- 1 Division of Biology, Kansas State University, Manhattan, KS, USA
- 2 Department of Horticulture, University of Kentucky, Lexington, KY, USA
- 3 Division of Glycoscience, Royal Institute of Technology, School of Biotechnology, AlbaNova University Centre, Stockholm, Sweden
Sterols play vital roles in plant growth and development, as components of membranes and as precursors to steroid hormones. Analysis of Arabidopsis mutants indicates that sterol composition is crucial for cellulose biosynthesis. Sterols are widespread in the plasma membrane (PM), suggesting a possible link between sterols and the multimeric cellulose synthase complex. In one possible scenario, molecular interactions in sterol-rich PM microdomains or another form of sterol-dependent membrane scaffolding may be critical for maintaining the correct subcellular localization, structural integrity and/or activity of the cellulose synthase machinery. Another possible link may be through steryl glucosides, which could act as primers for the attachment of glucose monomers during the synthesis of β−(1 → 4) glucan chains that form the cellulose microfibrils. This mini-review examines genetic and biochemical data supporting the link between sterols and cellulose biosynthesis in cell wall formation and explores potential approaches to elucidate the mechanism of this association.
Introduction
In plants, cellulose functions as a major component of the cell wall to provide mechanical support and structural integrity to various tissues. Cellulose consists of glucose molecules arranged in parallel hydrogen-bonded β-(1 → 4) glucan chains that form microfibrils of 2–4 nm in diameter and up to several micrometer in length depending on their origin (Mutwil et al., 2008; Fernandes et al., 2011). Cellulose microfibrils function in scaffolding other cell wall polymers such as hemicelluloses and pectins.
Cellulose is synthesized at the plasma membrane (PM) by terminal complexes organized as six-lobed rosettes in higher plants (Kimura et al., 1999). Current models suggest that the rosettes are organized as tetramers (Endler and Persson, 2011) or hexamers (Fernandes et al., 2011) consisting of multiple cellulose synthase catalytic subunits (CESA) arranged with a sixfold symmetry. Each CESA subunit has a predicted topology of eight transmembrane helices for anchorage in the PM (Delmer, 1999). Although much progress has been made in the identification of proteins involved in cellulose formation, our understanding of the biosynthetic process is far from complete (Guerriero et al., 2010). One major difficulty is to determine the precise composition of the cellulose synthase complex (CSC) and to solve its structure. This has proven extremely challenging using biochemical approaches because of the instability of the complex, and molecular genetics have provided only partial and essentially indirect answers to some of the most fundamental questions related to cellulose formation.
Several pieces of evidence suggest that the lipid environment of the CSC is crucial for its proper structural organization and function at the PM. In metazoans, sterols act as components of membranes, molecular ligands, or as precursors of steroid hormones. However, in plants, much less is known about the roles of sterols. They predominantly occur in the PM (Grebe et al., 2003) and are abundantly synthesized during the early stages of seed development, coincident with intense cell division and expansion (Schrick et al., 2011). Below we describe the genetic and biochemical evidence that support a link between sterols and cellulose biosynthesis, and we discuss experimental approaches to decipher the molecular mechanisms that underlie this connection.
The Roles of Sterols in Higher Plants
Compared to vertebrates, in which mainly cholesterol synthesis occurs, higher plants synthesize a complex mixture of sterols, commonly referred to as phytosterols (Benveniste, 2004). In Arabidopsis seedlings, sitosterol is the major sterol, followed by campesterol, stigmasterol, and over 20 minor sterols, many of which are biosynthetic intermediates (Schrick et al., 2000, 2002). Sitosterol and stigmasterol play major roles as PM components and are critical for membrane fluidity and permeability (Schuler et al., 1991; Grandmougin-Ferjani et al., 1997). Campesterol is a precursor of the brassinosteroids which stimulate stem elongation and cell division, and are the only plant steroids known to act as hormones (Clouse, 2011).
The first genetic evidence for the role of sterols in plant growth and development came from the identification of three Arabidopsis sterol biosynthesis mutants: fackel (fk; Jang et al., 2000; Schrick et al., 2000), cephalopod/sterol methyl transferase 1 (cph/smt1; Diener et al., 2000; Schrick et al., 2002), and hydra1 (hyd1; Schrick et al., 2002; Souter et al., 2002). These mutants exhibit cell division and expansion defects, as well as embryonic patterning defects, and correspond to sterol C-14 reductase, C-24 sterol methyltransferase, and sterol C-8,7 isomerase, respectively. The discovery that all three affect enzymes upstream of brassinosteroid DWARF (DWF) steps, coupled with the inability to rescue the mutants by brassinosteroids, led to the hypothesis that sterols participate in novel signaling pathways (Lindsey et al., 2003). Although such pathways have not been revealed to date, this idea is further supported by the characterization of smt2/cvp1 and smt3 double mutants of C-24 sterol methyltransferases that exhibit homeotic floral transformations and other developmental defects (Carland et al., 2010).
