Cardio-miRNAs and onco-miRNAs: circulating miRNA-based diagnostics for non-cancerous and cancerous diseases
- Department of Omics Network, National Cancer Center, Tokyo, Japan
Cardiovascular diseases and cancers are the leading causes of morbidity and mortality in the world. MicroRNAs (miRNAs) are short non-coding RNAs that primarily repress target mRNAs. Here, miR-24, miR-125b, miR-195, and miR-214 were selected as representative cardio-miRs that are upregulated in human heart failure. To bridge the gap between miRNA studies in cardiology and oncology, the targets and functions of these miRNAs in cardiovascular diseases and cancers will be reviewed. ACVR1B, BCL2, BIM, eNOS, FGFR3, JPH2, MEN1, MYC, p16, and ST7L are miR-24 targets that have been experimentally validated in human cells. ARID3B, BAK1, BCL2, BMPR1B, ERBB2, FGFR2, IL6R, MUC1, SITR7, Smoothened, STAT3, TET2, and TP53 are representative miR-125b targets. ACVR2A, BCL2, CCND1, E2F3, GLUT3, MYB, RAF1, VEGF, WEE1, and WNT7A are representative miR-195 targets. BCL2L2, ß-catenin, BIM, CADM1, EZH2, FGFR1, NRAS, PTEN, TP53, and TWIST1 are representative miR-214 targets. miR-125b is a good cardio-miR that protects cardiomyocytes; miR-195 is a bad cardio-miR that elicits cardiomyopathy and heart failure; miR-24 and miR-214 are bi-functional cardio-miRs. By contrast, miR-24, miR-125b, miR-195, and miR-214 function as oncogenic or tumor suppressor miRNAs in a cancer (sub)type-dependent manner. Circulating miR-24 is elevated in diabetes, breast cancer and lung cancer. Circulating miR-195 is elevated in acute myocardial infarction, breast cancer, prostate cancer and colorectal adenoma. Circulating miR-125b and miR-214 are elevated in some cancers. Cardio-miRs and onco-miRs bear some similarities in functions and circulation profiles. miRNAs regulate WNT, FGF, Hedgehog and other signaling cascades that are involved in orchestration of embryogenesis and homeostasis as well as pathogenesis of human diseases. Because circulating miRNA profiles are modulated by genetic and environmental factors and are dysregulated by genetic and epigenetic alterations in somatic cells, circulating miRNA association studies (CMASs) within several thousands of cases each for common non-cancerous diseases and major cancers are necessary for miRNA-based diagnostics.
Introduction
MicroRNAs (miRNAs) are short non-coding RNAs that primarily repress protein expression from target mRNAs with imperfect or perfect complementarity through mRNA degradation and translational inhibition or mRNA cleavage, respectively (Kasinski and Slack, 2011; van Rooij and Olson, 2012). For example, miR-15, miR-16, miR-20a, and miR-20b are anti-angiogenic miRNAs that repress VEGFA (VEGF) (Wang and Olson, 2009; Katoh, 2013b). miR-200 family members inhibit epithelial-to-mesenchymal transition (EMT) and self-renewal of stem cells through repression of ZEB1/2 and BMI1, respectively (Katoh and Katoh, 2008; Oishi et al., 2012; Feng et al., 2014). miRNAs regulate a variety of cellular processes, such as stemness, proliferation, senescence, apoptosis, inflammatory cytokine production, EMT, metastasis and drug resistance.
Cardiovascular diseases and cancers are the leading causes of morbidity and mortality in the world (Lozano et al., 2012). miRNAs involved in heart diseases (Divakaran and Mann, 2008), vascular diseases (Wang and Olson, 2009) and cancers (Croce, 2009) are designated cardio-miRs, angio-miRs, and onco-miRs, respectively, and the same miRNA can function as a cardio-miR, angio-miR or onco-miR in a context-dependent manner. Among 32,233 miRNA manuscripts in the PubMed database, 3334 and 15,740 manuscripts were extracted by using cardiovascular and oncological terms, respectively, and only 926 manuscripts were extracted by using both terms (Figure 1A), which indicates that the outcomes of miRNA studies might not be efficiently shared between the different disciplines. To bridge the gap between miRNA studies in cardiology and oncology, representative cardio-miRs upregulated in human heart failure were selected based on database screening. The targets and functions of these miRNAs in cardiovascular diseases and cancers are comprehensively reviewed, and then circulating miRNA-based diagnostics for non-cancerous and cancerous diseases are discussed with a focus on personal diversity related to genetic and environmental factors.
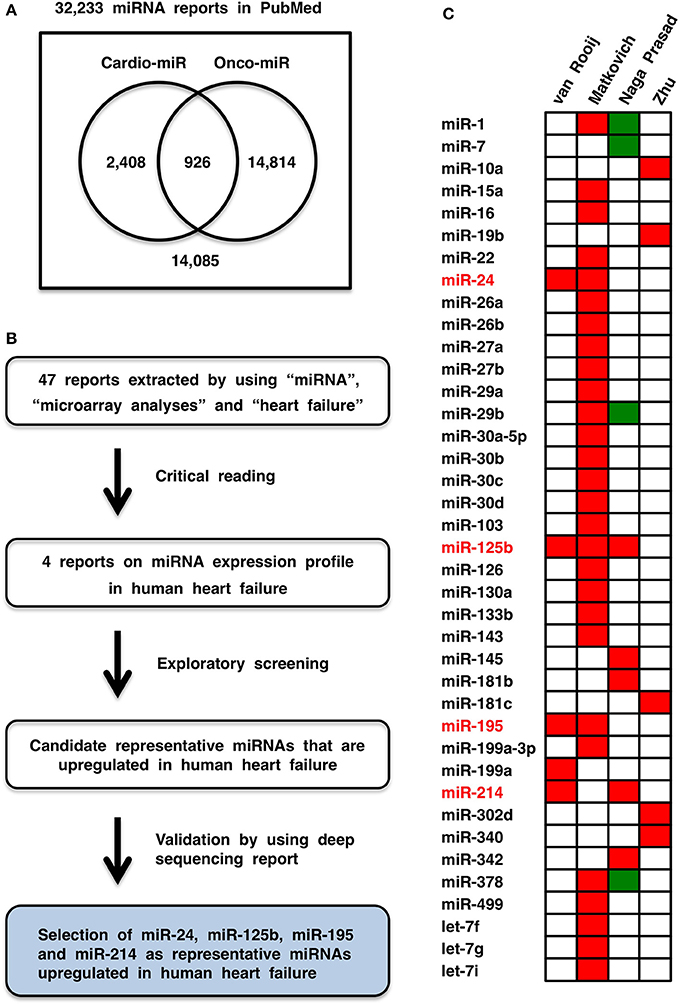
Figure 1. Cardio-miRNAs upregulated in human heart failure. (A) Bibliology of miRNAs, cardio-miRs, and onco-miRs. (B) Flowchart of representative cardio-miR selection. Based on exploratory screening of miRNA expression reports (van Rooij et al., 2006; Matkovich et al., 2009; Naga Prasad et al., 2009; Zhu et al., 2013) and validation process utilizing next-generation sequence report (Leptidis et al., 2013), miR-24, miR-125b, miR-195, and miR-214 were selected as representative cardio-miRs that are upregulated in human heart failure. (C) Heat map of miRNA expression profiles in human heart failure. Red, upregulated; green, downregulated.
Representative Cardio-miRs Upregulated in Heart Failure
Heart failure is a progressive decline in cardiac functions that occurs at the end stage of cardiovascular diseases, such as ischemic heart disease, hypertension and diabetes (Hill and Olson, 2008; Shah and Mann, 2011; Zhou et al., 2013b). Myocardial infarction is caused by coronary artery occlusion, which leads to the death of cardiomyocytes in the infarcted region owing to insufficient oxygen supply. Ischemic stress occurs in surviving cardiomyocytes in the surrounding or peripheral area of an infarcted region, and then hypertrophic growth of myocardiocytes and interstitial fibrosis occur in the non-infarcted region of the heart. By contrast, persistent pressure overload causes cardiac wall thickening of the left ventricle and hypertrophic growth of cardiomyocytes. Cardiac hypertrophy leads to maladaptive remodeling of the left ventricle and eventually results in patient death owing to fatal arrhythmia and/or heart failure.
Forty-seven reports were recovered by initial screening of the literature in the PubMed and Web of Science (WoS) databases by using “heart failure,” “miRNA or miRNAs,” and “microarray.” Then, four reports on microarray analyses (van Rooij et al., 2006; Matkovich et al., 2009; Naga Prasad et al., 2009; Zhu et al., 2013) were selected by critical reading (Figure 1B). Based on the criterion “miRNA that is upregulated in at least two reports on microarray analyses,” miR-24, miR-125b, miR-195, and miR-214 were selected as candidate representative cardio-miRs that are upregulated in human heart failure (Figure 1C). Because data obtained by using microarray analyses are not always correct, upregulation of miR-24, miR-125b, miR-195, and miR-214 in human heart failure were then validated by using a deep sequencing report on miRNA profiles in human heart failure (Leptidis et al., 2013). Based on the exploration and validation processes, miR-24, miR-125b, miR-195, and miR-214 were designated the representative cardio-miRs upregulated in human heart failure (Figure 1B).
miR-24
Human chromosomal loci of miR-24 genes
miR-24 is derived from the miR-23b/miR-27b/miR24-1 locus at human chromosome 9q22.32 and the miR-23a/miR-27a/miR-24-2 locus at human chromosome 19p13.13 (Figure 2).
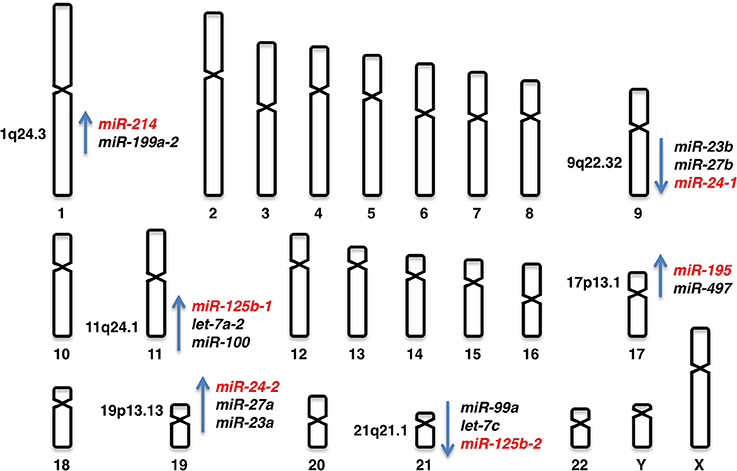
Figure 2. Human chromosomal loci of miR-24-1, miR-24-2, miR-125b-1, miR-125b-2, miR-195, and miR-214.
Targets of miR-24
miRNA targets demonstrated in rodents are not always conserved in humans owing to species divergence (Le et al., 2009), while putative miRNA targets predicted by using bioinformatics tools, such as TargetScan (http://www.targetscan.org), PicTar (http://pictar.mdc-berlin.de) and miRanda (http://www.microrna.org), are not always true. In this review, miRNA targets validated in human cells are listed up (Table 1).
ACVR1B (Activin receptor 1B) (Wang et al., 2008a), ARHGAP19 (Amelio et al., 2012), AURKB (Aurora kinase B) (Lal et al., 2009a), BCL2 (Srivastava et al., 2011), BCL2L11 (pro-apoptotic BIM) (Qian et al., 2011), CCNA2 (Cyclin A2) (Lal et al., 2009a), CDC2 (Lal et al., 2009a), CDK4 (Cyclin-dependent kinase 4) (Lal et al., 2009a), CDKN1B (p27 KIP1) (Giglio et al., 2013), CDKN2A (p16 INK4a) (Lal et al., 2008), DHFR (Dihydrofolate reductase) (Mishra et al., 2007), DIAPH1 (Diaphanos homolog 1) (Zhou et al., 2013a), DUSP16 (MKP7) (Zaidi et al., 2009), E2F2 (Lal et al., 2009a), eNOS (NOS3) (Meloni et al., 2013), FAF1 (Fas-associated factor 1) (Qin et al., 2010), FEN1 (Lal et al., 2009a), FGFR3 (FGF receptor 3) (Rio-Machin et al., 2013), GATA2 (Fiedler et al., 2011), H2AFX (Histone H2AX) (Lal et al., 2009b), HNF4A (HNF4α) (Takagi et al., 2010), JPH2 (Junctophilin 2) (Xu et al., 2012b), LIMK2 (LIM-domain kinase 2) (Zhou et al., 2013a), MEN1 (Luzi et al., 2012), MYC (c-Myc) (Lal et al., 2009a), NET1 (NET1A or ARHGEF8) (Papadimitriou et al., 2012), PAK4 (Fiedler et al., 2011), PTPN9 (Protein tyrosine phosphatase, non-receptor type 9) (Du et al., 2013), PTPRF (Protein tyrosine phosphatase, receptor type F) (Du et al., 2013), RASA1 (Ras GAP) (Fiedler et al., 2011), SH3PXD2A (TSK5) (Amelio et al., 2012), SLC4A1 (Anion exchanger 1) (Wu et al., 2010), SPRY2 (Sprouty homolog 2) (Li et al., 2013a), ST7L (Chen et al., 2013a), TRIB3 (Tribbles pseudokinase 3) (Chan et al., 2010), XIAP (X-linked inhibitor of apoptosis) (Xie et al., 2013), and ZNF217 (Zinc finger protein 217) (Szczyrba et al., 2013) are all validated targets of miR-24 (Table 1).
Involvement of miR-24 in cardiovascular diseases
miR-24 is upregulated in ischemic heart endothelial cells as a result of hypoxia-induced HIF-dependent transcription, but it is then transiently downregulated in adjacent surviving regions of acute myocardial infarction owing to the recovery of blood supply (Fiedler et al., 2011; Qian et al., 2011; Camps et al., 2014). miR-24 promotes cardiomyocyte survival through repression of pro-apoptotic Bim (Qian et al., 2011) and reduces cardiac fibrosis through repression of Furin protease that controls the activation of latent TGFβ (Wang et al., 2012b). On the other hand, miR-24 inhibits the survival, migration, proliferation and tube formation of endothelial cells (angiogenesis) through repression of eNOS and actin cytoskeleton regulators, such as DIAPH1, LIMK2, and PAK4 (Fiedler et al., 2011; Meloni et al., 2013; Zhou et al., 2013a). miR-24 is upregulated in the chronic phase after myocardial infarction and promotes hypertrophic growth of cardiomyocytes in mouse model experiments and disturbs cardiac contraction through repression of JPH2 that is involved in the excitation-contraction coupling process of the heart (van Rooij et al., 2006; Xu et al., 2012b). Because miR-24 protects cardiomyocytes themselves and reduces cardiac fibrosis but inhibits angiogenesis and deteriorates heart failure, miR-24 is a multi-functional cardio-miR that plays good and bad roles in heart failure (Figure 3A).
