- Department of Biology, Faculty of Arts and Science, Concordia University, Montreal, PQ, Canada
Studies employing the budding yeast Saccharomyces cerevisiae as a model organism have provided deep insights into molecular mechanisms of cellular and organismal aging in multicellular eukaryotes and have demonstrated that the main features of biological aging are evolutionarily conserved. Aging in S. cerevisiae is studied by measuring replicative or chronological lifespan. Yeast replicative aging is likely to model aging of mitotically competent human cell types, while yeast chronological aging is believed to mimic aging of post-mitotic human cell types. Emergent evidence implies that various organelle-organelle and organelle-cytosol communications play essential roles in chronological aging of S. cerevisiae. The molecular mechanisms underlying the vital roles of intercompartmental communications in yeast chronological aging have begun to emerge. The scope of this review is to critically analyze recent progress in understanding such mechanisms. Our analysis suggests a model for how temporally and spatially coordinated movements of certain metabolites between various cellular compartments impact yeast chronological aging. In our model, diverse changes in these key metabolites are restricted to critical longevity-defining periods of chronological lifespan. In each of these periods, a limited set of proteins responds to such changes of the metabolites by altering the rate and efficiency of a certain cellular process essential for longevity regulation. Spatiotemporal dynamics of alterations in these longevity-defining cellular processes orchestrates the development and maintenance of a pro- or anti-aging cellular pattern.
Introduction
Studies of the budding yeast Saccharomyces cerevisiae have been instrumental in discovering genes, signaling pathways, and chemical compounds that influence cellular and organismal aging in evolutionarily distant eukaryotes (Fontana et al., 2010; Kaeberlein, 2010; Longo et al., 2012; Arlia-Ciommo et al., 2014a,b). These studies have revealed that the key aspects of the aging process and mechanisms of its modulation by certain genetic, dietary, and pharmacological interventions have been conserved in the course of evolution (Eisenberg et al., 2009; Fontana et al., 2010; Arlia-Ciommo et al., 2014a; Denoth Lippuner et al., 2014; Medkour et al., 2016a). One paradigm of aging in yeast is replicative aging. It is believed to imitate aging of mitotic human cell types capable of dividing (Steinkraus et al., 2008; Kaeberlein, 2010; Longo et al., 2012; Denoth Lippuner et al., 2014; McCormick et al., 2015), although recent findings suggest that yeast replicative aging may also serve as a suitable model for the aging of post-mitotic tissues and for the aging of whole organism in the nematode C. elegans and humans (Ghavidel et al., 2015; McCormick et al., 2015; Janssens and Veenhoff, 2016). A body of evidence supports the notion that diverse interorganelle communications influence yeast replicative aging (Beach et al., 2012; Hughes and Gottschling, 2012; Henderson et al., 2014; Janssens et al., 2015; Hughes et al., 2016). This evidence has been comprehensively discussed elsewhere (Jazwinski, 2012, 2013, 2014, 2015; Jazwinski and Kriete, 2012). Another paradigm of aging in yeast is chronological aging. It is likely to mimic aging of post-mitotic human cell types incapable of dividing (Burtner et al., 2009, 2011; Kaeberlein, 2010; Longo et al., 2012; Arlia-Ciommo et al., 2014a), although there is evidence that yeast chronological aging may converge with yeast replicative aging into a single aging process (reviewed in Arlia-Ciommo et al., 2014b; see also Mirisola and Longo, 2012; Murakami et al., 2012; Polymenis and Kennedy, 2012; Delaney et al., 2013; Molon et al., 2015). Recent findings indicate that many organelle-organelle and organelle-cytosol communications impact yeast chronological aging (Goldberg et al., 2009a; Titorenko and Terlecky, 2011; Beach et al., 2012; Beach and Titorenko, 2013; Leonov and Titorenko, 2013). Mechanisms underlying the essential roles of such intercompartmental communications in yeast chronological aging have begun to emerge. Here, we critically analyze recent progress in understanding these mechanisms.
Spatiotemporal Dynamics of Intercompartmental Communications Define the Chronology of Cellular Aging in Yeast
Recent studies have revealed that various intercompartmental communications (i.e., organelle-organelle and organelle-cytosol) play essential roles in chronological aging of yeast cultured in media with glucose as the only carbon source (Beach and Titorenko, 2011; Beach et al., 2012, 2015a; Leonov and Titorenko, 2013; Medkour and Titorenko, 2016b). A model for how such communications impact yeast chronological aging is depicted schematically in Figure 1. Our model includes the notion that the longevity-defining intercompartmental communications involve unidirectional and bidirectional movements of a distinct set of metabolites between mitochondria and the cytosol, mitochondria and peroxisomes, mitochondria and the nucleus, peroxisomes and the nucleus, mitochondria and vacuoles, the endoplasmic reticulum (ER) and the plasma membrane (PM), the ER and the cytosol, the PM and the cytosol, the PM and vacuoles, the ER and lipid droplets (LD), and LD and peroxisomes (Figure 1). The intracellular concentrations of such metabolites and/or the rates of their movement between cellular compartments undergo age-related changes. In our model, different changes of the key metabolites are temporally restricted to several longevity-defining periods; the term “checkpoints” has been coined to describe these critical periods in yeast chronological lifespan (Burstein et al., 2012; Kyryakov et al., 2012; Arlia-Ciommo et al., 2014a; Beach et al., 2015a,b) (Figure 1). Most of these checkpoints occur early in life of chronologically aging yeast cells, during diauxic (D), and post-diauxic (PD) growth phases. Some of the checkpoints are late-life checkpoints that exists in the non-proliferative stationary (ST) phase of culturing. At each of these checkpoints, the changes of the key metabolites are detected by a distinct set of checkpoint-specific proteins called “master regulators” (Arlia-Ciommo et al., 2014a; Beach et al., 2015a). Our model further posits that each of these master regulators can respond to a change of the detected key metabolite by altering the rate and efficiency of a certain cellular process essential for longevity regulation (Figure 1). By establishing the rates and efficiencies of different longevity-defining cellular processes throughout chronological lifespan, the checkpoint-specific master regulators set up a pro- or anti-aging cellular pattern (Arlia-Ciommo et al., 2014a; Beach et al., 2015a).
