Genotyping markers used for multi locus VNTR analysis with ompA (MLVA-ompA) and multi sequence typing (MST) retain stability in Chlamydia trachomatis
- 1Molecular Microbiology and Infection, School of Medicine, University of Southampton, Southampton, Hampshire, UK
- 2Department of Clinical Microbiology, Malmö University Hospital, Malmö, Sweden
- 3Department of Obstetrics and Gynaecology, Malmö University Hospital, Malmö, Sweden
- 4Department of Medical Sciences, Section of Clinical Bacteriology, Uppsala University, Uppsala, Sweden
- 5Health Protection Agency, Microbiology Services, Public Health Laboratory Southampton, Southampton General Hospital, Southampton, Hampshire, UK
We aimed to evaluate the stability of the Chlamydia trachomatis multi locus VNTR analysis (MLVA-ompA) and multi sequence typing (MST) systems through multiple passages in tissue culture. Firstly, we analyzed the stability of these markers through adaptation of C. trachomatis to tissue culture and secondly, we examined the stability of a four-locus MLVA-ompA and a five-locus MST system after multiple passages in tissue culture. Marker sequences were monitored through successive chlamydial developmental cycles to evaluate the stability of the individual DNA markers through many bacterial divisions and this, in turn, informed us of the usefulness of using such typing systems for short and long-term molecular epidemiology. Southampton genitourinary medicine (GUM) clinic isolates from endocervical swabs collected from C. trachomatis positive women were passaged through tissue culture. MLVA-ompA typing was applied to primary swab samples and to the same samples after C. trachomatis had been passaged through cell culture (eight passages). Sequence data from time-zero and passage-eight isolates were aligned with reference sequences to determine the stability of the markers. The Swedish new variant (nvCT) underwent 72 passages in cell culture and the markers of the two schemes were similarly analyzed. Analysis of genetic markers of the MLVA-ompA typing system before and after the isolates were introduced to tissue culture showed no change in the dominant sequence. The nvCT that had been passaged 72 times over the duration of a year also showed no variation in the dominant sequence for both the genotyping schemes. MLVA-ompA and MST markers are stable upon adaptation of C. trachomatis to tissue culture following isolation of strains from primary endocervical swab samples. These markers remain stable throughout multiple rounds of cell-division in tissue culture, concomitant with the incubation period and appearance of symptoms normally associated with host-infection. Both genotyping schemes are, therefore, suitable for epidemiology of C. trachomatis.
Introduction
Chlamydia trachomatis, an intracellular bacterial pathogen is the leading cause of sexually transmitted infections (STIs) worldwide. The majority of diagnoses are in young people between the ages of 15 and 24. C. trachomatis is currently divided into at least 15 serotypes. Genital infections are caused by the non-invasive serotypes D to K and by the invasive lymphogranuloma venereum (LGV) which infects the lymphatic system. If left untreated, which is the case for up to 50% of men and 70% of women due to the asymptomatic nature of this STI, a range of complications may persist, although opportunistic screening and partner notification detect a lot of asymptomatic cases. Individuals infected with LGV can progress to develop proctitis, lymphadenopathy, and genital ulcerations, whereas disease outcome for individuals infected with serotypes D to K range from infertility, ectopic pregnancy, and pelvic inflammatory disease in women and epididymitis in men.