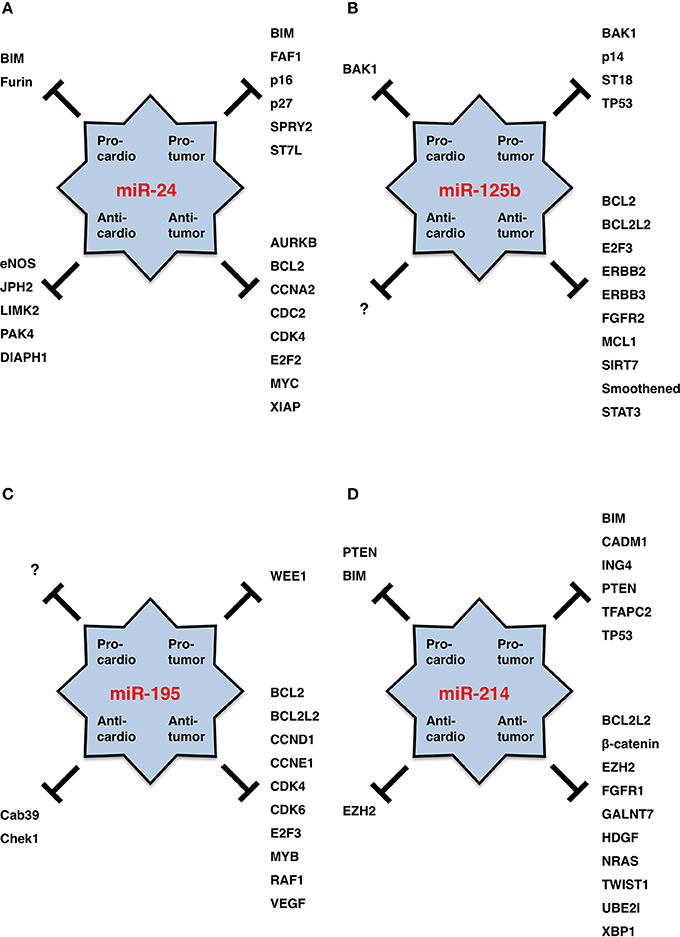
Figure 3. miRNA functions in cardiology and oncology. (A) miR-24. (B) miR-125b. (C) miR-195. (D) miR-214. Targets for pro-tumor, anti-tumor, pro-cardio and anti-cardio functions of each miRNA are shown. miR-125b is a good cardio-miR that protects cardiomyocytes. miR-195 is a bad cardio-miR that elicits cardiomyopathy and heart failure. miR-24 and miR-214 are bi-functional cardio-miRs. By contrast, miR-24, miR-125b, miR-195, and miR-214 function as oncogenic or tumor suppressor miRNAs in a cancer (sub)type-dependent manner.
Involvement of miR-24 in cancers
miR-24 is transcriptionally upregulated in acute myeloid leukemia (AML) with t(8;21) by the RUNX1-RUNX1T1 (AML1-ETO) fusion protein, which promotes proliferation and blocks differentiation of myeloid cells through repression of DUSP16 and subsequent activation of mitogen-activated protein kinase (MAKP) signaling (Zaidi et al., 2009). miR-24 is transcriptionally upregulated in breast cancer with lymph node metastasis in part by MYC (Li et al., 2013a), and overexpression of miR-24 in MCF-7 breast cancer cells promotes invasion and metastasis through repression of SPRY2 and subsequent MAPK activation (Li et al., 2013a). miR-24 is upregulated by the E6 and E7 oncoproteins of human papilloma virus type 16 (HPV16), which promotes proliferation through p27 repression (McKenna et al., 2014). Upregulation of miR-24 in glioblastoma promotes survival, proliferation and invasion through repression of tumor suppressor ST7L (Chen et al., 2013a). Upregulation of miR-24 in pancreatic endocrine tumors (Volinia et al., 2006) and parathyroid tumors (Luzi et al., 2012) can contribute to the progression of multiple endocrine neoplasia type 1 (MEN1) syndrome through repression of its causative gene product. miR-24 is also upregulated in colon cancer (Volinia et al., 2006), lung adenocarcinoma (Yanaihara et al., 2006), pancreatic ductal adenocarcinoma (Jamieson et al., 2012) and gastric cancer (Volinia et al., 2006; Bandres et al., 2009). Because pro-tumor miR-24 promotes survival, proliferation and invasion through repression of BIM, FAF1, p16, p27, SPRY2, and ST7L (Figure 3A), oncogenic miR-24 is upregulated in human cancers.
By contrast, miR-24 is downregulated in A549 and H1437 non-small-cell lung cancer cells owing to copy number loss of the miR-24-2 locus (Xie et al., 2013) (Table 2). miR-24 is also downregulated in prostate cancer (Volinia et al., 2006) and hepatocellular carcinoma (HCC) recurring after liver transplantation (Han et al., 2012). Because anti-tumor miR-24 promotes differentiation, growth arrest and apoptosis through repression of AURKB, BCL2, CCNA2, CDC2, CDK4, E2F2, MYC, and XIAP (Figure 3A), tumor suppressor miR-24 is downregulated in human cancers.
miR-24 functions as an oncogenic or tumor suppressor miRNA in a cancer (sub)type- or cell line-dependent manner (Figure 3A).
miR-125b
Human chromosomal loci of miR-125b genes
miR-125b is derived from the miR-125b-1 and miR-125b-2 loci. miR-125b-1 is clustered with let-7a-2 and miR-100 at human chromosome 11q24.1 and miR-125b-2 is clustered with let-7c and miR-99a at human chromosome 21q21.1 (Figure 2).
Targets of miR-125b
ARID3B (Akhavantabasi et al., 2012), BAK1 (BCL2-antagonist/killer 1) (Shi et al., 2007), BCL2 (Zhao et al., 2012a), BCL2L2 (anti-apoptotic BCL-W) (Gong et al., 2013), BMPR1B (Sætrom et al., 2009), CBFB (Core binding factor β) (Lin et al., 2011), CDH5 (VE-cadherin) (Muramatsu et al., 2013), CDKN2A (p14 ARF) (Amir et al., 2013), DICER1 (Klusmann et al., 2010), E2F3 (Huang et al., 2011a), EDN1 (Endothelin 1) (Li et al., 2010), EPO (Ferracin et al., 2013), EPOR (Ferracin et al., 2013), ERBB2 (Scott et al., 2007), ERBB3 (Scott et al., 2007), ETS1 (Zhang et al., 2011), FGFR2 (Xu et al., 2011), IL6R (Gong et al., 2013), IRF4 (Malumbres et al., 2009), JUN (c-Jun) (Kappelmann et al., 2013), LIN28A (Lin-28) (Wu and Belasco, 2005), LIN28B (Liang et al., 2010), MCL1 (Gong et al., 2013), MMP13 (Xu et al., 2012c), MUC1 (Rajabi et al., 2010), NCOR2 (Yang et al., 2012a), PGF (Placental growth factor) (Alpini et al., 2011), PRDM1 (BLIMP1) (Malumbres et al., 2009), SIRT7 (Kim et al., 2013a), Smoothened (SMO) (Ferretti et al., 2008), ST18 (Klusmann et al., 2010), STARD13 (Tang et al., 2012), STAT3 (Liu et al., 2011), TET2 (Cheng et al., 2013), TNF (TNF-α) (Rajaram et al., 2011), TNFSF4 (Smirnov and Cheung, 2008), and TP53 (Le et al., 2009) are representative targets of miR-25b (Table 1).
Involvement of miR-125b in cardiovascular diseases
miR-125b and LIN28A are human homologs of Caenorhabditis elegans (C. elegans) lin-4 and lin-28, respectively. C. elegans lin-4 is involved in the repression of lin-28 to orchestrate morphogenesis during larval stage, whereas human miR-125b is involved in the repression of LIN28A during the differentiation of embryonic stem cells (ESCs) into myocardial precursors and cardiomyocytes (Wu and Belasco, 2005; Wong et al., 2012).
miR-125b is physiologically expressed in perivascular stromal cells rather than cardiomyocytes of the developing mouse heart (Schneider et al., 2011) and in cardiac valves rather than myocardium of the adult rat heart (Vacchi-Suzzi et al., 2013). miR125b is upregulated in mouse cardiac endothelial cells during endothelial-to-mesenchymal transition (EndMT) induced by TGF-ß (Ghosh et al., 2012). In addition, mir-125b is upregulated in early-stage cardiac hypertrophy after aortic banding (Busk and Cirera, 2010) and also in late-stage cardiac hypertrophy and heart failure (van Rooij et al., 2006). Ectopic miR-125b expression by using adenovirus vector does not elicit cardiomyocyte hypertrophy in vitro (van Rooij et al., 2006), whereas ectopic miR-125b expression by using lentivirus reduces myocardial infarct size and preserves cardiac functions in a mouse experimental model of acute myocardial infarction (Wang et al., 2014b). miR-125b is a good cardio-miR that protects the heart from ischemia/reperfusion injury (Figure 3B).
Involvement of miR-125b in human cancers
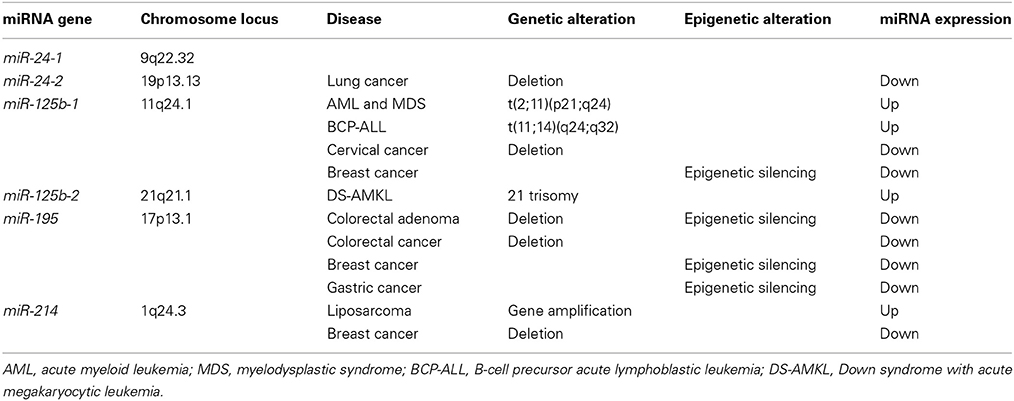
miR-125b is overexpressed in hematological malignancies owing to genetic alterations, such as chromosomal translocation and copy number gain (Table 2). miR-125b-1 at human chromosome 11q24.1 is upregulated as a result of chromosomal translocation in AML and myelodysplastic syndrome (MDS) with t(2;11)(p21;q24) (Bousquet et al., 2008; Thorsen et al., 2012) and B-cell precursor acute lymphoblastic leukemia (BCP-ALL) with t(11;14)(q24;q32) (Chapiro et al., 2010). miR-125b-2 at human chromosome 21q21.1 is upregulated as a result of copy number gain (21 trisomy) in Down syndrome with acute megakaryocytic leukemia (DS-AMKL) (Klusmann et al., 2010), which leads to the proliferation and self-renewal of hematopoietic progenitors of megakaryocytic and erythroid lineages in part through repression of DICER1 and ST18. miR-125b is also upregulated in childhood ALL with t(12;21)(p13.1;q22) (ETV6/RUNX1-ALL) (Gefen et al., 2010), pancreatic endocrine tumors (Volinia et al., 2006) and urothelial cancer at T2/T3 stages (Veerla et al., 2009). Upregulation of pro-tumor (oncogenic) miR-125b in human cancers promotes proliferation, survival and drug resistance of tumor cells through repression of BAK1, p14, ST18, and TP53 (Figure 3B).
miR-125b is repressed in solid tumors as a result of deletion and epigenetic silencing (Table 2). miR-125b is downregulated in in cervical cancer owing to a deletion of chromosome 11q24.1 that involves miR-125b-1 (Wilting et al., 2013) and in oral squamous cell carcinoma (OSCC) owing to deletions of chromosome 11q or 21 involving miR-125b-1 or miR-125b-2, respectively (Henson et al., 2009). miR-125b is downregulated in breast cancer (Zhang et al., 2011) and HCC (Alpini et al., 2011) owing to epigenetic silencing induced by CpG hypermetheylation of promoter region(s). miR-125b is also downregulated in prostate cancer (Porkka et al., 2007; Ozen et al., 2008) and colorectal cancer (Chen et al., 2009). Downregulation of anti-tumor (tumor suppressor) miR-125b in human cancers promotes survival, proliferation and invasion of tumor cells through de-repression of BCL2, BCL2L2, E2F3, ERBB2, FGFR2, MCL1, SIRT7, Smoothened and STAT3 (Figure 3B).
miR-125b also functions as an oncogenic or tumor suppressor miRNA in a context-dependent manner (Figure 3B).
miR-195
Human chromosomal locus of miR-195 gene
miR-195 is derived from the miR-497/miR-195 locus at human chromosome 17p13.1 (Figure 2).
Targets of miR-195
ACVR2A (Bai et al., 2012), ARL2 (ADP-ribosylation factor-like 2) (Zhou et al., 2013c), BCL2 (Liu et al., 2010), BCL2L2 (Yang et al., 2012b), BIRC5 (API4, Apoptosis inhibitor 4) (Itesako et al., 2014), CCND1 (Cyclin D1) (Xu et al., 2009), CCNE1 (Cyclin E1) (Hui et al., 2013), CDC42 (Wang et al., 2013c), CDK4 (Lin et al., 2012b), CDK6 (Xu et al., 2009), E2F3 (Xu et al., 2009), GLUT3 (SLC2A3) (Fei et al., 2012), IKKα (CHUK) (Ding et al., 2013), MYB (Zhou et al., 2014), RAF1 (Li et al., 2011), TAB3 (TAK1/MAP3K7-binding protein 3) (Ding et al., 2013), VAV2 (Wang et al., 2013c), VEGF (Wang et al., 2013c), WEE1 (Bhattacharya et al., 2013), and WNT7A (Itesako et al., 2014) are all validated targets of miR-195 (Table 1).
Involvement of miR-195 in cardiovascular diseases
During early post-natal development of mice, miR-195 is upregulated in cardiac ventricles and induces cell-cycle arrest in cardiomyocytes through repression of cell cycle regulators, such as Cdc2a, Chek1, Birc5, Nusap1, and Spag5 (Porrello et al., 2011). Overexpression of miR-195 in the developing heart of transgenic mice by using the β-myosin heavy chain (MHC) promoter gives rise to perinatal cardiomyopathy in one line and ventricular hypoplasia and ventricular septal defects in another line (Porrello et al., 2011). Overexpression of miR-195 in primary neonatal rat cardiomyocytes by using adenoviral vector leads to hypertrophic growth and sarcomeric assembly, and overexpression of miR-195 in the heart of post-natal transgemic mice by using the α-MHC promoter gives rise to cardiac hypertrophy and dilated cardiomyopathy (van Rooij et al., 2006). In transgenic mice with the α-MHC mutation R403Q, miR-195 upregulation and subsequent repression of Cab39 in the heart leads to hypertrophic cardiomyopathy owing to inhibition of Lkb1/Strad/Cab39-dependent AMPK signaling (Chen et al., 2012). Together these facts indicate that miR-195 is a bad cardio-miR that elicits hypertrophic cardiomyopathy, dilated cardiomyopathy and heart failure (Figure 3C).
Involvement of miR-195 in cancers
miR-195 is upregulated in metastatic melanoma (Bhattacharya et al., 2013) and some cases of lung cancer (Volinia et al., 2006), colorectal cancer (Ding et al., 2013), prostate cancer (Volinia et al., 2006), gastric cancer (Bandres et al., 2009; Ding et al., 2013) and HCC (Ding et al., 2013). miR-195 can function as an oncogenic miRNA through repression of WEE1 kinase (Figure 3C).
