Comparative genomics evidence that only protein toxins are tagging bad bugs
- Unité de Recherche en Maladies Infectieuses Tropical Emergentes (URMITE), CNRS–IRD UMR 6236-198, Université de la Méditerranée, Marseille, France
The term toxin was introduced by Roux and Yersin and describes macromolecular substances that, when produced during infection or when introduced parenterally or orally, cause an impairment of physiological functions that lead to disease or to the death of the infected organism. Long after the discovery of toxins, early genetic studies on bacterial virulence demonstrated that removing a certain number of genes from pathogenic bacteria decreases their capacity to infect hosts. Each of the removed factors was therefore referred to as a “virulence factor,” and it was speculated that non-pathogenic bacteria lack such supplementary factors. However, many recent comparative studies demonstrate that the specialization of bacteria to eukaryotic hosts is associated with massive gene loss. We recently demonstrated that the only features that seem to characterize 12 epidemic bacteria are toxin–antitoxin (TA) modules, which are addiction molecules in host bacteria. In this study, we investigated if protein toxins are indeed the only molecules specific to pathogenic bacteria by comparing 14 epidemic bacterial killers (“bad bugs”) with their 14 closest non-epidemic relatives (“controls”). We found protein toxins in significantly more elevated numbers in all of the “bad bugs.” For the first time, statistical principal components analysis, including genome size, GC%, TA modules, restriction enzymes, and toxins, revealed that toxins are the only proteins other than TA modules that are correlated with the pathogenic character of bacteria. Moreover, intracellular toxins appear to be more correlated with the pathogenic character of bacteria than secreted toxins. In conclusion, we hypothesize that the only truly identifiable phenomena, witnessing the convergent evolution of the most pathogenic bacteria for humans are the loss of metabolic activities, i.e., the outcome of the loss of regulatory and transcription factors and the presence of protein toxins, alone, or coupled as TA modules.
Introduction
The term toxin was introduced 123 years ago by Roux and Yersin to describe macromolecular substances that, when produced during infection or when introduced parenterally or orally, cause an impairment of physiological functions that leads to disease or to the death of the infected organism (Alouf, 2000). Since 1884, 323 toxins have been identified (Figure 1). Toxins are mainly classified as bacterial protein toxins/exotoxins or toxic lipopolysaccharide complexes (LPSs)/endotoxins. Exotoxins are usually secreted by living bacteria during exponential growth. The production of the toxin is generally specific to a particular bacterial species that produces the disease associated with the toxin. For example, only Clostridium tetani produces Tetanus toxin, and only Corynebacterium diphtheriae produces Diphtheria toxin. Toxins have a chemical, quantifiable action that establishes an aggressive frontal assault strategy during microbial pathogenesis. In contrast, LPSs establish a non-specific stealth assault that subverts the host’s immune response (Merrell and Falkow, 2004).
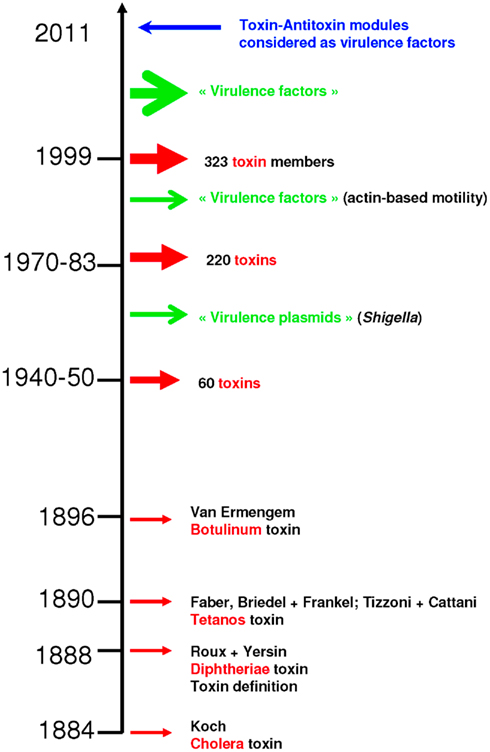
Figure 1. Discovery of toxins. The first toxin was identified in 1884, while toxin–antitoxin modules were considered as virulence factors for the first time in 2011. Chronological data were retrieved from Alouf (2000).
Long after the discovery of toxins, early genetic studies on bacterial virulence demonstrated that removing a certain number of genes from pathogenic bacteria decreases their capacity to infect hosts. Each of the removed factors was therefore referred to as a “virulence factor” (Lawrence, 1999a; Dobrindt et al., 2004; Ochman et al., 2005), and it was speculated that non-pathogenic bacteria lack such supplementary factors (Lawrence, 1999a; Ochman et al., 2005). However, many recent comparative studies demonstrate that specialization of bacteria to eukaryotic hosts is associated with massive gene loss (Nierman et al., 2004; Merhej et al., 2009) and even loss of identified “virulence factors” (Audic et al., 2007; Georgiades and Raoult, 2011a,b). Rickettsiae constitute the most representative example of this observation. Genomic analysis of rickettsial species reveals that a shift to pathogenicity does not require acquisition of new genes. Rather, gene loss is implicated in the emergence of their virulence (Andersson and Kurland, 1998; Andersson and Andersson, 1999; Moran, 2002; Blanc et al., 2007; Darby et al., 2007; Fournier et al., 2009; Merhej and Raoult, 2010). Furthermore, in a recent study in our laboratory comparing the 12 most dangerous epidemic bacteria for humans with their closest sequenced non-epidemic relatives, we demonstrated that the epidemic bacteria (“bad bugs”) have significantly smaller genomes, resulting from degraded recombination and repair systems, while no significant differences were observed concerning characteristics that were considered to play a role in pathogenesis (Georgiades and Raoult, 2011b). In the latter study, the only features that appear to characterize epidemic bacteria are, surprisingly, toxin–antitoxin (TA) modules (Figure 1; Georgiades and Raoult, 2011b). Other than their roles as addiction molecules in host bacteria, TA modules (Jensen and Gerdes, 1995; Fozo et al., 2010) seem to play a role in bacterial virulence, given that pathogenicity is initiated after attempts to limit their translation in bacteria (Kristoffersen et al., 2000; Yamamoto et al., 2002; Picardeau et al., 2003). Other than toxins and TA modules, some restriction enzymes have toxic activities (Arber, 1978). The actual role of restriction enzymes is bacterial defense against bacteriophage DNA by cleaving the DNA at specific sites. They are also considered to act as selfish elements (Bourniquel and Bickle, 2002). The toxic effects of restriction enzymes are mediated in the form of mutagenic effects on host cellular DNA (Arber, 1978; Kinashi et al., 1993; Price et al., 1995).
In this study, we investigated if toxins are indeed the only molecules specific to pathogenic bacteria. We supplemented the 12 “bad bugs” from our previous study with two additional “bad bugs” (Neisseria meningitidis and Staphylococcus aureus) and compared all 14 with their 14 closest non-epidemic relatives (“controls”). These 14 “bad bugs” are the most dangerous epidemic bacteria of all times and we chose to limit our study to them in order to achieve a neutral and not biased approach.