The availability of whole genome sequences for C. trachomatis has led to the development of several prototype genotyping systems. Multilocus sequence typing (MLST) is one of these typing techniques and it is usually based on 6–8 housekeeping genes which are specifically chosen because they are not under immune selection, and because of their role in cell survival. Therefore, MLST is useful when exploring long-term and global epidemiology and two such systems have been developed for Chlamydiacae (Pannekoek et al., 2008; Dean et al., 2009). To achieve a higher discriminating capacity a system based on five non-housekeeping genes was created (Klint et al., 2007). It is aimed for partner tracing and other short-term epidemiology. This scheme was originally named as MLST, but the term “MLST” strictly refers to the exclusive sequencing of small amplicons from non-selectable housekeeping genes. In this paper the designation of this scheme is reassigned to “multi sequence typing” (MST) as it encapsulates large and small amplicons, and includes selectable non-housekeeping genes (Wang et al., 2011). Pedersen et al. (2008) developed a high resolution typing scheme which encompassed multi locus variable number tandem repeat (VNTR) analysis and ompA (MLVA-ompA) (Pedersen et al., 2008, 2009; Wang et al., 2011). VNTR are defined as short regions of nucleotide repeats or motifs. There are several MLVA schemes practiced in bacteriology with various end-point measurement methods including fluorescent amplicon size-discrimination and sequence analysis of the VNTR loci (Pedersen et al., 2008; Schouls et al., 2009). For C. trachomatis Pedersen et al. (2008) described three VNTR regions; CT1335, CT1299, CT1291. Both the MST system of Klint et al. (2007) and the Pedersen et al. (2008) MLVA-ompA system provide a Simpson index of diversity between 0.94 and 0.96 as opposed to 0.64–0.83 when only ompA is typed (Ikryannikova et al., 2010; Bom et al., 2011; Wang et al., 2011). The guidelines outlined by the European Society of Clinical Microbiology and Infectious Disease (ESCMID) states that a typing system should have a discriminatory power of no less than 0.95 in order to be “ideal” for studying epidemiology (van Belkum et al., 2007). The MLVA DNA markers were selected for typing because of their variability therefore, it is also particularly important to assess the stability of these markers when the study involves sample collection by isolation of C. trachomatis in continuous cell culture. In the study by Wang et al. (2011) a sub panel of samples could not be typed initially and were subsequently assigned a MLVA-ompA type after culture (Wang et al., 2011).
Pedersen et al. (2008) have tested the stability of the three markers and ompA in vivo in 24 patients with persistent or recurrent infection. Their results show that the markers are stable between 70 and 394 days. However, this is very difficult to control as these patients were assessed under the assumption that it was the same isolate the individual was infected with and also that each of the 24 patients had not contracted chlamydia from a different sex partner and could have, therefore, been re-infected.
The aim of our study was to evaluate the stability of the C. trachomatis typing systems. The MLVA-ompA system was assessed after multiple passages in the controlled conditions of cell culture. By following potential changes through many developmental cycles it was possible to establish the stability of the individual markers within a numerical framework of bacterial divisions and this in turn informed us of the usefulness of using such a typing system for short-term and longer-term molecular epidemiology. In this study we analyzed firstly, the stability of these markers through adaptation of C. trachomatis to cell culture, in doing so the initial genetic impact on the C. trachomatis genome resulting from changing environmental conditions was analyzed. Secondly, the stability of the VNTR markers with ompA as well as the MST system (Klint et al., 2007) after multiple passages in cell culture was evaluated.
Methods
Cells and Culture of C. trachomatis
McCoy cells were seeded into wells or flasks 24 h pre-infection. Cells were grown overnight in Dulbecco's Modified Eagle medium (DMEM) containing 10% foetal calf serum (FCS) and then incubated at 37°C with 5% CO2. On the day of infection the medium was removed and replaced with inoculum containing the C. trachomatis isolates. This was then centrifuged at 754×g for 30 min. Subsequently the inoculum was replaced with DMEM containing cycloheximide (1 μg/mL), gentamicin (20 μg/mL), and vancomycin (10 μg/mL), and the cells incubated at 37°C for 48 h. After 48 h the inclusions were large enough and visible under a light microscope so that the extent of infection could also be determined visually to decide whether a larger surface area was required for the next passage.
