Stereochemical course of hydrolytic reaction catalyzed by alpha-galactosidase from cold adaptable marine bacterium of genus Pseudoalteromonas
- G.B. Elyakov Pacific Institute of Bioorganic Chemistry, Far Eastern Branch, Russian Academy of Sciences, Vladivostok, Russia
The recombinant α-galactosidase of the marine bacterium (α-PsGal) was synthesized with the use of the plasmid 40Gal, consisting of plasmid pET-40b (+) (Novagen) and the gene corresponding to the open reading frame of the mature α-galactosidase of marine bacterium Pseudoalteromonas sp. KMM 701, transformed into the Escherichia coli Rosetta(DE3) cells. In order to understand the mechanism of action, the stereochemistry of hydrolysis of 4-nitrophenyl α-D-galactopyranoside (4-NPGP) by α-PsGal was measured by 1H NMR spectroscopy. The kinetics of formation of α- and β-anomer of galactose showed that α-anomer initially formed and accumulated, and then an appreciable amount of β-anomer appeared as a result of mutarotation. The data clearly show that the enzymatic hydrolysis of 4-NPGP proceeds with the retention of anomeric configuration, probably, due to a double displacement mechanism of reaction.
Introduction
α-Galactosidase (α-D-galactoside galactohydrolase; EC 3.2.1.22) catalyzes the hydrolysis of terminal α-linked galactose residues in oligosaccharides and polymeric galactomannans. They have found a number of useful potential biotechnological and medical applications. An interesting application of α-galactosidase in biomedicine is the processing of B-red blood cells for conversion into universal O-red blood cells (Olsson et al., 2004). In medicine, it plays a crucial role in the treatment of Fabry's disease (Breunig et al., 2003) and for overcoming xenorejection for xenotransplantation (Ezzelarab and Cooper, 2005). α-Galactosidases are used in the food and feed industry to improve the quality of products containing oligosaccharides of raffinose series (Anisha and Prema, 2008).
α-Galactosidases are wide distributed in marine bacteria, especially among γ-proteobacteria (Ivanova et al., 1998) and Bacteroidetes (Bakunina et al., 2012), however, an α-galactosidase isolated from the marine bacterium Pseudoalteromonas sp. KMM 701 was the first biochemicaly characterized marine α-galactosidase. It catalyzes the hydrolysis of α-galactose residues from non-reducing end of B-trisaccharides and is capable for reducing a serological activity of B-red blood cells at neutral pH. Furthermore, the α-galactosidase is able to interrupt the adhesion of pathogens to human buccal epithelium (Balabanova et al., 2010). These properties showed the great therapeutic potential and opened up broad prospects for application of the enzyme in medicine. The α-galactosidase gene was isolated (GenBank DQ530422) and the amino acid sequence was reconstructed (UniProt Q19AX0). The marine bacterium Pseudoalteromonas sp. KMM 701 α-galactosidase has been found to belong to GH36 family in CAZy classification, according to its amino acids sequence (Balabanova et al., 2010).
The question of stereochemical outcome of enzymatic hydrolysis of glycosides is principal in the understanding of a catalytic mechanism and in classification of glycoside hydrolases. 1H NMR spectroscopy is a direct way for determining the stereochemical configuration of the product anomers.
The paper aims to produce the recombinant α-galactosidase of marine bacterium Pseudoalteromonas sp. KMM 701 (α-PsGal) and investigate stereochemistry of the glycoside hydrolysis for understanding the enzymatic mechanism of action.
Materials and Methods
Rcombinant α-Galactosidase Construction
For the α-PsGal gene amplification, Encyclo Taq Polymerase (Eurogene), genomic DNA of the marine bacterium Pseudoalteromonas sp. KMM 701 and the gene-specific the upstream primer, G2-NcoI-for40: 5′-ATTACCATGGATGACGACGACAAGGCCGACACTAAATCATTTTATCGATTAGACA-3′, and the downstream primer, G3-SalI-rev40: 5′-ACACGTCGACTTACGCTTTGTTGAGCTCAAATATAAGC-3′ were used. The resultant PCR products were purified with agarose gel (Helicon). PCR was carried out in automatic amplifier “Eppendorf.” Restriction endonucleases and T4 DNA ligase were purchased from “Thermo Scientific.” The restricted PCR product and plasmid pET-40b (+) were purified from agarose gel with the use of a “Qiagen” column. The recombinant insert of the resultant plasmid 40Gal was sequenced using the automated PE/ABI 310 DNA sequencer and the PE/ABI-ABI PRISM BigDye Terminator cycle sequencing Ready Reaction Kit (PE Applied Biosystems). Preparation of Escherichia coli competent cells and heat shock transformation were carried out according to the standard methods (Sambrook et al., 1989).
