the Hox/ParaHox and PBC/Meis Gene Families
Hox proteins belong to the ANTP (Antennapedia) class of HD-containing transcription factors. This class contains two large groups of sister gene families: (i) the non-Hox ANTP-class group, which includes the NK and Extended (Ext)-Hox gene families, and (ii) the Hox/ParaHox genes (Garcia-Fernàndez, 2005). The Hox gene family is usually found organized in clusters and contains several paralog groups (PGs) that are themselves classified into anterior (PG1-3), central (PG4-8), and posterior (PG9-14) (Duboule, 2007). The ParaHox family contains three clustered genes initially discovered in the cephalochordate amphioxus (Brooke et al., 1998), and named Gsx, Pdx/Xlox, and Cdx. ParaHox genes share common ancestors with specific Hox gene families, Gsx, and Pdx/Xlox with the anterior PG2/PG3, Cdx with the posterior PG9 (Quiquand et al., 2009).
Two different scenarios are proposed to explain the evolutionary history of the Hox/ParaHox gene family with regard to the other ANTP-class members. In the first one, the Hox/ParaHox family is specific to eumetazoans (which regroup Bilateria, Cnidaria, and Placozoa) and would have originated from duplications of a ProtoHox gene derived from NK genes and related to Evx/Mox (Ext-Hox family) (Gauchat et al., 2000; Minguillón and Garcia-Fernàndez, 2003; Larroux et al., 2007; Quiquand et al., 2009; Ryan et al., 2010) (Figure 1A). This scenario is supported by the presence of a Gsx ParaHox gene in the genome of the placozoan Trichoplax adhaerens (Schierwater and Kuhn, 1998; Schierwater et al., 2008b), the presence of several NK representatives and the absence of Hox/ParaHox genes in the genome of the ctenophores Mnemiopsis leidyi (Ryan et al., 2010) and Pleurobrachia bachei (Moroz et al., 2014) and the demosponge Amphimedon queenslandica (Srivastava et al., 2010). However, this scenario is challenged by the “ghost loci hypothesis”, which postulates that Hox/ParaHox genes were already present in the Last Common Ancestor (LCA) of metazoans and secondarily lost in Porifera over evolutionary times (Mendivil Ramos et al., 2012). In this second scenario, the Hox/ParaHox and NK families emerged independently from a common ProtoANTP ancestor gene (Figure 1B). The recent finding of a Cdx-like ParaHox gene in the genome of two calcareous sponges (Fortunato et al., 2014), now argues in favor of the ghost loci hypothesis.
In addition to ANTP, other classes of HD-containing transcription factors are also present in early branch phyla. These include the Paired-like, Pax, Pou, Lim, Six, and TALE classes (Galliot and de Vargas, 1999; Larroux et al., 2008; Srivastava et al., 2010; Holland, 2013; Fortunato et al., 2014). Members of the TALE class contain an atypical 63-residues long HD, due to the presence of three extra residues in between the helices 1 and 2 of the HD (Mukherjee and Bürglin, 2007). TALE class members are among the most ancient transcription factors in eukaryotes, with several of them present in unicellular organisms, plants and fungi (Bürglin, 1997, 1998), therefore predating the origin of animals. Interestingly, TALE-class members can interact with different types of HD-containing proteins in plants (Bellaoui et al., 2001; Hackbusch et al., 2005; Kanrar et al., 2006; Hay and Tsiantis, 2010), fungi (Keleher et al., 1989; Stark and Johnson, 1994; Carr et al., 2004), and animals (Bürglin, 1998).
The TALE class comprises five families (PBC, Meis, Iro, TGIF, and MKX), among which two, PBC and Meis are known to interact with ANTP members (Mukherjee and Bürglin, 2007). PBC and Meis were already present before multicellular organisms appeared (King et al., 2008; Clarke et al., 2013; Suga et al., 2013) and (Figure 1). Animal representatives of PBC and Meis include the Pbx1-4 or Extradenticle (Exd) and Meis1-3 or Homothorax (Hth) proteins, as named in mammals and in the fruit fly Drosophila melanogaster, respectively. PBC and Meis families originated from the duplication of a common ancestor gene named MEINOX, and this duplication was proposed to coincide with the apparition of the first Hox cluster in metazoans (Bürglin, 1998). Genome comparisons between early-branched metazoan species and unicellular organisms now establish that the PBC/Meis duplication predated the ANTP class and therefore the Hox/ParaHox family. Thomas Bürglin was however the first one to consider the partnership between Hox and TALE proteins as an informative molecular hallmark to trace the origin of the Hox patterning system in Metazoa (Bürglin, 1998).
The Ground State of Hox/TALE Interaction Networks in Bilateria: Role of the Hexapeptide (HX) Motif
The formation of Hox/PBC/Meis complexes in Bilateria is described to rely on Hox-PBC and PBC-Meis interactions (Figure 2A). Interaction between PBC and Meis involves the N-terminal PBC-A and Meis-A domains, respectively (Mann and Affolter, 1998). In the absence of Meis, the PBC-A domain is masking two nuclear localization signals located in the HD of PBC. The interaction with Meis relieves the masking activity of the PBC-A domain, allowing the nuclear translocation of PBC (Saleh et al., 2000; Stevens and Mann, 2007).
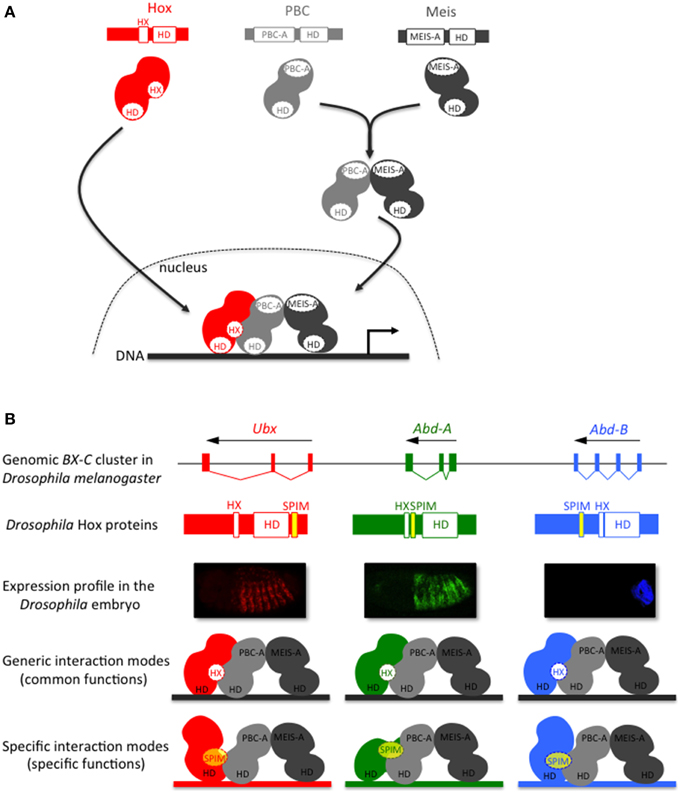
Figure 2. The Hox-TALE interaction network: role of generic (HX motif) and specific PBC interaction motifs (SPIMs). (A) Generic association mode between Hox and TALE proteins. The interaction between Meis and PBC allows the nuclear translocation of PBC. The hexapeptide (HX) motif, present in Hox proteins of all bilaterian lineages, is necessary and sufficient for the generic association mode of the Hox/TALE complex on DNA. (B) Model for the role of SPIMs in specifying patterning functions among Drosophila Hox proteins. The usage of SPIMs allows each Hox protein of the Bithorax complex (BX-C) to adopt different conformation modes with the TALE cofactors and regulate different target genes in vivo (as illustrated by the color code). The placement of SPIMs (highlighted in yellow) in Ultrabithorax (Ubx) and Abdominal-A (Abd-A) reflects the position of the UbdA and TDWM motifs, respectively (Hudry et al., 2012). The placement of the SPIM in Abdominal-B (Abd-B) is speculative.