Harvesting of C. trachomatis Isolates from Wells and Flasks
Two days after infection tissue culture flasks were harvested, for this a cell scraper was used to detach the cells from the plastic of the flask. The sample was then centrifuged at 2851×g for 10 min, the supernatant was then discarded and the pellet resuspended in 1:10 cold phosphate buffered saline (PBS). The sample was then added to glass beads and agitated for 1 min to release the elementary bodies from the cell, centrifuged at 110×g for 5 min to remove any cell debris and the supernatant was then added to an equal volume of 4 Sucrose Phosphate (4SP) and stored at −80°C. To harvest from wells the cells were scraped up into the medium using a sterilized pipette tip, the 10 min centrifugation step was omitted as was the addition of PBS, instead the harvest was immediately beaded and then the protocol for harvesting was continued as for harvesting from flasks.
Chlamydia trachomatis Isolates
In 2009, 157 endocervical swabs were collected over a six month period from women who had attended GUM clinics and GP surgeries in Southampton (Wang et al., 2011). The samples tested positive for C. trachomatis using a commercial real time PCR (Cobas Taqman 48, Roche), performed in the Southampton Health Protection Agency (HPA) Molecular Diagnostics Laboratory. To test the stability of ompA and the VNTR markers (CT1335, CT1299, and CT1291), seven of the archived isolates were chosen (Table 1). Locations of these loci within the chlamydial genome are shown in Table 2. The isolates were selected based on their ompA type, so that all available ompA types could be included in this study. The isolates were initially typed using the MLVA-ompA typing system i.e., before inoculation of the primary sample into tissue culture. All seven isolates were then taken through eight serial passages in cell culture and were subsequently re-typed.
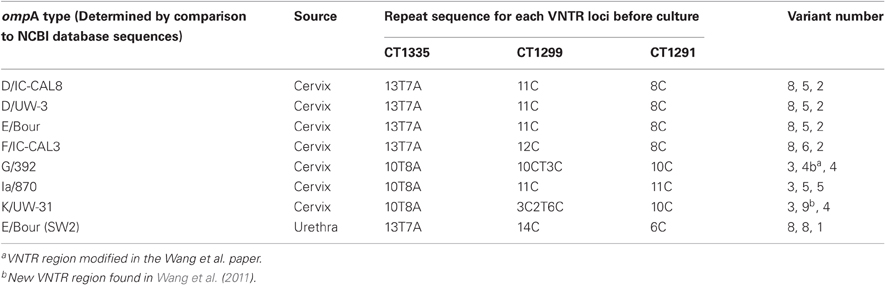
Table 1. Isolates included in all studies and their sites of isolation, also included are the repeat sequences for the three VNTR loci, their associated variant number, and the corresponding ompA NCBI database strain type for each isolate before culture.

Table 2. Location and putative function of the MLVA-ompA and MST markers within the genome, also included is the strand (negative or positive) in which the MST and ompA markers are located.
The stability of the Swedish new variant in cell culture was also determined, an isolate from Malmo in Sweden was sent to Southampton (Table 1), where it was passaged 72 times in cell culture over the period of a year. The sample was obtained from the urethra of a male patient who had contracted the new variant infection in Sweden in 2006. The Swedish new variant is a strain which has a 377 bp deletion in the cryptic plasmid and 44 bp duplication immediately upstream of CDS3, and is, therefore, a mutable strain and most suitable for studying marker stability (Ripa and Nilsson, 2006, 2007).
DNA Extraction
DNA was extracted from isolates that had been expanded to a level of infection were there was enough to infect 100% of the cells in a T75 tissue culture flask. DNA extraction was performed using the Promega Wizard genomic DNA purification kit (Promega, Southampton, UK) and was carried out according to manufacturer's instructions. The presence of DNA in the sample was verified on an agarose DNA gel. The concentration of DNA present in each isolate was determined using the Nanodrop 2000 (Thermo Scientific, Delaware, USA). PCR was conducted with high fidelity DNA polymerase, Phusion high fidelity DNA polymerase mastermix (New England Biolabs UK, Hitchin, UK), to ensure that the sequence data obtained was of the best quality possible.