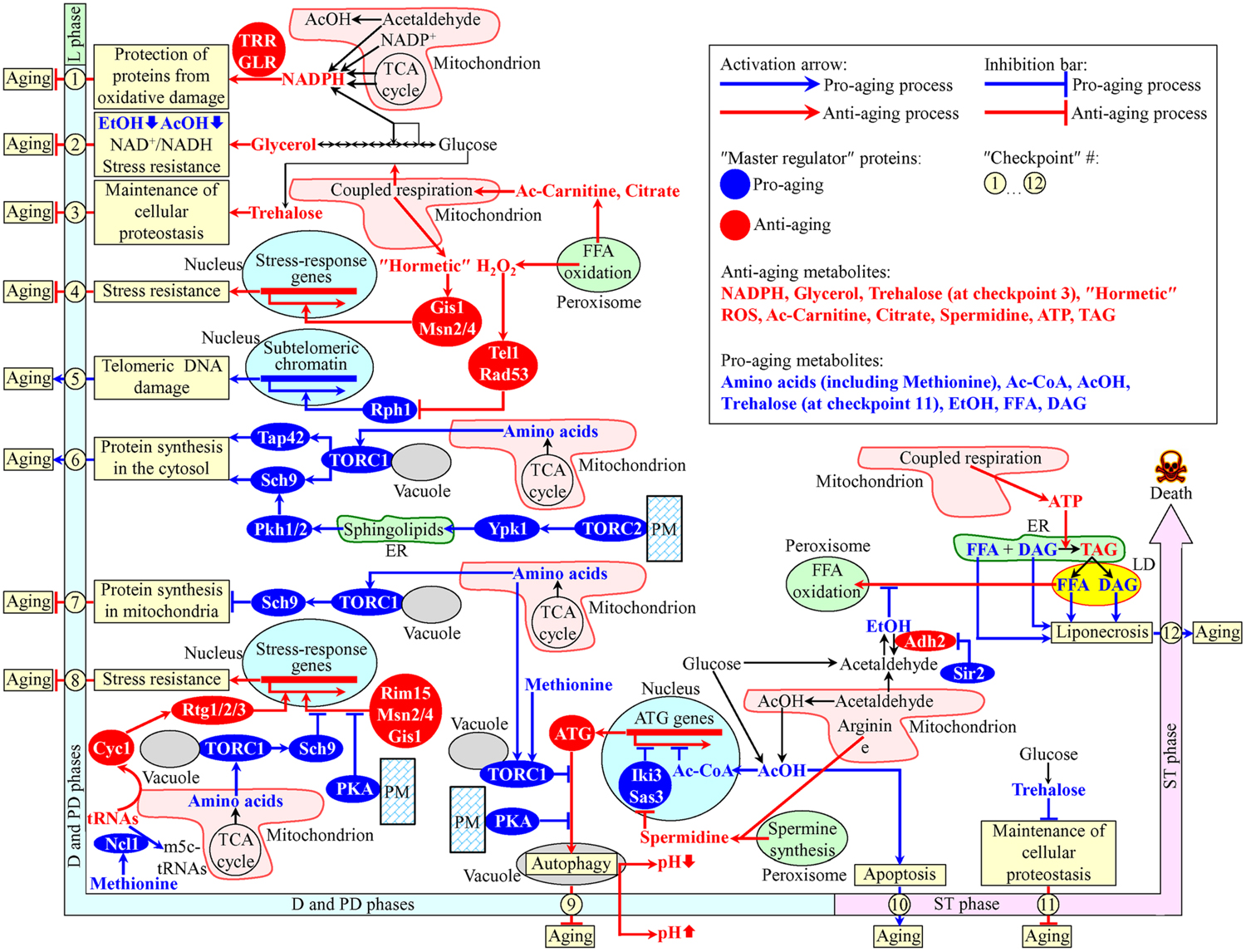
FIGURE 1. A model for how various organelle-organelle and organelle-cytosol communications impact yeast chronological aging. These communications involve movements of certain metabolites between various cellular compartments. Different changes in these metabolites are temporally restricted to longevity-defining periods called checkpoints. At each checkpoint, the changes of these metabolites are detected by certain master regulator proteins. Because each of the master regulator proteins modulates certain longevity-defining cellular processes, a coordinated in space and time action of these proteins orchestrates the development and maintenance of a pro- or anti-aging cellular pattern. See text for details. Ac-Carnitine, acetyl-carnitine; Ac-CoA, acetyl-CoA; AcOH, acetic acid; DAG, diacylglycerol; EtOH, ethanol; FFA, free (non-esterified) fatty acids; m5c-tRNAs, 5-methylcytosine tRNAs; PKA, protein kinase A; PM, the plasma membrane; ROS, reactive oxygen species; TAG, triacylglycerols; TCA, tricarboxylic; TORC, target of rapamycin complex; tRNAs, transfer RNAs.
At checkpoint 1, which exists early in D growth phase, two oxidative reactions of the pentose phosphate pathway in the cytosol and four enzymatic reactions in mitochondria create NADPH (Cai and Tu, 2012; Barral, 2013; Brandes et al., 2013; Arlia-Ciommo et al., 2014a) (Figure 1). NADPH provides reducing equivalents for the synthesis of amino acids, fatty acids, and sterols (Fraenkel, 2011; Cai and Tu, 2012). NADPH is also a donor of electrons for thioredoxin and glutathione reductase systems. Both these reductase systems contribute to the establishment and maintenance of an anti-aging cellular pattern because they protect many thiol-containing proteins from oxidative damage; such thiol-containing proteins reside in the nucleus, mitochondria and cytosol (Barral, 2013; Brandes et al., 2013) (Figure 1).
Glycerol is produced by glucose fermentation in the cytosol (Fraenkel, 2011). At checkpoint 2, glycerol plays an important role in the establishment and maintenance of an anti-aging cellular pattern by affecting the following cellular processes: (1) glucose fermentation to glycerol weakens its fermentation to ethanol and acetic acid, both known to be pro-aging metabolites in yeast; (2) glucose fermentation to glycerol enables to sustain the NAD+/NADH ratio that slows yeast chronological aging; and (3) glycerol increases resistance to acute oxidative, thermal, and osmotic stresses that accelerate yeast chronological aging (Burtner et al., 2009; Wei et al., 2009; Arlia-Ciommo et al., 2014a) (Figure 1).
