Bacterial toxin effector-membrane targeting: outside in, then back again
- Department of Microbiology-Immunology, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA
Pathogenic bacteria utilize multiple approaches to establish infection and mediate their toxicity to eukaryotic cells. Dedicated protein machines deposit toxic effectors directly inside the host, whereas secreted toxins must enter cells independently of other bacterial components. Regardless of how they reach the cytosol, these bacterial proteins must accurately identify their intracellular target before they can manipulate the host cell to benefit their associated bacteria. Within eukaryotic cells, post-translational modifications and individual targeting motifs spatially regulate endogenous host proteins. This review focuses on the strategies employed by bacterial effectors to associate with a frequently targeted location within eukaryotic cells, the plasma membrane.
Introduction
Pathogenic bacteria produce a wide array of virulence determinants that are either secreted into the extracellular matrix or “injected” directly into the target cell. These determinants can be classified as toxins or effectors, with the distinction depending on their mechanism of entry into the host cell, activity, and several other factors (reviewed in Galan, 2009). However, this nomenclature is further clouded by the existence of toxins that release effectors into the host upon toxin entry and protein cleavage (Egerer and Satchell, 2010). Regardless of their entrance mechanism or catalytic activity, toxic effectors must correctly identify the intracellular location of their target molecule once they reach the interior of the host cell.
Bacterial effectors have been shown to mediate their activity on many different targets within host cells, including proteins in the nucleus, mitochondria, endoplasmic reticulum, cytoskeleton, or plasma membrane (PM), as well as acting on the various host membrane phospholipids themselves. In addition to mediating cellular toxicity, numerous effectors combat host signaling pathways and clearance responses allowing for intracellular replication, host cell remodeling, intercellular spread, and immune evasion. Similar to how viruses hijack host cell processes utilizing molecular mimicry, many bacterial effectors also mimic the catalytic activities and targeting mechanisms of host proteins to ensure proper function within their host.
Although hundreds have been identified, much of the current research focus is on isolating new effectors as well as establishing catalytic activities and determining the roles in virulence played by known effectors. However, with the advent of advanced microscopic, genetic, bioinformatic, and biochemical tools the regions and mechanisms responsible for governing the intracellular location of these effectors are beginning to be identified.
Mechanisms of Translocating Bacterial Effectors into Host Cells
The most numerous and best-characterized effectors are those translocated through the type III and IV secretion systems (T3SS, T4SS). Proteins of the T3SS form a needle-like complex, or injectisome, that enables the bacteria to directly transport/inject effectors from the bacterial cytoplasm to the host cytosol. T3SS effectors are essential virulence factors for some of the most recognized human and insect pathogens including Pseudomonas aeruginosa, Yersinia pestis, Salmonella enterica, Shigella flexneri, Bordetella pertussis, Escherichia coli, non-O1 non-O139 Vibrio cholerae, and Photorhabdus luminescens. In addition, many plant pathogens including Xanthomonas spp., Erwinia spp., and Pseudomonas syringae utilize T3SS effectors to mediate their colonization of and toxicity to plant cells.