PCR Amplification of VNTR and ompA Sequences
MLVA-ompA analysis was carried out at Molecular Microbiology and Infection, School of Medicine, University of Southampton. VNTR and ompA sequences were amplified using PCR. OmpA was amplified using primers P1F (Frost et al., 1991) and CT5 (Rodriguez et al., 1991), whilst the three VNTR regions were amplified using primers outlined in Pedersen et al. (2008). PCR reactions were carried out in 20 μl volumes with the components consisting of 10 μl of Phusion high fidelity DNA polymerase mastermix (New England Biolabs UK, Hitchin, UK), 250 nM of the forward and reverse primers and 2 μl of DNA.
PCR conditions for the three VNTR sequences and ompA were as follows: 10 s at 98°C, and 35 cycles of 2 s at 98°C, 5 s at 59°C, and 10 s at 72°C, followed by an elongation step for 1 min at 72°C. The temperature was then maintained at 10°C. All strains amplified had a CT1291 band that was 225 bp in size.
Purification of the PCR products was carried out using Wizard SV gel and PCR clean-up system (Promega, Southampton, UK) and this was carried out using the manufacturer's instructions.
Sequencing of MLVA-ompA Markers and Determination of Sequence Stability
Sequencing of PCR products were carried out by a commercial company (Source Bioscience, Nottingham, UK). Sequencing primers are described in Wang et al. (2011). OmpA sequences were initially (before chlamydial passage) compared with sequences available on the NCBI database using a BLAST search and in doing so the ompA genotype was assigned to each isolate. The VNTR sequences were compared to the whole genome of D/UW-3/CX and thereafter assigned MLVA-types based on the dominant sequence in the chromatograms according to rules outlined in Pedersen et al. (2008). After the isolates were taken through eight passages, the chlamydial ompA and VNTR sequences were compared to sequence data received before the isolates had been passaged.
The Swedish new variant, which was passaged 72 times, was sequenced before passaging began and then every twelfth passage up to 72 passages. These sequences were compared to the database sequences and also to the sequence before the new variant had been passaged.
Sequencing of MST Markers and Determination of Sequence Stability
MST was carried out at the Section of Clinical Bacteriology, Department of Medical Sciences, Uppsala University. The MST scheme comprised five highly variable target regions and was performed as previously described (Klint et al., 2007) except that the pbpB region was amplified as two separate fragments according to Jurstrand et al. (2010), and for sequencing of the longer PCR products CT058 and pbpB internal sequence primers were used in the scheme (Herrmann, personal communication). DNA samples of chlamydia cell cultures passaged 72 times, sent from Southampton to Uppsala as part of a batch of samples that had been random coded so that sequence analysis was unbiased. Sequences were compared to the sequence at each locus to all known corresponding alleles available in the Uppsala University C. trachomatis MLST database (http://mlstdb.bmc.uu.se), 178 sequence types comprising 183 allele variants in March 2011. Sequences were thereafter compared to the original sequence of the C. trachomatis strain in question, as obtained before starting passage.
Results
Determination of the Stability of the MLVA-ompA Typing System
Seven endocervical C. trachomatis isolates from the Southampton women's study (Wang et al., 2011) were selected to analyse the stability of the MLVA-ompA markers in cell culture. The criteria for choosing these isolates were that they had to be capable of at least two passages in cell culture and were, therefore, capable of being further passaged for this study. Secondly all isolates selected for marker stability had to have different ompA genotypes and they all had to have gone through the same number of passages in order for the data to be comparable. The isolates included in this study comprised of seven ompA genotypes, which corresponded to the following NCBI database strains: D/IC-CAL8, D/UW-3, E/Bour, K/UW-31, F/IC-CAL3, G/392, Ia/870 (Table 1). For this study we wanted to include samples with shared MLVA types (i.e., samples that had the same MLVA types but different ompA types, these included local isolates which had identical ompA sequences to the database sequence of D/IC-CAL8, D/UW-3, and E/BOUR) (Table 1) so we could identify if there were any differences in stability of the MLVA markers between different ompA types.
