- 1Department of Environmental Health Science, Arnold School of Public Health, University of South Carolina, Columbia, SC, USA
- 2National Ocean Service, Hollings Marine Laboratory, Charleston, SC, USA
- 3Department of Chemistry and Biochemistry, University of South Carolina, Columbia, SC, USA
Aflatoxin is a mycotoxin and a secondary metabolite, and the most potent known liver carcinogen that contaminates several important crops, and represents a significant threat to public health and the economy. Available approaches reported thus far have been insufficient to eliminate this threat, and therefore provide the rational to explore novel methods for preventing aflatoxin accumulation in the environment. Many terrestrial plants and microbes that share ecological niches and encounter the aflatoxin producers have the ability to synthesize compounds that inhibit aflatoxin synthesis. However, reports of natural aflatoxin inhibitors from marine ecosystem components that do not share ecological niches with the aflatoxin producers are rare. Here, we show that a non-pathogenic marine bacterium, Vibrio gazogenes, when exposed to low non-toxic doses of aflatoxin B1, demonstrates a shift in its metabolic output and synthesizes a metabolite fraction that inhibits aflatoxin synthesis without affecting hyphal growth in the model aflatoxin producer, Aspergillus parasiticus. The molecular mass of the predominant metabolite in this fraction was also different from the known prodigiosins, which are the known antifungal secondary metabolites synthesized by this Vibrio. Gene expression analyses using RT-PCR demonstrate that this metabolite fraction inhibits aflatoxin synthesis by down-regulating the expression of early-, middle-, and late- growth stage aflatoxin genes, the aflatoxin pathway regulator, aflR and one global regulator of secondary metabolism, laeA. Our study establishes a novel system for generation of aflatoxin synthesis inhibitors, and emphasizes the potential of the under-explored Vibrio’s silent genome for generating new modulators of fungal secondary metabolism.
Introduction
Aflatoxins are a group of secondary metabolites that are synthesized primarily by food-borne fungi such as Aspergillus parasiticus and Aspergillus flavus. These Aspergilli contaminate a variety of economically important crops such as corn, wheat, peanuts, tree nuts, dried fruits, vegetables, and medicinal plants in tropical and subtropical areas worldwide (Trail et al., 1995; Bennett and Klich, 2003; Chanda et al., 2009; Georgianna and Payne, 2009). Aflatoxin B1 is the most potent liver carcinogen known and its contamination in food and feed is a significant risk factor of liver cancer risk in humans and animals (CAST, 2003; Liu and Wu, 2010). With liver carcinomas already being the third leading cause of cancer-related mortality worldwide, the global increase in prevalence of hepatitis B virus (HBV) and immunocompromised population has increased the risk of aflatoxin-induced liver cancer (Liu and Wu, 2010). The elimination of aflatoxin accumulation in food and feed, therefore, is of primary importance for reducing its global burden on public health and economy.
Common agricultural approaches used for prevention of aflatoxin contamination in crops include use of fungicides, biocontrol agents and fungi-resistant plants, crop rotation, choice of a plantation time that avoids the aflatoxin-conducive climatic conditions, and control of environmental factors during post-harvest (Kabak et al., 2006; Wu and Khlangwiset, 2010a,b; Cary et al., 2011). However, most of these strategies are expensive, time-consuming and have demonstrated limited success. To complement these conventional strategies, the use of compounds and extracts, collected from plants and microbes that share ecological niches with the aflatoxin producers, are becoming increasingly popular (Holmes et al., 2008). Examples of these natural compounds include a variety of naturally derived volatile compounds (Greene-McDowelle et al., 1999; Zeringue, 2000; Roze et al., 2004, 2007b, 2011). Despite the significant efforts in discovering aflatoxin biocontrol agents, over 55 billion people worldwide still suffer from uncontrolled exposure to aflatoxin (Strosnider et al., 2006), resulting in an est. 25,200 to 155,000 liver cancer cases globally (Liu and Wu, 2010). Chronic low-level exposure to aflatoxins and other carcinogenic mycotoxins remains a serious health threat in the USA (Kensler et al., 1992) and it is estimated that children in rural areas of the southern USA ingest ∼40 μg aflatoxin each day through contaminated food; a situation contributing to the significant rise in aflatoxin-induced liver cancer cases (Stoloff and Friedman, 1976; Van Rensburg, 1977). NIH statistics indicate that 16,600 new cases of aflatoxin-induced liver cancer annually in the USA (Kensler et al., 2011). Therefore, the aflatoxin monitoring programs and the destruction and/or decontamination of agricultural commodities, which are adopted to meet aflatoxin levels imposed by regulations from USA and Europe for food and feed, remain an expensive and time-consuming process. Hence, development of additional novel methodologies and compounds for aflatoxin elimination is essential.
Vibrio gazogenes is an estuarine Gram-negative bacterium that is well-known for its ability to synthesize industrially relevant proteins such as amylases and proteases (Ratcliffe et al., 1982) and bactericidal and fungicidal pigments, magnesidin A (Imamura et al., 1994), prodigiosins and cycloprodigiosins (Allen et al., 1983). Previous studies have also shown that random mutations in this bacterium with 1-methyl-3-nitro-l-nitrosoguanidine expanded its metabolic output and activated the synthesis of additional bactericidal prodigiosin-related pigments, norprodigiosin and propyl prodigiosin (Alihosseini et al., 2010). This prompted us to hypothesize that a portion of the bacterium’s metabolic potential remains silent under normal growth conditions, and can be activated by genetic and environmental perturbations. In this study, we conducted alterations of metabolism in V. gazogenes through exposures to non-toxic doses of the mycotoxin, aflatoxin. While aflatoxin B1 has been reported to bind to several probiotic bacteria (Kabak et al., 2009) and has also demonstrated the ability to alter bioluminescence responses in Vibrio fischeri (Li et al., 2011), there remains a lack of understanding on how interaction of aflatoxin B1 or other mycotoxins affect fundamental bacterial cell biology. To our surprise, aflatoxin exposure to V. gazogenes diminished prodigiosin release into the growth medium, but additionally resulted in the production of a new compound that demonstrated the ability to specifically inhibit aflatoxin synthesis in the model aflatoxin producer, A. parasiticus. Here we report the findings of this study. We establish a novel system for generation of aflatoxin-inhibitors and provide a new avenue in our fundamental understanding of Vibrio cell biology.
Materials and Methods
Ethics Statement
All experiments in this study were conducted in accordance with the general safety and biohazard protocols approved by the Envixronmental Health and Safety, University of South Carolina.
Strains, Media, and Growth Conditions
Aspergillus parasiticus, SU-1 (ATCC 56775), a wild-type aflatoxin producer. The strain was grown on 100 mm Petri dishes containing potato dextrose agar for 2 weeks. Fresh spores collected from these colonies were used for all the experiments in this study that involved the use of SU-1. In these experiments the fungus was grown in aflatoxin-inducing yeast-extract-sucrose (YES); a rich growth medium (containing 2% w/v yeast extract, 6% w/v sucrose, pH 5.8), by inoculation of 104 spores per mL of liquid medium and incubated in the dark (29°C; shaking at 150 rpm). A bacterial seawater isolate of V. gazogenes, having a 98% similarity of its 16S rRNA gene to ATCC 43942 (Farmer et al., 1988), was grown in Difco Marine Broth 2216 (BD Biosciences, Sparks, MD, USA) at 28°C in a shaking incubator (190 rpm). The bacterial isolate is currently being submitted to ATCC and NCBI.
