Cofactor engineering for enhancing the flux of metabolic pathways
- 1Department of Biochemical Engineering, University College London, London, UK
- 2Department of Life Sciences, Imperial College London, London, UK
The manufacture of a diverse array of chemicals is now possible with biologically engineered strains, an approach that is greatly facilitated by the emergence of synthetic biology. This is principally achieved through pathway engineering in which enzyme activities are coordinated within a genetically amenable host to generate the product of interest. A great deal of attention is typically given to the quantitative levels of the enzymes with little regard to their overall qualitative states. This highly constrained approach fails to consider other factors that may be necessary for enzyme functionality. In particular, enzymes with physically bound cofactors, otherwise known as holoenzymes, require careful evaluation. Herein, we discuss the importance of cofactors for biocatalytic processes and show with empirical examples why the synthesis and integration of cofactors for the formation of holoenzymes warrant a great deal of attention within the context of pathway engineering.
Introduction
Synthetic biology permits the engineering of biological devices or systems with novel or enhanced functions (Church et al., 2014). Such an approach has numerous applications, most notably in the manufacture of a diverse number of molecules including household chemicals, biofuels, and pharmaceutical drugs (Keasling, 2010). This is principally achieved through pathway engineering in which enzyme activities are carefully coordinated to generate the product of interest. Approaches based on the activities of isolated enzymes (in vitro) or whole cells (in vivo) can be employed for this purpose (Guterl et al., 2012; Stephanopoulos, 2012). The latter approach in particular offers a significant benefit with respect to complex, multi-step pathways that rely on secondary cellular factors, as well as additional pathway processing. Since the enzymes serve as the workhorse components of these biocatalytic systems, both their quantitative levels and qualitative states are important parameters to consider for pathway engineering. A great deal of focus is typically placed on the quantitative levels of the enzyme (Zelcbuch et al., 2013) with little attention given to their overall qualitative states. Thus, the assumption is usually made that these enzyme components are functioning at full capacity. However, this may not always be the case given that a large subset of enzymes depend on cofactors for functionality.
Significance of Cofactors in Biology