Materials and Methods
The following “bad bugs” were used: Mycobacterium leprae TN (NC_002677), Mycobacterium tuberculosis H37Rv (NC_000962), Rickettsia prowazekii Madrid E (NC_000963), Corynebacterium diphtheriae NCTC 13129 (NC_002935), Treponema pallidum SS14 (NC_010741), Yersinia pestis KIM (NC_004088), Bordetella pertussis Tohama 1 (NC_002929), Streptococcus pneumoniae G54 (NC_011072), Streptococcus pyogenes M1 GAS (NC_002737), Salmonella typhi CT18 (NC_003198), Shigella dysenteriae Sd197 (NC_007606), Vibrio cholerae O395 (NC_009457), Staphylococcus aureus N315 (NC_002745), and Neisseria meningitidis MC58 (NC_003112). For the “controls,” we constructed a 16s rRNA phylogenetic tree for each group of species and the non-pathogenic, or at least, non-epidemic species found as the closest relative to the “bad bug” was chosen as “control.” The following 14 related bacterial species were used: Mycobacterium avium 104 (NC_008595), Mycobacterium smegmatis MC2 155 (NC_008596), Rickettsia africae ESF-5 (NC_012633), Corynebacterium glutamicum R (NC_009342), Treponema denticola ATCC 35405 (NC_002967), Yersinia pseudotuberculosis IP 32953 (NC_006155), Bordetella bronchiseptica RB50 (NC_002927), Streptococcus agalactiae 2603V/R (NC_004116), Streptococcus suis 05ZYH33 (NC_009442), Salmonella schwarzengrund CVM19633 (NC_011094), Escherichia coli HS (NC_009800), Vibrio parahaemolyticus RIMD 2210633 (NC_004603), Staphylococcus haemolyticus JCSC1435 (NC_007168), and Neisseria cinerea (NZ_ACDY00000000).
Genomic Characteristics
All genomic characteristics used in this study [i.e., genome size, GC%, and number of open reading frames (ORFs)] were retrieved from the NCBI database. A graph was plotted and a χ2 statistical test was performed for each feature to determine if there were statistically significant differences between the “bad bugs” and the “controls.”
Restriction Enzymes
Restriction enzymes for each of the 28 bacterial species were retrieved from the REBASE database (Roberts et al., 2010). A graph was designed, and a χ2 statistical test was performed to conclude whether there were statistically significant differences between the “bad bugs” and the “controls.”
TA Modules
Text-mining searches were conducted in the GenBank protein database for the following seven type II TA families: VapB/C, RelE/B, ParE/D, MazE/F, phd/doc, ccdA/B, and higA/B. Each protein was used in a tBLASTN query, and hits were defined based on an e-value threshold of 10e-5 with >30% identity and at least 70% coverage. A graph was designed and a χ2 statistical test was performed to conclude whether there were statistically significant differences between the “bad bugs” and the “controls.”
Protein Toxins
Protein toxins for each of the 28 bacterial species were retrieved from the MvirDB database (Zhou et al., 2007). A graph was designed, and a χ2 statistical test was performed to conclude whether there were statistically significant differences between the “bad bugs” and the “controls.”
Phylogenies
Phylogenomic trees were constructed for all restriction enzymes, protein toxins, and all toxins and TA modules by generating a matrix of binary discrete characters (“0” and “1” for absence and presence, respectively) and implementing the neighbor-ioining (NJ) method in Phylogeny Inference Package (PHYLIP; Felsenstein, 1993). Phylogenetic trees were also separately constructed for each restriction enzyme family gene using three methods: NJ, maximum-parsimony (MP), and maximum-likelihood (ML). Alignments were performed with ClustalX2 (Larkin et al., 2007), and trees were constructed using Mega4 (Tamura et al., 2007).
Origins of the Toxins and TA Modules
A literature analysis allowed us to determine which of the toxins were transported to the bacterium by various mobile elements. Additionally, using phylogenies, we were able to determine whether toxins and TA modules were acquired by horizontal gene transfer (HGT) for each of the bacteria in our study. A tBLASTN query was performed for each toxin, and hits were defined based on an e-value threshold of 10e-5 with >30% identity and at least 70% coverage. Alignments were performed with ClustalX2 (Larkin et al., 2007), and trees were constructed using Mega4 (Tamura et al., 2007) via three methods: NJ, MP, and ML.
Principal Components Analyses
A first principal components analysis (PCA) was performed, including genome size, GC%, number of ORFs, protein toxins, restriction enzymes, and TA modules. The second PCA included protein toxins and TA modules, taking into consideration the lifestyle of the toxins (intracellular or extracellular). The PCAs were performed using R software (http://www.r-project.org/version 2.11.0).
Results
Confirming our previous study (Georgiades and Raoult, 2011b), our current results reveal significant differences in genome size and the number of ORFs between the “bad bugs” and “controls.” Indeed, “bad bugs” have smaller genomes and less ORFs than “controls” (Table 1; Figures A1A,B in Appendix). However, while most “bad bugs” have a lower GC%, the differences are not statistically significant (Table 1; Figure A1C in Appendix).
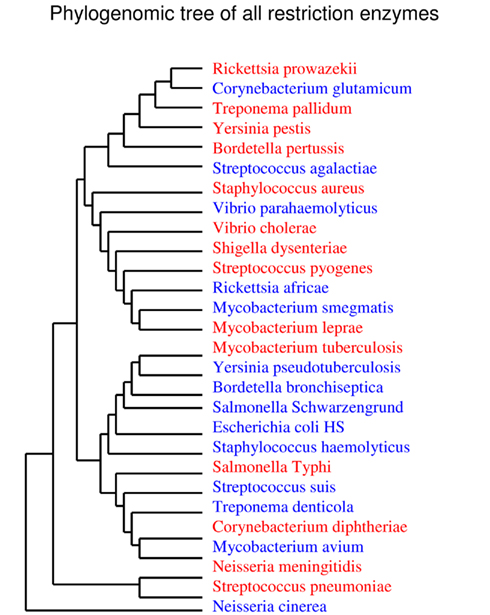
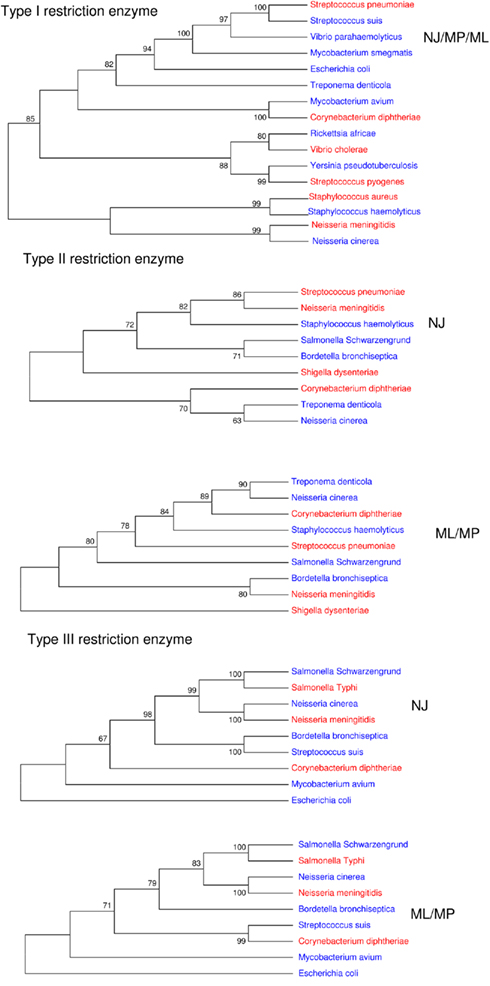
Furthermore, only five bad bugs have more restriction enzymes than their controls (M. tuberculosis, C. diphtheriae, S. pneumoniae, S. typhi, and N. meningitidis), and the differences are not statistically significant (p = 0.2857; Table 1; Figure A1D and Table A1 in Appendix). The phylogenomic tree for restriction enzymes displays a small cluster containing six species that do not encode restriction enzymes, of which four are “bad bugs” (Y. pestis, B. pertussis, T. pallidum, and R. prowazekii; Figure A2 in Appendix). The phylogenetic tree for type I restriction enzymes displays a cluster containing five “controls” (S. suis, V. parahaemolyticus, M. smegmatis, E. coli HS, and T. denticola) and only one “bad bug” (S. pneumoniae). The rest of the trees generally present topologies that are different from the topologies given by a 16S rRNA phylogenetic tree, with closely related species not clustering together. This result holds true for all of the methods used in this study (NJ, MP, and ML; Figure A3 in Appendix).