Impact of Mutations in Sterol Biosynthetic Enzymes on Cellulose Formation
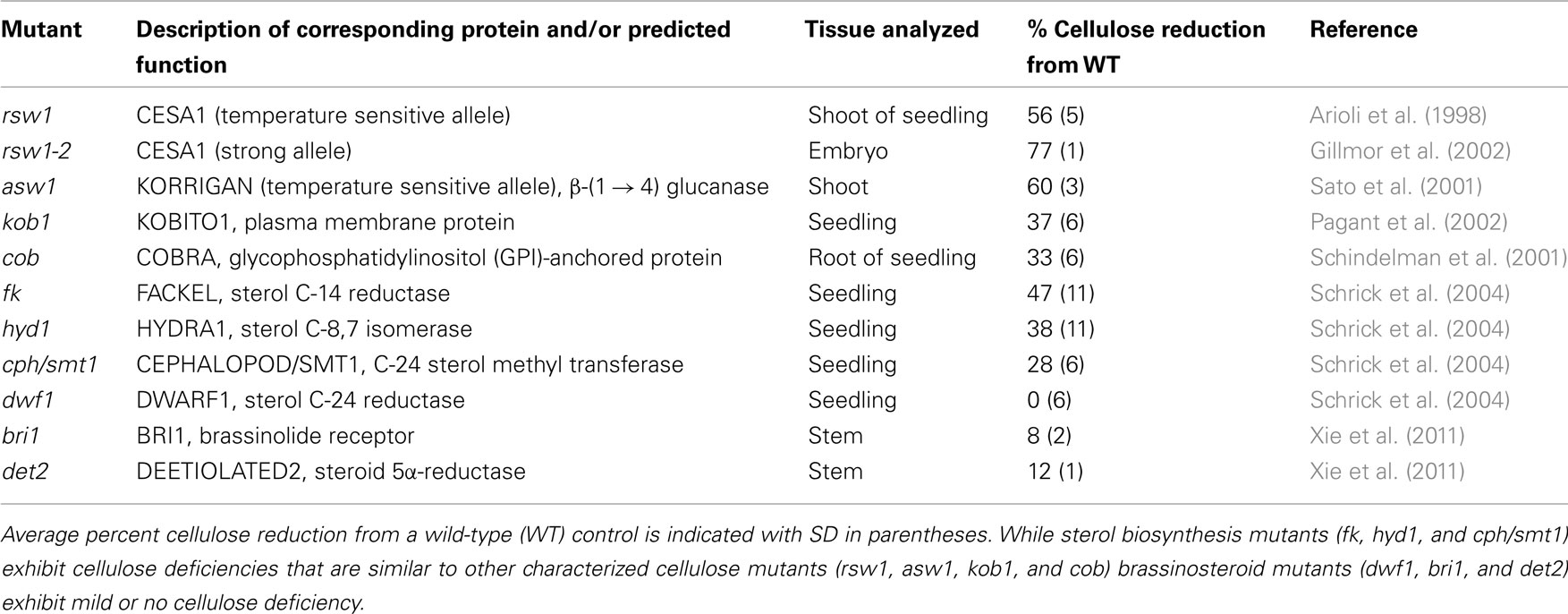
In addition to embryonic defects, fk, cph/smt1, and hyd1 mutants display a striking deficiency in cellulose content, a phenotype that can be mimicked by the sterol biosynthesis inhibitors fenpropimorph and 15-azasterol (Schrick et al., 2004). As is typical for cellulose deficiency, the mutants exhibit characteristic symptoms such as cell wall gaps, multiple nuclei, and aberrant cell wall thickenings with ectopic deposition of callose and lignin. Other cell wall components such as pectins are not reduced, arguing against a general defect in cell wall biogenesis. The decrease in cellulose content in the fk, cph/smt1, and hyd1 mutants is comparable to that observed in several cellulose biosynthesis mutants (Table 1). In contrast, brassinosteroid mutants exhibit mild or no cellulose deficiency (Table 1) although CESA genes are transcriptionally up-regulated by brassinolides (Xie et al., 2011). It is intriguing that dwf1/dim mutants which display normal cellulose content are deficient in campesterol, the precursor to brassinosteroids, while abnormally accumulating 24-methylenecholesterol, a membrane sterol (Klahre et al., 1998; Choe et al., 1999). Recently, a tomato mutant with a hyper-cracking fruit phenotype coupled with pericarp cell division and expansion defects and reduced cellulose levels was shown to correspond to a 3-β-hydroxysteroid dehydrogenase/C-4 decarboxylase (3-betaHSD/D) (Jocelyn Rose, personal communication). A multienzyme complex containing 3-betaHSD/D is required for removal of two methyl groups at C-4, rendering sterols functional as membrane constituents (Rahier et al., 2006). Taken together, these observations suggest that membrane sterols, and not brassinosteroids, are critical for cellulose accumulation.