All biological organisms possess a network of pathways that lead to the production of metabolites with an array of cellular functions (Feist et al., 2009). The synthesis and interconversion of these metabolites is made possible by the catalytic activities of countless enzymes. By lowering the activation energy barrier, enzymes catalyze reactions at considerably faster rates than their chemical counterparts. This characteristic property along with the relatively high degree of substrate selectivity, permit the use of enzymes as catalysts for industrial purposes. For those enzymes, which rely solely on amino acids for catalysis, the types of reactions are extremely narrow in scope and, for the most part, restricted to acid/base and electrophilic/nucleophilic reactions (Broderick, 2001). To further extend, the range of reactions, enzymes are commonly associated with non-protein moieties known as cofactors (Broderick, 2001).
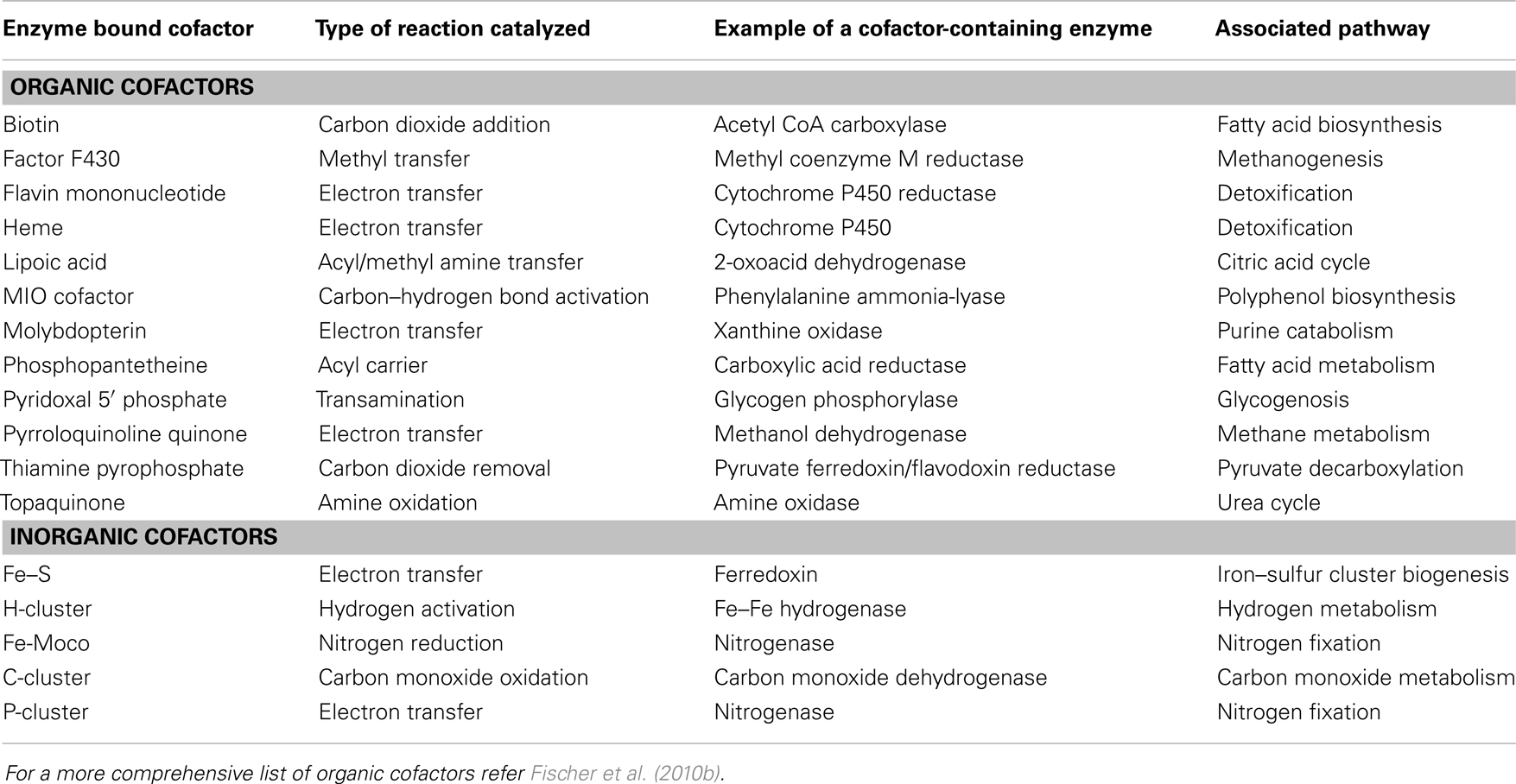
In the broadest sense of the term, cofactors are thought to be associated with well over half of known proteins (Fischer et al., 2010a). However, for this article, the term “cofactor” will refer specifically to those moieties, either organic or inorganic, which remain physically associated with the enzyme throughout the catalytic cycle (Fischer et al., 2010a). This excludes dissociable cosubstrates such as NADPH and glutathione. Additionally, since the primary focus of this article is on pathway engineering, only those cofactors that require de novo pathways for syntheses will be mentioned and sole metal entities such as calcium and selenium will also be excluded. Using this strict definition, a selection of common cofactors are listed in Table 1. These are categorized into two types: organic and inorganic (Rees, 2002; Fischer et al., 2010b). Members of the organic group of cofactors tend to be derivatives of vitamins and undertake numerous types of reactions, while the inorganic group is usually based on various arrangements of iron–sulfur (Fe–S) clusters.
In its cofactor-bound state, enzymes are referred to as holoenzymes while in the unbound state, they are known as apoenzymes (Figure 1A). Two discrete structural parts are required for the synthesis of a holoenzyme: a polypeptide chain and a cofactor moiety. The former is generated by the ubiquitous translational machinery while the latter, depending upon the cofactor, is synthesized by a defined metabolic pathway. In certain cases, subtle variations of the cofactor pathway may exist. For example, in animals, fungi, and α-proteobacteria, heme synthesis is initiated by 5-aminolevulinate synthase, via condensation of glycine and succinyl CoA, while in photosynthetic eukaryotes and some species of the α-proteobacterial group, it depends on glutamate, via the concerted actions of three enzymes (Layer et al., 2010). Once synthesized, the cofactor is integrated with the apoenzyme, either in a co-translational or post-translational manner, to form the holoenzyme. The nature of the association may be covalent and, in such cases, linkages are typically formed with serine, threonine, histidine, tyrosine, and lysine residues and catalyzed by a distinct maturation system. As an example, the heme c in cytochrome c is attached to a cysteine residue, via the vinyl group with the aid of Ccm (cytochrome c maturation) factors (Sanders et al., 2010). Alternatively, the interaction may be tight but non-covalent as in the case of the flavin-containing enzyme, acyl CoA dehydrogenase (Thorpe and Kim, 1995).
