Transcriptional regulators of Na,K-ATPase subunits
- Nemours Center for Childhood Cancer Research, Nemours/Alfred I. duPont Hospital for Children, Wilmington, DE, USA
The Na,K-ATPase classically serves as an ion pump creating an electrochemical gradient across the plasma membrane that is essential for transepithelial transport, nutrient uptake and membrane potential. In addition, Na,K-ATPase also functions as a receptor, a signal transducer and a cell adhesion molecule. With such diverse roles, it is understandable that the Na,K-ATPase subunits, the catalytic α-subunit, the β-subunit and the FXYD proteins, are controlled extensively during development and to accommodate physiological needs. The spatial and temporal expression of Na,K-ATPase is partially regulated at the transcriptional level. Numerous transcription factors, hormones, growth factors, lipids, and extracellular stimuli modulate the transcription of the Na,K-ATPase subunits. Moreover, epigenetic mechanisms also contribute to the regulation of Na,K-ATPase expression. With the ever growing knowledge about diseases associated with the malfunction of Na,K-ATPase, this review aims at summarizing the best-characterized transcription regulators that modulate Na,K-ATPase subunit levels. As abnormal expression of Na,K-ATPase subunits has been observed in many carcinoma, we will also discuss transcription factors that are associated with epithelial-mesenchymal transition, a crucial step in the progression of many tumors to malignant disease.
Introduction
Na,K-ATPase is an integral membrane protein that mainly functions as an ion pump, hydrolyzing one molecule of ATP to pump three Na+ out of the cell in exchange for two K+ entering the cell per pump cycle. This function is crucial to maintaining the ion gradient across the membrane and is critical for the resting membrane potential, electrical activity of muscle and nerve, Na+-coupled transport, and osmotic balance and cell volume regulation (Blanco et al., 1998; Kaplan, 2002). Besides being an ion pump, Na,K-ATPase acts as a signal transducer (Xie and Askari, 2002; Pierre and Xie, 2006; Rajasekaran and Rajasekaran, 2009; Reinhard et al., 2013). Both α- and β-subunit associate with various signaling molecules, including Src, phosphoinositide 3-kinase (PI3K), caveolin-1, protein phosphatase 2, and EGFR thereby activating a number of intracellular signaling pathways, including MAPK and Akt signaling, to modulate cell polarity, cell growth, cell motility and gene expression (Haas et al., 2002; Barwe et al., 2005; Cai et al., 2008; Kimura et al., 2011). Na,K-ATPase also functions as a receptor for the cardiac glycoside ouabain, a specific inhibitor of the pump. Studies with ouabain at concentrations that do not inhibit the Na,K-ATPase pump function have been particularly useful in elucidating the pump-independent signaling pathways associated with Na,K-ATPase (Xie and Askari, 2002; Silva and Soares-da-Silva, 2012; Reinhard et al., 2013). Na,K-ATPase also regulates the formation and stabilization of intercellular junctions and the β1 and β2-subunits act as cell adhesion molecules (Gloor et al., 1990; Rajasekaran et al., 2001a,b; Vagin et al., 2006, 2012).
Na,K-ATPase is an abundantly expressed protein and can use up to about 2/3 of the energy expenditure of a cell (Howarth et al., 2012) thus requiring tight regulation. Aberrant expression and/or function of Na,K-ATPase has been associated with many disorders, with cancer being one of the more recent ones (Litan and Langhans, 2015). This review will focus on the transcriptional regulation of Na,K-ATPase by hormones, transcription factors, growth factors, extracellular stimuli, and epigenetic modification in general; followed by the role of transcription factors involved in epithelial-mesenchymal transition (EMT) and carcinoma.
The Expression Profile of Na,K-ATPase
Na,K-ATPase is a heteromeric transmembrane protein composed of an essential α- and β-subunit (Lingrel and Kuntzweiler, 1994), and an optional third subunit belonging to the FXYD proteins which are more tissue specific regulatory subunits of the enzyme (Geering, 2006). The α-subunit is the catalytic subunit responsible for transport activities of the enzyme. It contains 10 transmembrane segments; a large intracellular domain and a simple extracellular domain. An ATP-binding site and a phosphorylation site locate within the large cytoplasmic loops. The extracellular domain and the transmembrane regions harbor the binding sites for cardiac glycosides such as digoxin and ouabain (Kaplan, 2002; Sandtner et al., 2011; Laursen et al., 2013). The β-subunit is a highly glycosylated transmembrane protein, which increases the translation efficiency and stability of the α-subunit and targets the α-subunit to the plasma membrane (Blanco et al., 1998; Rajasekaran et al., 2004; Benarroch, 2011). It also modulates the affinity of Na+ and K+ to the enzyme (Blanco et al., 1998) and functions as a cell adhesion molecule (Gloor et al., 1990; Rajasekaran et al., 2001b; Barwe et al., 2005; Kitamura et al., 2005; Shoshani et al., 2005; Vagin et al., 2006, 2012). Unlike the α-subunit, the β-subunit spans the plasma membrane only once and has a very short intracellular N-terminus and a large extracellular C-terminal domain that contains several N-glycosylation sites (Blanco et al., 1998; Benarroch, 2011). In recent years, the FXYD family has been identified as another subunit of Na,K-ATPase. They are a group of small, single-span membrane proteins that are characterized by the FXYD motif in the extracellular domain, two conserved glycine residues within the membrane, and a serine residue at the membrane-cytoplasm interface (Garty and Karlish, 2006; Geering, 2008). FXYD proteins modify the affinity for Na+, K+, and ATP, pump kinetics and transport properties and stabilize Na,K-ATPase (Garty and Karlish, 2006; Geering, 2006, 2008; Mishra et al., 2011).
The Na,K-ATPase subunits are highly conserved across species and among isoforms. Four isoforms of α-subunit (α1–α4) and three isoforms of β-subunit (β1–β3) have been identified so far, which are encoded by different genes (Blanco, 2005). In human, there is high identity in amino acid sequence among the α-subunit isoforms with 87% identity between α2 and α1 and 88% between α3 and α1. α4 is a little more divergent, but still sharing 79% identity with α1. The β-subunit isoforms are more diverse. Compared to β1, β2 shows 39% identity and β3 is 36%. Seven different FXYD proteins have been identified (FXYD1 to FXYD7), which show 43 ~ 51% identity in their amino acid sequence.
The α- and β-subunits are expressed in a tissue specific manner (Herrera et al., 1987; Orlowski and Lingrel, 1988; Sweadner, 1989; Blanco et al., 1998; Benarroch, 2011). The α1-subunit is present in most tissues and the highest expression is found in kidney, brain and heart. Compared to the broad tissue distribution of the α1-subunit, the expression of other α-subunit isoforms is more restricted. The α2-subunit predominates in skeletal muscle, brain and heart; the α3-subunit is abundant in nervous tissue and heart. The α4-subunit was discovered as a testis-specific subunit (Shamraj and Lingrel, 1994), but now has also been found in adult human and mouse skeletal muscle (Keryanov and Gardner, 2002). Within the nervous system, the α-subunit isoforms are expressed in a cell type specific manner. For example, the α1-subunit is expressed in both neurons and glial cells, the α2-subunit is primarily found in astrocytes and oligodendrocytes and the α3-subunit is abundant in neurons (Benarroch, 2011). Like the α1-subunit, the β1-subunit is ubiquitously expressed in almost all tissues with the greatest levels in brain and kidney (Orlowski and Lingrel, 1988). The β2-subunit is mainly found in nervous tissue, kidney and heart (Avila et al., 1998), whereas the β3-subunit predominates in kidney and adrenal gland but is also presents in brain, lung and liver (Malik et al., 1998). The FXYD proteins show a tissue-specific distribution as well. FXYD1 (phospholemman) is expressed in heart and skeletal muscle; FXYD2 (γ-subunit) in kidney; FXYD3 (MAT-8) in stomach and colon; FXYD4 (CHIF) in kidney and colon; FXYD5 (dysadherin) in intestine, lung and kidney; FXYD6 and FXYD7 in brain (Garty and Karlish, 2006; Geering, 2006). In addition, the expression levels of α- and β-subunit change during development. For example, in brain, all three α-subunit isoforms increase in abundance from fetus to adult (Orlowski and Lingrel, 1988). However, the α1-subunit is more abundant in fetal kidney and heart than in adult tissues (Herrera et al., 1987). In rat heart, the α3-subunit decreases sharply after birth and is replaced by α2-subunit in adults (Zahler et al., 1996). The β1-subunit increases in the early stage of embryonic development, but decreases to the basal level during embryo implantation (Deng et al., 2013). With these diverse tissue distributions of Na,K-ATPase subunit isoforms, multiple Na,K-ATPase isozymes can form, rendering cells able to respond more precisely to extracellular stimuli. For instance, while in kidney α1β1 is the major αβ complex, all combinations of αβ complexes have been found in heart and brain (Mobasheri et al., 2000).
The spatial and temporal control of Na,K-ATPase expression occurs at the transcriptional, post-transcriptional, translational and post-translational level. Here, we will review findings on the transcriptional regulation of Na,K-ATPase over the past decades (Table 1). Extracellular stimuli, such as growth factors play a key role in the regulation of Na,K-ATPase by activating intracellular signaling pathways including protein kinase A (PKA), protein kinase C (PKC), and other Ca2+-regulated signaling pathways. Numerous DNA-binding-site-specific transcription factors have been reported to be involved in the regulation of Na,K-ATPase expression, including hormone receptors, Snail1, specificity protein (Sp) and Zinc finger E-box-binding homeobox 1 (ZEB1). Recently, it has been shown that hypermethylation in promoter regions of human ATP1B1 gene is associated with reduced expression of β1-subunit (Selvakumar et al., 2014), indicating that epigenetic modification might be another mechanism of transcriptional regulation of Na,K-ATPase subunits.
Promoter Analysis of Na,K-ATPase α- and β-subunits
Transcription factors activate or repress Na,K-ATPase expression by binding to specific DNA elements on the promoter regions. There are numerous sequence-specific elements on the promoter regions of the various Na,K-ATPase subunits, of which β1 has been extensively studied (Figure 1).
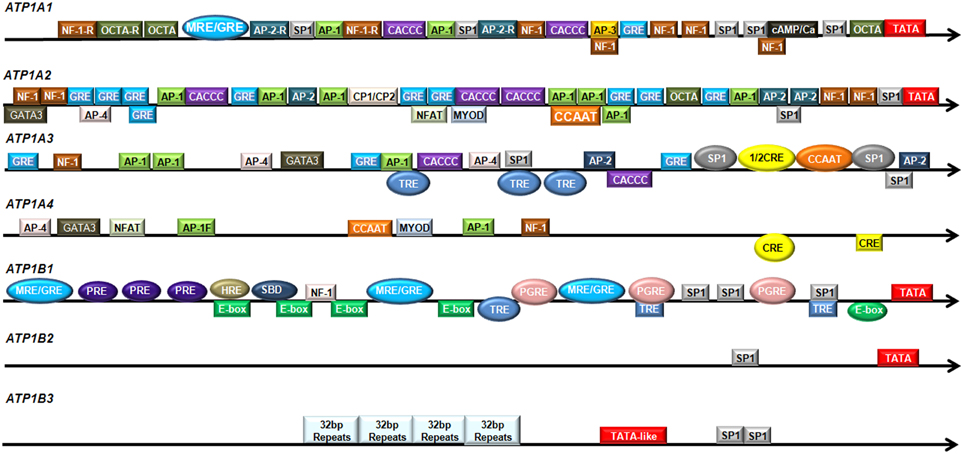
Figure 1. Regulatory elements in the promoter regions of human Na,K-ATPase subunits. The promoter regions of human Na,K-ATPase are shown in the direction from 5′ to 3′. The colored boxes indicate potential transcription factor binding sites. The colored ovals represent experimentally verified transcription factor binding sites.
α1-subunit
The α1-subunit is encoded by the gene ATP1A1. The ATP1A1 promoter has very high GC content, which is a common characteristic of all housekeeping genes. It contains a potential TATA box (−27 bp; Shull et al., 1990), two mineralocorticoid/glucocorticoid response element (MRE/GRE) sites (−598 to −574 bp and −252 bp; Kolla et al., 1999); potential binding sites for AP-1(−507 and −453 bp), AP-2 (−568 and −380 bp), AP-3 (−286 bp), and several CTF/NF-1 and potential Sp1 binding sites (Shull et al., 1990). The nucleotide sequence from around −100 bp to the transcription initiation site is highly conserved among the human and rat Atp1a1 genes (Kobayashi et al., 1997). An asymmetrical ATF/CRE site (−77 to −64 bp) is found in the rat Atp1a1 promoter (Kobayashi and Kawakami, 1995). In addition, the rat Atp1a1 promoter contains a Sp1 consensus sequence (−56 to −51 bp) and a Sp1 binding site (−120 to −106 bp; Suzuki-Yagawa et al., 1992).
α2-subunit
The α2-subunit is encoded by the gene ATP1A2. The promoter region of human ATP1A2 contains a potential TATA box, two potential Sp1 recognition sites, numerous potential AP-1, AP-2, and NF-1 binding sites, and several sequences that are similar to the glucocorticoid receptor binding site (Shull et al., 1989; Malyshev et al., 1991). The major positive regulatory elements are located at the position from −175 to −108 bp on the rat Atp1a2 gene, harboring two E boxes (−144 to −139 bp and −135 to −130 bp), a Spl binding consensus sequence (−123 to −118 bp) and a Sp1 binding site (−114 to −109 bp; Ikeda et al., 1993).