The passaging of the isolate initially began with each isolate being grown in a 24-well tray, then in a six-well tray and then into flasks (T25 and T75). By the eighth passage all isolates were now growing in a T75 flask where there were enough elementary bodies available to infect all the cells. DNA was then extracted from the elementary bodies and PCR reactions conducted for sequencing purposes. The chromatograms and sequence data received were aligned to sequences obtained before the isolates were taken through eight passages. Analysis of the alignments of the loci showed that the markers were stable upon adaptation of the chlamydia to cell culture as the sequence was unchanged from the sequence before the sample had been cultured in mouse cells and also that there is no change in the dominant sequence after eight passages in cell culture.
Passaging of the Swedish New Variant and Determination of the Stability of MLVA-ompA Markers in Cell Culture
The Swedish new variant (serotype E with a 377 bp deletion in the cryptic plasmid) isolated in Malmo, Sweden was taken through a further 72 passages in cell culture (Figure 1) in Southampton, UK, the process took one year to complete. The Swedish new variant was chosen because this isolate shows restricted cell tropism (Unemo et al., 2010) and there is also greater possibility that the MLVA-ompA markers are unstable in this “mutant” because the strain was evidently mutable based on the plasmid sequence. Sequences for markers from every twelfth passage were determined up to and including passage 72 (passage 0, 12, 24, 36, 48, 60, and 72). All the sequence data were aligned and there were no changes seen in the MLVA-ompA profile at any passage that had been sequenced.
Figure 1. Flow diagram showing the passage process of Sweden 2 and how the sample was processed to determine the stability of ompA and the VNTR markers.
Determination of the Stability of the MST Typing System in Cell Culture
The stability of the C. trachomatis MST markers in cell culture was also determined for the Swedish new variant which had been passaged 72 times (Figure 1). For this study five MST regions were analyzed these included; hctB, CT058, CT144, CT172, and pbpB and were first described by Klint et al. (2007). The five regions were sequenced and then aligned against corresponding sequences for each marker that was available from the sequence data of the Swedish new variant. The MST profile obtained was 21-19-1-2-1 and this was identical to the MST profile of the Swedish new variant. This data shows that these five MST markers are also stable in tissue culture up to 72 passages.
Discussion
There are currently five DNA typing systems published for genotyping C. trachomatis (Klint et al., 2007; Pannekoek et al., 2008; Pedersen et al., 2008; Dean et al., 2009; Bom et al., 2011). These typing systems use a number of regions within the C. trachomatis genome to assign distinct labels to clinical specimens containing C. trachomatis based upon the order of nucleotides in the sequence. The repeating sequences of VNTR loci build up due to an increased error rate during DNA replication at these sites, therefore, this raised error rate means that the stability of such markers over multiple bacterial cell-divisions must be verified to validate their use in epidemiology studies. To be useful for epidemiological studies the markers need to remain stable over the duration of the study allowing the data collected to be comparable (van Belkum et al., 2007). Use of genomic regions which are unstable may over represent the true distribution of bacterial genotypes within a community. It is, therefore, imperative to evaluate and understand the stability of the markers chosen for epidemiological studies.
Utilization of the gene for the major outer membrane protein A (ompA) has, for a long time been one of the main typing methods for distinguishing C. trachomatis. However, this method actually provides little discrimination (Pedersen et al., 2008; Bom et al., 2011) and masks the true phylogenetic relationships of the strains (Harris et al., 2012). Three “MLST” schemes have also been developed, two of which use seven housekeeping genes to determine overall population structure of the whole chlamydiae family (Pannekoek et al., 2008; Dean et al., 2009) and lastly a scheme which is based on five highly variable genes, some of which are under selection (which is designed to ensure high resolution) (Klint et al., 2007). Multi sequence typing (MST) best describes the typing scheme designed by Klint et al. (2007) and is the most discriminatory of the three “MLST” typing schemes available for C. trachomatis. The fifth typing system available for C. trachomatis is the MLVA-ompA scheme which uses three variable regions within the genome to assign types to each isolate (Pedersen et al., 2008). These DNA markers were selected for typing because of their variability, therefore, it is particularly important to assess the stability of these markers in continuous cell culture.