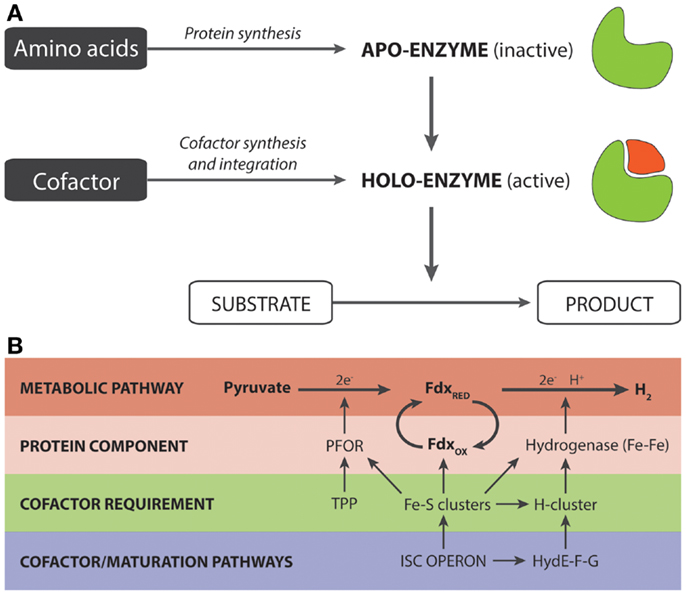
Figure 1. (A) Generalized overview of the synthesis of holoenzymes. (B) Significance of cofactor engineering for enhancing the output of holoenzyme-dependent pathways. The example (Akhtar and Jones, 2009) illustrates a synthetic pyruvate:H2-pathway that is heavily dependent on Fe–S clusters. These clusters are required for (i) the proteins/enzymes directly involved in the pathway for hydrogen production and (ii) the maturation factors that are responsible for the synthesis and integration of the H-cluster present in Fe–Fe hydrogenases.
Importance of Cofactor Synthesis for Both Enzyme and Pathway Functionality
As integral components of numerous holoenzymes, cofactors are required for the majority of metabolic pathways. To name but a few, these include: FAD in succinate dehydrogenase for the Krebs cycle; TPP in transketolase for the pentose phosphate pathway; pyridoxal phosphate in glycogen phosphorylase for glycogenolysis; H-clusters in hydrogenases for hydrogen metabolism; Fe–MoCo in nitrogenases for nitrogen fixation; biotin in acetyl CoA carboxylase for fatty acid biosynthesis; and haem in cytochrome P450 for various detoxification pathways. The most crucial point to consider is that the functional output of holoenzymes can only arise if the apoenzyme is correctly folded with its cofactor. Without the cofactor, such enzymes would be rendered inoperable and the associated pathways would become redundant. This situation, though undesirable, is likely to be encountered in a bottom-up approach to microbial engineering in which a genetically amenable and well-characterized organism, such as Escherichia coli and Saccharomyces cerevisiae may be completely devoid of cofactors necessary for heterologous enzyme activity. Alternatively, the capability of the host to synthesize the required cofactor exists but may be insufficient, resulting in a pool of enzymes with a high ratio of apo to holo form (see next section). The most obvious strategy for resolving these issues would be to resort to “cofactor engineering,” by genetically modifying the host so that the cofactor assembly system is present or that the level of native cofactor assembly is in sufficient supply.
Consider the expression of the clostridial Fe–Fe hydrogenase. This enzyme depends on an H-cluster that essentially is a di-iron arrangement with three carbon monoxide, two cyanide ligands, and one dithiolate bridge (Mulder et al., 2011). The formation of this cluster is catalyzed by three maturation enzymes; namely HydE, HydF, and HydG (Posewitz et al., 2004). Since E. coli is not naturally endowed with the Hyd maturation enzymes, E. coli invariably produces a non-functional Fe–Fe hydrogenase. This can be circumvented by simply complementing the expression of Fe–Fe hydrogenases with the maturation pathway for the H-cluster in order to form the active Fe–Fe hydrogenase (Posewitz et al., 2004; Akhtar and Jones, 2008b). Pyrroloquinoline (PQQ) is another example of a cofactor, which is not naturally synthesized in E. coli (Matsushita et al., 1997). This cofactor, present in a family of quinoproteins, has potential uses in biofuel cells, bioremediation, and biosensing (Matsushita et al., 2002). By incorporating the pqqABCDE gene cluster from Gluconobacter oxydansa, Yang et al. (2010) were able to successfully demonstrate the activity of a PQQ-requiring d-glucose dehydrogenase in E. coli. They noted, however, that the gene cluster was most likely complemented by the native tldD gene. A final example is tetrahydrobiopterin, which in itself is a desirable commodity for the treatment of mild and moderate forms of phenylketonuria (Perez-Duenas et al., 2004). It can be synthesized in vivo from GTP via a three-step pathway comprising GTP cyclohydrolase I, 6-pyruvoyl-tetrahydropterin synthase, and sepiapterin reductase (Yamamoto et al., 2003). By augmenting the pathway with expression of a GTP cyclohydrolase I sourced from Bacillus subtilis, a 1.5-fold improvement was observed with titers reaching as high as 4 g biopterin per liter of culture (Yamamoto et al., 2003).