For producing α-PsGal of marine bacterium Pseudoalteromonas sp. KMM 701 the recombinant plasmid 40Gal was transformed in to the E. coli Rosetta(DE3) cells. The obtained recombinant cells were grown on LB plate containing 25 μg/mL of kanamycin overnight at 37°C. A single colony was picked and grown at 200 rpt in 20 mL of LB, with 25 μg/mL of kanamycin at 37°C for 12 h. Overnight culture was transferred to 1 L of fresh LB with 25 μg/mL of kanamycin. When the cell density reached an OD600 of 0.6–0.8, 0.2 mM IPTG was added to induce the expression and the incubation was continued at 16°C up to 12 h at 200 rpt. The E. coli Rosetta(DE3) cells were transformed with the pET-40b (+) plasmid as a control.
Recombinant α-Galactosidase Purification
All purification steps were carried out at +6°C. After harvesting, the transgenic E. coli Rosetta (DE3)/40Gal cells were resuspended in 200 mL of buffer A (0.01 M NaH2PO4, pH 7.7, 0.01% NaN3) and sonificated by ultrasonic treatment, then centrifuged at 10,000 g for 30 min. The supernatant was applied to a column (2.5 × 30 cm) of Macro-Prep (BioRad). Elution of the protein was performed with the use a linear gradient of NaCl concentration (0.05–0.3 M) in the buffer A. The enzymatic active fractions were collected and applied to a column (1 × 2.5 cm) of Ni-NTA agarose (Qiagen). Elution of the protein was carried out in buffer B (0.01 M NaH2PO4, pH 7.7, 0.01% NaN3, 0.04 M EDTA). The active fractions were collected and desalted with the use of Bio-Scale™ Mini Macro-Prep® High Q 1 mL cartridge, then incubated with enterokinase (Invitrogen) at 21°C for 15 h to cleave N-terminal plasmid overhang from the chimeric protein (32.5 kDa). Then, the protein solution is applied to a gel filtration column (1.5 × 170 cm) of Superdex 200 (Sigma) equilibrated with the buffer C (0.01 M NaH2PO4, pH 7.7, 0.01% NaN3, 0.1 M NaCl). All purification steps were examined by SDS-PAGE. The concentration of the protein was determined according to Bredford (1976). The obtained recombinant polypeptide is determined by the first 10 amino acids on an automatic sequencer Procise model 492 (Applied Biosystems, USA). All biochemicals and reagents were from “Thermo scientific” and “Sigma-Aldrich.”
Enzyme Essay
Enzyme activity of the α-PsGal was determined with 3.3 mM of 4-NPGP in 50 mM sodium phosphate buffer pH 7.5, at 20°C. One unit of the α-galactosidase activity was defined as amount of the enzyme that hydrolyzed 1 μmol of 4-NPGP per min. Total quantity of p-nitrophenol was determined spectrophotometrically at 400 nm (ε400 = 18300 M−1 cm−1).
Molecular Mass Determination
The molecular size of the active α-PsGal after treatment of enterokinase was determined by gel filtration on the column of Superdex 200 (Sigma) (1.5 × 170 cm) in the buffer A at a flow of 0.16 mL/min at 6°C and calibrated using Bio-Rad standard molecular weight markers: Thyroglobulin bovine (670 kDa), γ-globulin bovine (158 kDa), Ovalbumin chicken (44 kDa), Mioglobin horse (17 kDa), Vitamin B12 (1.35 kDa). The molecular mass of the enzyme was determined by 12.5 % polyacrylamide gel electrophoresis (PAGE). The protein preparation was mixed with Laemmli sample buffer with heat treatment at 95°C, and then applied to PAGE. The SDS-PAGE gels were stained according to Laemmli (1970).
1H NMR Spectra Recording and Analysis
Determination of the anomeric center configuration of the product of the hydrolysis reaction was monitored by 1H NMR spectroscopy. The experiment was performed at 20°C with the use of NMR DRX-500 spectrometer (Bruker, Germany). 1H NMR spectra were acquired over 32,000 data points using a spectral width 5000 Hz.