Interactions between Hox and PBC have been extensively studied at the biochemical and structural levels. All these analyses converge to show a preponderant role for a short conserved motif present in Hox proteins, named hexapeptide (HX) (Mann et al., 2009). The HX motif lies upstream to the HD and contains a core Y/FPWM sequence in all but Abdominal B-group Hox proteins, which have a more divergent sequence (Merabet et al., 2009). More generally, the HX motif is defined as a PBC interaction motif (PIM) that contains an invariant Tryptophan residue located in a hydrophobic environment, followed by basic residues from +2 to +5 (In der Rieden et al., 2003). Crystal structures of vertebrate and invertebrate Hox/PBC complexes solved with anterior, central or posterior Hox proteins point to the critical role of the Tryptophan residue in maintaining strong interactions within the hydrophobic pocket formed in part by the three extra residues of the PBC HD (Passner et al., 1999; Piper et al., 1999; LaRonde-LeBlanc and Wolberger, 2003; Joshi et al., 2007). A recent structural analysis of the Hox/PBC complex bound on a physiological DNA-binding site further underlined that Hox paralog specific residues located in the N-terminal arm of the HD and in the linker region connecting the HX motif to the HD are important for recognizing a specific shape of the DNA minor groove in the presence of PBC (Joshi et al., 2007). SELEX-seq based approaches confirmed that Drosophila Hox/PBC complexes preferentially recognize different nucleotide sequences characterized by distinct minor groove topographies (Slattery et al., 2011). These results open new avenues for apprehending the molecular mechanisms underlying Hox and Hox/PBC DNA-binding specificity (Abe et al., 2015). Nevertheless, the systematic involvement of a unique Hox protein motif in the interaction with PBC does not easily explain the broad variety of functions that Hox/TALE complexes have in vivo (Hueber and Lohmann, 2008; Mann et al., 2009).
Specific PBC Interactions Motifs (SPIMs) as Versatile Complements to Diversify Hox/TALE Interaction Properties in Bilateria and Cnidaria
Our knowledge of Hox-TALE interaction properties results mostly from in vitro approaches. Along the same line, the duplication of Pbx and Meis genes in vertebrates could provide a supplementary layer of complexity. For example, direct Hox-Meis interactions are described with mouse proteins but their functional significance remains to be elucidated (Shen et al., 1997; Williams et al., 2005). The existence of alternative modes in Hox-PBC interaction came from the observation that the HX mutation does not obligatorily affect PBC-dependent functions of Hox proteins in the Drosophila embryo (Galant et al., 2002; Merabet et al., 2003). Additionally, several central and posterior Hox proteins from vertebrates and invertebrates interact with the TALE cofactors independently of the HX motif in vitro and in vivo (Hudry et al., 2012). Interestingly, HX-independent interactions between Hox and PBC are most often observed in the presence of Meis, and the involvement of Meis in such HX-independent interactions actually depends on its DNA-binding near the Hox/PBC binding site (Hudry et al., 2012). In other words, in acting at the level of target cis-regulatory sequences, Meis contributes to diversify the mode of Hox-PBC interactions and thus Hox functions (Merabet and Hudry, 2013).
The flexibility of Hox-TALE interaction properties is predicted to rely on Hox protein motifs that are more gene-specific than the generic HX motif. These motifs are named SPIMs [Specific PBC Interaction Motifs, see also Merabet and Hudry, 2013]. Like the HX motif, SPIMs belong to the so-called short linear motifs, which are classically 5–10 residues long and most often located within intrinsically disordered protein regions (Tompa et al., 2014). Two such motifs have been identified in the Drosophila Ultrabithorax (Ubx) and AbdominalA (AbdA) proteins (Merabet et al., 2007, 2011; Hudry et al., 2012). One of them is conserved in insect AbdA proteins, with a core TDWM sequence reminiscent of the HX motif. The other motif, named UbdA, is conserved between the protostome Ubx and AbdA proteins (Balavoine et al., 2002). Recent structural analyses showed that the UbdA motif constitutes a flexible extension of the HD that can establish direct contacts with the PBC partner (Foos et al., 2015). Altogether, studies with Ubx and AbdA confirm that Hox-TALE interactions and functions can rely on species- and/or paralog-specific motifs.
SPIMs remain to be identified in the majority of Hox proteins exerting HX-independent interactions with the TALE cofactors. Still, the usage of different SPIMs in Hox proteins constitutes an appealing molecular strategy for supporting the specific patterning functions of Hox/TALE complexes during development (Figure 2B). Moreover, the conservation of this property in vertebrate and invertebrate species (Hudry et al., 2012) strongly suggests that interaction flexibility is ancient in Bilateria. As a consequence, it is of upmost interest to trace its origin beyond Bilateria and assess its role in developmental and/or patterning functions.
Besides Bilateria, Cnidaria is the only other phylum that contains a bona fide Hox repertoire (Chourrout et al., 2006; Kamm et al., 2006). As mentioned previously, the role of cnidarian Hox genes in axis patterning is unclear. Furthermore, not all cnidarian Hox proteins contain an intact HX motif [(Hudry et al., 2014) and Table 1]. Nevertheless, as cnidarians express PBC and Meis genes (Matus et al., 2006; Hudry et al., 2014), a Hox/PBC/Meis network could potentially exist. The interaction properties of Hox, PBC and Meis proteins of the sea anemone Nematostella vectensis were recently tested, and as expected, these proteins form dimeric and trimeric complexes in vitro (Hudry et al., 2014). In addition, mutating the HX motif leads to the loss of the cnidarian Hox/PBC complex, but this loss is rescued in the presence of Meis. Hence, as observed in bilaterians, the Nematostella Meis allows Nematostella Hox proteins to use alternative modes of interaction with PBC. Thus, bilaterian and cnidarian Hox proteins share the property of using different interfaces for recruiting the TALE cofactors. We propose that these additional interfaces could correspond to SPIMs that remain to be identified in several instances (Figure 3). Moreover, with the exception of the HX motif, bilaterian and cnidarian Hox proteins do not share strong sequence similarities outside the HD, suggesting that those putative SPIMs could have evolved independently during eumetazoan evolution (see also below).
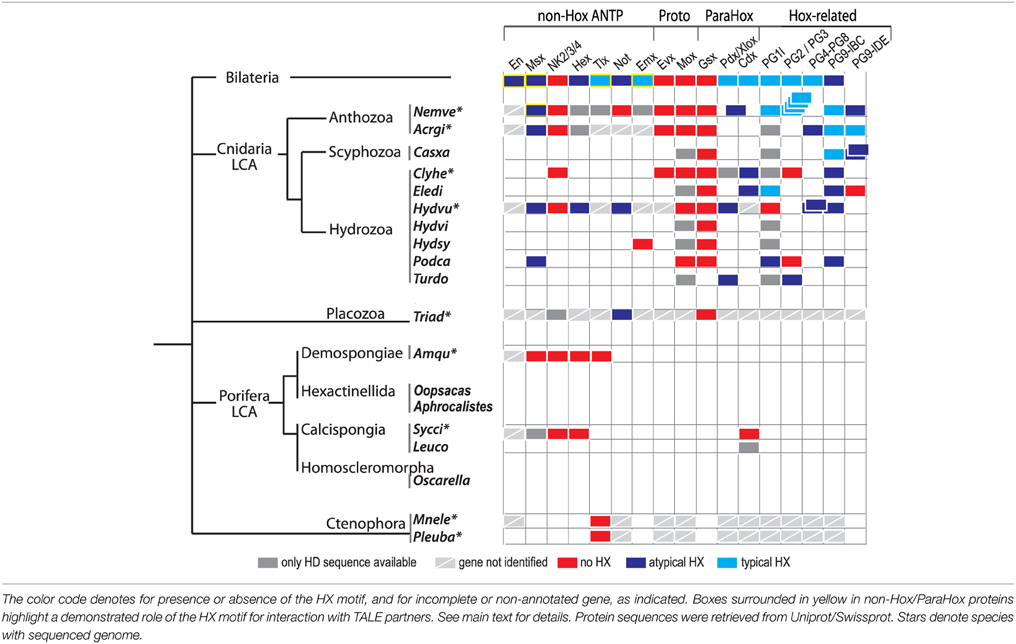
Table 1. Presence or absence of the HX motif among the Hox/ParaHox and non-Hox/ParaHox families across Metazoa.