α3-subunit
The α3-subunit is encoded by the gene ATP1A3. The human ATP1A3 promoter has high GC content, but no conventional TATA box (Malyshev et al., 1991). Sequence analysis reveals several potential Sp-1 binding sites and AP-1 sites, two AP-2 sites, two AP-4 sites, three GRE elements, two CACCC sequences, a NF-1 binding site, a couple of TRE elements (Pathak et al., 1990), and the binding sites for CREB/ATF and NF-Y/C-EBP (Benfante et al., 2005). These sites are well conserved between the human and rat Atp1a3 gene (Pathak et al., 1990). It was subsequently shown that two Sp1 binding sites (−110 to −100 bp; −59 to −47 bp), a CCAAT box (−64 to −61 bp) and a half CRE-like site (−87 to −83 bp) are functional. Mutations in the Sp1 binding site (−110 to −100 bp) or the CCAAT box significantly reduced the ATP1A3 promoter activity and these two sites worked synergistically in inducing ATP1A3 promoter activity. The CRE-like element itself did not affect ATP1A3 promoter activity, but cooperated with the upstream Sp1 site (Benfante et al., 2005). Three TRE elements (−636 to −457 bp; −218 to −106 bp; and −106 to −6 bp) showed strong and specific interaction with thyroid hormone receptor (TR) and modulated ATP1A3 promoter activity (Bajpai et al., 2001). In addition, the region from −210 bp to the transcription initiation site was sufficient to direct brain-specific expression of the rat α3-subunit (Pathak et al., 1994).
α4-subunit
The α4-subunit is encoded by the gene ATP1A4. In silicon sequence analysis of putative promoter regions of human and mouse ATP1A4 showed that they do not contain a TATA-box sequence, but have potential binding sites for transcription factors AP-1, AP-4, GATA3, NF-Y, MYOD, NF-1, NFAT (Keryanov and Gardner, 2002) and two consensus CRE sites (Rodova et al., 2006). However, ATP1A4 does not have any Sp1 binding site that is common on the promoter regions of other subunits (Keryanov and Gardner, 2002).
β1-subunit
The β1-subunit is encoded by the gene ATP1B1. It contains three potential MRE/GRE half sites (−1048 to −1027 bp; −650 to −630 bp; −276 to −271 bp; Derfoul et al., 1998), four E-boxes (−752 to −747 bp; −708 to −703 bp; −654 to −649 bp; −509 to −504 bp), a non-canonical E-box (−71 to −66 bp), a NF-1 binding site (−657 to −653 bp; Espineda et al., 2004), three prostaglandin response elements (PGRE: −445 to −438 bp; −226 to −216 bp; −100 to −92 bp; Matlhagela et al., 2005; Matlhagela and Taub, 2006), a putative hypoxia response element (HRE: −750 to −746 bp; Mony et al., 2013), a Smad binding domain (SBD: −728 and −721 bp; Mony et al., 2013), three thyroid hormone response elements (TRE; Feng et al., 1993), three progesterone element (PRE) half sites (Cochrane et al., 2012) and several Sp1 binding sites (Matlhagela et al., 2005; Matlhagela and Taub, 2006). Furthermore, the mouse Atp1b1 promoter region also contains a Cebpb binding site (Deng et al., 2013). The 5′ flanking region of rat Atp1b1 shows 68% homology with human ATP1B1. It contains a consensus TATA box sequence (−31 bp), two CAAT boxes, four GC boxes, five half-site TREs, several GRE and calcium and serum response elements (Liu and Gick, 1992).
β2-subunit
The β2-subunit is encoded by the gene ATP1B2. The region from −280 bp to the transcriptional initiation site on the promoter of ATP1B2 gene is highly conserved between rat, mouse and human. The human ATP1B2 promoter contains a consensus TATA sequence at position −94 bp and a Sp1 binding site at position −120 bp (Avila et al., 1998). The mouse Atp1b2 promoter has a TATA-like sequence (−23 to −28 bp), a putative CAAT box (−137 to −142 bp), and a GC box (−144 to −149 bp; Shyjan et al., 1991). Sequence analysis revealed a potential TATA box (−29 bp); two Sp1 binding sites (−55 and −147 bp) and a serum response element (SRE: −263 bp) on the rat Atp1b2 promoter region (Kawakami et al., 1990). In addition, an AMOG regulatory element (AMRE: GAGGCGGGG at position −87 to −79 bp) and a functional Sp1 binding site (−147 to −142 bp) were found in rat Atp1b2 promoter regions (Kawakami et al., 1992, 1993).
β3-subunit
The β3-subunit is encoded by the gene ATP1B3. The human ATP1B3 promoter contains a TATA-like sequence (−413 to −408 bp) and two Sp1-like elements (−323 to −315 bp and −312 to −304 bp). Moreover, the human ATP1B3 also has four identical 32-nucleotide-long repeats from position −760 to −633 bp. The sequence from −1 to −328 bp on the 5′ flanking region of human and mouse Atp1b3 shows high homology. The mouse Atp1b3 promoter has one Sp1-like sequence (−319 to −310 bp), but does not have any TATA-like sequence. No CAAT or GC boxes are present at the promoter region of human or mouse Atp1b3 (Malik et al., 1998).
Transcriptional Regulation of Na,K-ATPase by Hormones and Nuclear Receptors
Hormones are major regulators of Na,K-ATPase expression. Mineralocorticoids (Verrey et al., 1987, 1988, 1989; Ikeda et al., 1991; Oguchi et al., 1993; Farman et al., 1994; Muto et al., 1996; Tsuchiya et al., 1996; Grillo et al., 1997; Seok et al., 1999; Olivera et al., 2000; Blot-Chabaud et al., 2001; Phakdeekitcharoen et al., 2011), glucocorticoids (Orlowski and Lingrel, 1990; Bhutada et al., 1991; Celsi et al., 1991; González et al., 1994, 1996; Muto et al., 1996; Barquin et al., 1997; Ingbar et al., 1997; Chalaka et al., 1999; Dagenais et al., 2001; Hao et al., 2003a,b), thyroid hormone (McDonough et al., 1988; Gick and Ismail-Beigi, 1990; Orlowski and Lingrel, 1990; Melikian and Ismail-Beigi, 1991; Hensley et al., 1992; Kamitani et al., 1992; Azuma et al., 1993; Giannella et al., 1993; Ohara et al., 1993; Huang et al., 1994; Lin and Tang, 1997; Bajpai and Chaudhury, 1999; Shao et al., 2000; Phakdeekitcharoen et al., 2007), insulin (Russo and Sweadner, 1993; Tirupattur et al., 1993; Sweeney and Klip, 1998), progesterone (Cochrane et al., 2012; Deng et al., 2013), androgen (Blok et al., 1999), and vitamin D3 (Billecocq et al., 1997) have been reported to activate or repress Na,K-ATPase transcription. These hormones bind to and activate their receptors, which belong to the family of nuclear receptors with C4 zinc-fingers that then translocate into the nucleus to bind to the promoter regions of the target genes to regulate gene expression.
Mineralocorticoid Receptor (MR) and Glucocorticoid Receptor (GR)
MR and GR are members of the nuclear receptor family. They have three major functional domains: a N-terminal regulatory domain (NTD); a DNA-binding domain (DBD) and a ligand-binding domain (LBD; Kumar and Thompson, 2005). The MR is highly homologous to the GR in the DBD and LBD but not in the NTD (Kolla et al., 1999) and while mineralocorticoids only activate the MR, glucocorticoids activate both GR and MR. The activated MR and GR translocate into the nucleus and bind to a specific palindromic DNA sequence known as glucocorticoid/mineralocorticoid response element (GRE/MRE) located in the promoter regions of target genes, thus modulating gene transcription. The GRE is composed of imperfect inverted hexanucleotide repeats separated by three nucleotides (GGTACA NNN TGTTCT; Nordeen et al., 1990).
Glucocorticoids regulate the transcription of Na,K-ATPase in a variety of tissues including lung, kidney, liver, heart, spinal cord, cardiac muscle and smooth muscle (Orlowski and Lingrel, 1990; Bhutada et al., 1991; Celsi et al., 1991; Wang and Celsi, 1993; González et al., 1994, 1996; Wang et al., 1994; Muto et al., 1996; Barquin et al., 1997; Ingbar et al., 1997; Chalaka et al., 1999; Dagenais et al., 2001; Hao et al., 2003b). This regulation is specific to the cell type and the developmental stage. For example, dexamethasone, a synthetic potent glucocorticoid, increased the mRNA levels of α1- and β1-subunit in a rat liver cell line and in a fetal rat lung epithelial cell line (Bhutada et al., 1991; Chalaka et al., 1999). However, in cultured neonatal rat cardiac myocytes, dexamethasone only induced α2-subunit mRNA, and α1-, α3-, and β-subunit transcripts remained unchanged (Orlowski and Lingrel, 1990). Injection of betamethasone, another synthetic analog of glucocorticoid, increased α1- and β-subunit mRNA abundance in 10-day-old rats, but not in adult rats (Celsi et al., 1991).
Mineralocorticoids, like aldosterone, also regulate the mRNA levels of Na,K-ATPase in diverse tissues such as kidney, heart, brain, vascular smooth muscle and skeletal muscle (Verrey et al., 1987, 1988, 1989; Ikeda et al., 1991; Oguchi et al., 1993; Farman et al., 1994; Tsuchiya et al., 1996; Grillo et al., 1997; Seok et al., 1999; Olivera et al., 2000; Blot-Chabaud et al., 2001; Phakdeekitcharoen et al., 2011). Aldosterone increased both α1- and β1-subunit transcripts in kidney, heart and vascular muscles and this up-regulation of β1-subunit was much stronger than that of α1-subunit (Verrey et al., 1987, 1988, 1989; Ikeda et al., 1991; Oguchi et al., 1993). Aldosterone also increased α2- and β1-subunit mRNA in human skeletal muscle (Phakdeekitcharoen et al., 2011). In addition, aldosterone selectively up-regulated α3- and β1-subunit mRNA in some brain regions, such as hippocampus, gyrus dentatus and periventricular gray substance (Farman et al., 1994; Grillo et al., 1997). The transcriptional regulation of α1- and β1-subunit by aldosterone was mediated mostly by MR (Muto et al., 1996; Kolla et al., 1999), whereas dexamethasone-mediated induction of β1-subunit mRNA occurred through GR (Muto et al., 1996). Inhibiting GR and MR activities by their specific antagonists also reduced α1- and β1-subunit levels (Whorwood et al., 1994; Hao et al., 2003b). In monkey kidney CV-1 cells, which have no detectable MR and GR, activation of exogenous MR or GR alone by aldosterone or triamcinolone acetonide increased β1-subunit gene promoter activity. However, activation of both receptors inhibited transcription from the β1-subunit gene promoter (Derfoul et al., 1998) and the N-terminal region of MR mediated this inhibitory effect (Derfoul et al., 2000).
The promoter region of the ATP1B1 gene contains three potential MRE/GREs at nucleotide position −1048 to −1027 bp; −650 to −630 bp and −276 to −271 bp. The first two contain two half-binding sites separated by 10 and 9 base pairs, respectively, whereas the last one consists of a single half-site. Although all three MRE/GREs were involved in the induction of ATP1B1 promoter activity by corticoids, the one at position −650 to −630 bp was the strongest (Derfoul et al., 1998). Both MR and GR bound to the GRE/MRE (−650 to −630 bp) and activated the transcription from a heterologous neutral promoter in response to corticoids (Derfoul et al., 1998; Hao et al., 2003b). A similar element was found in the human ATP1A1 promoter region at position −598 to −574 bp in which two half sites are separated by 12 base pairs. This MRE/GRE bound both MR and GR and activated a heterologous neutral promoter activity (Kolla et al., 1999). In addition, a GRE (+434 to +448 bp) in the coding region of the rat Atp1b1 gene has been reported in a rat lung epithelial cell line. This element acted as an enhancer and increased the transcription from a heterologous promoter by dexamethasone (Hao et al., 2003a).
Progesterone Receptor
Progesterone receptor (PR) also belongs to the nuclear receptor family. PR's sequence and architecture is very similar to MR and GR (Leo et al., 2004). Like MR and GR, PR is activated by its ligand progesterone and then translocates into the nucleus, forms a homodimer and then binds to specific progesterone response elements (PREs) within the promoter of target genes to modulate transcription (Leonhardt et al., 2003). Progesterone increased the mRNA and protein levels of β1-subunit (Cochrane et al., 2012; Deng et al., 2013), and this up-regulation was enhanced by estradiol-17β (Deng et al., 2013). RU486, a PR antagonist, completely blocked the increase in β1-subunit mRNA by progesterone (Deng et al., 2013). In addition, human β1-subunit was up-regulated by both PR-A and PR-B, two different PR isoforms (Richer et al., 2002). Moreover, PR bound to three PRE half sites on the ATP1B1 promoter; Progestin, a synthetic progestogen with similar effect to progesterone, induced ATP1B1 promoter activity, an induction that was suppressed by RU486 (Cochrane et al., 2012).
Thyroid Hormone Receptor
Thyroid hormone receptor (TR) is a member of the nuclear receptor superfamily and is activated by thyroid hormone, triiodothyronine (T3) and its prohormone, thyroxine (T4). Although TR alone can bind as monomer or homodimer to specific DNA sequences in the gene promoter regions known as thyroid hormone response elements (TREs), it preferentially forms heterodimers with retinoid X receptor (RXR) and then binds to TREs, thus enhancing or inhibiting the expression of genes that are involved in development, homeostasis, cellular proliferation and differentiation (Wu and Koenig, 2000). T3 has been shown to regulate the abundance of Na,K-ATPase mRNA and protein in heart, kidney, liver, intestines, brain, and skeletal muscle among others (McDonough et al., 1988; Gick and Ismail-Beigi, 1990; Orlowski and Lingrel, 1990; Melikian and Ismail-Beigi, 1991; Hensley et al., 1992; Kamitani et al., 1992; Azuma et al., 1993; Giannella et al., 1993; Ohara et al., 1993; Huang et al., 1994; Lin and Tang, 1997; Bajpai and Chaudhury, 1999; Shao et al., 2000; Phakdeekitcharoen et al., 2007). In general, T3 up-regulated the transcription of Na,K-ATPase subunits. For example, T3 increased the mRNA levels of α1-, α2-, α3-, and β1-subunit in cultured neonatal rat cardiocytes (Kamitani et al., 1992) and α2-, β1-, and β2-subunit in skeletal muscle (Azuma et al., 1993; Phakdeekitcharoen et al., 2007). In hypothyroid rat brain, the mRNA level of α1-, α2-, and α3-subunit was reduced during the first 3 weeks of brain development (Chaudhury et al., 1996). The transcription of these mRNAs was decreased in nuclei from hypothyroid cerebra and pretreatment with T3 increased the rates of transcription (Bajpai and Chaudhury, 1999). The promoter regions of Atp1a1, Atp1a2, Atp1a3, and ATP1B1 genes have TRE sequences (Kamitani et al., 1992; Feng et al., 1993; Giannella et al., 1993). In addition, two negative thyroid receptor elements (−116 and −6 bp and −6 and +80 bp) were found in rat Atp1a3 promoter. The TR/RXR complex bound to these elements and inhibited T3-mediated gene expression in neonatal rat cardiac myocytes, which only occured with prolonged incubation with T3 (He et al., 1996).