By contrast, miR-195 is preferentially downregulated in breast cancer (Li et al., 2011), gastric cancer (Deng et al., 2013; Ding et al., 2013), colorectal cancer (Chen et al., 2009; Liu et al., 2010; Guo et al., 2013), HCC (Xu et al., 2009; Wang et al., 2013c), bladder cancer (Lin et al., 2012b; Itesako et al., 2014) and prostate cancer (Porkka et al., 2007). miR-195 is repressed in breast cancer (Li et al., 2011) and gastric cancer (Deng et al., 2013) owing to hypermethylation of CpG islands upstream of the miR-497/miR-195 locus. miR-195 is repressed in colorectal cancer owing to deletion of the miR-497/miR-195 locus (Guo et al., 2013), while miR-195 is repressed in colorectal adenoma mainly owing to epigenetic silencing and in part owing to deletion (Menigatti et al., 2013). miR-195 is downregulated in human cancers and pre-cancerous lesions as a result of epigenetic silencing and deletion (Table 2). Because miR-195 is involved in repression of cell cycle accelerators (CCND1, CCNE1, CDK4, CDK6, and E2F3) and anti-apoptotic factors (BCL2, BCL2L2, and BIRC5) (Figure 3C), miR-195 functions as a tumor suppressor miRNA in various types of human cancers.
miR-214
Human chromosomal locus of miR-214 gene
miR-214 is derived from the miR-199a-2/miR-214 locus at human chromosome 1q24.3 (Figure 2).
Targets of miR-214
ASF1B (Misiewicz-Krzeminska et al., 2013), BCL2L2 (Wang et al., 2013a), ß-catenin (CTNNB1) (Xia et al., 2012), BIM (Zhang et al., 2014), CADM1 (IGSF4A) (Momose et al., 2013), CCL5 (C-C motif ligand 5) (Mitra et al., 2012), CD276 (B7-H3) (Nygren et al., 2014), EZH2 (Derfoul et al., 2011), FGFR1 (Wang et al., 2013b), GALNT7 (N-acetylgalactosaminyltransferase 7) (Peng et al., 2012), HDGF (MGG1L2) (Shih et al., 2012b), ING4 (Zhang et al., 2010), ITGA3 (Integrin α3) (Penna et al., 2011), LTF (Lactoferrin) (Liao et al., 2010), LZTS1 (Xu and Wang, 2014), MAP2K3 (MEK3) (Yang et al., 2009), MAPK8 (JNK1) (Yang et al., 2009), NRAS (Huang et al., 2014a), PSMD10 (Misiewicz-Krzeminska et al., 2013), PTEN (Yang et al., 2008), TFAP2C (AP2γ) (Penna et al., 2011), TP53 (Xu et al., 2012a), TWIST1 (Twist) (Li et al., 2012), UBE2I (UBC9) (Zhao et al., 2012b), and XBP1 (Duan et al., 2012) are all validated targets of miR-214 (Table 1).
Involvement of miR-214 in cardiovascular diseases
miR-214 is upregulated as a result of cardiac ischemia and heart failure. In a mouse model of ischemic cardiac injury induced by permanent ligation of the left anterior descending coronary artery, miR-214 prevents cardiomyocyte death owing to Ca2+ overload, subsequent cardiac insufficiency and cardiac fibrosis through repression of Slc8a1 (Ncx1, sodium/calcium exchanger), which is the primary Ca2+ outflow pump in cardiomyocytes (Aurora et al., 2012). miR-214 protects primary neonatal rat cardiomyocytes from apoptosis induced by ischemia-reperfusion injury and represses Bim, Camk2d (Calmodulin kinase II delta) and Slc8a1 (Aurora et al., 2012). miR-214 also protects primary neonatal rat cardiomyocytes from apoptosis induced by H2O2 through PTEN repression (Lv et al., 2014). Overexpression of miR-214 in transgenic mice under control of the α-MHC promoter does not induce a deteriorating cardiac phenotype; however, adenovirus-mediated pri-miR-214 delivery and lentivirus-mediated miR-214 delivery induce hypertrophic growth of primary neonatal rat cardiomyocytes in part through Ezh2 repression (van Rooij et al., 2006; Yang et al., 2013). miR-214 is a bi-functional cardio-miR that plays good and bad roles (Figure 3D).
Involvement of miR-214 in cancers
Copy number gain of the 1q24.3 region around the miR-214 locus occurs in 35% of de-differentiated liposarcomas (Tap et al., 2011). miR-214 is upregulated in ovarian cancer (Yang et al., 2008; Xu et al., 2012a), gastric cancer (Volinia et al., 2006; Bandres et al., 2009), pancreatic cancer (Zhang et al., 2010; Jamieson et al., 2012), lung squamous cell carcinoma (Yanaihara et al., 2006), Sézary syndrome (Narducci et al., 2011), liposarcoma (Tap et al., 2011), osteosarcoma (Wang et al., 2014d) and nasopharyngeal cancer (Zhang et al., 2014). miR-214 upregulation in primary gastric cancer occurs as a result of its expression in mesenchymal stem cells (MSCs) rather than cancer cells (Wang et al., 2014a). Genetic alteration as well as tumor-stromal interaction are involved in miR-214 upregulation in human cancers.
Copy number loss of the miR-214 locus occurs in 24% of breast cancers (Derfoul et al., 2011). miR-214 is downregulated in cervical cancer (Peng et al., 2012; Wang et al., 2013a), HCC (Duan et al., 2012; Shih et al., 2012b), colorectal cancer (Chen et al., 2009), breast cancer (Derfoul et al., 2011), cholangiocarcinoma (Li et al., 2012), glioma (Zhao et al., 2012b), prostate cancer (Srivastava et al., 2013), and bladder cancer (Ratert et al., 2013).
Malignant phenotypes of cancer cells, such as proliferation, survival, drug resistance, invasion and metastasis, are induced by miR-214 upregulation through repression of BIM, CADM1, ING4, PTEN, TFAP2C, and TP53 and also by miR-214 downregulation through de-repression of BCL2L2, ß-catenin, EZH2, FGFR1, GALNT7, HDGF, NRAS, TWIST1, UBE2I, and XBP1 (Figure 3D). miR-214 performs oncogenic functions in some types/subtypes of human cancers and tumor-suppressor functions in other types/subtypes of human cancers.
Regulatory Signaling Networks and miRNA-Based Therapeutics
Regulatory signaling networks are defined as mutual interactions or cross-talks of receptor tyrosine kinase (RTK), G protein-coupled receptor (GPCR) and other receptor signaling cascades (Katoh, 2013a), which are involved in orchestration of fetal development and post-natal homeostasis as well as pathogenesis of non-cancerous and cancerous diseases. WNT, FGF Hedgehog, Notch, TGF-ß, BMP, Nodal, and Activin signaling cascades are major components of the regulatory signaling networks (Bailey et al., 2007; Katoh, 2007; Jayasena et al., 2008; Boulter et al., 2012; Nowell and Radtke, 2013; Coleman et al., 2014).
WNT signals are transduced through Frizzled receptors to the ß-catenin-dependent (canonical) and ß-catenin-independent (non-canonical) cascades (Cohen et al., 2007; Katoh and Katoh, 2007; Klaus and Birchmeier, 2008; Rao and Kühl, 2010). In the absence of canonical WNT signaling, β-catenin is phosphorylated by GSK-3β and is degraded in the proteasome system. By contrast, in the presence of canonical WNT signaling, β-catenin is released from the APC/AXIN degradation complex and activates transcription of canonical WNT target genes, such as CCND1, FGF20, JAG1, and MYC (Figure 4A). ß-catenin is a direct target of miR-200a (Saydam et al., 2009), miR-214 (Xia et al., 2012), miR-320a (Sun et al., 2012), and miR-1826 (Hirata et al., 2012). Downregulation of miR-200a, miR-214, miR-320a, and miR-1826 de-repress ß-catenin and activate the canonical WNT signaling cascade in human cancers.
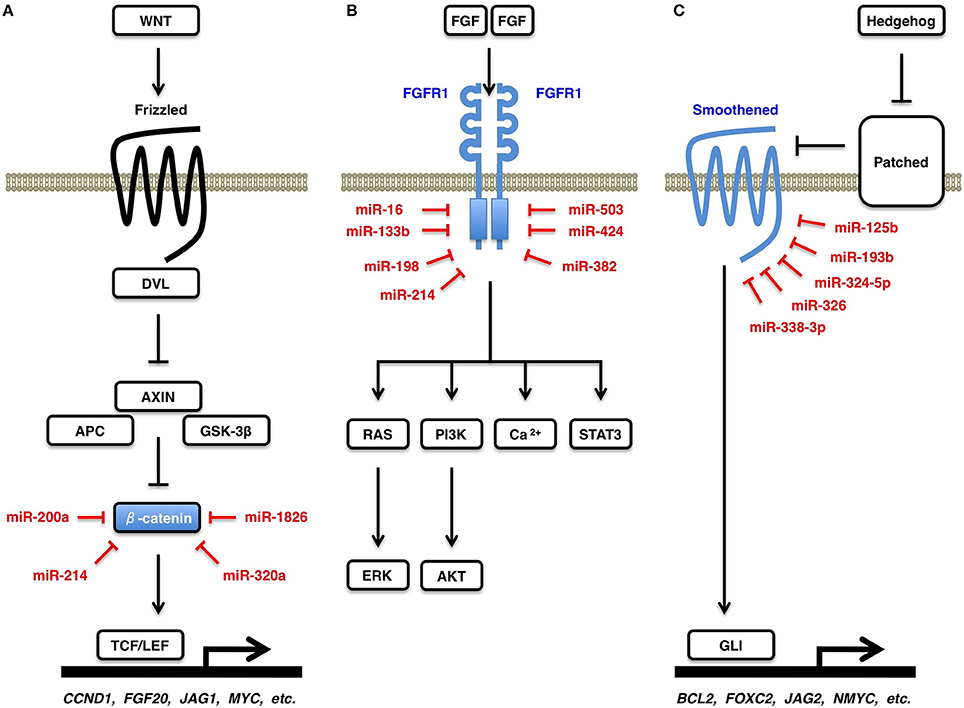
Figure 4. WNT, FGF, and Hedgehog signaling cascades. (A) Canonical WNT signaling cascade and β-catenin. Canonical WNT signaling activation releases β-catenin from its degradation complex of APC and AXIN, which results in nuclear translocation of β-catenin and transcriptional activation of TCF/LEF-target genes, such as CCND1 (Cyclin D1), FGF20, JAG1, and MYC (c-Myc). β-catenin is a direct target of miR-200a, miR-214, miR-320a, and miR-1826. (B) FGF signaling cascades and FGFR1. FGF signals induce dimerization and auto-phosphorylation of a receptor tyrosine kinase FGFR1, which activates downstream RAS-ERK, PI3K-AKT, STAT3, and Ca2+-release signaling cascades. FGFR1 is a direct target of miR-16, miR-133b, miR-198, miR-214, miR-382, miR-424, and miR-503. (C) Hedgehog signaling cascade and Smoothened. Hedgehog signals are transduced from Patched receptor to Smoothened signal transducer, which activates transcription of GLI-target genes, such as BCL2, FOXC2, JAG2, and MYCN (N-Myc). Smoothened is a direct target of miR-125b, miR-193b, miR-324-5p, miR-326, and miR-338-3p.
FGF signals are transduced to the RAS-ERK, PI3K-AKT, STAT3, and Ca2+-release signaling branches through FGFR1 (Figure 4B), FGFR2, FGFR3, and FGFR4 (Turner and Grose, 2010; Goetz and Mohammadi, 2013; Katoh and Nakagama, 2014). FGFR1 is a direct target of miR-16 (Chamorro-Jorganes et al., 2011), miR-133b (Wen et al., 2013), miR-198 (Yang et al., 2014), miR-214 (Wang et al., 2013b), miR-382 (Mor et al., 2013), miR-424 (Chamorro-Jorganes et al., 2011), and miR-503 (Kim et al., 2013b). Upregulation of miR-382 in olfactory neuroepithelium of schizophrenia patients repress FGFR1 (Mor et al., 2013). By contrast, downregulation of miR-133b and miR-214 in human cancers (Wen et al., 2013; Wang et al., 2013b) and that of miR-424 and miR-503 in pulmonary artery epithelial cells of patients with pulmonary arterial hypertension (Kim et al., 2013b) de-repress FGFR1 and promote proliferation of tumor cells and endothelial cells, respectively, through FGF signaling activation.
Hedgehog signals are transduced from Patched receptors to Smoothened signal transducer, which activates GLI-dependent transcription of target genes, such as BCL2, FOXC2, JAG2, and MYCN (N-Myc) (Figure 4C). Hedgehog-Smoothened-GLI signaling cascade is involved in the regulation of cellular survival, proliferation, motility and stemness (Jiang and Hui, 2008; Katoh and Katoh, 2009; Lin and Matsui, 2012a). Smoothened is a direct target of miR-125b (Ferretti et al., 2008), miR-193b (González-Gugel et al., 2013), miR-324-5p (Ferretti et al., 2008), miR-326 (Ferretti et al., 2008), and miR-338-3p (Huang et al., 2011b). Downregulation of miR-125b, miR-193b, miR-324-5p, miR-326, and miR-338-3p in human cancers de-repress Smoothened and promotes tumor proliferation and invasion through aberrant Hedgehog signaling activation.
miRNAs are therapeutic targets for non-cancerous diseases as well as cancers, because disease-related miRNAs dysregulate the regulatory signaling networks (Katoh and Katoh, 2008; Mo et al., 2013; Parpart and Wang, 2013; Katoh et al., 2013c). Reduction of elevated pro-disease miRNA and restoration of declined anti-disease miRNA are two major strategies of miRNA-based therapeutics. Locked-nucleic-acid-modified anti-miRNA oligonucleotides (LNA-antimiRs) are utilized for the reduction of pro-disease miRNAs, while adenovirus and letivirus vectors are utilized for the restoration of anti-disease miRNAs (Ji et al., 2008; Kasinski and Slack, 2011; Shi et al., 2011; van Rooij and Olson, 2012). Reduction of FGFR1-targeting miRNAs for cancer therapy deteriorate diabetes and cardiac functions, because the FGFR1-PI3K-AKT signaling cascade is involved in cancer promotion (Katoh et al., 2013c) as well as diabetes control (Suh et al., 2014). By contrast, restoration of miRNA targeting BAK1, BIM, or PTEN for cardiomyocyte protection promotes survival of tumor cells (Figure 3). miRNA-based therapy is at the risk of adverse effects owing to repression of verified targets in different disciplines. In addition, because multiple miRNAs repress the same target (Figure 4) and each miRNA represses multiple targets (Table 1), miRNA-based therapy is also at the risk of adverse effects owing to repression of unidentified targets in individual patients. There are many obstacles before clinical application of miRNA-based therapeutics.
Circulating miR-24, miR-125b, miR-195, and miR-214
miRNAs function within the cell where they were produced as well as in other cells that receive miRNAs secreted or released from the cell of their origin (Valadi et al., 2007; Skog et al., 2008). Extracellular miRNAs are detected in various types of body fluids, such as blood, tears, saliva, urine, vitreous humor, cerebro-spinal fluid, pleural fluid, peritoneal fluid, seminal fluid, breast milk, and amniotic fluid (Mitchell et al., 2008; Weber et al., 2010; Ragusa et al., 2013). Extracellular miRNAs are classified into miRNAs in the blood (circulating miRNAs) and those in other body fluids. Because circulating miRNAs within exosomes (Taylor and Gercel-Taylor, 2008), microvesicles (Hunter et al., 2008) and high-density lipoprotein (Vickers et al., 2011) or those conjugated with AGO2 protein (Arroyo et al., 2011) are stable, circulating miRNAs are going to be utilized as diagnostics and prognostic biomarkers (Table 3).