In the cytosol of chronologically “young” yeast progressing through D and PD growth phases, the non-reducing disaccharide trehalose is synthesized from glucose (Goldberg et al., 2009b; Kyryakov et al., 2012). The rate of such synthesis sustains cellular trehalose homeostasis and is modulated by the efficiency of coupled mitochondrial respiration (Ocampo et al., 2012). The efficiency of such respiration is, in turn, modulated by the rate of peroxisome-to-mitochondria transfer of citrate and acetyl-carnitine (Epstein et al., 2001; Traven et al., 2001; Hiltunen et al., 2003; Titorenko and Terlecky, 2011; Arlia-Ciommo et al., 2014a). At checkpoint 3, trehalose is essential for maintaining an anti-aging pattern of cellular proteostasis because it attenuates the misfolding, aggregation and oxidative damage of newly synthesized polypeptides (Goldberg et al., 2009b; Kyryakov et al., 2012; Arlia-Ciommo et al., 2014a) (Figure 1).
During D and PD growth phases, the intracellular concentration of hydrogen peroxide (H2O2) in chronologically aging yeast depends on the efficiencies with which this major reactive oxygen species (ROS) is produced by and released from mitochondria and peroxisomes (Goldberg et al., 2010; Mirisola and Longo, 2013; Schroeder et al., 2013; Schroeder and Shadel, 2014). If the concentration of H2O2 at checkpoint 4 is sustained at a sub-lethal (“hormetic”) level, it elicits the establishment of an anti-aging cellular pattern by stimulating the master regulators Gis1, Msn2, and Msn4. In the nucleus, these three transcriptional factors activate expression of genes that encode proteins involved in heat-shock and DNA-damage responses, ROS decomposition, cell cycle progression and transition to quiescence, autophagy, maintenance of cell wall integrity, trehalose synthesis and degradation, glycogen synthesis and degradation, glycolysis and gluconeogenesis, the pentose phosphate pathway, glycerol and amino acid synthesis, ergosterol synthesis, maintenance of glutathione and thioredoxin homeostasis, methylglyoxal detoxification, maintenance of heavy metal ion homeostasis, potassium transport, and mitochondrial electron transport; these proteins are needed for resistance to thermal, oxidative, osmotic, low pH, carbon source starvation, sorbic acid, high ethanol concentration, and DNA-damage stresses (Martínez-Pastor et al., 1996; Schmitt and McEntee, 1996; Boy-Marcotte et al., 1998; Causton et al., 2001; Fabrizio et al., 2001) (Figure 1).
At checkpoint 5, H2O2 produced by and released from mitochondria and peroxisomes modulates a signaling pathway which includes the DNA damage response kinases Tel1 and Rad53 (both of which are anti-aging master regulators) and the histone demethylase Rph1 (a pro-aging master regulator) (Mirisola and Longo, 2013). If the concentration of H2O2 at this checkpoint is sustained at a hormetic level, it stimulates the Tel1-depenent phosphorylation/activation of Rad53, which in response phosphorylates and inactivates Rph1 (Schroeder et al., 2013; Schroeder and Shadel, 2014). The resulting inactivation of Rph1 establishes an anti-aging cellular pattern because it allows to attenuate the Rph1-dependent transcription of subtelomeric chromatin regions in the nucleus, thereby lessening the extent of telomeric DNA damage (Mirisola and Longo, 2013; Schroeder et al., 2013; Schroeder and Shadel, 2014) (Figure 1).
During D and PD growth phases, the amino acids aspartate, asparagine, glutamate, and glutamine are synthesized from intermediates of the TCA cycle in mitochondria (Fraenkel, 2011; Cai and Tu, 2012). After being released into the cytosol, these amino acids stimulate protein kinase (PK) activity of the TOR (target of rapamycin) complex 1 (TORC1) at the surface of vacuoles (Crespo et al., 2002; Powers et al., 2006; Jewell et al., 2013; Conrad et al., 2014; Shimobayashi and Hall, 2014; Swinnen et al., 2014). Following its activation, TORC1 acts as a pro-aging master regulator at checkpoint 6 by phosphorylating the nutrient-sensory PK Sch9 and the Tap42 protein. Once phosphorylated, Sch9 and Tap42 accelerate the pro-aging process of protein synthesis in the cytosol by stimulating ribosome biogenesis and augmenting translation initiation (Hinnebusch, 2005; Huber et al., 2009; Conrad et al., 2014; Swinnen et al., 2014; Eltschinger and Loewith, 2016) (Figure 1). The TOR complex 2 (TORC2) at the PM also functions as a pro-aging master regulator at checkpoint 6. If activated, TORC2 phosphorylates the PK Ypk1. After being phosphorylated, Ypk1 stimulates the synthesis of complex sphingolipids in the ER. These sphingolipids then stimulate the PKs Pkh1 and Pkh2, both of which in response phosphorylate Sch9 to intensify the pro-aging process of protein synthesis in the cytosol (Roelants et al., 2004; Liu et al., 2005; Urban et al., 2007; Huang et al., 2014; Eltschinger and Loewith, 2016; Teixeira and Costa, 2016) (Figure 1).
At checkpoint 7, the amino acids aspartate, asparagine, glutamate, and glutamine are released from mitochondria and activate TORC1 at the surface of vacuoles. Active TORC1 sets off a pro-aging cellular pattern by phosphorylating Sch9, which then attenuates the anti-aging process of protein synthesis in mitochondria (Bonawitz et al., 2007; Pan and Shadel, 2009; Conrad et al., 2014; Shimobayashi and Hall, 2014; Swinnen et al., 2014) (Figure 1).