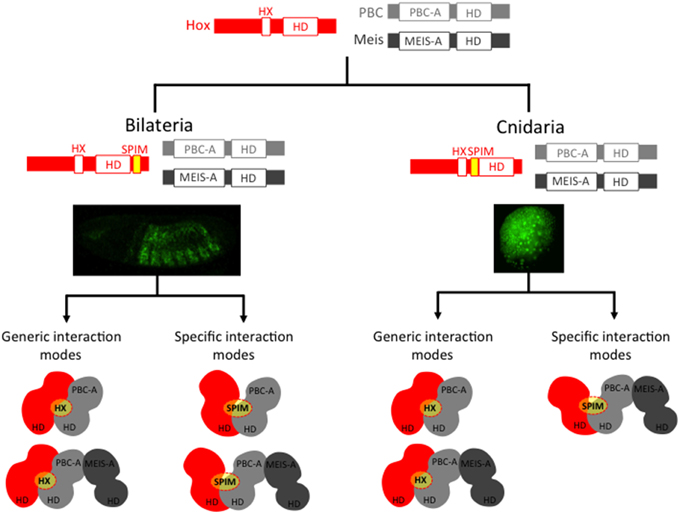
Figure 3. Cnidarian and bilaterian Hox/TALE networks display similar interaction properties. Ancestral Hox/TALE networks were strictly relying on the HX motif. The apparition of SPIMs in bilaterian and cnidarian lineages allowed Hox proteins to diversify their interaction modes with the TALE partners. Pictures depict in vivo interaction between Hox and PBC proteins in a live Drosophila (right) or Nematostella embryo, as described in Hudry et al. (2011, 2014). Compared to Drosophila, the usage of SPIMs in Nematostella Hox proteins is strictly dependent on the presence of Meis (Hudry et al., 2014). The absence of identical SPIMs between bilaterian and cnidarian Hox proteins suggests that these motifs emerged independently in these two groups (see also Figure 4).
Genesis of ANTP-TALE Networks during Early Metazoan Evolution
Molecular analyses underline that the HX motif is a generic interaction platform for recruiting the TALE partners. We therefore analyzed a large number of available protein sequences for assessing the presence of a putative HX motif in ANTP class members. A peptide sequence was considered as a putative HX motif when containing the consensus Y/FPWM (typical HX motif) or a single W (atypical/divergent HX motif) residue followed by a basic residue (R or K) from +2 to +6 and not localized more than 30 residues away from the HD (Table 1). In Bilateria, the HX motif is found in almost all Hox/ParaHox members, and in several individual representatives of non-Hox/ParaHox protein families, including Engrailed (En), Msx, Hex, Tlx, Not, and Emx proteins (Table 1). The HX motif is found in cnidarian Hox/ParaHox members among early-branched animal phyla. It is however less conserved when compared to Bilateria, being lost or divergent in several cnidarian lineages (Table 1). Atypical HX motifs are also found in Msx and Hex members of Cnidaria, and in Not members of Cnidaria and Placozoa (Table 1). Interestingly, the Evx, Mox, and Gsx proteins, which likely represent the most ancestral ProtoHox and Hox/ParaHox family members (Minguillón and Garcia-Fernàndez, 2003; Quiquand et al., 2009) all lack the HX motif (Table 1).
PBC-recruiting functions have been assigned to few non-Hox proteins among the ANTP class so far. Among them are the mammalian Tlx, Drosophila En and Nematostella Msx proteins, which do interact in a fully HX-dependent manner with the TALE cofactors (Rhee et al., 2004; Brendolan et al., 2005; Fujioka et al., 2012; Hudry et al., 2014). Still, these proteins display subtle differences in their TALE interaction properties. For example, the Drosophila En protein interacts with PBC or PBC/Meis in a HX-dependent manner (Hudry et al., 2014). By comparison, the Msx protein from Nematostella interacts in a HX-dependent manner with PBC, but only in the presence of Meis (Hudry et al., 2014). These observations highlight that the role of the PBC/Meis partnership in HX-dependent interactions can be different depending on the protein family and animal lineage considered.
We propose two different evolutionary scenarios to explain the presence of the HX motif in several ANTP family members among metazoan lineages: (i) either the HX motif was already present in the ProtoANTP ancestor, constituting the first molecular interface for recruiting the TALE cofactors (Figure 4A), or (ii) it emerged multiple times independently in the different ANTP families across animal evolution (Figure 4B). The position of the HX motif systematically located in the upstream vicinity of the HD supports the first scenario. As a corollary, the absence of any HX-like motif in all but one (Not) ANTP members of Placozoa, Porifera and Ctenophora would be attributed to repeated secondary losses. Although more sequences are needed in these three early-branched animal phyla, this apparently global and systematic loss of HX motif sequences is intriguing. This could argue in favor of the second scenario, whereby the HX motif would have appeared sporadically by convergent evolution in the different protein families. This second scenario does not exclude additional secondary losses, as observed in cnidarian Hox/ParaHox proteins (Table 1). Moreover, evolution by convergence is not atypical for short motifs in general (Van Roey et al., 2013), and has for example already been proposed for another motif widely found in ANTP-class members (including Gsx, En, Emx, and several NK) and other non-homeoproteins (Williams and Holland, 2000). In the case of the HX motif, it seemingly appeared later during evolution in bilaterian Tlx, Emx and En proteins (Table 1), suggesting a mechanism of convergent evolution. Of note, these bilaterian proteins are known to interact and/or participate with TALE cofactors in the context of tissue-specific functions (Brendolan et al., 2005; Capellini et al., 2010). Along the same line, an HX motif is also present in non-ANTP class proteins, including LIM and several myogenic bHLH proteins (see In der Rieden et al., 2003, for a more complete list of HX-containing proteins). In the case of bHLH proteins the HX motif was further shown to be involved in the interaction and function with TALE cofactors during skeletal muscle differentiation in vertebrates (Knoepfler et al., 1999; Maves et al., 2007, 2009; Yao et al., 2013). Together these observations highlight the strong evolutionary plasticity of the HX motif for providing a TALE-recruiting activity to highly divergent protein families.