It is striking that sterol composition differs considerably between the fk, cph/smt1, and hyd1 mutants despite their similar cellulose deficiencies. In particular, fk mutants accumulate Δ8,14 sterols and exhibit a reduction in both sitosterol and campesterol (Schrick et al., 2000), while hyd1 mutants are similarly reduced in these sterols, but they accumulate stigmasta-monoen-3β-ol (Schrick et al., 2002). In contrast, cph/smt1 mutants abnormally accumulate both cycloartenol and cholesterol, and are reduced in sitosterol but not in campesterol (Schrick et al., 2002). Consistent with the possibility that accumulation of abnormal sterols, such as biosynthetic intermediates, contributes to cellulose deficiency, fk mutants are not rescued by exogenous application of sterol end-products (Schrick et al., 2000). Even subtle changes in membrane sterol composition may disrupt the functional requirements for cellulose biosynthesis, since simple structural variations in the acyl chain alter membrane protein function in vitro (Litman and Mitchell, 1996). In mouse, the accumulation of cholesterol precursors results in embryonic defects although the cholesterol content is normal, suggesting that the build-up of precursors interferes with sterol function (Engelking et al., 2006). In this system, the drug lovastatin, which inhibits an early step leading to sterol biosynthesis (HMG-CoA reductase), was used to effectively block the accumulation of sterol intermediates and restore function. Therefore, it may be informative to prevent the accumulation of abnormal sterols in fk, hyd1, or cph/smt1 by application of lovastatin, and then to observe whether the cellulose defects can be complemented by application of sterol end-products.
Other Effects of Sterol Biosynthesis Mutations in Plants
In addition to effects on cellulose synthesis, abnormal sterol composition is associated with other physiological and cellular processes related to membranes, such as plastid biogenesis (Babiychuk et al., 2008) and the regulation of reactive oxygen species (Pose et al., 2009). Sterol biosynthesis takes place at the endoplasmic reticulum (Benveniste, 2004). At steady state, sterols are found in the Golgi membranes and endocytic compartments, although they accumulate mostly at the PM (Grebe et al., 2003). Several studies incorporating sterol biosynthesis mutants have shown that sterols play critical roles in cell polarity through their requirement for clathrin-mediated endocytosis of PIN proteins, which are presumptive auxin efflux carriers (Boutte and Grebe, 2009). The molecular mechanism is not clear nor is it known how specific this effect is, since the cytokinesis-specific syntaxin KNOLLE was shown to also require sterol-dependent endocytosis for proper localization (Boutte et al., 2010). More recent work suggests that isoprenoids and sterols are crucial for microRNA processing in providing the correct membrane composition for ARGONAUTE1 membrane association and function (Brodersen et al., 2012). Collectively, these findings indicate that sterol biosynthesis mutations can perturb membrane structure and protein trafficking.
Sterol-Rich PM Microdomains may be Critical for Maintaining the Structural Integrity and Activity of Cellulose Synthase
As for many integral membrane complexes, biochemical analysis of cellulose synthase has proven to be a major challenge. The enzyme complex is highly unstable, and PM extractions typically result in loss of cellulose synthase activity (Delmer, 1999; Bessueille and Bulone, 2008). In vitro synthesis of cellulose from plant cell-free extracts is sensitive to detergents that facilitate the isolation of intact complexes (Lai-Kee-Him et al., 2002; Colombani et al., 2004; Cifuentes et al., 2010), suggesting that cellulose synthase requires a specific lipid environment. Consistent with this idea, its activity has been identified in detergent-resistant membranes (DRMs) that exhibit biochemical properties similar to sterol-rich PM microdomains (Bessueille et al., 2009).
In the past 15 years, a model for the organization of the PM has emerged in which sterol and sphingolipid rich microdomains also known as lipid rafts coexist with more fluid domains containing phospholipids and unsaturated hydrocarbon chains (Simons and Ikonen, 1997). Lipid rafts, to which specific classes of proteins are associated, play a role in many biological processes, including cell polarity, protein trafficking, and signal transduction. While lipid rafts are defined by their composition and distribution in vivo, DRMs are biochemically characterized by their insolubility in the non-ionic detergent Triton X-100 at 4°C (Mongrand et al., 2004; Borner et al., 2005). The relationship between DRMs and lipid rafts is debated since the experimental conditions used for DRM isolation may artificially induce their formation (Lichtenberg et al., 2005; Tanner et al., 2011). Despite this consideration, extractions of DRMs could reflect differential affinities of resident membrane proteins to various lipid environments.