Insulin
Insulin is an important regulator of Na,K-ATPase. It binds to insulin receptor and activates various cellular signaling pathways, thus regulating cell functions. Insulin modulates Na,K-ATPase activity by increasing Na+ influx; increasing the affinity of Na,K-ATPase for intracellular Na+; altering phosphorylation status of Na,K-ATPase and facilitating the translocation of intracellular Na,K-ATPase to the plasma membrane. In addition, insulin also regulates the transcription, translation and stability of Na,K-ATPase (Ewart and Klip, 1995; Lavoie et al., 1996; Sweeney and Klip, 1998; Therien and Blostein, 2000; Clausen, 2003). The transcriptional regulation of Na,K-ATPase by insulin occurs in an isoform specific manner. For example, insulin selectively increased α2, but not α1 mRNA level in vascular smooth muscle cells (VSMC) and 3T3-L1 cells (Russo and Sweadner, 1993; Tirupattur et al., 1993; Sweeney and Klip, 1998). However, it reduced the β1 mRNA level (Russo and Sweadner, 1993).
Transcription Factors Regulating Na,K-ATPase
Specificity Protein
Specificity protein (Sp) is a small family of zinc-finger transcription factors with four members: Sp1, Sp2, Sp3, and Sp4. Sp1, Sp2, and Sp3 are ubiquitously expressed, while Sp4 is expressed mainly in neurons and testes. The Sp family members contain three zinc fingers close to the C-terminus, which is DBD, and preferentially bind to the GC box [(G/T)(G/A)GGC(G/T)(G/A)(G/A)(G/T)] on the promoter regions of target genes to enhance gene expression. Sp1, Sp3, and Sp4 have a highly conserved DBDs and they recognize the same consensus GC rich element with identical affinities. However, Sp2, in which the important histidine residue within the first zinc finger is replaced by a leucine residue, does not bind to the GC box but instead binds to a GT rich element (Suske, 1999). Potential Sp1 binding sites were found on the promoter regions of almost all Na,K-ATPase subunits, including Atp1a1, Atp1a2, Atp1a3, Atp1b1, and Atp1b2 (Shull et al., 1989, 1990; Kawakami et al., 1990; Yagawa et al., 1990; Malyshev et al., 1991; Suzuki-Yagawa et al., 1992; Ikeda et al., 1993; Yu et al., 1996; Avila et al., 1998; Benfante et al., 2005). It has been shown that Sp1 and Sp3 bind to GC boxes on the promoter regions of rat Atp1a1 (Kobayashi et al., 1997) and Atp1b1 (Murakami et al., 1997). Mutations in the GC box sequences decreased basal Atp1b1 transcription in neonatal rat cardiac myocytes (Zhuang et al., 2000). Sp1, Sp3 and Sp4 also bind to two Sp1 binding sites (−110 to −100 bp; −59 to −47 bp) on human ATP1A3 promoter. Mutations in Sp1 binding site (−110 to −100 bp) significantly reduced the promoter activity (Benfante et al., 2005). Recently, Sp1, Sp3, and Sp4 had been shown to increase α1-, α3-, and β1-subunit transcription in murine neurons by binding to their promoter regions (Johar et al., 2012). Mutating the Sp binding sites on Atp1a1, Atp1a3, and Atp1b1 promoters led to a significant decrease in their promoter activity. Silencing Sp1, Sp3, or Sp4 significantly decreased the mRNA levels of α1-, α3-, and β1-subunit, while overexpressing Sp1, Sp3, and Sp4 increased their transcription (Johar et al., 2012). Sp1 also binds to two Sp binding sites located near its TATA box on rat Atp1b2 promoter and enhanced the expression of β2-subunit. In addition, the functional Sp-binding site on Atp1a1 promoter is well conserved in rat brain, kidney and liver (Suzuki-Yagawa et al., 1992; Kobayashi et al., 1997; Nemoto et al., 1997), indicating that it might contribute to the constitutional expression of α1-subunit. The Sp binding sites are also required for the regulation of Na,K-ATPase transcription in response to extracellular stimuli. For example, hyperoxia selectively up-regulated β1-subunit transcription in MDCK cells (Wendt et al., 1998). Hyperoxia increased the binding of Sp1 and Sp3 to the Atp1b1 promoter and deletion of a GC rich element resulted in loss of both basal and hyperoxia-activated transcription (Wendt et al., 2000). Sp might also augment transcriptional activation of Na,K-ATPase through other transcription factors. For example, Sp1 significantly enhanced MR and GR expression (Kolla et al., 1999) and elevated MR and GR might synergize with Sp1 to upregulate Na,K-ATPase transcription.
Snail
Snail1 is the first identified member of the Snail family, which belongs to the zinc-finger transcription factors and has three members in vertebrates: Snail1, Snail2, and Snail3. The Snail family has a highly conserved C-terminal region, which is the DBD containing four to six zinc fingers; and a divergent N-terminal region, which contains a SNAG (Snail/Gfi) domain and recruits the transcription repressor complex (such as sin3A/HDAC). The Snail family functions as a transcriptional repressor by binding to the E-box (CAGGTG) on the promoter regions of target genes which are important for embryonic development as well as tumor progression (Nieto, 2002). Four E-boxes and a non-canonical E-box are present on the promoter region of the ATP1B1 gene. Snail1 binds to the non-canonical E-box located at position −71 to −66 bp and selectively repressed the expression of β1-subunit without affecting the α1-subunit level (Espineda et al., 2004). Overexpression of Snail1 repressed ATP1B1 promoter activity in a dose dependent manner and markedly reduced the level of β1-subunit in MCF7 and MDCK cells. Knock-down of Snail by RNA interference increased the abundance of β1-subunit mRNA (Espineda et al., 2004).
Activating Transcription Factor 1 (ATF-1)/cAMP Response Element Binding Protein (CREB) Family
ATF/CREB belongs to the basic leucine zipper factor (bZIP) family, which bind to DNA as dimers and function as activators. The ATF/CREB family is composed of different ATFs, CREB, CREM (cAMP response element modulator) and related proteins. This family contains an N-terminal DBD and a C-terminal bZIP domain that binds other bZIP transcription factors to form homo- and heterodimers. Both ATF and CREB bind to the cAMP response element (CRE: GTGACGT A/C A/G). Phosphorylation of ATF-1 and CREB by multiple kinases enhances the binding to CRE and activates the transcription of target genes which play an important role in cell survival and cell growth (Hai and Hartman, 2001). Dibutyryl cAMP (db-cAMP), a cell-permeable cAMP analog that activates cAMP dependent protein kinase (PKA) and phosphorylates the CREB family, induced the mRNA of α1-subunit but not β1-subunit (Dagenais et al., 2001). The rat Atp1a1 promoter has an asymmetrical ATF/CRE site at position −70 to −66 bp. The ATF-1/CREB heterodimer bound to this site and induced constitutive expression of αl-subunit. Blocking the binding of ATF-1/CREB to the CRE site reduced α1-subunit transcription (Kobayashi and Kawakami, 1995). Phosphorylated ATF-1 increased rat α1-subunit transcription and dephosphorylation of the ATF-1/CREB heterodimer by alkaline phosphatase reduced its binding to the CRE site on rat Atp1a1 gene (Kobayashi et al., 1997). In addition, human ATP1A4 promoter region has consensus CRE sites. db-cAMP and ectopic expression of CREMtau, a testis specific splice variant of CREM activated the ATP1A4 promoter activity. This activation required the CRE site located at −263 bp relative to the transcription initiation site (Rodova et al., 2006).
Zinc Finger E-Box Binding Homeobox 1 (ZEB1)
The ZEB family has two members in vertebrates: ZEB1 (also known as Areb6) and ZEB2. They contain five major domains: two zinc-finger clusters separated by a central homeodomain, a Smad interacting domain (SID) and a CtBP interacting domain (CID). Each zinc finger cluster contains 3 or 4 zinc fingers and is responsible for DNA binding. ZEB binds to E-boxes (CACCT and CACCTG) on the promoter of target genes and recruits co-activators (PCAF or p300), thus activating gene transcription (Vandewalle et al., 2009). The rat Atp1a1 promoter has a consensus Areb6 (ZEB1) binding sequence and Areb6 (ZEB1) had been shown to activate gene transcription in a cell-specific manner (Watanabe et al., 1993). High-fat diet also induced the binding of ZEB to rat Atp1α1 promoter and increased the mRNA and protein level of α1-subunit in nuclear extracts from gastrocnemius muscle (Galuska et al., 2009). Moreover, C-peptide increased the binding of ZEB to rat Atp1α1 promoter region. Partial silencing of ZEB markedly reduced the effect of C-peptide on the increase in α1-subunit expression (Galuska et al., 2011). In addition, ZEB binding sites were also found on FXYD1 promoter at positions −270 to −295 bp and −329 to −354 bp (Galuska et al., 2009).
CCAAT Box Binding Transcription Factors (CBFs)
CCAAT box is one of the most common elements in eukaryotic promoters, which is often found between -60 to -100 bp relative to the transcription initiation site. Several transcription factors have been reported to bind to the CCAAT box, including NF-Y, NF-1, and CCAAT/enhancer-binding protein (C/EBP) isoforms (CEBP-α, CEBP-β, CEBP-γ, CEBP-δ, and CEBP-ε), which are essential for optimal transcriptional activation of target genes (Mantovani, 1998). CCAAT boxes have been found in Atp1a2, Atp1a3, and Atp1b1 promoter regions (Pathak et al., 1990; Malyshev et al., 1991; Benfante et al., 2005; Deng et al., 2013). The CCAAT box (−61 to −64 bp) on human ATP1A3 promoter is well conserved between rat and human and NF-YB had been shown to bind to this CCAAT box. The presence of the CCAAT box significantly increased ATP1A3 promoter activity in both neuron and non-neuron cell lines, while mutations in the CCAAT box reduced the promoter activity (Benfante et al., 2005). In addition, the CCAAT/enhancer binding protein beta (Cebpb) might be involved in regulating β1-subunit expression in human luminal epithelial cells. The active isoform of Cebpb significantly increased Atp1b1 promoter activity, but the truncated isoform of Cebpb had no effects on Atp1b1 promoter activity (Deng et al., 2013).
Growth Factors and Transcriptional Regulation of Na,K-ATPase
Transforming Growth Factor-β
Transforming growth factor-β (TGF-β) is a secreted protein that regulates numerous physiological processes, including cell differentiation, proliferation, development and survival. TGF-β has three isoforms in mammals: TGF-β1, TGF-β2, and TGF-β3, which are encoded by different genes. Aberrant TGF-β signaling has been linked to cancer initiation, invasion and metastasis (Lamouille et al., 2014). TGF-β treatment reduced mRNA and protein levels of Na,K-ATPase α- and β-subunits in primary cultures of renal proximal tubule cells (Tang et al., 1995). However, TGF-β1 seems to regulate Na,K-ATPase subunit expression in a cell type specific manner. Some studies showed that TGF-β1 reduced β1-subunit as well as α1-, α2-, and α3-subunit mRNA levels in young FRTL-5 rat thyroid cells (Pekary et al., 1997). Studies from our group found that although TGF-β1 decreased the surface levels of β1-subunit in kidney cells, this seemed to occur at the post-translational level since no significant changes in mRNA levels were observed between control and TGF-β1-treated cells (Rajasekaran et al., 2010). TGF-β1 also prevented the increase in α1-subunit mRNA induced by steroid hormones (Husted et al., 2000). Nevertheless, TGF-β2 selectively decreased the transcription of β1-subunit in ARPE-19 cells, a human retinal pigmented epithelial cell line, involving two transcription factors: hypoxia inducible factor (HIF) and Smad3 (Mony et al., 2013). HIF belongs to the basic helix-loop-helix (bHLH) superfamily and is a heterodimer composed of a hypoxia induced HIF-α subunit and a constitutively expressed HIF-1β subunit. Three HIF-α subunits have been identified so far: HIF-1α, HIF-2α, and HIF-3α. HIF-1α contains three major domains: a N-terminal domain, which is a DBD containing a bHLH; an oxygen-dependent degradation domain (ODDD) that mediates oxygen-regulated protein stability and a C-terminal domain, which recruits transcriptional coactivators such as CBP/p300 to activate gene transcription. The HIF complex associates with hypoxia response elements (HREs) in the regulatory regions of target genes and binds the transcriptional coactivators to induce gene expression. Besides hypoxia, HIF-1 is also regulated in an oxygen-independent manner (Ke and Costa, 2006). TGF-β2 increased HIF-1α expression in ARPE-19 cells and blocking the binding of HIF-1α to HREs with echinomycin prevented the TGF-β2-induced decrease of the β1-subunit (Mony et al., 2013). Since HIF-1α generally functions as a transcriptional activator, this observation raised the possibility that other transcription repressors may antagonize the function of HIF-1α in the regulation of Na,K-ATPase expression. The ATP1B1 promoter region also contains a SBD in close proximity to a putative HRE. Smads are transcription modulators usually activated by TGF-β signaling that can inhibit gene expression by preventing the action of transcriptional activators (Massague et al., 2005). Studies in ARPE19 cells indeed support the idea that both Smad3 and HIF-1α cooperated in regulating the expression of β1-subunit expression by TGF-β2 (Mony et al., 2013).