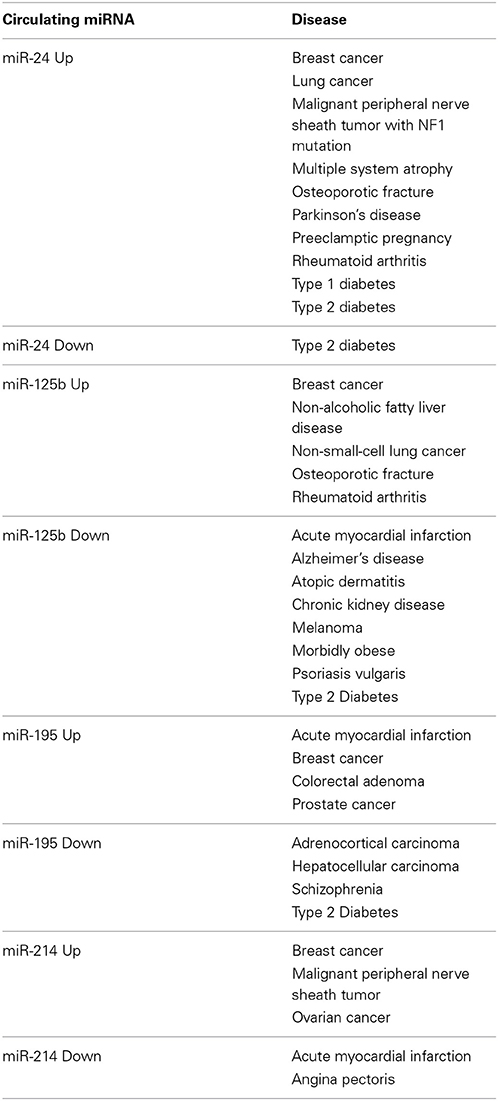
Circulating miR-24 is elevated in patients with breast cancer (Wu et al., 2012b; Sochor et al., 2014), lung cancer (Le et al., 2012), malignant peripheral nerve sheath tumor with the NF1 mutation (Weng et al., 2013), multiple system atrophy (Vallelunga et al., 2014), osteoporotic fracture (Seeliger et al., 2014), Parkinson's disease (Vallelunga et al., 2014), preeclamptic pregnancy (Wu et al., 2012a), rheumatoid arthritis (Murata et al., 2013) and type 1 diabetes (Nielsen et al., 2012). Wang et al. reported elevated miR-24 in type 2 diabetes patients (Wang et al., 2014c), whereas Zampetaki et al. reported reduced miR-24 in type 2 diabetes patients (Zampetaki et al., 2010).
Circulating miR-125b is elevated in patients with breast cancer (Wang et al., 2012a; Mar-Aguilar et al., 2013), non-alcoholic fatty liver disease (Pirola et al., 2014), non-small-cell lung cancer (Yuxia et al., 2012; Cui et al., 2013), osteoporotic fracture (Seeliger et al., 2014) and rheumatoid arthritis (Duroux-Richard et al., 2014), whereas circulating miR-125b is reduced in patients with acute myocardial infarction (Huang et al., 2014b), Alzheimer's disease (Tan et al., 2014), atopic dermatitis (Koga et al., 2014), chronic kidney disease (Chen et al., 2013b), melanoma (Alegre et al., 2014), morbidly obese (Ortega et al., 2013), psoriasis vulgaris (Koga et al., 2014), and type 2 diabetes (Ortega et al., 2014).
Circulating miR-195 is elevated in patients with acute myocardial infarction (Long et al., 2012), breast cancer (Heneghan et al., 2010), colorectal adenoma (Kanaan et al., 2013), and prostate cancer (Mahn et al., 2011), whereas circulating miR-195 is reduced in adrenocortical carcinoma (Chabre et al., 2013), HCC (Qu et al., 2011), schizophrenia (Shi et al., 2012), and type 2 diabetes (Ortega et al., 2014).
Circulating miR-214 is elevated in patients with breast cancer (Schwarzenbach et al., 2012), malignant peripheral nerve sheath tumor (Weng et al., 2013) and ovarian cancer (Taylor and Gercel-Taylor, 2008), whereas circulating miR-214 is reduced in patients with acute myocardial infarction and angina pectoris (Lu et al., 2013).
These facts clearly indicate that circulating miRNAs reported as cancer biomarkers are also dysregulated in non-cancerous diseases, and that miRNAs reported as biomarkers of non-cancerous diseases are also dysregulated in cancers (Table 3).
miRNA Regulation by Genetic and Environmental Factors
Genetic factors are associated with individual traits and disease susceptibility (Lichtenstein et al., 2000; Zimmet et al., 2001; Milne et al., 2009). Single nucleotide polymorphisms (SNPs) and copy number variations (CNVs) are major germ-line variations. The SNP rs1434536 is located in the miR-125b-binding site within the 3′-untranslated region (UTR) of BMPR1B. The C and T alleles of the rs1434536 SNP are sensitive and resistant to BMPR1B repression by miR-125b, respectively (Sætrom et al., 2009). The homozygous T genotype of rs1434536 is associated with increased risk of breast cancer (Sætrom et al., 2009) and decreased risk of endometriosis (Chang et al., 2013). Copy number loss of the miR-195 locus occurs in autism patients (Vaishnavi et al., 2013). Copy number gain of the miR-125b-2 locus occurs in Down syndrome patients as a result of trisomy 21, which leads to elevated circulating miR-125b in pregnant women with a Down syndrome fetus (Kotlabova et al., 2013) and causes acute megakaryocytic leukemia in Down syndrome patients (Klusmann et al., 2010). Genetic factors directly affect expression profiles and functions of miRNAs (Figure 5, upper left).
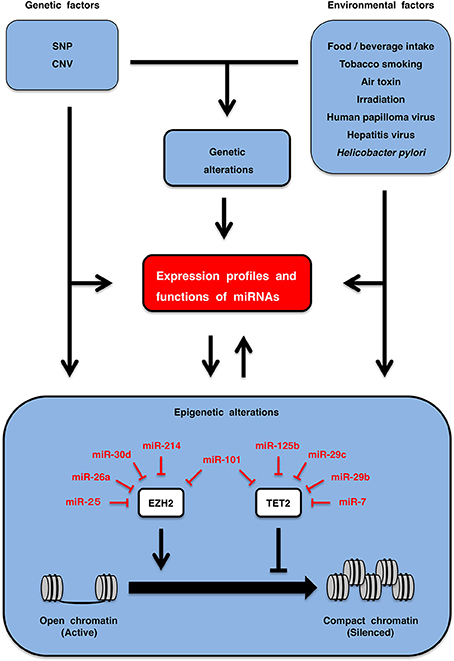
Figure 5. Regulation of circulating miRNAs. Genetic factors, such as single nucleotide polymorphism (SNP) and copy number variation (CNV), as well as environmental factors, including life style (food/beverage intake, tobacco smoking, air toxin, irradiation, etc.) and chronic infection (human papilloma virus, hepatitis virus, Helicobacter pylori, etc.) are involved in the regulation of expression profiles and functions of miRNAs (upper part). EZH2 and TET2 are epigenetic regulators that are involved in inactivation and activation of genes through repressive histone marking and CpG-island de-methylation, respectively. miRNA expression is downregulated by epigenetic silencing, while epigenetic regulators are repressed by multiple miRNAs. miRNAs and epigenetics are in the relationship of mutual regulation (lower part). Genetic and environmental factors regulate circulation miRNA profiles directly as well as indirectly through genetic and epigenetic alterations.
Environmental factors are also associated with disease susceptibility (Lichtenstein et al., 2000; Zimmet et al., 2001). Life style (food/beverage intake, tobacco smoking, air toxin, irradiation, etc.) and chronic infection (papilloma virus, hepatitis virus, Helicobater pylori, etc.) are environmental factors affecting individuals. Human miR-125b is downregulated in the bronchial epithelium of current smokers compared with never smokers (Schembri et al., 2009), and rat miR-125b is downregulated in the lungs of rats that were exposed to environmental smoke for 28 days (Izzotti et al., 2009). The expression profile of miRNAs in airway epithelial cells is altered by air toxins, such as diesel exhaust particles and formaldehyde (Jardim et al., 2009; Rager et al., 2011), whereas that in breast cancer cells is altered by endocrine disruptors, such as o,p'-dichlorodiphenyltrichloroethane (DDT), bisphenol A (BPA), fenhexamid and fludioxonil (Tilghman et al., 2012; Teng et al., 2013). The circulating miRNA landscape is altered by total-body γ-irradiation (Jacob et al., 2013) and by uptake of dietary polyphenols, such as quercetin, hesperidin, naringenin, anthocyanin, catechin, proanthocyanin, caffeic acid, ferulic acid and curcumin (Milenkovic et al., 2012), in model animal experiments. miRNA expression profile is altered by chronic infection with human papilloma virus, hepatitis virus and Helicobacter pylori, which are involved in pathogenesis of cervical cancer (Wang et al., 2008b), HCC (Ladeiro et al., 2008; Arzumanyan et al., 2013), and gastric cancer (Zhang et al., 2008), respectively. Environmental factors directly alter circulating or tissue levels of miRNAs (Figure 5, upper right).
Genetic alterations, such as gene amplification, deletion, translocation, point mutation or single nucleotide variation (SNV), occur in tumor cells during multi-stage carcinogenesis owing to mutual interactions of genetic and environmental factors (Lichtenstein et al., 2000; Katoh et al., 2013c; Katoh and Nakagama, 2014). SNVs in diffuse large B-cell lymphomas that disrupt the miR-125b-binding site within the 3′-UTR of TP53 are associated with better prognosis of patients owing to de-repression of a tumor suppressor TP53 (Li et al., 2013b). Effects of gene amplification, deletion and translocation on expression profiles of miR-24, miR-125b, miR-195, and miR-214 have been described above (Table 2). Genetic alterations play a key role for the regulation of miRNA profiles in somatic cells (Figure 5, upper middle).
Epigenetics is chromatin-based genomic regulations that are involved in the modulation of expression landscapes of mRNAs and miRNAs during fetal development, post-natal homeostasis and pathogenesis of human diseases (Datta et al., 2008; Kulis and Esteller, 2010; Ordovás and Smith, 2010; Baylin and Jones, 2011; Dawson and Kouzarides, 2012). EZH2 and TET2 are representative epigenetic regulators that are repressed by miR-214 and miR-125b, respectively (Table 1). EZH2 is a human homolog of Drosophiula Enhancer of zeste, which is a component of the Polycomb repressive complex 2 (PRC2) and PRC2-like complex (Sparmann and van Lohuizen, 2006). EZH2 is involved in epigenetic silencing of PRC target genes through trimethylation of histone H3 lysine 27 (H3K27me3) and CpG hypermethylation of promoters (Figure 5, lower part). Because EZH2 is a target of miR-25 (Esposito et al., 2012), miR-26a (Sander et al., 2008), miR-30d (Esposito et al., 2012), miR-101 (Varambally et al., 2008), and miR-214 (Derfoul et al., 2011), downregulation of miR-25, miR-26a, miR-30d, miR-101, and miR-214 in human cancers are associated with EZH2 upregulation and malignant phenotypes. TET2 is involved in promoter de-methylation through enzymatic conversion of 5-methylcytosine (5mC) to 5-hydroxylmethyl-cytosine (5hmC) (Ito et al., 2010). Loss-of-function TET2 mutations occur in patients with myeloproliferative neoplasms, MDS and AML (Shih et al., 2012a), while upregulation of TET2-targeting miRNAs, such as miR-7, miR-29b, miR-29c, miR-101, and miR-125b, occur in AML patients with wild-type TET2 (Cheng et al., 2013). miRNAs targeting EZH2 and TET2 alter epigenetic regulations of disease-associated genes. By contrast, disease-associated miRNAs are epigenetically silenced owing to promoter CpG hypermethylation in human diseases (Table 2). Epigenetic alterations also play a key role for the regulation of miRNA profiles in somatic cells (Figure 5, lower part).
Genetic and environmental factors dynamically alter expression profiles of miRNAs in individuals and also indirectly alter miRNA profiles through genetic and epigenetic alterations in patients with non-cancerous diseases and cancers (Figure 5).
Circulating miRNA-Based Diagnostics
Circulating miR-195 is upregulated in colorectal adenoma (Kanaan et al., 2013); however, miR-195 in colorectal adenoma tissues is repressed owing to epigenetic silencing and deletion (Menigatti et al., 2013). Circulating miR-125b, miR-195, and miR-214 are upregulated in breast cancer patients (Table 2), whereas these miRNAs in breast cancer tissues are downregulated owing to epigenetic silencing or deletion (Table 2). These facts clearly indicate that circulating miRNAs in cancer patients are not always derived from tumor tissues.
Because circulating miRNA profiles are dynamically regulated by genetic and environmental factors (Figure 5), circulating miRNA profiles of cancer patients reflect co-existing non-cancerous diseases or individual whole-body conditions. Therefore, circulating miRNA association studies (CMASs) within several thousands of cases each for common non-cancerous diseases as well as major cancers (Figure 6) should be carried out to establish a reliable and robust platform of miRNA-based diagnostics.
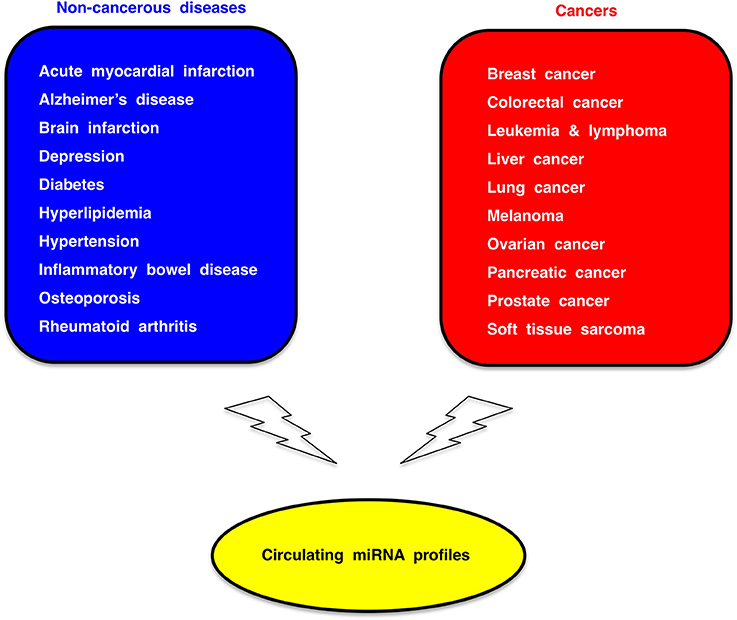
Figure 6. Circulating miRNA association study (CMAS). Dysregulation of circulating miRNAs occur in a variety of human disease. Circulating miRNA profiles in several thousands of cases each for non-cancerous common diseases (blue box) and major cancers (red box) should be investigated for the establishment of miRNA-based diagnostic platform.
Conclusion
Cardio-miRs and onco-miRs bear some similarities in functions and circulation profiles. miRNAs modulate the regulatory signaling networks that are involved in orchestration of embryogenesis and homeostasis as well as pathogenesis of human diseases. Circulating miRNA profiles within several thousands of cases each for non-cancerous and cancerous diseases are necessary for the establishment of miRNA-based diagnostics.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported in part by a grant-in-aid for the Knowledgebase Project from the Masaru Katoh's Fund.