Plant lipid rafts are thought to exhibit unique structural features, owing to the greater molecular diversity of plant sterols and sphingolipids compared to animal or yeast counterparts. Plant-derived DRMs contain multiple sterol molecules such as 24-methylcholesterol, sitosterol, and stigmasterol (Mongrand et al., 2004), instead of primarily cholesterol as in mammals, and two distinct classes of sphingolipids, inositol phosphorylceramides, and glucosylceramides (Markham et al., 2006). Methylcyclodextrin, a chaotropic agent that extracts sterols from membranes, can serve as a tool to purify DRM sub-populations that rely on sterol function (Kierszniowska et al., 2009). In oomycetes, β-(1 → 3)-glucan synthase is released from DRMs by methylcyclodextrin, while chitin synthase is not (Briolay et al., 2009). Analogously, methylcyclodextrin might be applied to study the dependency of cellulose synthase activity on sterol-rich membrane environments in DRM preparations.
It is intriguing that lipid rafts and the CSC are formed in the same compartment before they emerge at the PM. In yeast and mammalian cells, it was shown that sterols and sphingolipids are enriched at the trans-Golgi network from where they are sorted via Golgi-derived vesicles to the PM (Klemm et al., 2009). Although rosettes have been visualized in the Golgi apparatus (Haigler and Brown, 1986), they appear to be non-functional in this compartment. Rosettes could assemble on the Golgi stacks followed by transport to the PM on Golgi-derived vesicles.
Sterols might Provide Direct Structural Scaffolding for Cellulose Synthase
Alternatively or in addition to their function in lipid rafts, sterols may interact directly with cellulose synthase by providing a scaffold to assist in proper structural conformation of the enzyme or to stabilize the complex (Figure 1). According to the original fluid-mosaic model (Singer and Nicolson, 1972), a small fraction of lipids may interact specifically with membrane proteins. It was recently shown that specific structural lipids are bound to the intact integral membrane complex ATPase within the membrane rotors (Zhou et al., 2011). Sterols could similarly directly bind the CESA subunits and/or other proteins associated with the CSC, thus enabling and/or stabilizing the function of the complex.
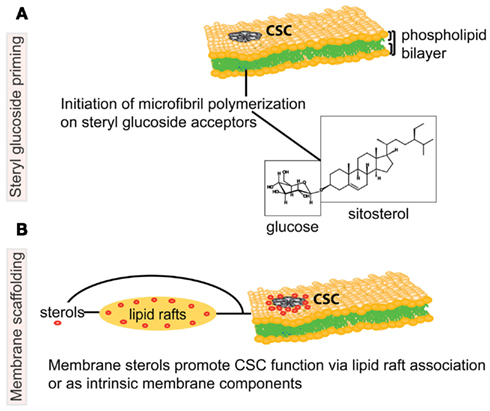
Figure 1. Schematic representation of potential associations between sterols and the cellulose biosynthetic machinery in higher plants. The cellulose synthase complex (CSC) is embedded in the phospholipid bilayer of the plasma membrane (PM), and catalyzes production of a cellulose microfibril, a major constituent of the plant cell wall. In the upper model (A), steryl glucosides (SGs) comprised of sitosterol (molecule shown in boxes) prime the synthesis of glucan chains as proposed by Peng et al. (2002). The lower model (B) depicts molecular interactions whereby sterols (in red) are necessary for the correct targeting, structural integrity and/or catalytic activity of the CSC. The proposed molecular interactions between sterols and the CSC may occur in PM microdomains, also known as lipid rafts, or via another form of molecular scaffolding. Note that the proposed models are not mutually exclusive. Figure is not drawn to scale.
Probing the Function of Sterols in Cellulose Biosynthesis
The cellulose-deficient sterol biosynthesis mutants and/or pharmacological inhibitors may serve as tools to investigate the role of sterols and plasma PM microdomains in subcellular targeting, structural integrity and/or activity of the CSC. In leek seedlings treated with the sterol biosynthesis inhibitor fenpropimorph, recovery of DRMs from the Golgi, but not from the PM, has been reported (Laloi et al., 2007). Moreover, raft protein contents are altered in mammalian cells by the sterol biosynthesis inhibitor AY9944 (Keller et al., 2004). These observations suggest that sterol composition is critical for the transport and organization of membrane microdomains.
Live imaging of fluorescently tagged CESA subunits in sterol biosynthesis mutants may be used to determine whether normal sterol composition is needed for correct localization of the CSC. In wild-type Arabidopsis, subcellular localization of CESA6 and CESA3 subunits reveals their presence at the Golgi and in the PM (Paradez et al., 2006; Crowell et al., 2009; Gutierrez et al., 2009). In sterol biosynthesis mutants, this localization might be shifted toward the Golgi if the translocation of DRMs to the PM is altered. If it is found that the CSC is properly targeted to the PM, the next step will be to investigate whether the DRMs and/or lipid rafts of sterol biosynthesis mutants lack cellulose synthase activity. Such experiments could resolve the possible mechanism by which sterols and/or sterol-rich PM microdomains or another type of sterol-dependent membrane scaffolding affects CSC function.