Fibroblast Growth Factor
Fibroblast growth factors (FGFs) control a wide range of biological functions and regulate cellular proliferation, survival, migration, and differentiation. FGFs are a family of glyocoproteins that are generally sequestered by the extracellular matrix and cell surface heparan sulfate proteoglycans. Activation of FGF signaling occurs upon release of the factor and binding to one of the four highly conserved FGF receptors, FGR1, FGR2, FGR3, and FGR4 (Turner and Grose, 2010). Incubation of cells in serum rapidly increased α1- and β1-subunit mRNA levels in VSMC and rat liver cells and induced Atp1a1 and Atp1b1 promoter activity, suggesting that serum regulates Na,K-ATPase gene expression at the transcriptional level (Kirtane et al., 1994; Nemoto et al., 1997). Serum differentially regulated α1- and β1-subunit mRNA induction and activation of PKC and tyrosine kinase activity was required for up-regulation of α1-subunit mRNA but not for β1-subunit. Further studies revealed that FGF treatment stimulated Atp1a1 and Atp1b1 promoter activities, similar to the observation in serum treated VSMC (Nemoto et al., 1997). Furthermore, keratinocyte growth factor (KGF), a member of the FGF family and a polypeptide mitogen secreted by fibroblasts and endothelial cells that acts primarily on epithelial cells, increased the abundance of α1-subunit, but not β1-subunit mRNA in alveolar type II cells (Borok et al., 1998).
Prostaglandin and Na,K-ATPase Transcription
Prostaglandin E1 (PGE1) is a member of the prostaglandin family, which are lipid mediators produced from arachidonic acid by cyclooxygenase and prostaglandin synthases. Prostaglandins exert their effects by activating G protein-coupled receptors and four PGE receptors have been reported: EP1 (E prostanoid receptor 1), EP2, EP3, and EP4 (Sugimoto and Narumiya, 2007). Prostaglandins are important regulators of ion transport in the kidney (Bonvalet et al., 1987) and PGE1 increased the transcription of α- and β-subunit of Na,K-ATPase in MDCK cells (Taub et al., 1992, 2004; Matlhagela and Taub, 2006). The increase in the transcription of β1-subunit induced by PGE1 was mediated by EP1 and EP2 receptors and PGE receptor antagonists inhibited the promoter activity of human ATP1B1 induced by PGE1 (Matlhagela and Taub, 2006). Three prostaglandin response elements (PGRE) have been identified within the human ATP1B1 promoter, which are located at position −445 to −438 bp (PGRE1: TGACCTTC); −226 to −216 bp (PGRE2: GTCCCTCA); and −92 to −100 bp (PGRE3: AGTCCCTGC; Matlhagela et al., 2005; Matlhagela and Taub, 2006) and PGRE1 is well conserved among species (Matlhagela and Taub, 2006). The PGRE sequence is similar to the consensus CRE, and CREB had been shown to bind to these sequences indicating that PGREs might be cAMP response elements. The binding affinity of CREB to three PGREs was varied with PGRE3 > PGRE1 > PGRE2 (Matlhagela et al., 2005). Sp1, Sp3 as well as CREB all bound to PGRE (Matlhagela and Taub, 2006) and the increase in transcription of β1-subunit was mediated through CREB binding to PGRE1 and PGRE3 as well as Sp1 binding to an adjacent Sp1 site (Matlhagela et al., 2005; Matlhagela and Taub, 2006). Mutations in PGRE3 or the GC box (−118 to −112 bp) immediately downstream of PGRE3 failed to mediate the increase in ATP1B1 promoter activity induced by PGE1 (Matlhagela and Taub, 2006). Besides PGE1, PGE2 and PGF2α also have been shown to induce ATP1B1 promoter activity in primary renal proximal tubule (RPT) cells (Herman et al., 2010).
Na,K-ATPase Activity and Transcriptional Regulation
Chronic inhibition of Na,K-ATPase activity by ouabain leads to an increase in the abundance of Na,K-ATPase. For example, ouabain increased the mRNA levels of α1- and β1-subunit in cultured rat astrocytes (Hosoi et al., 1997; Muto et al., 2000), which was abolished by actinomycin D, a transcription inhibitor acting by interfering with mRNA synthesis (Muto et al., 2000). Ouabain also regulated the transcription of α3- and β1-subunit in cultured neonatal rat cardiac myocytes. Ouabain augmented the Atp1b1 promoter activity and the effect of ouabain on the regulation of Na,K-ATPase subunits was dependent on extracellular Ca2+ and calmodulin (Kometiani et al., 2000). The inhibition of Na,K-ATPase activity by ouabain can be mimicked by incubating cells in a low K+ environment and low extracellular K+ increased α1- and β1-subunit mRNA levels in rat cardiac myocytes (Qin et al., 1994; Zhuang et al., 2000; Wang et al., 2007a). Sp and CREB family transcription factors were required for the up-regulation of Na,K-ATPase subunits induced by low K+. Rat Atp1a1 promoter region has a CRE/ATF site (at −70 to −63 bp) and a GC box motif (at −57 to −48 bp). Mutations in the CRE/ATF site or GC box substantially reduced low K+-mediated Atp1a1 promoter activity. Low K+ increased the expression of Sp1, Sp3 and CREB-1 and enhanced the binding of these transcription factors to the GC box and, to a lesser extent, to the CRE/ATF site on rat Atp1a1 promoter (Wang et al., 2007b). The sequences between -102 to +151 bp on rat Atp1b1 were required for low K+-induced trans-activation of reporter gene expression (Qin et al., 1994). Low K+ enhanced the binding of Sp1 and Sp3 to GC box elements on the Atp1b1 promoter (Zhuang et al., 2000). Mutations in potential GC box sequences decreased basal and low K+-mediated up-regulation of β1-subunit transcription in neonatal rat cardiac myocytes (Zhuang et al., 2000). Inhibiting the binding of Sp1 and Sp3 to GC boxes by mithramycin, which has high affinity for GC-rich DNA sequences (Matlhagela et al., 2005), blocked low K+-mediated up-regulation of Atp1a1 and Atp1b1 promoter activity. Multiple cellular signaling pathways were involved in this low K+-induced increase in the transcription of α1- and β1-subunits. Protein kinase A (PKA), ERK1/2, and histone deacetylase (HDAC) were required for up-regulation of α1-subunit transcription, whereas the transcription of β1-subunit was dependent on protein kinase C (PKC), c-Jun-N-terminal kinase (JNK) and p38 mitogen-activated protein kinase (MAPK; Wang et al., 2007a). However, it is important to point out that inhibition of Na,K-ATPase and therefore an increase in intracellular sodium regulates Na,K-ATPase on multiple levels including transcription, transport, endocytosis and degradation (Vinciguerra et al., 2003, 2004; Wang et al., 2014).
Neuronal Activity and Na,K-ATPase Transcriptional Regulation
α1-, α3-, and β1-subunits are abundantly expressed in most neurons in the central nervous system and neuronal activity modulates the transcription of Na,K-ATPase subunits. Depolarization by KCl increased the promoter activities as well as the mRNA levels of α1-, α3- and β1-subunits; however, impulse blockade induced by tetrodotoxin (TTX), a voltage-dependent Na+ channel blocker, significantly reduced α1-, α3-, and β1-subunit transcripts in murine neurons (Johar et al., 2012, 2014). At least two transcription factors were involved in this activity-dependent transcriptional regulation in neurons, nuclear respiratory factor 1 (NRF1) and the Sp family (Johar et al., 2012, 2014).
NRF1 is a transcription factor which was first identified as an activator of cytochrome C (Evans and Scarpulla, 1989). NRF1 increases the expression of nuclear genes required for mitochondrial biogenesis and function, but also binds to the promoter region of target genes which control cell cycle, cell growth, cell adhesion, migration, and tumor invasion (Okoh et al., 2011). Increased NRF1 levels have been observed in hepatoma, thyroid oncocytoma and breast cancers (Dong et al., 2002; Savagner et al., 2003; Okoh et al., 2011). The consensus NRF1 binding site is (T/C)GCGCA(C/T)GCGC(A/G), which is a GC-rich element. In addition, NRF1 binds to sequences with an invariant GCA core flanked by GC-rich regions, which can be found in 5′ flanking regions of Atp1a1, Atp1a3, and Atp1b1 genes. It has been reported that NRF1 binds to the promoter regions of Atp1a1 and Atp1b1 in murine neurons at binding sites that are conserved among mice, rats and humans. Mutations in the NRF1 binding sites significantly decreased Atp1b1 promoter activity, but increased Atp1a1 promoter activity, which indicates that NRF1 differentially regulates Na,K-ATPase subunits. Indeed, silencing of NRF1 significantly decreased β1-subunit transcript but increased the mRNA level of α1-subunit and overexpression of NRF1 increased the mRNA level of the β1-subunit but decreased the α1-subunit transcript. NRF1 itself is regulated by neuronal activity (Yang et al., 2006). Depolarization by KCl increased NRF1 expression, and TTX reduced the NRF1 expression (Johar et al., 2012) and knockdown of NRF1 with small interference RNA blocked the up-regulation of the β1-subunit and down-regulation of the α1-subunit induced by KCl, whereas overexpression of NRF1 rescued the down-regulation of the β1-subunit and up-regulation of the α1-subunit by TTX. The inverse regulation of Na,K-ATPase α1- and β1-subunit by NRF1 in response to KCl and TTX points to additional transcription factors and an intricate network regulating Na,K-ATPase expression by neuronal activity.
Sp transcription factors are regulated by neuronal activity and depolarization by KCl increased Sp1 and Sp4 expression, while TTX reduced the expression of Sp1 and Sp4 (Johar et al., 2014). Sp1, Sp3, and Sp4 bound to the promoter regions of Atp1a1, Atp1a3, and Atp1b1 in murine neurons, and Sp4 showed the highest binding affinity. Mutations in the Sp binding sites of Atp1a1, Atp1a3, and Atp1b1 promoters significantly decreased the promoter activity. Silencing Sp1, Sp3, and Sp4 decreased α1-, α3-, and β1-subunit mRNA levels; while overexpression of Sp1, Sp3, and Sp4 increased α1-, α3-, and β1-subunit transcripts. Silencing Sp1 or Sp4 also blocked the KCl-induced up-regulation of Na,K-ATPase subunits and overexpression of Sp1, Sp3, or Sp4 rescued the down-regulation of Na,K-ATPase subunits by TTX (Johar et al., 2014).
Epigenetic Mechanisms in the Regulation of Na,K-ATPase Transcription
DNA methylation is an important epigenetic mechanism in the regulation of gene expression. Methylation generally occurs at the cytosine bases of CpG sequences throughout the entire genome, with the exception of CpG islands usually found in gene promoter regions (Cedar and Bergman, 2009). Methylation patterns often depend on tissue type and developmental stage, correlating with transcriptional activity. Aberrant DNA methylation patterns have been linked to altered gene expression in various genetic diseases and tumors (Bird, 2002). Higher methylated CpGs were found in the first exon of Atp1a3, which correlated with the lower expression of this gene in pig liver (Henriksen et al., 2013). Manganese exposure led to sustained Atp1a3 promoter hypermethylation and downregulation of its transcript level in mice (Wang et al., 2013). CpG islands are also found in ATP1B1 and Atp1b2 promoter regions (Alvarez de la Rosa et al., 2002; Selvakumar et al., 2014), which are conserved between human and mice and remain unmethylated (Alvarez de la Rosa et al., 2002). However, ATP1B1 promoter is hypermethylated in tumor samples of clear cell renal cell carcinoma, which showed reduced β1-subunit levels (Selvakumar et al., 2014). Knock-down of the tumor suppressor gene von Hippel-Lindau (VHL) enhanced ATP1B1 promoter methylation and decreased β1-subunit expression, while inhibition of methyltransferase by 5-Aza-2′-deoxycytidine rescued β1-subunit mRNA expression in VHL-knockdown cells (Selvakumar et al., 2014).
Methylation is also involved in transcriptional regulation and differential distribution of FXYD1. The FXYD1 promoter contains methylated cytosines and a predicted CpG island. 5′-aza-cytidine markedly increased FXYD1 mRNA levels. Mouse heart showed lower methylation in fxyd1 promoter and higher fxyd1 expression; while brain had a higher methylation of the fxyd1 promoter and lower fxyd1 expression (Deng et al., 2007). In brain, the fxyd1 mRNA level is lower in the frontal cortex than in cerebellum, which correlates with the more frequent promoter methylation found in frontal cortex (Banine et al., 2011). The fxyd1 promoter is a target of the Methyl CpG binding protein 2 (MeCP2), a common transcriptional modulator. MeCP2 directly bound to methylated CpG in the fxyd1 promoter and repressed fxyd1 transcription (Deng et al., 2007). On the other hand, activating histones, such as histone 3 acetylated at lysines 9 and 14 (H3K9/14ac) and histone 3 methylated at lysine 4 (H3K4me3), disassociated with the fxyd1 promoter to augment the inhibitory effect (Banine et al., 2011).
Conclusions
The expression of Na,K-ATPase is transcriptionally regulated by hormones, growth factors, lipid mediators and other extracellular stimuli through mediating transcription factor binding to promoter regions of Na,K-ATPase subunits. Many regulators of Na,K-ATPase are involved in the response to demands of cellular activity and ion homeostasis. Together with the spatial and temporal regulation of Na,K-ATPase subunits and their individual isoforms, this allows for a complex and controlled regulation of Na,K-ATPase in response to physiological demands in a tissue-specific manner. Interestingly, various recently identified transcriptional regulators of Na,K-ATPase are transcription factors that are also activated during development, in stem cells and during epithelial-mesenchymal transition (EMT). EMT is a switch in which epithelial cells undergo a shift from a well-differentiated polarized epithelial phenotype to a fibroblastic, mesenchymal phenotype and is a natural developmental process in tissue and organ formation. EMT can also be activated during wound healing, in tissue fibrosis, and is a major factor in cancer developing into a malignant disease (Kalluri and Weinberg, 2009). Vice versa, EMT activated transcription factors regulate the expression of Na,K-ATPase. For example, TGF-β and FGF up-regulate the expression of ZEB1, ZEB2, and Snail; steroid hormones, IGF-1 and PGE2 also induce ZEB1 and ZEB2. And ZEB1 and Snail1 regulate the transcription of Na,K-ATPase. Most recently, we identified the β1-subunit as a target of the Sonic hedgehog (Shh) signaling pathway (Lee et al., 2015). Shh is a critical morphogen involved in patterning of the early embryo and organogenesis, including the neural tube and limb system. The correlation between these signaling pathways and the transcriptional regulation of Na,K-ATPase is intriguing as elucidating the cross-regulation network among these factors may help in our understanding of the role of Na,K-ATPase with its pump-dependent and pump-independent functions in normal development, stem cell biology, and disease.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Support was provided by the National Institute of General Medical Sciences of the National Institutes of Health Awards Number NIGMS-P20GM103464, the American Cancer Society Grant Number RSG-09-021-01-CNE and The Nemours Foundation.