Growth Measurements of A. parasiticus and V. gazogenes
All fungal growth quantifications were performed using dry weight measurements. Briefly, the mycelia were filtered out of the growth media using a miracloth (Millipore, Billerica, MA, USA) and dried at 75°C for 6 h and the final weight was recorded. All Vibrio growth measurements were performed using absorbance readings of growth media at 600 nm.
Aflatoxin Exposure Experiments, Extraction, and Analysis of Vibrio Metabolites
Aflatoxin B1 was commercially obtained (Sigma). Three different doses (0.1, 0.2, or 0.3 μg/mL) of aflatoxin B1 were added to the Vibrio growth medium at the start of the culture. In the control flask only the vehicle (70% Methanol) was added. To extract the metabolites from V. gazogenes the cells were first harvested by centrifugation and extracted with 60 mL acetone. A portion of the filtrate was concentrated by evaporation under N2 gas. The concentrate was loaded onto a silica gel column (1.2 cm × 15 cm) and eluted with dichloromethane: methanol (80:1.5). The fractions were then purified on a silica gel column using chloroform and methanol (50:2). After purification the fractions were concentrated by evaporation under N2 gas and re-suspended in 1 mL methanol for spectral analysis.
ARMs Exposure Experiments and Aflatoxin Comparisons
Comparative semi-quantitative estimations of accumulation of aflatoxin in growth medium was performed using thin layer chromatography (TLC) of the growth medium as described previously (Banerjee et al., 2014).
Total RNA Purification and Transcript Analysis
Isolation of total RNA from fungal cells exposed to aflatoxin response metabolites from V. gazogenes was performed using 30 h old cultures. This is a time point that corresponds to the activation of secondary metabolism (hence the expression of aflatoxin genes in A. parasiticus) under the growth conditions adopted in this study (Roze et al., 2007a). Purification of total RNA and preparation of complementary DNA was performed as described previously (Chanda et al., 2009). Transcript levels were quantified by performing quantitative real-time PCR assays using SsoAdvanced universal SYBR Green supermix (BioRad Laboratories, Hercules, CA, USA) and gene-specific forward and reverse primers (Table 1) that were designed using Primer3 online software1. Reactions were performed in a CFX96 thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). As described for previous gene expression studies in A. parasiticus (Roze et al., 2007a; Chanda et al., 2009), expression value of each gene was obtained from the threshold cycle values were normalized against β-tubulin (the house keeping gene) in each sample. All RT-PCRs were performed in triplicates for each gene per sample. Data analyses were performed using CFX Manager software (Bio-Rad Laboratories).
Statistical Analyses
All statistical tests were performed using GraphPad Prism Software (GraphPad, La Jolla, CA, USA). Statistical analyses to determine for statistical significance of differences between control versus experimental groups were determined using one-way ANOVA (with sample size 3). An unpaired t-test was used to determine the gene expression effects of ARMs on A. parasiticus compared to the untreated samples. Significance was set at p < 0.05.
Results
Aflatoxin B1 Exposures Do Not Inhibit V. gazogenes Growth
As a first step in understanding how V. gazogenes, responds to aflatoxin B1, we investigated the effect of three different doses of aflatoxin B1 on the growth of V. gazogenes. The doses, 10, 30, and 50 ppb were either below, approximately equal to or fivefold higher than the highest-allowed aflatoxin level (20 ppb) in food and feed (Mazumder and Sasmal, 2001; CAST, 2003; Liu and Wu, 2010). Time-course absorbance readings were recorded to compare the growth rates of V. gazogenes, in presence of aflatoxin B1, with untreated-controls. As shown in Figure 1, none of the aflatoxin B1 doses demonstrated any significant effect on the growth of V. gazogenes.
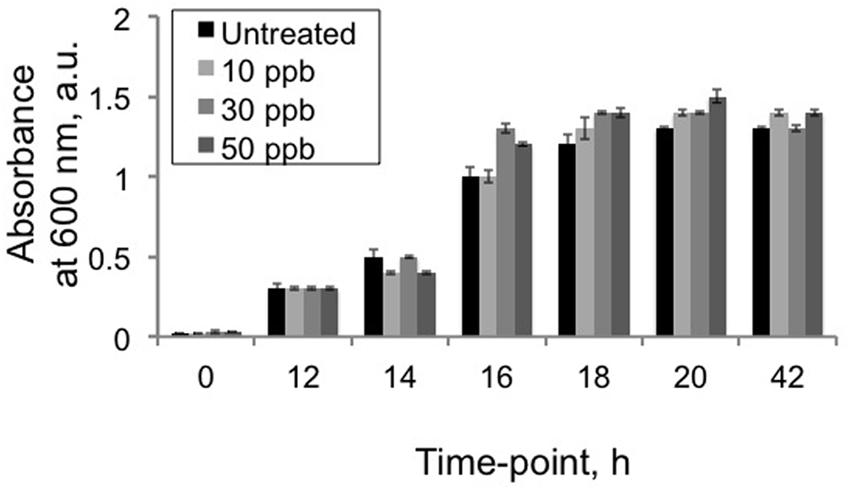
FIGURE 1. Effect of aflatoxin B1 exposure on Vibrio gazogenes growth. Growth comparisons were performed using comparisons of 600 nm absorbance values between untreated V. gazogenes cultures and cultures were supplemented with 10, 30, and 50 ppb of aflatoxin B1. Statistical significance of two-tailed p-values were determined using an unpaired t-test with sample size of 3 and significance set as p < 0.05.
Aflatoxin B1 Exposures Do Not Inhibit Prodigiosin Synthesis
Next, we investigated the effect of aflatoxin B1 exposures on the production of prodigiosins by V. gazogenes. The prodigiosin fraction was obtained from cells (either untreated control cells or cells exposed to aflatoxin B1) using our optimized laboratory protocol (see Materials and Methods). Since, the prodigiosins exhibit an absorbance peak at 530 nm (Figure 2A), this wavelength was used to compare prodigiosin levels between experimental treatments and controls at three different time-points of growth (12, 18, and 42 h). Our results (Figure 2B) demonstrated that although cells exposed to aflatoxin B1 showed a minor increase in absorbance values compared to the untreated samples, the difference was not statistically significant.