Similar to the activity and translocation of T3SS effectors, T4SS deliver virulence proteins from their associated pathogens from bacteria into host cells via an injectisome-like structure (Cascales and Christie, 2003). However unlike T3SS, T4SS effectors are not always directly deposited inside the host cytosol through the T4SS, but can also be secreted into the extracellular milieu or released into vacuoles within the cell depending on where their associated pathogens are found in relation to the cell (Backert and Meyer, 2006; Fronzes et al., 2009). T4 secretion has been shown to deliver toxins and effectors from pathogens that are strictly extracellular including Helicobacter pylori and B. pertussis as well as bacteria that replicate inside host cells including Legionella pneumophila, Bartonella spp., and Coxiella burnetii. Therefore T4SS substrates must contain specialized mechanisms to correctly identify their surface-bound, extravacuolar, or otherwise intracellular targets following secretion.
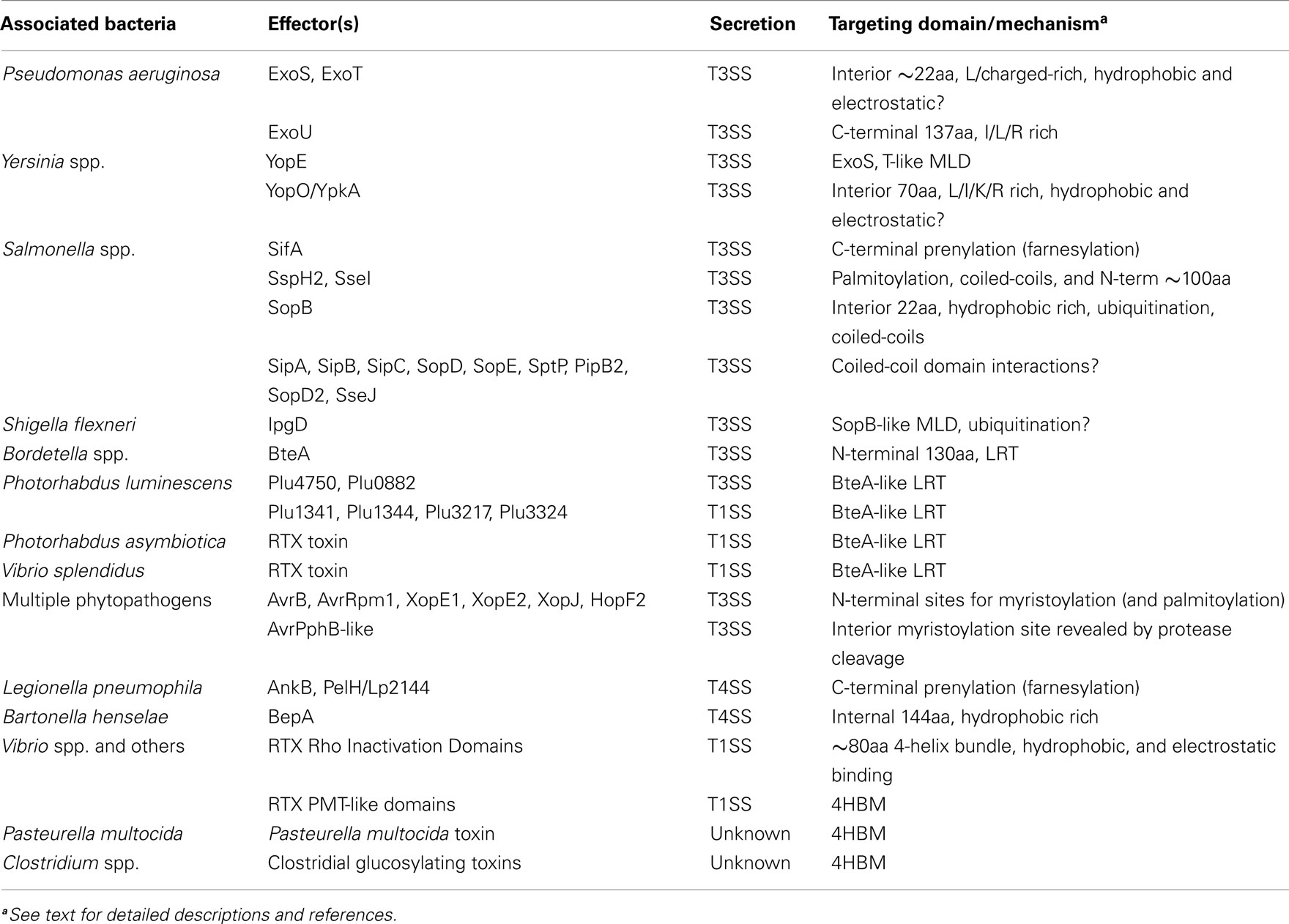
Similar to the array of functions performed by T3 and T4SS effectors, bacterial protein toxins display a huge diversity in the mechanisms they employ to intoxicate and manipulate host cells. A large number of these toxins utilize proteolysis within the host to release effectors as well as, reveal, enhance, and/or activate their catalytic activities; a variety of secreted multifunctional autoprocessing RTX toxins (MARTX) have recently been shown to utilize autoprocessing to release multiple individual catalytic effector domains into the host cytosol (Egerer and Satchell, 2010). MARTX toxins are produced by many species of Vibrio as well as an assortment of human and insect pathogens. These large toxins are generally composed of a central region containing multiple effector domains that is flanked by N- and C-terminal repeat regions (Satchell, 2011). While these repeat regions are thought to mediate host cell binding and translocation, the effector domains are believed to carry out the actual cytotoxicity of their associated toxins. Despite all of the MARTX toxins sharing this organization, the defining feature of this family of toxins is the presence of a cysteine protease domain (CPD), which is responsible for cleaving the full-length toxin into individual effector domains upon binding host cell inositol hexakisphosphate (recently reviewed in Egerer and Satchell, 2010). These cleavage events lead to the effector domains being released into the cytosol where they can then identify their intracellular target and carry out their activities as individual effectors. In this manner, the liberation of the MARTX toxin effector domains into the cell mimics that of the delivery of effectors through translocation by T3 and T4 secretion systems (Figure 1).
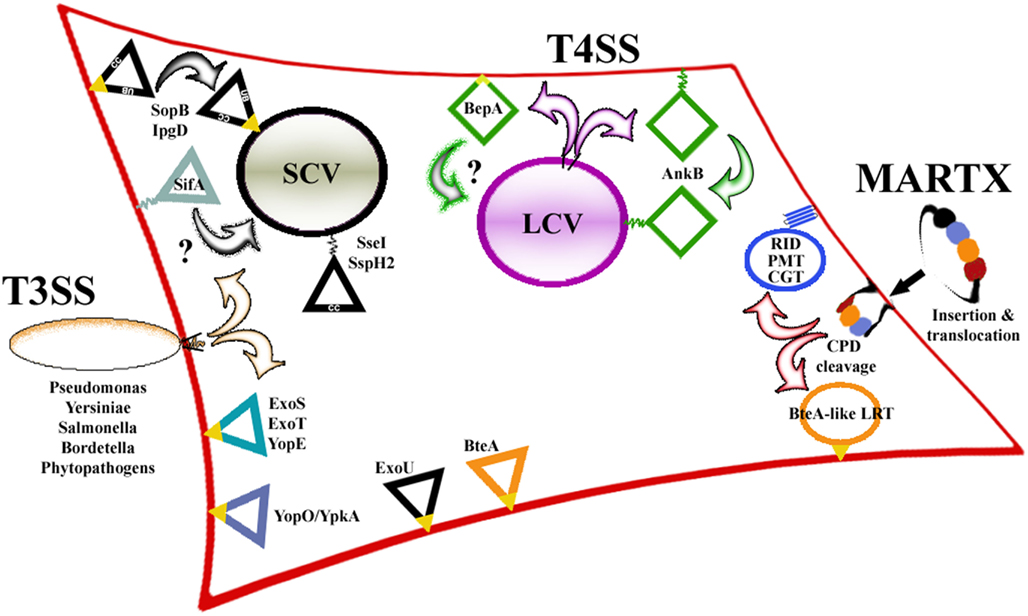
Figure 1. Schematic summary of the toxic bacterial effectors that target host cell plasma membranes. Shown are three central pathways (T3SS, T4SS, MARTX) responsible for effector translocation into the host cell and the intracellular locations of the effectors with lipidation motifs (jagged lines at either end of an effector) or dedicated membrane localization domains (triangles, T3SS; squares, T4SS; circles, MARTX). Potential hydrophobic interactions with the membrane are indicated as yellow patches on the effector. CC, coiled-coil domain; UB, ubiquitination site; SCV, Salmonella-containing vacuole; LCV, Legionella-containing vacuole. See text for details.