Abbreviations
CRE, cAMP response element; EMT, epithelial-mesenchymal transition; FGFs, fibroblast growth factors; GR, glucocorticoid receptor; GRE, glucocorticoid response element; HRE, hypoxia response element; MR, mineralocorticoid receptor; MRE, mineralocorticoid response element; Na,K-ATPase, sodium, potassium-adenosine triphosphatase; NRF1, nuclear respiratory factor 1; PGE1, prostaglandin E1; PGRE, prostaglandin response element; PR, progesterone receptor; PRE, progesterone response element; SBD, Smad binding domain; Sp, specificity protein; SRE, serum response element; TGF-β, transforming growth factor-β; TR, thyroid hormone receptor; TRE, thyroid hormone response element; ZEB1, zinc finger E-box binding homeobox.
References
Adachi, S., Zelenin, S., Matsuo, Y., and Holtback, U. (2004). Cellular response to renal hypoxia is different in adolescent and infant rats. Pediatr. Res. 55, 485–491. doi: 10.1203/01.PDR.0000106805.54926.2C
Alvarez de la Rosa, D., Avila, J., and Martín-Vasallo, P. (2002). Chromatin structure analysis of the rat Na,K-ATPase beta2 gene 5'-flanking region. Int. J. Biochem. Cell Biol. 34, 632–644. doi: 10.1016/S1357-2725(02)00006-7
Avila, J., Alvarez de la Rosa, D., González-Martínez, L. M., Lecuona, E., and Martín-Vasallo, P. (1998). Structure and expression of the human Na,K-ATPase beta 2-subunit gene. Gene 208, 221–227. doi: 10.1016/S0378-1119(97)00661-6
Azuma, K. K., Hensley, C. B., Tang, M. J., and McDonough, A. A. (1993). Thyroid hormone specifically regulates skeletal muscle Na(+)-K(+)-ATPase alpha 2- and beta 2-isoforms. Am. J. Physiol. 265(3 Pt 1), C680–C687.
Bajpai, M., and Chaudhury, S. (1999). Transcriptional and post-transcriptional regulation of Na+,K(+)-ATPase alpha isoforms by thyroid hormone in the developing rat brain. Neuroreport 10, 2325–2328. doi: 10.1097/00001756-199908020-00019
Bajpai, M., Mandal, S. K., and Chaudhury, S. (2001). Identification of thyroid regulatory elements in the Na-K-ATPase alpha3 gene promoter. Mol. Biol. Rep. 28, 1–7. doi: 10.1023/A:1011986418897
Banine, F., Matagne, V., Sherman, L. S., and Ojeda, S. R. (2011). Brain region-specific expression of Fxyd1, an Mecp2 target gene, is regulated by epigenetic mechanisms. J. Neurosci. Res. 89, 840–851. doi: 10.1002/jnr.22608
Barquin, N., Ciccolella, D. E., Ridge, K. M., and Sznajder, J. I. (1997). Dexamethasone upregulates the Na-K-ATPase in rat alveolar epithelial cells. Am. J. Physiol. 273(4 Pt 1), L825–L830.
Barwe, S. P., Anilkumar, G., Moon, S. Y., Zheng, Y., Whitelegge, J. P., Rajasekaran, S. A., et al. (2005). Novel role for Na,K-ATPase in phosphatidylinositol 3-kinase signaling and suppression of cell motility. Mol. Biol. Cell 16, 1082–1094. doi: 10.1091/mbc.E04-05-0427
Benarroch, E. E. (2011). Na+, K+-ATPase: functions in the nervous system and involvement in neurologic disease. Neurology 76, 287–293. doi: 10.1212/WNL.0b013e3182074c2f
Benfante, R., Antonini, R. A., Vaccari, M., Flora, A., Chen, F., Clementi, F., et al. (2005). The expression of the human neuronal alpha3 Na+,K+-ATPase subunit gene is regulated by the activity of the Sp1 and NF-Y transcription factors. Biochem. J. 386(Pt 1), 63–72. doi: 10.1042/BJ20041294
Bhutada, A., Wassynger, W. W., and Ismail-Beigi, F. (1991). Dexamethasone markedly induces Na,K-ATPase mRNA beta 1 in a rat liver cell line. J. Biol. Chem. 266, 10859–10866.
Billecocq, A., Horne, W. C., Chakraborty, M., Takeyasu, K., Levenson, R., and Baron, R. (1997). 1,25-Dihydroxyvitamin D3 selectively induces increased expression of the Na,K-ATPase beta 1 subunit in avian myelomonocytic cells without a concomitant change in Na,K-ATPase activity. J. Cell. Physiol. 172, 221–229.
Bird, A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. doi: 10.1101/gad.947102
Blanco, G. (2005). Na,K-ATPase subunit heterogeneity as a mechanism for tissue-specific ion regulation. Semin. Nephrol. 25, 292–303. doi: 10.1016/j.semnephrol.2005.03.004
Blanco, G., Sánchez, G., and Mercer, R. W. (1998). Differential regulation of Na,K-ATPase isozymes by protein kinases and arachidonic acid. Arch. Biochem. Biophys. 359, 139–150. doi: 10.1006/abbi.1998.0904
Blok, L. J., Chang, G. T., Steenbeek-Slotboom, M., van Weerden, W. M., Swarts, H. G., De Pont, J. J., et al. (1999). Regulation of expression of Na+,K+-ATPase in androgen-dependent and androgen-independent prostate cancer. Br. J. Cancer 81, 28–36. doi: 10.1038/sj.bjc.6690647
Blot-Chabaud, M., Djelidi, S., Courtois-Coutry, N., Fay, M., Cluzeaud, F., Hummler, E., et al. (2001). Coordinate control of Na,K-atpase mRNA expression by aldosterone, vasopressin and cell sodium delivery in the cortical collecting duct. Cell. Mol. Biol. (Noisy-le-grand) 47, 247–253.
Bonilla, S., Goecke, I. A., Bozzo, S., Alvo, M., Michea, L., and Marusic, E. T. (1991). Effect of chronic renal failure on Na,K-ATPase alpha 1 and alpha 2 mRNA transcription in rat skeletal muscle. J. Clin. Invest. 88, 2137–2141. doi: 10.1172/JCI115544
Bonvalet, J. P., Pradelles, P., and Farman, N. (1987). Segmental synthesis and actions of prostaglandins along the nephron. Am. J. Physiol. 253(3 Pt 2), F377–F387.
Borok, Z., Danto, S. I., Dimen, L. L., Zhang, X. L., and Lubman, R. L. (1998). Na(+)-K(+)-ATPase expression in alveolar epithelial cells: upregulation of active ion transport by KGF. Am. J. Physiol. 274(1 Pt 1), L149–L158.
Cai, T., Wang, H., Chen, Y., Liu, L., Gunning, W. T., Quintas, L. E., et al. (2008). Regulation of caveolin-1 membrane trafficking by the Na/K-ATPase. J. Cell Biol. 182, 1153–1169. doi: 10.1083/jcb.200712022
Cedar, H., and Bergman, Y. (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304. doi: 10.1038/nrg2540
Celsi, G., Nishi, A., Akusjärvi, G., and Aperia, A. (1991). Abundance of Na(+)-K(+)-ATPase mRNA is regulated by glucocorticoid hormones in infant rat kidneys. Am. J. Physiol. 260(2 Pt 2), F192–F197.
Chalaka, S., Ingbar, D. H., Sharma, R., Zhau, Z., and Wendt, C. H. (1999). Na(+)-K(+)-ATPase gene regulation by glucocorticoids in a fetal lung epithelial cell line. Am. J. Physiol. 277(1 Pt 1), L197–L203.
Chaudhury, S., Bajpai, M., and Bhattacharya, S. (1996). Differential effects of hypothyroidism on Na-K-ATPase mRNA alpha isoforms in the developing rat brain. J. Mol. Neurosci. 7, 229–234. doi: 10.1007/BF02736843
Clausen, T. (2003). Na+-K+ pump regulation and skeletal muscle contractility. Physiol. Rev. 83, 1269–1324. doi: 10.1152/physrev.00011.2003
Cochrane, D. R., Jacobsen, B. M., Connaghan, K. D., Howe, E. N., Bain, D. L., and Richer, J. K. (2012). Progestin regulated miRNAs that mediate progesterone receptor action in breast cancer. Mol. Cell. Endocrinol. 355, 15–24. doi: 10.1016/j.mce.2011.12.020
Dagenais, A., Denis, C., Vives, M. F., Girouard, S., Massé, C., Nguyen, T., et al. (2001). Modulation of alpha-ENaC and alpha1-Na+-K+-ATPase by cAMP and dexamethasone in alveolar epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 281, L217–L230.
Danto, S. I., Borok, Z., Zhang, X. L., Lopez, M. Z., Patel, P., Crandall, E. D., et al. (1998). Mechanisms of EGF-induced stimulation of sodium reabsorption by alveolar epithelial cells. Am. J. Physiol. 275(1 Pt 1), C82–C92.
Deng, V., Matagne, V., Banine, F., Frerking, M., Ohliger, P., Budden, S., et al. (2007). FXYD1 is an MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum. Mol. Genet. 16, 640–650. doi: 10.1093/hmg/ddm007
Deng, W. B., Tian, Z., Liang, X. H., Wang, B. C., Yang, F., and Yang, Z. M. (2013). Progesterone regulation of Na/K-ATPase beta1 subunit expression in the mouse uterus during the peri-implantation period. Theriogenology 79, 1196–1203. doi: 10.1016/j.theriogenology.2013.02.018
Derfoul, A., Robertson, N. M., Hall, D. J., and Litwack, G. (2000). The N-terminal domain of the mineralocorticoid receptor modulates both mineralocorticoid receptor- and glucocorticoid receptor-mediated transactivation from Na/K ATPase beta1 target gene promoter. Endocrine 13, 287–295. doi: 10.1385/ENDO:13:3:287
Derfoul, A., Robertson, N. M., Lingrel, J. B., Hall, D. J., and Litwack, G. (1998). Regulation of the human Na/K-ATPase beta1 gene promoter by mineralocorticoid and glucocorticoid receptors. J. Biol. Chem. 273, 20702–20711. doi: 10.1074/jbc.273.33.20702
Dong, X., Ghoshal, K., Majumder, S., Yadav, S. P., and Jacob, S. T. (2002). Mitochondrial transcription factor A and its downstream targets are up-regulated in a rat hepatoma. J. Biol. Chem. 277, 43309–43318. doi: 10.1074/jbc.M206958200
Espineda, C. E., Chang, J. H., Twiss, J., Rajasekaran, S. A., and Rajasekaran, A. K. (2004). Repression of Na,K-ATPase beta1-subunit by the transcription factor snail in carcinoma. Mol. Biol. Cell 15, 1364–1373. doi: 10.1091/mbc.E03-09-0646
Evans, M. J., and Scarpulla, R. C. (1989). Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J. Biol. Chem. 264, 14361–14368.
Ewart, H. S., and Klip, A. (1995). Hormonal regulation of the Na(+)-K(+)-ATPase: mechanisms underlying rapid and sustained changes in pump activity. Am. J. Physiol. 269(2 Pt 1), C295–C311.
Farman, N., Bonvalet, J. P., and Seckl, J. R. (1994). Aldosterone selectively increases Na(+)-K(+)-ATPase alpha 3-subunit mRNA expression in rat hippocampus. Am. J. Physiol. 266(2 Pt 1), C423–C428.
Feng, J., Orlowski, J., and Lingrel, J. B. (1993). Identification of a functional thyroid hormone response element in the upstream flanking region of the human Na,K-ATPase beta 1 gene. Nucleic Acids Res. 21, 2619–2626. doi: 10.1093/nar/21.11.2619
Galuska, D., Kotova, O., Barrès, R., Chibalina, D., Benziane, B., and Chibalin, A. V. (2009). Altered expression and insulin-induced trafficking of Na+-K+-ATPase in rat skeletal muscle: effects of high-fat diet and exercise. Am. J. Physiol. Endocrinol. Metab. 297, E38–E49. doi: 10.1152/ajpendo.90990.2008
Galuska, D., Pirkmajer, S., Barrès, R., Ekberg, K., Wahren, J., and Chibalin, A. V. (2011). C-peptide increases Na,K-ATPase expression via PKC- and MAP kinase-dependent activation of transcription factor ZEB in human renal tubular cells. PLoS ONE 6:e28294. doi: 10.1371/journal.pone.0028294
Garty, H., and Karlish, S. J. (2006). Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 68, 431–459. doi: 10.1146/annurev.physiol.68.040104.131852
Geering, K. (2006). FXYD proteins: new regulators of Na-K-ATPase. Am. J. Physiol. Renal Physiol. 290, F241–F250. doi: 10.1152/ajprenal.00126.2005
Geering, K. (2008). Functional roles of Na,K-ATPase subunits. Curr. Opin. Nephrol. Hypertens. 17, 526–532. doi: 10.1097/MNH.0b013e3283036cbf
Giannella, R. A., Orlowski, J., Jump, M. L., and Lingrel, J. B. (1993). Na(+)-K(+)-ATPase gene expression in rat intestine and Caco-2 cells: response to thyroid hormone. Am. J. Physiol. 265(4 Pt 1), G775–G782.
Gick, G. G., and Ismail-Beigi, F. (1990). Thyroid hormone induction of Na(+)-K(+)-ATPase and its mRNAs in a rat liver cell line. Am. J. Physiol. 258(3 Pt 1), C544–C551.