References
Akhavantabasi, S., Sapmaz, A., Tuna, S., and Erson-Bensan, A. E. (2012). miR-125b targets ARID3B in breast cancer cells. Cell Struct. Funct. 37, 27–38. doi: 10.1247/csf.11025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Alegre, E., Sanmamed, M. F., Rodriguez, C., Carranza, O., Martín-Algarra, S., and González, A. (2014). Study of circulating microRNA-125b levels in serum exosomes in advanced melanoma. Arch. Pathol. Lab. Med. 138, 828–832. doi: 10.5858/arpa.2013-0134-OA
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Alpini, G., Glaser, S. S., Zhang, J. P., Francis, H., Han, Y., Gong, J., et al. (2011). Regulation of placenta growth factor by microRNA-125b in hepatocellular cancer. J. Hepatol. 55, 1339–1345. doi: 10.1016/j.jhep.2011.04.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Amelio, I., Lena, A. M., Viticchiè, G., Shalom-Feuerstein, R., Terrinoni, A., Dinsdale, D., et al. (2012). miR-24 triggers epidermal differentiation by controlling actin adhesion and cell migration. J. Cell Biol. 199, 347–363. doi: 10.1083/jcb.201203134
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Amir, S., Ma, A. H., Shi, X. B., Xue, L., Kung, H. J., and deVere White, R. W. (2013). Oncomir miR-125b suppresses p14(ARF) to modulate p53-dependent and p53-independent apoptosis in prostate cancer. PLoS ONE 8:e61064. doi: 10.1371/journal.pone.0061064
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arroyo, J. D., Chevillet, J. R., Kroh, E. M., Ruf, I. K., Pritchard, C. C., Gibson, D. F., et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008. doi: 10.1073/pnas.1019055108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arzumanyan, A., Reis, H. M., and Feitelson, M. A. (2013). Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat. Rev. Cancer 13, 123–135. doi: 10.1038/nrc3449
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Aurora, A. B., Mahmoud, A. I., Luo, X., Johnson, B. A., van Rooij, E., Matsuzaki, S., et al. (2012). MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca2+ overload and cell death. J. Clin. Invest. 122, 1222–1232. doi: 10.1172/JCI59327
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bai, Y., Yang, W., Yang, H. X., Liao, Q., Ye, G., Fu, G., et al. (2012). Downregulated miR-195 detected in preeclamptic placenta affects trophoblast cell invasion via modulating ActRIIA expression. PLoS ONE 7:e38875. doi: 10.1371/journal.pone.0038875
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bailey, J. M., Singh, P. K., and Hollingsworth, M. A. (2007). Cancer metastasis facilitated by developmental pathways: sonic hedgehog, Notch, and bone morphogenic proteins. J. Cell. Biochem. 102, 829–839. doi: 10.1002/jcb.21509
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bandres, E., Bitarte, N., Arias, F., Agorreta, J., Fortes, P., Agirre, X., et al. (2009). microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin. Cancer Res. 15, 2281–2290. doi: 10.1158/1078-0432.CCR-08-1818
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baylin, S. B., and Jones, P. A. (2011). A decade of exploring the cancer epigenome: biological and translational implications. Nat. Rev. Cancer 11, 726–734. doi: 10.1038/nrc3130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhattacharya, A., Schmitz, U., Wolkenhauer, O., Schönherr, M., Raatz, Y., and Kunz, M. (2013). Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene 32, 3175–3183. doi: 10.1038/onc.2012.324
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boulter, L., Govaere, O., Bird, T. G., Radulescu, S., Ramachandran, P., Pellicoro, A., et al. (2012). Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat. Med. 18, 572–579. doi: 10.1038/nm.2667
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bousquet, M., Quelen, C., Rosati, R., Mansat-De Mas, V., La Starza, R., Bastard, C., et al. (2008). Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J. Exp. Med. 205, 2499–2506. doi: 10.1084/jem.20080285
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Busk, P. K., and Cirera, S. (2010). MicroRNA profiling in early hypertrophic growth of the left ventricle in rats. Biochem. Biophys. Res. Commun. 396, 989–993. doi: 10.1016/j.bbrc.2010.05.039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Camps, C., Saini, H. K., Mole, D. R., Choudhry, H., Reczko, M., Guerra-Assunção, J. A., et al. (2014). Integrated analysis of microRNA and mRNA expression and association with HIF binding reveals the complexity of microRNA expression regulation under hypoxia. Mol. Cancer 13:28. doi: 10.1186/1476-4598-13-28
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chabre, O., Libé, R., Assie, G., Barreau, O., Bertherat, J., Bertagna, X., et al. (2013). Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocr. Relat. Cancer 20, 579–594. doi: 10.1530/ERC-13-0051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chamorro-Jorganes, A., Araldi, E., Penalva, L. O., Sandhu, D., Fernández-Hernando, C., and Suárez, Y. (2011). MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler. Thromb. Vasc. Biol. 31, 2595–2606. doi: 10.1161/ATVBAHA.111.236521
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chan, M. C., Hilyard, A. C., Wu, C., Davis, B. N., Hill, N. S., Lal, A., et al. (2010). Molecular basis for antagonism between PDGF and the TGFβ family of signalling pathways by control of miR-24 expression. EMBO J. 29, 559–573. doi: 10.1038/emboj.2009.370
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, C. Y., Chen, Y., Lai, M. T., Chang, H. W., Cheng, J., Chan, C., et al. (2013). BMPR1B up-regulation via a miRNA binding site variation defines endometriosis susceptibility and CA125 levels. PLoS ONE 8:e80630. doi: 10.1371/journal.pone.0080630
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chapiro, E., Russell, L. J., Struski, S., Cavé, H., Radford-Weiss, I., Valle, V. D., et al. (2010). A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia 24, 1362–1364. doi: 10.1038/leu.2010.93
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, H., Untiveros, G. M., McKee, L. A., Perez, J., Li, J., Antin, P. B., et al. (2012). Micro-RNA-195 and -451 regulate the LKB1/AMPK signaling axis by targeting MO25. PLoS ONE 7:e41574. doi: 10.1371/journal.pone.0041574
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, L., Zhang, A., Li, Y., Zhang, K., Han, L., Du, W., et al. (2013a). MiR-24 regulates the proliferation and invasion of glioma by ST7L via β-catenin/Tcf-4 signaling. Cancer Lett. 329, 174–180. doi: 10.1016/j.canlet.2012.10.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, N. X., Kiattisunthorn, K., O'Neill, K. D., Chen, X., Moorthi, R. N., Gattone, V. H. 2nd., et al. (2013b). Decreased microRNA is involved in the vascular remodeling abnormalities in chronic kidney disease (CKD). PLoS ONE 8:e64558. doi: 10.1371/journal.pone.0064558
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, X., Guo, X., Zhang, H., Xiang, Y., Chen, J., Yin, Y., et al. (2009). Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28, 1385–1392. doi: 10.1038/onc.2008.474
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, J., Guo, S., Chen, S., Mastriano, S. J., Liu, C., D'Alessio, A. C., et al. (2013). An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell Rep. 5, 471–481. doi: 10.1016/j.celrep.2013.08.050
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cohen, E. D., Wang, Z., Lepore, J. J., Lu, M. M., Taketo, M. M., Epstein, D. J., et al. (2007). Wnt/β-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J. Clin. Invest. 117, 1794–1804. doi: 10.1172/JCI31731
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coleman, S. J., Bruce, C., Chioni, A., Kocher, H. M., and Grose, R. P. (2014). The ins and outs of fibroblast growth factor signaling. Clin. Sci. 127, 217–231. doi: 10.1042/CS20140100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Croce, C. M. (2009). Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714. doi: 10.1038/nrg2634
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cui, E. H., Li, H. J., Hua, F., Wang, B., Mao, W., Feng, X. R., et al. (2013). Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta Pharmacol. Sin. 34, 309–313. doi: 10.1038/aps.2012.125
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Datta, J., Kutay, H., Nasser, M. W., Nuovo, G. J., Wang, B., Majumder, S., et al. (2008). Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 68, 5049–5058. doi: 10.1158/0008-5472.CAN-07-6655
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dawson, M. A., and Kouzarides, T. (2012). Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27. doi: 10.1016/j.cell.2012.06.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deng, H., Guo, Y., Song, H., Xiao, B., Sun, W., Liu, Z., et al. (2013). MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene 518, 351–359. doi: 10.1016/j.gene.2012.12.103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Derfoul, A., Juan, A. H., Difilippantonio, M. J., Palanisamy, N., Ried, T., and Sartorelli, V. (2011). Decreased microRNA-214 levels in breast cancer cells coincides with increased cell proliferation, invasion and accumulation of the Polycomb Ezh2 methyltransferase. Carcinogenesis 32, 1607–1614. doi: 10.1093/carcin/bgr184
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ding, J., Huang, S., Wang, Y., Tian, Q., Zha, R., Shi, H., et al. (2013). Genome-wide screening reveals that miR-195 targets the TNF-α/NF-κB pathway by down-regulating IκB kinase α and TAB3 in hepatocellular carcinoma. Hepatology 58, 654–666. doi: 10.1002/hep.26378
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Divakaran, V., and Mann, D. L. (2008). The emerging role of microRNAs in cardiac remodeling and heart failure. Circ. Res. 103, 919–928. doi: 10.1161/CIRCRESAHA.108.183087
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Du, W. W., Fang, L., Li, M., Yang, X., Liang, Y., Peng, C., et al. (2013). MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J. Cell Sci. 126, 1440–1453. doi: 10.1242/jcs.118299
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duan, Q., Wang, X., Gong, W., Ni, L., Chen, C., He, X., et al. (2012). ER stress negatively modulates the expression of the miR-199a/214 cluster to regulates tumor survival and progression in human hepatocellular cancer. PLoS ONE 7:e31518. doi: 10.1371/journal.pone.0031518
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duroux-Richard, I., Pers, Y. M., Fabre, S., Ammari, M., Baeten, D., Cartron, G., et al. (2014). Circulating miRNA-125b is a potential biomarker predicting response to rituximab in rheumatoid arthritis. Mediators Inflamm. 2014:342524. doi: 10.1155/2014/342524
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Esposito, F., Tornincasa, M., Pallante, P., Federico, A., Borbone, E., Pierantoni, G. M., et al. (2012). Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J. Clin. Endocrinol. Metab. 97, E710–E718. doi: 10.1210/jc.2011-3068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fei, X., Qi, M., Wu, B., Song, Y., Wang, Y., and Li, T. (2012). MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 586, 392–397. doi: 10.1016/j.febslet.2012.01.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feng, X., Wang, Z., Fillmore, R., and Xi, Y. (2014). MiR-200, a new star miRNA in human cancer. Cancer Lett. 344, 166–173. doi: 10.1016/j.canlet.2013.11.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ferracin, M., Bassi, C., Pedriali, M., Pagotto, S., D'Abundo, L., Zagatti, B., et al. (2013). miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol. Cancer 12:130. doi: 10.1186/1476-4598-12-130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ferretti, E., De Smaele, E., Miele, E., Laneve, P., Po, A., Pelloni, M., et al. (2008). Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 27, 2616–2627. doi: 10.1038/emboj.2008.172
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fiedler, J., Jazbutyte, V., Kirchmaier, B. C., Gupta, S. K., Lorenzen, J., Hartmann, D., et al. (2011). MicroRNA-24 regulates vascularity after myocardial infarction. Circulation 124, 720–730. doi: 10.1161/CIRCULATIONAHA.111.039008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gefen, N., Binder, V., Zaliova, M., Linka, Y., Morrow, M., Novosel, A., et al. (2010). Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia 24, 89–96. doi: 10.1038/leu.2009.208
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ghosh, A. K., Nagpal, V., Covington, J. W., Michaels, M. A., and Vaughan, D. E. (2012). Molecular basis of cardiac endothelial-to-mesenchymal transition (EndMT): differential expression of microRNAs during EndMT. Cell. Signal. 24, 1031–1036. doi: 10.1016/j.cellsig.2011.12.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giglio, S., Cirombella, R., Amodeo, R., Portaro, L., Lavra, L., and Vecchione, A. (2013). MicroRNA miR-24 promotes cell proliferation by targeting the CDKs inhibitors p27Kip1 and p16INK4a. J. Cell. Physiol. 228, 2015–2023. doi: 10.1002/jcp.24368
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Goetz, R., and Mohammadi, M. (2013). Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 14, 166–180. doi: 10.1038/nrm3528
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gong, J., Zhang, J. P., Li, B., Zeng, C., You, K., Chen, M. X., et al. (2013). MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 32, 3071–3079. doi: 10.1038/onc.2012.318
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
González-Gugel, E., Villa-Morales, M., Santos, J., Bueno, M. J., Malumbres, M., Rodríguez-Pinilla, S. M., et al. (2013). Down-regulation of specific miRNAs enhances the expression of the gene Smoothened and contributes to T-cell lymphoblastic lymphoma development. Carcinogenesis 34, 902–908. doi: 10.1093/carcin/bgs404
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guo, S. T., Jiang, C. C., Wang, G. P., Li, Y. P., Wang, C. Y., Guo, X. Y., et al. (2013). MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene 32, 1910–1920. doi: 10.1038/onc.2012.214
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Han, Z. B., Zhong, L., Teng, M. J., Fan, J. W., Tang, H. M., Wu, J. Y., et al. (2012). Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol. Oncol. 6, 445–457. doi: 10.1016/j.molonc.2012.04.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heneghan, H. M., Miller, N., Lowery, A. J., Sweeney, K. J., Newell, J., and Kerin, M. J. (2010). Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 251, 499–505. doi: 10.1097/SLA.0b013e3181cc939f
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Henson, B. J., Bhattacharjee, S., O'Dee, D. M., Feingold, E., and Gollin, S. M. (2009). Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer 48, 569–582. doi: 10.1002/gcc.20666
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hill, J. A., and Olson, E. N. (2008). Cardiac plasticity. N. Engl. J. Med. 358, 1370–1380. doi: 10.1056/NEJMra072139
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hirata, H., Hinoda, Y., Ueno, K., Nakajima, K., Ishii, N., and Dahiya, R. (2012). MicroRNA-1826 directly targets β-catenin (CTNNB1) and MEK1 (MAP2K1) in VHL-inactivated renal cancer. Carcinogenesis 33, 501–508. doi: 10.1093/carcin/bgr302
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, H. J., Liu, J., Hua, H., Li, S. E., Zhao, J., Yue, S., et al. (2014a). MiR-214 and N-ras regulatory loop suppresses rhabdomyosarcoma cell growth and xenograft tumorigenesis. Oncotarget 5, 2161–2175.