Modifying Membranes for Intracellular Survival: Salmonella and Legionella Effectors
Many bacterial pathogens utilize effectors for initiation of intracellular survival and replication. S. enterica serovar Typhimurium stimulates host cell invasion and spread through translocation of at least 39 effectors by two T3SS (T3SS1 and T3SS2). These effectors modulate aspects of internalization, virulence, as well as the production of a membrane-bound compartment where Salmonella reside within the host cell. This compartment, the Salmonella-containing vacuole (SCV), is a host cell derived membrane-bound vacuole that, through the action of multiple T3SS effectors, provides a protected intracellular home for Salmonella to replicate and evade degradation. In addition to establishing their connection to infection dynamics, several specific functions have been attributed to many of these effectors (for a recent comprehensive review of Salmonella effectors and their activities and roles in disease, see Agbor and McCormick, 2011). Many of these effectors function at different stages of the Salmonella infection cycle, therefore necessitating stringent regulatory mechanisms to ensure proper timing of effector activity. To control this, in addition to having their expression and translocation regulated temporally, the effectors utilize host cell modifications and intrinsic motifs to control their intracellular locations. Several of these spatial determinants have been identified, particularly from effectors targeting the host cell PM; SifA, SspH2, SseI, SopB, PipB2, and SseJ have all been found associated with the PM, albeit through diverse means (Figure 1; Table 1).
The most well characterized T4SS involved in translocation of virulence factors, the Legionella Dot/Icm system, is also involved in modifying host cell membranes. Similar to Salmonella, Legionella evade the host immune response by modifying phagosomes to prevent fusion with host lysosomes, thereby permitting replication inside the cell within Legionella-containing vacuoles (LCV; Isberg et al., 2009). Several groups have shown that L. pneumophila utilizes the Dot/Icm T4SS and >275 effectors to modulate nearly every step of the infection process, including formation of the LCV from host cell membranes (Segal et al., 1998; Vogel et al., 1998; Nagai and Roy, 2003; Ensminger and Isberg, 2009; Isberg et al., 2009; Zhu et al., 2011). Also like Salmonella, many of these Dot/Icm effectors are directed to the host PM and/or the LCV through host cell modifications or by PM targeting motifs (Figure 1; Table 1).
Although they possess different methods of entry into the host cell, bacterial effectors play diverse roles in the pathogenesis of their cognate organisms, with many acting on host membranes and membrane-associated proteins that are often the first potential target upon entry. Accordingly, these effectors have evolved an array of mechanisms to identify and interact with the PM. Therefore, the focus of this review will be on the bacterial effectors that target the host PM and will discuss the two main strategies employed by these effectors to mediate these associations: localization via host cell modifications and possession of dedicated structural motifs sufficient for PM localization (summarized in Figure 1; Table 1).
Host Cell Modifications that Mediate PM Localization
Intracellular targeting in eukaryotic cells is often determined through post-translational modifications, with lipidation serving as the major means to deliver proteins to the PM. Although host cell enzymes carry out these modifications, bacterial effectors have evolved mechanisms to mimic the recognition sites of these enzymes in order to utilize these modifications for their intracellular targeting.
Prenylation of C-Terminal Cysteine Residues
The Salmonella effector SifA is a GTPase involved in SCV membrane maintenance and contains a C-terminal CAAX-motif that is necessary and sufficient for targeting it to the PM of host cells (Figure 2; Boucrot et al., 2003). Many eukaryotic proteins containing CAAX-motifs, including several Rho-GTPases, are modified by host cell enzymes that attach a lipid isoprenyl moiety to the conserved Cys residue (Gao et al., 2009). This post-translational modification serves to enhance membrane localization by increasing the overall hydrophobicity of the protein thereby influencing the intracellular targeting of CAAX-motif-containing proteins. Studies on transfected HEK 293T cells show that SifA is prenylated within the cell, while in vitro studies confirm that prenylation occurs on the Cys residue within the SifA CAAX-motif by protein geranylgeranyl transferase I (Reinicke et al., 2005). Intriguingly, although deletion of SifA resulted in significantly attenuated survival and growth of Salmonella in macrophages, the infection dynamics of Salmonella expressing SifA with a mutated CAAX-motif were nearly indistinguishable from the wild-type SifA expressing parental strain (Reinicke et al., 2005) despite the SifA CAAX mutant showing decreased activity (Boucrot et al., 2003). These results indicate that either the reduced levels of SifA activity are sufficient to maintain a typical infection or that there are multiple overlapping mechanisms in place to ensure SifA reaches its membrane-associated intracellular target during infection.