At checkpoint 8, the efflux of the amino acids aspartate, asparagine, glutamate and glutamine from mitochondria, resulting activation of TORC1 at the vacuolar surface and subsequent phosphorylation of Sch9 cause a retention of the nutrient-sensory PK Rim15 in the cytosol (Wanke et al., 2008; Smets et al., 2010). Because under such conditions Rim15 cannot enter the nucleus, it is unable to stimulate Msn2, Msn4, and Gis1; these three transcriptional activators can orchestrate an anti-aging transcriptional program in the nucleus only if they are stimulated by Rim15 (Wanke et al., 2008; Smets et al., 2010; Conrad et al., 2014; Shimobayashi and Hall, 2014; Swinnen et al., 2014) (Figure 1). Furthermore, protein kinase A (PKA) activity at the cytosolic leaflet of the PM also contributes to the establishment of a pro-aging cellular pattern at checkpoint 8. This PK activity inhibits nuclear import of Msn2 and Msn4, thus turning off an anti-aging transcriptional program driven – in a Rim15-dependent manner – by these two transcriptional activators (Medvedik et al., 2007; Lee et al., 2008; Smets et al., 2010; Conrad et al., 2014) (Figure 1). Moreover, a study on a methionine restriction-induced delay of yeast chronological aging implies that the excess of methionine can elicit a pro-aging cellular pattern at checkpoint 8 by activating the tRNA methyltransferase Ncl1 in the cytosol (Johnson and Johnson, 2014). This decreases the concentration of non-methylated tRNAs, attenuates the efflux of cytochrome C (Cyc1) from mitochondria and mitigates nuclear import of the cytosolic Rtg1/Rtg2/Rtg3 heterotrimeric transcriptional factor, which is required for the stimulation of an anti-aging transcriptional program in the nucleus (Johnson and Johnson, 2014) (Figure 1).
If TORC1 at the surface of vacuoles is activated by the release of the amino acids aspartate, asparagine, glutamate, and glutamine from mitochondria at checkpoint 9, active TORC1 phosphorylates the autophagy-initiating protein Atg13 (Laplante and Sabatini, 2012; Conrad et al., 2014; Shimobayashi and Hall, 2014; Swinnen et al., 2014). At this checkpoint, Atg13 can also be phosphorylated by PKA kinase activity confined to the cytosolic face of the PM (Yorimitsu et al., 2007; Stephan et al., 2009, 2010). The TORC1- and PKA-driven phosphorylation of Atg13 at checkpoint 9 inhibits autophagosome formation in the cytosol, thus suppressing the anti-aging process of autophagy (Yorimitsu et al., 2007; Stephan et al., 2009, 2010; Shimobayashi and Hall, 2014) (Figure 1). Furthermore, a study on the methionine restriction-induced delay of yeast chronological aging revealed that the excess of methionine in the cytosol can trigger a pro-aging cellular pattern at checkpoint 9 because it weakens autophagy, either by stimulating TORC1 at the vacuolar surface or by attenuating autophagosome formation in the cytosol (Ruckenstuhl et al., 2014). Such methionine-driven weakening of autophagy accelerates aging by decreasing the extent of vacuolar acidification and by increasing acetic acid accumulation in cultural medium (Wu et al., 2013; Johnson and Johnson, 2014) (Figure 1). Moreover, mitochondria, peroxisomes and the cytosol house individual reactions for the synthesis of the polyamine spermidine (Minois et al., 2011; Beach and Titorenko, 2013; Minois, 2014). At checkpoint 9, spermidine inhibits the histone acetyltransferases Iki3 and Sas3 (Eisenberg et al., 2009). Although such spermidine-driven inhibition of Iki3 and Sas3 causes global decline in the acetylation of histone H3 and silencing of numerous genes in the nucleus, histones in the promoter regions of several ATG (autophagy) genes get acetylated under these conditions (Eisenberg et al., 2009; Morselli et al., 2009, 2011; Madeo et al., 2010). The resulting selective activation of transcription of these genes at checkpoint 9 promotes the anti-aging process of autophagy (Figure 1). Also, a fraction of acetic acid in the cytosol can be imported into the nucleus and then converted into acetyl-CoA in the Acs2-dependent reaction (Eisenberg et al., 2014). At checkpoint 9, this acetyl-CoA selectively represses transcription of nuclear ATG genes, thus suppressing the anti-aging process of autophagy (Eisenberg et al., 2014; Schroeder et al., 2014) (Figure 1).
Chronologically aging yeast cells produce acetic acid as follows: (1) it is generated as the alternative product of glucose fermentation in the cytosol; and (2) it is formed in the Ald4-dependent reaction in mitochondria, from which acetic acid can be released into the cytosol (Burtner et al., 2009; Fraenkel, 2011; Longo et al., 2012; Arlia-Ciommo et al., 2014a). At the late-life checkpoint 10 in ST phase, a pool of acetic acid in the cytosol accelerates yeast chronological aging because it elicits an age-related form of apoptotic programmed death (Burtner et al., 2009, 2011; Longo et al., 2012; Murakami et al., 2012; Arlia-Ciommo et al., 2014a) (Figure 1).
While at the early-life checkpoint 3 trehalose is essential for maintaining an anti-aging pattern of cellular proteostasis (see above), this non-reducing disaccharide sets off a pro-aging cellular pattern at the late-life checkpoint 11 in ST phase. This is because in chronologically “old” yeast cells, which do not grow or divide, trehalose covers hydrophobic amino acid side chains of misfolded and unfolded proteins (Goldberg et al., 2009b; Kyryakov et al., 2012; Arlia-Ciommo et al., 2014a). Such side chains are needed to be recognized by a group of molecular chaperones that help to refold these misfolded and unfolded proteins (Chen et al., 2011; Lindquist and Kelly, 2011; Taylor and Dillin, 2011; Kim et al., 2013). By competing with molecular chaperones for binding to such clusters of hydrophobic amino acids, trehalose attenuates the anti-aging process of maintaining cellular proteostasis (Goldberg et al., 2009b; Kyryakov et al., 2012; Arlia-Ciommo et al., 2014a) (Figure 1).
