- 1 Department of Microbiology, University of Texas Southwestern Medical Center, Dallas, TX, USA
- 2 Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, CA, USA
Many infections start with microbial invasion of mucosal surfaces, which are typically colonized by a community of resident microbes. A growing body of literature demonstrates that the resident microbiota plays a significant role in host susceptibility to pathogens. Recent work has largely focused on the considerable effect that the intestinal microbiota can have upon bacterial pathogenesis. These studies reveal many significant gaps in our knowledge about the mechanisms by which the resident community impacts pathogen invasion and the nature of the ensuing host immune response. It is likely that as viral pathogens become the focus of studies that examine microbiota–host interaction, substantial effects of resident communities exerted via diverse mechanisms will be elucidated. Here we provide a perspective of the exciting emerging field that examines how the intestinal microbiota influences host susceptibility to viruses.
The Intestinal Microbiota
A staggering number of microbes reside in and upon the human body (Dethlefsen et al., 2007). Our co-evolution with this microbiota has resulted in the integration of microbially derived developmental cues and metabolic capacities into human biology (Backhed et al., 2005). The vast majority of the 10–100 trillion microbial cells that inhabit the human body are found within the distal digestive tract. An altered intestinal microbiota composition has been linked to numerous pathologic states such as inflammatory bowel diseases and obesity in mouse models and in humans (Ley et al., 2005, 2006; Frank et al., 2007). In some cases an etiological relationship between the microbiota and disease state has been established (Turnbaugh et al., 2006; Garrett et al., 2007; Vijay-Kumar et al., 2010). Thus, human biology and health are intimately intertwined with the biology of our microbial inhabitants.
Ongoing large-scale sequencing efforts are providing a comprehensive sequence-based definition of this intestinal microbiota (Qin et al., 2010). Typically two bacterial phyla dominate the healthy Western adult intestine, the Firmicutes and Bacteroidetes compose >90% of the bacterial cells. Proteobacterium and Bifidobacterium and a handful of other bacterial phyla make up the remainder of the community, along with members of Archaea and Eukaryota. Despite the limited representation of bacterial phyla, at finer scales of phylogenetic resolution, such as species and strain, the intestinal microbiota is highly diverse and exhibits substantial compositional variability between people, and thus represents an “individualized fingerprint.” In addition to inter-individual variability, the microbiota exhibits temporal variability, likely due to the numerous factors that continually perturb this dynamic microbial ecosystem, such as changes in host diet and introduction of orally acquired pathogens.
The tremendous foundation of microbiome sequencing data is propelling human microbiota studies into a second phase focused on function and mechanism. Recently published metagenomic data highlight the overlap and conservation of core functionalities within the intestinal microbiome of different individuals (Turnbaugh et al., 2007, 2009). This similarity embedded within the combined genomes of different consortia reveals that each person’s gut is endowed with a core set of genes (a core microbiome) that carry out functions common to the human intestine. This new phase of investigation, which is focused on the emergent properties of the host–microbial super-organism, is attracting biomedical scientists from diverse fields, including virologists.
A Well-Supported Case for Microbiota Impact on Pathogen Susceptibility
The ability of an orally acquired pathogen to cause disease is dependant upon multiple steps that the microbiota may influence. A pathogen must navigate through the dense community of microbes, gain access to the epithelial surface, and manage the ensuing immune response. Commensal microbes present significant competition for nutrients (Sonnenburg et al., 2006), secrete microbicidal proteins (Corr et al., 2007), and elicit host responses that are fundamental to the development and maintenance of the mucosal innate and adaptive immune system (Cebra, 1999; Hooper et al., 2001; Ivanov et al., 2009). Despite this seemingly inhospitable environment, pathogens can gain access to host tissue and cause disease.
The role of the intestinal microbiota in reducing host susceptibility to enteric bacterial pathogens is commonly referred to as “colonization resistance,” although the underlying mechanisms are poorly understood. The increased susceptibility of oral antibiotic-treated mice or humans to enteric infection nicely illustrates the potential impact of microbiota perturbation on pathogen emergence (Pavia et al., 1990; Barthel et al., 2003; Doorduyn et al., 2006; Lawley et al., 2009). Furthermore, treating infectious diarrheas with live microbial supplements (i.e., probiotics) has proven successful in decreasing the duration of symptoms, but the mechanisms of action and whether such treatment is more effective for certain causative agents (e.g., bacterial vs. viral) remain obscure (Servin, 2004; Vandenplas et al., 2007; Guandalini, 2008; Allen et al., 2010). Together, these data support that alterations in host-associated microbial communities can impact an ensuing interaction with a pathogen.