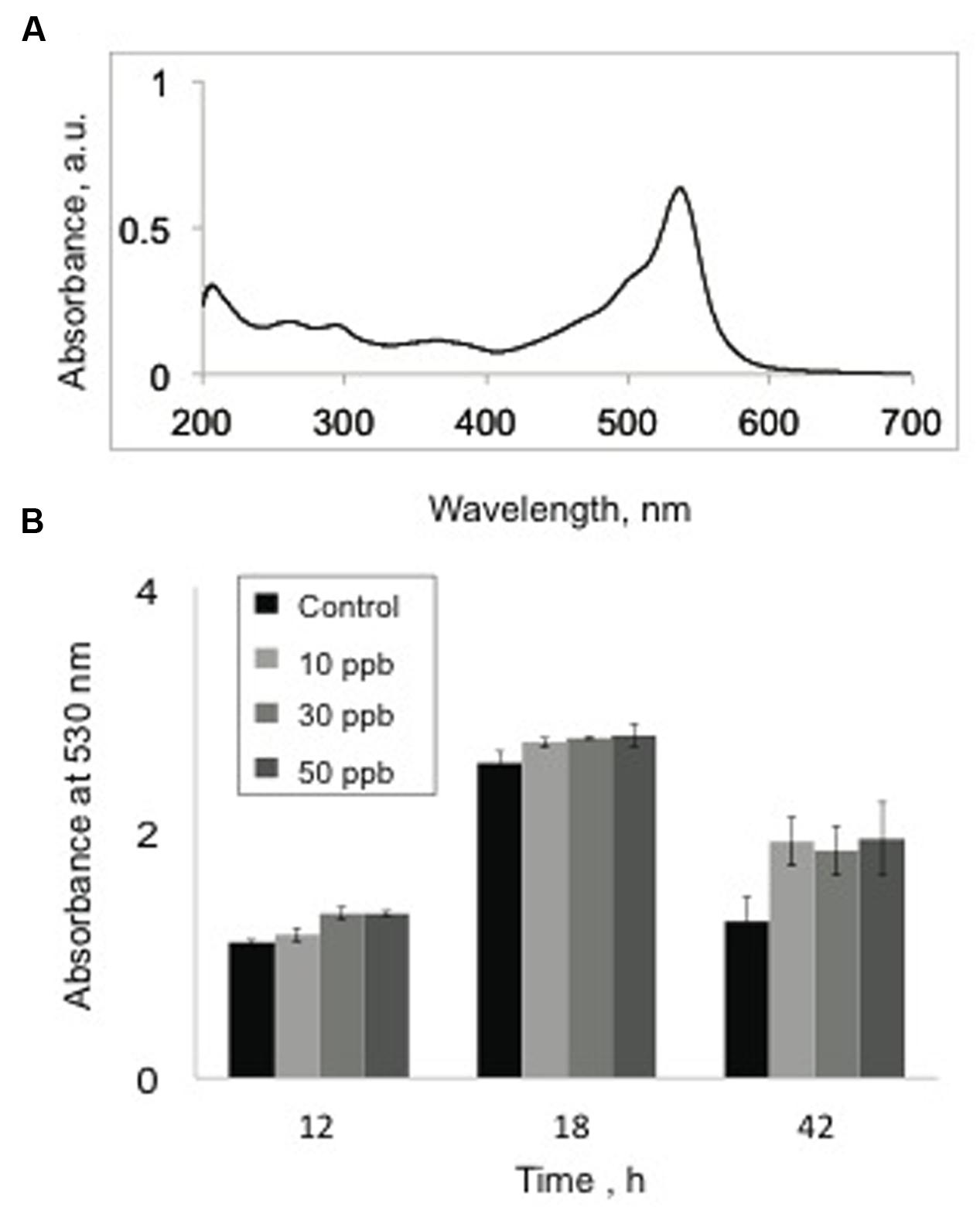
FIGURE 2. Effect of aflatoxin B1 exposure on prodigiosin production. (A) UV-Vis spectral profile of a prodigiosin-rich metabolite fraction demonstrating peak absorbance at 530 nm. (B) Comparison of absorbance values at 530 nm, of methanol extracts from untreated V. gazogenes cultures and cultures were supplemented with 10, 30, and 50 ppb of aflatoxin B1. Statistical significance of two-tailed p-values were determined using an unpaired t-test, with n = 3, and p < 0.05 as significance level.
Additional V. gazogenes Metabolite Fraction Obtained by Bacterial Exposure to Aflatoxin B1: Aflatoxin Response Metabolites (ARMs)
While growth and prodigiosin production by V. gazogenes was not affected in presence of aflatoxin B1, we observed that exposure to aflatoxin B1 resulted in a distinct alteration of color in the growth medium (Figure 3A) suggesting the presence of a different metabolite compared to untreated cells. Based on the ‘blue-shift’ in color of the growth medium (bright red to orange) upon addition of aflatoxin B1, we hypothesized that the bacterium synthesizes an additional metabolite fraction under these conditions with a corresponding absorbance lower than that of the prodigiosin fraction. To test this, we performed UV-Vis spectral analysis on the metabolite fractions of aflatoxin B1-treated samples. The Vibrio metabolite fractions obtained from aflatoxin B1 treated samples demonstrated a new absorbance peak at 240 nm (Figure 3B). This suggested that aflatoxin B1 exposure affects the cellular metabolism of V. gazogenes resulting in a different metabolite profile, compared to the untreated control. Here, we denote this additional metabolite fraction in response to aflatoxin B1 exposure as ‘aflatoxin response metabolites (ARMs).’
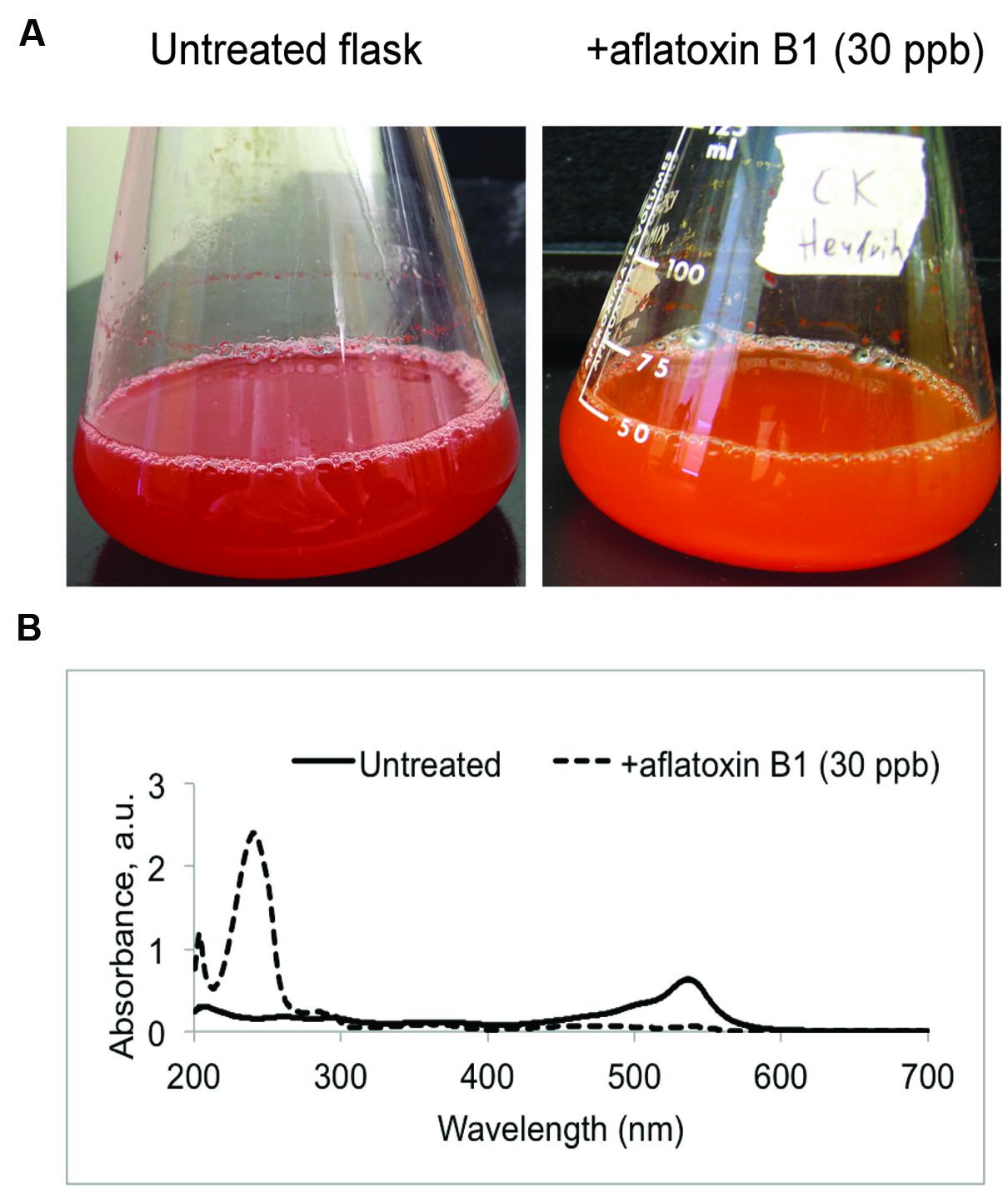
FIGURE 3. Aflatoxin-response metabolites (ARMs) produced by the bacterium V. gazogenes during exposure to aflatoxin B1 (AFB1). (A) Representative flasks demonstrating the differences in appearance of untreated V. gazogenes cultures and the aflatoxin B1 supplemented cultures. (B) Comparison of UV-Vis profiles of the methanol extracts from untreated and supplemented V. gazogenes cultures.