Huang, L., Luo, J., Cai, Q., Pan, Q., Zeng, H., Guo, Z., et al. (2011a). MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int. J. Cancer 128, 1758–1769. doi: 10.1002/ijc.25509
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, S., Chen, M., Li, L., He, M., Hu, D., Zhang, X., et al. (2014b). Circulating MicroRNAs and the occurrence of acute myocardial infarction in Chinese populations. Circ. Cardiovasc. Genet. 7, 189–198. doi: 10.1161/CIRCGENETICS.113.000294
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, X. H., Chen, J. S., Wang, Q., Chen, X. L., Wen, L., Chen, L. Z., et al. (2011b). miR-338-3p suppresses invasion of liver cancer cell by targeting smoothened. J. Pathol. 225, 463–472. doi: 10.1002/path.2877
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hui, W., Yuntao, L., Lun, L., WenSheng, L., ChaoFeng, L., HaiYong, H., et al. (2013). MicroRNA-195 inhibits the proliferation of human glioma cells by directly targeting cyclin D1 and cyclin E1. PLoS ONE 8:e54932. doi: 10.1371/journal.pone.0054932
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hunter, M. P., Ismail, N., Zhang, X., Aguda, B. D., Lee, E. J., Yu, L., et al. (2008). Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 3:e3694. doi: 10.1371/journal.pone.0003694
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Itesako, T., Seki, N., Yoshino, H., Chiyomaru, T., Yamasaki, T., Hidaka, H., et al. (2014). The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS ONE 9:e84311. doi: 10.1371/journal.pone.0084311
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ito, S., D'Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi: 10.1038/nature09303
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Izzotti, A., Calin, G. A., Arrigo, P., Steele, V. E., Croce, C. M., and De Flora, S. (2009). Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 23, 806–812. doi: 10.1096/fj.08-121384
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacob, N. K., Cooley, J. V., Yee, T. N., Jacob, J., Alder, H., Wickramasinghe, P., et al. (2013). Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PLoS ONE 8:e57603. doi: 10.1371/journal.pone.0057603
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jamieson, N. B., Morran, D. C., Morton, J. P., Ali, A., Dickson, E. J., Carter, C. R., et al. (2012). MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin. Cancer Res. 18, 534–545. doi: 10.1158/1078-0432.CCR-11-0679
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jardim, M. J., Fry, R. C., Jaspers, I., Dailey, L., and Diaz-Sanchez, D. (2009). Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ. Health Perspect. 117, 1745–1751. doi: 10.1289/ehp.0900756
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jayasena, C. S., Ohyama, T., Segil, N., and Groves, A. K. (2008). Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development 135, 2251–2261. doi: 10.1242/dev.017905
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ji, Q., Hao, X., Meng, Y., Zhang, M., Desano, J., Fan, D., et al. (2008). Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 8:266. doi: 10.1186/1471-2407-8-266
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiang, J., and Hui, C. C. (2008). Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812. doi: 10.1016/j.devcel.2008.11.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kanaan, Z., Roberts, H., Eichenberger, M. R., Billeter, A., Ocheretner, G., Pan, J., et al. (2013). A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann. Surg. 258, 400–408. doi: 10.1097/SLA.0b013e3182a15bcc
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kappelmann, M., Kuphal, S., Meister, G., Vardimon, L., and Bosserhoff, A. K. (2013). MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 32, 2984–2991. doi: 10.1038/onc.2012.307
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kasinski, A. L., and Slack, F. J. (2011). MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer 11, 849–864. doi: 10.1038/nrc3166
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katoh, M. (2007). Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev. 3, 30–38. doi: 10.1007/s12015-007-0006-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katoh, M. (2013a). Great challenges in molecular medicine: toward personalized medicine. Front. Cell Dev. Biol. 1:1. doi: 10.3389/fcell.2013.00001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katoh, M. (2013b). Therapeutics targeting angiogenesis: genetics and epigenetics, extracellular miRNAs and signaling networks. Int. J. Mol. Med. 32, 763–767. doi: 10.3892/ijmm.2013.1444
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katoh, M., Igarashi, M., Fukuda, H., Nakagama, H., and Katoh, M. (2013c). Cancer genetics and genomics of human FOX family genes. Cancer Lett. 328, 198–206. doi: 10.1016/j.canlet.2012.09.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katoh, M., and Katoh, M. (2007). WNT signaling pathway and stem cell signaling network. Clin. Cancer Res. 13, 4042–4045. doi: 10.1158/1078-0432.CCR-06-2316
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katoh, M., and Nakagama, H. (2014). FGF receptors: cancer biology and therapeutics. Med. Res. Rev. 34, 280–300. doi: 10.1002/med.21288
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katoh, Y., and Katoh, M. (2008). Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA. Int. J. Mol. Med. 22, 271–275. doi: 10.3892/ijmm_00000019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Katoh, Y., and Katoh, M. (2009). Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med. 9, 873–886. doi: 10.2174/156652409789105570
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J., Kang, Y., Kojima, Y., Lighthouse, J. K., Hu, X., Aldred, M. A., et al. (2013b). An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat. Med. 19, 74–82. doi: 10.1038/nm.3040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J. K., Noh, J. H., Jung, K. H., Eun, J. W., Bae, H. J., Kim, M. G., et al. (2013a). Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors miR-125a-5p and miR-125b. Hepatology 57, 1055–1067. doi: 10.1002/hep.26101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Klaus, A., and Birchmeier, W. (2008). Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 8, 387–398. doi: 10.1038/nrc2389
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Klusmann, J. H., Li, Z., Böhmer, K., Maroz, A., Koch, M. L., Emmrich, S., et al. (2010). miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 24, 478–490. doi: 10.1101/gad.1856210
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koga, Y., Jinnin, M., Ichihara, A., Fujisawa, A., Moriya, C., Sakai, K., et al. (2014). Analysis of expression pattern of serum microRNA levels in patients with psoriasis. J. Dermatol. Sci. 74, 170–171. doi: 10.1016/j.jdermsci.2014.01.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kotlabova, K., Doucha, J., Chudoba, D., Calda, P., Dlouha, K., and Hromadnikova, I. (2013). Extracellular chromosome 21-derived microRNAs in euploid and aneuploid pregnancies. Indian J. Med. Res. 138, 935–943.
Kulis, M., and Esteller, M. (2010). DNA methylation and cancer. Adv. Genet. 70, 27–56. doi: 10.1016/B978-0-12-380866-0.60002-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ladeiro, Y., Couchy, G., Balabaud, C., Bioulac-Sage, P., Pelletier, L., Rebouissou, S., et al. (2008). MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology 47, 1955–1963. doi: 10.1002/hep.22256
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lal, A., Kim, H. H., Abdelmohsen, K., Kuwano, Y., Pullmann, R. Jr., Srikantan, S., et al. (2008). p16(INK4a) translation suppressed by miR-24. PLoS ONE 3:e1864. doi: 10.1371/journal.pone.0001864
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lal, A., Navarro, F., Maher, C. A., Maliszewski, L. E., Yan, N., O'Day, E., et al. (2009a). miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol. Cell 35, 610–625. doi: 10.1016/j.molcel.2009.08.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lal, A., Pan, Y., Navarro, F., Dykxhoorn, D. M., Moreau, L., Meire, E., et al. (2009b). miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 16, 492–498. doi: 10.1038/nsmb.1589
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Le, H. B., Zhu, W. Y., Chen, D. D., He, J. Y., Huang, Y. Y., Liu, X. G., et al. (2012). Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med. Oncol. 29, 3190–3197. doi: 10.1007/s12032-012-0303-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Le, M. T., Teh, C., Shyh-Chang, N., Xie, H., Zhou, B., Korzh, V., et al. (2009). MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 23, 862–876. doi: 10.1101/gad.1767609
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leptidis, S., El Azzouzi, H., Lok, S. I., de Weger, R., Olieslagers, S., Kisters, N., et al. (2013). A deep sequencing approach to uncover the miRNOME in the human heart. PLoS ONE 8:e57800. doi: 10.1371/journal.pone.0057800
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, B., Han, Q., Zhu, Y., Yu, Y., Wang, J., and Jiang, X. (2012). Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. FEBS J. 279, 2393–2398. doi: 10.1111/j.1742-4658.2012.08618.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, D., Yang, P., Xiong, Q., Song, X., Yang, X., Liu, L., et al. (2010). MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells. J. Hypertens. 28, 1646–1654. doi: 10.1097/HJH.0b013e32833a4922
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, D., Zhao, Y., Liu, C., Chen, X., Qi, Y., Jiang, Y., et al. (2011). Analysis of miR-195 and miR-497 expression, regulation and role in breast cancer. Clin. Cancer Res. 17, 1722–1730. doi: 10.1158/1078-0432.CCR-10-1800
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, X., Liu, X., Xu, W., Zhou, P., Gao, P., Jiang, S., et al. (2013a). c-MYC-regulated miR-23a/24-2/27a cluster promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty2. J. Biol. Chem. 288, 18121–18133. doi: 10.1074/jbc.M113.478560
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, Y., Gordon, M. W., Xu-Monette, Z. Y., Visco, C., Tzankov, A., Zou, D., et al. (2013b). Single nucleotide variation in the TP53 3′ untranslated region in diffuse large B-cell lymphoma treated with rituximab-CHOP: a report from the International DLBCL rituximab-CHOP consortium program. Blood 121, 4529–4540. doi: 10.1182/blood-2012-12-471722
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liang, L., Wong, C. M., Ying, Q., Fan, D. N., Huang, S., Ding, J., et al. (2010). MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology 52, 1731–1740. doi: 10.1002/hep.23904
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liao, Y., Du, X., and Lönnerdal, B. (2010). miR-214 regulates lactoferrin expression and pro-apoptotic function in mammary epithelial cells. J. Nutr. 140, 1552–1556. doi: 10.3945/jn.110.124289
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lichtenstein, P., Holm, N. V., Verkasalo, P. K., Iliadou, A., Kaprio, J., Koskenvuo, M., et al. (2000). Environmental and heritable factors in the causation of cancer: analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343, 78–85. doi: 10.1056/NEJM200007133430201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lin, K. Y., Zhang, X. J., Feng, D. D., Zhang, H., Zeng, C. W., Han, B. W., et al. (2011). miR-125b, a target of CDX2, regulates cell differentiation through repression of the core binding factor in hematopoietic malignancies. J. Biol. Chem. 286, 38253–38263. doi: 10.1074/jbc.M111.269670
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lin, T. L., and Matsui, W. (2012a). Hedgehog pathway as a drug target: smoothened inhibitors in development. Onco. Targets Ther. 5, 47–58. doi: 10.2147/OTT.S21957
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lin, Y., Wu, J., Chen, H., Mao, Y., Liu, Y., Mao, Q., et al. (2012b). Cyclin-dependent kinase 4 is a novel target in micoRNA-195-mediated cell cycle arrest in bladder cancer cells. FEBS Lett. 586, 442–447. doi: 10.1016/j.febslet.2012.01.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, L., Chen, L., Xu, Y., Li, R., and Du, X. (2010). microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem. Biophys. Res. Commun. 400, 236–240. doi: 10.1016/j.bbrc.2010.08.046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, L. H., Li, H., Li, J. P., Zhong, H., Zhang, H. C., Chen, J., et al. (2011). miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem. Biophys. Res. Commun. 416, 31–38. doi: 10.1016/j.bbrc.2011.10.117
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Long, G., Wang, F., Duan, Q., Yang, S., Chen, F., Gong, W., et al. (2012). Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS ONE 7:e50926. doi: 10.1371/journal.pone.0050926
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lozano, R., Naghavi, M., Foreman, K., Lim, S., Shibuya, K., Aboyans, V., et al. (2012). Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128. doi: 10.1016/S0140-6736(12)61728-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lu, H. Q., Liang, C., He, Z. Q., Fan, M., and Wu, Z. G. (2013). Circulating miR-214 is associated with the severity of coronary artery disease. J. Geriatr. Cardiol. 10, 34–38. doi: 10.3969/j.issn.1671-5411.2013.01.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luzi, E., Marini, F., Giusti, F., Galli, G., Cavalli, L., and Brandi, M. L. (2012). The negative feedback-loop between the oncomir Mir-24-1 and menin modulates the Men1 tumorigenesis by mimicking the “Knudson's second hit.” PLoS ONE 7:e39767. doi: 10.1371/journal.pone.0039767
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lv, G., Shao, S., Dong, H., Bian, X., Yang, X., and Dong, S. (2014). MicroRNA-214 protects cardiac myocytes against H2O2-induced injury. J. Cell. Biochem. 115, 93–101. doi: 10.1002/jcb.24636
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mahn, R., Heukamp, L. C., Rogenhofer, S., von Ruecker, A., Müller, S. C., and Ellinger, J. (2011). Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology 77:1265.e9–16. doi: 10.1016/j.urology.2011.01.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Malumbres, R., Sarosiek, K. A., Cubedo, E., Ruiz, J. W., Jiang, X., Gascoyne, R. D., et al. (2009). Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood 113, 3754–3764. doi: 10.1182/blood-2008-10-184077
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mar-Aguilar, F., Mendoza-Ramírez, J. A., Malagón-Santiago, I., Espino-Silva, P. K., Santuario-Facio, S. K., Ruiz-Flores, P., et al. (2013). Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis. Markers 34, 163–169. doi: 10.1155/2013/259454
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matkovich, S. J., van Booven, D. J., Youker, K. A., Torre-Amione, G., Diwan, A., Eschenbacher, W. H., et al. (2009). Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation 119, 1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McKenna, D. J., Patel, D., and McCance, D. J. (2014). miR-24 and miR-205 expression is dependent on HPV onco-protein expression in keratinocytes. Virology 448, 210–216. doi: 10.1016/j.virol.2013.10.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meloni, M., Marchetti, M., Garner, K., Littlejohns, B., Sala-Newby, G., Xenophontos, N., et al. (2013). Local inhibition of microRNA-24 improves reparative angiogenesis and left ventricle remodeling and function in mice with myocardial infarction. Mol. Ther. 21, 1390–1402. doi: 10.1038/mt.2013.89
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Menigatti, M., Staiano, T., Manser, C. N., Bauerfeind, P., Komljenovic, A., Robinson, M., et al. (2013). Epigenetic silencing of monoallelically methylated miRNA loci in precancerous colorectal lesions. Oncogenesis 2:e56. doi: 10.1038/oncsis.2013.21
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Milenkovic, D., Deval, C., Gouranton, E., Landrier, J. F., Scalbert, A., Morand, C., et al. (2012). Modulation of miRNA expression by dietary polyphenols in apoE deficient mice: a new mechanism of the action of polyphenols. PLoS ONE 7:e29837. doi: 10.1371/journal.pone.0029837