The Roles of Sterol Glycosides in Plant Cells
In probing the potential functions of sterols in cellulose biosynthesis, the role of conjugated sterols should not be overlooked. An abundant portion of sterols in plant membranes form steryl glycoside (SG) conjugates and the most commonly observed SG contains glucose as a sugar moiety (Grille et al., 2010). It is not clear why some sterols are glucosylated in vivo while others remain as free sterols, except that glucosylation requires end-product sterols and does not occur on intermediates. SGs have been found to be prevalent in DRM preparations from Arabidopsis and leek, suggesting their presence in lipid rafts (Laloi et al., 2007).
Steryl glycosides are synthesized by UDP-glucose:sterol glucosyltransferases (UGTs) that catalyze the glycosylation of the C3-hydroxyl of the sterol (Warnecke et al., 1997, 1999). In Arabidopsis, two genes code for the related UGT80 enzymes, UGT80A2 and UGT80B1 (DeBolt et al., 2009). Consistent with their predicted UGT functions, ugt80A2,B1 T-DNA insertion double mutants exhibit a significant reduction in SGs in various plant tissues examined (DeBolt et al., 2009) including seeds (Schrick et al., 2012). While ugt80A2 mutants display only minor effects on growth, ugt80B1 mutants exhibit an array of phenotypes in the seed, such as transparent testa, defects in flavonoid deposition, loss of the cuticle, and a decrease in aliphatic suberin and cutin-like polymers (DeBolt et al., 2009). The findings suggest a role for SGs in trafficking lipid polyester precursors in seeds.
A Proposed Function for Sitosteryl Glucoside in Cellulose Biosynthesis
Sitosteryl glucoside (SSG) has been proposed to act as a primer for cellulose biosynthesis based on the evidence that SSGs can be used by cellulose synthase as a glucose acceptor in vitro (Figure 1; Peng et al., 2002). Using crude membranes from cotton fibers, radioactive sterol cellodextrins were synthesized upon incubation of sitosterol-[14C]glucoside and non-radioactive UDP-glucose. Membranes from yeast heterologously expressing a cotton CESA subunit were shown to catalyze the same reaction. These data provide evidence that SSGs can be used by cellulose synthase as an acceptor in vitro, but whether SSGs are primers for cellulose biosynthesis in vivo remains an open question. The idea that in higher plants polymerization of cellulose chains may be initiated on lipid acceptors by cellulose synthase is consistent with models proposed for bacterial systems in which polyisoprenes and their phosphorylated forms are acceptor molecules for a transglycosylase enzyme (Matthysse et al., 1995).
To test the hypothesis that SGs play a critical role in cellulose biosynthesis in vivo, cellulose content was examined in the Arabidopsis ugt80 mutants (DeBolt et al., 2009). Although the ugt80A2,B1 double mutants exhibit a slow growth phenotype coupled with elongation defects in embryos, no cellulose deficiency was observed (DeBolt et al., 2009). It is possible that the low levels of SGs in the ugt80A2,B1 mutants can fulfill a role in priming cellulose biosynthesis. In ugt80A2,B1, residual SG levels indicate that additional plant enzymes are able to catalyze steryl glucosylation. One candidate for this function is glucosylceramide synthase (GCS), encoded by At2g19880 in Arabidopsis, which shares a low level of sequence similarity with UGT80 enzymes. A predicted GCS enzyme from cotton was shown to produce SGs in yeast (Hillig et al., 2003), consistent with the possibility that it catalyzes steryl glucosylation in planta. BLAST searches with UGT80 protein sequences reveal another candidate enzyme encoded by At5g24750, which has thus far not been characterized. Investigations of mutants corresponding to these and other candidate enzymes are expected to reveal the putative functions of SGs in cellulose biosynthesis.
Concluding Remarks
Several studies point toward an involvement of sterols in cellulose biosynthesis of higher plants, but investigations on this specific fundamental question are still at their infancy. In particular, the mode of interaction of sterols with the cellulose synthase machinery, either direct or indirect, remains to be determined (Figure 1). The in-depth analysis of cellulose formation in mutants affected in sterol biosynthesis represents a promising strategy to address this basic question. Multiple experimental approaches need to be developed to (i) determine the impact of mutations in sterol biosynthesis enzymes, or other manipulations of sterol content, on the catalytic activity of cellulose synthase, both in vitro and in vivo; (ii) analyze the impact of these perturbations on cellulose synthase subcellular localization, lipid and protein composition of membrane microdomains, and stability of the cellulose synthase machinery; (iii) quantify the effect of sterol depletion on the structural properties of cellulose microfibrils; and (iv) evaluate the importance of SGs in priming cellulose biosynthesis or in any other step of cellulose formation. This emerging topic of research in the plant cell wall field is poised to advance our mechanistic understanding of cellulose biosynthesis, and may lead to novel insights in the dynamic interplay between sterols and proteins in membranes systems.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the National Science Foundation (MCB-1122016) and the Swedish Center for Biomimetic Fiber Engineering for funding support. Contribution no. 12-387-J from the Kansas Agricultural Experiment Station.