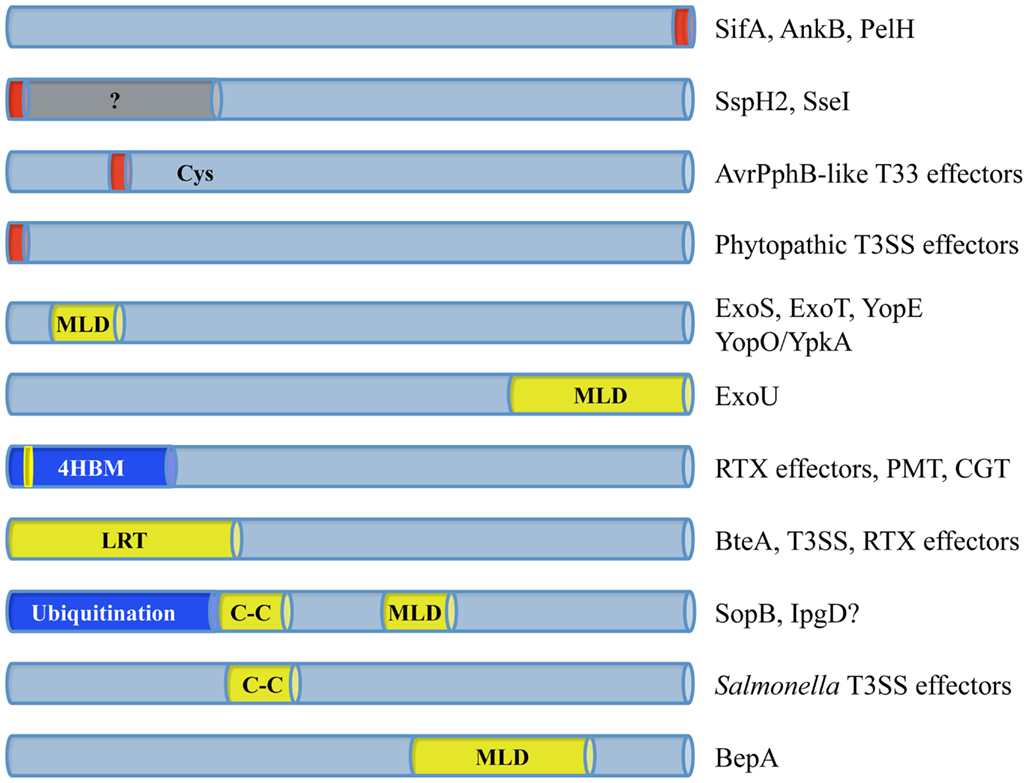
Figure 2. Plasma membrane targeting motifs are not restricted to specific locations within the various effectors. The dedicated membrane localization domains (MLD) that have been identified are found at both termini and in the interior of their effectors. Many of these MLDs contain stretches of Ile/Leu/Val residues (yellow) to mediate hydrophobic membrane associations. In addition, regions rich in basic residues (dark blue) can enhance membrane binding by generating electrostatic interactions or serving as sites for host cell ubiquitination. Sites for host cell lipidation (red) are also found at both termini as well as on the interior of certain effectors, which is exposed following cysteine protease cleavage. Several prenylated effectors contain motifs that provide additional targeting information (gray) to identify specific locations within the PM.
Similar to SifA, many L. pneumophila effectors translocated into the host via T4SS are prenylated, which delivers them to the PM, LCV, or both, as in the case of AnkB. AnkB is a L. pneumophila effector involved in binding polyubiquitinated bacterial proteins to the LCV (Price et al., 2009). These polyubiquitinated proteins are essential for proper formation and maintenance of the LCV, and accordingly, deletion of AnkB prevents L. pneumophila replication within the host as well as disease (Price et al., 2009, 2010a). When ectopically expressed in either mammalian cells or the amebae Dictyostelium discoideum, AnkB was found exclusively at the cell periphery. Peripheral localization was subsequently shown to be dependent on a CAAX-motif (CVLC) at the extreme C-terminus of AnkB, because mutation of this motif abolished localization. Further, when these residues were fused to a cytosolic protein, they were sufficient to target the PM (Figure 2; Price et al., 2010a). AnkB was also confirmed to be prenylated by a host cell farnesyltransferase and that this modification was necessary for PM targeting. Besides these lipidation events, Ivanov et al. (2010) showed that AnkB contains a stretch of positively charged residues that although not required to maintain membrane association, were necessary for PM localization suggesting that additional targeting information might determine membrane specificity. Interestingly, despite its exclusively peripheral localization when ectopically expressed, AnkB is predominantly associated with the outside of the LCV when translocated by the T4SS during infection (Price et al., 2009, 2010a,b). This alternate localization could indicate that AnkB is associating with another protein on the LCV or that the changes in the LCV membrane caused by additional Legionella proteins drive AnkB to the LCV (Yeung et al., 2009). In addition to studies looking at specific phospholipids and proteins present on each, these localization results show that the outer membrane of the LCV retains sufficient similarity to the PM for lipidation to be employed by bacterial effectors to target the LCV.
In silico Identification of Prenylated Effectors
Apart from SifA and AnkB, several additional bacterial effectors have been shown to contain putative CAAX-motifs responsible for their intracellular targeting. Two recent bioinformatic analyses looking at the sequenced genomes of four different Legionella isolates for CAAX-motifs both identified 11 potentially farnesylated effectors (Ivanov et al., 2010; Price et al., 2010b). Both studies transiently expressed these effectors in mammalian cells and found that only PelH localization appeared similar to that of AnkB, with all of the others displaying various patterns of peri-nuclear localization and co-localization with Golgi markers. Importantly, the CAAX-motifs and host modification were necessary for all patterns of localization, as mutation of the Cys residue or treatment with a prenylation inhibitor decreased membrane association for all of the effectors (Table 1; Ivanov et al., 2010; Price et al., 2010b). In further support of the overall importance of lipidation to the targeting of Dot/Icm effectors, co-localization studies in D. discoideum show that the host cell enzymes responsible for protein farnesylation are associated with the LCV during infection (Al-Quadan and Kwaik, 2011). Therefore, effector lipidation can either occur within the cytoplasm before the LCV is formed or locally on the surface of the LCV once the effectors are released from the interior of the LCV. These results show that many Dot/Icm proteins are localized to various membranes within the cell through host-mediated post-translational modification and that targeting influences the proper formation and maturation of the LCV during disease.