Prior to analysis by NMR, 0.6 mL 50 mM sodium phosphate buffer, pH 7.5, and substrate were dried on rotary evaporator and dissolved in 0.6 mL D2O. The deuterium-exchanged α-PsGal was obtained on vivaspin turbo 10 K MWCO (Sartorius, Germany). Chemical shifts were measured relative to an external standard—acetone (trace) in D2O. CH3—signal acetone was set to δ = 2.22 ppm. After recording of the original spectrum at τ = 0 min 6.0 mmol of the deuterium-exchanged p-nitrophenyl galactopyranoside in 0.6 mL of D2O, 0.1 mL of the deuterium-exchanged α-PsGal containing 1.14 U was added for initiation of the reaction. 1H NMR spectra were recorded automatically at intervals of 3 min for 60 min from the start of the reaction.
1H NMR anomer signals (αH-1) of α-4-NPGP, α-galactopyranose, and (β H-1) of β-galactopyranose, as well as proton signals of free 4-nitrophenol ring were analyzed and integrated by the standard software “TopSpin 3.2.” Integral intensities of signals of each anomer were calculated as a percentage of the total integral intensities of all signals of anomers and were plotted depend on time. The degree of substrate hydrolysis (4-NPGP) was defined as a percent ratio of the 1H NMR integral intensities of proton signals of free 4-nitrophenol ring at 8.109 ppm to proton signals of 4-NPGP at 8.275 ppm and also was plotted depend on time.
Results
Recombinant α-Galactosidase Production and Characterization
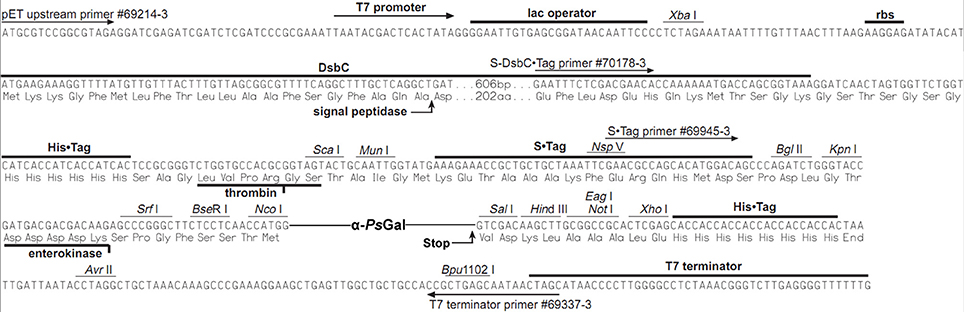
The recombinant plasmid 40Gal of 8306 base pairs (bp) for the synthesis of the recombinant protein α-PsGal consisted of the NcoI/SalI-fragment of plasmid pET-40b (+) (Novagen) and the gene of 2130 bp corresponding to the open reading frame of the mature α-galactosidase of marine bacterium Pseudoalteromonas sp. KMM 701 (Figure 1).
The IPTG concentration of 0.2 mM, strain cultivation temperature of 16°C and duration of the cultivation of 12 h at 200 rpt were optimal conditions for the α-PsGal expression. The induced α-PsGal expression in 1 L culture of E.coli Rosetta(DE3)/40Gal cells yielded up to 10 mg of the functionally active protein with the specific activity 160 U/mg at the final purification step. α-PsGal has been found to provide the complete inactivation of serological activity of B-red blood cells and its conversion into the O-red blood cells in the same condition as for the native enzyme (data not shown).
PAGE-SDS showed a single band of the purified recombinant protein with a molecular mass of approximately 80 kDa corresponding to the one subunit of α-PsGal (Figure 2).
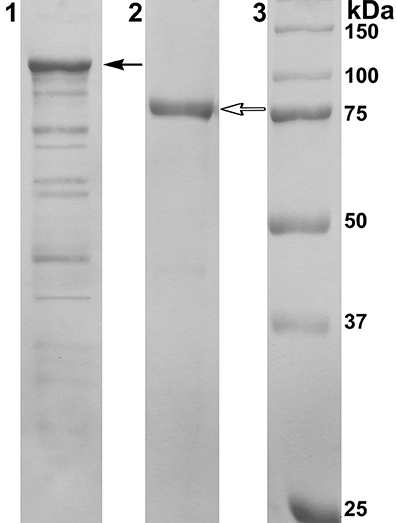
Figure 2. Results of 12.5% SDS-PAGE electrophoresis. Line 1: Cell extract of E. coli Rosetta(DE3)/40Gal; Line 2: α-PsGal after final purification step; Line 3: MW-standards. Filled arrow indicates the band corresponding to the chimeric protein with DsbC overhang (112.5 kDa); empty arrow indicates the band corresponding to the mature protein after enterokinase treatment (80 kDa).