References
Arioli, T., Peng, L., Betzner, A. S., Burn, J., Wittke, W., Herth, W., Camilleri, C., Hofte, H., Plazinski, J., Birch, R., Cork, A., Glover, J., Redmond, J., and Williamson, R. E. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720.
Babiychuk, E., Bouvier-Nave, P., Compagnon, V., Suzuki, M., Muranaka, T., Van, M. M., Kushnir, S., and Schaller, H. (2008). Allelic mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3163–3168.
Benveniste, P. (2004). Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 55, 429–457.
Bessueille, L., and Bulone, V. (2008). A survey of cellulose biosynthesis in higher plants. Plant Biotechnol. 25, 315–322.
Bessueille, L., Sindt, N., Guichardant, M., Djerbi, S., Teeri, T. T., and Bulone, V. (2009). Plasma membrane microdomains from hybrid aspen cells are involved in cell wall polysaccharide biosynthesis. Biochem. J. 420, 93–103.
Borner, G. H., Sherrier, D. J., Weimar, T., Michaelson, L. V., Hawkins, N. D., Macaskill, A., Napier, J. A., Beale, M. H., Lilley, K. S., and Dupree, P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104–116.
Boutte, Y., Frescatada-Rosa, M., Men, S., Chow, C. M., Ebine, K., Gustavsson, A., Johansson, L., Ueda, T., Moore, I., Jürgens, G., and Grebe, M. (2010). Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J. 29, 546–558.
Boutte, Y., and Grebe, M. (2009). Cellular processes relying on sterol function in plants. Curr. Opin. Plant Biol. 12, 705–713.
Briolay, A., Bouzenzana, J., Guichardant, M., Deshayes, C., Sindt, N., Bessueille, L., and Bulone, V. (2009). Cell wall polysaccharide synthases are located in detergent-resistant membrane microdomains in oomycetes. Appl. Environ. Microbiol. 75, 1938–1949.
Brodersen, P., Sakvarelidze-Achard, L., Schaller, H., Khafif, M., Schott, G., Bendahmane, A., and Voinnet, O. (2012). Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 1778–1783.
Carland, F., Fujioka, S., and Nelson, T. (2010). The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 153, 741–756.
Choe, S., Dilkes, B. P., Gregory, B. D., Ross, A. S., Yuan, H., Noguchi, T., Fujioka, S., Takatsuto, S., Tanaka, A., Yoshida, S., Tax, F. E., and Feldmann, K. (1999). The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 119, 897–907.
Cifuentes, C., Bulone, V., and Emons, A. M. (2010). Biosynthesis of callose and cellulose by detergent extracts of tobacco cell membranes and quantification of the polymers synthesized in vitro. J. Integr. Plant Biol. 52, 221–233.
Clouse, S. D. (2011). “Brassinosteroids,” in The Arabidopsis Book 9, eds N. Winchester, and D. McCauley (Rockville, MD: American Society of Plant Biologists), e0151.
Colombani, A., Djerbi, S., Bessueille, L., Blomqvist, K., Ohlsson, A., Berglund, T., Teeri, T. T., and Bulone, V. (2004). In vitro synthesis of (1(3)-beta-D-glucan (callose) and cellulose by detergent extracts of membranes from cell suspension cultures of hybrid aspen. Cellulose 11, 313–327.
Crowell, E. F., Bischoff, V., Desprez, T., Rolland, A., Stierhof, Y. D., Schumacher, K., Gonneau, M., Hofte, H., and Vernhettes, S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21, 1141–1154.
DeBolt, S., Scheible, W.-R., Schrick, K., Auer, M., Beisson, F., Bischoff, V., Bouvier-Navé, P., Carroll, A., Hematy, K., Li, Y., Milne, J., Nair, M., Schaller, H., Zemla, M., and Somerville, C. (2009). Mutations in UDP-glucose:sterol glucosyltransferase in Arabidopsis cause transparent testa phenotype and suberization defect in seeds. Plant Physiol. 151, 78–87.
Delmer, D. P. (1999). Cellulose biosynthesis: exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 245–276.
Diener, A. C., Li, H., Zhou, W., Whoriskey, W. J., Nes, W. D., and Fink, G. R. (2000). Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12, 853–870.
Endler, A., and Persson, S. (2011). Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 4, 199–211.
Engelking, L. J., Evers, B. M., Richardson, J. A., Goldstein, J. L., Brown, M. S., and Liang, G. (2006). Severe facial clefting in Insig-deficient mouse embryos caused by sterol accumulation and reversed by lovastatin. J. Clin. Invest. 116, 2356–2365.
Fernandes, A. N., Thomas, L. H., Altaner, C. M., Callow, P., Forsyth, V. T., Apperley, D. C., Kennedy, C. J., and Jarvis, M. C. (2011). Nanostructure of cellulose microfibrils in spruce wood. Proc. Natl. Acad. Sci. U.S.A. 108, E1195–E1203.
Gillmor, C. S., Poindexter, P., Lorieau, J., Palcic, M. M., and Somerville, C. (2002). Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156, 1003–1013.