Gloor, S., Antonicek, H., Sweadner, K. J., Pagliusi, S., Frank, R., Moos, M., et al. (1990). The adhesion molecule on glia (AMOG) is a homologue of the beta subunit of the Na,K-ATPase. J. Cell Biol. 110, 165–174. doi: 10.1083/jcb.110.1.165
González, S., Grillo, C., De Nicola, A. G., Piroli, G., Angulo, J., McEwen, B. S., et al. (1994). Dexamethasone increases adrenalectomy-depressed Na+,K(+)-ATPase mRNA and ouabain binding in spinal cord ventral horn. J. Neurochem. 63, 1962–1970. doi: 10.1046/j.1471-4159.1994.63051962.x
González, S., Grillo, C., Gonzalez Deniselle, M. C., Lima, A., McEwen, B. S., and De Nicola, A. F. (1996). Dexamethasone up-regulates mRNA for Na+,K(+)-ATPase in some spinal cord neurones after cord transection. Neuroreport 7, 1041–1044. doi: 10.1097/00001756-199604100-00017
Grillo, C., Piroli, G., Lima, A., McEwen, B. S., and De Nicola, A. F. (1997). Aldosterone up-regulates mRNA for the alpha3 and beta1 isoforms of (Na,K)-ATPase in several brain regions from adrenalectomized rats. Brain Res. 767, 120–127. doi: 10.1016/S0006-8993(97)00541-6
Guerrero, C., Lecuona, E., Pesce, L., Ridge, K. M., and Sznajder, J. I. (2001). Dopamine regulates Na-K-ATPase in alveolar epithelial cells via MAPK-ERK-dependent mechanisms. Am. J. Physiol. Lung Cell. Mol. Physiol. 281, L79–L85.
Haas, M., Wang, H., Tian, J., and Xie, Z. (2002). Src-mediated inter-receptor cross-talk between the Na+/K+-ATPase and the epidermal growth factor receptor relays the signal from ouabain to mitogen-activated protein kinases. J. Biol. Chem. 277, 18694–18702. doi: 10.1074/jbc.M111357200
Hai, T., and Hartman, M. G. (2001). The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273, 1–11. doi: 10.1016/S0378-1119(01)00551-0
Hao, H., Rhodes, R., Ingbar, D. H., and Wendt, C. H. (2003a). Dexamethasone responsive element in the rat Na,K-ATPase beta1 gene coding region. Biochim. Biophys. Acta 1630, 55–63. doi: 10.1016/j.bbaexp.2003.09.003
Hao, H., Wendt, C. H., Sandhu, G., and Ingbar, D. H. (2003b). Dexamethasone stimulates transcription of the Na+-K+-ATPase beta1 gene in adult rat lung epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 285, L593–L601. doi: 10.1152/ajplung.00037.2003
He, H., Chin, S., Zhuang, K., Hartong, R., Apriletti, J., and Gick, G. (1996). Negative regulation of the rat Na-K-ATPase alpha 3-subunit gene promoter by thyroid hormone. Am. J. Physiol. 271(5 Pt 1), C1750–C1756.
Henriksen, C., Kjaer-Sorensen, K., Einholm, A. P., Madsen, L. B., Momeni, J., Bendixen, C., et al. (2013). Molecular cloning and characterization of porcine Na(+)/K(+)-ATPase isoforms alpha1, alpha2, alpha3 and the ATP1A3 promoter. PLoS ONE 8:e79127. doi: 10.1371/journal.pone.0079127
Hensley, C. B., Azuma, K. K., Tang, M. J., and McDonough, A. A. (1992). Thyroid hormone induction of rat myocardial Na(+)-K(+)-ATPase: alpha 1-, alpha 2-, and beta 1-mRNA and -protein levels at steady state. Am. J. Physiol. 262(2 Pt 1), C484–C492.
Herman, M. B., Rajkhowa, T., Cutuli, F., Springate, J. E., and Taub, M. (2010). Regulation of renal proximal tubule Na-K-ATPase by prostaglandins. Am. J. Physiol. Renal Physiol. 298, F1222–F1234. doi: 10.1152/ajprenal.00467.2009
Herrera, V. L., Emanuel, J. R., Ruiz-Opazo, N., Levenson, R., and Nadal-Ginard, B. (1987). Three differentially expressed Na,K-ATPase alpha subunit isoforms: structural and functional implications. J. Cell Biol. 105, 1855–1865. doi: 10.1083/jcb.105.4.1855
Hosoi, R., Matsuda, T., Asano, S., Nakamura, H., Hashimoto, H., Takuma, K., et al. (1997). Isoform-specific up-regulation by ouabain of Na+,K+-ATPase in cultured rat astrocytes. J. Neurochem. 69, 2189–2196. doi: 10.1046/j.1471-4159.1997.69052189.x
Howarth, C., Gleeson, P., and Attwell, D. (2012). Updated energy budgets for neural computation in the neocortex and cerebellum. J. Cereb. Blood Flow Metab. 32, 1222–1232. doi: 10.1038/jcbfm.2012.35
Huang, F., He, H., and Gick, G. (1994). Thyroid hormone regulation of Na,K-ATPase alpha 2 gene expression in cardiac myocytes. Cell. Mol. Biol. Res. 40, 41–52.
Husted, R. F., Sigmund, R. D., and Stokes, J. B. (2000). Mechanisms of inactivation of the action of aldosterone on collecting duct by TGF-beta. Am. J. Physiol. Renal Physiol. 278, F425–F433.
Ikeda, K., Nagano, K., and Kawakami, K. (1993). Anomalous interaction of Sp1 and specific binding of an E-box-binding protein with the regulatory elements of the Na,K-ATPase alpha 2 subunit gene promoter. Eur. J. Biochem. 218, 195–204. doi: 10.1111/j.1432-1033.1993.tb18365.x
Ikeda, U., Hyman, R., Smith, T. W., and Medford, R. M. (1991). Aldosterone-mediated regulation of Na+, K(+)-ATPase gene expression in adult and neonatal rat cardiocytes. J. Biol. Chem. 266, 12058–12066.
Ingbar, D. H., Duvick, S., Savick, S. K., Schellhase, D. E., Detterding, R., Jamieson, J. D., et al. (1997). Developmental changes of fetal rat lung Na-K-ATPase after maternal treatment with dexamethasone. Am. J. Physiol. 272(4 Pt 1), L665–L672.
Isenovic, E. R., Jacobs, D. B., Kedees, M. H., Sha, Q., Milivojevic, N., Kawakami, K., et al. (2004). Angiotensin II regulation of the Na+ pump involves the phosphatidylinositol-3 kinase and p42/44 mitogen-activated protein kinase signaling pathways in vascular smooth muscle cells. Endocrinology 145, 1151–1160. doi: 10.1210/en.2003-0100
Johar, K., Priya, A., and Wong-Riley, M. T. (2012). Regulation of Na(+)/K(+)-ATPase by nuclear respiratory factor 1: implication in the tight coupling of neuronal activity, energy generation, and energy consumption. J. Biol. Chem. 287, 40381–40390. doi: 10.1074/jbc.M112.414573
Johar, K., Priya, A., and Wong-Riley, M. T. (2014). Regulation of Na(+)/K(+)-ATPase by neuron-specific transcription factor Sp4: implication in the tight coupling of energy production, neuronal activity and energy consumption in neurons. Eur. J. Neurosci. 39, 566–578. doi: 10.1111/ejn.12415
Kalluri, R., and Weinberg, R. A. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428. doi: 10.1172/JCI39104
Kamitani, T., Ikeda, U., Muto, S., Kawakami, K., Nagano, K., Tsuruya, Y., et al. (1992). Regulation of Na,K-ATPase gene expression by thyroid hormone in rat cardiocytes. Circ. Res. 71, 1457–1464. doi: 10.1161/01.RES.71.6.1457
Kaplan, J. H. (2002). Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 71, 511–535. doi: 10.1146/annurev.biochem.71.102201.141218
Karitskaya, I., Aksenov, N., Vassilieva, I., Zenin, V., and Marakhova, I. (2010). Long-term regulation of Na,K-ATPase pump during T-cell proliferation. Pflugers Arch. 460, 777–789. doi: 10.1007/s00424-010-0843-z
Kawakami, K., Okamoto, H., Yagawa, Y., and Nagano, K. (1990). Regulation of Na+,K(+)-ATPase. II. Cloning and analysis of the 5'-flanking region of the rat NKAB2 gene encoding the beta 2 subunit. Gene 91, 271–274. doi: 10.1016/0378-1119(90)90099-D
Kawakami, K., Suzuki-Yagawa, Y., Watanabe, Y., and Nagano, K. (1992). Identification and characterization of the cis-elements regulating the rat AMOG (adhesion molecule on glia)/Na,K-ATPase beta 2 subunit gene. J. Biochem. 111, 515–522.
Kawakami, K., Watanabe, Y., Araki, M., and Nagano, K. (1993). Sp1 binds to the adhesion-molecule-on-glia regulatory element that functions as a positive transcription regulatory element in astrocytes. J. Neurosci. Res. 35, 138–146. doi: 10.1002/jnr.490350204
Ke, Q., and Costa, M. (2006). Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 70, 1469–1480. doi: 10.1124/mol.106.027029
Keryanov, S., and Gardner, K. L. (2002). Physical mapping and characterization of the human Na,K-ATPase isoform, ATP1A4. Gene 292, 151–166. doi: 10.1016/S0378-1119(02)00647-9
Kimura, T., Han, W., Pagel, P., Nairn, A. C., and Caplan, M. J. (2011). Protein phosphatase 2A interacts with the Na,K-ATPase and modulates its trafficking by inhibition of its association with arrestin. PLoS ONE 6:e29269. doi: 10.1371/journal.pone.0029269
Kirtane, A., Ismail-Beigi, N., and Ismail-Beigi, F. (1994). Role of enhanced Na+ entry in the control of Na,K-ATPase gene expression by serum. J. Membr. Biol. 137, 9–15. doi: 10.1007/BF00234994
Kitamura, N., Ikekita, M., Sato, T., Akimoto, Y., Hatanaka, Y., Kawakami, H., et al. (2005). Mouse Na+/K+-ATPase beta1-subunit has a K+-dependent cell adhesion activity for beta-GlcNAc-terminating glycans. Proc. Natl. Acad. Sci. U.S.A. 102, 2796–2801. doi: 10.1073/pnas.0409344102
Kobayashi, M., and Kawakami, K. (1995). ATF-1CREB heterodimer is involved in constitutive expression of the housekeeping Na,K-ATPase alpha 1 subunit gene. Nucleic Acids Res. 23, 2848–2855. doi: 10.1093/nar/23.15.2848
Kobayashi, M., Shimomura, A., Hagiwara, M., and Kawakami, K. (1997). Phosphorylation of ATF-1 enhances its DNA binding and transcription of the Na,K-ATPase alpha 1 subunit gene promoter. Nucleic Acids Res. 25, 877–882. doi: 10.1093/nar/25.4.877
Kolla, V., Robertson, N. M., and Litwack, G. (1999). Identification of a mineralocorticoid/glucocorticoid response element in the human Na/K ATPase alpha1 gene promoter. Biochem. Biophys. Res. Commun. 266, 5–14. doi: 10.1006/bbrc.1999.1765
Kometiani, P., Tian, J., Li, J., Nabih, Z., Gick, G., and Xie, Z. (2000). Regulation of Na/K-ATPase beta1-subunit gene expression by ouabain and other hypertrophic stimuli in neonatal rat cardiac myocytes. Mol. Cell. Biochem. 215, 65–72. doi: 10.1023/A:1026503319387
Kone, B. C., and Higham, S. (1999). Nitric oxide inhibits transcription of the Na+-K+-ATPase alpha1-subunit gene in an MTAL cell line. Am. J. Physiol. 276(4 Pt 2), F614–F621.
Kumar, R., and Thompson, E. B. (2005). Gene regulation by the glucocorticoid receptor: structure:function relationship. J. Steroid Biochem. Mol. Biol. 94, 383–394. doi: 10.1016/j.jsbmb.2004.12.046
Lamouille, S., Xu, J., and Derynck, R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196. doi: 10.1038/nrm3758
Laursen, M., Yatime, L., Nissen, P., and Fedosova, N. U. (2013). Crystal structure of the high-affinity Na+K+-ATPase-ouabain complex with Mg2+ bound in the cation binding site. Proc. Natl. Acad. Sci. U.S.A. 110, 10958–10963. doi: 10.1073/pnas.1222308110
Lavoie, L., Roy, D., Ramlal, T., Dombrowski, L., Martín-Vasallo, P., Marette, A., et al. (1996). Insulin-induced translocation of Na+-K+-ATPase subunits to the plasma membrane is muscle fiber type specific. Am. J. Physiol. 270(5 Pt 1), C1421–C1429.
Lee, J., Ha, J. H., Kim, S., Oh, Y., and Kim, S. W. (2002). Caffeine decreases the expression of Na+/K+-ATPase and the type 3 Na+/H+ exchanger in rat kidney. Clin. Exp. Pharmacol. Physiol. 29, 559–563. doi: 10.1046/j.1440-1681.2002.03697.x
Lee, S. J., Litan, A., Li, Z., Graves, B., Lindsey, S., Barwe, S. P., et al. (2015). Na,K-ATPase beta1-subunit is a target of sonic hedgehog signaling and enhances medulloblastoma tumorigenicity. Mol. Cancer 14, 159. doi: 10.1186/s12943-015-0430-1
Leo, J. C., Guo, C., Woon, C. T., Aw, S. E., and Lin, V. C. (2004). Glucocorticoid and mineralocorticoid cross-talk with progesterone receptor to induce focal adhesion and growth inhibition in breast cancer cells. Endocrinology 145, 1314–1321. doi: 10.1210/en.2003-0732
Leonhardt, S. A., Boonyaratanakornkit, V., and Edwards, D. P. (2003). Progesterone receptor transcription and non-transcription signaling mechanisms. Steroids 68, 761–770 doi: 10.1016/S0039-128X(03)00129-6
Lin, H. H., and Tang, M. J. (1997). Thyroid hormone upregulates Na,K-ATPase alpha and beta mRNA in primary cultures of proximal tubule cells. Life Sci. 60, 375–382. doi: 10.1016/S0024-3205(96)00661-3
Litan, A., and Langhans, S. A. (2015). Cancer as a channelopathy: ion channels and pumps in tumor development and progression. Front. Cell. Neurosci. 9:86. doi: 10.3389/fncel.2015.00086
Liu, B., and Gick, G. (1992). Characterization of the 5′ flanking region of the rat Na+/K(+)-ATPase beta 1 subunit gene. Biochim. Biophys. Acta 1130, 336–338. doi: 10.1016/0167-4781(92)90449-A
Malik, N., Canfield, V., Sanchez-Watts, G., Watts, A. G., Scherer, S., Beatty, B. G., et al. (1998). Structural organization and chromosomal localization of the human Na,K-ATPase beta 3 subunit gene and pseudogene. Mamm. Genome 9, 136–143. doi: 10.1007/s003359900704
Malyshev, I. V., Bessarab, D. A., Orlova, M. Yu., Petrukhin, K. E., Volik, S. V., Broude, N. E., et al. (1991). A comparative analysis of the putative regulatory regions in human genes for the alpha-subunit family of Na(+)-K+ ATPase. Biomed. Sci. 2, 54–62.