ARMs Do Not Inhibit A. parasiticus Growth but Inhibit Aflatoxin Synthesis
Next we proceeded to investigate whether ARMs affect the aflatoxin synthesis in the model aflatoxin B1 producer, A. parasticus. The activation of ARM production by Vibrio occurred upon addition of aflatoxin B1 to their growth medium. Therefore, we envisioned this alteration of metabolite profiles as a defensive response from Vibrio cells. We hypothesized that ARMs will have a specific inhibitory effect on aflatoxin synthesis in the producer cells. To test this we studied the growth and aflatoxin production by A. parasiticus in presence of two different doses of the ARMs metabolite fraction (1 and 2 μg per mL of growth medium); the doses were chosen arbitrarily. To compare the levels of aflatoxin biosynthesis in A. parasiticus exposed to ARMs exposed with the untreated cells, we adopted a semi-quantitative approach in which we compared the intensities of aflatoxin 1 and aflatoxin B2 bands on the TLC plates (see Materials and Methods). As predicted, our TLC results generated from 40 h cultures of A. parasiticus, demonstrated that ARMs applied at the concentration of 2 μg per mL of growth medium inhibited both aflatoxin B1 and aflatoxin B2 by approximately twofold (Figure 4A). Since, the drop in aflatoxin synthesis could also have resulted from the inhibition of A. parasiticus growth, we next compared the dry-weights of the A. parasiticus mycelia exposed to 1 and 2 μg per mL of ARMs extract with the untreated control mycelia. As shown in Figure 4B, addition of ARMs to the growth medium did not result any significant change in A. parasiticus dry weight, suggesting that inhibition of aflatoxin synthesis in A. parasiticus by ARMs was a direct effect and not a growth dependent effect.
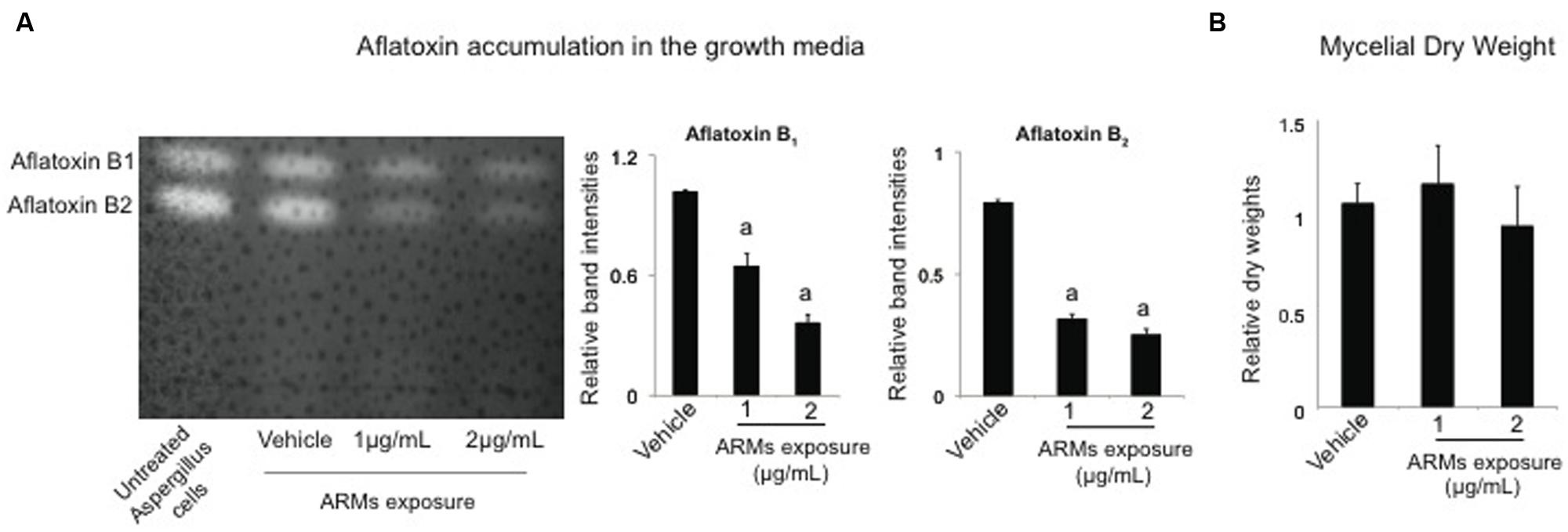
FIGURE 4. Effect of ARMs on aflatoxin biosynthesis and fungal growth. (A) Effect on aflatoxin accumulation in the growth media. Left panel, a representative TLC plate providing a qualitative comparison of aflatoxin accumulation in the untreated culture and cultures that were supplemented with 1 and 2 μg/mL ARMs extract and the vehicle (DMSO). Right panel, semi-quantitative comparative comparisons of band intensities of aflatoxin B1 and aflatoxin B2. a, significant difference in band intensity compared to the vehicle control. (B) Effect on growth. Comparison of dry-weight measurements. Bars represent measurements relative to the dry-weight of untreated cells. Statistical significance of two-tailed p-values were determined using an unpaired t-test, with sample size of n = 3 and p < 0.05 set as level of significance.