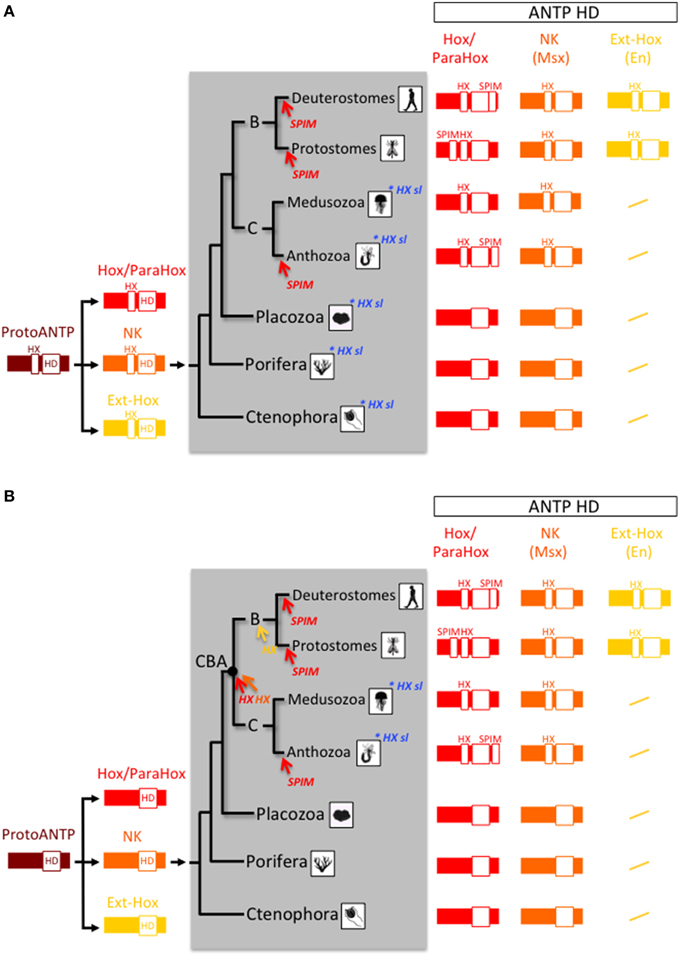
Figure 4. Two evolutionary scenarios for the origin and early evolution of Hox-TALE interaction properties. (A) In the first scenario, the HX motif arose in the ProtoANTP gene of the metazoan LCA. There were multiple secondary losses in the Hox/ParaHox (sl; highlighted in blue) and other families (not indicated) in ctenophores, porifers, placozoans and cnidarians. (B) In the second scenario, the HX motif appeared independently several times during evolution, acquired in the Hox/ParaHox (red arrow), and NK (Msx, orange arrow) families of the last common ancestor of Cnidaria and Bilateria (CBA), or in Ext-Hox members (as exemplified with En, yellow arrow) of the bilaterian ancestor. The absence of the HX motif in Hox/ParaHox members of several cnidarian species indicates secondary lost events (highlighted in blue; see also Table 1). In both scenarios, the HX motif served as a molecular template for diversifying TALE interaction properties only in the Hox/ParaHox family. This was achieved by the emergence of SPIMs. These motifs were independently acquired (highlighted in red) in Bilateria and Cnidaria, coinciding with strong morphological radiation in these two phyla.
Genesis of the Hox-TALE Patterning System during Metazoan Evolution
The evolutionarily conserved PBC-A and Meis-A domains in PBC and Meis proteins are restricted to Bilateria, Cnidaria and Placozoa, suggesting that a Hox/TALE network exists only in these three phyla (Figure 1). Like all cnidarian and bilaterian Gsx proteins, the unique ParaHox Gsx representative of Trichoplax adhaerens has no HX motif (Table 1) and cannot interact with PBC and Meis (Hudry et al., 2014). By contrast, the two other ParaHox and the Hox-related proteins have retained an HX motif in most cnidarians and bilaterians (Table 1). Thus, Cnidaria and Bilateria are the only phyla where a Hox-TALE interaction network is effective.
Since interaction with TALE proteins is not a specific feature of Hox proteins, the next question is “When did Hox proteins acquire their patterning functions linked to the interaction with the TALE cofactors?” We postulate here that the acquisition of differential patterning functions was tightly linked to the emergence of diversified interaction properties between Hox and TALE proteins. Then the question could be reformulated as: “When did alternative TALE interaction motifs appear in addition to the HX motif in the Hox/ParaHox family?”
Recent work with Nematostella Hox and TALE proteins (Hudry et al., 2014) suggests that SPIMs co-evolved with the specification of embryonic axes. As SPIMs are specific to a given Hox family or to a given species, they likely emerged independently several times during evolution (Figure 4). We propose that the original HX-dependent interaction mode served as an initial molecular template for experiencing these novel HX-independent interaction properties with the TALE partners. It is tempting to speculate that SPIMs were a molecular prerequisite for allowing Hox proteins to acquire patterning functions during early eumetazoan evolution. In this model, the acquisition of SPIMs in Hox proteins likely happened in parallel to mechanisms regulating their expression, allocating Hox genes to specific spatio-temporal domains along the longitudinal axis (Figure 5).
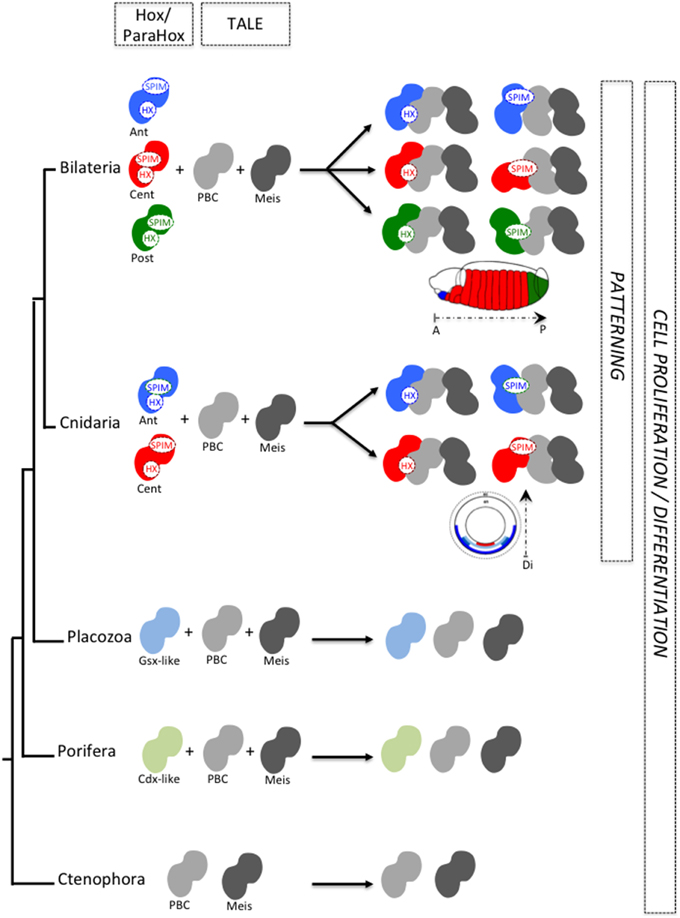
Figure 5. SPIMs as molecular markers of a Hox/TALE patterning system during animal evolution. The acquisition of SPIMs in Cnidaria (Nematostella) and Bilateria (Drosophila, mammals) allowed Hox proteins to diversify their interaction modes with the TALE partners. This molecular diversification was essential for providing differential activities to Hox proteins along the longitudinal axis. Illustrative examples are provided along the anterior-posterior axis of the Drosophila embryo or along the directive axis of the Nematostella embryo. Anterior (ant), central (cent), and posterior (post) Hox proteins are depicted by a different color. In Placozoa and Porifera, ParaHox-like members are present but these proteins do not contain any HX motif. Along the same line, placozoan and poriferan PBC and Meis representatives lack the PBC-A and MEIS-A domains (see Figure 1) and thus cannot interact together. As a consequence, TALE interaction networks do likely not exist in those two phyla. We postulate that Hox/ParaHox transcription factors were initially dedicated to cell proliferation/cell differentiation with no patterning function, whereas Hox/TALE interactions co-evolved with patterning functions.
Finally, SPIMs do not necessarily correspond to related peptide sequences, as already noticed for the TDWM and UbdA motifs in Drosophila (Merabet and Hudry, 2013), making their identification difficult. Additional SPIMs need however to be identified to validate our model. Several tools are now available for predicting the presence of short interaction motifs in protein sequences, based on the analysis of amino acid chemical properties and the classification of hundreds of characterized short motifs in databases (Tompa et al., 2014). Interestingly, these tools predict a number of short motifs in several regions of bilaterian (Merabet and Dard, 2014) and cnidarian (Baëza et al., 2015) Hox proteins. These regions are often involved in the interaction with different TFs (Baëza et al., 2015), and could therefore contain good candidate SPIMs to test in the future.