The molecular mass of the mature protein determined by gel filtration was to be 160 kDa, suggesting the two-subunit molecular organization of the active α-PsGal.
1H NMR Spectra Recording and Analysis
1H NMR spectra of deuterium-exchanged 4-NPGP and products of it's hydrolysis under the action of the recombinant α-PsGal are shown on Figure 3.
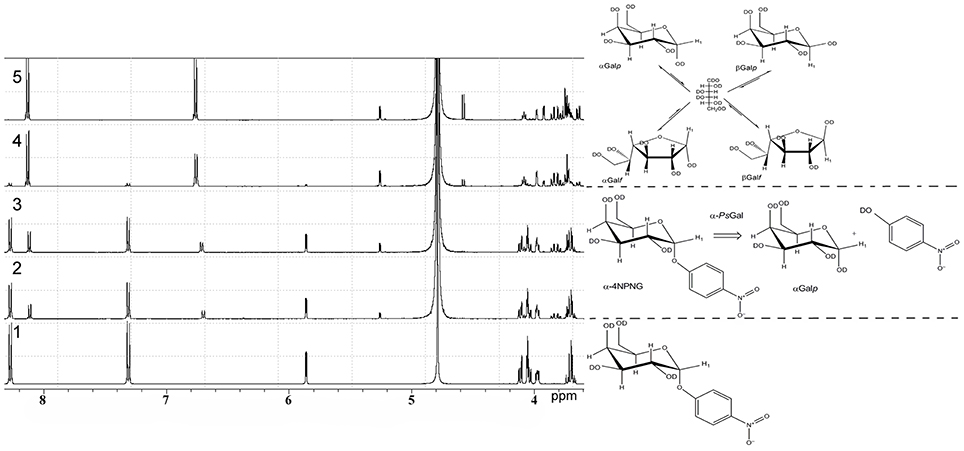
Figure 3. 1H NMR analysis of the stereo selectivity for the recombinant α-PsGal catalyzed reaction. One-dimensional 1H NMR spectra of the deuterium-exchanged 4-NPGP prior to enzyme addition, at τ = 0 min (spectrum 1), at τ = 3 min (spectrum 2); at τ = 6 min (spectrum 3); at τ = 24 min (spectrum 4); at τ = 60 min (spectrum 5) after initiation of the reaction. (in 50 mM sodium phosphate buffer pH 7.5, at 20°C, D2O).
In 1H NMR spectrum 4-NPGP (Figure 3, spectrum 1, formula of the compound is shown on the right) one signal was a doublet at 5.84 ppm [J(1, 2) = 3.43 Hz] and two groups signals at 8.28 and 7.31 ppm corresponded to αH-1 anomer and 4-nitrophenol ring protons in 4-NPGP, respectively. Moreover, signals in region of resonance from 4.13 to 3.96 ppm are observed. These signals corresponded to H-2, H-3, H-4, H-5, and H-6 protons of α-galactopyranose in 4-NPGP (Figure 3, spectrum 1).
After addition of the enzyme, the doublet signal was appeared at 5.26 ppm [J(1, 2) = 3.8 Hz] during the first 3 min (Figure 3, spectrum 2, reaction equation is shown on the right). The doublet was identical in chemical shift to the αH-1 resonance of free α-D-galactopyranose (Angyal and Pickles, 1972). Signals arising at 8.11 and 6.68 ppm related to protons of free 4-nitrophenol formed through the reaction. Decrease in the amplitude of signals at 8.28 and 7.31 ppm indicates disappearance the substrate. A doublet signal at 4.57 ppm [J(1, 2) = 7.8 Hz], which was identical in chemical shift to the H-1 resonance of free β-D-galactopyranose (β H-1) (Angyal and Pickles, 1972), appeared after 10 min of the reaction in accordance with the time of spontaneous mutarotation (Figure 3, spectrum 4). Signals of protons H-2, H-3, H-4, H-5, and H-6 of free α- and β-D-galactopyranose occurred at 3.40–4.20 ppm, minor signals at 5.21 ppm [J(1, 2) = 3.30] rising at late time of reaction corresponded to β-D-galactofuranose (Angyal and Pickles, 1972) (Figure 3, spectra 4 and 5, equation of equilibrium of D-galactose is shown on the right Zhu et al., 2001). Signals in the anomeric region, which could indicate the emergence of new products of the reaction or formation of O-glycosidic bond in the spectra were not found.