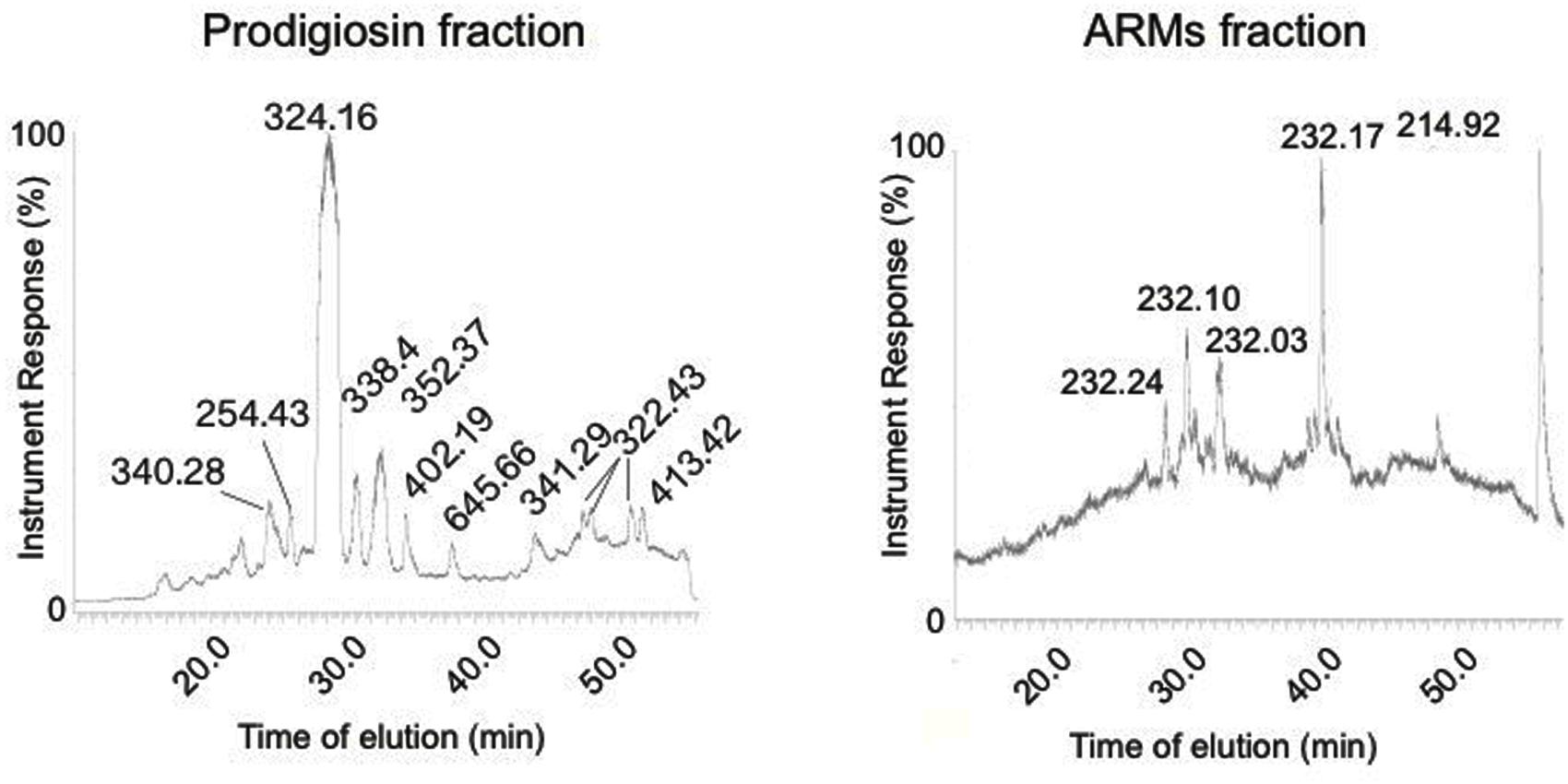
ARMs Metabolite Fraction Displays a Different HPLC Trace Compared to Prodigiosin Fraction
Our UV-Vis spectral analysis suggested that ARMs were synthesized by V. gazogenes upon exposure to aflatoxin. We then proceeded to confirm that this fraction (peak absorbance at 240 nm) was composed of metabolites of molecular masses that are different from the Vibrio’s prodigiosin fraction (peak absorbance at 530 nm). As shown in Figure 5, HPLC traces showed that the prodigiosin fraction predominantly demonstrated the expected molecular weight of 324 D, corresponding to the known prodigiosin. The HPLC trace of ARMs on the contrary was clearly different, with a predominantly displayed molecular mass 232 D, which demonstrate that the metabolite fraction of ARMs was chemically different from the Vibrio’s prodigiosin fraction. These results suggest that the differential metabolite profile in response to aflatoxin exposure can occur either due to synthesis of new metabolites by V. gazogenes or due to breakdown of prodigiosins resulting in novel smaller molecules with aflatoxin synthesis inhibitory activity.
ARMs Inhibit A. parasiticus Aflatoxin Biosynthesis at the Level of Transcript Accumulation
The fungal growth and aflatoxin results then prompted us to investigate whether aflatoxin biosynthesis was inhibited at the level of transcript accumulation of aflatoxin genes. To conduct this analysis we performed a quantitative comparison of transcript accumulation of two genes nor-1, and ver-1 that encode two enzymes, Nor-1, Vbs, and Ver-1, respectively, involved in the aflatoxin biosynthetic pathway (Chanda et al., 2009). Activation of these genes in A. parasiticus occurs at 24 h, and transcripts of all aflatoxin enzymes accumulate by 30 h, when the fungus is grown in YES growth medium (Roze et al., 2007a). Hence, we chose to examine the effects of ARMs extract on A. parasiticus at three different time-points, 24, 30 and 40 h, a time-point when aflatoxin is synthesized by the fungus at peak levels (Roze et al., 2007a). In addition to these genes, we also compared the transcript accumulation of the aflatoxin pathway regulator, aflR, at the same time points. As shown in Figure 6, nor-1, ver-1 as well as the aflR genes transcript levels demonstrated ≥5 fold reduction in presence of ARMs extract compared to the vehicle control by 30 h. Hence, our results suggest that ARMs extract reduces aflatoxin synthesis at the level of transcript accumulation.
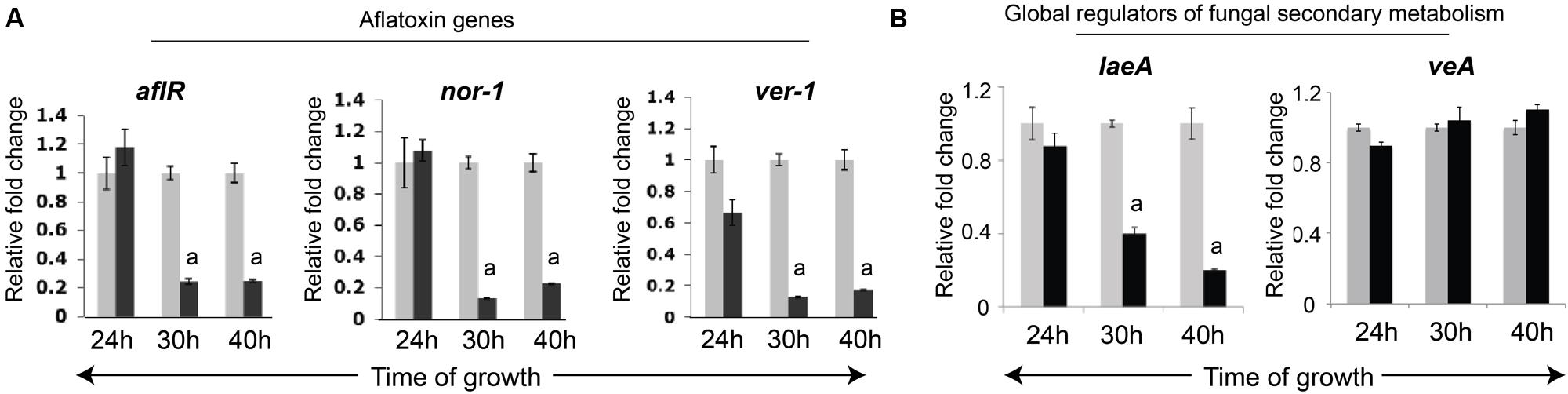
FIGURE 6. Effects of ARMs on Aspergillus parasiticus gene expression. (A) Comparison of transcript accumulation of aflatoxin-synthesis regulatory genes in A. parasiticus. mRNA levels for each gene were observed at 24 h (aflatoxin synthesis start point), 30 and 40 h time points (aflatoxin synthesis is activated and reaches peak levels by 40 h). Black bars, cells grown in presence of ARMs (2 μg/mL), Gray Bars, DMSO (vehicle) control. (B) Comparison of transcript accumulation of two global regulators of secondary metabolism, veA and laeA at the same time-points. Statistical significance of difference in transcript accumulation between control and ARMs-treated cells were determined using an unpaired t-test with sample size of 3 and two tailed p < 0.05 set as level of significance. a, p < 0.05.
