Optimization of the IPP precursor supply for the production of lycopene, decaprenoxanthin and astaxanthin by Corynebacterium glutamicum
- Faculty of Biology and Center for Biotechnology (CeBiTec), Bielefeld University, Bielefeld, Germany
The biotechnologically relevant bacterium Corynebacterium glutamicum, currently used for the million ton-scale production of amino acids for the food and feed industries, is pigmented due to synthesis of the rare cyclic C50 carotenoid decaprenoxanthin and its glucosides. The precursors of carotenoid biosynthesis, isopenthenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate, are synthesized in this organism via the methylerythritol phosphate (MEP) or non-mevalonate pathway. Terminal pathway engineering in recombinant C. glutamicum permitted the production of various non-native C50 and C40 carotenoids. Here, the role of engineering isoprenoid precursor supply for lycopene production by C. glutamicum was characterized. Overexpression of dxs encoding the enzyme that catalyzes the first committed step of the MEP-pathway by chromosomal promoter exchange in a prophage-cured, genome-reduced C. glutamicum strain improved lycopene formation. Similarly, an increased IPP supply was achieved by chromosomal integration of two artificial operons comprising MEP pathway genes under the control of a constitutive promoter. Combined overexpression of dxs and the other six MEP pathways genes in C. glutamicum strain LYC3-MEP was not synergistic with respect to improving lycopene accumulation. Based on C. glutamicum strain LYC3-MEP, astaxanthin could be produced in the milligrams per gram cell dry weight range when the endogenous genes crtE, crtB, and crtI for conversion of geranylgeranyl pyrophosphate to lycopene were coexpressed with the genes for lycopene cyclase and β-carotene hydroxylase from Pantoea ananatis and carotene C(4) oxygenase from Brevundimonas aurantiaca.
Introduction
Carotenoids are ubiquitous natural pigments with colors ranging from yellow to red. They are composed of isoprene units and belong to the family of terpenoids. These pigments do not only play important and versatile roles in their biological hosts, but are also suggested to have a beneficial effect on human health. Furthermore, they are intensively applied for food and beverage coloration (Downham and Collins, 2000; Gassel et al., 2013). Hence, carotenoids have received extensive considerable attention and especially the interest for an efficient and environmental-friendly production by microbial hosts is increasing (Lee and Schmidt-Dannert, 2002; Das et al., 2007; Harada and Misawa, 2009; Cutzu et al., 2013). In order to compete with already existing production processes, such as chemical synthesis or extraction from organic material, the large-scale production in microbial hosts requires process as well as strain optimization. One of the most common strategies for enhanced production is the efficient supply of precursor molecules as all carotenoids derive from the universal C5 precursor molecule IPP and its isomer DMAPP. IPP and DMAPP can be synthesized via two independent pathways, the mevalonate (MVA) and the 2-methylerythritol 4-phosphate (MEP) pathway (Rodriguez-Concepcion and Boronat, 2002). The MVA pathway starts from acetyl-CoA and operates mainly in eukaryotes (mammals, fungi, in the cytoplasm of plant cells), archaea, and a limited number of bacteria. The MEP pathway that starts from pyruvate and glyceraldehyde 3-phosphate and proceeds via the eponymous intermediate MEP was identified much later (Rohmer et al., 1993) and is found in most bacteria as well as in plant plastids (Rohmer, 1999; Lange et al., 2000; Lee and Schmidt-Dannert, 2002). Both pathways also differ regarding redox and energy requirements (Steinbüchel, 2003). As the MEP pathway is present in several pathogens such as Plasmodium falciparum and Mycobacterium tuberculosis, but not in mammals, it is considered a drug target (Jomaa et al., 1999; Testa and Brown, 2003).
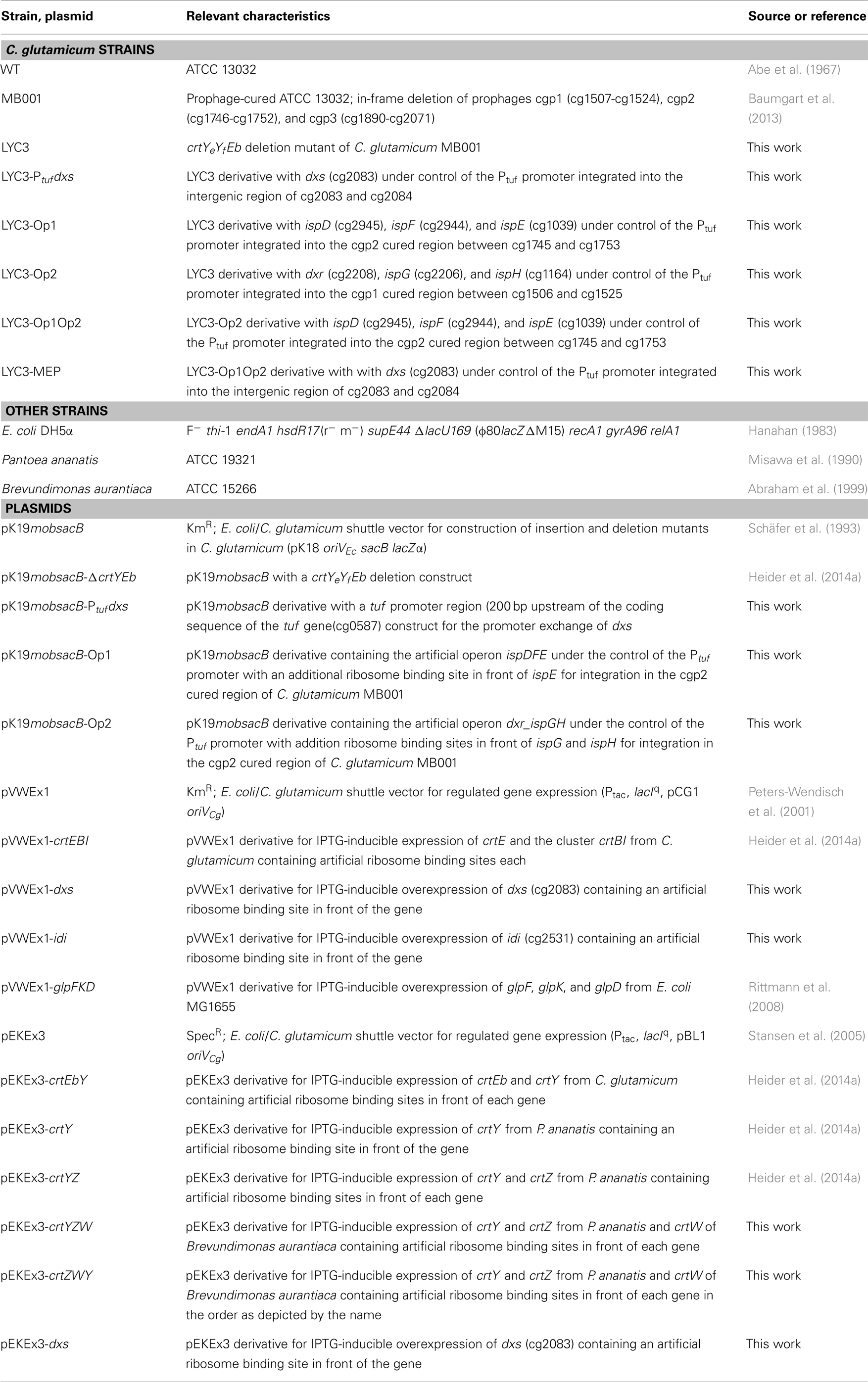
The MEP pathway consists of nine reactions catalyzed by eight enzymes (Figure 1) starting with the transfer of an acetaldehyde group derived from pyruvate to GAP, forming 1-deoxy-d-xylulose 5-phosphate (DXP), in the reaction of DXP synthase Dxs (EC 2.2.1.7). The intermediate DXP is also the precursor for thiamine (vitamine B1) (Begley et al., 1999) and pyridoxol (vitamine B6) (Hill et al., 1996) biosynthesis. Subsequently, DXP reductoisomerase Dxr (EC 1.1.1.267) converts DXP to MEP using NADPH as cofactor. MEP is then converted to the cyclic diphosphate 2C-methyl-d-erythritol-2,4-cyclodiphosphate (ME-cPP) by the three enzymes IspD, IspE, and IspF (Gräwert et al., 2011). ME-cPP is then converted to IPP and DMAPP by a reduction and elimination reaction catalyzed by the two iron–sulfur proteins IspG and IspH (Rohdich et al., 2004). It is proposed that flavodoxin is an essential redox partner for one of the enzymes (Adam et al., 2002; Gräwert et al., 2004; Puan et al., 2005). IPP and DMAPP can be synthesized independently by IspH (Gräwert et al., 2004). IPP and DMAPP often do not occur in the same ratio as for example in Escherichia coli IPP is synthesized in a 5:1 proportion to DMAPP (Rohdich et al., 2002; Gräwert et al., 2004; Xiao et al., 2008). The IPP:DMAPP isomerase Idi (EC 5.3.3.2) facilitates the isomerization between IPP and DMAPP. In the case of microorganisms using the MVA pathway produce/synthesize IPP exclusively, isomerases are essential enzymes, whereas in bacteria possessing the MEP pathway idi is not essential for the survival of the cells (Hahn et al., 1999; Julsing et al., 2007).