Intestinal Microbiota Influences Immune Function and Pathogen Susceptibility
The gut microbiota influences the status of the host immune system during development and throughout life. The host typically maintains a dynamic and attenuated physiological state of inflammation in the mucosa that is tuned to the membership of the adjacent microbial community (Cebra, 1999). Disruption of the microbiota-dependent homeostasis can be deleterious to the host. For example, in an experimental model of chemically induced intestinal epithelial injury, cell damage is exacerbated in the absence of a microbiota or if the mice are deficient in Myd88, an adaptor protein critical for toll-like, receptor-mediated recognition of the commensal microbiota (Rakoff-Nahoum et al., 2004). Similarly, Salmonella-induced inflammation becomes more severe in the absence of an intestinal microbial community (Stecher et al., 2005). Antimicrobial protein secretion (Hooper et al., 2001), induction of secretory IgA (Moreau et al., 1978; Shroff et al., 1995), Th1:Th2 bias (Mazmanian et al., 2005), and recruitment of the myeloid and lymphoid cell lineages to the gut (Cebra et al., 1998; Zaph et al., 2008) are impacted by microbial signals derived from the microbiota.
Therefore, aspects of mucosal and systemic immunity are similar in concept to a rheostat, with the microbiota playing a significant role in modulating the set-point. The concept that microbial modulation of the immune system can alter pathogen susceptibility is illustrated in a mouse model of latent herpesvirus infection. In this model, the latent virus is capable of elevating steady state levels of IFN-γ, effectively altering the set-point of the immune system and decreasing the host susceptibility to intracellular bacterial pathogens (Barton et al., 2007). The increased resistance to Listeria monocytogenes and Yersinia pestis of mice harboring latent herpesvirus implies that such dormant viruses (most humans possess several such herpesviruses) may be viewed as symbionts with mutualistic potential. These data are also suggestive that these same principles may apply to the microbiota: specifically, host immune status may be modulated by commensal microbes prior to pathogen exposure to alter host resistance to infection.
Since the vast majority of infections begin at the mucosal surface, it is not surprising that pathogen susceptibility may be impacted by the presence or absence of resident microbes that are capable of influencing local and systemic aspects of immunity. Segmented filamentous bacteria (SFB), an intestinal resident of mice, were recently identified as necessary and sufficient to induce Th17 development in the gut (Ivanov et al., 2009). In this same study, the presence of SFB proved critical in minimizing the inflammation caused by a colitis-inducing bacterial pathogen, Citrobacter rodentium. Similarly, alteration in host susceptibility to respiratory infection following intestinal exposure to a specific microbe has been reported. In an experimental model of Cryptococcus neoformans-induced pulmonary eosinophilia, the Th2-driven pathology can be abrogated by prior introduction of C. rodentium into the digestive tract (Williams et al., 2006). C. rodentium inflammation induces a Th1-response, which reduces the Th2-skewing and C. neoformans-associated pulmonary inflammation. This protective effect is lost if mice are colonized with a mutant strain of C. rodentium that is deficient in the intimin receptor, and therefore is incapable of attaching to the host colonic epithelium and unable to induce a Th1 response. Similarly, multiple studies have illustrated the ability of either chronic or acute parasitic infection in the intestine to influence immune responses in the lung, specifically decreasing allergic airway inflammation (Wilson et al., 2005; Kitagaki et al., 2006; Gibbons et al., 2009). The ability of a group of Clostridium species to induce regulatory T-cells in the colon and decrease the severity of colitis and IgE responses in a mouse model further affirms the role of gut microbes in modulating aspects of mucosal and systemic immune function (Atarashi et al., 2010).