The stability of the DNA markers used in the two typing methods with the highest discriminatory power was assessed; the Pedersen et al. MLVA-ompA typing system Pedersen et al. (2008) and the Klint et al. (2007) MST typing system. Although ompA had previously been shown to be stable through adaptation of C. trachomatis to cell culture (Stothard et al., 1998), more recent epidemiological studies suggest that within a single clone (e.g., nvCT—according to the MST scheme), minor single base substitutions occasionally appear (Herrmann et al., 2008); a similar trend has been observed in LGV isolates (Christerson et al., 2010).
Our hypothesis was that adaptation of C. trachomatis to cell culture conditions could affect marker stability, however, on the contrary our data shows that the dominant MLVA sequences are stable on adaptation of the C. trachomatis to cell culture. The MLVA-ompA sequences were analyzed from DNA extracts taken from primary swab samples (Wang et al., 2011) and after eight passages in cell culture which is equivalent to approximately 80 divisions. To ensure our study covered a wide spread of C. trachomatis types all the isolates chosen were selected on the basis that they had different MLVA-ompA types. Three isolates with different ompA types but the same MLVA profile were also included. This could be indicative of recombination of the ompA gene. Choosing isolates with different ompA types also acted as a verification that no contamination between strains had occurred during passaging of the isolates. Although the in vivo conditions in which C. trachomatis persists are much more complex (involves the immune system etc.) than that of the in vitro system that is described here, the basis for taking the samples through eight passages to analyse the initial impact in cell culture was to mimic the actual time it takes for symptoms to appear within an individual. Symptoms usually arise in individuals seven to twenty-one days after infection occurs (Black, 1997), therefore, the isolates were taken through twenty-four days from infection to harvesting in cell culture to ensure the total time from being infected to symptoms persisting in individuals was covered over the eight passages.
A review by Bjorn-Arne Lindstedt states that it is imperative that the VNTR loci selected for MLVA typing are thoroughly checked before being applied to real-life situations, this includes long-term passaging and re-typing to ensure the stability and the reproducibility of each locus (Lindstedt, 2005). Pedersen et al. (2008) previously assessed the markers in patients with recurrent or persistent C. trachomatis. Others have also evaluated the MLVA system and found that some VNTR-markers may vary with replication of single clones and cause difficulties in interpretation (Bom et al., 2011). Analysis of the stability in a controlled environment by long term passaging of isolates was, therefore, important thus the second part of our study was performed to determine the stability of the MLVA-ompA and MST markers (Klint et al., 2007) over seventy two passages using the Swedish new variant. This isolate had already been subject to change in vivo as illustrated by both a deletion and duplication within the plasmid and may, therefore, be an unstable strain (or perhaps a strain which has encountered an unknown selective pressure causing the mutation), making this a useful isolate to test stability over a long duration. Three studies carried out in Sweden have shown that the Swedish new variant is clonal. The first study by Herrmann et al. (2008) typed 48 specimens from different regions in Sweden and three other countries. The study not only showed that all isolates were genotype E according to the ompA typing system but also that 96% of the isolates had identical sequence types when typed using a highly discriminatory MST method (Klint et al., 2007). Later studies by Jurstrand et al. (2010) also showed that all specimens typed (n = 41) had the same ompA type (genotype E) and the same MST sequence types. The sequence type assigned to all samples was unique for the Swedish new variant. The third study included nvCT cases from 2009 and although only 13 specimens were analyzed it supports the stability of the MST target regions over time (Klint et al., 2011). Although this evidence suggested that the Swedish new variant is clonal and may be stable in vivo the transmission of the new variant from patient to patient in these studies remains unknown and, therefore, it is also unknown as to how many passages the new variant has been through. Our in vitro studies were conducted under carefully controlled conditions so the number of continuous passages were defined.