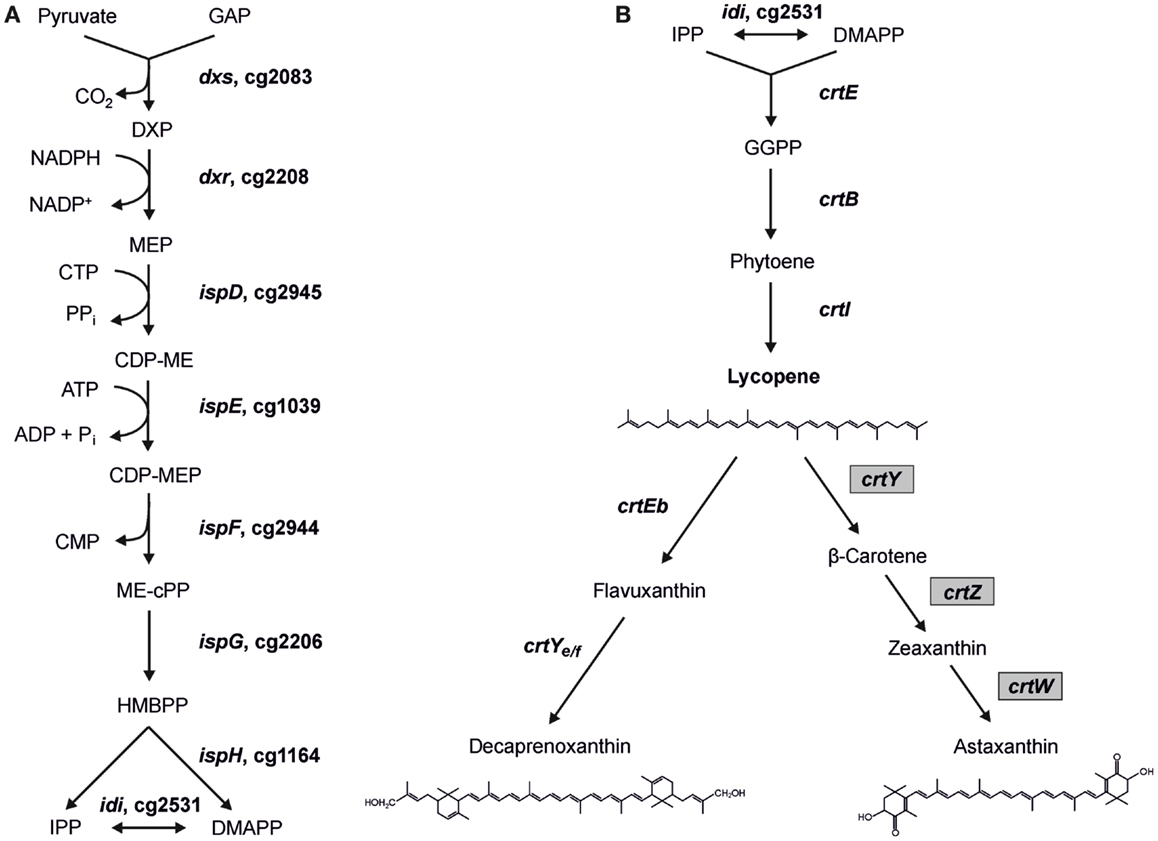
Figure 1. Scheme of the MEP pathway (A) and of decaprenoxanthin biosynthesis in C. glutamicum (B) with heterologous astaxanthin biosynthesis. Gene names from C. glutamicum (and gene IDs for MEP pathway genes) as well as gene names from Pantoea ananatis and Brevundimonas aurantiaca (gray boxes) are indicated. The structures of the endogenous C50 carotenoid decaprenoxanthin and the heterologous C40 carotenoid astaxanthin are given (GAP, glyceraldehyde 3-phosphate; DXP, 1-deoxy-d-xylulose 5-phosphate; MEP, 2-methylerythritol 4-phosphate; CDP-ME, 4-diphosphocytidyl-2-methylerythritol; CDP-MEP, 4-diphosphocytidyl-2-methylerythritol 2-phosphate; ME-cPP, 2-methylerythritol 2,4-cyclopyrophosphate; HMBPP, 4-hydroxy-3-methyl-but-2-enyl pyrophosphate; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate).
Corynebacterium glutamicum is a pigmented Gram-positive bacterium with a long and safe history in the food and feed sector as it is used for the fermentative production of amino acids. Annually, about 2.6 million tons of l-glutamate and about 1.95 million tons of l-lysine are produced biotechnologically worldwide (Ajinomoto, Food Products Business. Available from http://www.ajinomoto.com/en/ir/pdf/Food-Oct2012.pdf and /Feed-useAA-Oct2013.pdf, Cited 18 March 2014). Besides amino acids, the diamines cadaverine and putrescine (Mimitsuka et al., 2007; Schneider and Wendisch, 2010) and the alcohols ethanol and isobutanol (Sakai et al., 2007; Blombach and Eikmanns, 2011), among others, can be produced from sugars by recombinant C. glutamicum strains. Furthermore, access of C. glutamicum to alternative feed stocks like glycerol from the biodiesel process (Meiswinkel et al., 2013), pentoses from lignocellulosics (Gopinath et al., 2011), amino sugars (Uhde et al., 2013; Matano et al., 2014), starch (Seibold et al., 2006), and β-glucans (Tsuchidate et al., 2011) has been engineered.
Recently, the potential of C. glutamicum for production of carotenoids has been explored. C. glutamicum synthesizes the cyclic C50 carotenoid decaprenoxanthin and its glucosides (Figure 1). Its carotenogenic pathway and the respective genes have been elucidated (Krubasik et al., 2001; Heider et al., 2012, 2014a) and overproduction of the C50 carotenoids decaprenoxanthin, sarcinaxanthin, and C.p. 450 in the milligrams per gram cell dry weight (DCW) range by C. glutamicum was achieved by metabolic engineering of the terminal carotenoid pathway (Heider et al., 2014a). Moreover, the heterologous production of the C40 carotenoids β-carotene and zeaxanthin could be established (Heider et al., 2014a) and hydroxylated carotenoids could be produced either as aglycons or as di-glucosides (Heider et al., 2014a). Engineering of C. glutamicum for the production of a sesquiterpene, (+)-valencene, was possible as well (Frohwitter et al., 2014).
Based on its genome sequence, all genes of the MEP pathway of C. glutamicum have been putatively assigned. However, neither have the respective genes or enzymes of the MEP pathway been functionally analyzed nor has engineering for an increased IPP supply been reported. The MEP pathway genes are distributed over the genome of C. glutamicum. The MEP pathway genes dxs (cg2083), ispH (cg1164), and idi (cg2531) are monocistronic, while dxr (cg2208), ispD (cg2945), ispE (cg1039), ispF (cg2944), and ispG (cg2206) belong to operons. IspE is the third gene of the operon cg1037-ksgA-ispE-cg1040-pdxK with genes for a putative resuscitation-promoting factor (cg1037), putative dimethyladenosine transferase KsgA, and putative pyridoxamine kinase PdxK. IspD and ispF are encoded in the cg2946-ispDF operon with cg2946, which codes for a CarD-like transcriptional regulator. Dxr and ispG are organized in a transcriptional unit separated by an uncharacterized gene (cg2207) putatively encoding a membrane-embedded Zn-dependent protease. In bacteria, two bottlenecks in the MEP pathway were proposed. On the one hand, DXP synthase, which catalyzes the first reaction is claimed to be rate-limiting (Sprenger et al., 1997; Xiang et al., 2007) and is essential in E. coli (Sauret-Gueto et al., 2003) and Bacillus subtilis (Julsing et al., 2007) and possibly further bacteria. On the other hand, overproduction of Idi, which is not essential in bacteria possessing the MEP pathway (Hahn et al., 1999; Julsing et al., 2007), improved carotenoid production (Harker and Bramley, 1999; Kim and Keasling, 2001).
In this study, two synthetic operons (ispDFE and dxr-ispGH) under control of the strong promoter Ptuf of the C. glutamicum translation elongation factor EF-Tu gene were integrated into the prophage-cured, genome-reduced C. glutamicum strain MB001 (Baumgart et al., 2013). Furthermore, dxs was overexpressed from the chromosome by exchanging the endogenous promoter with the Ptuf promoter. Finally, idi was overexpressed from an IPTG-inducible plasmid. The genome-reduced strain overexpressing all of the eight MEP pathway genes was then shown to be suitable for production of lycopene and endogenous decaprenoxanthin as well as for production of the non-native astaxanthin.
Materials and Methods
Bacterial Strains, Media and Growth Conditions
The strains and plasmids used in this work are listed in Table 1. C. glutamicum ATCC13032 was used as wild type (WT), for metabolic engineering the prophage-cured C. glutamicum MB001 (Baumgart et al., 2013) was used as platform strain. Precultivation of C. glutamicum strains was performed in LB medium or LB with glucose. For cultivation in CGXII medium (Eggeling and Reyes, 2005), precultivated cells were washed once with CGXII medium without carbon source and inoculated to an initial OD600 of 1. Glucose was added as carbon and energy source to a concentration of 100 mM. Standard cultivations of C. glutamicum were performed at 30°C in a volume of 50 ml in 500 ml flasks with two baffles shaking at 120 rpm. The OD600 was measured in dilutions using a Shimadzu UV-1202 spectrophotometer (Duisburg, Germany). Alternatively, cultivations were performed in 1 ml volume in microtiterplates at 1100 rpm at 30°C using Biolector® micro fermentation system (m2p-labs GmbH, Baesweiler, Germany). For cloning, E. coli DH5α was used as host and cultivated in LB medium at 37°C. When appropriate, kanamycin or spectinomycin was added to concentrations of 25 and 100 μg ml−1, respectively. Gene expression was induced by adding 50 μM and 1 mM IPTG, respectively, at inoculation of the main culture.