Cofactor Insertion is Key to Maximizing Total and Specific Holoenzyme Activity
To maximize the specific activity of a holoenzyme, cofactor synthesis would need to be complemented and/or coupled with cofactor insertion. For enzymes, which bind to cofactors in a non-covalent fashion, the process of cofactor insertion is somewhat of an enigma. It is still not known, even for the well-known heme b cofactor, whether this process is facilitated by dedicated in vivo components or is a spontaneous process (Thöny-Meyer, 2009). Though overwhelming in vitro data show that heme b insertion can be a spontaneous event, recent evidence with in vivo model systems have implicated the involvement of cellular factors that have yet to be elucidated (Waheed et al., 2010; Correia et al., 2011). For those enzymes that have covalently attached cofactors, specialized maturation systems have evolved to catalyze both insertion and covalent linkage of the cofactor. Induction of the maturation system can increase holoenzyme activity, as in the case of a carboxylic acid reductase (CAR), which was recently employed for the production of a broad range of chemical commodities (Akhtar et al., 2013). Venkitasubramanian et al. (2007) had verified that CAR requires a cofactor known as phosphopantetheine. This cofactor, during its synthesis, is concomitantly integrated with the enzyme, via a phosphodiester bond, by a maturation enzyme known as phosphopantetheinyl transferase. The sole expression of CAR in E. coli leads to an observable, but exceedingly poor, activity. However, with coexpression of the phosphopantetheinyl transferase Sfp from Bacillus subtilis, the specific activity of CAR can be enhanced several-fold to a level that is on par with one that is purified from the native organism.
In addition to stimulating the specific enzyme activity, increasing the intracellular levels of cofactors is also known to improve the overall production levels of holoenzymes, suggesting a relationship between holo/apo-forms and degradation. This is a phenomenon that has been frequently observed for hemoproteins (Harnastai et al., 2006; Lu et al., 2010, 2013; Michener et al., 2012). By elevating heme levels, via supplementation of the media with δ-aminolevulinic acid, the expression levels of hemoglobin and cytochrome b5 can be significantly improved (Gallagher et al., 1992; Liu et al., 2014). Likewise, increasing Fe–S levels, via overexpression of the isc (Fe–S cluster) operon, also leads to similar effects (Nakamura et al., 1999; Akhtar and Jones, 2008a). An explanation for the increased holoenzyme levels may be gleaned from studies of proteins, which utilize divalent metal ions as cofactors (Wilson et al., 2004; Bushmarina et al., 2006; Palm-Espling et al., 2012). Evidence from these published reports suggests that cofactors may aid in the folding of the polypeptide chain and, in turn, accelerate the formation of a functional protein (Goedken et al., 2000). In the case of ribonuclease HI, metal cofactor integration was found to impart a greater degree of rigidity on the final native conformational state of the protein, in addition to improving the refolding rate of the protein (Wittung-Stafshede, 2002). Further insights on the importance of cofactors in protein folding can be gained with the S-adenosylmethionine-containing biotin synthase. This enzyme relies on an intact Fe–S cluster for the addition of sulfur to dethiobiotin to form the biotin thiophane ring. Reyda et al. (2008) noticed that the loss of the Fe–S cluster destabilized the protein, which led to transient unfolding of specific regions, as well as increased proteolysis. Proteolytic degradation was found to proceed by an apparent ATP-dependent proteolysis mechanism, via sequential cleavage of small C-terminal fragments. Interestingly, it was also speculated that since the activity of the protein is generally well maintained under high-iron conditions, a repair process, possibly mediated by the Isc and/or Suf (Sulfur mobilization) machinery, may be active under conditions of destabilization (Reyda et al., 2008).