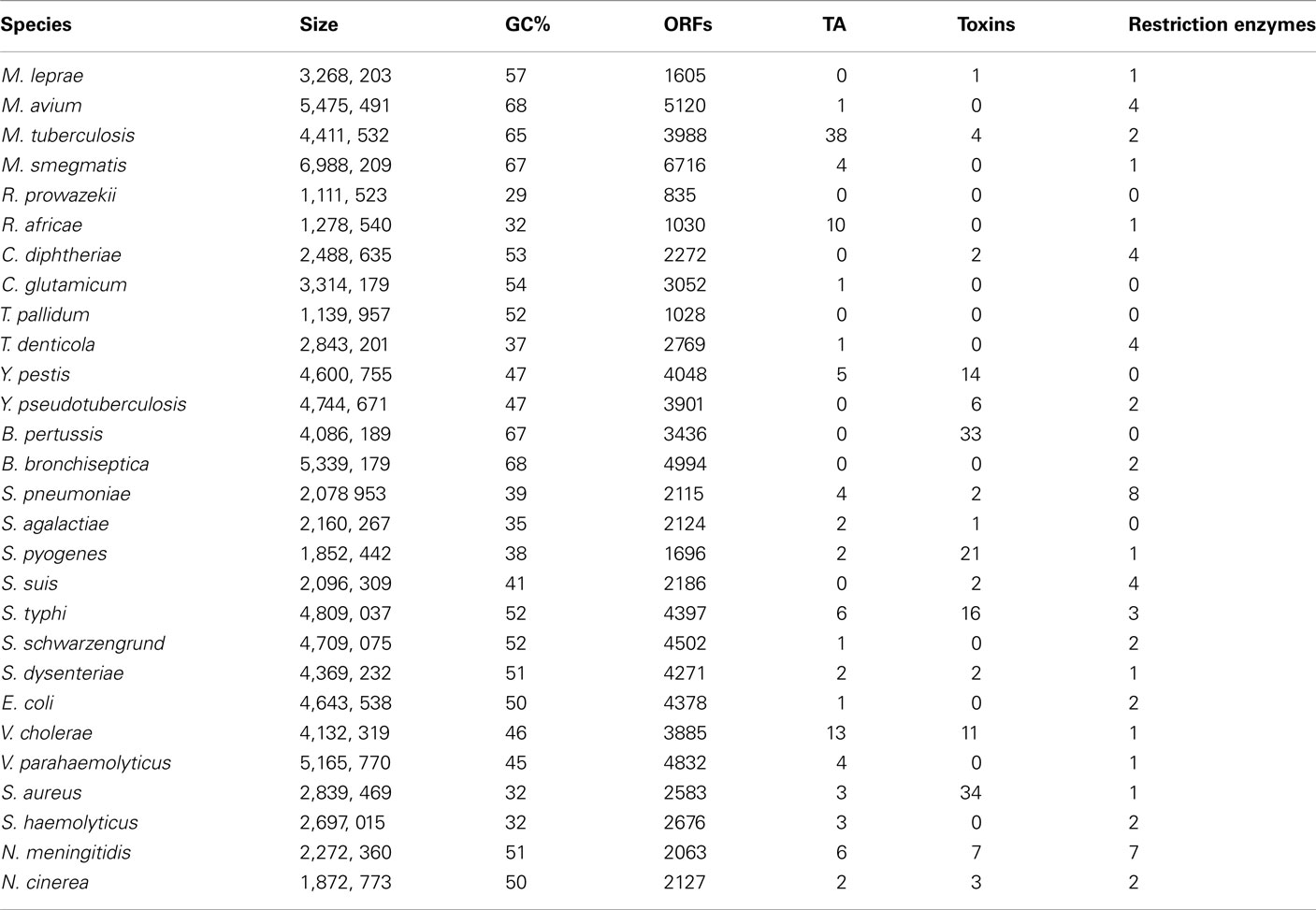
Table 1. Genomic characteristics studied for each of the 28 bacterial species. “Bad bugs” are in red and “controls” in blue.
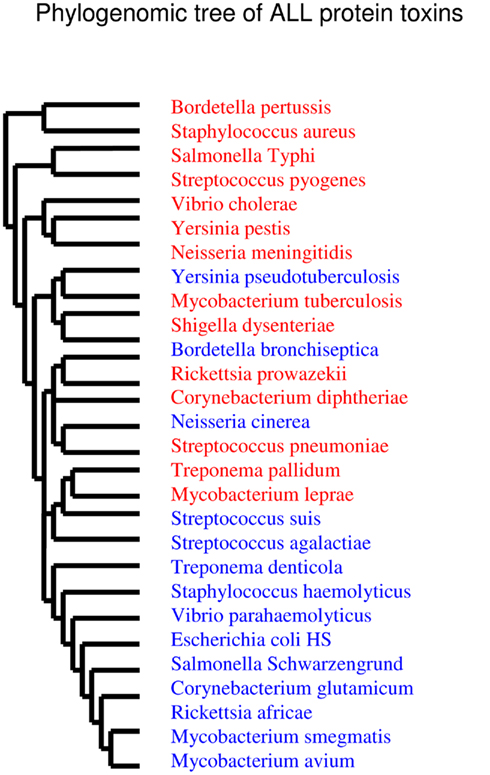
Concerning the protein toxins, all of the “bad bugs” contain more proteins, and the difference is statistically significant (p = 0.0002; Table 1; Figure A1E and Table A2 in Appendix). The phylogenomic tree of all toxins includes a cluster containing 12 species, of which nine are “controls” missing toxins (T. denticola, S. haemolyticus, V. parahaemolyticus, E. coli HS, S. schwarzengrund, C. glutamicum, R. africae, M. smegmatis, and M. avium). Moreover, a cluster of seven “bad bugs” containing toxins is also present. “Bad bugs” or “controls” tend to cluster together and not with their phylogenetically closest relatives (Figure A4 in Appendix). Finally, we searched the 28 genomes for members of the seven known TA modules and found that eight “bad bugs” (M. tuberculosis, Y. pestis, S. pneumoniae, S. pyogenes, S. typhi, S. dysenteriae, V. cholerae, and N. meningitidis) contain significantly more TA modules than their controls (p = 0.0445; Tables 1 and 2, Figure A1F in Appendix). Our results agree with the findings of previous studies (Pandey and Gerdes, 2005; Goulard et al., 2010; Georgiades and Raoult, 2011b) and the data in the toxin–antitoxin database (TADB; Shao et al., 2011).