NMR spectroscopic monitoring of the hydrolysis of 4-NPGP by α-PsGal is shown on Figure 4.
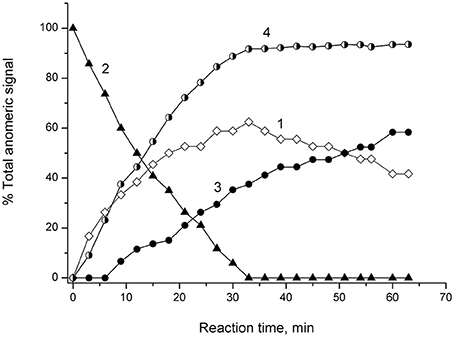
Figure 4. 1H NMR Monitoring of reaction of α-4-NPGP hydrolysis catalyzed by α-PsGal in 50 mM sodium phosphate buffer pH 7.5, at 20°C, D2O. Experimental time-dependent changes in integral intensity of anomer signals. Curve 1: αH-1 of reaction product of galatopyranose at 5.26 ppm; Curve 2: αH-1 of 4-NPGP at 5.84 ppm; Curve 3: β H-1 mutarotation product of α-galatopyranose at 4.57 ppm; Curve 4: shows the change in the degree of hydrolysis of the substrate vs. time.
Signal at 5.26 ppm the α-D-galactopyranose αH-1 appeared at the first minutes after the addition of α-PsGal (Figure 4, curve 1). Disappearance of doublet at 5.84 ppm of αH-1 of 4-NPGP (Figure 4, curve 2) simultaneously with appearance of the signal at 5.26 ppm of αH-1 of α-D-galactopyranose increasing in intensity (Figure 4, curve 1) were observed within next 30 min. On curve of the time-dependent changes in signal amplitude of β H-1 at 4.57 ppm lag-period were observed during 9 min (Figure 4, curve 3), when degree of the substrate hydrolysis reached up to ~30% (Figure 4, curve 4). After 33 min, the degree of substrate hydrolysis reached up to 98–100% (Figure 4, curve 4). After 60 min, the ratio of integral intensities of αH-1 and β H-1 signal was 40:60. Typical equilibrium ratio 31:69 between αH-1 and β H-1 D-galactopyranose anomers (Angyal and Pickles, 1972; Zhu et al., 2001) was formed for 180 min from the start of the reaction in experimental conditions pH 7.5 and 20°C.
Discussion
Marine bacteria have been found to be a good source for α-galatosidases, which are stable at low temperatures and hence find use in medical and food industry applications. These enzymes appear to be also distinct from their terrestrial counterparts in substrate preference and catalytic efficiency. The first example was the enzyme from the marine bacterium Pseudoalteromonas sp. KMM 701 that displayed a good stability at low temperatures (20°C for 24 h) and neutral pH (pH 6.7–7.7) and was 4-fold more efficient than the α-galactosidase from green coffee beans for the B-red blood cells conversion into O-red blood cells (Bakunina et al., 1998). Furthermore, a single cold-active α-galactosidase isolated from the Antarctic Bacillus sp. LX-1 was reported to be a promising biocatalyst for soybean processing in the food and feed industry (Lee et al., 2012).
Here, we have firstly developed a method for the production of the completely soluble highly active recombinant α-galactosidase of the psychrotrophic marine bacterium Pseudoalteromonas sp. KMM 701 (α-PsGal) in E. coli. The use of E. coli signal peptide included in the expression plasmid pET40b(+) resulted in overproduction of α-PsGal. It should be noted that the step of the hybrid protein 6xHis-Dsb-α-PsGal incubation with enterokinase could be optionally due to the absence of any effect of 32.5 kDa plasmid fusion protein including N-terminal His-tag on the α-PsGal activity (Figures 1, 2 plasmid scheme and electrophoregram, respectively).