Grandmougin-Ferjani, A., Schuler-Muller, I., and Hartmann, M. A. (1997). Sterol modulation of the plasma membrane H+-ATPase activity from corn roots reconstituted into soybean lipids. Plant Physiol. 113, 163–174.
Grebe, M., Xu, J., Mobius, W., Ueda, T., Nakano, A., Geuze, H. J., Rook, M. B., and Scheres, B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13, 1378–1387.
Grille, S., Zaslawski, A., Thiele, S., Plat, J., and Warnecke, D. (2010). The functions of steryl glycosides come to those who wait: recent advances in plants, fungi, bacteria and animals. Prog. Lipid Res. 49, 262–288.
Guerriero, G., Fugelstad, J., and Bulone, V. (2010). What do we really know about cellulose biosynthesis in higher plants? J. Integr. Plant Biol. 52, 161–175.
Gutierrez, R., Lindeboom, J. J., Paredez, A. R., Emons, A. M., and Ehrhardt, D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806.
Haigler, C. H., and Brown, R. M. (1986). Transport of rosettes from the Golgi apparatus to the plasma membrane in isolated mesophyll cells of Zinnia elegans during differentiation to tracheary elements in suspension culture. Protoplasma 134, 111–120.
Hillig, I., Leipelt, M., Ott, C., Zahringer, U., Warnecke, D., and Heinz, E. (2003). Formation of glucosylceramide and sterol glucoside by a UDP-glucose-dependent glucosylceramide synthase from cotton expressed in Pichia pastoris. FEBS Lett. 553, 365–369.
Jang, J. C., Fujioka, S., Tasaka, M., Seto, H., Takatsuto, S., Ishii, A., Aida, M., Yoshida, S., and Sheen, J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14, 1485–1497.
Keller, R. K., Arnold, T. P., and Fliesler, S. J. (2004). Formation of 7-dehydrocholesterol-containing membrane rafts in vitro and in vivo, with relevance to the Smith-Lemli-Opitz syndrome. J. Lipid Res. 45, 347–355.
Kierszniowska, S., Seiwert, B., and Schulze, W. X. (2009). Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Mol. Cell. Proteomics 8, 612–623.
Kimura, S., Laosinchai, W., Itoh, T., Cui, X., Linder, C. R., and Brown, R. M. Jr. (1999). Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11, 2075–2086.
Klahre, U., Noguchi, T., Fujioka, S., Takatsuto, S., Yokota, T., Nomura, T., Yoshida, S., and Chua, N. H. (1998). The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10, 1677–1690.
Klemm, R. W., Ejsing, C. S., Surma, M. A., Kaiser, H. J., Gerl, M. J., Sampaio, J. L., de Robillard, Q., Ferguson, C., Proszynski, T. J., Shevchenko, A., and Simons, K. (2009). Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell Biol. 185, 601–612.
Lai-Kee-Him, J., Chanzy, H., Muller, M., Putaux, J. L., Imai, T., and Bulone, V. (2002). In vitro versus in vivo cellulose microfibrils from plant primary wall synthases: structural differences. J. Biol. Chem. 277, 36931–36939.
Laloi, M., Perret, A. M., Chatre, L., Melser, S., Cantrel, C., Vaultier, M. N., Zachowski, A., Bathany, K., Schmitter, J. M., Vallet, M., Lessire, R., Hartmann, M. A., and Moreau, P. (2007). Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiol. 143, 461–472.
Lichtenberg, D., Goni, F. M., and Heerklotz, H. (2005). Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem. Sci. 30, 430–436.
Lindsey, K., Pullen, M. L., and Topping, J. F. (2003). Importance of plant sterols in pattern formation and hormone signalling. Trends Plant Sci. 8, 521–525.
Litman, B. J., and Mitchell, D. C. (1996). A role for phospholipid polyunsaturation in modulating membrane protein function. Lipids 31(Suppl.) S193–S197.
Markham, J. E., Li, J., Cahoon, E. B., and Jaworski, J. G. (2006). Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281, 22684–22694.
Matthysse, A. G., Thomas, D. L., and White, A. R. (1995). Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177, 1076–1081.
Mongrand, S., Morel, J., Laroche, J., Claverol, S., Carde, J. P., Hartmann, M. A., Bonneu, M., Simon-Plas, F., Lessire, R., and Bessoule, J. J. (2004). Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279, 36277–36286.
Mutwil, M., Debolt, S., and Persson, S. (2008). Cellulose synthesis: a complex complex. Curr. Opin. Plant Biol. 11, 252–257.
Pagant, S., Bichet, A., Sugimoto, K., Lerouxel, O., Desprez, T., McCann, M., Lerouge, P., Vernhettes, S., and Hofte, H. (2002). KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14, 2001–2013.
Paradez, A., Wright, A., and Ehrhardt, D. W. (2006). Microtubule cortical array organization and plant cell morphogenesis. Curr. Opin. Plant Biol. 9, 571–578.