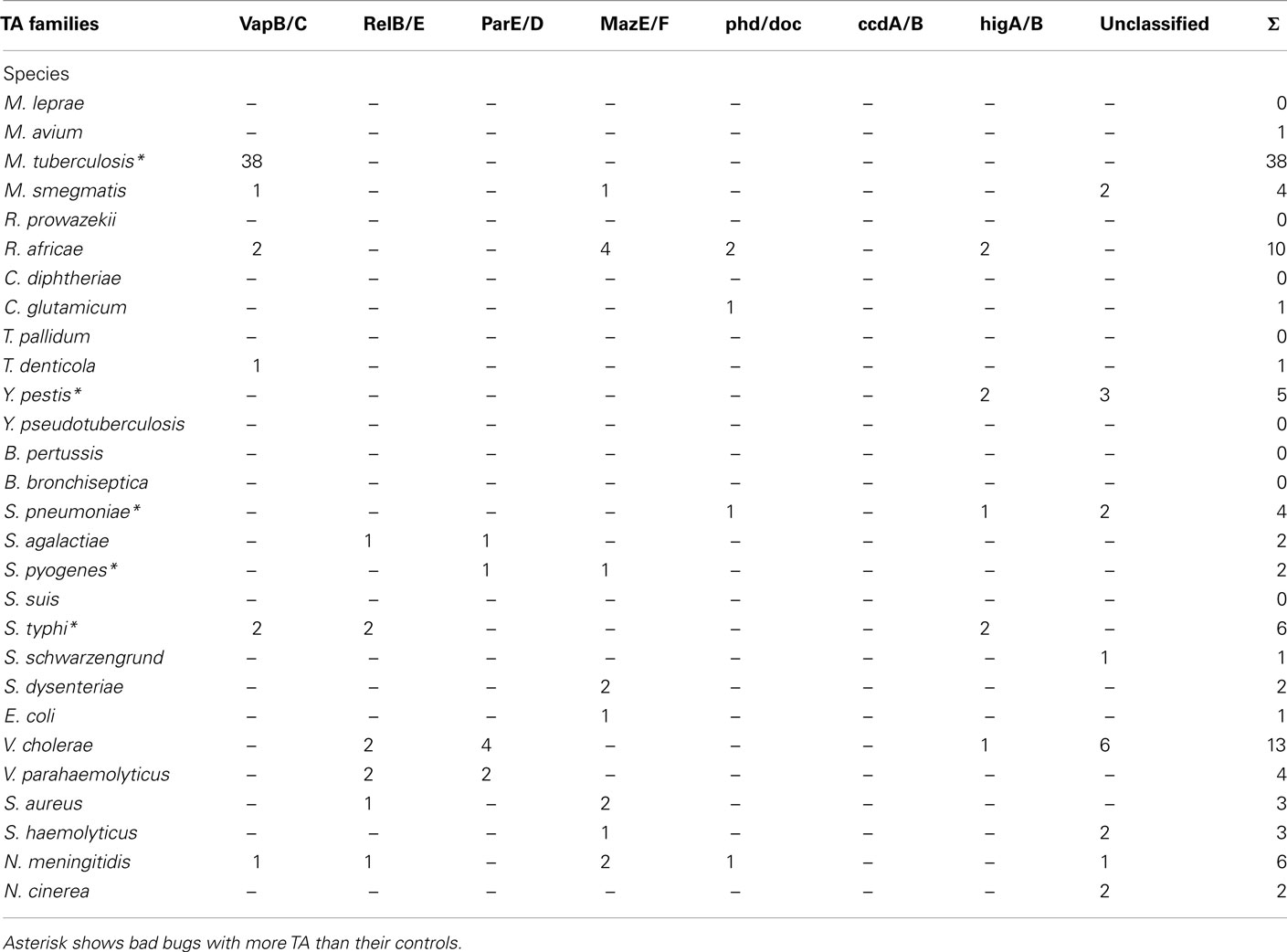
Table 2. Number of TA modules in each of the TA families. “Bad bugs” are in red and “controls” in blue.
We constructed a phylogenomic tree based on the presence/absence of toxins and TA modules that presents three clusters. One of these clusters contains seven “controls” with no toxins or TA modules (M. avium, R. africae, C. glutamicum, S. schwarzengrund, E. coli, V. parahaemolyticus, and S. haemolyticus), whereas “bad bugs” that encode many toxic elements are grouped together (M. tuberculosis, Y. pestis, S. pneumoniae, S. pyogenes, S. typhi, S. dysenteriae, V. cholerae, S. aureus, and N. meningitidis; Figure 2).
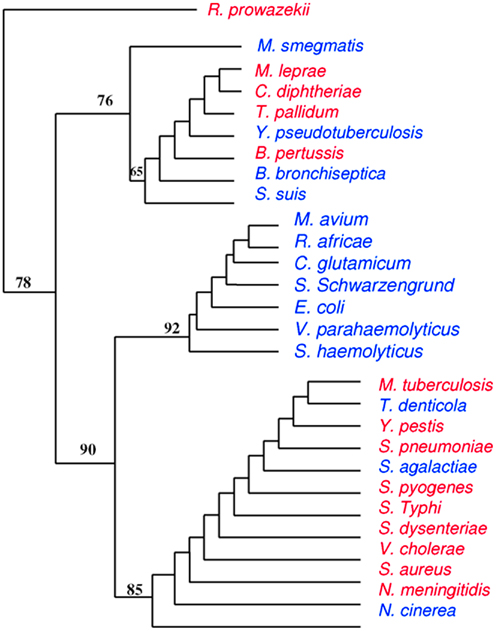
Figure 2. Phylogenomic tree based on presence/absence of toxin–antitoxin modules and toxins. Closely related species do not cluster together while totally unrelated species do, meaning that highly divergent species may have common evolutionary histories. “Bad bugs” are in red, and “controls” are in blue.