Recombinant DNA Work
Plasmids were constructed in E. coli DH5α from PCR-generated fragments (KOD, Novagen, Darmstadt, Germany) and isolated with the QIAprep spin miniprep kit (QIAGEN, Hilden, Germany). Oligonucleotides used in this study were obtained from Eurofins MWG Operon (Ebersberg, Germany) and are listed in Table 2. Standard reactions like restriction, ligation, and PCR were performed as described previously (Sambrook and Russell, 2001). Besides the common ligation reaction, the Gibson assembly has been applied for the construction of plasmids (Gibson et al., 2009). If applicable, PCR products were purified using the PCR purification kit or MinElute PCR purification kit (QIAGEN, Hilden, Germany). For transformation of E. coli, the RbCl method was used (Hanahan, 1983) and C. glutamicum was transformed via electroporation (van der Rest et al., 1999) at 2.5 kV, 200 Ω, and 25 μF. All cloned DNA fragments were shown to be correct by sequencing.
Deletion of Carotenogenic Genes in C. glutamicum MB001
For deletion of the carotenogenic genes crtYe/f and crtEb, encoding the C45/C50 carotenoid ε-cyclase and the lycopne elongase, respectively, the suicide vector pK19mobsacB was used (Schäfer et al., 1994). Genomic regions flanking the crtYEb cluster were amplified from genomic DNA of C. glutamicum WT using primer pairs crtY-A/crtY-B and crtEb-C/crtEb-D (Table 2), respectively. The PCR products were purified and linked by crossover PCR using the primer pair crtY-A/crtEb-D (Table 2). The purified PCR product was cloned into pK19mobsacB resulting in the construction of deletion vector pK19mobsacB-δcrtYEb (Table 1). The targeted deletion of crtYEb via two-step homologous recombination as well as the selection for the first and second recombination events were carried out as described previously (Eggeling and Bott, 2005). Deletion of crtYEb was verified by PCR analysis of the constructed mutant using primer pair crtY-E/crtEb-F (Table 2).
Construct Design of the Synthetic MEP Operons and Their Integration into the Genome of C. glutamicum LYC3
The integration of the synthetic operons Op1 and Op2 was conducted by using the suicide vector pK19mobsacB (Schäfer et al., 1994). Op1 consists of the MEP-pathway genes ispD, ispF, and ispE under the control of the constitutive Ptuf promoter. IspD and ispF form a transcription unit and were amplified as such from genomic DNA from C. glutamicum WT using the oligonucleotides 5 and 6. The primer pair 7/8 was used to amplify ispE from C. glutamicum WT, introducing an artificial ribosome binding site (RBS) in front of the gene. The promoter region was amplified using the oligonucleotides 3 and 4. In Op2 dxr, ispG and ispH were combined, by amplification from the C. glutamicum WT genome using the primer pairs 15/16, 17/18, and 19/20, respectively. An artificial RBS in front of ispG and ispH each was introduced by the oligonucleotides 17 and 19, respectively. Also the genes of Op2 were put under the control of the Ptuf promoter, amplified from genomic DNA using the primers 13 and 14. Genomic regions flanking the selected insertion region were amplified from genomic DNA of C. glutamicum LYC3 using primer pairs 1/2 and 9/10 for integration in the cgp2 cured region in the case of Op1, or 11/12 and 20/22 for integration of Op2 in the cgp1 cured region (Table 2), respectively. The purified PCR products were either linked by crossover PCR or were directly combined together with the plasmid by Gibson assembly (Gibson et al., 2009). The final assembly of the insert with linearized pK19mobsacB led to the construction of the respective integration vectors pK19mobsacB-Op1 and pK19mobsacB-Op2 (Table 1). The following integration of the operon by two-step homologous recombination was performed according to the deletion of genes. The integration of operon1 and 2 was verified by PCR using the primers 29/30 and 31/32, respectively.
Promoter Exchange of the dxs Gene in C. glutamicum LYC3
The plasmid pK19mobsacB-Ptufdxs was constructed to replace the native dxs promoter with the tuf promoter region from C. glutamicum WT. For this purpose, the upstream region of dxs (483 bp), the 3′ part of dxs and the tuf promoter region [200 bp upstream of the coding sequence of the tuf gene(cg0587)] were amplified from chromosomal DNA of C. glutamicum LYC3 using the oligonucleotide pairs 27/28, 23/24, and 25/26, respectively (Table 2). By crossover PCR, the dxs 3′ fragment and the tuf promoter region were fused with oligonucleotides 23/26. Afterward, the dxs upstream region was fused to this 644 bp long fragment using oligonucleotides 27/26. The final purified PCR product was cloned into pK19mobsacB resulting in the vector pK19mobsacB-Ptufdxs (Table 1). The following process for the promoter exchange by two-step homologous recombination was performed as described earlier for the deletion of genes. The promoter exchange was verified by PCR using the primers dxs_E and 33, and sequencing of the PCR product.
Overexpression of Carotenogenic Genes
Plasmids harboring a carotenogenic gene (general abbreviation crt), pEKEx3-crt or pVWEx1-crt allowed an IPTG-inducible overexpression of crt. They were constructed on the basis of pEKEx3 (Stansen et al., 2005) or pVWEx1 (Peters-Wendisch et al., 2001), respectively. Amplification of crt by polymerase chain reaction (PCR) from genomic DNA of C. glutamicum WT, P. ananatis and B. aurentiaca, which was prepared as described (Eikmanns et al., 1995), was carried out using the respective primers (Table 2). The amplification of the crt genes from was based on genomic DNA as template. The amplified products were cloned into the appropriately restricted pEKEx3 or pVWEx1 plasmid DNA.
Extraction Analysis of Carotenoids
To extract carotenoids from the C. glutamicum strains 15 ml aliquots of the cell cultures were centrifuged at 10,000 × g for 15 min and the pellets were washed with deionized H2O. The pigments were extracted with 10 ml methanol:acetone mixture (7:3) at 60°C for 30 min with thorough vortexing every 10 min. When necessary, several extraction cycles were performed to remove all visible colors from the cell pellet (Heider et al., 2012).
The extraction mixture was centrifuged 10,000 × g for 15 min and the supernatant was transferred to a new tube. The carotenoid content in the extracts was quantified through absorbance at 470 nm by HPLC analysis (see below) and the concentrations were calculated using a standard curve and appropriate dilutions. High performance liquid chromatography (HPLC) analyses of the C. glutamicum extracts were performed like described earlier (Heider et al., 2014a) on an Agilent 1200 series HPLC system (Agilent Technologies Sales & Services GmbH & Co., KG, Waldbronn), including a diode array detector (DAD) for UV/visible (Vis) spectrum recording. For separation, a column system consisting of a precolumn (10 mm × 4 mm MultoHigh 100 RP18-5, CS Chromatographie Service GmbH, Langerwehe, Germany) and a main column (ProntoSIL 200-5 C30, 250 mm × 4 mm, CS Chromatographie Service GmbH, Langerwehe, Germany) was used. Quantification of carotenoids was performed using the extracted wavelength chromatogram at 470 nm for decaprenoxanthin and carotenoids with corresponding UV/Vis profiles as well as for lycopene and corresponding carotenoids. Lycopene from tomato (Sigma, Steinheim, Germany), astaxanthin (Ehrenstorfer GmbH, Augsburg, Germany), and β-carotene (Merck, Darmstadt, Germany) were used as standards. The carotenoids were dissolved in chloroform according to its solubility and diluted in methanol:acetone (7:3). Due to the lack of appropriate standards decaprenoxanthin and zeaxanthin quantification was calculated based on a β-carotene standard and reported as β-carotene equivalents. The HPLC protocol comprised a gradient elution for 10 min and a mobile phase composition of (A) methanol and (B) methanol/methyl tert-butyl ether/ethyl acetate (5:4:1) starting from 10 to 100% eluent B followed by 20 min of isocratic elution with 100% B. After that, the eluent composition is set back to 10% B for 3 min. The injection volume was 50 μl and the flow rate was kept constant at 1.4 ml/min.
DXS Activity Assay
The DXS activity of C. glutamicum crude extracts was determined using an endpoint assay adopted from Xiang et al. (2007), which is based on the measurement of the remaining pyruvate level in the reaction mixture. The assays were carried out at 30°C in total volume of 1 ml containing 50 mM Tris (pH 7.5), 60 μM pyruvate, 60 μM GAP, 10 mM dithiothreitol (DTT), 5 mM MgCl2, and 600 μM TPP. Reactions were stopped after 5, 15, 30, and +60 min of incubation by heat inactivation (5 min at 95°C). Subsequent the leftover pyruvate was converted to lactate with lactate dehydrogenase and the concomitant consumption of NADH was determined by fluorescence. Therefore, the reaction was allowed to proceed for 60 min at room temperature. Then, 2.5 U ml−1 lactate dehydrogenase and 0.1 mM NADH was added to the reaction mixture and incubated for 30 min at 37°C. The NADH diminution was determined photometrically at 340 nm.