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Milne, A. N., Carneiro, F., O'Morain, C., and Offerhaus, G. J. (2009). Nature meets nurture: molecular genetics of gastric cancer. Hum. Genet. 126, 615–628. doi: 10.1007/s00439-009-0722-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mishra, P. J., Humeniuk, R., Mishra, P. J., Longo-Sorbello, G. S., Banerjee, D., and Bertino, J. R. (2007). A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc. Natl. Acad. Sci. U.S.A. 104, 13513–13518. doi: 10.1073/pnas.0706217104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Misiewicz-Krzeminska, I., Sarasquete, M. E., Quwaider, D., Krzeminski, P., Ticona, F. V., Paíno, T., et al. (2013). Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica 98, 640–648. doi: 10.3324/haematol.2012.070011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518. doi: 10.1073/pnas.0804549105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mitra, A. K., Zillhardt, M., Hua, Y., Tiwari, P., Murmann, A. E., Peter, M. E., et al. (2012). MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2, 1100–1108. doi: 10.1158/2159-8290.CD-12-0206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mo, Y. Y., Tang, H., and Miele, L. (2013). Notch-associated microRNAs in cancer. Curr. Drug Targets 14, 1157–1166. doi: 10.2174/13894501113149990188
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Momose, K., Minami, A., Shimono, Y., Mizutani, K., Nobutani, K., Azuma, T., et al. (2013). miR-214 and hypoxia down-regulate Necl-2/CADM1 and enhance ErbB2/ErbB3 signaling. Genes Cells 18, 195–202. doi: 10.1111/gtc.12027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mor, E., Kano, S., Colantuoni, C., Sawa, A., Navon, R., and Shomron, N. (2013). MicroRNA-382 expression is elevated in the olfactory neuroepithelium of schizophrenia patients. Neurobiol. Dis. 55, 1–10. doi: 10.1016/j.nbd.2013.03.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muramatsu, F., Kidoya, H., Naito, H., Sakimoto, S., and Takakura, N. (2013). microRNA-125b inhibits tube formation of blood vessels through translational suppression of VE-cadherin. Oncogene 32, 414–421. doi: 10.1038/onc.2012.68
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murata, K., Furu, M., Yoshitomi, H., Ishikawa, M., Shibuya, H., Hashimoto, M., et al. (2013). Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS ONE 8:e69118. doi: 10.1371/journal.pone.0069118
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naga Prasad, S. V. N., Duan, Z. H., Gupta, M. K., Surampudi, V. S., Volinia, S., Calin, G. A., et al. (2009). Unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. J. Biol. Chem. 284, 27487–27499. doi: 10.1074/jbc.M109.036541
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Narducci, M. G., Arcelli, D., Picchio, M. C., Lazzeri, C., Pagani, E., Sampogna, F., et al. (2011). MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sézary syndrome. Cell Death Dis. 2, e151. doi: 10.1038/cddis.2011.32
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nielsen, L. B., Wang, C., Sørensen, K., Bang-Berthelsen, C. H., Hansen, L., Andersen, M. L., et al. (2012). Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual β-cell function and glycaemic control during disease progression. Exp. Diabetes Res. 2012:896362. doi: 10.1155/2012/896362
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nowell, C., and Radtke, F. (2013). Cutaneous Notch signaling in health and disease. Cold Spring Harb. Perspect. Med. 3:a017772. doi: 10.1101/cshperspect.a017772
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nygren, M. K., Tekle, C., Ingebrigtsen, V. A., Mäkelä, R., Krohn, M., Aure, M. R., et al. (2014). Identifying microRNAs regulating B7-H3 in breast cancer: the clinical impact of microRNA-29c. Br. J. Cancer 110, 2072–2080. doi: 10.1038/bjc.2014.113
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oishi, N., Kumar, M. R., Roessler, S., Ji, J., Forgues, M., Budhu, A., et al. (2012). Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology 56, 1792–1803. doi: 10.1002/hep.25890
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ordovás, J. M., and Smith, C. E. (2010). Epigenetics and cardiovascular disease. Nat. Rev. Cardiol. 7, 510–519. doi: 10.1038/nrcardio.2010.104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ortega, F. J., Mercader, J. M., Catalán, V., Moreno-Navarrete, J. M., Pueyo, N., Sabater, M., et al. (2013). Targeting the circulating microRNA signature of obesity. Clin. Chem. 59, 781–792. doi: 10.1373/clinchem.2012.195776
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ortega, F. J., Mercader, J. M., Moreno-Navarrete, J. M., Rovira, O., Guerra, E., Esteve, E., et al. (2014). Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care 37, 1375–1383. doi: 10.2337/dc13-1847
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ozen, M., Creighton, C. J., Ozdemir, M., and Ittmann, M. (2008). Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 27, 1788–1793. doi: 10.1038/sj.onc.1210809
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Papadimitriou, E., Vasilaki, E., Vorvis, C., Iliopoulos, D., Moustakas, A., Kardassis, D., et al. (2012). Differential regulation of the two RhoA-specific GEF isoforms Net1/Net1A by TGF-β and miR-24: role in epithelial-to-mesenchymal transition. Oncogene 31, 2862–2875. doi: 10.1038/onc.2011.457
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parpart, S., and Wang, X. W. (2013). microRNA regulation and its consequences in cancer. Curr. Pathobiol. Rep. 1, 71–79. doi: 10.1007/s40139-012-0002-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peng, R. Q., Wan, H. Y., Li, H. F., Liu, M., Li, X., and Tang, H. (2012). MicroRNA-214 suppresses growth and invasiveness of cervical cancer cells by targeting UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7. J. Biol. Chem. 287, 14301–14309. doi: 10.1074/jbc.M111.337642
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Penna, E., Orso, F., Cimino, D., Tenaglia, E., Lembo, A., Quaglino, E., et al. (2011). microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 30, 1990–2007. doi: 10.1038/emboj.2011.102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pirola, C. J., Fernández Gianotti, T., Castaño, G. O., Mallardi, P., San Martino, J., Mora Gonzalez Lopez Ledesma, M., et al. (2014). Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. doi: 10.1136/gutjnl-2014-306996. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Porkka, K. P., Pfeiffer, M. J., Waltering, K. K., Vessella, R. L., Tammela, T. L., and Visakorpi, T. (2007). MicroRNA expression profiling in prostate cancer. Cancer Res. 67, 6130–6135. doi: 10.1158/0008-5472.CAN-07-0533
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Porrello, E. R., Johnson, B. A., Aurora, A. B., Simpson, E., Nam, Y. J., Matkovich, S. J., et al. (2011). MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 109, 670–679. doi: 10.1161/CIRCRESAHA.111.248880
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qian, L., Van Laake, L. W., Huang, Y., Liu, S., Wendland, M. F., and Srivastava, D. (2011). miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J. Exp. Med. 208, 549–560. doi: 10.1084/jem.20101547
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qin, W., Shi, Y., Zhao, B., Yao, C., Jin, L., Ma, J., et al. (2010). miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS ONE 5:e9429. doi: 10.1371/journal.pone.0009429
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qu, K. Z., Zhang, K., Li, H., Afdhal, N. H., and Albitar, M. (2011). Circulating microRNAs as biomarkers for hepatocellular carcinoma. J. Clin. Gastroenterol. 45, 355–360. doi: 10.1097/MCG.0b013e3181f18ac2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rager, J. E., Smeester, L., Jaspers, I., Sexton, K. G., and Fry, R. C. (2011). Epigenetic changes induced by air toxics: formaldehyde exposure alters miRNA expression profiles in human lung cells. Environ. Health Perspect. 119, 494–500. doi: 10.1289/ehp.1002614
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ragusa, M., Caltabiano, R., Russo, A., Puzzo, L., Avitabile, T., Longo, A., et al. (2013). MicroRNAs in vitreus humor from patients with ocular diseases. Mol. Vis. 19, 430–440.
Rajabi, H., Jin, C., Ahmad, R., McClary, C., Joshi, M. D., and Kufe, D. (2010). Mucin 1 oncoprotein expression is suppressed by the miR-125b oncomir. Genes Cancer 1, 62–68. doi: 10.1177/1947601909357933
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rajaram, M. V., Ni, B., Morris, J. D., Brooks, M. N., Carlson, T. K., Bakthavachalu, B., et al. (2011). Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc. Natl. Acad. Sci. U.S.A. 108, 17408–17413. doi: 10.1073/pnas.1112660108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rao, T. P., and Kühl, M. (2010). An updated overview on Wnt signaling pathways: a prelude for more. Circ. Res. 106, 1798–1806. doi: 10.1161/CIRCRESAHA.110.219840
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ratert, N., Meyer, H. A., Jung, M., Lioudmer, P., Mollenkopf, H. J., Wagner, I., et al. (2013). miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J. Mol. Diagn. 15, 695–705. doi: 10.1016/j.jmoldx.2013.05.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rio-Machin, A., Ferreira, B. I., Henry, T., Gómez-López, G., Agirre, X., Alvarez, S., et al. (2013). Downregulation of specific miRNAs in hyperdiploid multiple myeloma mimics the oncogenic effect of IgH translocations occurring in the non-hyperdiploid subtype. Leukemia 27, 925–931. doi: 10.1038/leu.2012.302
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sætrom, P., Biesinger, J., Li, S. M., Smith, D., Thomas, L. F., Majzoub, K., et al. (2009). A risk variant in an miR-125b binding site in BMPR1B is associated with breast cancer pathogenesis. Cancer Res. 69, 7459–7465. doi: 10.1158/0008-5472.CAN-09-1201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sander, S., Bullinger, L., Klapproth, K., Fiedler, K., Kestler, H. A., Barth, T. F., et al. (2008). MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 112, 4202–4212. doi: 10.1182/blood-2008-03-147645
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saydam, O., Shen, Y., Würdinger, T., Senol, O., Boke, E., James, M. F., et al. (2009). Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/β-catenin signaling pathway. Mol. Cell Biol. 29, 5923–5940. doi: 10.1128/MCB.00332-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schembri, F., Sridhar, S., Perdomo, C., Gustafson, A. M., Zhang, X., Ergun, A., et al. (2009). MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 106, 2319–2324. doi: 10.1073/pnas.0806383106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schneider, M., Andersen, D. C., Silahtaroglu, A., Lyngbæk, S., Kauppinen, S., Hansen, J. L., et al. (2011). Cell-specific detection of microRNA expression during cardiomyogenesis by combined in situ hybridization and immunohistochemistry. J. Mol. Histol. 42, 289–299. doi: 10.1007/s10735-011-9332-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schwarzenbach, H., Milde-Langosch, K., Steinbach, B., Müller, V., and Pantel, K. (2012). Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res. Treat. 134, 933–941. doi: 10.1007/s10549-012-1988-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scott, G. K., Goga, A., Bhaumik, D., Berger, C. E., Sullivan, C. S., and Benz, C. C. (2007). Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 282, 1479–1486. doi: 10.1074/jbc.M609383200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Seeliger, C., Karpinski, K., Haug, A., Vester, H., Schmitt, A., Bauer, J., et al. (2014). Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. 29, 1718–1728. doi: 10.1002/jbmr.2175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shah, A. M., and Mann, D. L. (2011). In search of new therapeutic targets and strategies for heart failure: recent advances in basic science. Lancet 378, 704–712. doi: 10.1016/S0140-6736(11)60894-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shi, W., Du, J., Qi, Y., Liang, G., Wang, T., Li, S., et al. (2012). Aberrant expression of serum miRNAs in schizophrenia. J. Psychiatr. Res. 46, 198–204. doi: 10.1016/j.jpsychires.2011.09.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shi, W., Gerster, K., Alajez, N. M., Tsang, J., Waldron, L., Pintilie, M., et al. (2011). MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 71, 2926–2937. doi: 10.1158/0008-5472.CAN-10-3369
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shi, X. B., Xue, L., Yang, J., Ma, A. H., Zhao, J., Xu, M., et al. (2007). An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 104, 19983–19988. doi: 10.1073/pnas.0706641104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shih, A. H., Abdel-Wahab, O., Patel, J. P., and Levine, R. L. (2012a). The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer 12, 599–612. doi: 10.1038/nrc3343
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shih, T. C., Tien, Y. J., Wen, C. J., Yeh, T. S., Yu, M. C., Huang, C. H., et al. (2012b). MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J. Hepatol. 57, 584–591. doi: 10.1016/j.jhep.2012.04.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Skog, J., Würdinger, T., van Rijn, S., Meijer, D. H., Gainche, L., Sena-Esteves, M., et al. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476. doi: 10.1038/ncb1800
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smirnov, D. A., and Cheung, V. G. (2008). ATM gene mutations result in both recessive and dominant expression phenotypes of genes and microRNAs. Am. J. Hum. Genet. 83, 243–253. doi: 10.1016/j.ajhg.2008.07.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sochor, M., Basova, P., Pesta, M., Dusilkova, N., Bartos, J., Burda, P., et al. (2014). Oncogenic MicroRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 14:448. doi: 10.1186/1471-2407-14-448
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sparmann, A., and van Lohuizen, M. (2006). Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6, 846–856. doi: 10.1038/nrc1991
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Srivastava, A., Goldberger, H., Dimtchev, A., Ramalinga, M., Chijioke, J., Marian, C., et al. (2013). MicroRNA profiling in prostate cancer—the diagnostic potential of urinary miR-205 and miR-214. PLoS ONE 8:e76994. doi: 10.1371/journal.pone.0076994
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Srivastava, N., Manvati, S., Srivastava, A., Pal, R., Kalaiarasan, P., Chattopadhyay, S., et al. (2011). miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: a potential for therapeutic intervention. Breast Cancer Res. 13:R39. doi: 10.1186/bcr2861
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suh, J. M., Jonker, J. W., Ahmadian, M., Goetz, R., Lackey, D., Osborn, O., et al. (2014). Endocrinization of FGF1 produces a neomorphic and potent insulin sensitizer. Nature 513, 436–439. doi: 10.1038/nature13540
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, J. Y., Huang, Y., Li, J. P., Zhang, X., Wang, L., Meng, Y. L., et al. (2012). MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem. Biophys. Res. Commun. 420, 787–792. doi: 10.1016/j.bbrc.2012.03.075
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Szczyrba, J., Nolte, E., Hart, M., Döll, C., Wach, S., Taubert, H., et al. (2013). Identification of ZNF217, hnRNP-K, VEGF-A and IPO7 as targets for microRNAs that are downregulated in prostate carcinoma. Int. J. Cancer 132, 775–784. doi: 10.1002/ijc.27731
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takagi, S., Nakajima, M., Kida, K., Yamaura, Y., Fukami, T., and Yokoi, T. (2010). MicroRNAs regulate human hepatocyte nuclear factor 4α, modulating the expression of metabolic enzymes and cell cycle. J. Biol. Chem. 285, 4415–4422. doi: 10.1074/jbc.M109.085431
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tan, L., Yu, J. T., Liu, Q. Y., Tan, M. S., Zhang, W., Hu, N., et al. (2014). Circulating miR-125b as a biomarker of Alzheimer's disease. J. Neurol. Sci. 336, 52–56. doi: 10.1016/j.jns.2013.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tang, F., Zhang, R., He, Y., Zou, M., Guo, L., and Xi, T. (2012). MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PLoS ONE 7:e35435. doi: 10.1371/journal.pone.0035435
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tap, W. D., Eilber, F. C., Ginther, C., Dry, S. M., Reese, N., Barzan-Smith, K., et al. (2011). Evaluation of well-differentiated/de-differentiated liposarcomas by high-resolution oligonucleotide array-based comparative genomic hybridization. Genes Chromosomes Cancer 50, 95–112. doi: 10.1002/gcc.20835
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Taylor, D. D., and Gercel-Taylor, C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21. doi: 10.1016/j.ygyno.2008.04.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Teng, Y., Manavalan, T. T., Hu, C., Medjakovic, S., Jungbauer, A., and Klinge, C. M. (2013). Endocrine disruptors fludioxonil and fenhexamid stimulate miR-21 expression in breast cancer cells. Toxicol. Sci. 131, 71–83. doi: 10.1093/toxsci/kfs290
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thorsen, J., Aamot, H. V., Roberto, R., Tjønnfjord, G. E., Micci, F., and Heim, S. (2012). Myelodysplastic syndrome with a t(2;11)(p21;q23-24) and translocation breakpoint close to miR-125b-1. Cancer Genet. 205, 528–532. doi: 10.1016/j.cancergen.2012.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tilghman, S. L., Bratton, M. R., Segar, H. C., Martin, E. C., Rhodes, L. V., Li, M., et al. (2012). Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PLoS ONE 7:e32754. doi: 10.1371/journal.pone.0032754