In conclusion MLVA-ompA markers are stable through adaptation of C. trachomatis to growth in vitro and are also stable over eight passages in cell culture. We have also cultured a mutable isolate (the Swedish new variant of C. trachomatis) through 72 passages and showed that the MLVA-ompA and MST markers are stable, however, complete definition of evolutionary adaptation requires more isolates and extensive further passaging.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the financial support from the HPA PhD Fund for Clare Labiran. We would like to thank Prof. Nicholas Thomson and Dr. Helena Seth-Smith of The Wellcome Trust Sanger Institute for their expert advice. We also acknowledge Dr. Stuart C. Clarke for his help in securing funding for the PhD project.
References
Black, C. M. (1997). Current methods of laboratory diagnosis of Chlamydia trachomatis infections. Clin. Microbiol. Rev. 10, 160–184.
Bom, R. J., Christerson, L., Schim van der Loeff, M. F., Coutinho, R. A., Herrmann, B., and Bruisten, S. M. (2011). Evaluation of high-resolution typing methods for Chlamydia trachomatis in samples from heterosexual couples. J. Clin. Microbiol. 49, 2844–2853.
Christerson, L., de Vries, H. J., de Barbeyrac, B., Gaydos, C. A., Henrich, B., Hoffmann, S., Schachter, J., Thorvaldsen, J., Vall-Mayans, M., Klint, M., Herrmann, B., and Morre, S. A. (2010). Typing of lymphogranuloma venereum Chlamydia trachomatis strains. Emerg. Infect. Dis. 16, 1777–1779.
Dean, D., Bruno, W. J., Wan, R., Gomes, J. P., Devignot, S., Mehari, T., de Vries, H. J. C., Morre, S. A., Myers, G., Read, T. D., and Spratt, B. G. (2009). Predicting phenotype and emerging strains among Chlamydia trachomatis infections. Emerg. Infect. Dis. 15, 1385–1394.
Frost, E. H., Deslandes, S., Veilleux, S., and Bourgaux-Ramoisy, D. (1991). Typing Chlamydia trachomatis by detection of restriction fragment length polymorphism in the gene encoding the major outer membrane protein. J. Infect. Dis. 163, 1103–1107.
Harris, S. R., Clarke, I. N., Seth-Smith, H. M., Solomon, A. W., Cutcliffe, L. T., Marsh, P., Skilton, R. J., Holland, M. J., Mabey, D., Peeling, R. W., Lewis, D. A., Spratt, B. G., Unemo, M., Persson, K., Bjartling, C., Brunham, R., de Vries, H. J., Morre, S. A., Speksnijder, A., Bebear, C. M., Clerc, M., de Barbeyrac, B., Parkhill, J., and Thomson, N. R. (2012). Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat. Genet. 44, 413–419.
Herrmann, B., Torner, A., Low, N., Klint, M., Nilsson, A., Velicko, I., Soderblom, T., and Blaxhult, A. (2008). Emergence and spread of Chlamydia trachomatis variant, Sweden. Emerg. Infect. Dis. 14, 1462–1465.
Ikryannikova, L. N., Shkarupeta, M. M., Shitikov, E. A., Il'ina, E. N., and Govorun, V. M. (2010). Comparative evaluation of new typing schemes for urogenital Chlamydia trachomatis isolates. FEMS Immunol. Med. Microbiol. 59, 188–196.
Jurstrand, M., Christerson, L., Klint, M., Fredlund, H., Unemo, M., and Herrmann, B. (2010). Characterisation of Chlamydia trachomatis by ompA sequencing and multilocus sequence typing in a Swedish county before and after identification of the new variant. Sex. Transm. Infect. 86, 56–60.
Klint, M., Fuxelius, H. H., Goldkuhl, R. R., Skarin, H., Rutemark, C., Andersson, S. G. E., Persson, K., and Herrmann, B. (2007). High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J. Clin. Microbiol. 45, 1410–1414.
Klint, M., Hadad, R., Christerson, L., Lor, B., Anagrius, C., Osterlund, A., Larsson, I., Sylvan, S., Fredlund, H., Unemo, M., and Herrmann, B. (2011). Prevalence trends in Sweden for the new variant of Chlamydia trachomatis. Clin. Microbiol. Infect. 17, 683–689.