Stimulating Cofactor Synthesis Can Enhance the Flux of Synthetic Pathways
With regard to the actual impact that cofactor engineering can have on the metabolic performance of synthetic pathways, two studies are particularly worthy of mention. In the first study relating to the production of the vitamin C precursor, 2-keto-l-gulonic acid, Gao et al. (2013) utilized two PQQ-dependent dehydrogenases with d-sorbitol as the starting substrate. The authors noted that induced expression did not improve titers beyond a certain threshold and hypothesized that PQQ was the limiting factor. This was proven to be correct since induction of a pathway for PQQ synthesis resulted in a 20% increase in overall titer. A later refinement of the work in which the two pathway enzymes were incorporated as a fusion protein in Ketogulonigenium vulgare also resulted in a similar improvement in titer (Gao et al., 2014).
The second study relates to a synthetic pathway for hydrogen production consisting of a pyruvate:ferredoxin oxidoreductase (PFOR, also known as YdbK in the literature), Fdx and Fe–Fe hydrogenase (Akhtar and Jones, 2009). The design of this pathway was based on the observation that pyruvate, rather than NAD(P)H, was a metabolically superior source of electrons for hydrogen synthesis (Veit et al., 2008). Initial structural analysis of the PFOR-based pathway, in addition to the maturation enzymes, revealed that each protein component was associated with at least one Fe–S cluster that essentially provides the route of electron transfer from pyruvate to the H-cluster of the Fe–Fe hydrogenase (Figure 1B). Up to a total of 12 Fe–S clusters were found to be required for the pathway. In E. coli, the formation of Fe–S clusters is undertaken by the isc operon, though an analogous suf operon is also present (Fontecave et al., 2005; Roche et al., 2013). Since the isc operon is controlled by the negative IscR transcriptional regulator, our initial work using a ΔiscR background strain had shown that the levels of holo-ferredoxin and the in vitro hydrogenase activity could be improved two and threefold, respectively, relative to the wild-type strain (Akhtar and Jones, 2008a). Based on this insight and given the heavy demand for Fe–S clusters, we reasoned that the ΔiscR strain may improve the pathway flux toward hydrogen production by increasing the availability of Fe–S clusters. In accordance with our prediction, we observed a twofold improvement in hydrogen yield relative to the wild-type, resulting in an overall yield of 1.5 moles of H2 per mole of glucose. Even more remarkably with addition of TPP, which serves as a cofactor for the PFOR component, the hydrogen yield was further increased to give a final yield of 1.9, out of a theoretical yield of two moles per mole of glucose (Akhtar and Jones, 2009). Given also that both the specific and total in vitro activity of PFOR was improved, this suggests that the availability of the TPP cofactor may well be another potential limiting cofactor for hydrogen production (Akhtar and Jones, 2009).
Although cofactor biosynthesis and integration under native control can be limiting and unresponsive to high-expression levels of the apoform, presumably to conserve cellular resources, cofactor engineering can quite clearly be advantageous for the assembly of metabolic pathways that depend on the activity of heterologous holoenzymes. This benefit presumably arises from the improved holoenzyme activity, via increased structural stability and/or folding rates in conjunction with reduced protein degradation and/or protein unfolding (explained in the previous section). Interestingly, data from our work on the in vitro activity of Fe–S proteins also suggests that, under certain conditions, a steady and constant supply of cofactors may well aid the restoration of damaged or inactivated enzymes (Akhtar and Jones, 2008a).
Concluding Remarks
Achieving a balanced production of polypeptide and cofactor for optimal holoenzyme activity would be an iterative process involving the (i) modulated induction of genes, (ii) monitoring of cofactor levels, (iii) evaluation of enzyme activity, and (iv) evaluation of whole-system productivity. Genetic modulation can be controlled at the transcriptional and translational levels by varying the strengths of promoter and ribosomal binding sites, while cofactor levels can be monitored and quantified using suitable analytical methods, e.g., mass spectrometry, high-performance liquid chromatography (HPLC). Combining this with information relating to specific enzyme activity should allow provide profound insights into the intracellular cofactor levels required to achieve optimal holoenzyme activity. Furthermore, if a product reporter system is available, it may also be possible to screen for optimal cofactor metabolism, for example using RBS-variation libraries (Zelcbuch et al., 2013).