Results
Overexpression of dxs Increased Lycopene Yield
The first and often rate-limiting reaction in the MEP pathway is the condensation of pyruvate and GAP to DXP catalyzed by Dxs (Harker and Bramley, 1999; Kim and Keasling, 2001). To test if Dxs is a bottleneck in carotenoid biosynthesis in C. glutamicum, dxs was overexpressed in C. glutamicum LYC3, a mutant derived from the genome-reduced C. glutamicum strain MB001 (Baumgart et al., 2013) that accumulates lycopene due to deletion of the lycopene elongase and C45/C50 carotenoid ε-cyclase genes crtEb and crtYe/f. To exchange the native dxs promoter by the strong constitutive promoter of tuf (cg0587), which encodes for the elongation factor EF-Tu (Fukui et al., 2011), the replacement vector pK19mobsacB-Ptuf dxs was constructed and C. glutamicum LYC3-Ptuf dxs was obtained. Dxs activities measured in crude extracts were about twofold higher in C. glutamicum LYC3-Ptuf dxs (16 ± 1 mU mg−1) than in the control strain C. glutamicum LYC3 (Table 3). As consequence of enhanced Dxs activity, lycopene production doubled (0.08 ± 0.01 mg g−1 DCW as compared to 0.04 ± 0.01 mg g−1 DCW) (Table 3). Thus, increased Dxs activity improved lycopene production by C. glutamicum. Increased specific Dxs activities were also observed when a plasmid-borne copy of dxs was overexpressed from an IPTG-inducible promoter in LYC3, but lycopene production was only slightly improved (Table 3). Hence, chromosomal overexpression proved better and was therefore chosen for subsequent metabolic engineering of the MEP pathway.
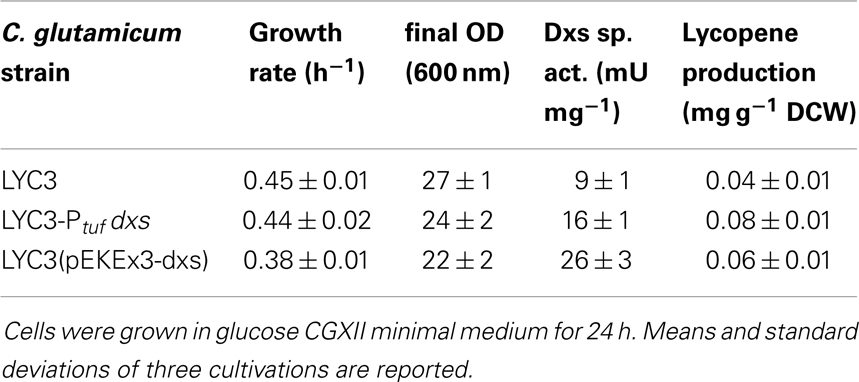
Table 3. Influence of chromosomal promoter exchange of the 1-deoxy-d-xylulose 5-phosphate synthase gene dxs on Dxs actitivities, growth rates, and lycopene production.
Overproduction of Enzymes Converting DXP to IPP Using Two Synthetic Operons Integrated into the C. glutamicum Chromosome
For overproduction of the six MEP pathway enzymes catalyzing the conversion of DXP to IPP, two synthetic operons were constructed and integrated into the chromosome of C. glutamicum LYC3. Operon 1 was constructed to drive expression of ispDF, which are cotranscribed naturally, fused to ispE from Ptuf. The RBS of the tuf gene was inserted upstream of ispD, while the endogenous RBS of ispF and a perfect C. glutamicum RBS upstream of ispE were used. To construct operon 2, dxr, ispG, and ispH were fused for expression from Ptuf and perfect C. glutamicum RBS were inserted upstream of ispG and ispH while the RBS of the tuf gene was used upstream of dxr. Both operons were integrated by homologous recombination into the chromosome of C. glutamicum LYC3, which lacks prophages cgp1 and cgp2. Operon 1 was integrated into the chromosome of C. glutamicum LYC3 between cg1506 and cg1525, i.e., at the position that harbors prophage cgp2 in the C. glutamicum WT, but which is absent from LYC3, and the resulting strain was named LYC3-Op1. Similarly, C. glutamicum LYC3-Op2 was obtained by integrating operon 2 into the chromosome of C. glutamicum LYC3 at the position (between cg1745 and cg1753) that in C. glutamicum WT harbors prophage cgp1, but which is absent from LYC3. The constructed C. glutamicum strain LYC3-Op1Op2 contains both operons in the chromosome instead of prophages cgp1 and cgp2. C. glutamicum LYC3-Op1 showed slightly higher lycopene accumulation than C. glutamicum strains LYC3 and LYC3-Op2. C. glutamicum LYC3-Op2 grew slower than LYC3 and LYC3-Op1. C. glutamicum LYC3-Op1Op2 that harbors both operons also grew slower, but accumulated almost threefold more lycopene than LYC3. Thus, overexpression of MEP pathway genes from two chromosomally integrated synthetic operons improved lycopene production (Figure 2).
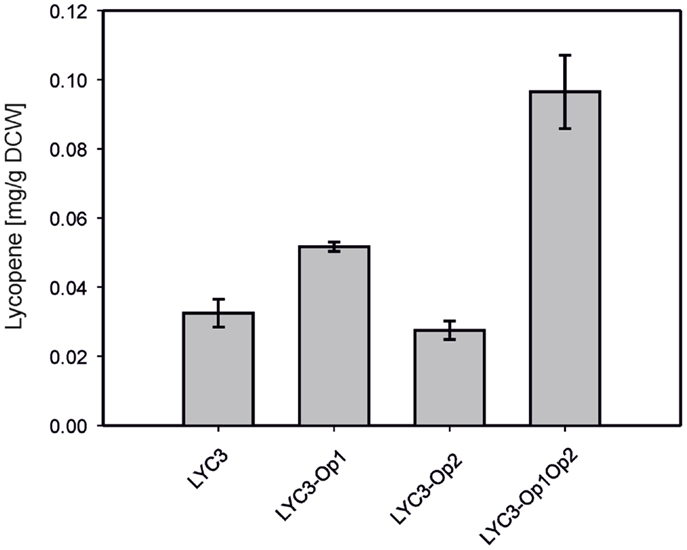
Figure 2. Lycopene production by C. gluamicum LYC3 and derived strains expressing the synthetic ispDFE operon (LYC3-Op1), the synthetic dxr-ispGH operon (LYC3-Op2) or both operons (LYC3-Op1Op2) for overproduction of MEP pathway enzymes. Cells were grown in glucose CGXII minimal medium. Means and standard deviations of three cultivations are shown.
Improved IPP Supply by Chromosome-Based Enhancement of MEP Pathway Gene Expression
To combine chromosome-based overexpression of the genes necessary for conversion of DXP to IPP with overproduction of Dxs, the first enzyme of the MEP pathway, the endogenous promoter of chromosomal dxs was exchanged by Ptuf in C. glutamicum LYC3-Op1Op2 and the resulting strain was named C. glutamicum LYC3-MEP. Surprisingly, LYC3-MEP showed slower growth on solid as well as in liquid medium. Poor growth in liquid glucose medium was accompanied by little lycopene production, although LYC3-MEP colonies appeared well pigmented on plates. Since the central carbon metabolites pyruvate and GAP are the immediate precursors of the MEP pathway, it was tested if lycopene production by C. glutamicum LYC3-MEP was affected by the carbon source. To this end, pyruvate and glycerol were tested as carbon sources. Since glycerol is no carbon source for C. glutamicum WT, glpFKD from E. coli encoding the enzyme for conversion of glycerol to GAP were expressed from plasmid pVWEx1-glpFKD (Rittmann et al., 2008) in C. glutamicum LYC3-MEP. Growth by C. glutamicum LYC3-MEP(pVWEx1-glpFKD) on glycerol, glycerol + glucose, or glycerol + pyruvate was still impaired, but about twofold more lycopene (around 0.07 ± 0.01 mg g−1 DCW) accumulated than with glucose as sole carbon source (Figure 3).
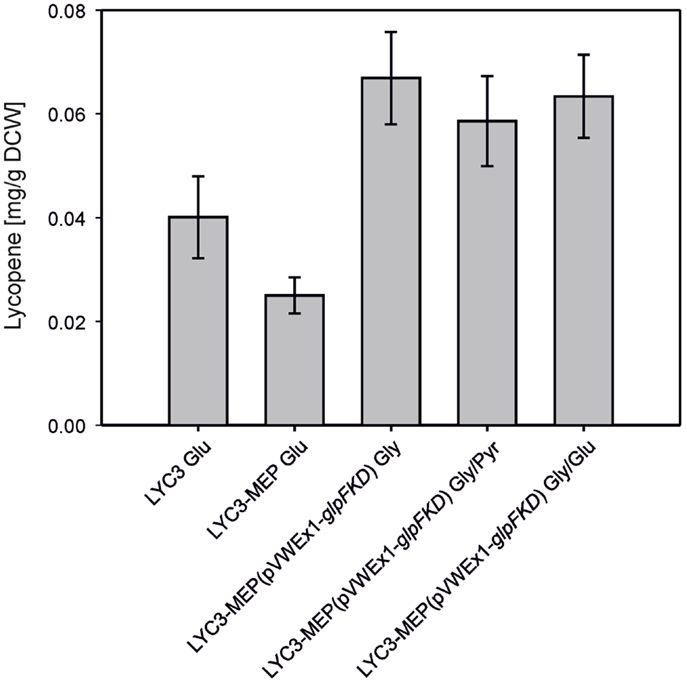
Figure 3. Lyopene production by C. gluamicum LYC3-MEP(pVWEx1- glpFKD) on glycerol as sole and combined carbon source. LYC3-MEP (pVWEx1-glpFKD) cells were grown in CGXII minimal medium with 200 mM glycerol (Gly), 100 mM glycerol + 100 mM pyruvate (Gly/Pyr), or 100 mM glycerol + 50 mM glucose (Gly/Glu), respectively. Expression of glpFKD was induced by 50 μM IPTG. As reference, lycopene production of the strains LYC3 and LYC3-MEP grown in CGXII minimal medium with 100 mM glucose (Glu) is given. Means and standard deviations of three cultivations are reported.