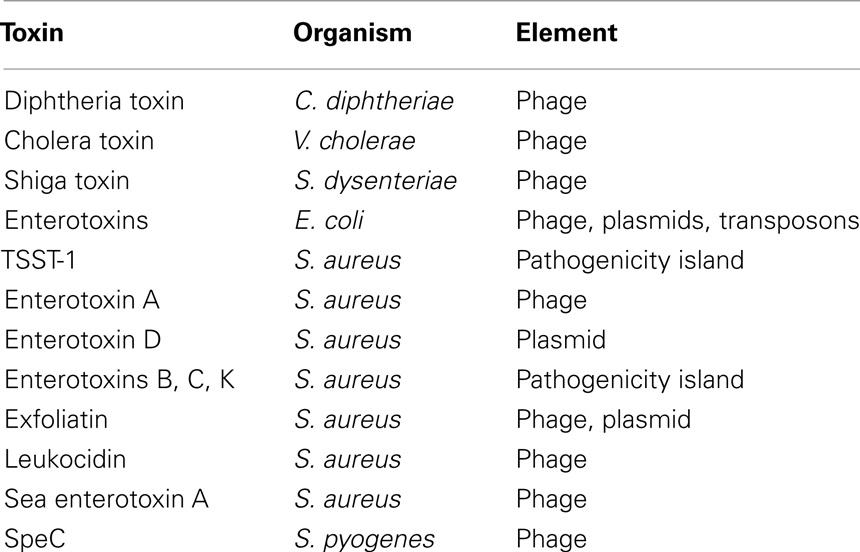
It has been hypothesized and widely accepted that bacterial toxins are encoded by mobile and variable elements (Novick, 2003). Surveying the literature, we reviewed the origins of some of these protein toxins (Table 3). Most bacterial toxins, including Cholera toxin, Streptococcal pyrogenic exotoxin (SpeC), Shiga (and Shiga-like) toxins, Diphtheria toxin, most staphylococcal and pathogenic E. coli Enterotoxins, and the Leukocidin toxin of S. aureus, are phage-associated, encoded on bacteriophages and inserted into the bacterial chromosomes (Waldor and Mekalanos, 1996; Nataro and Kaper, 1998; Boudy et al., 2001; Muniesca et al., 2003; Novick, 2003; Tinsley et al., 2006). Similarly, other mobile elements encode bacterial toxins, such as transposons for some E. coli enterotoxins (Nataro and Kaper, 1998), plasmids that carry staphylococcal enterotoxins and exfoliatins (Novick, 2003), and staphylococcal pathogenicity islands (SaPIs) carrying enterotoxins B, C, and K, and the S. aureus toxin shock syndrome toxin (TSST-1; Novick, 2003).
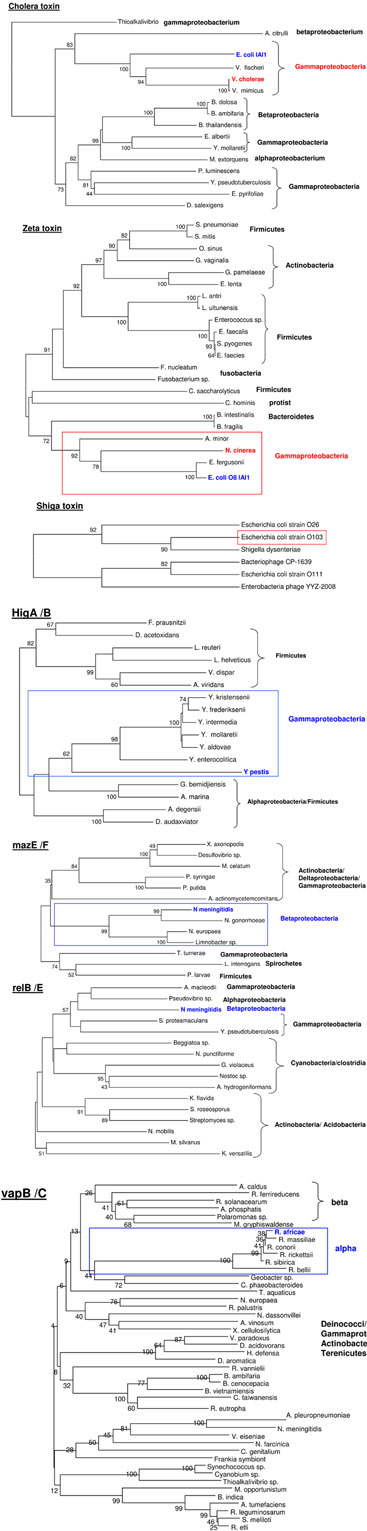
We also searched for possible HGT events, which would be located in the genomic origins of toxin and TA module sequences present in the genomes of the bacteria of interest. We did not find strong evidence for such events, except in three cases: cholera toxin seems to have been gained by V. cholerae from E. coli, and Zeta toxin of N. cinerea, and Shiga toxin also grouped with E. coli strains (the commensal strain O8 IAI1 and strain O103, respectively). As for TA modules, they appear to have different origins; HigA/B were likely transferred by Firmicutes, MazE/F likely came from different proteobacteria, RelB/E may have been transferred by Gammaproteobacteria, and VapB/C were most likely transferred from Betaproteobacteria (Figure A5 in Appendix).
Our initial PCA revealed that other than TA modules, only toxins appear to be correlated with pathogenicity (Figure 3). “Bad bugs” and “controls” are separated on the analysis plot when these two features are taken into consideration. Furthermore, as discovered in the second PCA, when we considered the lifestyles of the toxins (i.e., whether they are intracellular or extracellular), the intracellular toxins, including TA modules, are most related to the pathogenic capacity of the bacteria studied and not the extracellular toxins, as would be expected (Figure 4). Other than TA modules, which are intracellular toxins, the toxins of Salmonella, Shigella, V. cholerae, Yersinia, and B. pertussis are also intracellular. This is why their pathogenic capacities were so difficult to understand and why the TA modules were never considered important for bacterial pathogenesis.
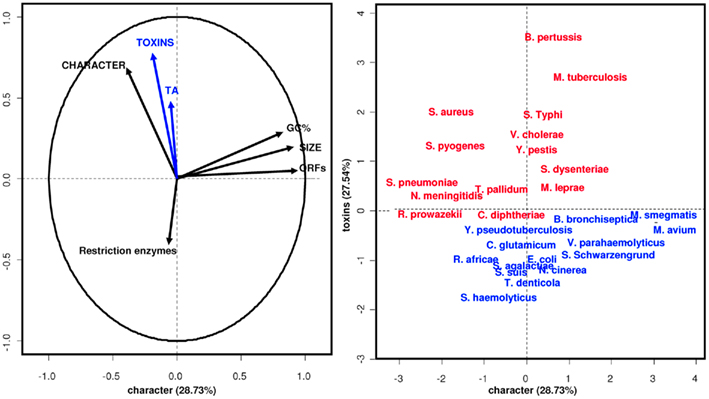
Figure 3. Principal components analysis. TA modules and toxins characterize epidemic bacteria. “Bad bugs” are in red, and “controls” are in blue.
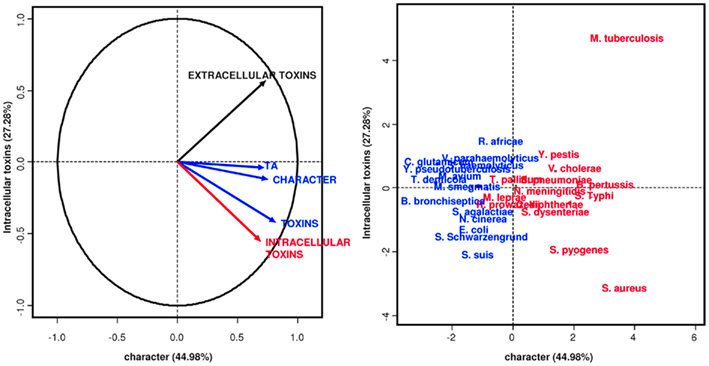
Figure 4. Principal components analysis. Intracellular toxins and TA modules characterize epidemic bacteria. “Bad bugs” are in red, and “controls” are in blue.