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Turner, N., and Grose, R. (2010). Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 10, 116–129. doi: 10.1038/nrc2780
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vacchi-Suzzi, C., Hahne, F., Scheubel, P., Marcellin, M., Dubost, V., Westphal, M., et al. (2013). Heart structure-specific transcriptomic atlas reveals conserved microRNA-mRNA interactions. PLoS ONE 8:e52442. doi: 10.1371/journal.pone.0052442
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vaishnavi, V., Manikandan, M., Tiwary, B. K., and Munirajan, A. K. (2013). Insights on the functional impact of microRNAs present in autism-associated copy number variants. PLoS ONE 8:e56781. doi: 10.1371/journal.pone.0056781
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J. J., and abd Lötvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. doi: 10.1038/ncb1596
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vallelunga, A., Ragusa, M., Di Mauro, S., Iannitti, T., Pilleri, M., Biundo, R., et al. (2014). Identification of circulating microRNAs for the differential diagnosis of Parkinson's disease and Multiple System Atrophy. Front. Cell. Neurosci. 8:156. doi: 10.3389/fncel.2014.00156
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Rooij, E., and Olson, E. N. (2012). MicroRNA therapeutics for cardiovascular disease: opportunity and obstacles. Nat. Rev. Drug Disc. 11, 860–872. doi: 10.1038/nrd3864
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Rooij, E., Sutherland, L. B., Liu, N., Williams, A. H., McAnally, J., Gerard, R. D., et al. (2006). A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. U.S.A. 103, 18255–18260. doi: 10.1073/pnas.0608791103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Varambally, S., Cao, Q., Mani, R. S., Shankar, S., Wang, X., Ateeq, B., et al. (2008). Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699. doi: 10.1126/science.1165395
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Veerla, S., Lindgren, D., Kvist, A., Frigyesi, A., Staaf, J., Persson, H., et al. (2009). MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int. J. Cancer 124, 2236–2242. doi: 10.1002/ijc.24183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433. doi: 10.1038/ncb2210
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Volinia, S., Calin, G. A., Liu, C. G., Ambs, S., Cimmino, A., Petrocca, F., et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261. doi: 10.1073/pnas.0510565103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, F., Liu, M., Li, X., and Tang, H. (2013a). MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 587, 488–495. doi: 10.1016/j.febslet.2013.01.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H., Tan, G., Dong, L., Cheng, L., Li, K., Wang, Z., et al. (2012a). Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS ONE 7:e34210. doi: 10.1371/journal.pone.0034210
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J., Huang, W., Xu, R., Nie, Y., Cao, X., Meng, J., et al. (2012b). MicroRNA-24 regulates cardiac fibrosis after myocardial infarction. J. Cell. Mol. Med. 16, 2150–2160. doi: 10.1111/j.1582-4934.2012.01523.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J., Li, J., Wang, X., Zheng, C., and Ma, W. (2013b). Downregulation of microRNA-214 and overexpression of FGFR-1 contribute to hepatocellular carcinoma metastasis. Biochem. Biophys. Res. Commun. 439, 47–53. doi: 10.1016/j.bbrc.2013.08.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, M., Zhao, C., Shi, H., Zhang, B., Zhang, L., Zhang, X., et al. (2014a). Deregulated microRNAs in gastric cancer tissue-derived mesenchymal stem cells: novel biomarkers and a mechanism for gastric cancer. Br. J. Cancer 110, 1199–1210. doi: 10.1038/bjc.2014.14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, Q., Huang, Z., Xue, H., Jin, C., Ju, X. L., Han, J. D., et al. (2008a). MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood 111, 588–595. doi: 10.1182/blood-2007-05-092718
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, R., Zhao, N., Li, S., Fang, J. H., Chen, M. X., Yang, J., et al. (2013c). MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology 58, 642–653. doi: 10.1002/hep.26373
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, S., and Olson, E. N. (2009). AngiomiRs—key regulators of angiogenesis. Curr. Opin. Genet. Dev. 19, 205–211. doi: 10.1016/j.gde.2009.04.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X., Ha, T., Zou, J., Ren, D., Liu, L., Zhang, X., et al. (2014b). MicroRNA-125b protects against myocardial ischaemia/reperfusion injury via targeting p53-mediated apoptotic signalling and TRAF6. Cardiovasc. Res. 102, 385–395. doi: 10.1093/cvr/cvu044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X., Sundquist, J., Zöller, B., Memon, A. A., Palmér, K., Sundquist, K., et al. (2014c). Determination of 14 circulating microRNAs in Swedes and Iraqis with and without diabetes mellitus type 2. PLoS ONE 9:e86792. doi: 10.1371/journal.pone.0086792
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X., Tang, S., Le, S. Y., Lu, R., Rader, J. S., Meyers, C., et al. (2008b). Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE 3:e2557. doi: 10.1371/journal.pone.0002557
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, Z., Cai, H., Lin, L., Tang, M., and Cai, H. (2014d). Upregulated expression of microRNA-214 is linked to tumor progression and adverse prognosis in pediatric osteosarcoma. Pediatr. Blood Cancer 61, 206–210. doi: 10.1002/pbc.24763
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weber, J. A., Baxter, D. H., Zhang, S., Huang, D. Y., Huang, K. H., Lee, M. J., et al. (2010). The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741. doi: 10.1373/clinchem.2010.147405
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wen, D., Li, S., Ji, F., Cao, H., Jiang, W., Zhu, J., et al. (2013). miR-133b acts as a tumor suppressor and negatively regulates FGFR1 in gastric cancer. Tumour Biol. 34, 793–803. doi: 10.1007/s13277-012-0609-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weng, Y., Chen, Y., Chen, J., Liu, Y., and Bao, T. (2013). Identification of serum microRNAs in genome-wide serum microRNA expression profiles as novel noninvasive biomarkers for malignant peripheral nerve sheath tumor diagnosis. Med. Oncol. 30:531. doi: 10.1007/s12032-013-0531-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wilting, S. M., Snijders, P. J. F., Verlaat, W., Jaspers, A., van de Wiel, M. A., van Wieringen, W. N., et al. (2013). Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis. Oncogene 32, 106–116. doi: 10.1038/onc.2012.20
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wong, S. S., Ritner, C., Ramachandran, S., Aurigui, J., Pitt, C., Chandra, P., et al. (2012). miR-125b promotes early germ layer specification through Lin28/let-7d and preferential differentiation of mesoderm in human embryonic stem cells. PLoS ONE 7:e36121. doi: 10.1371/journal.pone.0036121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, J., Zhang, Y. C., Suo, W. H., Liu, X. B., Shen, W. W., Tian, H., et al. (2010). Induction of anion exchanger-1 translation and its opposite roles in the carcinogenesis of gastric cancer cells and differentiation of K562 cells. Oncogene 29, 1987–1996. doi: 10.1038/onc.2009.481
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, L., and Belasco, J. G. (2005). Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell Biol. 25, 9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, L., Zhou, H., Lin, H., Qi, J., Zhu, C., Gao, Z., et al. (2012a). Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction 143, 389–397. doi: 10.1530/REP-11-0304
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, Q., Wang, C., Lu, Z., Guo, L., and Ge, Q. (2012b). Analysis of serum genome-wide microRNAs for breast cancer detection. Clin. Chim. Acta 413, 1058–1065. doi: 10.1016/j.cca.2012.02.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xia, H., Ooi, L. L., and Hui, K. M. (2012). MiR-214 targets β-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS ONE 7:e44206. doi: 10.1371/journal.pone.0044206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xie, Y., Tobin, L. A., Camps, J., Wangsa, D., Yang, J., Rao, M., et al. (2013). MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells. Oncogene 32, 2442–2451. doi: 10.1038/onc.2012.258
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, C. X., Xu, M., Tan, L., Yang, H., Permuth-Wey, J., Kruk, P. A., et al. (2012a). MicroRNA miR-214 regulates ovarian cancer cell stemness by targeting p53/Nanog. J. Biol. Chem. 287, 34970–34978. doi: 10.1074/jbc.M112.374611
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, M., Wu, H. D., Li, R. C., Zhang, H. B., Wang, M., Tao, J., et al. (2012b). Mir-24 regulates junctophilin-2 expression in cardiomyocytes. Circ. Res. 111, 837–841. doi: 10.1161/CIRCRESAHA.112.277418
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, N., Brodin, P., Wei, T., Meisgen, F., Eidsmo, L., Nagy, N., et al. (2011). MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J. Invest. Dermatol. 131, 1521–1529. doi: 10.1038/jid.2011.55
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, N., Zhang, L., Meisgen, F., Harada, M., Heilborn, J., Homey, B., et al. (2012c). MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 287, 29899–29908. doi: 10.1074/jbc.M112.391243
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, T., Zhu, Y., Xiong, Y., Ge, Y. Y., Yun, J. P., and Zhuang, S. M. (2009). MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 50, 113–121. doi: 10.1002/hep.22919
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, Z., and Wang, T. (2014). miR-214 promotes the proliferation and invasion of osteosarcoma cells through direct suppression of LZTS1. Biochem. Biophys. Res. Commun. 449, 190–195. doi: 10.1016/j.bbrc.2014.04.140
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yanaihara, N., Caplen, N., Bowman, E., Seike, M., Kumamoto, K., Yi, M., et al. (2006). Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198. doi: 10.1016/j.ccr.2006.01.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, H., Kong, W., He, L., Zhao, J. J., O'Donnell, J. D., Wang, J., et al. (2008). MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 68, 425–433. doi: 10.1158/0008-5472.CAN-07-2488
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, J., Zhao, H., Xin, Y., and Fan, L. (2014). MicroRNA-198 inhibits proliferation and induces apoptosis of lung cancer cells via targeting FGFR1. J. Cell. Biochem. 115, 987–995. doi: 10.1002/jcb.24742
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, T., Zhang, G. F., Chen, X. F., Gu, H. H., Fu, S. Z., Xu, H. F., et al. (2013). MicroRNA-214 provokes cardiac hypertrophy via repression of EZH2. Biochem. Biophys. Res. Commun. 436, 578–584. doi: 10.1016/j.bbrc.2013.05.079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, X., Bemis, L., Su, L. J., Gao, D., and Flaig, T. W. (2012a). miR-125b regulation of androgen receptor signaling via modulation of the receptor complex co-repressor NCOR2. Biores. Open Access 1, 55–62. doi: 10.1089/biores.2012.9903
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, X., Yin, J., Yu, J., Xiang, Q., Liu, Y., Tang, S., et al. (2012b). miRNA-195 sensitizes human hepatocellular carcinoma cells to 5-FU by targeting BCL-w. Oncol. Rep. 27, 250–257. doi: 10.3892/or.2011.1472
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, Z., Chen, S., Luan, X., Li, Y., Liu, M., Li, X., et al. (2009). MicroRNA-214 is aberrantly expressed in cervical cancers and inhibits the growth of HeLa cells. IUBMB Life 61, 1075–1082. doi: 10.1002/iub.252
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yuxia, M., Zhennan, T., and Wei, Z. (2012). Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J. Cancer Res. Clin. Oncol. 138, 2045–2050. doi: 10.1007/s00432-012-1285-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zaidi, S. K., Dowdy, C. R., van Wijnen, A. J., Lian, J. B., Raza, A., Stein, J. L., et al. (2009). Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network. Cancer Res. 69, 8249–8255. doi: 10.1158/0008-5472.CAN-09-1567
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zampetaki, A., Kiechl, S., Drozdov, I., Willeit, P., Mayr, U., Prokopi, M., et al. (2010). Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 107, 810–817. doi: 10.1161/CIRCRESAHA.110.226357
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, X. J., Ye, H., Zeng, C. W., He, B., Zhang, H., and Chen, Y. Q. (2010). Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J. Hematol. Oncol. 3:46. doi: 10.1186/1756-8722-3-46
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Yan, L. X., Wu, Q. N., Du, Z. M., Chen, J., Liao, D. Z., et al. (2011). miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 71, 3552–3562. doi: 10.1158/0008-5472.CAN-10-2435
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Z., Li, Z., Gao, C., Chen, P., Chen, J., Liu, W., et al. (2008). miR-21 plays pivotal role in gastric cancer pathogenesis and progression. Lab. Invest. 88, 1358–1366. doi: 10.1038/labinvest.2008.94
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, Z. C., Li, Y. Y., Wang, H. Y., Fu, S., Wang, X. P., Zeng, M. S., et al. (2014). Knockdown of miR-214 promotes apoptosis and inhibits cell proliferation in nasopharyngeal carcinoma. PLoS ONE 9:e86149. doi: 10.1371/journal.pone.0086149
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, A., Zeng, Q., Xie, X., Zhou, J., Yue, W., Li, Y., et al. (2012a). MicroRNA-125b induces cancer cell apoptosis through suppression of Bcl-2 expression. J. Genet. Genomics 39, 29–35. doi: 10.1016/j.jgg.2011.12.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, Z., Tan, X., Zhao, A., Zhu, L., Yin, B., Yuan, J., et al. (2012b). microRNA-214-mediated UBC9 expression in glioma. BMB Rep. 45, 641–646. doi: 10.5483/BMBRep.2012.45.11.097
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, Q., Anderson, C., Zhang, H., Li, X., Inglis, F., Jayagopal, A., et al. (2013a). Repression of choroidal neovascularization through actin cytoskeleton pathways by microRNA-24. Mol. Ther. 22, 378–389. doi: 10.1038/mt.2013.243
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, S., Liu, Y., Prater, K., Zheng, Y., and Cai, L. (2013b). Roles of microRNAs in pressure overload- and ischemia-related myocardial remodeling. Life Sci. 93, 855–862. doi: 10.1016/j.lfs.2013.08.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, Y., Jiang, H., Gu, J., Tang, Y., Shen, N., and Jin, Y. (2013c). MicroRNA-195 targets ADP-ribosylation factor-like protein 2 to induce apoptosis in human embryonic stem cell-derived neural progenitor cells. Cell Death Dis. 4:e695. doi: 10.1038/cddis.2013.195
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, Y., Tian, L., Wang, X., Ye, L., Zhao, G., et al. (2014). MicroRNA-195 inhibits non-small cell lung cancer cell proliferation, migration and invasion by targeting MYB. Cancer Lett. 347, 65–74. doi: 10.1016/j.canlet.2014.01.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, X., Wang, H., Liu, F., Chen, L., Luo, W., Su, P., et al. (2013). Identification of micro-RNA networks in end-stage heart failure because of dilated cardiomyopathy. J. Cell. Mol. Med. 17, 1173–1187. doi: 10.1111/jcmm.12096
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zimmet, P., Alberti, K. G., and Shaw, J. (2001). Global and societal implications of the diabetes epidemic. Nature 414, 782–787. doi: 10.1038/414782a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Alzheimer's disease, early diagnosis, gastric cancer, hypertension, pancreatic cancer, personalize medicine, rheumatoid arthritis, stem cells
Citation: Katoh M (2014) Cardio-miRNAs and onco-miRNAs: circulating miRNA-based diagnostics for non-cancerous and cancerous diseases. Front. Cell Dev. Biol. 2:61. doi: 10.3389/fcell.2014.00061
Received: 25 August 2014; Accepted: 29 September 2014;
Published online: 16 October 2014.
Edited by:
Frank Emmert-Streib, Queen's University Belfast, UKReviewed by:
Guozhang Zou, National Center for Nanoscience and Technology, ChinaWen-Shu Wu, University of Illinois at Chicago, USA
Shephali Bhatnagar, University of Louisville, USA
Copyright © 2014 Katoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masaru Katoh, Department of Omics Network, National Cancer Center, 5-1-1 Tsukiji, Chuo-ward, Tokyo 104-0045, Japan e-mail: mkatoh-kkr@umin.ac.jp
 Masaru Katoh
Masaru Katoh