Since IspH synthesizes both IPP and DMAPP, but typically not in equimolar amounts (Rohdich et al., 2002; Gräwert et al., 2004; Xiao et al., 2008), it is possible that unbalanced biosynthesis of IPP and DMAPP in C. glutamicum LYC3-MEP impairs growth and carotenogenesis. To test this hypothesis, isopentenyl pyrophosphate isomerase Idi was overproduced. Indeed, C. glutamicum LYC3-MEP(pVWEx1-idi)(pEKEx3) produced twofold more lycopene (0.08 ± 0.02 mg g−1 DCW) than C. glutamicum strains LYC3, LYC3-MEP, and the empty vector control strain, but still showed impaired growth (Table 4). Thus, a lycopene producing C. glutamicum strain with improved IPP supply overexpressing all MEP pathway genes and idi could be constructed. However, lycopene production by this strain (Table 4) was comparable to that by C. glutamicum strains LYC3-Ptuf dxs (Table 3) and LYC3-Op1Op2 (Figure 2) indicating that the positive effects did not act synergistically. This was also observed when the strains were grown in LB medium supplemented with 100 mM glucose; however, they grew faster (data not shown). Taken together, C. glutamicum strains with improved IPP and DMAPP supply showed higher lycopene production than the respective parental strains.
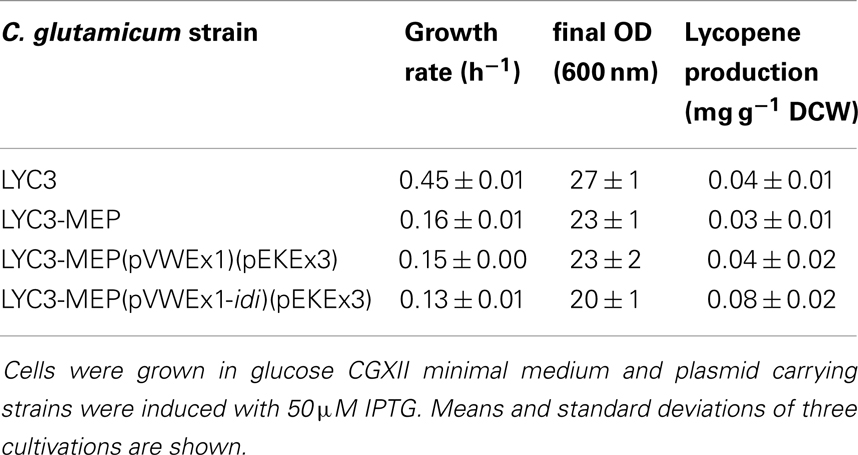
Table 4. Growth rates and lycopene production by prophage-cured, MEP pathway genes overexpressing C. glutamicum strain LYC3-MEP.
Application of C. glutamicum with Improved IPP Supply for Production of Decaprenoxanthin and Astaxanthin
To test if C. glutamicum LYC3-MEP overexpressing idi is suitable for production of the endogenous C50 carotenoid decaprenoxanthin, this strain was transformed with plasmid pEKEx3-crtEbY. Expression of lycopene elongase gene crtEb and of carotenoid ε-cyclase gene crtYe/f from this plasmid complements the lycopene producing C. glutamicum LYC3-MEP, which carries chromosomal crtEb and crtYe/f deletions allowing for decaprenoxanthin biosynthesis. The resulting strain LYC3-MEP(pVWEX1-idi)(pEKEx3-crtEbY) overproduces all enzymes of endogenous carotenogenesis except crtE, crtB, and crtI (Figure 1). Although it grew slowly, LYC3-MEP (pVWEX1-idi)(pEKEx3-crtEbY) produced 0.35 ± 0.02 mg g−1 DCW (Table 5) and, thus, is a genome-reduced strain with improved IPP supply suitable for the overproduction of the endogenous C50 carotenoid decaprenoxanthin.
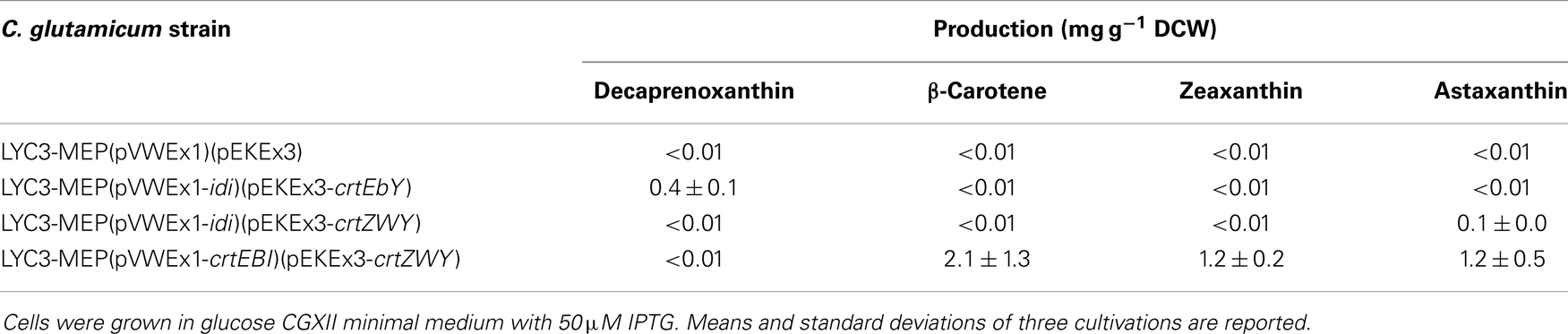
Table 5. Astaxanthin and decaprenoxanthin production by recombinant C. glutamicum strains with improved IPP supply.
C. glutamicum has previously been engineered for the production of the non-native C40 carotenoids β-carotene and zeaxanthin (Heider et al., 2014a). When crtYPa (PANA_4160) encoding lycopene cyclase from Pantoea ananatis was expressed, β-carotene accumulated. Additional expression of crtZPa (PANA_4163), which encodes β-carotene hydroxylase, resulted in partial conversion of β-carotene to zeaxanthin (Heider et al., 2014a). To enable astaxanthin production, crtWBa encoding carotene C(4) oxygenase from Brevundimonas aurantiaca, which oxidizes zeaxanthin to yield astaxanthin, was expressed in addition to crtYPa and crtZPa. The resulting plasmid pEKEx3-crtZWY was used to transform LYC3-MEP(pVWEX1-idi). C. glutamicum LYC3-MEP(pVWEX1-idi)(pEKEx3-crtZWY) produced 0.14 ± 0.01 mg g−1 DCW astaxanthin and neither β-carotene nor zeaxanthin accumulated (Table 5). Thus, to the best of our knowledge, this is the documentation of astaxanthin production by recombinant C. glutamicum. Although levels were low, LYC3-MEP(pVWEX1-idi)(pEKEx3-crtZWY) produced astaxanthin as only carotenoid.
Based on our previous findings that overexpression of the genes crtE, crtB, and crtI (Figure 1) strongly increased lycopene production (Heider et al., 2012), as well as decaprenoxanthin production (Heider et al., 2014a); these genes were overexpressed from plasmid pVWEx3-crtEBI. The resulting strain C. glutamicum LYC3-MEP(pVWEx3-crtEBI)(pEKEx3-crtZWY) produced 2.1 ± 1.3 mg g−1 DCW β-carotene and 1.2 ± 0.2 mg g−1 DCW zeaxanthin (Table 5), but also ninefold more astaxanthin (1.2 ± 0.5 mg g−1 DCW) than LYC3-MEP(pVWEx1-idi)(pEKEx3-crtZWY). Thus, it was shown that astaxanthin can be produced by recombinant C. glutamicum in the milligrams per gram DCW range.