Perspective: the HX Motif and SPIMs as Molecular Markers of Patterning Functions in the ParaHox Family?
ParaHox genes share several common features with the Hox genes. For example, they are organized in clusters and display spatial-temporal constraints for their expression during embryogenesis of several bilaterian species (Garstang and Ferrier, 2013). The expression profile of ParaHox genes in Cnidaria is also reminiscent of important functions during embryogenesis, regeneration or budding, as seen in the solitary polyp Hydra (Schummer et al., 1992; Miljkovic-Licina et al., 2007), the coral Acropora (Hayward et al., 2001), the jellyfish Podocoryne (Yanze et al., 2001), the sea anemone Nematostella (Finnerty et al., 2003), or the colonial polyp Hydractinia (Cartwright et al., 2006). Moreover, ParaHox genes, and more particularly Gsx, could be more representative of the ProtoHox ancestor gene than any other Hox gene (Quiquand et al., 2009). Although Gsx does not contain any HX motif, it has a conserved role for the specification of neuroblast lineages in bilaterians (Weiss et al., 1998; Waclaw et al., 2009; Winterbottom et al., 2010; López-Juárez et al., 2013) and cnidarians, with a fine regulation along the body axis (Hayward et al., 2001; Miljkovic-Licina et al., 2007). Along the same line, Pdx-1 plays a crucial role in pancreatic beta-cell differentiation (Kaneto et al., 2007). These observations suggest that a primordial ParaHox (and Hox) function was dedicated to the emergence of novel cell types along the body axis, possibly in a TALE-independent manner (Figure 5) (De Jong et al., 2006; Miljkovic-Licina et al., 2007; Quiquand et al., 2009). This role could then have been deployed in several Hox/ParaHox members and in different tissues, requiring the acquisition of additional molecular features such as the HX motif and SLIMs for diversifying the novel patterning functions. In agreement with this hypothesis, in Bilateria Pdx/Xlox and Cdx transcription factors are required for the patterning of endodermal derivatives (Cole et al., 2009; Beck and Stringer, 2010; Annunziata et al., 2013; Ikuta et al., 2013) or during axis elongation with the Hox genes (Moreno and Morata, 1999; Van den Akker et al., 2002; Shinmyo et al., 2005; Young et al., 2009). The impact of TALE cofactors in those patterning functions remains to be investigated. The role of Pdx/Xlox and Cdx is also largely unknown in cnidarians. Testing their interaction properties with TALE cofactors could undoubtedly provide new insightful information into the origin and evolvability of the Hox/TALE patterning system in Metazoa. Ultimately, such studies should tell us whether the combination of one HX motif plus several SPIMs in the ParaHox proteins was necessary and sufficient to promote a spatial organization of cell differentiation along the body axis and thus the emergence of patterning functions in different tissues.
Conclusion
Hox proteins are TFs displaying highly similar DNA binding properties in vitro. Still, each Hox protein will dictate a specific developmental program with the same set of TALE cofactors. We proposed here that the apparition of a functional Hox/TALE patterning system during metazoan evolution was tightly linked to the acquisition of different short motifs named SPIMs. The usage of different SPIMs in Hox proteins constitutes an appealing molecular strategy for explaining the specific and various developmental functions of Hox/TALE complexes. Due to their small size, SPIMs present the advantage of being highly dynamic during evolution, allowing diversifying the molecular code between Hox and TALE proteins. This model supposes that interaction flexibility is an important feature of the Hox/TALE patterning system. Whether this molecular strategy could more widely apply to other key patterning networks constitutes a major issue to investigate in the future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We warmly thank Daniel Chourrout and Maja Adamska for helpful comments and criticisms on early versions of this review. Work in the laboratory of SM is supported by Association Française contre les Myopathies (AFM), Fondation pour la recherché Médicale (FRM), Association pour la Recherche contre le Cancer (ARC), Ligue Régionale contre le Cancer, Centre National de la Recherche Scientifique (CNRS), and Ecole Normale Supérieure (ENS) de Lyon. Work in the Galliot lab is supported by the State of Geneva, the Swiss National Fonds for Research (SNF-31003A-149630). We apologize to colleagues whose work was not cited due to space constraints.
Abbreviations
ANTP, Antennapedia; AP, anterior posterior; HD, Homeodomain; HX, Hexapeptide; PG, Paralog Group; SPIM, Specific PBC Interaction Motif; TALE, Three Amino acid Loop Extension; TF, Transcription factor.
References
Abe, N., Dror, I., Yang, L., Slattery, M., Zhou, T., Bussemaker, H. J., et al. (2015). Deconvolving the recognition of DNA shape from sequence. Cell 161, 307–318. doi: 10.1016/j.cell.2015.02.008
Annunziata, R., Martinez, P., and Arnone, M. I. (2013). Intact cluster and chordate-like expression of ParaHox genes in a sea star. BMC Biol. 11:68. doi: 10.1186/1741-7007-11-68
Arthur, W. (2002). The emerging conceptual framework of evolutionary developmental biology. Nature 415, 757–764. doi: 10.1038/415757a
Baëza, M., Viala, S., Heim, M., Dard, A., Hudry, B., Duffraisse, M., et al. (2015). Inhibitory activities of short linear motifs underlie Hox interactome specificity in vivo. Elife 4, 1–28. doi: 10.7554/eLife.06034
Balavoine, G., de Rosa, R., and Adoutte, A. (2002). Hox clusters and bilaterian phylogeny. Mol. Phylogenet. Evol. 24, 366–373. doi: 10.1016/S1055-7903(02)00237-3
Beck, F., and Stringer, E. J. (2010). The role of Cdx genes in the gut and in axial development. Biochem. Soc. Trans. 38, 353–357. doi: 10.1042/BST0380353
Bellaoui, M., Pidkowich, M. S., Samach, A., Kushalappa, K., Kohalmi, S. E., Modrusan, Z., et al. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13, 2455–2470. doi: 10.1105/tpc.13.11.2455
Brendolan, A., Ferretti, E., Salsi, V., Moses, K., Quaggin, S., Blasi, F., et al. (2005). A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development 132, 3113–3126. doi: 10.1242/dev.01884
Brooke, N. M., Garcia-Fernàndez, J., and Holland, P. W. (1998). The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392, 920–922. doi: 10.1038/31933
Bürglin, T. R. (1997). Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25, 4173–4180. doi: 10.1093/nar/25.21.4173
Bürglin, T. R. (1998). The PBC domain contains a MEINOX domain: coevolution of Hox and TALE homeobox genes? Dev. Genes Evol. 208, 113–116. doi: 10.1007/s004270050161
Capellini, T. D., Vaccari, G., Ferretti, E., Fantini, S., He, M., Pellegrini, M., et al. (2010). Scapula development is governed by genetic interactions of Pbx1 with its family members and with Emx2 via their cooperative control of Alx1. Development 137, 2559–2569. doi: 10.1242/dev.048819
Carr, E. A., Mead, J., and Vershon, A. K. (2004). Alpha1-induced DNA bending is required for transcriptional activation by the Mcm1-alpha1 complex. Nucleic Acids Res. 32, 2298–2305. doi: 10.1093/nar/gkh560