Mantovani, R. (1998). A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26, 1135–1143. doi: 10.1093/nar/26.5.1135
Massagué, J., Seoane, J., and Wotton, D. (2005). Smad transcription factors. Genes Dev. 19, 2783–2810. doi: 10.1101/gad.1350705
Matlhagela, K., Borsick, M., Rajkhowa, T., and Taub, M. (2005). Identification of a prostaglandin-responsive element in the Na,K-ATPase beta 1 promoter that is regulated by cAMP and Ca2+. Evidence for an interactive role of cAMP regulatory element-binding protein and Sp1. J. Biol. Chem. 280, 334–346. doi: 10.1074/jbc.M411415200
Matlhagela, K., and Taub, M. (2006). Regulation of the Na-K-ATPase beta(1)-subunit promoter by multiple prostaglandin-responsive elements. Am. J. Physiol. Renal Physiol. 291, F635–F646. doi: 10.1152/ajprenal.00452.2005
McDonough, A. A., Brown, T. A., Horowitz, B., Chiu, R., Schlotterbeck, J., Bowen, J., et al. (1988). Thyroid hormone coordinately regulates Na+-K+-ATPase alpha- and beta-subunit mRNA levels in kidney. Am. J. Physiol. 254(2 Pt 1), C323–C329.
Melikian, J., and Ismail-Beigi, F. (1991). Thyroid hormone regulation of Na,K-ATPase subunit-mRNA expression in neonatal rat myocardium. J. Membr. Biol. 119, 171–177. doi: 10.1007/BF01871416
Mishra, N. K., Peleg, Y., Cirri, E., Belogus, T., Lifshitz, Y., Voelker, D. R., et al. (2011). FXYD proteins stabilize Na,K-ATPase: amplification of specific phosphatidylserine-protein interactions. J. Biol. Chem. 286, 9699–9712. doi: 10.1074/jbc.M110.184234
Mobasheri, A., Avila, J., Cózar-Castellano, I., Brownleader, M. D., Trevan, M., Francis, M. J., et al. (2000). Na+, K+-ATPase isozyme diversity; comparative biochemistry and physiological implications of novel functional interactions. Biosci. Rep. 20, 51–91. doi: 10.1023/A:1005580332144
Mony, S., Lee, S. J., Harper, J. F., Barwe, S. P., and Langhans, S. A. (2013). Regulation of Na,K-ATPase beta1-subunit in TGF-beta2-mediated epithelial-to-mesenchymal transition in human retinal pigmented epithelial cells. Exp. Eye Res. 115, 113–122. doi: 10.1016/j.exer.2013.06.007
Murakami, Y., Ikeda, U., Shimada, K., and Kawakami, K. (1997). Promoter of the Na,K-ATPase alpha3 subunit gene is composed of cis elements to which NF-Y and Sp1/Sp3 bind in rat cardiocytes. Biochim. Biophys. Acta 1352, 311–324. doi: 10.1016/S0167-4781(97)00032-8
Muto, S., Nemoto, J., Ebata, S., Kawakami, K., and Asano, Y. (1998a). Corticosterone and 11-dehydrocorticosterone stimulate Na,K-ATPase gene expression in vascular smooth muscle cells. Kidney Int. 54, 492–508. doi: 10.1046/j.1523-1755.1998.00033.x
Muto, S., Nemoto, J., Ohtaka, A., Watanabe, Y., Yamaki, M., Kawakami, K., et al. (1996). Differential regulation of Na+-K+-ATPase gene expression by corticosteriods in vascular smooth muscle cells. Am. J. Physiol. 270(3 Pt 1), C731–C739.
Muto, S., Nemoto, J., Okada, K., Miyata, Y., Kawakami, K., Saito, T., et al. (2000). Intracellular Na+ directly modulates Na+,K+-ATPase gene expression in normal rat kidney epithelial cells. Kidney Int. 57, 1617–1635. doi: 10.1046/j.1523-1755.2000.00006.x
Muto, S., Ohtaka, A., Nemoto, J., Kawakami, K., and Asano, Y. (1998b). Effects of hyperosmolality on Na,K-ATPase gene expression in vascular smooth muscle cells. J. Membr. Biol. 162, 233–245. doi: 10.1007/s002329900361
Nemoto, J., Muto, S., Ohtaka, A., Kawakami, K., and Asano, Y. (1997). Serum transcriptionally regulates Na(+)-K(+)-ATPase gene expression in vascular smooth muscle cells. Am. J. Physiol. 273(3 Pt 1), C1088–C1099.
Nieto, M. A. (2002). The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 3, 155–166. doi: 10.1038/nrm757
Nordeen, S. K., Suh, B. J., Kühnel, B., and Hutchison, C. A. III. (1990). Structural determinants of a glucocorticoid receptor recognition element. Mol. Endocrinol. 4, 1866–1873. doi: 10.1210/mend-4-12-1866
Oguchi, A., Ikeda, U., Kanbe, T., Tsuruya, Y., Yamamoto, K., Kawakami, K., et al. (1993). Regulation of Na-K-ATPase gene expression by aldosterone in vascular smooth muscle cells. Am. J. Physiol. 265(4 Pt 2), H1167–H1172.
Ohara, T., Ikeda, U., Muto, S., Oguchi, A., Tsuruya, Y., Yamamoto, K., et al. (1993). Thyroid hormone stimulates Na(+)-K(+)-ATPase gene expression in cultured rat mesangial cells. Am. J. Physiol. 265(3 Pt 2), F370–F376.
Okoh, V., Deoraj, A., and Roy, D. (2011). Estrogen-induced reactive oxygen species-mediated signalings contribute to breast cancer. Biochim. Biophys. Acta 1815, 115–133. doi: 10.1016/j.bbcan.2010.10.005
Olivera, W. G., Ciccolella, D. E., Barquin, N., Ridge, K. M., Rutschman, D. H., Yeates, D. B., et al. (2000). Aldosterone regulates Na,K-ATPase and increases lung edema clearance in rats. Am. J. Respir. Crit. Care Med. 161(2 Pt 1), 567–573. doi: 10.1164/ajrccm.161.2.9808050
Orlowski, J., and Lingrel, J. B. (1988). Tissue-specific and developmental regulation of rat Na,K-ATPase catalytic alpha isoform and beta subunit mRNAs. J. Biol. Chem. 263, 10436–10442.
Orlowski, J., and Lingrel, J. B. (1990). Thyroid and glucocorticoid hormones regulate the expression of multiple Na,K-ATPase genes in cultured neonatal rat cardiac myocytes. J. Biol. Chem. 265, 3462–3470.
Pathak, B. G., Neumann, J. C., Croyle, M. L., and Lingrel, J. B. (1994). The presence of both negative and positive elements in the 5'-flanking sequence of the rat Na,K-ATPase alpha 3 subunit gene are required for brain expression in transgenic mice. Nucleic Acids Res. 22, 4748–4755. doi: 10.1093/nar/22.22.4748
Pathak, B. G., Pugh, D. G., and Lingrel, J. B. (1990). Characterization of the 5'-flanking region of the human and rat Na,K-ATPase alpha 3 gene. Genomics 8, 641–647. doi: 10.1016/0888-7543(90)90250-X
Pekary, A. E., Levin, S. R., Johnson, D. G., Berg, L., and Hershman, J. M. (1997). Tumor necrosis factor-alpha (TNF-alpha) and transforming growth factor-beta 1 (TGF-beta 1) inhibit the expression and activity of Na+/K(+)-ATPase in FRTL-5 rat thyroid cells. J. Interferon Cytokine Res. 17, 185–195. doi: 10.1089/jir.1997.17.185
Phakdeekitcharoen, B., Kittikanokrat, W., Kijkunasathian, C., and Chatsudthipong, V. (2011). Aldosterone increases Na+ -K+ -ATPase activity in skeletal muscle of patients with Conn's syndrome. Clin. Endocrinol. (Oxf). 74, 152–159. doi: 10.1111/j.1365-2265.2010.03912.x
Phakdeekitcharoen, B., Phudhichareonrat, S., Pookarnjanamorakot, C., Kijkunasathian, C., Tubtong, N., Kittikanokrat, W., et al. (2007). Thyroid hormone increases mRNA and protein expression of Na+-K+-ATPase alpha2 and beta1 subunits in human skeletal muscles. J. Clin. Endocrinol. Metab. 92, 353–358. doi: 10.1210/jc.2006-0552
Pierre, S. V., and Xie, Z. (2006). The Na,K-ATPase receptor complex: its organization and membership. Cell Biochem. Biophys. 46, 303–316. doi: 10.1385/CBB:46:3:303
Planès, C., Escoubet, B., Blot-Chabaud, M., Friedlander, G., Farman, N., and Clerici, C. (1997). Hypoxia downregulates expression and activity of epithelial sodium channels in rat alveolar epithelial cells. Am. J. Respir. Cell Mol. Biol. 17, 508–518. doi: 10.1165/ajrcmb.17.4.2680
Qin, X., Liu, B., and Gick, G. (1994). Low external K+ regulates Na,K-ATPase alpha 1 and beta 1 gene expression in rat cardiac myocytes. Am. J. Hypertens. 7, 96–99.
Rajasekaran, S. A., Gopal, J., Willis, D., Espineda, C., and Twiss, J. L. Rajasekaran, A. K. (2004). Na,K-ATPase beta1-subunit increases the translation efficiency of the alpha1-subunit in MSV-MDCK cells. Mol. Biol. Cell 15, 3224–3232. doi: 10.1091/mbc.E04-03-0222
Rajasekaran, S. A., Huynh, T. P., Wolle, D. G., Espineda, C. E., Inge, L. J., Skay, A., et al. (2010). Na,K-ATPase subunits as markers for epithelial-mesenchymal transition in cancer and fibrosis. Mol. Cancer Ther. 9, 1515–1524. doi: 10.1158/1535-7163.MCT-09-0832
Rajasekaran, S. A., Palmer, L. G., Moon, S. Y., Peralta Soler, A., Apodaca, G. L., Harper, J. F., et al. (2001a). Na,K-ATPase activity is required for formation of tight junctions, desmosomes, and induction of polarity in epithelial cells. Mol. Biol. Cell 12, 3717–3732. doi: 10.1091/mbc.12.12.3717
Rajasekaran, S. A., Palmer, L. G., Quan, K., Harper, J. F., Ball, W. J. Jr., Bander, N. H., et al. (2001b). Na,K-ATPase beta-subunit is required for epithelial polarization, suppression of invasion, and cell motility. Mol. Biol. Cell 12, 279–295. doi: 10.1091/mbc.12.2.279
Rajasekaran, S. A., and Rajasekaran, A. K. (2009). Na,K-ATPase and epithelial tight junctions. Front. Biosci. 14, 2130–2148. doi: 10.2741/3367
Rayson, B. M. (1991). [Ca2+]i regulates transcription rate of the Na+/K(+)-ATPase alpha 1 subunit. J. Biol. Chem. 266, 21335–21338.
Reinhard, L., Tidow, H., Clausen, M., and Nissen, P. (2013). Na+,K+-ATPase as a docking station: protein–protein complexes of the Na+,K+-ATPase. Cell. Mol. Life Sci. 70, 205–222. doi: 10.1007/s00018-012-1039-9
Richer, J. K., Jacobsen, B. M., Manning, N. G., Abel, M. G., Wolf, D. M., and Horwitz, K. B. (2002). Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J. Biol. Chem. 277, 5209–5218. doi: 10.1074/jbc.M110090200
Rodova, M., Nguyen, A. N., and Blanco, G. (2006). The transcription factor CREMtau and cAMP regulate promoter activity of the Na,K-ATPase alpha4 isoform. Mol. Reprod. Dev. 73, 1435–1447. doi: 10.1002/mrd.20518
Russo, J. J., and Sweadner, K. J. (1993). Na(+)-K(+)-ATPase subunit isoform pattern modification by mitogenic insulin concentration in 3T3-L1 preadipocytes. Am. J. Physiol. 264(2 Pt 1), C311–C316.
Sandtner, W., Egwolf, B., Khalili-Araghi, F., Sánchez-Rodríguez, J. E., Roux, B., Bezanilla, F., et al. (2011). Ouabain binding site in a functioning Na+/K+ ATPase. J. Biol. Chem. 286, 38177–38183. doi: 10.1074/jbc.M111.267682
Savagner, F., Mirebeau, D., Jacques, C., Guyetant, S., Morgan, C., Franc, B., et al. (2003). PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem. Biophys. Res. Commun. 310, 779–784. doi: 10.1016/j.bbrc.2003.09.076
Selvakumar, P., Owens, T. A., David, J. M., Petrelli, N. J., Christensen, B. C., Lakshmikuttyamma, A., et al. (2014). Epigenetic silencing of Na,K-ATPase beta 1 subunit gene ATP1B1 by methylation in clear cell renal cell carcinoma. Epigenetics 9, 579–586. doi: 10.4161/epi.27795
Seok, J. H., Hong, J. H., Jeon, J. R., Hur, G. M., Sung, J. Y., and Lee, J. H. (1999). Aldosterone directly induces Na,K-ATPase alpha 1-subunit mRNA in the renal cortex of rat. Biochem. Mol. Biol. Int. 47, 251–254.
Sevieux, N., Alam, J., and Songu-Mize, E. (2001). Effect of cyclic stretch on alpha-subunit mRNA expression of Na+-K+-ATPase in aortic smooth muscle cells. Am. J. Physiol. Cell Physiol. 280, C1555–C1560.