Discussion
Recently, C. glutamicum has been engineered for production of diverse lycopene-derived carotenoids (Heider et al., 2014a) and of a sesquiterpene (Frohwitter et al., 2014). There is an increasing demand for efficient, low-cost, and natural production of terpenoids (Zhu et al., 2014) as they have many applications, e.g., in the medicinal and nutraceutical industries or as fuels (Martin et al., 2003; Ajikumar et al., 2010; Peralta-Yahya et al., 2011). Besides terminal terpenoid pathway engineering, an efficient supply of the prenyl pyrophosphate precursors is important (Heider et al., 2014b). It could be shown here that MEP pathway engineering to improve IPP supply in C. glutamicum improved lycopene production. However, as observed in similar studies of MEP pathway engineering in other bacteria individual bottlenecks may be overcome, but the individual beneficial effects do not necessarily add up (Kim and Keasling, 2001; Martin et al., 2003; Rodriguez-Villalon et al., 2008). Overexpressing the initial MEP pathway gene, dxs improved lycopene production by C. glutamicum (see Figure 1) and by other bacteria (Harker and Bramley, 1999; Matthews and Wurtzel, 2000). However, optimal overexpression levels need to be established since, e.g., chromosomal overexpression proved better than overexpression from a multy-copy plasmid (Yuan et al., 2006). Similarly, when dxs was overexpressed in C. glutamicum by exchanging the native promoter of dxs with the strong constitutive tuf promoter more lycopene accumulated than when plasmid-borne dxs overexpression, which led to higher Dxs activities, was tested (Table 3). The complex interplay of MEP pathway enzymes is also reflected by the fact that overexpression of dxr, ispG, and ispH in LYC3-Op2 only improved lycopene accumulation when combined with overexpression of ispDF and ispE (Op1) (Figure 2). Although lycopene titers obtained with C. glutamicum LYC3-Op1Op2 were comparable to the dxs overexpressing strain LYC3-Ptufdxs (Figure 2 and Table 3), their combination in strain LYC3-MEP was not synergistic and even perturbed growth. This may be explained by accumulation of inhibitory MEP pathway intermediates as shown for B. subtilis (Sivy et al., 2011) and E. coli (Martin et al., 2003; Zou et al., 2013), from an excessive drain of central metabolic intermediates (Kim and Keasling, 2001) and/or from an imbalance between IPP and DMAPP (Kajiwara et al., 1997). In C. glutamicum, improved lycopene production as consequence of overexpression of IPP isomerase gene idi was observed in LYC3-MEP (Table 4). However, lycopene production by LYC3-MEP overexpressing idi was not higher than by LYC3-Ptufdxs or by LYC3-Op1Op2. Moreover, when dxs was overexpressed in the WT-derived strain ΔcrtEb lycopene production increased from about 0.04 to about 0.12 mg g−1 DCW, but combined overexpression of dxs and idi did not further increase lycopene production (data not shown). Thus, the perturbed growth may not only be due to an imbalance between IPP and DMAPP.
It remains to be shown if combinatorial approaches to optimize multiple gene expression levels (Zelcbuch et al., 2013; Nowroozi et al., 2014) would improve the IPP precursor supply in C. glutamicum. Fine-tuning of gene expression in recombinant C. glutamicum by varying promoters (Holátko et al., 2009; van Ooyen et al., 2011; Schneider et al., 2012), RBSs (Schneider et al., 2012), translational start codons (Schneider et al., 2012), or translational stop codons (Jensen and Wendisch, 2013) improved production of amino acids and diamines. In addition, overexpression of heterologous instead of endogenous genes may be beneficial, e.g., as shown for improving isoprene production by E. coli via overexpression of two MEP pathway genes dxs and dxr from B. subtilis (Zhao et al., 2011) or by combining overexpression of xylA from Xanthomonas campestris with endogenous xylB to accelerate xylose utilization of C. glutamicum (Meiswinkel et al., 2013).
Besides fine-tuning of MEP pathway gene overexpression, growth, and terpenoid production by recombinant C. glutamicum with increased IPP supply could be improved by metabolic pull, i.e., by overexpression of genes of the downstream terpenoid pathway (Table 5). Similarly, amorphadiene synthase overexpression prevented accumulation of inhibitory isoprenoid pathway intermediates in E. coli (Martin et al., 2003). Overcoming the toxicity of accumulating IPP and DMAPP was successfully used as screening method for the identification of genes that are involved in isoprenoid biosynthesis (Withers et al., 2007). Accumulation of the MEP pathway intermediate ME-cPP inhibits growth and isoprenoid production by recombinant E. coli. To abolish its accumulation overexproducing the two enzymes downstream of ME-cPP (ispG and ispH) needed to be combined with overexpressing an operon for iron–sulfur cluster assembly since both IspG and IspH are containing iron–sulfur clusters (Zou et al., 2013).
To the best of our knowledge, production of astaxanthin by recombinant C. glutamicum was shown here for the first time. Astaxanthin is the third most important carotenoid after β-carotene and lutein and its global market amounted to about 230 million US$ in 2010 (BBC Research, 2011). The economically most significant application of astaxanthin is its use as feed additive in aquaculture industry (Lorenz and Cysewski, 2000; Higuera-Ciapara et al., 2006; Schmidt et al., 2011), but it also exhibits high potential as a nutraceutical and as an approved ingredient for cosmetics due to its remarkably high antioxidative activity (Miki, 1991; Schmidt et al., 2011). Astaxanthin is mainly produced by marine bacteria and microalgae, but only the green freshwater microalga Haematococcus pluvialis and the red yeasts Xanthophyllomyces dendrohous/Phaffia rhodozyma are established as hosts for commercial production (Bhosale and Bernstein, 2005; Rodriguez-Saiz et al., 2010). Algae-based production of astaxanthin is still more costly than chemical synthesis (Jackson et al., 2008), but markets more and more demand naturally produced carotenoids. The astaxanthin titers by recombinant C. glutamicum reported here are in the milligrams per gram DCW range and, thus, they are comparable to yields described for P. rhodozyma (ranging from 0.16 to 6.6 mg g−1 DCW (Cruz and Parajo, 1998; Jacobson et al., 1999). The highest product titer of 9.7 mg g−1 DCW is reported for a P. rhodozyma strain improved by metabolic engineering and classical mutagenesis (Gassel et al., 2013), while the highest titer in a recombinant bacterium, i.e., E. coli strain was 5.8 mg g−1 DCW astaxanthin (Zelcbuch et al., 2013). Thus, the astaxanthin titers reported for C. glutamicum are comparable and it is conceivable that they may be improved further by combining metabolic engineering with classical mutagenesis as in P. rhodozyma (Gassel et al., 2013), by combinatorial approaches to gene expression (Zelcbuch et al., 2013), or by high-cell density cultivation since biomass concentrations of up to 95 g DCW/l have been reported for C. glutamicum (Riesenberg and Guthke, 1999).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abe, S., Takayarna, K., and Kinoshita, S. (1967). Taxonomical studies on glutamic acid producing bacteria. J. Gen. Appl. Microbiol. 13, 279–301. doi: 10.2323/jgam.13.279
Abraham, W. R., Strömpl, C., Meyer, H., Lindholst, S., Moore, E. R., Christ, R., et al. (1999). Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int. J. Syst. Bacteriol. 49(Pt 3), 1053–1073. doi:10.1099/00207713-49-3-1053
Adam, P., Hecht, S., Eisenreich, W., Kaiser, J., Grawert, T., Arigoni, D., et al. (2002). Biosynthesis of terpenes: studies on 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Proc. Natl. Acad. Sci. U.S.A. 99, 12108–12113. doi:10.1073/pnas.182412599
Ajikumar, P. K., Xiao, W. H., Tyo, K. E., Wang, Y., Simeon, F., Leonard, E., et al. (2010). Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330, 70–74. doi:10.1126/science.1191652
Baumgart, M., Unthan, S., Ruckert, C., Sivalingam, J., Grunberger, A., Kalinowski, J., et al. (2013). Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 79, 6006–6015. doi:10.1128/AEM.01634-13
BBC Research. (2011). The Global Market for Carotenoids. Available from: http://www.bccresearch.com/market-research/food-and-beverage/carotenoids-global-market-fod025d.html
Begley, T. P., Downs, D. M., Ealick, S. E., Mclafferty, F. W., Van Loon, A. P., Taylor, S., et al. (1999). Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171, 293–300. doi:10.1007/s002030050713
Bhosale, P., and Bernstein, P. S. (2005). Microbial xanthophylls. Appl. Microbiol. Biotechnol. 68, 445–455. doi:10.1007/s00253-005-0032-8
Blombach, B., and Eikmanns, B. J. (2011). Current knowledge on isobutanol production with Escherichia coli, Bacillus subtilis and Corynebacterium glutamicum. Bioeng. Bugs 2, 346–350. doi:10.4161/bbug.2.6.17845
Cruz, J. M., and Parajo, J. C. (1998). Improved astaxanthin production by Xanthophyllomyces dendrorhous growing on enzymatic wood hydrolysates containing glucose and cellobiose. Food Chem. 63, 479–484. doi:10.1016/S0308-8146(98)00061-2
Cutzu, R., Coi, A., Rosso, F., Bardi, L., Ciani, M., Budroni, M., et al. (2013). From crude glycerol to carotenoids by using a Rhodotorula glutinis mutant. World J. Microbiol. Biotechnol. 29, 1009–1017. doi:10.1007/s11274-013-1264-x
Das, A., Yoon, S. H., Lee, S. H., Kim, J. Y., Oh, D. K., and Kim, S. W. (2007). An update on microbial carotenoid production: application of recent metabolic engineering tools. Appl. Microbiol. Biotechnol. 77, 505–512. doi:10.1007/s00253-007-1206-3
Downham, A., and Collins, P. (2000). Colouring our foods in the last and next millennium. Int. J. Food Sci. Technol. 35, 5–22. doi:10.1046/j.1365-2621.2000.00373.x
Eggeling, L., and Bott, M. (eds) (2005). Handbook of Corynebacterium glutamicum. Boca Raton: CRC Press.