Discussion
In this study, we investigated whether restriction enzymes and protein toxins are the only features, other than TA modules (Georgiades and Raoult, 2011b), that can be considered natural virulence factors for bacteria. To maintain an unbiased analysis, we assessed the 14 most dangerous epidemic bacteria for humans and compared them to their closest non-epidemic related species.
In a previous study, we demonstrated that 12 epidemic bacteria have reduced-size genomes and more selfish elements, i.e., TA modules, which have evolved such that the host cells become addicted to them (Lawrence, 1999b; Makarova et al., 2009; Georgiades and Raoult, 2011b). Other studies also report a high number of TA modules in pathogenic bacteria, such as Y. pestis (Goulard et al., 2010), but their role in pathogenesis is not evident and was never considered previously. TA modules are mostly encoded on plasmids or within prophages and were initially identified as plasmid stabilization factors. When they were also found in multiple copies on chromosomes, it was hypothesized that they might also have a role in the stabilization of integrons in bacterial chromosomes (Szekeres et al., 2007). Other systems that possess properties of selfish elements are restriction enzymes, which may also constitute a toxic danger for bacterial host cells. It has been demonstrated that restriction enzymes can be toxic for mammalian cells by promoting DNA mutations (Kinashi et al., 1993; Price et al., 1995). However, our study did not reveal a significant difference in the numbers of restriction enzymes in “bad bugs.”
Since Diphtheria toxin was isolated by Roux and Yersin in 1888, bacterial toxins have been recognized as the primary virulence factors for pathogenic bacteria. These toxins are defined as “soluble substances that alter the normal metabolism of host cells with deleterious effects on the host,” are considered as one of the most powerful human poisons known, and can retain high activity when very dilute. Indeed, the major symptoms associated with diseases caused by C. diphtheriae, B. pertussis, V. cholerae, Bacillus anthracis, Clostridium botulinum, and enterohemorrhagic E. coli are all related to the activities of the toxins produced by these organisms (Alouf, 2000; Merrell and Falkow, 2004). Thus, we hypothesize that toxins have a direct measurable effect on cells. Injecting toxins in an animal or a cell leads to its death, whereas administration of antibodies against the toxin in question offers protection against future infections. This is what constitutes the principle of vaccinations against Diphtheria and Tetanus and the principle of preventive passive immunization (Stiehm, 1998). Recent experimental and cellular models testing different individual genes from bacterial genomic repertoires demonstrate that some glycoproteins have similar toxic roles (Merrell and Falkow, 2004). These endotoxins may be released in a soluble form by young cultures grown in the laboratory; however they act in a way that does not reflect bacterial virulence in experimental models (Wesselink et al., 1978). For the most part, endotoxins remain associated with the cell wall until disintegration of the organisms. In vivo, this is the result of autolysis, external lysis mediated by complement and lysozyme, and phagocytic digestion of bacterial cells. Both the toxic component of LPS and the immunogenic portion of LPS act as determinants of pathogenicity for bacteria (Merrell and Falkow, 2004). Furthermore, bacterial pathogenicity in humans is also associated with the epidemic capacity of the bacteria, about which there are few hypotheses, except in the case of vector-borne diseases (Gubler, 1997), whose multiplication in the blood and levels of bacterial load are critical. Such a considerable multiplication causes deleterious effects to the host that may lead to its death, and events such as high multiplication results in a de-regulation associated with the disappearance of transcription regulators observed in pathogenic species (Merhej et al., 2009; Georgiades and Raoult, 2011b). This likely causes the persistence of multiplication in the blood, even if nutrients decrease, and the appearance of a less favorable condition that breaks the balance with the host. In our study, we found significantly elevated numbers of protein toxins in all of the “bad bugs,” while most of the “controls” only possess two or less toxins. The statistical PCA, including genome size, GC%, TA modules, restriction enzymes, and toxins, revealed that other than TA modules, toxins are the only proteins linked to the pathogenicity of bacteria (Figure 3). Moreover, intracellular toxins appear to be more related to the pathogenicity of bacteria than secreted toxins (Figure 4). Of course, this hypothesis needs to be confirmed by experimentations and possible mechanisms should be proposed. Phylogenomic trees, constructed based on the presence/absence of restriction enzyme genes, toxins, and TA modules, display different topologies compared to 16S rRNA phylogenetic trees, meaning that closely related species are not found in sister taxa. “Bad bugs” or “controls” tend to cluster together rather than with their phylogenetic neighbors. This demonstrates that specialized epidemic bacteria have congruent evolutionary histories, resulting in a virulent gene repertoire defined by both present and absent genes. The gene repertoires of closely related species can possess different histories, while distant species can have a similar gene repertoire due to related evolutionary events. In particular, TA modules are found in “bad bugs” as a result of HGT events from other bacteria, including Firmicutes, Gammaproteobacteria, Betaproteobacteria, and other proteobacteria (Figure A5 in Appendix). Furthermore, most toxins arrive in bacterial genomes by various mobile elements, especially bacteriophages. This is the case for the streptococcal and staphylococcal toxins, Cholera toxin, Shiga toxin, and E. coli Enterotoxins. Modifications of bacterial virulence by bacteriophages were initially brought to light in 1951 (Freeman, 1951). This first demonstration showed that avirulent strains of C. diphtheriae infected with a bacteriophage yielded virulent lysogens producing Diphtheria toxin (Freeman, 1951). The outbreak of bloody diarrhea and hemolytic uremic syndrome (HUS) due to an E. coli O104:H4 strain in Germany in May and June 2011 illustrates the capacity of bacterial species to produce new combinations of genes, leading to the emergence of highly aggressive strains. The O104:H4 strain acquired the phage-mediated Shiga toxin and resistance to numerous antibiotics. However, antibiotic selective pressure has nothing to do with specialization (Werner et al., 2004; Furuya and Lowy, 2006). As in the HUS case, antibiotics are not always indicated in the treatment of the human disease because they may worsen the symptoms when the toxin is released (Denamur, 2011).
In conclusion, the only truly identifiable phenomena, witnessing a convergent evolution in the most pathogenic bacteria for humans, are the loss of metabolic activities (Georgiades and Raoult, 2011b) that occur via the loss of regulatory and transcription factors and the acquisition of protein toxins, alone, or coupled as TA modules (Figure 5). For example, in the case of Shigella, the loss of the lysine decarboxylase activity is a major event in the acquisition of its virulence capacity (Maurelli et al., 1998). TA modules stabilize and protect neighboring genes, which explains the fact that massive gene loss in some specialized bacteria seems to be decreased, while in some others (e.g., R. prowazekii) that do not contain TA modules, gene loss is much more extreme and on-going.