The configuration of the anomeric center of galactose, which was a reaction product of the of 4-NPGP hydrolysed of by α-PsGal, was monitored by 1H NMR spectroscopy. Upon occurrence of signals of anomeric protons at the initial time of reaction one can be appreciate stereochemical configuration of the anomeric center of the formed product. In 1H NMR spectra of the initial time of the reaction the resonance of the α-anomeric center of galactose is clearly observed (Figure 3). β-Anomeric as a result of galactose mutarotation appeared only after 10 min from the beginning of hydrolysis reaction of 4-NPGP under the action of α-PsGal (Figure 4). Mutarotation proceeded until the anomer ratio of 40% α- and 60% β-galactopyranose was established after 60 min, when the reaction was stopped (Figure 4). These observations indicate that the primary product must be α-D-galactopyranose, and the enzyme acts on a mechanism leading to retaining of the anomer configuration of the substrate. Based on these data, we conclude that α-PsGal catalyzes the hydrolysis of glycosidic bound with double displacement mechanism. This mechanism involves two catalytic residues; one responsible for the protonation of the glycosidic oxygen, and the other for stabilization of a carbocationic intermediate. The glycosyl-enzyme intermediate is decomposed by the transfer of a glycosyl moiety to an acceptor molecule, which is in the case of hydrolysis a molecule of water (Sinnott, 1990). The absence of signals corresponding to the anomeric protons involved in the formation of O-glycosidic bonds indicates the lack of transglycosylation in the conditions of this experiment. It is known that initiation of transglycosylation reaction of galactosidases requires high concentrations of substrates (donors and acceptors) and enzymes, low temperatures and the presence of special solvents (Matsuo et al., 1997; Nieder et al., 2003; Goulas et al., 2009).
Considering the postulation that glycoside hydrolases grouped into the same family shared a common catalytic mechanism (Henrissat and Bairoch, 1993, 1996), all enzymes of GH36 family are retaining glycoside hydrolases. At first the technique of 1H NMR experiments was applied for investigation of GH36 enzyme reaction stereochemistry of α-galactosidase from thermopile bacterium Thermotoga maritima (Comfort et al., 2007). It was unequivocally shown, that α-galactosidase from T. maritima was a retaining glycoside hydrolase. However, this thermophile enzyme has low identity (19.1%) of amino acids sequence with psychrophillic marine Pseudoalteromonas sp. KMM 701 (Balabanova et al., 2006). The related catalytic mechanism was shown for GH27 α-galactosidases from Streptomyces griseoalbus (Anisha et al., 2011), fungus Thermomyces lanuginosus (Puchart et al., 2000), Phanerochaete chrysosporium (Brumer et al., 1999), Trichoderma reesei (Shabalin et al., 2002). Catalytic mechanism of human α-galactosidase was also studied with 1H NMR experiments (Guce et al., 2010).
1H NMR experiments were successfully applied for direct detecting of catalytic mechanism of novel inverting GH110 family α-galactosidase from Bacteroides fragilis (Liu et al., 2008), as well as for detecting a retaining and inverting properties of GH97 α-galactosidase and α-glucosidase from Bacteroides thetaiotaomicron (Gloster et al., 2008).
Conclusion
For the first time the completely soluble and functional recombinant cold-active α-galactosidase α-PsGal of marine bacterium was successfully produced in E. coli with a high level of yield and specific activity. Based on the results of 1H NMR experiments, it was demonstrated that α-PsGal catalyzes the hydrolysis of substrate with retaining of the anomeric configuration. It is evident that the marine α-galactosidase catalyzes the substrate hydrolyses with double displacement mechanism as all classical glycoside hydrolases of GH36 family of clan-D GH (Comfort et al., 2007; Cantarel et al., 2009).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by grants from RFBR 13-04-00806, FEB RAS, and the RAS Presidium under the project “Molecular and Cellular Biology” 12-II6-10.