The Microbiota’s Direct Influence on Pathogens
In addition to acting through the immune system, the intestinal microbiota may impact pathogens more directly. The emergence of bacterial pathogens, such as Clostridium difficile and Salmonella spp., during or shortly after antibiotic treatment, suggests the loss of inhibitory effects of the microbiota upon antibiotic-induced disruption. Several bacterial pathogens, such as Salmonella and Citrobacter, appear to subvert the host immune response within the intestine causing persistent microbiota disruption thus aiding their own proliferation (Lupp et al., 2007; Stecher et al., 2007). Competing for nutrients, physically obstructing access to the mucosa, and producing antimicrobial proteins have all been proposed as mechanisms that contribute to colonization resistance (Stecher and Hardt, 2008). The bacteriocin, Abp118, produced by a Lactobacillus salivarius strain, is capable of killing L. monocytogenes, and its production in vivo is required to protect the host in a murine model of oral Listeria infection (Corr et al., 2007). This example provides a glimpse of the type of warfare that can take place amongst and between permanent and transient gut residents. Alternatively, egg hatching of Trichuris muris, a nematode that invades the intestinal epithelium of mice, is enhanced in the presence of the microbiota-derived microbes in vitro, and T. muris burden is decreased after antibiotic knock-down of the microbiota (Hayes et al., 2010). The ability of multiple pathogens to infect in the absence of apparent microbiota disruption (i.e., not all orally acquired pathogens require antibiotic treatment to emerge and cause disease) suggests that several infectious agents may be adapted to and benefit from the microbiota in a normal state.
Potential Impact of the Microbiota on Viral Infections
In general, the microbiota may enhance viral infection, reduce viral infection, or have no effect on viral infection. In instances when the microbiota influences viral infection, it may be via direct mechanisms, such as virion modification, or indirect mechanisms, such as host cell modification. Direct effects of the microbiota on viruses are likely to occur locally, whereas indirect effects may occur locally or systemically.
Several studies have shown that the microbiota has the potential to reduce viral infection and disease. First, germ-free mice have enhanced susceptibility to influenza A virus (Dolowy and Muldoon, 1964), coxsackie B virus (Schaffer et al., 1963), and Friend virus (Mirand and Grace, 1963), which suggests that the microbiota directly or indirectly limits disease in mice with a conventional flora. Second, while dengue virus is a blood-borne pathogen in mammals, it initiates mosquito infection through the enteric route. Interestingly, dengue virus replication is enhanced in the midguts of antibiotic-treated mosquitoes, which suggests that the insect microbiota limits viral replication. These effects may occur indirectly via microbiota-mediated anti-viral immune activation through the Toll pathway (Xi et al., 2008). Third, adenovirus replication is inhibited by defensins, which are antimicrobial peptides produced by the host in response to the microbiota (Gropp et al., 1999). Defensins directly bind adenovirus virions and limit replication in vitro (Smith and Nemerow, 2008). Therefore, the microbiota may limit adenovirus infection through induction of host defense mechanisms, although the effects of defensins on enteric viruses in vivo is unknown.
Conversely, several studies have shown that microbial products can indirectly enhance viral replication and disease. First, persistence of mouse mammary tumor virus is enhanced by lipopolysaccharide (LPS), a component of Gram-negative bacteria. LPS activates the innate immune pathways culminating in IL-10-mediated tolerance and viral persistence in mice (Jude et al., 2003). Second, Theiler’s murine encephalitis virus-induced replication and disease in the central nervous system is enhanced by treatment with LPS. These effects likely occur through indirect mechanisms involving increased inflammation in the central nervous system, which promotes viral replication (Pullen et al., 1995). In future work, it will be important to determine whether native microbiota-derived LPS, rather than experimentally administered LPS, is sufficient to promote viral infection. Third, human immunodeficiency virus (HIV) pathogenesis and progression to acquired immunodeficiency syndrome (AIDS) is enhanced by LPS-mediated chronic immune activation from microbial translocation through the intestinal wall (Brenchley et al., 2006). Therefore, the microbiota may indirectly enhance progression to AIDS through indirect immune activation mechanisms.
When considering effects of the microbiota on viruses, it is important to be mindful of the evolutionary potential of viruses, particularly RNA viruses. Viruses have error frequencies up to a million-fold higher than bacteria or mammals and likely evolved to replicate in their respective niches, including niches containing a microbiota. Thus, the hypothesis that some viruses will generally fare worse in the absence of the microbiota with which they have evolved will be important to test using microbiota-deficient mice.
Gaps that Need to be Addressed for the Field to Move Forward
Clearly, it is important to understand how the microbiota influences viral infections, but what are the major questions to address? The questions will be shaped by virus type. For example, the intestinal microbiota may have direct and indirect effects for enteric viruses that directly encounter them in the intestinal lumen (enterovirus, rotavirus, Norwalk virus, astrovirus, etc.). However, the intestinal microbiota may also impact non-enteric viruses through indirect mechanisms. Several viruses encounter host–microbiota at other body sites, such as skin and the genital tract (HIV, papilloma virus, herpes simplex virus, etc.). Therefore, microbiota may affect a wide variety of viruses through diverse mechanisms (Figure 1).