At the late-life checkpoint 12 in ST phase, the excessive accumulation of free (non-esterified) fatty acids (FFA) and diacylglycerol (DAG) in cellular membranes accelerates yeast chronological aging because it triggers an age-related form of programmed cell death called liponecrosis (Goldberg et al., 2009a,b; Arlia-Ciommo et al., 2014a, 2016; Richard et al., 2014) (Figure 1). ATP, which is produced mainly in mitochondria, slows age-related liponecrosis by providing energy needed for the detoxification of FFA in the ER through the incorporation of FFA into triacylglycerols (TAG) and other neutral lipids (Arlia-Ciommo et al., 2014a; Richard et al., 2014; Sheibani et al., 2014). Ethanol, a product of glucose fermentation, accelerates age-related liponecrosis by suppressing peroxisomal oxidation of FFA that are generated in LD due to lipolysis of TAG and other neutral lipids (Goldberg et al., 2009a,b; Arlia-Ciommo et al., 2014a; Beach et al., 2015a). The sirtuin deacetylase Sir2 promotes ethanol accumulation by inactivating the Adh2 isoform of alcohol dehydrogenase, which is required for ethanol catabolism (Fabrizio et al., 2005) (Figure 1).
Conclusion
In this review, we analyzed mechanisms through which temporally and spatially coordinated organelle-organelle and organelle-cytosol communications impact yeast chronological aging. Our analysis indicates that these communications are integrated into a convoluted network involving unidirectional and bidirectional movements of certain metabolites between cellular compartments. Different changes in the intracellular concentrations and the rates of movement of these metabolites are restricted to critical longevity-defining periods of chronological lifespan called checkpoints. Certain proteins known as master regulators can detect the changes of the key metabolites at each of these checkpoints. The checkpoint-specific master regulator proteins contribute to setting up a pro- or anti-aging cellular pattern because each of these proteins modulates certain longevity-defining cellular processes. Future work will aim at understanding how certain dietary and pharmacological interventions known to delay aging can modulate information flow within the intricate network of intercompartmental communications.
Author Contributions
PD and VT wrote the text. VT prepared the figure.
Funding
This work was supported by grant RGPIN 2014-04482 from the NSERC of Canada. PD was supported by the Concordia University Graduate Fellowship Award.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to all laboratory members for stimulating discussions. We apologize to those whose work has not been cited owing to space limitations. VT is a Concordia University Research Chair in Genomics, Cell Biology and Aging.
References
Arlia-Ciommo, A., Leonov, A., Piano, A., Svistkova, V., and Titorenko, V. I. (2014a). Cell- autonomous mechanisms of chronological aging in the yeast Saccharomyces cerevisiae. Microbial Cell 1, 164–178. doi: 10.15698/mic2014.06.152
Arlia-Ciommo, A., Piano, A., Leonov, A., Svistkova, V., and Titorenko, V. I. (2014b). Quasi-programmed aging of budding yeast: a trade-off between programmed processes of cell proliferation, differentiation, stress response, survival and death defines yeast lifespan. Cell Cycle 13, 3336–3349. doi: 10.4161/15384101.2014.965063
Arlia-Ciommo, A., Svistkova, V., Mohtashami, S., and Titorenko, V. I. (2016). A novel approach to the discovery of anti-tumor pharmaceuticals: searching for activators of liponecrosis. Oncotarget 7, 5204–5225. doi: 10.18632/oncotarget.6440
Beach, A., Burstein, M. T., Richard, V. R., Leonov, A., Levy, S., and Titorenko, V. I. (2012). Integration of peroxisomes into an endomembrane system that governs cellular aging. Front. Physiol. 3:283. doi: 10.3389/fphys.2012.00283
Beach, A., Leonov, A., Arlia-Ciommo, A., Svistkova, V., Lutchman, V., and Titorenko, V. I. (2015a). Mechanisms by which different functional states of mitochondria define yeast longevity. Int. J. Mol. Sci. 16, 5528–5554. doi: 10.3390/ijms16035528
Beach, A., Richard, V. R., Bourque, S., Boukh-Viner, T., Kyryakov, P., Gomez-Perez, A., et al. (2015b). Lithocholic bile acid accumulated in yeast mitochondria orchestrates a development of an anti-aging cellular pattern by causing age-related changes in cellular proteome. Cell Cycle 14, 1643–1656. doi: 10.1080/15384101.2015.1026493
Beach, A., and Titorenko, V. I. (2011). In search of housekeeping pathways that regulate longevity. Cell Cycle 10, 3042–3044. doi: 10.4161/cc.10.18.16947
Beach, A., and Titorenko, V. I. (2013). Essential roles of peroxisomally produced and metabolized biomolecules in regulating yeast longevity. Subcell. Biochem. 69, 153–167. doi: 10.1007/978-94-007-6889-5_9
Bonawitz, N. D., Chatenay-Lapointe, M., Pan, Y., and Shadel, G. S. (2007). Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 5, 265–277. doi: 10.1016/j.cmet.2007.02.009
Boy-Marcotte, E., Perrot, M., Bussereau, F., Boucherie, H., and Jacquet, M. (1998). Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180, 1044–1052.
Brandes, N., Tienson, H., Lindemann, A., Vitvitsky, V., Reichmann, D., Banerjee, R., et al. (2013). Time line of redox events in aging postmitotic cells. Elife 2:e00306. doi: 10.7554/eLife.00306
Burstein, M. T., Kyryakov, P., Beach, A., Richard, V. R., Koupaki, O., Gomez-Perez, A., et al. (2012). Lithocholic acid extends longevity of chronologically aging yeast only if added at certain critical periods of their lifespan. Cell Cycle 11, 3443–3462. doi: 10.4161/cc.21754
Burtner, C. R., Murakami, C. J., Kennedy, B. K., and Kaeberlein, M. (2009). A molecular mechanism of chronological aging in yeast. Cell Cycle 8, 1256–1270. doi: 10.4161/cc.8.8.8287
Burtner, C. R., Murakami, C. J., Olsen, B., Kennedy, B. K., and Kaeberlein, M. (2011). A genomic analysis of chronological longevity factors in budding yeast. Cell Cycle 10, 1385–1396. doi: 10.4161/cc.10.9.15464
Cai, L., and Tu, B. P. (2012). Driving the cell cycle through metabolism. Annu. Rev. Cell Dev. Biol. 28, 59–87. doi: 10.1146/annurev-cellbio-092910-154010
Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., et al. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323–337. doi: 10.1091/mbc.12.2.323
Chen, B., Retzlaff, M., Roos, T., and Frydman, J. (2011). Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 3:a004374. doi: 10.1101/cshperspect.a004374
Conrad, M., Schothorst, J., Kankipati, H. N., Van Zeebroeck, G., Rubio-Texeira, M., and Thevelein, J. M. (2014). Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38, 254–299. doi: 10.1111/1574-6976.12065
Crespo, J. L., Powers, T., Fowler, B., and Hall, M. N. (2002). The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. U.S.A. 99, 6784–6789.