References
Angyal, S. J., and Pickles, V. A. (1972). Equilibria between pyranoses and furanoses. II. Aust. J. Chem. 25, 1695–1710. doi: 10.1071/CH9721695
Anisha, G. S., John, R. P., and Prema, P. (2011). Substrate specificities and mechanism of action of multiple α-galactosidases from Streptomyces griseoloalbus. Food Chem. 124, 349–353. doi: 10.1016/j.foodchem.2010.06.001
Anisha, G. S., and Prema, P. (2008). Reduction of non-digestible oligosaccharides in horse gram and green gram flours using crude α-galactosidase from Streptomyces griseoloalbus. Food Chem. 106, 1175–1179. doi: 10.1016/j.foodchem.2007.07.058
Bakunina, I. Y., Nedashkovskaya, O. I., Kim, S. B., Zvyagintseva, T. N., and Mikhailov, V. V. (2012). Diversity of glycosidase activities in the bacteria of the phylum bacteroidetes isolated from marine algae. Microbiology 81, 688–695. doi: 10.1134/S0026261712060033
Bakunina, I. Y., Sova, V. V., Nedashkovskaya, O. I., Kuhlmann, R. A., Likhosherstov, L. M., Martynova, M. D., et al. (1998). α-galactosidase of the marine bacterium Pseudoalteromonas sp. KMM 701. Biochemistry (Mosc) 63, 1209–1215.
Balabanova, L. A., Bakunina, I. Y., Nedashkovskaya, O. I., Makarenkova, I. D., Zaporozhets, T. S., Besednova, N. N., et al. (2010). Molecular characterization and therapeutic potential of a marine bacterium Pseudoalteromonas sp. KMM 701 α-galactosidase. Mar. Biotechnol. 12, 111–120. doi: 10.1007/s10126-009-9205-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Balabanova, L. A., Likshatskaya, G. N., Bakunina, I. Y., Petruk, S. V., Kozhemyako, V. B., and Rasskazov, V. A. (2006). Primary structure and homology modeling of the novel α-galactosidase from marine bacterium. FEBS J. 273, 262–263. doi: 10.1111/j.1742-4658.2006.05277.x
Bredford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Breunig, F., Knoll, A., and Wanner, C. (2003). Enzyme replacement therapy in Fabry disease: clinical implications. Curr. Opin. Nephrol. Hypertens. 12, 491–495. doi: 10.1097/00041552-200309000-00002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brumer, H., Sims, P. F. G., and Sinnott, M. L. (1999). Lignocellulose degradation by Phanerochaete chrysosporium: purification and characterization of the main α-galactosidase. Biochem. J. 339, 43–53. doi: 10.1042/0264-6021:3390043
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Comfort, D. A., Bobrov, K. S., Ivanen, D. R., Shabalin, K. A., Harris, J. M., Kulminskaya, A. A., et al. (2007). Biochemical analysis of Thermotoga maritima GH-36 α-galactosidase (TmGalA) confirms the mechanistic commonality of clan GH-D glycoside hydrolases. Biochemistry 46, 3319–3330. doi: 10.1021/bi061521n
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ezzelarab, M., and Cooper, D. K. C. (2005). Reducing Gal expression on the pig organ – a retrospective review. Xenotransplantation 12, 278–285. doi: 10.1111/j.1399-3089.2005.00236.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gloster, T. M., Turkenburg, J. P., Potts, J. R., Henrissat, B., and Davies, G. J. (2008). Divergence of catalytic mechanism within a glycosidase family provides insight into evolution of carbohydrate metabolism by human gut flora. Chem. Biol. 15, 1058–1067. doi: 10.1016/j.chembiol.2008.09.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Goulas, T., Goulas, A., Tzortzis, G., and Gibson, G. R. (2009). A novel α-galactosidase from Bifidobacterium bifidum with transgalactosylating properties: gene molecular cloning and heterologous expression. Appl. Microbiol. Biotechnol. 82, 471–477. doi: 10.1007/s00253-008-1750-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guce, A. I., Clark, N. E., Salgado, E. N., Ivanen, D. R., Kulminskaya, A. A., Brumer, H., et al. (2010). Catalytic mechanism of human α-galactosidase. J. Biol. Chem. 285, 3625–3632. doi: 10.1074/jbc.M109.060145
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Henrissat, B., and Bairoch, A. (1993). New families in the classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem. J. 293, 781–788.
Henrissat, B., and Bairoch, A. (1996). Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316, 695–696.