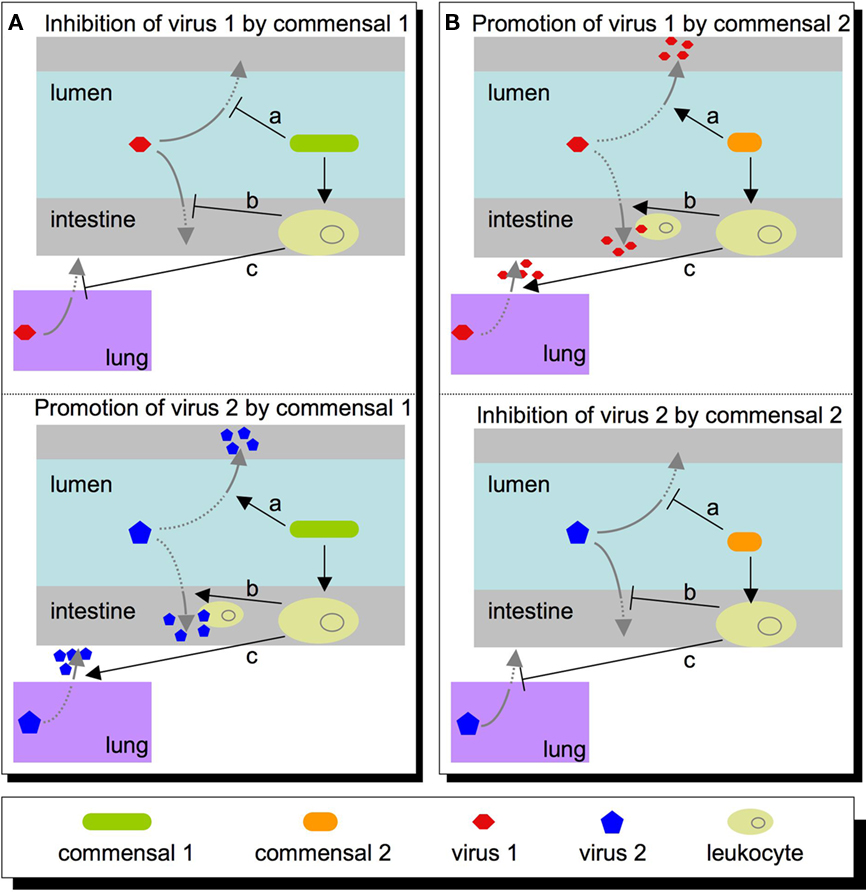
Figure 1. Potential roles that commensal microbes within the intestine may play in host susceptibility to virus. (A) A commensal may inhibit a virus (top left panel) directly (a), or indirectly at a local site (b), or at a peripheral site such as the lung (c). Conversely, this same commensal may promote viral infectivity (lower left panel) via these same mechanisms, such as directly activating the virus (a), recruiting leukocytes that promote/permit viral replication (b), or setting systemic immune parameters that are permissive for viral replication at a distant site (c). (B) Other commensals may have opposite effects on the same viruses, and alterations in microbiota composition or functional state may shift the balance in favor of the first or second scenario.
The first step is to determine the impact of the microbiota on various viruses by depleting the microbiota and quantifying viral replication and pathogenesis. Mouse models will be very useful for these experiments, provided that there is an appropriate mouse model for the virus of interest. Mice are naturally resistant to many pathogens, including viruses, often requiring the use of immunodeficient or neonatal mice for pathogenesis studies. Therefore, diverse systems may be required to overcome this “species barrier.” Microbiota depletion can be accomplished with broad-spectrum antibiotic treatment or the use of germ-free animals. Antibiotic treatment is an attractive option since it is relatively simple and inexpensive and can be used with any animal model. However, in antibiotic-treated animals, microbiota clearance is incomplete, the ratios of various species are altered, and the antibiotics can indirectly alter the host via microbiota-independent effects (Rakoff-Nahoum et al., 2004). Another negative is that antibiotic-mediated microbiota clearance requires 1 week or more, and since neonatal mice are required for some viral pathogenesis models, the antibiotic model is not ideal. Using germ-free mice overcomes many of these obstacles. Germ-free mice are completely microbiologically sterile, eliminating background and allowing recolonization with specific microbes to examine specificity of microbiota effects. However, germ-free mice require highly specialized facilities, are expensive, and a limited number of germ-free mouse strains are available. Additionally, germ-free mice have immature immune systems and altered intestinal physiology, which may complicate interpretation of experiments (Smith et al., 2007). A combination of experiments with antibiotic-treated mice and germ-free mice may be necessary to paint a complete picture of microbiota effects on viruses. Retrospective and/or prospective studies on humans treated with antibiotics and their incidence of viral infections may also be useful in evaluating microbiota effects on viruses.