ARMs Inhibit Transcript Accumulation of the Secondary Metabolism Global Regulator, laeA but not veA
Since the regulatory network of the aflatoxin biosynthesis pathway is integral to the global network of secondary metabolism in A. parasiticus (Chanda et al., 2009), we also proceeded to investigate whether, ARMs target the global regulation of secondary metabolism. One key global regulatory complex of fungal secondary metabolism is the Velvet complex which consists of proteins VeA, LaeA and other regulators (Bayram et al., 2008). Central to this complex is the cross-talk between the two global regulatory proteins, LaeA, a methyltransferase that is key to the epigenetic regulation of aflatoxin biosynthetic pathway (Bok and Keller, 2004), and VeA, a light responsive regulatory protein that migrates from cytoplasm to the nucleus in absence of light to particpate in the VeA complex (Bayram et al., 2008). In this study we investigated whether ARMs affect transcript accumulation of either laeA or veA genes. To our surprise we found that, while no significant changes occurred in veA transcripts, the laeA transcript accumulation was reduced by ∼2 fold by 30 h and ∼4 fold by 40 h (Figure 7), suggesting that ARMs inhibit aflatoxin biosynthesis at least in part, through inhibition of laeA gene activation.
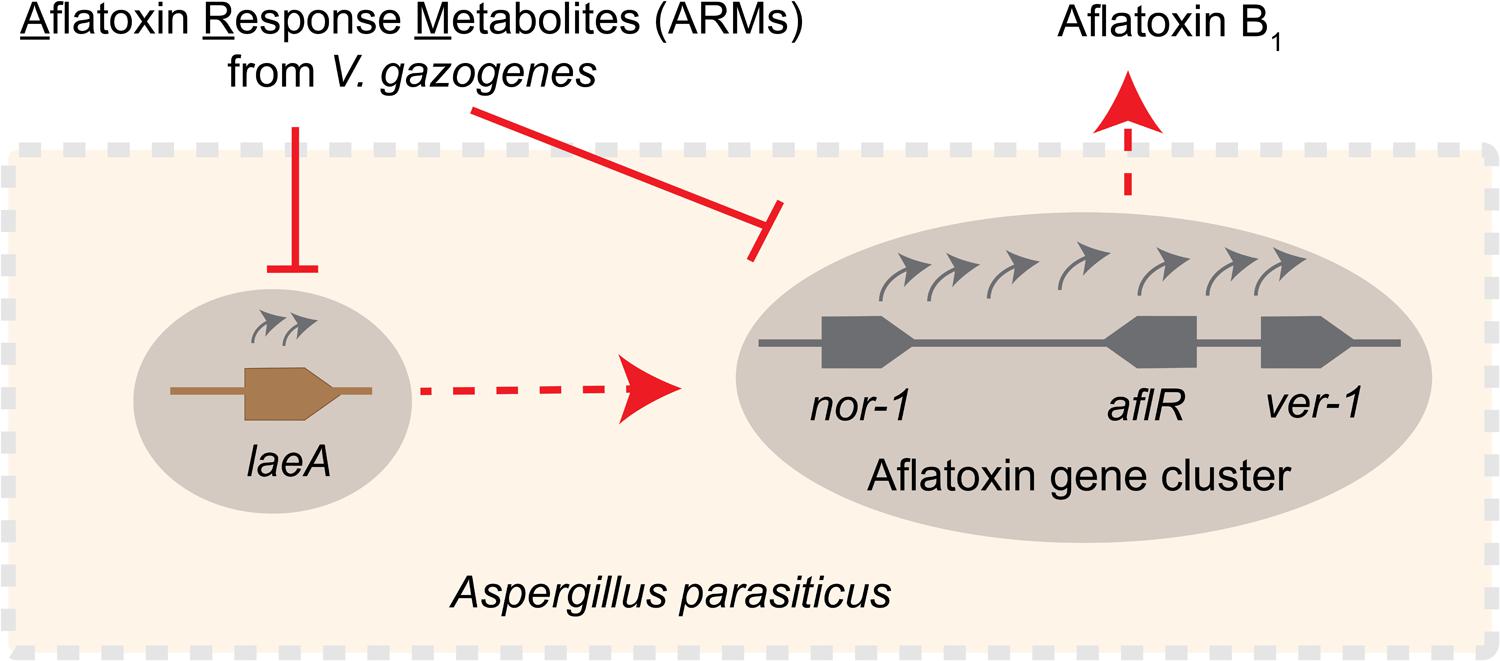
FIGURE 7. Schematic representation of the inhibitory effect of ARMs on aflatoxin biosynthesis. Current study demonstrates that ARMs inhibit aflatoxin biosynthesis in A. parasiticus at the level of gene expression. We hypothesize that the inhibition of aflatoxin genes as exemplified by the decreased nor-1, ver-1, and aflR transcripts in presence of ARMs can be the effect of one or both of the following: (1) inhibition of laeA expression, which in turn can have inhibitory impact on the activation of the aflatoxin genes, or (2) a dual inhibition caused by direct inhibition on aflatoxin gene cluster activation along with a laeA mediated inhibition. Red dotted arrows, regulatory roles established in previous studies, red solid lines, inhibitory effect, gray curved arrows, gene activation, gray solid line, schematic of the aflatoxin gene cluster showing relative positions of nor-1, ver-1, and aflR in the cluster, brown solid line, laeA gene.
Discussion
Here, we demonstrate the feasibility of a novel system for generation of aflatoxin biosynthesis inhibitors, a concept that is analogous to the generation of antibodies upon antigen exposure. Our data reveal that the estuarine bacterium V. gazogenes, upon aflatoxin exposure, produces a metabolite profile that is chemically different from untreated-cells. Upon isolation of the ARMs and applying them on the aflatoxin producer cells, we found that the metabolites inhibit aflatoxin biosynthesis at the level of transcript accumulation of aflatoxin biosynthesis regulatory genes. Based on our current study we propose two possible explanations underlying this inhibition (illustrated in the schematic in Figure 7). One possible mechanism of inhibition is through the regulation of the laeA gene activation. The laeA transcripts dropped by 2–4 fold during 30–40 h time points suggesting that ARMs inhibit the formation of the Velvet complex, a protein complex comprising LaeA protein that regulate fungal secondary metabolism (Bayram et al., 2008). Alternatively, it is also possible that in addition to laeA mediated inhibition ARMs inhibit the activation of aflatoxin genes directly. Fungal growth was not inhibited during the ARMs-mediated inhibition of aflatoxin biosynthesis, suggesting that the metabolites target secondary metabolism specifically. Future studies will identify the molecule(s) within ARMs that results in the aflatoxin inhibition. From our current preliminary studies, we postulate that two or more compounds generated in response to aflatoxin exposure act either complementarily or synergistically to inhibit aflatoxin synthesis inhibition. These collaborative effects will be determined in those functional characterization studies with the purified compounds.