Given the importance of cofactor synthesis and integration for holoenzyme activity, a few key points need to be considered when assembling pathways involving holoenzymes. Firstly, holoenzyme activity will only be possible within a host that is metabolically equipped to synthesize the necessary cofactor, otherwise a complementary pathway for cofactor production would also need to be implemented. This is particularly relevant for synthetic pathways that employ holoenzymes from diverse origins. Secondly, to ensure maximal activity of the holoenzyme, the apoenzyme needs to be sufficiently coupled with the synthesis and insertion of its respective cofactor. An imbalance between the two will lead to poor enzyme activity, one that would most likely be inadequate for catalytic purposes. Even native cofactor biosynthesis may not be optimally tuned or responsive to demand from an over-expressed apoenzyme, as would be required in a biocatalytic system where the objective function has shifted from biomass to metabolite producer. Thirdly, since cofactor stimulation is known to improve the stability and activity of holoenzymes, cofactor engineering is likely to be a useful strategy for enhancing the total activity of all holoenzymes in the engineered pathway, and maximizing chances for high flux toward the product of interest. As it currently stands, the impact of cofactor engineering on the activity of holoenzymes is still very much a poorly studied area and certainly warrants more attention given its potential impact on the success of engineered biocatalytic systems.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work that spurred our views, reflected within this perspective, was supported in part by the Academy of Finland project no. 25369 and European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/European Research Council Grant Agreement 260661 (Patrik R. Jones).
References
Akhtar, M., and Jones, P. (2008a). Deletion of iscR stimulates recombinant clostridial Fe-Fe hydrogenase activity and H2-accumulation in Escherichia coli BL21(DE3). Appl. Microbiol. Biotechnol. 78, 853–862. doi: 10.1007/s00253-008-1377-6
Akhtar, M., and Jones, P. (2008b). Engineering of a synthetic hydF-hydE-hydG-hydA operon for biohydrogen production. Anal. Biochem. 373, 170–172. doi:10.1016/j.ab.2007.10.018
Akhtar, M., and Jones, P. (2009). Construction of a synthetic YdbK-dependent pyruvate: H2 pathway in Escherichia coli BL21(DE3). Metab. Eng. 11, 139–147. doi:10.1016/j.ymben.2009.01.002
Akhtar, M. K., Turner, N. J., and Jones, P. R. (2013). Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. U.S.A. 110, 87–92. doi:10.1073/pnas.1216516110
Bushmarina, N. A., Blanchet, C. E., Vernier, G., and Forge, V. (2006). Cofactor effects on the protein folding reaction: acceleration of alpha-lactalbumin refolding by metal ions. Protein Sci. 15, 659–671. doi:10.1110/ps.051904206
Church, G. M., Elowitz, M. B., Smolke, C. D., Voigt, C. A., and Weiss, R. (2014). Realizing the potential of synthetic biology. Nat. Rev. Mol. Cell Biol. 15, 289–294. doi:10.1038/nrm3767
Correia, M. A., Sinclair, P. R., and De Matteis, F. (2011). Cytochrome P450 regulation: the interplay between its heme and apoprotein moieties in synthesis, assembly, repair, and disposal. Drug Metab. Rev. 43, 1–26. doi:10.3109/03602532.2010.515222
Feist, A., Herrgård, M., Thiele, I., Reed, J., and Palsson, B. (2009). Reconstruction of biochemical networks in microorganisms. Nat. Rev. Microbiol. 7, 129–143. doi:10.1038/nrmicro1949
Fischer, J. D., Holliday, G. L., Rahman, S. A., and Thornton, J. M. (2010a). The structures and physicochemical properties of organic cofactors in biocatalysis. J. Mol. Biol. 403, 803–824. doi:10.1016/j.jmb.2010.09.018
Fischer, J. D., Holliday, G. L., and Thornton, J. M. (2010b). The CoFactor database: organic cofactors in enzyme catalysis. Bioinformatics 26, 2496–2497. doi:10.1093/bioinformatics/btq442
Fontecave, M., Choudens, S. O., Py, B., and Barras, F. (2005). Mechanisms of iron-sulfur cluster assembly: the SUF machinery. J. Biol. Inorg. Chem. 10, 713–721. doi:10.1007/s00775-005-0025-1
Gallagher, J., Kaderbhai, N., and Kaderbhai, M. A. (1992). Gene-dose-dependent expression of soluble mammalian cytochrome b5 in Escherichia coli. Appl. Microbiol. Biotechnol. 38, 77–83. doi:10.1007/BF00169423
Gao, L., Du, G., Zhou, J., Chen, J., and Liu, J. (2013). Characterization of a group of pyrroloquinoline quinone-dependent dehydrogenases that are involved in the conversion of l-sorbose to 2-Keto-l-gulonic acid in Ketogulonicigenium vulgare WSH-001. Biotechnol. Prog. 29, 1398–1404. doi:10.1002/btpr.1803