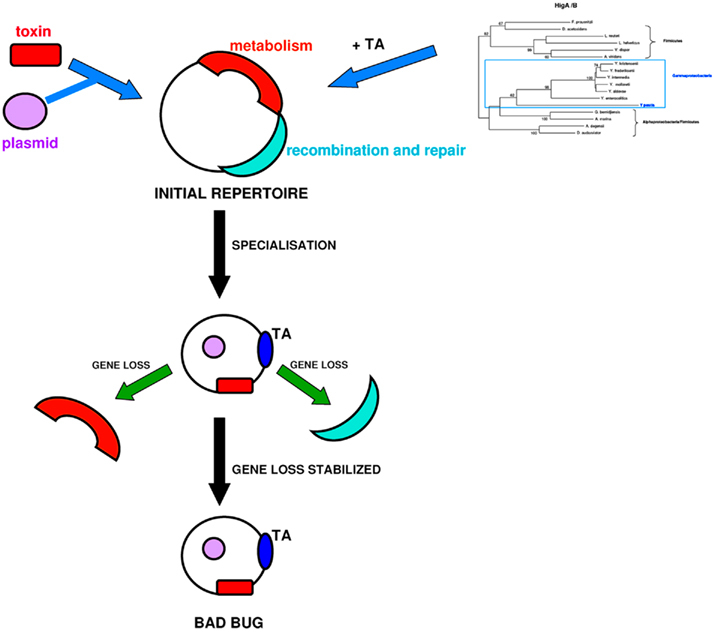
Figure 5. The “bad bug” creation scenario. Initially, bacteria contain metabolic functions and recombination machinery. TA modules are gained by gene transfer, and toxin genes arrive in bacterial genomes by various mobile elements, such as plasmids. After specialization, the bacterial recombination systems are degraded, and metabolic functions are lost. “Bad bugs” are characterized by toxins and TA modules that stabilize neighboring genes and limit massive gene loss.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Andersson, J. O., and Andersson, S. G. E. (1999). Genome degradation is an ongoing process in Rickettsia. Mol. Biol. Evol. 16, 1178–1191.
Andersson, S. G. E., and Kurland, C. G. (1998). Reductive evolution of resident genomes. Trends Microbiol. 6, 263–268.
Arber, W. (1978). Autobiography. Nobelprize.org 2011. Available at: http://nobelprize.org/nobel_prizes/medicine/laureates/1978-arber-autobiography.html
Audic, S., Robert, C., Campagna, B., Parinello, H., Claverie, J. M., Raoult, D., and Drancourt, M. (2007). Genome analysis of Minibacterium massiliensis highlights the convergent evolution of water-living bacteria. PLoS Genet. 3, e138.
Blanc, G., Ogata, H., Robert, C., Audic, S., Suhre, K., Vestris, G., Claverie, J. M., and Raoult, D. (2007). Reductive genome evolution from the mother of Rickettsia. PLoS Genet. 3, e14.
Boudy, T. B., Pancholi, V., and Fischetti, V. A. (2001). Induction of lysogenic bacteriophage and phage-associated toxin from Group A Streptococci during co-culture with human pharyngeal cells. Infect. Immun. 69, 1440–1443.
Bourniquel, A. A., and Bickle, T. A. (2002). Complex restriction enzymes: NTP-driven molecular motors. Biochimie 84, 1074–1059.
Darby, A. C., Cho, N. H., Fuxelius, H. H., Westberg, J., and Andersson, S. G. E. (2007). Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 23, 511–520.
Denamur, E. (2011). The 2011 Shiga toxin-producing Escherichia coli O104:H4 German outbreak: a lesson of genomic plasticity. Clin. Microbiol. Infect.
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 214–424.
Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package). Distributed by the author, Departement of Genetics, University of Washington, Seattle.
Fournier, P. E., Karkouri, El K., Leroy, Q., Robert, C., Giumelli, B., Renesto, P., Socolovschi, C., Parola, P., Audic, S., and Raoult, D. (2009). Analysis of the Rickettsia africae genome reveals that virulence acquisition in Rickettsia species may be explained by genome reduction. BMC Genomics 10, 166.
Fozo, E. M., Makarova, K. S., Shabalina, S. A., Yutin, N., Koonin, E. V., and Storz, G. (2010). Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 38, 3743–3759.
Freeman, V. J. (1951). Studies on the virulence of bacteriophages-infected strains of Corynebacterium diphtheriae. J. Bacteriol. 61, 675–688.
Furuya, E. Y., and Lowy, F. D. (2006). Antimicrobial-resistance bacteria in the community setting. Nat. Rev. Microbiol. 4, 36–45.
Georgiades, K., and Raoult, D. (2011a). Defining pathogenic bacterial species in the genomic era. Front Microbiol. 1:151.
Georgiades, K., and Raoult, D. (2011b). Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PLoS ONE 6, e17962.
Goulard, C., Langrand, S., Carniel, E., and Chauvaux, S. (2010). The Yersinia pestis chromosome encodes active addiction toxins. J. Bacteriol. 192, 3669–3677.
Gubler, D. J. (1997). Resurgent vector-borne diseases as a global health problem. Emerging Infect. Dis. 4, 442–450.
Jensen, R. B., and Gerdes, K. (1995). Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17, 205–210.
Kinashi, Y., Nagasama, H., and Little, B. J. (1993). Mutagenic effects of restriction enzymes in Chinese hamster cells: evidence for high mutagenicity of Sau3AI and the hprt locus. Mutat. Res. 285, 251–257.
Kristoffersen, P., Jensen, G. B., Gerdes, K., and Piskur, J. (2000). Bacterial toxin-antitoxin gene system as containment control in yeast cells. Appl. Environ. Microbiol. 66, 5524–5526.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0 . Bioinformatics 23, 2947–2948.
Lawrence, J. (1999a). Gene transfer, speciation and the evolution of bacterial genomes. Curr. Opin. Microbiol. 2, 519–523.
Lawrence, J. (1999b). Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 9, 642–648.
Makarova, K. S., Wolf, Y. I., van der Oost, J. E., and Koonin, E. V. (2009). Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 4, 29.
Maurelli, A. T., Fernandez, R. E., Bloch, C. A., Rode, C. K., and Fasano, A. (1998). Black holes and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 3943–3948.
Merhej, V., and Raoult, D. (2010). Rickettsial evolution in the light of comparative genomics. Biol. Rev. 86, 379–405.
Merhej, V., Royer-Carenzi, M., Pontarotti, P., and Raoult, D. (2009). Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol. Direct 4, 13.
Merrell, D. S., and Falkow, S. (2004). Frontal and stealth attack strategies in microbial pathogenesis. Nature 430, 250–256.
Moran, N. A. (2002). Microbial minimalism: genome reduction in bacterial pathogens. Cell 108, 583–586.
Muniesca, M., de Simon, M., Prats, G., Ferrer, D., Panella, H., and Jofre, J. (2003). Shiga toxin 2-converting bacteriophage associated with clonal valiability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71, 4554–4562.
Nataro, J. P., and Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201.
Nierman, W. C., DeShazer, D., Kim, H. S., Tettelin, H., Nelson, K. E., Feldblyum, T., Ulrich, R. L., Ronning, C. M., Brinkac, L. M., Daugherty, S. C., Davidsen, T. D., Deboy, R. T., Dimitrov, G., Dodson, R. J., Durkin, A. S., Gwinn, M. L., Haft, D. H., Khouri, H., Kolonay, J. F., Madupu, R., Mohammoud, Y., Nelson, W. C., Radune, D., Romero, C. M., Sarria, S., Selengut, J., Shamblin, C., Sullivan, S. A., White, O., Yu, Y., Zafar, N., Zhou, L., and Fraser, C. M. (2004). Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. U.S.A. 101, 14246–14251.