Peng, L., Kawagoe, Y., Hogan, P., and Delmer, D. (2002). Sitosterol-beta-glucoside as primer for cellulose synthesis in plants. Science 295, 147–150.
Pose, D., Castanedo, I., Borsani, O., Nieto, B., Rosado, A., Taconnat, L., Ferrer, A., Dolan, L., Valpuesta, V., and Botella, M. A. (2009). Identification of the Arabidopsis dry2/sqe1-5 mutant reveals a central role for sterols in drought tolerance and regulation of reactive oxygen species. Plant J. 59, 63–76.
Rahier, A., Darnet, S., Bouvier, F., Camara, B., and Bard, M. (2006). Molecular and enzymatic characterizations of novel bifunctional 3beta-hydroxysteroid dehydrogenases/C-4 decarboxylases from Arabidopsis thaliana. J. Biol. Chem. 281, 27264–27277.
Sato, S., Kato, T., Kakegawa, K., Ishii, T., Liu, Y. G., Awano, T., Takabe, K., Nishiyama, Y., Kuga, S., Nakamura, Y., Tabata, S., and Shibata, D. (2001). Role of the putative membrane-bound endo-1,4-beta-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol. 42, 251–263.
Schindelman, G., Morikami, A., Jung, J., Baskin, T. I., Carpita, N. C., Derbyshire, P., McCann, M. C., and Benfey, P. N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127.
Schrick, K., Cordova, C., Li, G., Murray, L., and Fujioka, S. (2011). A dynamic role for sterols in embryogenesis of Pisum sativum. Phytochemistry 72, 465–475.
Schrick, K., Fujioka, S., Takatsuto, S., Stierhof, Y. D., Stransky, H., Yoshida, S., and Jürgens, G. (2004). A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J. 38, 227–243.
Schrick, K., Mayer, U., Horrichs, A., Kuhnt, C., Bellini, C., Dangl, J., Schmidt, J., and Jürgens, G. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14, 1471–1484.
Schrick, K., Mayer, U., Martin, G., Bellini, C., Kuhnt, C., Schmidt, J., and Jürgens, G. (2002). Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J. 31, 61–73.
Schrick, K., Shiva, S., Arpin, J. C., Delimont, N., Isaac, G., Tamura, P., and Welti, R. (2012). Steryl glucoside and acyl steryl glucoside analysis of Arabidopsis seeds by electrospray ionization tandem mass spectrometry. Lipids 47, 185–193.
Schuler, I., Milon, A., Nakatani, Y., Ourisson, G., Albrecht, A. M., Benveniste, P., and Hartman, M. A. (1991). Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phosphatidylcholine bilayers. Proc. Natl. Acad. Sci. U.S.A. 88, 6926–6930.
Singer, S. J., and Nicolson, G. L. (1972). The fluid mosaic model of the structure of cell membranes. Science 175, 720–731.
Souter, M., Topping, J., Pullen, M., Friml, J., Palme, K., Hackett, R., Grierson, D., and Lindsey, K. (2002). Hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14, 1017–1031.
Tanner, W., Malinsky, J., and Opekarova, M. (2011). In plant and animal cells, detergent-resistant membranes do not define functional membrane rafts. Plant Cell 23, 1191–1193.
Warnecke, D., Erdmann, R., Fahl, A., Hube, B., Muller, F., Zank, T., Zahringer, U., and Heinz, E. (1999). Cloning and functional expression of UGT genes encoding sterol glucosyltransferases from Saccharomyces cerevisiae, Candida albicans, Pichia pastoris, and Dictyostelium discoideum. J. Biol. Chem. 274, 13048–13059.
Warnecke, D. C., Baltrusch, M., Buck, F., Wolter, F. P., and Heinz, E. (1997). UDP-glucose:sterol glucosyltransferase: cloning and functional expression in Escherichia coli. Plant Mol. Biol. 35, 597–603.
Xie, L., Yang, C., and Wang, X. (2011). Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 62, 4495–4506.
Keywords: sterols, steryl glucosides, cellulose, plasma membrane microdomains, lipid rafts, cell wall
Citation: Schrick K, DeBolt S and Bulone V (2012) Deciphering the molecular functions of sterols in cellulose biosynthesis. Front. Plant Sci. 3:84. doi: 10.3389/fpls.2012.00084
Received: 20 March 2012; Paper pending published: 02 April 2012;
Accepted: 15 April 2012; Published online: 03 May 2012.
Edited by:
Jose Manuel Estevez, University of Buenos Aires and CONICET, ArgentinaReviewed by:
Andrew Carroll, Stanford University, USACharles T. Anderson, The Pennsylvania State University, USA
Copyright: © 2012 Schrick, DeBolt and Bulone. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Kathrin Schrick, Division of Biology, Kansas State University, Ackert Hall 116, Manhattan, KS 66506, USA. e-mail: kschrick@ksu.edu