Gao, L., Hu, Y., Liu, J., Du, G., Zhou, J., and Chen, J. (2014). Stepwise metabolic engineering of gluconobacter oxydans WSH-003 for the direct production of 2-keto-l-gulonic acid from d-sorbitol. Metab. Eng. 24, 30–37. doi:10.1016/j.ymben.2014.04.003
Goedken, E. R., Keck, J. L., Berger, J. M., and Marqusee, S. (2000). Divalent metal cofactor binding in the kinetic folding trajectory of Escherichia coli ribonuclease HI. Protein Sci. 9, 1914–1921. doi:10.1110/ps.9.10.1914
Guterl, J. K., Garbe, D., Carsten, J., Steffler, F., Sommer, B., Reisse, S., et al. (2012). Cell-free metabolic engineering: production of chemicals by minimized reaction cascades. ChemSusChem 5, 2165–2172. doi:10.1002/cssc.201200365
Harnastai, I. N., Gilep, A. A., and Usanov, S. A. (2006). The development of an efficient system for heterologous expression of cytochrome P450s in Escherichia coli using hemA gene co-expression. Protein Expr. Purif. 46, 47–55. doi:10.1016/j.pep.2005.07.006
Keasling, J. D. (2010). Manufacturing molecules through metabolic engineering. Science 330, 1355–1358. doi:10.1126/science.1193990
Layer, G., Reichelt, J., Jahn, D., and Heinz, D. W. (2010). Structure and function of enzymes in heme biosynthesis. Protein Sci. 19, 1137–1161. doi:10.1002/pro.405
Liu, L., Martinez, J. L., Liu, Z., Petranovic, D., and Nielsen, J. (2014). Balanced globin protein expression and heme biosynthesis improve production of human hemoglobin in Saccharomyces cerevisiae. Metab. Eng. 21, 9–16. doi:10.1016/j.ymben.2013.10.010
Lu, H., Ma, J., Liu, N., and Wang, S. (2010). Effects of heme precursors on CYP1A2 and POR expression in the baculovirus/Spodoptera frugiperda system. J. Biomed. Res. 24, 242–249. doi:10.1016/S1674-8301(10)60034-6
Lu, H. Y., Qiu, L. L., Yang, X. J., Zhang, X. M., Zhang, Z., and Wang, S. L. (2013). Optimization of heme precursors for the expression of human cytochrome P450 2A13 and its co-expression with oxidoreductase in baculovirus/sf9 system. J. Biochem. 153, 555–563. doi:10.1093/jb/mvt018
Matsushita, K., Arents, J. C., Bader, R., Yamada, M., Adachi, O., and Postma, P. W. (1997). Escherichia coli is unable to produce pyrroloquinoline quinone (PQQ). Microbiology 143, 3149–3156. doi:10.1099/00221287-143-10-3149
Matsushita, K., Toyama, H., Yamada, M., and Adachi, O. (2002). Quinoproteins: structure, function, and biotechnological applications. Appl. Microbiol. Biotechnol. 58, 13–22. doi:10.1007/s00253-001-0851-1
Michener, J. K., Nielsen, J., and Smolke, C. D. (2012). Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 109, 19504–19509. doi:10.1073/pnas.1212287109
Mulder, D. W., Shepard, E. M., Meuser, J. E., Joshi, N., King, P. W., Posewitz, M. C., et al. (2011). Insights into [FeFe]-hydrogenase structure, mechanism, and maturation. Structure 19, 1038–1052. doi:10.1016/j.str.2011.06.008
Nakamura, M., Saeki, K., and Takahashi, Y. (1999). Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hs cA-fdx-ORF3 gene cluster. J. Biochem. 126, 10–18. doi:10.1093/oxfordjournals.jbchem.a022409
Palm-Espling, M. E., Niemiec, M. S., and Wittung-Stafshede, P. (2012). Role of metal in folding and stability of copper proteins in vitro. Biochim. Biophys. Acta 1823, 1594–1603. doi:10.1016/j.bbamcr.2012.01.013
Perez-Duenas, B., Vilaseca, M. A., Mas, A., Lambruschini, N., Artuch, R., Gomez, L., et al. (2004). Tetrahydrobiopterin responsiveness in patients with phenylketonuria. Clin. Biochem. 37, 1083–1090. doi:10.1016/j.clinbiochem.2004.09.005
Posewitz, M. C., King, P. W., Smolinski, S. L., Zhang, L., Seibert, M., and Ghirardi, M. L. (2004). Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279, 25711–25720. doi:10.1074/jbc.M403206200
Rees, D. C. (2002). Great metalloclusters in enzymology. Annu. Rev. Biochem. 71, 221–246. doi:10.1146/annurev.biochem.71.110601.135406
Reyda, M. R., Dippold, R., Dotson, M. E., and Jarrett, J. T. (2008). Loss of iron-sulfur clusters from biotin synthase as a result of catalysis promotes unfolding and degradation. Arch. Biochem. Biophys. 471, 32–41. doi:10.1016/j.abb.2007.12.001
Roche, B., Aussel, L., Ezraty, B., Mandin, P., Py, B., and Barras, F. (2013). Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta 1827, 455–469. doi:10.1016/j.bbabio.2012.12.010
Sanders, C., Turkarslan, S., Lee, D. W., and Daldal, F. (2010). Cytochrome c biogenesis: the Ccm system. Trends Microbiol. 18, 266–274. doi:10.1016/j.tim.2010.03.006
Stephanopoulos, G. (2012). Synthetic biology and metabolic engineering. ACS Synth. Biol. 1, 514–525. doi:10.1021/sb300094q
Thöny-Meyer, L. (2009). “Heme transport and incorporation into proteins,” in Tetrapyrroles: Birth, Life and Death, eds M. J. Warren and A. G. Smith (Landes Bioscience and Springer Science+Business Media), 149–159.