Delaney, J. R., Murakami, C., Chou, A., Carr, D., Schleit, J., Sutphin, G. L., et al. (2013). Dietary restriction and mitochondrial function link replicative and chronological aging in Saccharomyces cerevisiae. Exp. Gerontol. 48, 1006–1013. doi: 10.1016/j.exger.2012.12.001
Denoth Lippuner, A., Julou, T., and Barral, Y. (2014). Budding yeast as a model organism to study the effects of age. FEMS Microbiol. Rev. 38, 300–325. doi: 10.1111/1574-6976.12060
Eisenberg, T., Knauer, H., Schauer, A., Büttner, S., Ruckenstuhl, C., Carmona-Gutierrez, D., et al. (2009). Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11, 1305–1314. doi: 10.1038/ncb1975
Eisenberg, T., Schroeder, S., Andryushkova, A., Pendl, T., Küttner, V., Bhukel, A., et al. (2014). Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme A stimulates autophagy and prolongs lifespan. Cell Metab. 19, 431–444. doi: 10.1016/j.cmet.2014.02.010
Eltschinger, S., and Loewith, R. (2016). TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol. 26, 148–159. doi: 10.1016/j.tcb.2015.10.003
Epstein, C. B., Waddle, J. A., Hale, W., Davé, V., Thornton, J., Macatee, T. L., et al. (2001). Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 12, 297–308. doi: 10.1091/mbc.12.2.297
Fabrizio, P., Gattazzo, C., Battistella, L., Wei, M., Cheng, C., McGrew, K., et al. (2005). Sir2 blocks extreme life-span extension. Cell 123, 655–667. doi: 10.1016/j.cell.2005.08.042
Fabrizio, P., Pozza, F., Pletcher, S. D., Gendron, C. M., and Longo, V. D. (2001). Regulation of longevity and stress resistance by Sch9 in yeast. Science 292, 288–290. doi: 10.1126/science.1059497
Fontana, L., Partridge, L., and Longo, V. D. (2010). Extending healthy life span – from yeast to humans. Science 328, 321–326. doi: 10.1126/science.1172539
Fraenkel, D. G. (2011). Yeast Intermediary Metabolism. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Ghavidel, A., Baxi, K., Ignatchenko, V., Prusinkiewicz, M., Arnason, T. G., Kislinger, T., et al. (2015). A genome scale screen for mutants with delayed exit from mitosis: Ire1-independent induction of autophagy integrates ER homeostasis into mitotic lifespan. PLoS Genet. 11:e1005429. doi: 10.1371/journal.pgen.1005429
Goldberg, A. A., Bourque, S. D., Kyryakov, P., Boukh-Viner, T., Gregg, C., Beach, A., et al. (2009a). A novel function of lipid droplets in regulating longevity. Biochem. Soc. Trans. 37, 1050–1055. doi: 10.1042/BST0371050
Goldberg, A. A., Bourque, S. D., Kyryakov, P., Gregg, C., Boukh-Viner, T., Beach, A., et al. (2009b). Effect of calorie restriction on the metabolic history of chronologically aging yeast. Exp. Gerontol. 44, 555–571. doi: 10.1016/j.exger.2009.06.001
Goldberg, A. A., Richard, V. R., Kyryakov, P., Bourque, S. D., Beach, A., Burstein, M. T., et al. (2010). Chemical genetic screen identifies lithocholic acid as an anti-aging compound that extends yeast chronological life span in a TOR-independent manner, by modulating housekeeping longevity assurance processes. Aging (Albany NY). 2, 393–414. doi: 10.18632/aging.100168
Henderson, K. A., Hughes, A. L., and Gottschling, D. E. (2014). Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. Elife 3:e03504. doi: 10.7554/eLife.03504
Hiltunen, J. K., Mursula, A. M., Rottensteiner, H., Wierenga, R. K., Kastaniotis, A. J., and Gurvitz, A. (2003). The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 27, 35–64. doi: 10.1016/S0168-6445(03)00017-2
Hinnebusch, A. G. (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450. doi: 10.1146/annurev.micro.59.031805.133833
Huang, X., Withers, B. R., and Dickson, R. C. (2014). Sphingolipids and lifespan regulation. Biochim. Biophys. Acta 1841, 657–664. doi: 10.1016/j.bbalip.2013.08.006
Huber, A., Bodenmiller, B., Uotila, A., Stahl, M., Wanka, S., Gerrits, B., et al. (2009). Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23, 1929–1943. doi: 10.1101/gad.532109
Hughes, A. L., and Gottschling, D. E. (2012). An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261–265. doi: 10.1038/nature11654
Hughes, A. L., Hughes, C. E., Henderson, K. A., Yazvenko, N., and Gottschling, D. E. (2016). Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. Elife 5:e13943. doi: 10.7554/eLife.13943
Janssens, G. E., Meinema, A. C., González, J., Wolters, J. C., Schmidt, A., Guryev, V., et al. (2015). Protein biogenesis machinery is a driver of replicative aging in yeast. Elife 4: e08527. doi: 10.7554/eLife.08527