Ivanova, E. P., Bakunina, I. Y., Nedashkovskaya, O. I., Gorshkova, N. M., Mikhailov, V. V., and Elyakova, L. A. (1998). Incidence of marine microorganisms producing β-N-acetylglucosaminidases, α-galactosidases and α-N-acetylgalactosaminidases. Russ. J. Mar. Biol. 24, 365–372.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, J., Park, I., and Cho, J. (2012). Production and partial characterization of α-galactosidase activity from an Antarctic bacterial isolate, Bacillus sp. LX-1. Afr. J. Biotechnol. 11, 12396–12405. doi: 10.5897/AJB12.1219
Liu, Q. P., Yuan, H., Bennett, E. P., Levery, S. B., Nudelman, E., Spence, J., et al. (2008). Identification of a GH110 Subfamily of α-1,3-galactosidases novel enzymes for removal of the α3Gal xenotransplantation antigen. J. Biol. Chem. 283, 8545–8554. doi: 10.1074/jbc.M709020200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matsuo, I., Fujimoto, H., Isomura, M., and Katsumi, A. (1997). Chemoenzymatic synthesis of Gal-α–1,3-gal, Gal-α-1,3-β 1,4GlcNAc, and their PEG-conjugates. Bioorg. Med. Chem. 7, 255–258. doi: 10.1016/S0960-894X(97)00017-6
Nieder, V., Marx, S. P., Gutiérrez Gallego, R., Kamerling, J. P., Johannes, F. G., Vliegenthart, J. F. G., et al. (2003). Synthesis of nucleotide-activated disaccharides with β-galactosidase from Bacillus circulans and α-galactosidase from Bifidobacterium adolescentis. J Mol. Cat. B: Enzym. 21, 157–166. doi: 10.1016/S1381-1177(02)00123-6
Olsson, M. L., Hilla, C. A., de laVega, H., Liu, P. Q., Stroud, M. R., Valdinocci, J., et al. (2004). Universal red blood cells-enzymatic conversion of blood group A and B antigens. Transfus. Clin. Biol. 11, 33–39. doi: 10.1016/j.tracli.2003.12.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Puchart, V., Vrsanska, M., Bhat, M. K., and Biely, P. (2000). Purification and characterization of α-galactosidase from a thermophilic fungus Thermomyces lanuginosus. Biochim. Biophys. Acta 1524, 27–37. doi: 10.1016/S0304-4165(00)00138-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 253.
Shabalin, K. A., Kulminskaya, A. A., Savel'ev, A. N., Shishlyannikov, S. M., and Neustroev, K. N. (2002). Enzymatic properties of α-galactosidase from Trichoderma reesei in the hydrolysis of galactooligosaccharides. Enzyme Microb. Technol. 30, 231–239. doi: 10.1016/S0141-0229(01)00482-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sinnott, M. L. (1990). Catalytic mechanisms of enzymatic glycosyl transfer. Chem. Rev. 90, 1171–1202. doi: 10.1021/cr00105a006
Zhu, Y., Zajicek, J., and Serianni, A. S. (2001). Acyclic forms of [1-13C] aldohexoses in aqueous solution: quantitation by 13C NMR and deuterium isotope effects on tautomeric equilibria. J. Org. Chem. 66, 6244–6251. doi: 10.1021/jo010541m
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Recombinant retaining α-galactosidase, marine bacterium, 1H NMR spectroscopy
Citation: Bakunina IY, Balabanova LA, Golotin VA, Slepchenko LV, Isakov VV and Rasskazov VA (2014) Stereochemical course of hydrolytic reaction catalyzed by alpha-galactosidase from cold adaptable marine bacterium of genus Pseudoalteromonas. Front. Chem. 2:89. doi: 10.3389/fchem.2014.00089
Received: 30 June 2014; Accepted: 25 September 2014;
Published online: 13 October 2014.
Edited by:
Ali Tavassoli, University of Southampton, UKReviewed by:
Rahman M. Mizanur, US Army Medical Research Institute of Infectious Diseases, USARui Zhao, Chinese Academy of Sciences, China
Copyright © 2014 Bakunina, Balabanova, Golotin, Slepchenko, Isakov and Rasskazov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Irina Y. Bakunina, Laboratory of Enzyme Chemistry, G.B. Elyakov Pacific Institute of Bioorganic Chemistry, Far Eastern Branch, Russian Academy of Sciences, Prospect 100 let Vladivostoku, 159, Vladivostok 690022, Russia e-mail: bakun@list.ru
 Irina Y. Bakunina
Irina Y. Bakunina Larissa A. Balabanova
Larissa A. Balabanova Vasiliy A. Golotin
Vasiliy A. Golotin Lyubov V. Slepchenko
Lyubov V. Slepchenko  Valeriy A. Rasskazov
Valeriy A. Rasskazov