Lindstedt, B. A. (2005). Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 26, 2567–2582.
Pannekoek, Y., Morelli, G., Kusecek, B., Morre, S. A., Ossewaarde, J. M., Langerak, A. A., and van der Ende, A. (2008). Multi locus sequence of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol. 8, 42.
Pedersen, L. N., Herrmann, B., and Moller, J. K. (2009). Typing Chlamydia trachomatis: from egg yolk to nanotechnology. FEMS Immunol. Med. Microbiol. 55, 120–130.
Pedersen, L. N., Podenphant, L., and Moller, J. K. (2008). Highly discriminative genotyping of Chlamydia trachomatis using omp1 and a set of variable number tandem repeats. Clin. Microbiol. Infect. 14, 644–652.
Ripa, T., and Nilsson, P. (2006). A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro Surveill. 11, E061109.
Ripa, T., and Nilsson, P. A. (2007). A Chlamydia trachomatis strain with a 377-bp deletion in the cryptic plasmid causing false-negative nucleic acid amplification tests. Sex. Transm. Dis. 34, 255–256.
Rodriguez, P., Vekris, A., de, B. B., Dutilh, B., Bonnet, J., and Bebear, C. (1991). Typing of Chlamydia trachomatis by restriction endonuclease analysis of the amplified major outer membrane protein gene. J. Clin. Microbiol. 29, 1132–1136.
Schouls, L. M., Spalburg, E. C., van Luit, M., Huijsdens, X. W., Pluister, G. N., van Santen-Verheuvel, M. G., and de Neeling, A. J. (2009). Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS ONE 4:e5082. doi: 10.1371/journal.pone.0005082
Stothard, D. R., van der Pol, B., Smith, N. J., and Jones, R. B. (1998). Effect of serial passage in tissue culture on sequence of omp1 from Chlamydia trachomatis clinical isolates. J. Clin. Microbiol. 36, 3686–3688.
Unemo, M., Seth-Smith, H. M., Cutcliffe, L. T., Skilton, R. J., Barlow, D., Goulding, D., Persson, K., Harris, S. R., Kelly, A., Bjartling, C., Fredlund, H., Olcen, P., Thomson, N. R., and Clarke, I. N. (2010). The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization. Microbiology 156, 1394–1404.
van Belkum, A., Tassios, P. T., Dijkshoorn, L., Haeggman, S., Cookson, B., Fry, N. K., Fussing, V., Green, J., Feil, E., Gerner-Smidt, P., Brisse, S., and Struelens, M. (2007). Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13 (Suppl 3), 1–46.
Keywords: Chlamydia trachomatis, MLVA-ompA, multi sequence typing, marker stability, tissue culture
Citation: Labiran C, Clarke IN, Cutcliffe LT, Wang Y, Skilton RJ, Persson K, Bjartling C, Herrmann B, Christerson L and Marsh P (2012) Genotyping markers used for multi locus VNTR analysis with ompA (MLVA-ompA) and multi sequence typing (MST) retain stability in Chlamydia trachomatis. Front. Cell. Inf. Microbio. 2:68. doi: 10.3389/fcimb.2012.00068
Received: 20 January 2012; Accepted: 30 April 2012;
Published online: 17 May 2012.
Edited by:
Yasuko Rikihisa, Ohio State University, USAReviewed by:
Ted Hackstadt, Rocky Mountain Laboratories/NIAID/NIH, USAYajun Song, Beijing Institute of Microbiology and Epidemiology, China
João P. Gomes, National Institute of Health, Portugal
Copyright: © 2012 Labiran, Clarke, Cutcliffe, Wang, Skilton, Persson, Bjartling, Herrmann, Christerson and Marsh. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Peter Marsh, Health Protection Agency, Microbiology Services, Public Health Laboratory Southampton, Southampton General Hospital, Tremona Road, Southampton, SO16 6YD, Hampshire, UK. e-mail: peter.marsh@uhs.nhs.uk