Due to the success of this bioinformatic approach in identifying L. pneumophila prenylated effectors, further computational methods were used to predict additional similarly targeted effectors (Al-Quadan et al., 2011). Analogous sequence and structure-based approaches had previously been employed in order to establish a defined prenylation motif and identify putative farnesylated substrates (Maurer-Stroh and Eisenhaber, 2005; London et al., 2011). An initial in silico analysis of 17 sequenced mammalian or plant pathogens using these techniques revealed the presence of 54 proteins from 14 species containing a putative secretion system that have a predicted CAAX-motif (Al-Quadan et al., 2011). While this list contains known and putative effectors delivered by multiple different pathways, 38 proteins were identified that were previously unclassified as effectors and are likely targeted to the PM via their associated CAAX-motifs. Given the commonality of prenylation as a PM targeting mechanism used by bacterial and eukaryotic proteins as well as the volume of genome sequence data available, applying these sorts of predictions to larger data sets will undoubtedly be useful in the preliminary stages of identifying novel effectors and PM-targeted proteins.
N-terminal Cysteine Palmitoylation
Similar to prenylation by geranylgeranyl transferases, other host enzymes catalyze the reversible fusion of palmitic acid or similar lipids to N-terminal Cys residues through amide or thioester linkages. These reactions, termed palmitoylation, are mediated by palmitoyl acyl transferases and have been shown to enhance the PM association of both eukaryotic and bacterial proteins (Resh, 1994). Two Salmonella T3SS effectors, SspH2 and SseI, have recently been shown to require palmitoylation to target the host PM (Figure 2; Hicks et al., 2011). SspH2 and SseI contain N-terminal consensus palmitoylation motifs and require the Cys residue for lipidation, PM localization, and effector activity (SseI; Hicks et al., 2011). Interestingly, both SspH2 and SseI contain additional information required for proper lipidation and localization (Figure 2). Fusion of the palmitoylation motif of SspH2 to a non-palmitoylated or PM-associated Salmonella effector did not result in PM localization or palmitoylation. Also, although full-length SseI fused to GFP localized to the basolateral side of the PM of polarized cells, a truncated version of SseI-GFP was found at the apical PM (Hicks et al., 2011). Therefore, palmitoylation alone was not sufficient to properly target either effector to its correct intracellular destination indicating that further study is necessary to fully understand their mechanisms of targeting.
Myristoylation Mediated Targeting
Besides attachment of lipid moieties to Cys residues during prenylation or palmitoylation, host cell enzymes can also lipidate Gly residues to enhance the hydrophobicity of proteins. This lipidation, myristoylation, is catalyzed by N-myristoyltransferase and involves the covalent linkage of C14 fatty acid (myristate) to Gly residues at the extreme N-terminus of a protein (Boutin, 1997; for a more detailed description of the consensus recognition motif see Martin et al., 2011). Similar to palmitoylation, myristoylation can also influence the intracellular localization of proteins and the two modifications are often combined to ensure targeting and association with the PM (Resh, 1994). Many Ras family members, frequent targets of T3SS effectors, utilize myristoylation and/or palmitoylation to enhance their membrane binding (Roy et al., 2000). Comparisons of the extreme N-terminal regions of several plant pathogen-secreted effectors with their targets show the presence of lipidation sites on both effector and target, suggesting that analogous modifications deliver each to shared sites within the cell.
Sequence homology searches have identified a large number of effectors (>20) produced by phytopathic species of P. syringae and Xanthomonas that contain putative myristoylation sites (Nimchuk et al., 2000; Robert-Seilaniantz et al., 2006; Thieme et al., 2007). Subsequent studies looking at the importance of the Gly within the myristoylation site of many of these proteins (including, but not limited to, AvrB, AvrRpm1, XopE1, XopE2, XopJ, or HopF2) found that mutants lacking this Gly were no longer PM-associated, functioned in plants, or lipidated in vitro (Figure 2; Nimchuk et al., 2000; Robert-Seilaniantz et al., 2006; Thieme et al., 2007). Interestingly, putative palmitoylation sites are found immediately adjacent to the myristoylation sites within most of these effectors. Mutation of the palmitoylated Cys within these sites on P. syringae AvrB or AvrRpm1 did not abolish PM association or activity like removal of the myristoylated Gly, although the Cys mutants displayed partially reduced activity and membrane association (Nimchuk et al., 2000). These varying effects on activity can be explained by the observation that myristoylation often precedes palmitoylation, therefore myristoylation is required for complete palmitoylation of these effectors and that, while myristoylation is sufficient for some activity, both modifications are needed for full membrane association and activity. Importantly, the targets of several of these effectors have been identified and are found at the PM, thus explaining the requirement for lipidation (Axtell and Staskawicz, 2003; Shao et al., 2003; Bosis et al., 2011; Wu et al., 2011) (see Block and Alfano, 2011 for a review of all P. syringae effector targets).