Cartwright, P., Schierwater, B., and Buss, L. W. (2006). Expression of a Gsx parahox gene, Cnox-2, in colony ontogeny in Hydractinia (Cnidaria: Hydrozoa). J. Exp. Zool. Part B Mol. Dev. Evol. 306, 460–469. doi: 10.1002/jez.b.21106
Chiori, R., Jager, M., Denker, E., Wincker, P., Da Silva, C., Le Guyader, H., et al. (2009). Are Hox genes ancestrally involved in axial patterning? Evidence from the hydrozoan Clytia hemisphaerica (cnidaria). PLoS ONE 4:4231. doi: 10.1371/journal.pone.0004231
Chourrout, D., Delsuc, F., Chourrout, P., Edvardsen, R. B., Rentzsch, F., Renfer, E., et al. (2006). Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature 442, 684–687. doi: 10.1038/nature04863
Clarke, M., Lohan, A. J., Liu, B., Lagkouvardos, I., Roy, S., Zafar, N., et al. (2013). Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 14:R11. doi: 10.1186/gb-2013-14-2-r11
Cole, A. G., Rizzo, F., Martinez, P., Fernandez-Serra, M., and Arnone, M. I. (2009). Two ParaHox genes, SpLox and SpCdx, interact to partition the posterior endoderm in the formation of a functional gut. Development 136, 541–549. doi: 10.1242/dev.029959
Davidson, E. H., and Erwin, D. H. (2006). Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800. doi: 10.1126/science.1113832
de Jong, D. M., Hislop, N. R., Hayward, D. C., Reece-Hoyes, J. S., Pontynen, P. C., Ball, E. E., et al. (2006). Components of both major axial patterning systems of the Bilateria are differentially expressed along the primary axis of a “radiate” animal, the anthozoan cnidarian Acropora millepora. Dev. Biol. 298, 632–643. doi: 10.1016/j.ydbio.2006.07.034
Duboule, D. (2007). The rise and fall of Hox gene clusters. Development 134, 2549–2560. doi: 10.1242/dev.001065
Erwin, D. H. (2009). Early origin of the bilaterian developmental toolkit. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 364, 2253–2261. doi: 10.1098/rstb.2009.0038
Finnerty, J. R., Pang, K., Burton, P., Paulson, D., and Martindale, M. Q. (2004). Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science 304, 1335–1337. doi: 10.1126/science.1091946
Finnerty, J. R., Paulson, D., Burton, P., Pang, K., and Martindale, M. Q. (2003). Early evolution of a homeobox gene: the parahox gene Gsx in the Cnidaria and the Bilateria. Evol. Dev. 5, 331–345. doi: 10.1046/j.1525-142X.2003.03041.x
Foos, N., Maurel-Zaffran, C., Maté, M. J., Vincentelli, R., Hainaut, M., Berenger, H., et al. (2015). A flexible extension of the Drosophila ultrabithorax homeodomain defines a novel Hox/PBC interaction mode. Structure 23, 270–279. doi: 10.1016/j.str.2014.12.011
Fortunato, S. A. V., Adamski, M., Ramos, O. M., Leininger, S., Liu, J., Ferrier, D. E. K., et al. (2014). Calcisponges have a ParaHox gene and dynamic expression of dispersed NK homeobox genes. Nature 514, 620–623. doi: 10.1038/nature13881
Fujioka, M., Gebelein, B., Cofer, Z. C., Mann, R. S., and Jaynes, J. B. (2012). Engrailed cooperates directly with Extradenticle and Homothorax on a distinct class of homeodomain binding sites to repress sloppy paired. Dev. Biol. 366, 382–392. doi: 10.1016/j.ydbio.2012.04.004
Galant, R., Walsh, C. M., and Carroll, S. B. (2002). Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development 129, 3115–3126.
Galliot, B., and de Vargas, C., Miller, D. (1999). Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. Dev. Genes Evol. 209, 186–197. doi: 10.1007/s004270050243
Garcia-Fernàndez, J. (2005). The genesis and evolution of homeobox gene clusters. Nat. Rev. Genet. 6, 881–892. doi: 10.1038/nrg1723
Garstang, M., and Ferrier, D. E. K. (2013). Time is of the essence for ParaHox homeobox gene clustering. BMC Biol. 11:72. doi: 10.1186/1741-7007-11-72
Gauchat, D., Mazet, F., Berney, C., Schummer, M., Kreger, S., Pawlowski, J., et al. (2000). Evolution of Antp-class genes and differential expression of Hydra Hox/paraHox genes in anterior patterning. Proc. Natl. Acad. Sci. U.S.A. 97, 4493–4498. doi: 10.1073/pnas.97.9.4493
Gould, S. J. (1992). Ontogeny and phylogeny—revisited and reunited. Bioessays 4, 275–279. doi: 10.1002/bies.950140413
Hackbusch, J., Richter, K., Müller, J., Salamini, F., and Uhrig, J. F. (2005). A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 4908–4912. doi: 10.1073/pnas.0501181102
Hay, A., and Tsiantis, M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. doi: 10.1242/dev.030049
Hayward, D. C., Catmull, J., Reece-Hoyes, J. S., Berghammer, H., Dodd, H., Hann, S. J., et al. (2001). Gene structure and larval expression of cnox-2Am from the coral Acropora millepora. Dev. Genes Evol. 211, 10–19. doi: 10.1007/s004270000112
Heffer, A., Xiang, J., and Pick, L. (2013). Variation and constraint in Hox gene evolution. Proc. Natl. Acad. Sci. U.S.A. 110, 2211–2216. doi: 10.1073/pnas.1210847110
Holland, P. W. H. (2013). Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2, 31–45. doi: 10.1002/wdev.78
Hudry, B., Remacle, S., Delfini, M.-C., Rezsohazy, R., Graba, Y., and Merabet, S. (2012). Hox proteins display a common and ancestral ability to diversify their interaction mode with the PBC class cofactors. PLoS Biol. 10:e1001351. doi: 10.1371/journal.pbio.1001351
Hudry, B., Thomas-Chollier, M., Volovik, Y., Duffraisse, M., Dard, A., Frank, D., et al. (2014). Molecular insights into the origin of the Hox-TALE patterning system. Elife 3:e01939. doi: 10.7554/eLife.01939
Hudry, B., Viala, S., Graba, Y., and Merabet, S. (2011). Visualization of protein interactions in living Drosophila embryos by the bimolecular fluorescence complementation assay. BMC Biol. 9:5. doi: 10.1186/1741-7007-9-5
Hueber, S. D., and Lohmann, I. (2008). Shaping segments: Hox gene function in the genomic age. Bioessays 30, 965–979. doi: 10.1002/bies.20823
Ikuta, T., Chen, Y.-C., Annunziata, R., Ting, H.-C., Tung, C. H., Koyanagi, R., et al. (2013). Identification of an intact ParaHox cluster with temporal colinearity but altered spatial colinearity in the hemichordate Ptychodera flava. BMC Evol. Biol. 13:129. doi: 10.1186/1471-2148-13-129
In der Rieden, P. M., der, Mainguy, G., Woltering, J. M., and Durston, A. J. (2003). Homeodomain to hexapeptide or PBC-interactiondomain distance: size apparently matters. Trends Genet. 20, 76–79. doi: 10.1016/j.tig.2003.12.001
Joshi, R., Passner, J. M., Rohs, R., Jain, R., Sosinsky, A., Crickmore, M., et al. (2007). Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131, 530–543. doi: 10.1016/j.cell.2007.09.024
Kamm, K., Schierwater, B., Jakob, W., Dellaporta, S. L., and Miller, D. J. (2006). Axial patterning and diversification in the cnidaria predate the Hox system. Curr. Biol. 16, 920–926. doi: 10.1016/j.cub.2006.03.036
Kaneto, H., Miyatsuka, T., Kawamori, D., and Matsuoka, T. A. (2007). Pleiotropic roles of PDX-1 in the pancreas. Rev. Diabet. Stud. 4, 209–225. doi: 10.1900/RDS.2007.4.209
Kanrar, S., Onguka, O., and Smith, H. M. S. (2006). Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224, 1163–1173. doi: 10.1007/s00425-006-0298-9
Keleher, C. A., Passmore, S., and Johnson, A. D. (1989). Yeast repressor alpha 2 binds to its operator cooperatively with yeast protein Mcm1. Mol. Cell. Biol. 9, 5228–5230.