Novick, R. P. (2003). Mobile genetic elements and bacterial toxinoses: the superantigens-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 49, 93–105.
Ochman, H., Lerat, E., and Daubin, V. (2005). Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl. Acad. Sci. U.S.A. 102, 6595–6599.
Pandey, P. D., and Gerdes, K. (2005). Toxin-antitoxin loci are highly adundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976.
Picardeau, M., Le Dantec, C., Richard, G. F., and Saint, G. I. (2003). The spirochetal chpK chromosomal toxin-antitoxin locus induces growth inhibition of yeast and Mycobacteria. FEMS Microbiol. Lett. 229, 277–281.
Price, R. C., Margison, G. P., Hendry, J. H., and West, C. M. (1995). Relationships between the cytotoxic effects of restriction endonucleases and radiation on mammalian cells. Int. J. Radiat. Biol. 67, 327–334.
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2010). REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38, 234–236.
Shao, Y., Harrison, E. M., Bi, D., Tai, C., He, X., Ou, H. Y., Rajakumar, K., and Deng, Z. (2011). TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and Archaea. Nucleic Acids Res. 39, D606–D611.
Stiehm, E. R. (1998). “Passive immunization,” in Textbook of Paediatric Infectious Diseases, 4th Edn, eds R. D. Feigin, and J. D. Cherry (Philadelphia, PA: The WB Sanders Co.), 2769–2802.
Szekeres, S., Dauti, M., Wilde, C., Mazel, D., and Rowe-Magnus, D. A. (2007). Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 63, 1588–1605.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0 . Mol. Biol. Evol. 24, 1596–1599.
Tinsley, C. R., Bille, E., and Nassif, X. (2006). Bacteriophages and pathogenicity: more than just providing a toxin? Microbes Infect. 8, 1365–1371.
Waldor, M. K., and Mekalanos, J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914.
Werner, C. A., Monnet, L. D., and Harbath, S. (2004). Antibiotic selection pressure and resistance in Streptococcus pneumoniae and Streptococcus pyogenes. Emerging Infect. Dis. 10, 514–517.
Wesselink, P. R., Thoden van Velzen, S. K., and Makkes, P. C. (1978). Release of endotoxins in an experimental model simulating the dental root canal. Oral Surg. Oral Med. Oral Pathol. 45, 789–795.
Yamamoto, T. A., Gerdes, K., and Tunnacliffe, A. (2002). Bacterial toxin RelE induces apoptosis in human cells. FEBS Lett. 519, 191–194.
Zhou, C. E., Smith, J., Lam, M., Zemla, A., Dyer, M. D., and Slezak, T. (2007). MvirDB-a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res. 35, 391–394.
Appendix
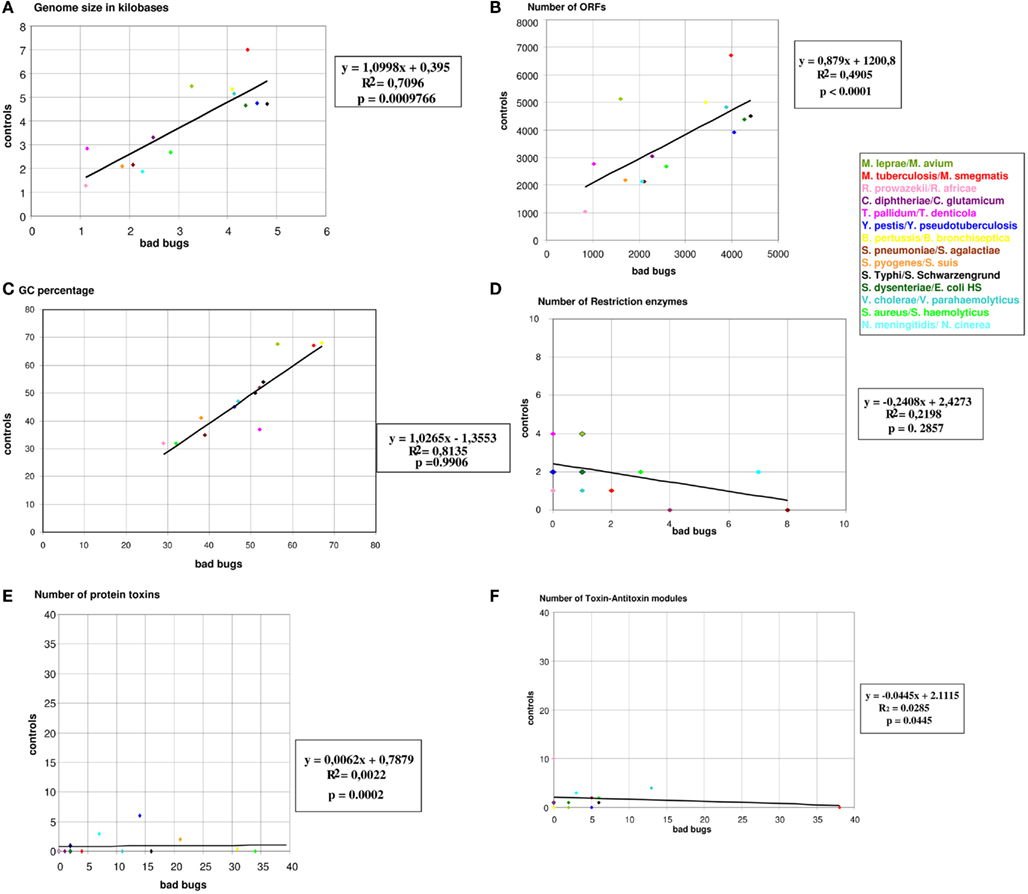
Figure A1. Graphical representations of the genomic characteristics of “bad bugs” and “controls.”(A) Genome sizes in kilobases; (B) numbers of ORFs; (C) numbers of restriction enzymes; (D) GC%; (E) numbers of toxin–antitoxin modules; (F) numbers of protein toxins. All colored dots represent “bad bug”/“control” couples.
Keywords: protein toxins, toxin–antitoxin modules, bad bugs, restriction enzymes, comparative genomics
Citation: Georgiades K and Raoult D (2011) Comparative genomics evidence that only protein toxins are tagging bad bugs. Front. Cell. Inf. Microbio. 1:7. doi: 10.3389/fcimb.2011.00007
Received: 12 July 2011;
Accepted: 07 October 2011;
Published online: 25 October 2011.
Edited by:
Jimmy Ballard, University of Oklahoma Health Sciences Center, USAReviewed by:
Nisheeth Agarwal, Translational Health Science and Technology Institute, IndiaJason Larabee, University of Oklahoma Health Science Center, USA
Copyright: © 2011 Georgiades and Raoult. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Didier Raoult, Unité des Rickettsies URMITE, UMR CNRS 6236, IRD 198, 27, boulevard Jean Moulin 13385, Marseille Cedex 05, France. e-mail: didier.raoult@gmail.com