In addition to the effectors that possess “classical” N-terminal sequences for lipidation, several classes of phytopathic effectors have been identified that contain internal myristoylation sites (Figure 2). These motifs are exposed following autocatalytic cleavage by CPDs attached to the effector (Nimchuk et al., 2000). Homologs of the most well characterized of these effectors, AvrPphB, can be identified within other plant-associated bacteria (Dowen et al., 2009). In vitro biochemical studies on 7 of the 13 homologs showed three unexpected findings: (1) not all of the AvrPphB family members displayed autoprocessing; (2) of those that were autoprocessed, not all of the cleavage events led to exposure of lipidation sites; and (3) two homologs displaying putative N-terminal myristoylation and palmitoylation sites following cleavage were not lipidated (Dowen et al., 2009; Fotiadis et al., 2012). Despite these discrepancies, mutation of the Gly residue adjacent to the cleavage site within the AvrPphB-like effectors abrogated PM localization and activity (Nimchuk et al., 2000; Dowen et al., 2009; Wu et al., 2011). Thus, although many of these effectors initially mask their targeting motifs, they ultimately behave similarly to other “classically” lipidated effectors and identify the PM through enhanced N-terminal hydrophobicity.
Altogether, despite significant differences in the composition of plant and mammalian cell membranes, an array of effectors utilize multiple types of lipidation by host cell enzymes in order to modulate their delivery to the host PM where they can mediate their function and benefit their cognate bacterial pathogens.
Proteins with Dedicated Domains for Membrane Localization
Similar to the diversity of lipidation-mediated targeting mechanisms employed within the host cell, several classes of post-translational modification independent PM targeting domains have been identified within eukaryotic, viral, and bacterial proteins. Homologous to the variety of host cell modifications utilized by bacterial effectors, several different dedicated PM targeting structural motifs are present within effectors released by MARTX toxins, T3SS, and T4SS. It is important to note that, in addition to the effectors and domains described below, a number of effectors and toxins are directly inserted into the PM as integral membrane proteins through trans-membrane domains (e.g., Tir, diphtheria toxin, etc.; Kenny et al., 1997; Zhao and London, 2005). However because of their prevalence and diversity, this review focuses on the mechanisms utilized by effectors to mediate peripheral attachment to the PM.
Small, Hydrophobic Domains for Membrane Binding
Many bacterial effectors target host cell Rho family GTPases that are responsible for modulating numerous intracellular signaling pathways. These effectors function to either activate or inactivate the Rho proteins, thereby altering host signaling to benefit the associated bacteria. Active Rho-GTPases are typically membrane-associated; therefore effectors have had to devise strategies to accurately target these membranes to subvert Rho signaling activity. P. aeruginosa ExoS and ExoT and Yersinia spp. YopE have been shown to act as GTPase-activating proteins (GAPs) on substrates within the ER-Golgi network in the peri-nuclear region of the cell (Black and Bliska, 2000; Kazmierczak and Engel, 2002; Krall et al., 2002, 2004). A shared membrane localization domain (MLD) has been identified within all three of these effectors that is responsible for mediating their initial association with the PM immediately following translocation, then with the ER-Golgi following trafficking (Krall et al., 2004; Isaksson et al., 2009). Each of these ∼22-residue conserved MLDs are rich in Leu residues, contain multiple charged residues (R/K, D/E), and possess at least one highly hydrophobic residue (Phe; Figure 2; Table 2). Deletion or mutation of any of these MLDs significantly reduced the virulence of either P. aeruginosa (Zhang and Barbieri, 2005) or Y. pseudotuberculosis (Isaksson et al., 2009), emphasizing the importance of these domains for effector activity.
Although direct membrane association has not been shown, mutagenesis studies and alpha-helical predictions on ExoS combined with structural evidence from YopE suggest how these interactions may occur. The crystal structure of YopE shows that the MLD is surface exposed, largely unstructured, and contains a short (five residues) β-sheet (β3) and a portion of an α-helix (α2; Birtalan et al., 2002). Mutagenesis studies on ExoS show that substitution of the conserved Leu residues in MLDExoS for Asn had the same effect on membrane binding as deletion of the entire MLD, whereas substitutions of the two Arg residues for Asn abolished PM localization but did not affect the overall percentage of membrane-associated ExoS (Zhang and Barbieri, 2005; Zhang et al., 2007). In addition, helical wheel predictions on ExoS show that the MLD forms an amphipathic helix with the Leu residues found on one face, whereas the charged residues are on another (Zhang et al., 2007). These findings indicate that the Leu residues possibly mediate binding to all cellular membranes while the charged residues act to enhance PM interactions, possibly through electrostatic contacts. Although MLDYopE has been crystallized, because it is bound to its chaperone in the crystal the in vivo orientation is not discernable (Birtalan et al., 2002). Although it is unlikely, in the absence of evidence showing direct membrane association and because amphipathic helices are often involved in protein–protein interactions, it is also possible that these MLDs could be responsible for binding a host membrane-associated protein.
The Salmonella T3SS effector, SopB, incorporates both a structural motif and host cell modification to regulate its intracellular location. SopB is a phosphoinositol phosphatase whose activity controls both the initial stages of SCV formation and maturation through hydrolysis of different host cell phospholipids. Hydrolysis of the phospholipids decorating the SCV leads to alterations in SCV membrane charge and host signaling cascades (Hernandez et al., 2004; Mason et al., 2007; Mallo et al., 2008; Bakowski et al., 2010). Immediately following translocation into the cell, SopB is found at the PM where it is ubiquitinated by host enzymes (Patel et al., 2009). Mutation of the residues required for SopB ubiquitination blocked delivery to the SCV leading to PM-retention, indicating that ubiquitination, unlike lipidation-mediated delivery to the PM, is likely responsible for removing SopB from the PM and that another mechanism was involved in PM targeting. In silico searches of SopB for trans-membrane domains that might mediate membrane binding led to the discovery of a 22-residue long region required for PM association following translocation into the cell (Figure 2). This region, in addition to sharing the same size as the MLDs from ExoS, ExoT, and YopE, is also rich in Leu/Val/Ser residues and is thought to mediate its attachment to the membrane through strong hydrophobic interactions (Table 2). Similar to ExoS, ExoT, and YopE, deletion of this domain abrogated PM targeting and ubiquitination of SopB (SopBΔ288–309). Further, Salmonella expressing SopBΔ288–309 were unable to invade or replicate in host cells and were unable to modulate intracellular signaling. Importantly, purified SopBΔ288–309 retained near wild-type phosphatase activity in in vitro assays, indicating that intracellular targeting and catalytic activity are separable. These data show that SopB utilizes a short hydrophobic sequence for PM localization and that this domain is necessary for Salmonella infection (Patel et al., 2009). Interestingly, a functionally homologous effector translocated by Shigella, IpgD, contains a highly similar sequence, suggesting that this is a shared motif employed for targeting phosphoinositol phosphatases to vacuoles generated by intracellular pathogens.