King, N. (2004). The unicelllular ancestry of animal development. Dev. Cell 7, 313–325. doi: 10.1016/j.devcel.2004.08.010
King, N., Hittinger, C. T., and Carroll, S. B. (2003). Evolution of key cell signaling and adhesion protein families predates animal origins. Science 301, 361–363. doi: 10.1126/science.1083853
King, N., Westbrook, M. J., Young, S. L., Kuo, A., Abedin, M., Chapman, J., et al. (2008). The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788. doi: 10.1038/nature06617
Kmita, M., and Duboule, D. (2003). Organizing axes in time and space; 25 years of colinear tinkering. Science 301, 331–333. doi: 10.1126/science.1085753
Knoepfler, P. S., Bergstrom, D. A., Uetsuki, T., Dac-korytko, I., Sun, Y. H., Wright, W. E., et al. (1999). A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with Pbx – Meis1/Prep1. 27, 3752–3761. doi: 10.1093/nar/27.18.3752
LaRonde-LeBlanc, N. A., and Wolberger, C. (2003). Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 17, 2060–2072. doi: 10.1101/gad.1103303
Larroux, C., Fahey, B., Degnan, S. M., Adamski, M., Rokhsar, D. S., and Degnan, B. M. (2007). The NK Homeobox Gene Cluster Predates the Origin of Hox Genes. Curr. Biol. 17, 706–710. doi: 10.1016/j.cub.2007.03.008
Larroux, C., Luke, G. N., Koopman, P., Rokhsar, D. S., Shimeld, S. M., and Degnan, B. M. (2008). Genesis and expansion of metazoan transcription factor gene classes. Mol. Biol. Evol. 25, 980–996. doi: 10.1093/molbev/msn047
Lewis, E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. doi: 10.1038/276565a0
López-Juárez, A., Howard, J., Ullom, K., Howard, L., Grande, A., Pardo, A., et al. (2013). Gsx2 controls region-specific activation of neural stem cells and injury-induced neurogenesis in the adult subventricular zone. Genes Dev. 27, 1272–1287. doi: 10.1101/gad.217539.113
Mann, R. S., and Affolter, M. (1998). Hox proteins meet more partners. Curr. Opin. Genet. Dev. doi: 10.1016/s0959-437x(98)80113-5
Mann, R. S., and Carroll, S. B. (2002). Molecular mechanisms of selector gene function and evolution. Curr. Opin. Genet. Dev. 12, 592–600. doi: 10.1016/S0959-437X(02)00344-1
Mann, R. S., Lelli, K. M., and Joshi, R. (2009). Chapter 3 Hox specificity. Unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 88, 63–101. doi: 10.1016/S0070-2153(09)88003-4
Martindale, M. Q. (2005). The evolution of metazoan axial properties. Nat. Rev. Genet. 6, 917–927. doi: 10.1038/nrg1725
Matus, D. Q., Pang, K., Marlow, H., Dunn, C. W., Thomsen, G. H., and Martindale, M. Q. (2006). Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc. Natl. Acad. Sci. U.S.A. 103, 11195–11200. doi: 10.1073/pnas.0601257103
Maves, L., Tyler, A., Moens, C. B., and Tapscott, S. J. (2009). Pbx acts with Hand2 in early myocardial differentiation. Dev. Biol. 333, 409–418. doi: 10.1016/j.ydbio.2009.07.004
Maves, L., Waskiewicz, A. J., Paul, B., Cao, Y., Tyler, A., Moens, C. B., et al. (2007). Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development 134, 3371–3382. doi: 10.1242/dev.003905
McGinnis, W., Krumlauf, R. (1992). Homeobox genes and axial patterning. Cell 68, 283–302. doi: 10.1016/0092-8674(92)90471-N
Mendivil Ramos, O., Barker, D., and Ferrier, D. E. K. (2012). Ghost loci imply Hox and parahox existence in the last common ancestor of animals. Curr. Biol. 22, 1951–1956. doi: 10.1016/j.cub.2012.08.023
Merabet, S., and Dard, A. (2014). Tracking context-specific transcription factors regulating Hox activity. Dev. Dyn. 243, 16–23. doi: 10.1002/dvdy.24002
Merabet, S., and Hudry, B. (2013). Hox transcriptional specificity despite a single class of cofactors: are flexible interaction modes the key? Plasticity in Hox/PBC interaction modes as a common molecular strategy for shaping Hox transcriptional activities. Bioessays 35, 88–92. doi: 10.1002/bies.201200146
Merabet, S., Hudry, B., Saadaoui, M., and Graba, Y. (2009). Classification of sequence signatures: a guide to Hox protein function. Bioessays 31, 500–511. doi: 10.1002/bies.200800229
Merabet, S., Kambris, Z., Capovilla, M., Bérenger, H., Pradel, J., and Graba, Y. (2003). The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev. Cell 4, 761–768. doi: 10.1016/S1534-5807(03)00126-6
Merabet, S., Litim-Mecheri, I., Karlsson, D., Dixit, R., Saadaoui, M., Monier, B., et al. (2011). Insights into Hox protein function from a large scale combinatorial analysis of protein domains. PLoS Genet. 7:e1002302. doi: 10.1371/journal.pgen.1002302
Merabet, S., Saadaoui, M., Sambrani, N., Hudry, B., Pradel, J., Affolter, M., et al. (2007). A unique Extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax. Proc. Natl. Acad. Sci. U.S.A. 104, 16946–16951. doi: 10.1073/pnas.0705832104
Miljkovic-Licina, M., Chera, S., Ghila, L., and Galliot, B. (2007). Head regeneration in wild-type hydra requires de novo neurogenesis. Development 134, 1191–1201. doi: 10.1242/dev.02804
Minguillón, C., and Garcia-Fernàndez, J. (2003). Genesis and evolution of the Evx and Mox genes and the extended Hox and ParaHox gene clusters. Genome Biol. 4:R12. doi: 10.1186/gb-2003-4-2-r12
Moens, C. B., and Selleri, L. (2006). Hox cofactors in vertebrate development. Dev. Dyn. 291, 193–206. doi: 10.1016/j.ydbio.2005.10.032
Moreno, E., and Morata, G. (1999). Caudal is the Hox gene that specifies the most posterior Drosophile segment. Nature 400, 873–877. doi: 10.1038/23709
Moroz, L. L., Kocot, K. M., Citarella, M. R., Dosung, S., Norekian, T. P., Povolotskaya, I. S., et al. (2014). The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. doi: 10.1038/nature13400
Mukherjee, K., and Bürglin, T. R. (2007). Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J. Mol. Evol. 65, 137–153. doi: 10.1007/s00239-006-0023-0
Ogishima, S., and Tanaka, H. (2007). Missing link in the evolution of Hox clusters. Gene 387, 21–30. doi: 10.1016/j.gene.2006.08.011
Passner, J. M., Ryoo, H. D., Shen, L., Mann, R. S., and Aggarwal, A. K. (1999). Structure of a DNA-bound Ultrabithorax–Extradenticle homeodomain complex. Nature 397, 714–719. doi: 10.1038/17833
Pearson, J. C., Lemons, D., and McGinnis, W. (2005). Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6, 893–904. doi: 10.1038/nrg1726
Piper, D. E., Batchelor, A. H., Chang, C. P., Cleary, M. L., and Wolberger, C. (1999). Structure of a HoxB1–Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96, 587–97. doi: 10.1016/S0092-8674(00)80662-5
Quiquand, M., Yanze, N., Schmich, J., Schmid, V., Galliot, B., and Piraino, S. (2009). More constraint on ParaHox than Hox gene families in early metazoan evolution. Dev. Biol. 328, 173–187. doi: 10.1016/j.ydbio.2009.01.022
Rhee, J. W., Arata, A., Selleri, L., Jacobs, Y., Arata, S., Onimaru, H., et al. (2004). Pbx3 deficiency results in central hypoventilation. Am. J. Pathol. 165, 1343–1350. doi: 10.1016/S0002-9440(10)63392-5
Ryan, J. F., and Baxevanis, A. D. (2007). Hox, Wnt, and the evolution of the primary body axis: insights from the early-divergent phyla. Biol. Direct 2, 37–48. doi: 10.1186/1745-6150-2-37
Ryan, J. F., Mazza, M. E., Pang, K., Matus, D. Q., Baxevanis, A. D., Martindale, M. Q., et al. (2007). Pre-bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis. PLoS ONE 2:e153. doi: 10.1371/journal.pone.0000153
Ryan, J. F., Pang, K., Mullikin, J. C., Martindale, M. Q., and Baxevanis, A. D. (2010). The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa. Evodevo 1:9. doi: 10.1186/2041-9139-1-9
Saleh, M., Rambaldi, I., Yang, X. J., and Featherstone, M. S. (2000). Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20, 8623–33. doi: 10.1128/MCB.20.22.8623-8633.2000
Schierwater, B., Kamm, K., Srivastava, M., Rokhsar, D., Rosengarten, R. D., and Dellaporta, S. L. (2008a). The early ANTP gene repertoire: insights from the placozoan genome. PLoS ONE 3:2457. doi: 10.1371/journal.pone.0002457
Schierwater, B., Kamm, K., Srivastava, M., Rokhsar, D., Rosengarten, R. D., and Dellaporta, S. L. (2008b). The early ANTP gene repertoire: insights from the placozoan genome. PLoS ONE 3:e2457. doi: 10.1371/journal.pone.0002457
Schierwater, B., and Kuhn, K. (1998). Homology of Hox genes and the zootype concept in early metazoan evolution. Mol. Phylogenet. Evol. 9, 375–381. doi: 10.1006/mpev.1998.0489
Schummer, M., Scheurlen, I., Schaller, C., and Galliot, B. (1992). HOM/HOX homeobox genes are present in hydra (Chlorohydra viridissima) and are differentially expressed during regeneration. EMBO J. 11, 1815–1823.