Eggeling, L., and Reyes, O. (2005). “Experiments,” in Handbook of Corynebacterium glutamicum, eds L. Eggeling and M. Bott (Boca Raton: CRC Press), 3535–3566.
Eikmanns, B. J., Rittmann, D., and Sahm, H. (1995). Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J. Bacteriol. 177, 774–782.
Frohwitter, J., Heider, S., Peters-Wendisch, P., Beekwilder, J., and Wendisch, V. (2014). Production of the sesquiterpene (+)-valencene by metabolically engineered Corynebacterium glutamicum. J. Biotechnol. doi:10.1016/j.jbiotec.2014.05.032
Fukui, K., Koseki, C., Yamamoto, Y., Nakamura, J., Sasahara, A., Yuji, R., et al. (2011). Identification of succinate exporter in Corynebacterium glutamicum and its physiological roles under anaerobic conditions. J. Biotechnol. 154, 25–34. doi:10.1016/j.jbiotec.2011.03.010
Gassel, S., Schewe, H., Schmidt, I., Schrader, J., and Sandmann, G. (2013). Multiple improvement of astaxanthin biosynthesis in Xanthophyllomyces dendrorhous by a combination of conventional mutagenesis and metabolic pathway engineering. Biotechnol. Lett. 35, 565–569. doi:10.1007/s10529-012-1103-4
Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A. III, and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi:10.1038/nmeth.1318
Gopinath, V., Meiswinkel, T. M., Wendisch, V. F., and Nampoothiri, K. M. (2011). Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose-utilizing Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 92, 985–996. doi:10.1007/s00253-011-3478-x
Gräwert, T., Groll, M., Rohdich, F., Bacher, A., and Eisenreich, W. (2011). Biochemistry of the non-mevalonate isoprenoid pathway. Cell. Mol. Life Sci. 68, 3797–3814. doi:10.1007/s00018-011-0753-z
Gräwert, T., Kaiser, J., Zepeck, F., Laupitz, R., Hecht, S., Amslinger, S., et al. (2004). IspH protein of Escherichia coli: studies on iron-sulfur cluster implementation and catalysis. J. Am. Chem. Soc. 126, 12847–12855. doi:10.1021/ja0471727
Hahn, F. M., Hurlburt, A. P., and Poulter, C. D. (1999). Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J. Bacteriol. 181, 4499–4504.
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi:10.1016/S0022-2836(83)80284-8
Harada, H., and Misawa, N. (2009). Novel approaches and achievements in biosynthesis of functional isoprenoids in Escherichia coli. Appl. Microbiol. Biotechnol. 84, 1021–1031. doi:10.1007/s00253-009-2166-6
Harker, M., and Bramley, P. M. (1999). Expression of prokaryotic 1-deoxy-d-xylulose-5-phosphatases in Escherichia coli increases carotenoid and ubiquinone biosynthesis. FEBS Lett. 448, 115–119.
Heider, S. A., Peters-Wendisch, P., Netzer, R., Stafnes, M., Brautaset, T., and Wendisch, V. F. (2014a). Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 98, 1223–1235. doi:10.1007/s00253-013-5359-y
Heider, S. A., Peters-Wendisch, P., Wendisch, V. F., Beekwilder, J., and Brautaset, T. (2014b). Metabolic engineering for the microbial production of carotenoids and related products with a focus on the rare C50 carotenoids. Appl. Microbiol. Biotechnol. 98, 4355–4368. doi:10.1007/s00253-014-5693-8
Heider, S. A., Peters-Wendisch, P., and Wendisch, V. F. (2012). Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol. 12:198. doi:10.1186/1471-2180-12-198
Higuera-Ciapara, I., Felix-Valenzuela, L., and Goycoolea, F. M. (2006). Astaxanthin: a review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 46, 185–196. doi:10.1080/10408690590957188
Hill, R. E., Himmeldirk, K., Kennedy, I. A., Pauloski, R. M., Sayer, B. G., Wolf, E., et al. (1996). The biogenetic anatomy of vitamin B6. A 13C NMR investigation of the biosynthesis of pyridoxol in Escherichia coli. J. Biol. Chem. 271, 30426–30435. doi:10.1074/jbc.271.48.30426
Holátko, J., Elisáková, V., Prouza, M., Sobotka, M., Nesvera, J., and Pátek, M. (2009). Metabolic engineering of the l-valine biosynthesis pathway in Corynebacterium glutamicum using promoter activity modulation. J. Biotechnol. 139, 203–210. doi:10.1016/j.jbiotec.2008.12.005
Jackson, H., Braun, C. L., and Ernst, H. (2008). The chemistry of novel xanthophyll carotenoids. Am. J. Cardiol. 101, 50D–57D. doi:10.1016/j.amjcard.2008.02.008
Jacobson, G., Jolly, S., Sedmak, J., Skatrud, T., Wasileski, J., Inventors. (1999). Astaxanthin Over-Producing Strains of Phaffia rhodozyma, Methods for Their Cultivation, and Their Use in Animal Feeds. Assignee: Archer Daniels Midland Company, Decatur, IL. United States Patent Application 08/967,034.
Jensen, J. V., and Wendisch, V. F. (2013). Ornithine cyclodeaminase-based proline production by Corynebacterium glutamicum. Microb. Cell Fact. 12, 63. doi:10.1186/1475-2859-12-63
Jomaa, H., Wiesner, J., Sanderbrand, S., Altincicek, B., Weidemeyer, C., Hintz, M., et al. (1999). Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285, 1573–1576. doi:10.1126/science.285.5433.1573
Julsing, M. K., Rijpkema, M., Woerdenbag, H. J., Quax, W. J., and Kayser, O. (2007). Functional analysis of genes involved in the biosynthesis of isoprene in Bacillus subtilis. Appl. Microbiol. Biotechnol. 75, 1377–1384. doi:10.1007/s00253-007-0953-5
Kajiwara, S., Fraser, P. D., Kondo, K., and Misawa, N. (1997). Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 324(Pt 2), 421–426.
Kim, S. W., and Keasling, J. D. (2001). Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 72, 408–415. doi:10.1002/1097-0290(20000220)72:4<408::AID-BIT1003>3.0.CO;2-H
Krubasik, P., Takaichi, S., Maoka, T., Kobayashi, M., Masamoto, K., and Sandmann, G. (2001). Detailed biosynthetic pathway to decaprenoxanthin diglucoside in Corynebacterium glutamicum and identification of novel intermediates. Arch. Microbiol. 176, 217–223. doi:10.1007/s002030100315
Lange, B. M., Rujan, T., Martin, W., and Croteau, R. (2000). Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. U.S.A. 97, 13172–13177. doi:10.1073/pnas.240454797
Lee, P. C., and Schmidt-Dannert, C. (2002). Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl. Microbiol. Biotechnol. 60, 1–11. doi:10.1007/s00253-002-1101-x
Lorenz, R. T., and Cysewski, G. R. (2000). Commercial potential for Haematococcus microalgae as a natural source of astaxanthin. Trends Biotechnol. 18, 160–167. doi:10.1016/S0167-7799(00)01433-5
Martin, V. J., Pitera, D. J., Withers, S. T., Newman, J. D., and Keasling, J. D. (2003). Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21, 796–802. doi:10.1038/nbt833
Matano, C., Uhde, A., Youn, J. W., Maeda, T., Clermont, L., Marin, K., et al. (2014). Engineering of Corynebacterium glutamicum for growth and l-lysine and lycopene production from N-acetyl-glucosamine. Appl. Microbiol. Biotechnol. 98, 5633–5643. doi:10.1007/s00253-014-5676-9
Matthews, P. D., and Wurtzel, E. T. (2000). Metabolic engineering of carotenoid accumulation in Escherichia coli by modulation of the isoprenoid precursor pool with expression of deoxyxylulose phosphate synthase. Appl. Microbiol. Biotechnol. 53, 396–400. doi:10.1007/s002530051632
Meiswinkel, T. M., Rittmann, D., Lindner, S. N., and Wendisch, V. F. (2013). Crude glycerol-based production of amino acids and putrescine by Corynebacterium glutamicum. Bioresour. Technol. 145, 254–258. doi:10.1016/j.biortech.2013.02.053
Miki, W. (1991). Biological functions and activities of animal carotenoids. Pure Appl. Chem. 63, 141–146. doi:10.1351/pac199163010141
Mimitsuka, T., Sawai, H., Hatsu, M., and Yamada, K. (2007). Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci. Biotechnol. Biochem. 71, 2130–2135. doi:10.1271/bbb.60699
Misawa, N., Nakagawa, M., Kobayashi, K., Yamano, S., Izawa, Y., Nakamura, K., et al. (1990). Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172, 6704–6712.
Nowroozi, F. F., Baidoo, E. E., Ermakov, S., Redding-Johanson, A. M., Batth, T. S., Petzold, C. J., et al. (2014). Metabolic pathway optimization using ribosome binding site variants and combinatorial gene assembly. Appl. Microbiol. Biotechnol. 98, 1567–1581. doi:10.1007/s00253-013-5361-4
Peralta-Yahya, P. P., Ouellet, M., Chan, R., Mukhopadhyay, A., Keasling, J. D., and Lee, T. S. (2011). Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun. 2, 483. doi:10.1038/ncomms1494
Peters-Wendisch, P. G., Schiel, B., Wendisch, V. F., Katsoulidis, E., Mockel, B., Sahm, H., et al. (2001). Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3, 295–300.