Janssens, G. E., and Veenhoff, L. M. (2016). Evidence for the hallmarks of human aging in replicatively aging yeast. Microbial Cell 3, 263–274. doi: 10.15698/mic2016.07.510
Jazwinski, S. M. (2012). The retrograde response and other pathways of interorganelle communication in yeast replicative aging. Subcell. Biochem. 57, 79–100. doi: 10.1007/978-94-007-2561-4_4
Jazwinski, S. M. (2013). The retrograde response: when mitochondrial quality control is not enough. Biochim. Biophys. Acta 1833, 400–409. doi: 10.1016/j.bbamcr.2012.02.010
Jazwinski, S. M. (2014). The retrograde response: a conserved compensatory reaction to damage from within and from without. Prog. Mol. Biol. Transl. Sci. 127, 133–154. doi: 10.1016/B978-0-12-394625-6.00005-2
Jazwinski, S. M. (2015). Mitochondria to nucleus signaling and the role of ceramide in its integration into the suite of cell quality control processes during aging. Ageing Res. Rev. 23, 67–74. doi: 10.1016/j.arr.2014.12.007
Jazwinski, S. M., and Kriete, A. (2012). The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Front. Physiol. 3:139. doi: 10.3389/fphys.2012.00139
Jewell, J. L., Russell, R. C., and Guan, K. L. (2013). Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14, 133–139. doi: 10.1038/nrm3522
Johnson, J. E., and Johnson, F. B. (2014). Methionine restriction activates the retrograde response and confers both stress tolerance and lifespan extension to yeast, mouse and human cells. PLoS ONE 9:e97729. doi: 10.1371/journal.pone.0097729
Kaeberlein, M. (2010). Lessons on longevity from budding yeast. Nature 464, 513–519. doi: 10.1038/nature08981
Kim, Y. E., Hipp, M. S., Bracher, A., Hayer-Hartl, M., and Hartl, F. U. (2013). Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355. doi: 10.1146/annurev-biochem-060208-092442
Kyryakov, P., Beach, A., Richard, V. R., Burstein, M. T., Leonov, A., Levy, S., et al. (2012). Caloric restriction extends yeast chronological lifespan by altering a pattern of age-related changes in trehalose concentration. Front. Physiol. 3:256. doi: 10.3389/fphys.2012.00256
Laplante, M., and Sabatini, D. M. (2012). mTOR signaling in growth control and disease. Cell 274–293. doi: 10.1016/j.cell.2012.03.017
Lee, P., Cho, B. R., Joo, H. S., and Hahn, J. S. (2008). Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 70, 882–895. doi: 10.1111/j.1365-2958.2008.06450.x
Leonov, A., and Titorenko, V. I. (2013). A network of interorganellar communications underlies cellular aging. IUBMB Life 65, 665–674. doi: 10.1002/iub.1183
Lindquist, S. L., and Kelly, J. W. (2011). Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: progress and prognosis. Cold Spring Harb. Perspect. Biol. 3:a004507. doi: 10.1101/cshperspect.a004507
Liu, K., Zhang, X., Lester, R. L., and Dickson, R. C. (2005). The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 280, 22679–22687. doi: 10.1074/jbc.M502972200
Longo, V. D., Shadel, G. S., Kaeberlein, M., and Kennedy, B. (2012). Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 16, 18–31. doi: 10.1016/j.cmet.2012.06.002
Madeo, F., Tavernarakis, N., and Kroemer, G. (2010). Can autophagy promote longevity? Nat. Cell Biol. 12, 842–846. doi: 10.1038/ncb0910-842
Martínez-Pastor, M. T., Marchler, G., Schüller, C., Marchler-Bauer, A., Ruis, H., and Estruch, F. (1996). The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15, 2227–2235.
McCormick, M. A., Delaney, J. R., Tsuchiya, M., Tsuchiyama, S., Shemorry, A., Sim, S., et al. (2015). A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 22, 895–906. doi: 10.1016/j.cmet.2015.09.008
Medkour, Y., Svistkova, V., and Titorenko, V. I. (2016a). Cell-nonautonomous mechanisms underlying cellular and organismal aging. Int. Rev. Cell Mol. Biol. 321, 259–297. doi: 10.1016/bs.ircmb.2015.09.003
Medkour, Y., and Titorenko, V. I. (2016b). Mitochondria operate as signaling platforms in yeast aging. Aging (Albany NY). 8, 212–213.
Medvedik, O., Lamming, D. W., Kim, K. D., and Sinclair, D. A. (2007). MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 5:e261. doi: 10.1371/journal.pbio.0050261
Minois, N. (2014). Molecular basis of the ‘anti-aging’ effect of spermidine and other natural polyamines – a mini-review. Gerontology 60, 319–326. doi: 10.1159/000356748
Minois, N., Carmona-Gutierrez, D., and Madeo, F. (2011). Polyamines in aging and disease. Aging (Albany NY). 3, 716–732. doi: 10.18632/aging.100361
Mirisola, M. G., and Longo, V. D. (2012). Acetic acid and acidification accelerate chronological and replicative aging in yeast. Cell Cycle 11, 3532–3533.