Thorpe, C., and Kim, J. J. (1995). Structure and mechanism of action of the acyl-CoA dehydrogenases. FASEB J. 9, 718–725.
Veit, A., Akhtar, M., Mizutani, T., and Jones, P. (2008). Constructing and testing the thermodynamic limits of synthetic NAD(P)H:H2 pathways. Microb. Biotechnol. 1, 382–394. doi:10.1111/j.1751-7915.2008.00033.x
Venkitasubramanian, P., Daniels, L., and Rosazza, J. P. (2007). Reduction of carboxylic acids by Nocardia aldehyde oxidoreductase requires a phosphopantetheinylated enzyme. J. Biol. Chem. 282, 478–485. doi:10.1074/jbc.M607980200
Waheed, S. M., Ghosh, A., Chakravarti, R., Biswas, A., Haque, M. M., Panda, K., et al. (2010). Nitric oxide blocks cellular heme insertion into a broad range of heme proteins. Free Radic. Biol. Med. 48, 1548–1558. doi:10.1016/j.freeradbiomed.2010.02.038
Wilson, C. J., Apiyo, D., and Wittung-Stafshede, P. (2004). Role of cofactors in metalloprotein folding. Q. Rev. Biophys. 37, 285–314. doi:10.1017/S003358350500404X
Wittung-Stafshede, P. (2002). Role of cofactors in protein folding. Acc. Chem. Res. 35, 201–208. doi:10.1021/ar010106e
Yamamoto, K., Kataoka, E., Miyamoto, N., Furukawa, K., Ohsuye, K., and Yabuta, M. (2003). Genetic engineering of Escherichia coli for production of tetrahydrobiopterin. Metab. Eng. 5, 246–254. doi:10.1016/S1096-7176(03)00046-6
Yang, X. P., Zhong, G. F., Lin, J. P., Mao, D. B., and Wei, D. Z. (2010). Pyrroloquinoline quinone biosynthesis in Escherichia coli through expression of the gluconobacter oxydans pqqABCDE gene cluster. J. Ind. Microbiol. Biotechnol. 37, 575–580. doi:10.1007/s10295-010-0703-z
Keywords: cofactors, metabolic pathway engineering, Fe–S clusters, enzymatic activity, synthetic biology
Citation: Akhtar MK and Jones PR (2014) Cofactor engineering for enhancing the flux of metabolic pathways. Front. Bioeng. Biotechnol. 2:30. doi: 10.3389/fbioe.2014.00030
Received: 12 June 2014; Accepted: 12 August 2014;
Published online: 28 August 2014.
Edited by:
Pablo Carbonell, University of Evry, FranceReviewed by:
Juan Manuel Pedraza, Universidad de los Andes, ColombiaGary Sawers, Martin-Luther University Halle-Wittenberg, Germany
Copyright: © 2014 Akhtar and Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Kalim Akhtar, Department of Biochemical Engineering, University College London, Torrington Place, London, WC1E 7JE, UK e-mail: kalim.akhtar@ucl.ac.uk;
Patrik R. Jones, Department of Life Sciences, Imperial College London, Sir Alexander Fleming building, London, SW7 2AZ, UK e-mail: p.jones@imperial.ac.uk
 M. Kalim Akhtar
M. Kalim Akhtar Patrik R. Jones
Patrik R. Jones