Puan, K. J., Wang, H., Dairi, T., Kuzuyama, T., and Morita, C. T. (2005). fldA is an essential gene required in the 2-C-methyl-d-erythritol 4-phosphate pathway for isoprenoid biosynthesis. FEBS Lett. 579, 3802–3806. doi:10.1016/j.febslet.2005.05.047
Riesenberg, D., and Guthke, R. (1999). High-cell-density cultivation of microorganisms. Appl. Microbiol. Biotechnol. 51, 422–430. doi:10.1007/s002530051412
Rittmann, D., Lindner, S. N., and Wendisch, V. F. (2008). Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 74, 6216–6222. doi:10.1128/AEM.00963-08
Rodriguez-Concepcion, M., and Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130, 1079–1089. doi:10.1104/pp.007138
Rodriguez-Saiz, M., De La Fuente, J. L., and Barredo, J. L. (2010). Xanthophyllomyces dendrorhous for the industrial production of astaxanthin. Appl. Microbiol. Biotechnol. 88, 645–658. doi:10.1007/s00253-010-2814-x
Rodriguez-Villalon, A., Perez-Gil, J., and Rodriguez-Concepcion, M. (2008). Carotenoid accumulation in bacteria with enhanced supply of isoprenoid precursors by upregulation of exogenous or endogenous pathways. J. Biotechnol. 135, 78–84. doi:10.1016/j.jbiotec.2008.02.023
Rohdich, F., Bacher, A., and Eisenreich, W. (2004). Perspectives in anti-infective drug design. The late steps in the biosynthesis of the universal terpenoid precursors, isopentenyl diphosphate and dimethylallyl diphosphate. Bioorg. Chem. 32, 292–308. doi:10.1016/j.bioorg.2004.05.012
Rohdich, F., Hecht, S., Gartner, K., Adam, P., Krieger, C., Amslinger, S., et al. (2002). Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. U.S.A. 99, 1158–1163. doi:10.1073/pnas.032658999
Rohmer, M. (1999). The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 16, 565–574. doi:10.1039/a709175c
Rohmer, M., Knani, M., Simonin, P., Sutter, B., and Sahm, H. (1993). Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 295(Pt 2), 517–524.
Sakai, S., Tsuchida, Y., Okino, S., Ichihashi, O., Kawaguchi, H., Watanabe, T., et al. (2007). Effect of lignocellulose-derived inhibitors on growth of and ethanol production by growth-arrested Corynebacterium glutamicum R. Appl. Environ. Microbiol. 73, 2349–2353. doi:10.1128/AEM.01858-07
Sambrook, J., and Russell, D. (2001). Molecular Cloning. A Laboratory Manual, 3rd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratoy Press.
Sauret-Gueto, S., Ramos-Valdivia, A., Ibanez, E., Boronat, A., and Rodriguez-Concepcion, M. (2003). Identification of lethal mutations in Escherichia coli genes encoding enzymes of the methylerythritol phosphate pathway. Biochem. Biophys. Res. Commun. 307, 408–415. doi:10.1016/S0006-291X(03)01211-7
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Puhler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi:10.1016/0378-1119(94)90324-7
Schäfer, T., Selig, M., and Schönheit, P. (1993). Acetyl-CoA synthetase (ADP forming) in archaea, a novle enzyme involved in acetate formation and ATP synthesis. Arch. Microbiol. 159, 72–83. doi:10.1007/BF00244267
Schmidt, I., Schewe, H., Gassel, S., Jin, C., Buckingham, J., Humbelin, M., et al. (2011). Biotechnological production of astaxanthin with Phaffia rhodozyma/Xanthophyllomyces dendrorhous. Appl. Microbiol. Biotechnol. 89, 555–571. doi:10.1007/s00253-010-2976-6
Schneider, J., Eberhardt, D., and Wendisch, V. F. (2012). Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system. Appl. Microbiol. Biotechnol. 95, 169–178. doi:10.1007/s00253-012-3956-9
Schneider, J., and Wendisch, V. F. (2010). Putrescine production by engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 88, 859–868. doi:10.1007/s00253-010-2778-x
Seibold, G., Auchter, M., Berens, S., Kalinowski, J., and Eikmanns, B. J. (2006). Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J. Biotechnol. 124, 381–391. doi:10.1016/j.jbiotec.2005.12.027
Sivy, T. L., Fall, R., and Rosenstiel, T. N. (2011). Evidence of isoprenoid precursor toxicity in Bacillus subtilis. Biosci. Biotechnol. Biochem. 75, 2376–2383. doi:10.1271/bbb.110572
Sprenger, G. A., Schorken, U., Wiegert, T., Grolle, S., De Graaf, A. A., Taylor, S. V., et al. (1997). Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. U.S.A. 94, 12857–12862. doi:10.1073/pnas.94.24.12857
Stansen, C., Uy, D., Delaunay, S., Eggeling, L., Goergen, J. L., and Wendisch, V. F. (2005). Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 71, 5920–5928. doi:10.1128/AEM.71.10.5920-5928.2005
Steinbüchel, A. (2003). Production of rubber-like polymers by microorganisms. Curr. Opin. Microbiol. 6, 261–270. doi:10.1016/S1369-5274(03)00061-4
Testa, C. A., and Brown, M. J. (2003). The methylerythritol phosphate pathway and its significance as a novel drug target. Curr. Pharm. Biotechnol. 4, 248–259. doi:10.2174/1389201033489784
Tsuchidate, T., Tateno, T., Okai, N., Tanaka, T., Ogino, C., and Kondo, A. (2011). Glutamate production from beta-glucan using endoglucanase-secreting Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 90, 895–901. doi:10.1007/s00253-011-3116-7
Uhde, A., Youn, J. W., Maeda, T., Clermont, L., Matano, C., Kramer, R., et al. (2013). Glucosamine as carbon source for amino acid-producing Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 97, 1679–1687. doi:10.1007/s00253-012-4313-8
van der Rest, M. E., Lange, C., and Molenaar, D. (1999). A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52, 541–545. doi:10.1007/s002530051557
van Ooyen, J., Emer, D., Bussmann, M., Bott, M., Eikmanns, B. J., and Eggeling, L. (2011). Citrate synthase in Corynebacterium glutamicum is encoded by two gltA transcripts which are controlled by RamA, RamB, and GlxR. J. Biotechnol. 154, 140–148. doi:10.1016/j.jbiotec.2010.07.004
Withers, S. T., Gottlieb, S. S., Lieu, B., Newman, J. D., and Keasling, J. D. (2007). Identification of isopentenol biosynthetic genes from Bacillus subtilis by a screening method based on isoprenoid precursor toxicity. Appl. Environ. Microbiol. 73, 6277–6283. doi:10.1128/AEM.00861-07
Xiang, S., Usunow, G., Lange, G., Busch, M., and Tong, L. (2007). Crystal structure of 1-deoxy-d-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J. Biol. Chem. 282, 2676–2682. doi:10.1074/jbc.M610235200
Xiao, Y., Zhao, Z. K., and Liu, P. (2008). Mechanistic studies of IspH in the deoxyxylulose phosphate pathway: heterolytic C-O bond cleavage at C4 position. J. Am. Chem. Soc. 130, 2164–2165. doi:10.1021/ja710245d
Yuan, L. Z., Rouviere, P. E., Larossa, R. A., and Suh, W. (2006). Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab. Eng. 8, 79–90. doi:10.1016/j.ymben.2005.08.005
Zelcbuch, L., Antonovsky, N., Bar-Even, A., Levin-Karp, A., Barenholz, U., Dayagi, M., et al. (2013). Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 41, e98. doi:10.1093/nar/gkt151
Zhao, Y., Yang, J., Qin, B., Li, Y., Sun, Y., Su, S., et al. (2011). Biosynthesis of isoprene in Escherichia coli via methylerythritol phosphate (MEP) pathway. Appl. Microbiol. Biotechnol. 90, 1915–1922. doi:10.1007/s00253-011-3199-1
Zhu, F., Zhong, X., Hu, M., Lu, L., Deng, Z., and Liu, T. (2014). In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli. Biotechnol. Bioeng. 111, 1396–1405. doi:10.1002/bit.25198
Keywords: carotenoid production, genome-reduced Corynebacterium glutamicum, MEP pathway, synthetic operons, astaxanthin
Citation: Heider SAE, Wolf N, Hofemeier A, Peters-Wendisch P and Wendisch VF (2014) Optimization of the IPP precursor supply for the production of lycopene, decaprenoxanthin and astaxanthin by Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2:28. doi: 10.3389/fbioe.2014.00028
Received: 02 May 2014; Accepted: 31 July 2014;
Published online: 20 August 2014.
Edited by:
Jean Marie François, Laboratoire d’Ingénierie des Systèmes Biologiques et des Procédés UMR-CNRS 5504, FranceReviewed by:
Klaas J. Jan Hellingwerf, University of Amsterdam, NetherlandsTiangang Liu, Wuhan University, China
Copyright: © 2014 Heider, Wolf, Hofemeier, Peters-Wendisch and Wendisch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Volker F. Wendisch, Faculty of Biology and CeBiTec, Bielefeld University, Universitätsstr. 25, Bielefeld 33615, Germany e-mail: volker.wendisch@uni-bielefeld.de
 Sabine A. E. Heider
Sabine A. E. Heider Natalie Wolf
Natalie Wolf
 Volker F. Wendisch
Volker F. Wendisch