Mirisola, M. G., and Longo, V. D. (2013). A radical signal activates the epigenetic regulation of longevity. Cell Metab. 17, 812–813. doi: 10.1016/j.cmet.2013.05.015
Molon, M., Zadrag-Tecza, R., and Bilinski, T. (2015). The longevity in the yeast Saccharomyces cerevisiae: a comparison of two approaches for assessment the lifespan. Biochem. Biophys. Res. Commun. 460, 651–656. doi: 10.1016/j.bbrc.2015.03.085
Morselli, E., Galluzzi, L., Kepp, O., Criollo, A., Maiuri, M. C., Tavernarakis, N., et al. (2009). Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY). 1, 961–970. doi: 10.18632/aging.100110
Morselli, E., Mariño, G., Bennetzen, M. V., Eisenberg, T., Megalou, E., Schroeder, S., et al. (2011). Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 192, 615–629. doi: 10.1083/jcb.201008167
Murakami, C., Delaney, J. R., Chou, A., Carr, D., Schleit, J., Sutphin, G. L., et al. (2012). pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast. Cell Cycle 11, 3087–3096. doi: 10.4161/cc.21465
Ocampo, A., Liu, J., Schroeder, E. A., Shadel, G. S., and Barrientos, A. (2012). Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 16, 55–67. doi: 10.1016/j.cmet.2012.05.013
Pan, Y., and Shadel, G. S. (2009). Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging (Albany NY). 1, 131–145. doi: 10.18632/aging.100016
Polymenis, M., and Kennedy, B. K. (2012). Chronological and replicative lifespan in yeast: do they meet in the middle? Cell Cycle 11, 3531–3532. doi: 10.4161/cc.22041
Powers, R. W., Kaeberlein, M., Caldwell, S. D., Kennedy, B. K., and Fields, S. (2006). Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20, 174–184. doi: 10.1101/gad.1381406
Richard, V. R., Beach, A., Piano, A., Leonov, A., Feldman, R., Burstein, M. T., et al. (2014). Mechanism of liponecrosis, a distinct mode of programmed cell death. Cell Cycle 13, 3707–3726. doi: 10.4161/15384101.2014.965003
Roelants, F. M., Torrance, P. D., and Thorner, J. (2004). Differential roles of PDK1- and PDK2- phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology 150, 3289–3304. doi: 10.1099/mic.0.27286-0
Ruckenstuhl, C., Netzberger, C., Entfellner, I., Carmona-Gutierrez, D., Kickenweiz, T., Stekovic, S., et al. (2014). Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 10:e1004347. doi: 10.1371/journal.pgen.1004347
Schmitt, A. P., and McEntee, K. (1996). Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 93, 5777–5782. doi: 10.1073/pnas.93.12.5777
Schroeder, E. A., Raimundo, N., and Shadel, G. S. (2013). Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 17, 954–964. doi: 10.1016/j.cmet.2013.04.003
Schroeder, E. A., and Shadel, G. S. (2014). Crosstalk between mitochondrial stress signals regulates yeast chronological lifespan. Mech. Ageing Dev. 135, 41–49. doi: 10.1016/j.mad.2013.12.002
Schroeder, S., Pendl, T., Zimmermann, A., Eisenberg, T., Carmona-Gutierrez, D., Ruckenstuhl, C., et al. (2014). Acetyl-coenzyme A: a metabolic master regulator of autophagy and longevity. Autophagy 10, 1335–1337. doi: 10.4161/auto.28919
Sheibani, S., Richard, V. R., Beach, A., Leonov, A., Feldman, R., Khelghatybana, L., et al. (2014). Macromitophagy, neutral lipids synthesis and peroxisomal fatty acid oxidation protect yeast from “liponecrosis”, a previously unknown form of programmed cell death. Cell Cycle 13, 138–147. doi: 10.4161/cc.26885
Shimobayashi, M., and Hall, M. N. (2014). Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 15, 155–162. doi: 10.1038/nrm3757
Smets, B., Ghillebert, R., De Snijder, P., Binda, M., Swinnen, E., De Virgilio, C., et al. (2010). Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 56, 1–32. doi: 10.1007/s00294-009-0287-1
Steinkraus, K. A., Kaeberlein, M., and Kennedy, B. K. (2008). Replicative aging in yeast: the means to the end. Annu. Rev. Cell Dev. Biol. 24, 29–54. doi: 10.1146/annurev.cellbio.23.090506.123509
Stephan, J. S., Yeh, Y. Y., Ramachandran, V., Deminoff, S. J., and Herman, P. K. (2009). The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. U.S.A. 106, 17049–17054. doi: 10.1073/pnas.0903316106
Stephan, J. S., Yeh, Y. Y., Ramachandran, V., Deminoff, S. J., and Herman, P. K. (2010). The Tor and cAMP-dependent protein kinase signaling pathways coordinately control autophagy in Saccharomyces cerevisiae. Autophagy 6, 294–295. doi: 10.4161/auto.6.2.11129
Swinnen, E., Ghillebert, R., Wilms, T., and Winderickx, J. (2014). Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation in Saccharomyces cerevisiae. FEMS Yeast Res. 14, 17–32. doi: 10.1111/1567-1364.12097
Taylor, R. C., and Dillin, A. (2011). Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3:a004440. doi: 10.1101/cshperspect.a004440
Teixeira, V., and Costa, V. (2016). Unraveling the role of the Target of Rapamycin signaling in sphingolipid metabolism. Prog. Lipid Res. 61, 109–133. doi: 10.1016/j.plipres.2015.11.001
Titorenko, V. I., and Terlecky, S. R. (2011). Peroxisome metabolism and cellular aging. Traffic 12, 252–259. doi: 10.1111/j.1600-0854.2010.01144.x
Traven, A., Wong, J. M., Xu, D., Sopta, M., and Ingles, C. J. (2001). Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J. Biol. Chem. 276, 4020–4027.
Urban, J., Soulard, A., Huber, A., Lippman, S., Mukhopadhyay, D., Deloche, O., et al. (2007). Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674. doi: 10.1016/j.molcel.2007.04.020
Wanke, V., Cameroni, E., Uotila, A., Piccolis, M., Urban, J., Loewith, R., et al. (2008). Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 69, 277–285. doi: 10.1111/j.1365-2958.2008.06292.x
Wei, M., Fabrizio, P., Madia, F., Hu, J., Ge, H., Li, L. M., et al. (2009). Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 5:e1000467. doi: 10.1371/journal.pgen.1000467
Wu, Z., Song, L., Liu, S. Q., and Huang, D. (2013). Independent and additive effects of glutamic acid and methionine on yeast longevity. PLoS ONE 8:e79319. doi: 10.1371/journal.pone.0079319
Keywords: yeast, chronological aging, interorganelle communications, macromolecular damage, cellular proteostasis, hormesis, programmed cell death, signal transduction
Citation: Dakik P and Titorenko VI (2016) Communications between Mitochondria, the Nucleus, Vacuoles, Peroxisomes, the Endoplasmic Reticulum, the Plasma Membrane, Lipid Droplets, and the Cytosol during Yeast Chronological Aging. Front. Genet. 7:177. doi: 10.3389/fgene.2016.00177
Received: 02 August 2016; Accepted: 16 September 2016;
Published: 27 September 2016.
Edited by:
S. Michal Jazwinski, Tulane University, USAReviewed by:
Peter William Piper, University of Sheffield, UKSean Patrick Curran, University of Southern California, USA
Mark A. McCormick, Buck Institute for Research on Aging, USA
Copyright © 2016 Dakik and Titorenko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vladimir I. Titorenko, vladimir.titorenko@concordia.ca
 Pamela Dakik
Pamela Dakik Vladimir I. Titorenko
Vladimir I. Titorenko