Once the impact of the microbiota has been established for a given virus, the second step is to determine whether effects occur through direct or indirect mechanisms. The microbiota may directly alter the infectivity of viral particles through virion modification or processing. Microbial components or products may interact with virions to enhance or reduce their infectivity. It is also possible that virions may “hitchhike” on motile bacteria to access host cells. Conversely, the microbiota may indirectly influence viral infection by altering the host. For example, the microbiota may promote or limit viral replication by altering innate and/or adaptive immune responses, changing the susceptibility of host cells, or altering viral receptor expression. A variety of experimental systems will be useful to determine the nature of microbiota effects on viruses. For example, direct effects of microbiota on virions can be examined through in vitro mixing experiments and infectivity assays. Indirect microbiota effects on viruses mediated by innate immune responses can be examined in mouse strains lacking components of the innate response. The specificity of microbiota effects can be examined by colonizing germ-free mice with specific types of bacteria to determine whether a particular type of bacteria is sufficient to affect the virus of interest. Together, these types of experiments will reveal the basics of microbiota–virus interactions and will provide information for detailed mechanistic studies in the future.
Implications for Human Health
It is clear that the microbiota provides a major source of biological variation between individuals, and it is a contributor to numerous biological processes. Microbiota plasticity, which is significantly greater than that of our own genome, may be harnessed to modulate aspects of human health, such as optimizing resistance to certain pathogens. The link between microbiota perturbation and enteric pathogen emergence illustrates that investigation into the factors that govern microbiota stability could have great significance for humans. Alternatively, if certain members of the microbiota, or certain functions encoded within the microbiota, are co-opted by viruses and are required for invasion of host tissue, this set of interactions presents new therapeutic targets.
The possibility of leveraging the microbiota’s role in infectious disease as a platform to alter the course of infections presents many important questions. Can we rationally and reproducibly manipulate the composition and function of our intestinal microbiota? Can this manipulation be performed in ways that prevent or ameliorate human disease? To address these questions, it is critical to lay a foundation of basic research that will establish the fundamental principles that govern this complex set of interactions. Highly controlled studies in model system are needed to establish how the gut microbiota may be manipulated rationally through the use of orally administered microbes (probiotics), dietary supplementation (prebiotics), and/or microbiota-targeted small molecules to optimize host resistance to infectious disease.
As we expand our understanding of pathogens that possess diverse routes of invasion, tropisms, and/or toxins, and we investigate how this biology intersects with the host–microbiota interaction, strategies for using this information to improve human health will become evident. For example, interferon (IFN) signaling is an important part of the immune response to many viral infections; however, this system is a target for inhibition by many viruses. Identifying a microbiota manipulation (e.g., dietary change or probiotic administration) that induces a mild anti-viral-like IFN response suggests a strategy for pre-conditioning the host immune response to provide increased resistance during periods of heightened vulnerability (e.g., in infancy or during epidemics). Tuning the immune rheostat to desired set-points using microbial exposure (i.e., colonization status) provides a promising avenue that may have broad applicability. Identification of biomarkers of these host responses, which can be surveyed in feces, urine, or blood, will enable these responses to be tracked and appropriately altered.
The ability to modulate the status of the mucosal immune system has implications for improving the efficacy of oral vaccination campaigns. One such example is the global effort to eradicate wild poliovirus, which has stalled in recent years due to the failure of oral polio vaccines to achieve seroconversion of children in developing nations. In the Indian states of Uttar Pradesh and Bihar, less than 71% of children are protected against type 1 poliovirus despite an average of 15 oral doses of trivalent vaccine per child before 5 years of age; 85% protection is achieved with 10 average doses across the rest of India (Grassly et al., 2006). This failure in protection is seen in several developing regions of the world with poor sanitation and has been linked to diarrhea and the presence of enteric viruses (Myaux et al., 1996). These studies lend further evidence that the status of the mucosal immune system and/or stability of the microbiota dictates responsiveness to newly encountered agents (whether vaccines or pathogens) and places additional importance on pursuing a mechanistic understanding of how the relationship between the microbiota and mucosal immune system may be optimized to reduce viral disease and to improve the efficacy of oral vaccines.