Shen, W. F., Montgomery, J. C., Rozenfeld, S., Moskow, J. J., Lawrence, H. J., Buchberg, A. M., et al. (1997). AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol. Cell. Biol. 17, 6448–58.
Shinmyo, Y., Mito, T., Matsushita, T., Sarashina, I., Miyawaki, K., Ohuchi, H., et al. (2005). Caudal is required for gnathal and thoracic patterning and for posterior elongation in the intermediate-germband cricket Gryllus bimaculatus. Mech. Dev. 122, 231–239. doi: 10.1016/j.mod.2004.10.001
Slattery, M., Riley, T., Liu, P., Abe, N., Gomez-Alcala, P., Dror, I., et al. (2011). Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270–1282. doi: 10.1016/j.cell.2011.10.053
Srivastava, M., Simakov, O., Chapman, J., Fahey, B., Gauthier, M. E. A, Mitros, T., et al. (2010). The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726. doi: 10.1038/nature09201
Stark, M. R., and Johnson, A. D. (1994). Interaction between two homeodomain proteins is specified by a short C-terminal tail. Nature 371, 429–432. doi: 10.1038/371429a0
Steele, R. E., David, C. N., and Technau, U. (2011). A genomic view of 500 million years of cnidarian evolution. Trends Genet. 27, 7–13. doi: 10.1016/j.tig.2010.10.002
Stevens, K. E., and Mann, R. S. (2007). A balance between two nuclear localization sequences and a nuclear export sequence governs extradenticle subcellular localization. Genetics 175, 1625–1636. doi: 10.1534/genetics.106.066449
Suga, H., Chen, Z., de Mendoza, A., Sebé-Pedrós, A., Brown, M. W., Kramer, E., et al. (2013). The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 4, 2325. doi: 10.1038/ncomms3325
Tompa, P., Davey, N. E., Gibson, T. J., and Babu, M. M. (2014). A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169. doi: 10.1016/j.molcel.2014.05.032
True, J. R., and Carroll, S. B. (2002). Gene co-option in physiological and morphological evolution. Annu. Rev. Cell Dev. Biol. 18, 53–80. doi: 10.1146/annurev.cellbio.18.020402.140619
Van den Akker, E., Forlani, S., Chawengsaksophak, K., de Graaff, W., Beck, F., Meyer, B. I., et al. (2002). Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development 129, 2181–2193.
Van Roey, K., Dinkel, H., Weatheritt, R. J., Gibson, T. J., and Davey, N. E. (2013). The switches.ELM resource: a compendium of conditional regulatory interaction interfaces. Sci. Signal. 6, rs7. doi: 10.1126/scisignal.2003345
Waclaw, R. R., Wang, B., Pei, Z., Ehrman, L. A., and Campbell, K. (2009). Distinct temporal requirements for the homeobox gene Gsx2 in specifying striatal and olfactory bulb neuronal fates. Neuron 63, 451–465. doi: 10.1016/j.neuron.2009.07.015
Wagner, G. P. (2007). The developmental genetics of homology. Nat. Rev. Genet. 8, 473–479. doi: 10.1038/nrg2099
Weiss, J. B., Von Ohlen, T., Mellerick, D. M., Dressler, G., Doe, C. Q., and Scott, M. P. (1998). Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes Dev. 12, 3591–3602. doi: 10.1101/gad.12.22.3591
Wenger, Y., and Galliot, B. (2013). Punctuated emergences of genetic and phenotypic innovations in eumetazoan, bilaterian, euteleostome, and hominidae ancestors. Genome Biol. Evol. 5, 1949–1968. doi: 10.1093/gbe/evt142
Williams, N. A., and Holland, P. W. (2000). An amphioxus Emx homeobox gene reveals duplication during vertebrate evolution. Mol. Biol. Evol. 17, 1520–1528. doi: 10.1093/oxfordjournals.molbev.a026251
Williams, T. M., Williams, M. E., and Innis, J. W. (2005). Range of HOX/TALE superclass associations and protein domain requirements for HOXA13:MEIS interaction. Dev. Biol. 277, 457–471. doi: 10.1016/j.ydbio.2004.10.004
Winterbottom, E. F., Illes, J. C., Faas, L., and Isaacs, H., V (2010). Conserved and novel roles for the Gsh2 transcription factor in primary neurogenesis. Development 137, 2623–2631. doi: 10.1242/dev.047159
Yanze, N., Spring, J., Schmidli, C., and Schmid, V. (2001). Conservation of Hox/ParaHox-related genes in the early development of a cnidarian. Dev. Biol. 236, 89–98. doi: 10.1006/dbio.2001.0299
Yao, Z., Farr, G. H., Tapscott, S. J., and Maves, L. (2013). Pbx and Prdm1a transcription factors differentially regulate subsets of the fast skeletal muscle program in zebrafish. Biol. Open 2, 546–555. doi: 10.1242/bio.20133921
Keywords: Hox, PBC, Meis, Metazoa, patterning, early-branching phyla, HX, SPIMs
Citation: Merabet S and Galliot B (2015) The TALE face of Hox proteins in animal evolution. Front. Genet. 6:267. doi: 10.3389/fgene.2015.00267
Received: 14 June 2015; Accepted: 31 July 2015;
Published: 18 August 2015.
Edited by:
Sylvain Marcellini, University of Concepcion, ChileReviewed by:
Ingo Braasch, University of Oregon, USAPedro Martinez, Universitat de Barcelona, Spain
Copyright © 2015 Merabet and Galliot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samir Merabet, Centre National de Recherche Scientifique, Ecole Normale Supérieure de Lyon, UMR5242, 46 Allée d'Italie, Lyon 69007, France, samir.merabet@ens-lyon.fr
 Samir Merabet
Samir Merabet Brigitte Galliot
Brigitte Galliot