It is important to emphasize that specific aflatoxin inhibitory natural products that have been characterized to-date were reported primarily from terrestrial organisms whose ecological domains likely overlap with those of the aflatoxin producers. Examples include natural products and volatiles from plants (Cleveland et al., 2009; Roze et al., 2011; Chitarrini et al., 2014), fungi (Ono et al., 1997; Yoshinari et al., 2010; Hua et al., 2014) and bacteria (Jermnak et al., 2013; Wang et al., 2013; Kong et al., 2014). Our study provides the first evidence, to the best of our knowledge, of an organism that demonstrates the ability of synthesizing aflatoxin inhibitors, while not sharing ecological niches with aflatoxin producers at all. Also this is the first report, to the best of our knowledge, of a Vibrio-producing metabolite(s) that specifically inhibit aflatoxin biosynthesis without affecting fungal growth. It is possible that mycotoxin triggered synthesis of mycotoxin inhibitors is a phenomenon that is conserved in the Vibrio species. Alternatively, it is also possible that V. gazogenes is a chemically gifted organism that has genetically evolved with the rising mycotoxin levels in the environment with global changes in climate (Kolpin et al., 2014; Rangel et al., 2015).
The effect of ARMs mediated down-regulation of laeA gene, but not veA gene suggests that the metabolites target cellular signaling receptors that specifically regulate laeA gene expression. Since LaeA is a global regulator of secondary metabolism and influences several mycotoxin biosynthetic pathways (Keller et al., 2005), we anticipate that aflatoxin inhibitor within ARMs will inhibit other mycotoxins as well. Hence, for our follow-up studies we will categorize these as secondary metabolism specific inhibitors instead denoting these as specific inhibitors against aflatoxin biosynthesis.
Current investigations in our laboratory reveal that other fungal secondary metabolites trigger synthesis of metabolite fractions in V. gazogenes that demonstrate different HPLC traces compared to either prodigiosins or ARMs fractions. These results implicate the need to examine the regulation of Vibrio genes under different environmental signals. It appears from our studies that many areas of the Vibrio genome remain silent under standard laboratory growth conditions and can be activated as needed to generate metabolites that are relevant to the public health. Our future studies will shed light on these silent areas of the V. gazogenes genome that encode the biosynthesis of the secondary metabolism modulatory metabolites; the knowledge will enable us to clone these areas on plasmids and engineer them as needed with the goal of purifying these compounds in large quantities.
Author Contributions
AC: conceived the research concept, PG, YPC, KB, KM, CM, NB, and MV-M: collected data, PG, YPC, PM, JF, AD, and AC: analyzed and interpreted the data and PG, AD, and AC: wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by funding from the Department of Environmental Health Sciences, University of South Carolina (Lab start-up funds to AC), National institute of health (R01AI120987-01 to AD) and the Office of Undergraduate Research, University of South Carolina (Magellan Scholarship and Magellan Minigrant to NB).
Footnotes
References
Alihosseini, F., Lango, J., Ju, K. S., Hammock, B. D., and Sun, G. (2010). Mutation of bacterium Vibrio gazogenes for selective preparation of colorants. Biotechnol. Prog. 26, 352–360. doi: 10.1002/btpr.346
Allen, G. R., Reichelt, J. L., and Gray, P. P. (1983). Influence of environmental factors and medium composition on Vibrio gazogenes growth and prodigiosin production. Appl. Environ. Microbiol. 45, 1727–1732.
Banerjee, S., Gummadidala, P. M., Rima, R. A., Ahmed, R. U., Kenne, G. J., Mitra, C., et al. (2014). Quantitative acoustic contrast tomography reveals unique multiscale physical fluctuations during aflatoxin synthesis in Aspergillus parasiticus. Fungal Genet. Biol. 73, 61–68. doi: 10.1016/j.fgb.2014.10.006
Bayram, O., Krappmann, S., Ni, M., Bok, J. W., Helmstaedt, K., Valerius, O., et al. (2008). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. doi: 10.1126/science.1155888
Bennett, J. W., and Klich, M. (2003). Mycotoxins. Clin. Microbiol. Rev. 16, 497–516. doi: 10.1128/CMR.16.3.497-516.2003
Bok, J. W., and Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3, 527–535. doi: 10.1128/EC.3.2.527-535.2004
Cary, J. W., Rajasekaran, K., Brown, R. L., Luo, M., Chen, Z. Y., and Bhatnagar, D. (2011). Developing resistance to aflatoxin in maize and cottonseed. Toxins (Basel) 3, 678–696. doi: 10.3390/toxins3060678
CAST (2003). Mycotoxins: Risks in Plant, Animal, and Human Systems. Task Force Report No. 139. Ames, IA: Council of Agricultural Science and Technology, 199.
Chanda, A., Roze, L. V., Kang, S., Artymovich, K. A., Hicks, G. R., Raikhel, N. V., et al. (2009). A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 106, 19533–19538. doi: 10.1073/pnas.0907416106
Chitarrini, G., Nobili, C., Pinzari, F., Antonini, A., De Rossi, P., Del Fiore, A., et al. (2014). Buckwheat achenes antioxidant profile modulates Aspergillus flavus growth and aflatoxin production. Int. J. Food Microbiol. 189, 1–10. doi: 10.1016/j.ijfoodmicro.2014.07.029
Cleveland, T. E., Carter-Wientjes, C. H., De Lucca, A. J., and Bou, S. M. (2009). Effect of soybean volatile compounds on Aspergillus flavus growth and aflatoxin production. J. Food Sci. 74, H83–H87. doi: 10.1111/j.1750-3841.2009.01078.x
Farmer, J. J. III, Hickman-Brenner, F. W., Fanning, G. R., Gordon, C. M., and Brenner, D. J. (1988). Characterization of Vibrio metschnikovii and Vibrio gazogenes by DNA-DNA hybridization and phenotype. J. Clin. Microbiol. 26, 1993–2000.
Georgianna, D. R., and Payne, G. A. (2009). Genetic regulation of aflatoxin biosynthesis: from gene to genome. Fungal Genet. Biol. 46, 113–125. doi: 10.1016/j.fgb.2008.10.011
Greene-McDowelle, D. M., Ingber, B., Wright, M. S., Zeringue, H. J. Jr., Bhatnagar, D., and Cleveland, T. E. (1999). The effects of selected cotton-leaf volatiles on growth, development and aflatoxin production of Aspergillus parasiticus. Toxicon 37, 883–893. doi: 10.1016/S0041-0101(98)00209-8
Holmes, R. A., Boston, R. S., and Payne, G. A. (2008). Diverse inhibitors of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 78, 559–572. doi: 10.1007/s00253-008-1362-0
Hua, S. S., Beck, J. J., Sarreal, S. B., and Gee, W. (2014). The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 30, 71–78. doi: 10.1007/s12550-014-0189-z
Imamura, N., Adachi, K., and Sano, H. (1994). Magnesidin A, a component of marine antibiotic magnesidin, produced by Vibrio gazogenes ATCC29988. J. Antibiot. (Tokyo) 47, 257–261. doi: 10.7164/antibiotics.47.257
Jermnak, U., Chinaphuti, A., Poapolathep, A., Kawai, R., Nagasawa, H., and Sakuda, S. (2013). Prevention of aflatoxin contamination by a soil bacterium of Stenotrophomonas sp. that produces aflatoxin production inhibitors. Microbiology 159, 902–912. doi: 10.1099/mic.0.065813-0
Kabak, B., Brandon, E. F., Var, I., Blokland, M., and Sips, A. J. (2009). Effects of probiotic bacteria on the bioaccessibility of aflatoxin B(1) and ochratoxin A using an in vitro digestion model under fed conditions. J. Environ. Sci. Health B 44, 472–480. doi: 10.1080/03601230902935154
Kabak, B., Dobson, A. D., and Var, I. (2006). Strategies to prevent mycotoxin contamination of food and animal feed: a review. Crit. Rev. Food Sci. Nutr. 46, 593–619. doi: 10.1080/10408390500436185
Keller, N. P., Turner, G., and Bennett, J. W. (2005). Fungal secondary metabolism – from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. doi: 10.1038/nrmicro1286
Kensler, T. W., Groopman, J. D., Eaton, D. L., Curphey, T. J., and Roebuck, B. D. (1992). Potent inhibition of aflatoxin-induced hepatic tumorigenesis by the monofunctional enzyme inducer 1,2-dithiole-3-thione. Carcinogenesis 13, 95–100. doi: 10.1093/carcin/13.1.95
Kensler, T. W., Roebuck, B. D., Wogan, G. N., and Groopman, J. D. (2011). Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol. Sci. 120, S28–S48. doi: 10.1093/toxsci/kfq283
Kolpin, D. W., Schenzel, J., Meyer, M. T., Phillips, P. J., Hubbard, L. E., Scott, T. M., et al. (2014). Mycotoxins: diffuse and point source contributions of natural contaminants of emerging concern to streams. Sci. Total Environ. 47, 669–676. doi: 10.1016/j.scitotenv.2013.09.062
Kong, Q., Chi, C., Yu, J., Shan, S., Li, Q., Guan, B., et al. (2014). The inhibitory effect of Bacillus megaterium on aflatoxin and cyclopiazonic acid biosynthetic pathway gene expression in Aspergillus flavus. Appl. Microbiol. Biotechnol. 98, 5161–5172. doi: 10.1007/s00253-014-5632-8
Li, X., Pan, L., and Wang, B. (2011). Influence of aflatoxin on Vibrio fischeri luminescence. Acta Microbiol. Sin. 51, 1669–1674.