A logical and likely extension of personal genomes is the personal microbiome, elucidating the genes encoded by the core species or taxa within an individual’s intestine. With ever the increasing understanding of how the biology of host and microbiota integrate, these two sets of DNA sequences will offer insight into current or looming pathologies by identifying potential deleterious combinations of polymorphisms in the genes of host and microbiota. The microbiome offers a set of genes much more amenable to manipulation compared to our human genome. A prerequisite for incorporating the microbiota into personalized, preventative medicine is to attain a mechanistic understanding of microbiota function and to understand how microbes within the intestine affect the resistance and susceptibility to pathogens (Nicholson et al., 2005; Zaneveld et al., 2008). The rational manipulation of our “microbial self” will soon be a reality, and it is imperative to understand which manipulations will promote human health.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Terry Dermody, Lora Hooper, and Harry Greenberg for helpful discussions and Sara Fisher for editing the manuscript. This work was funded in part by grants from the NIH-NIDDK R01DK085025 (to Justin L. Sonnenburg), and NIH-NIAID R01AI74668 and a Pew Scholar award (to Julie K. Pfeiffer).
References
Allen, S. J., Martinez, E. G., Gregorio, G. V., and Dans, L. F. (2010). Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst. Rev. 11, CD003048.
Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., Cheng, G., Yamasaki, S., Saito, T., Ohba, Y., Taniguchi, T., Takeda, K., Hori, S., Ivanov, I. I., Umesaki, Y., Itoh, K., and Honda, K. (2010). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341.
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920.
Barthel, M., Hapfelmeier, S., Quintanilla-Martinez, L., Kremer, M., Rohde, M., Hogardt, M., Pfeffer, K., Russmann, H., and Hardt, W. D. (2003). Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858.
Barton, E. S., White, D. W., Cathelyn, J. S., Brett-McClellan, K. A., Engle, M., Diamond, M. S., Miller, V. L., and Virgin, H. W. t. (2007). Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447, 326–329.
Brenchley, J. M., Price, D. A., Schacker, T. W., Asher, T. E., Silvestri, G., Rao, S., Kazzaz, Z., Bornstein, E., Lambotte, O., Altmann, D., Blazar, B. R., Rodriguez, B., Teixeira-Johnson, L., Landay, A., Martin, J. N., Hecht, F. M., Picker, L. J., Lederman, M. M., Deeks, S. G., and Douek, D. C. (2006). Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12, 1365–1371.
Cebra, J. J. (1999). Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69, 1046S–1051S.
Cebra, J. J., Periwal, S. B., Lee, G., Lee, F., and Shroff, K. E. (1998). Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev. Immunol. 6, 13–18.
Corr, S. C., Li, Y., Riedel, C. U., O’Toole, P. W., Hill, C., and Gahan, C. G. (2007). Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U.S.A. 104, 7617–7621.
Dethlefsen, L., McFall-Ngai, M., and Relman, D. A. (2007). An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449, 811–818.
Dolowy, W. C., and Muldoon, R. L. (1964). Studies of germfree animals. I. Response of mice to infection with influenza A virus. Proc. Soc. Exp. Biol. Med. 116, 365–371.
Doorduyn, Y., Van Den Brandhof, W. E., Van Duynhoven, Y. T., Wannet, W. J., and Van Pelt, W. (2006). Risk factors for Salmonella Enteritidis and Typhimurium (DT104 and non-DT104) infections in The Netherlands: predominant roles for raw eggs in Enteritidis and sandboxes in Typhimurium infections. Epidemiol. Infect. 134, 617–626.
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785.
Garrett, W. S., Lord, G. M., Punit, S., Lugo-Villarino, G., Mazmanian, S. K., Ito, S., Glickman, J. N., and Glimcher, L. H. (2007). Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45.
Gibbons, D. L., Haque, S. F., Copestake, S. L., Wells, J. W., Noble, A., Smith, A. L., and Hayday, A. C. (2009). Suppression of airway inflammation by a natural acute infection of the intestinal epithelium. Mucosal Immunol. 2, 144–155.