The T4SS Effector BepA Contains an MLD
Apart from lipidation, other T4SS effectors contain dedicated targeting motifs to mediate phospholipid binding and their intracellular location. While they are not found at the PM, the L. pneumophila effectors SetA, SidM/DrrA, SidC, LidA, and LpnE all bind specific phosphatidylinositols within the cell in order to associate with the LCV (Yeung et al., 2009; Jank et al., 2012). In addition to these and the prenylated L. pneumophila T4SS effectors that are discussed above, a 142aa region of the Bartonella henselae T4SS effector BepA (305–446) has been shown to be necessary and sufficient for PM-binding, indicating that it is a dedicated MLD (Figure 2; Schmid et al., 2006). Like the previously described T3SS effector MLDs that associate with the PM, MLDBepA contains a high percentage (25%) of highly hydrophobic (I, L, V) residues suggesting that membrane binding may be mediated through hydrophobic contacts (Table 2). Interestingly, BepA is responsible for preventing endothelial cell apoptosis that likely permits chronic B. henselae infection, and BepA PM localization is necessary for this activity (Schmid et al., 2006). Determining the mechanism of PM association by BepA will therefore be important in further characterizing how this intracellular pathogen evades clearance.
The ExoU MLD Affects Catalysis
In addition to those found in its other primary effectors, a dedicated MLD was recently identified within the P. aeruginosa effector ExoU (Rabin et al., 2006). Like ExoS and ExoT, ExoU requires an intact and functional MLD to retain full toxicity as strains secreting ExoU with mutations in the MLD show decreased cytotoxicity to HeLa cells and are less virulent in a mouse model of pneumonia (Veesenmeyer et al., 2010). ExoU does not target Rho-GTPases like ExoS and ExoT, but is a phospholipase (Phillips et al., 2003). Therefore, the MLD likely directs the catalytic domain of ExoU to the interior face of the PM after translocation where its phospholipase activity induces rapid cell lysis (Rabin et al., 2006). The final 87 residues of ExoU encompass the MLD and have been shown to be necessary and sufficient to target GFP to the PM, although inclusion of an additional 50 residues (550–687) enhances PM localization (Figure 2; Rabin et al., 2006). Similar to the other P. aeruginosa and Yersinia spp. effector MLDs, MLDExoU contains I/L and Arg residues that are required for proper localization (Table 2; Veesenmeyer et al., 2010). Interestingly, numerous mutations within the MLD that decrease PM association also decrease the catalytic activity of ExoU, indicating that MLDExoU is not only responsible for membrane association but affects catalysis directly, possibly through substrate or cofactor binding (Veesenmeyer et al., 2010). Further in vitro biochemical analysis and structural determinations will be required to determine the exact mechanism of how these activities are intertwined.
Conservation of a Four-Helix Bundle MLD in Three Toxin Families
In an effort to further characterize the mechanism of action of several MARTX toxin effectors, a deletion analysis revealed the presence of a dedicated MLD within the N-terminal portion of the V. cholerae Rho inactivation domain (RID; Figure 2; Geissler et al., 2010). PSI-BLAST searches performed on this 85aa portion of RID (MLDRID) identified 24 homologs of this sub-domain: nine MARTX RIDs; Pasteurella multocida mitogenic toxin (PMT) and nine PMT-like MARTX effector domains; and five Clostridial glucosylating toxins (CGTs; Figure 3A; Geissler et al., 2010, 2012). These homologs were therefore found associated with three different toxin families from a diverse collection of bacteria (Figure 3B) and while the mechanisms of intoxication employed by these families are different, the intracellular targets of all three types of toxin are found at the membrane; PMT acts on several Gα-subunits of different G-protein coupled receptors (Orth et al., 2009), the CGTs inactivate PM-bound Rho-GTPases (Just et al., 1995), and the RIDs likely act on an as yet unidentified target in the PM to inactivate RhoA (Sheahan and Satchell, 2007; Geissler et al., 2010). Interestingly, RIDΔMLD retained its cell-rounding ability when expressed at high levels during transient transfection, but was no longer toxic when delivered at more physiological levels (Geissler et al., 2010). Furthermore, despite retaining their full catalytic activities, disrupting the MLD from either PMT or TcsL abolished toxicity to tissue culture cells (Mesmin et al., 2004; Kamitani et al., 2010). These data show that while the catalytic site and enzymatic activities of each family of toxin are not linked to the MLDs, as in the case of ExoU, enhancing the ability of each toxin to identify its target is essential for toxicity.