Liu, Y., and Wu, F. (2010). Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ. Health Perspect. 118, 818–824. doi: 10.1289/ehp.0901388
Mazumder, P. M., and Sasmal, D. (2001). Mycotoxins – limits and regulations. Anc. Sci. Life 20, 1–19.
Ono, M., Sakuda, S., Suzuki, A., and Isogai, A. (1997). Aflastatin A, a novel inhibitor of aflatoxin production by aflatoxigenic fungi. J. Antibiot. (Tokyo) 50, 111–118. doi: 10.7164/antibiotics.50.111
Rangel, D. E., Alder-Rangel, A., Dadachova, E., Finlay, R. D., Kupiec, M., Dijksterhuis, J., et al. (2015). Fungal stress biology: a preface to the Fungal Stress Responses special edition. Curr. Genet. 61, 231–238. doi: 10.1007/s00294-015-0500-3
Ratcliffe, C., Sanders, R. L., Tittel, L., and O’Brien, R. W. (1982). Amylase and protease secretion by the marine bacterium Vibrio gazogenes. Aust. J. Biol. Sci. 35, 457–467.
Roze, L. V., Arthur, A. E., Hong, S. Y., Chanda, A., and Linz, J. E. (2007a). The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol. Microbiol. 66, 713–726. doi: 10.1111/j.1365-2958.2007.05952.x
Roze, L. V., Beaudry, R. M., Arthur, A. E., Calvo, A. M., and Linz, J. E. (2007b). Aspergillus volatiles regulate aflatoxin synthesis and asexual sporulation in Aspergillus parasiticus. Appl. Environ. Microbiol. 73, 7268–7276. doi: 10.1128/AEM.00801-07
Roze, L. V., Calvo, A. M., Gunterus, A., Beaudry, R., Kall, M., and Linz, J. E. (2004). Ethylene modulates development and toxin biosynthesis in Aspergillus possibly via an ethylene sensor-mediated signaling pathway. J. Food Prot. 67, 438–447.
Roze, L. V., Koptina, A. V., Laivenieks, M., Beaudry, R. M., Jones, D. A., Kanarsky, A. V., et al. (2011). Willow volatiles influence growth, development, and secondary metabolism in Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 92, 359–370. doi: 10.1007/s00253-011-3339-7
Stoloff, L., and Friedman, L. (1976). Information bearing on the hazard to man from aflatoxin ingestion. PAG Bull. 6, 21–32.
Strosnider, H., Azziz-Baumgartner, E., Banziger, M., Bhat, R. V., Breiman, R., Brune, M. N., et al. (2006). Workgroup report: public health strategies for reducing aflatoxin exposure in developing countries. Environ. Health Perspect. 114, 1898–1903.
Trail, F., Mahanti, N., and Linz, J. (1995). Molecular biology of aflatoxin biosynthesis. Microbiology 141(Pt 4), 755–765. doi: 10.1099/13500872-141-4-755
Van Rensburg, S. J. (1977). “Role of epidemiology in the elucidation of mycotoxin health risks,” in Mycotoxins in Human and Animal Health, eds J. V. Rodricks, C. W. Hesseltine, and M. A. Mehlman (Park Forest South, IL: Pathotox Publishers), 699–711.
Wang, K., Yan, P. S., Ding, Q. L., Wu, Q. X., Wang, Z. B., and Peng, J. (2013). Diversity of culturable root-associated/endophytic bacteria and their chitinolytic and aflatoxin inhibition activity of peanut plant in China. World J. Microbiol. Biotechnol. 29, 1–10. doi: 10.1007/s11274-012-1135-x
Wu, F., and Khlangwiset, P. (2010a). Evaluating the technical feasibility of aflatoxin risk reduction strategies in Africa. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 27, 658–676. doi: 10.1080/19440041003639582
Wu, F., and Khlangwiset, P. (2010b). Health economic impacts and cost-effectiveness of aflatoxin-reduction strategies in Africa: case studies in biocontrol and post-harvest interventions. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 27, 496–509. doi: 10.1080/19440040903437865
Yoshinari, T., Noda, Y., Yoda, K., Sezaki, H., Nagasawa, H., and Sakuda, S. (2010). Inhibitory activity of blasticidin A, a strong aflatoxin production inhibitor, on protein synthesis of yeast: selective inhibition of aflatoxin production by protein synthesis inhibitors. J. Antibiot. (Tokyo) 63, 309–314. doi: 10.1038/ja.2010.36
Keywords: aflatoxins, Vibrio gazogenes, Aspergillus parasiticus, aflatoxin response metabolites (ARMs), natural products
Citation: Gummadidala PM, Chen YP, Beauchesne KR, Miller KP, Mitra C, Banaszek N, Velez-Martinez M, Moeller PDR, Ferry JL, Decho AW and Chanda A (2016) Aflatoxin-Exposure of Vibrio gazogenes as a Novel System for the Generation of Aflatoxin Synthesis Inhibitors. Front. Microbiol. 7:814. doi: 10.3389/fmicb.2016.00814
Received: 23 February 2016; Accepted: 13 May 2016;
Published: 03 June 2016.
Edited by:
Vijai Kumar Gupta, NUI Galway, IrelandReviewed by:
Nicolas Toro, Estación Experimental del Zaidín – Consejo Superior de Investigaciones Científicas, SpainRam Prasad, Amity University, India
Copyright © 2016 Gummadidala, Chen, Beauchesne, Miller, Mitra, Banaszek, Velez-Martinez, Moeller, Ferry, Decho and Chanda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anindya Chanda, achanda@mailbox.sc.edu
†Co-primary authors
 Phani M. Gummadidala
Phani M. Gummadidala Yung Pin Chen1†
Yung Pin Chen1† Kristen P. Miller
Kristen P. Miller Chandrani Mitra
Chandrani Mitra John L. Ferry
John L. Ferry Alan W. Decho
Alan W. Decho Anindya Chanda
Anindya Chanda