Grassly, N. C., Fraser, C., Wenger, J., Deshpande, J. M., Sutter, R. W., Heymann, D. L., and Aylward, R. B. (2006). New strategies for the elimination of polio from India. Science 314, 1150–1153.
Gropp, R., Frye, M., Wagner, T. O., and Bargon, J. (1999). Epithelial defensins impair adenoviral infection: implication for adenovirus-mediated gene therapy. Hum. Gene Ther. 10, 957–964.
Guandalini, S. (2008). Probiotics for children with diarrhea: an update. J. Clin. Gastroenterol. 42(Suppl. 2), S53–S57.
Hayes, K. S., Bancroft, A. J., Goldrick, M., Portsmouth, C., Roberts, I. S., and Grencis, R. K. (2010). Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science 328, 1391–1394.
Hooper, L. V., Wong, M. H., Thelin, A., Hansson, L., Falk, P. G., and Gordon, J. I. (2001). Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884.
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., Wei, D., Goldfarb, K. C., Santee, C. A., Lynch, S. V., Tanoue, T., Imaoka, A., Itoh, K., Takeda, K., Umesaki, Y., Honda, K., and Littman, D. R. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498.
Jude, B. A., Pobezinskaya, Y., Bishop, J., Parke, S., Medzhitov, R. M., Chervonsky, A. V., and Golovkina, T. V. (2003). Subversion of the innate immune system by a retrovirus. Nat. Immunol. 4, 573–578.
Kitagaki, K., Businga, T. R., Racila, D., Elliott, D. E., Weinstock, J. V., and Kline, J. N. (2006). Intestinal helminths protect in a murine model of asthma. J. Immunol. 177, 1628–1635.
Lawley, T. D., Clare, S., Walker, A. W., Goulding, D., Stabler, R. A., Croucher, N., Mastroeni, P., Scott, P., Raisen, C., Mottram, L., Fairweather, N. F., Wren, B. W., Parkhill, J., and Dougan, G. (2009). Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77, 3661–3669.
Ley, R. E., Backhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075.
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023.
Lupp, C., Robertson, M. L., Wickham, M. E., Sekirov, I., Champion, O. L., Gaynor, E. C., and Finlay, B. B. (2007). Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 204.
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118.
Mirand, E. A., and Grace, J. T. Jr. (1963). Responses of germ-free mice to friend virus. Nature 200, 92–93.
Moreau, M. C., Ducluzeau, R., Guy-Grand, D., and Muller, M. C. (1978). Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect. Immun. 21, 532–539.
Myaux, J. A., Unicomb, L., Besser, R. E., Modlin, J. F., Uzma, A., Islam, A. M., and Santosham, M. (1996). Effect of diarrhea on the humoral response to oral polio vaccination. Pediatr. Infect. Dis. J. 15, 204–209.
Nicholson, J. K., Holmes, E., and Wilson, I. D. (2005). Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. 3, 431–438.
Pavia, A. T., Shipman, L. D., Wells, J. G., Puhr, N. D., Smith, J. D., McKinley, T. W., and Tauxe, R. V. (1990). Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. J. Infect. Dis. 161, 255–260.
Pullen, L. C., Park, S. H., Miller, S. D., Dal Canto, M. C., and Kim, B. S. (1995). Treatment with bacterial LPS renders genetically resistant C57BL/6 mice susceptible to Theiler’s virus-induced demyelinating disease. J. Immunol. 155, 4497–4503.
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., Nielsen, T., Pons, N., Levenez, F., Yamada, T., Mende, D. R., Li, J., Xu, J., Li, S., Li, D., Cao, J., Wang, B., Liang, H., Zheng, H., Xie, Y., Tap, J., Lepage, P., Bertalan, M., Batto, J. M., Hansen, T., Le Paslier, D., Linneberg, A., Nielsen, H. B., Pelletier, E., Renault, P., Sicheritz-Ponten, T., Turner, K., Zhu, H., Yu, C., Li, S., Jian, M., Zhou, Y., Li, Y., Zhang, X., Li, S., Qin, N., Yang, H., Wang, J., Brunak, S., Doré, J., Guarner, F., Kristiansen, K., Pedersen, O., Parkhill, J., Weissenbach, J., MetaHIT Consortium, Bork, P., Ehrlich, S. D., and Wang, J. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65.