Shamraj, O. I., and Lingrel, J. B. (1994). A putative fourth Na+,K(+)-ATPase alpha-subunit gene is expressed in testis. Proc. Natl. Acad. Sci. U.S.A. 91, 12952–12956. doi: 10.1073/pnas.91.26.12952
Shao, Y., Ojamaa, K., Klein, I., and Ismail-Beigi, F. (2000). Thyroid hormone stimulates Na,K-ATPase gene expression in the hemodynamically unloaded heterotopically transplanted rat heart. Thyroid 10, 753–759. doi: 10.1089/thy.2000.10.753
Shoshani, L., Contreras, R. G., Roldán, M. L., Moreno, J., Lázaro, A., Balda, M. S., et al. (2005). The polarized expression of Na+,K+-ATPase in epithelia depends on the association between beta-subunits located in neighboring cells. Mol. Biol. Cell 16, 1071–1081. doi: 10.1091/mbc.E04-03-0267
Shull, M. M., Pugh, D. G., and Lingrel, J. B. (1989). Characterization of the human Na,K-ATPase alpha 2 gene and identification of intragenic restriction fragment length polymorphisms. J. Biol. Chem. 264, 17532–17543.
Shull, M. M., Pugh, D. G., and Lingrel, J. B. (1990). The human Na,K-ATPase alpha 1 gene: characterization of the 5'-flanking region and identification of a restriction fragment length polymorphism. Genomics 6, 451–460. doi: 10.1016/0888-7543(90)90475-A
Shyjan, A. W., Canfield, V. A., and Levenson, R. (1991). Evolution of the Na,K- and H,K-ATPase beta subunit gene family: structure of the murine Na,K-ATPase beta 2 subunit gene. Genomics 11, 435–442. doi: 10.1016/0888-7543(91)90152-5
Silva, E., and Soares-da-Silva, P. (2012). New insights into the regulation of Na+,K+-ATPase by ouabain. Int. Rev. Cell Mol. Biol. 294, 99–132. doi: 10.1016/B978-0-12-394305-7.00002-1
Sugimoto, Y., and Narumiya, S. (2007). Prostaglandin E receptors. J. Biol. Chem. 282, 11613–11617. doi: 10.1074/jbc.R600038200
Suske, G. (1999). The Sp-family of transcription factors. Gene 238, 291–300. doi: 10.1016/S0378-1119(99)00357-1
Suzuki-Yagawa, Y., Kawakami, K., and Nagano, K. (1992). Housekeeping Na,K-ATPase alpha 1 subunit gene promoter is composed of multiple cis elements to which common and cell type-specific factors bind. Mol. Cell. Biol. 12, 4046–4055. doi: 10.1128/MCB.12.9.4046
Sweadner, K. J. (1989). Isozymes of the Na+/K+-ATPase. Biochim. Biophys. Acta 988, 185–220. doi: 10.1016/0304-4157(89)90019-1
Sweeney, G., and Klip, A. (1998). Regulation of the Na+/K+-ATPase by insulin: why and how? Mol. Cell. Biochem. 182, 121–133. doi: 10.1023/A:1006805510749
Tang, M. J., and McDonough, A. A. (1992). Low K+ increases Na(+)-K(+)-ATPase alpha- and beta-subunit mRNA and protein abundance in cultured renal proximal tubule cells. Am. J. Physiol. 263(2 Pt 1), C436–C442.
Tang, M. J., Wang, Y. K., and Lin, H. H. (1995). Butyrate and TGF-beta downregulate Na,K-ATPase expression in cultured proximal tubule cells. Biochem. Biophys. Res. Commun. 215, 57–66. doi: 10.1006/bbrc.1995.2433
Taub, M., Borsick, M., Geisel, J., Matlhagela, K., Rajkhowa, T., and Allen, C. (2004). Regulation of the Na,K-ATPase in MDCK cells by prostaglandin E1: a role for calcium as well as cAMP. Exp. Cell Res. 299, 1–14. doi: 10.1016/j.yexcr.2004.04.046
Taub, M. L., Wang, Y., Yang, I. S., Fiorella, P., and Lee, S. M. (1992). Regulation of the Na,K-ATPase activity of Madin-Darby canine kidney cells in defined medium by prostaglandin E1 and 8-bromocyclic AMP. J. Cell. Physiol. 151, 337–346. doi: 10.1002/jcp.1041510215
Therien, A. G., and Blostein, R. (2000). Mechanisms of sodium pump regulation. Am. J. Physiol. Cell Physiol. 279, C541–C566.
Tirupattur, P. R., Ram, J. L., Standley, P. R., and Sowers, J. R. (1993). Regulation of Na+,K(+)-ATPase gene expression by insulin in vascular smooth muscle cells. Am. J. Hypertens. 6(7 Pt 1), 626–629. doi: 10.1093/ajh/6.7.626
Tsuchiya, K., Giebisch, G., and Welling, P. A. (1996). Aldosterone-dependent regulation of Na-K-ATPase subunit mRNA in the rat CCD: competitive PCR analysis. Am. J. Physiol. 271(1 Pt 2), F7–F15.
Turner, N., and Grose, R. (2010). Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 10, 116–129. doi: 10.1038/nrc2780
Vagin, O., Dada, L. A., Tokhtaeva, E., and Sachs, G. (2012). The Na-K-ATPase alpha(1)beta(1) heterodimer as a cell adhesion molecule in epithelia. Am. J. Physiol. Cell Physiol. 302, C1271–C1281. doi: 10.1152/ajpcell.00456.2011
Vagin, O., Tokhtaeva, E., and Sachs, G. (2006). The role of the beta1 subunit of the Na,K-ATPase and its glycosylation in cell-cell adhesion. J. Biol. Chem. 281, 39573–39587. doi: 10.1074/jbc.M606507200
Van Why, S. K., Mann, A. S., Ardito, T., Siegel, N. J., and Kashgarian, M. (1994). Expression and molecular regulation of Na(+)-K(+)-ATPase after renal ischemia. Am. J. Physiol. 267(1 Pt 2), F75–F85.
Vandewalle, C., Van Roy, F., and Berx, G. (2009). The role of the ZEB family of transcription factors in development and disease. Cell. Mol. Life Sci. 66, 773–787. doi: 10.1007/s00018-008-8465-8
Verrey, F., Kraehenbuhl, J. P., and Rossier, B. C. (1989). Aldosterone induces a rapid increase in the rate of Na,K-ATPase gene transcription in cultured kidney cells. Mol. Endocrinol. 3, 1369–1376. doi: 10.1210/mend-3-9-1369
Verrey, F., Schaerer, E., Fuentes, P., Kraehenbuhl, J. P., and Rossier, B. C. (1988). Effects of aldosterone on Na,K-ATPase transcription, mRNAs, and protein synthesis, and on transepithelial Na+ transport in A6 cells. Prog. Clin. Biol. Res. 268B, 463–468.
Verrey, F., Schaerer, E., Zoerkler, P., Paccolat, M. P., Geering, K., Kraehenbuhl, J. P., et al. (1987). Regulation by aldosterone of Na+,K+-ATPase mRNAs, protein synthesis, and sodium transport in cultured kidney cells. J. Cell Biol. 104, 1231–1237. doi: 10.1083/jcb.104.5.1231
Vinciguerra, M., Arnaudeau, S., Mordasini, D., Rousselot, M., Bens, M., Vandewalle, A., et al. (2004). Extracellular hypotonicity increases Na,K-ATPase cell surface expression via enhanced Na+ influx in cultured renal collecting duct cells. J. Am. Soc. Nephrol. 15, 2537–2547. doi: 10.1097/01.ASN.0000139931.81844.10
Vinciguerra, M., Deschênes, G., Hasler, U., Mordasini, D., Rousselot, M., Doucet, A., et al. (2003). Intracellular Na+ controls cell surface expression of Na,K-ATPase via a cAMP-independent PKA pathway in mammalian kidney collecting duct cells. Mol. Biol. Cell 14, 2677–2688. doi: 10.1091/mbc.E02-11-0720
Wang, G., Kawakami, K., and Gick, G. (2007a). Divergent signaling pathways mediate induction of Na,K-ATPase alpha1 and beta1 subunit gene transcription by low potassium. Mol. Cell. Biochem. 294, 73–85. doi: 10.1007/s11010-006-9247-y
Wang, G., Kawakami, K., and Gick, G. (2007b). Regulation of Na,K-ATPase alpha1 subunit gene transcription in response to low K(+): role of CRE/ATF- and GC box-binding proteins. J. Cell. Physiol. 213, 167–176. doi: 10.1002/jcp.21107
Wang, L., Shiraki, A., Itahashi, M., Akane, H., Abe, H., Mitsumori, K., et al. (2013). Aberration in epigenetic gene regulation in hippocampal neurogenesis by developmental exposure to manganese chloride in mice. Toxicol. Sci. 136, 154–165. doi: 10.1093/toxsci/kft183
Wang, Y. B., Leroy, V., Maunsbach, A. B., Doucet, A., Hasler, U., Dizin, E., et al. (2014). Sodium transport is modulated by p38 kinase-dependent cross-talk between ENaC and Na,K-ATPase in collecting duct principal cells. J. Am. Soc. Nephrol. 25, 250–259. doi: 10.1681/ASN.2013040429
Wang, Z. M., and Celsi, G. (1993). Glucocorticoids differentially regulate the mRNA for Na+,K(+)-ATPase isoforms in infant rat heart. Pediatr. Res. 33, 1–4. doi: 10.1203/00006450-199301000-00001
Wang, Z. M., Yasui, M., and Celsi, G. (1994). Glucocorticoids regulate the transcription of Na(+)-K(+)-ATPase genes in the infant rat kidney. Am. J. Physiol. 267(2 Pt 1), C450–C455.
Watanabe, Y., Kawakami, K., Hirayama, Y., and Nagano, K. (1993). Transcription factors positively and negatively regulating the Na,K-ATPase alpha 1 subunit gene. J. Biochem. 114, 849–855.
Wendt, C. H., Gick, G., Sharma, R., Zhuang, Y., Deng, W., and Ingbar, D. H. (2000). Up-regulation of Na,K-ATPase beta 1 transcription by hyperoxia is mediated by SP1/SP3 binding. J. Biol. Chem. 275, 41396–41404. doi: 10.1074/jbc.M004759200
Wendt, C. H., Towle, H., Sharma, R., Duvick, S., Kawakami, K., Gick, G., et al. (1998). Regulation of Na-K-ATPase gene expression by hyperoxia in MDCK cells. Am. J. Physiol. 274(2 Pt 1), C356–C364.
Whorwood, C. B., and Stewart, P. M. (1995). Transcriptional regulation of Na/K-ATPase by corticosteroids, glycyrrhetinic acid and second messenger pathways in rat kidney epithelial cells. J. Mol. Endocrinol. 15, 93–103. doi: 10.1677/jme.0.0150093
Whorwood, C. B., Ricketts, M. L., and Stewart, P. M. (1994). Regulation of sodium-potassium adenosine triphosphate subunit gene expression by corticosteroids and 11 beta-hydroxysteroid dehydrogenase activity. Endocrinology 135, 901–910.
Wu, Y., and Koenig, R. J. (2000). Gene regulation by thyroid hormone. Trends Endocrinol. Metab. 11, 207–211. doi: 10.1016/S1043-2760(00)00263-0
Xie, Z., and Askari, A. (2002). Na(+)/K(+)-ATPase as a signal transducer. Eur. J. Biochem. 269, 2434–2439. doi: 10.1046/j.1432-1033.2002.02910.x
Xue, Z., Li, B., Gu, L., Hu, X., Li, M., Butterworth, R. F., et al. (2010). Increased Na,K-ATPase alpha2 isoform gene expression by ammonia in astrocytes and in brain in vivo. Neurochem. Int. 57, 395–403. doi: 10.1016/j.neuint.2010.04.014
Yagawa, Y., Kawakami, K., and Nagano, K. (1990). Cloning and analysis of the 5′-flanking region of rat Na+/K(+)-ATPase alpha 1 subunit gene. Biochim. Biophys. Acta 1049, 286–292. doi: 10.1016/0167-4781(90)90099-N
Yamamoto, K., Ikeda, U., Okada, K., Saito, T., Kawakami, K., and Shimada, K. (1994). Sodium ion mediated regulation of Na/K-ATPase gene expression in vascular smooth muscle cells. Cardiovasc. Res. 28, 957–962. doi: 10.1093/cvr/28.7.957
Yang, S. J., Liang, H. L., and Wong-Riley, M. T. (2006). Activity-dependent transcriptional regulation of nuclear respiratory factor-1 in cultured rat visual cortical neurons. Neuroscience 141, 1181–1192. doi: 10.1016/j.neuroscience.2006.04.063
Yu, H. Y., Nettikadan, S., Fambrough, D. M., and Takeyasu, K. (1996). Negative transcriptional regulation of the chicken Na+/K(+)-ATPase alpha 1-subunit gene. Biochim. Biophys. Acta 1309, 239–252. doi: 10.1016/S0167-4781(96)00130-3
Yu, L., and Hales, C. A. (2011). Long-term exposure to hypoxia inhibits tumor progression of lung cancer in rats and mice. BMC Cancer 11:331. doi: 10.1186/1471-2407-11-331
Zahler, R., Sun, W., Ardito, T., and Kashgarian, M. (1996). Na-K-ATPase alpha-isoform expression in heart and vascular endothelia: cellular and developmental regulation. Am. J. Physiol. 270(1 Pt 1), C361–C371.
Keywords: Na,K-ATPase α-subunit, Na,K-ATPase β-subunit, transcription, promoter analysis, cancer, epigenetics
Citation: Li Z and Langhans SA (2015) Transcriptional regulators of Na,K-ATPase subunits. Front. Cell Dev. Biol. 3:66. doi: 10.3389/fcell.2015.00066
Received: 19 July 2015; Accepted: 05 October 2015;
Published: 26 October 2015.
Edited by:
Ali Mobasheri, University of Surrey, UKReviewed by:
Diego Alvarez De La Rosa, Universidad de La Laguna, SpainPablo Martin-Vasallo, Universidad de La Laguna, Spain
Copyright © 2015 Li and Langhans. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sigrid A. Langhans, langhans@nemoursresearch.org
 Zhiqin Li
Zhiqin Li Sigrid A. Langhans
Sigrid A. Langhans