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S., and Medzhitov, R. (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241.
Schaffer, J., Beamer, P. R., Trexler, P. C., Breidenbach, G., and Walcher, D. N. (1963). Response of germ-free animals to experimental virus monocontamination. I. Observation on coxsackie B virus. Proc. Soc. Exp. Biol. Med. 112, 561–564.
Servin, A. L. (2004). Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28, 405–440.
Shroff, K. E., Meslin, K., and Cebra, J. J. (1995). Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 63, 3904–3913.
Smith, J. G., and Nemerow, G. R. (2008). Mechanism of adenovirus neutralization by human alpha-defensins. Cell Host Microbe 3, 11–19.
Smith, K., McCoy, K. D., and Macpherson, A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69.
Sonnenburg, J. L., Chen, C. T., and Gordon, J. I. (2006). Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4, e413. doi: 10.1371/journal.pbio.0040413
Stecher, B., and Hardt, W. D. (2008). The role of microbiota in infectious disease. Trends Microbiol. 16, 107–114.
Stecher, B., Macpherson, A. J., Hapfelmeier, S., Kremer, M., Stallmach, T., and Hardt, W. D. (2005). Comparison of Salmonella enterica serovar Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect. Immun. 73, 3228–3241.
Stecher, B., Robbiani, R., Walker, A. W., Westendorf, A. M., Barthel, M., Kremer, M., Chaffron, S., Macpherson, A. J., Buer, J., Parkhill, J., Dougan, G., von Mering, C., and Hardt, W. D. (2007). Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189. doi: 10.1371/journal.pbio.0050244
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., Sogin, M. L., Jones, W. J., Roe, B. A., Affourtit, J. P., Egholm, M., Henrissat, B., Heath, A. C., Knight, R., and Gordon, J. I. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484.
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. I. (2007). The human microbiome project. Nature 449, 804–810.
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031.
Vandenplas, Y., Salvatore, S., Vieira, M., Devreker, T., and Hauser, B. (2007). Probiotics in infectious diarrhoea in children: are they indicated? Eur. J. Pediatr. 166, 1211–1218.
Vijay-Kumar, M., Aitken, J. D., Carvalho, F. A., Cullender, T. C., Mwangi, S., Srinivasan, S., Sitaraman, S. V., Knight, R., Ley, R. E., and Gewirtz, A. T. (2010). Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231.
Williams, A. E., Edwards, L., and Hussell, T. (2006). Colonic bacterial infection abrogates eosinophilic pulmonary disease. J. Infect. Dis. 193, 223–230.
Wilson, M. S., Taylor, M. D., Balic, A., Finney, C. A., Lamb, J. R., and Maizels, R. M. (2005). Suppression of allergic airway inflammation by helminth-induced regulatory T cells. J. Exp. Med. 202, 1199–1212.
Xi, Z., Ramirez, J. L., and Dimopoulos, G. (2008). The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 4, e1000098. doi:10.1371/journal.ppat.1000098
Zaneveld, J., Turnbaugh, P. J., Lozupone, C., Ley, R. E., Hamady, M., Gordon, J. I., and Knight, R. (2008). Host-bacterial coevolution and the search for new drug targets. Curr. Opin. Chem. Biol. 12, 109–114.
Keywords: intestinal microbiota, virus, pathogen, mucosal immune system, infection
Citation: Pfeiffer JK and Sonnenburg JL (2011) The intestinal microbiota and viral susceptibility. Front. Microbio. 2:92. doi: 10.3389/fmicb.2011.00092
Received: 22 January 2011;
Paper pending published: 12 February 2011;
Accepted: 14 April 2011;
Published online: 27 April 2011
Edited by:
Peter J. Turnbaugh, Harvard University, USAReviewed by:
Alain Stintzi, Ottawa Institute of Systems Biology, CanadaDennis L. Kasper, Harvard Medical School, USA
Copyright: © 2011 Pfeiffer and Sonnenburg. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Julie K. Pfeiffer, Department of Microbiology, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX 75390-9048, USA. e-mail: julie.pfeiffer@utsouthwestern.edu; Justin L. Sonnenburg, Department of Microbiology and Immunology, Stanford University School of Medicine, 299 Campus Drive, Stanford, CA 94305-5124, USA. e-mail: jsonnenburg@stanford.edu
 Julie K. Pfeiffer1*
Julie K. Pfeiffer1*