Figure 3. Despite the shared overall structure for membrane associations by the four helical bundle membrane localization domains, their primary amino acid sequences show significant diversity. (A) Pairwise sequence alignment of all 24 of the currently identified 4HBMs organized according to their sequence identity. A schematic representation of the structure of the domains is depicted below alignment. To highlight their locations on either loops or helices, only basic (blue), hydrophobic (yellow), and Phe (magenta) residues are boxed. (B) Neighbor joining tree illustrating the phylogenetic relationships of the 4HBMs; tree is based on sequence pairwise identity. (C) Structural alignment of five different crystallized and modeled 4HBMs shows nearly complete overlap for all of the homologs, regardless of their primary amino acid sequence or associated catalytic domain.
The homology of MLDRID with PMT is confined to a region that is necessary and sufficient for PMT membrane targeting (Kitadokoro et al., 2007) and the initial 15aa of the homologous regions from TcsL and TcdB are required for full toxicity and in vitro liposome binding, suggesting that they were involved in PM associations (Mesmin et al., 2004). In support of these findings and similar to other MLDs that have been discussed herein, GFP-fusions to putative MLDs from each toxin family displayed significant PM localization and were found in the membrane fraction following cellular fractionation (Kitadokoro et al., 2007; Geissler et al., 2010, 2012; Kamitani et al., 2010).
While the amino acid sequence similarities between the MLD homologs from the different toxin families is relatively low (∼40%), each of these domains likely shares a conserved overall structure (Figures 3A,B; Table 2). The crystal structures of the catalytic C-terminal portion of PMT and the glucosyltransferase domains from TcdA, TcdB, TcsL, and TcnA show that each MLD is composed of four closely packed α-helices that are connected by short loop regions (Reinert et al., 2005; Kitadokoro et al., 2007; Ziegler et al., 2008; Pruitt et al., 2012). In addition, structural predictions performed on many of the Vibrio MARTX effector MLDs showed similar four helical bundles (Figure 3C; Geissler et al., 2010). Therefore, to distinguish these MLDs from those produced by T3 and T4SS effectors, the members of this family of domains are referred to as 4-helical bundle MLDs (4HBM; Geissler et al., 2012). Several studies using structure- and homology-based mutagenesis and structural modeling have provided insight into the mechanism of PM-binding by the 4HBMs. These studies show that individual residues involved in either maintaining the 4-helix bundle or generating a positively charged surface are essential for proper localization (Geissler et al., 2010, 2012; Kamitani et al., 2010). While maintenance of the bundle likely contributes to the overall structure of the protein, individual surface exposed residues likely play direct roles in protein-membrane association. In support of this, mutation of basic residues within the Loop1–Loop3 region (L1–L3) of several 4HBMs significantly reduced their ability to localize to the PM (Figure 3C; Geissler et al., 2010, 2012).
Positively charged surfaces of proteins often mediate electrostatic interactions with negatively charged host cell membranes; these interactions are often enhanced by the insertion of hydrophobic residues into the lipid bilayer (Arbuzova et al., 2000; Zhang et al., 2003; Mulgrew-Nesbitt et al., 2006). That basic residues within L1–L3 were required for proper PM localization and the observation that highly hydrophobic residues (I, L, F) were found in L1 of those 4HBMs that showed strong PM localization, suggested that this region might be involved in mediating membrane binding. Transfection and in vitro studies showed that both basic and hydrophobic residues within L1 are required for the 4HBMs to fully associate with the host PM and negatively charged phospholipids (Geissler et al., 2012). Therefore, those 4HBMs that contain an L1–L3 region rich in basic and hydrophobic residues are more capable of associating with the PM than those 4HBMs lacking either type of residue (Geissler et al., 2012).
The BteA-Like Family of Lipid Raft-Targeting MLDs
In contrast to the numerous effectors delivered by MARTX toxins, T3 and T4SS, human pathogenic Bordetella species only secrete one effector via their T3SS (Panina et al., 2005). This effector, BteA, is a 658aa cytotoxin produced by all three of the human pathogenic Bordetella species (B. bronchiseptica, B. pertussis, and B. parapertussis). Upon translocation into a variety of host cells, BteA induces non-apoptotic cell death and is thought to be a central virulence factor because deletion of BteA abolishes cytotoxicity and results in a phenotype indistinguishable from deletion of the T3SS itself (Panina et al., 2005). In an effort to characterize the intracellular localization of BteA, the N-terminal 130aa was found to contain an MLD and was necessary and sufficient to target GFP to the lipid raft portion of the HeLa cell membrane (Figure 2; French et al., 2009). Since it targets a specific portion of the PM, which distinguishes it from other known MLDs, the BteA MLD was therefore termed the lipid raft-targeting domain (LRT).
Further examination of the sequence of the BteA LRT domain shows that it has a similar composition to other identified MLDs: BteA is rich in highly hydrophobic (∼20% are Ile, Leu, Val) and positively charged (>10% are Arg, Lys) residues (Table 2). BLAST searches revealed that the central 78aa of this region (aa34–112) were homologous to 8 other proteins in the database (French et al., 2009). Two of these homologs are found within predicted T3SS effectors from P. luminescens, while the other 6 are found inside MARTX toxins. GFP-fusions to two other BteA-like LRT homologs displayed comparable localization patterns as BteA LRT-GFP, indicating that there is functional conservation amongst the domains and that these are a new family of MLDs that target eukaryotic membrane rafts (French et al., 2009).
Despite having a novel MLD, BteA displays many similarities to other effectors that possess dedicated MLDs. Expression of BteA lacking its targeting domain displayed toxicity indistinguishable from full-length BteA when it was expressed in the cell at high levels via transfection. However, BteA ΔLRT-GFP was unable to be detected in the membrane fraction (French et al., 2009). These results show that the LRT and catalytic domains of BteA are separable and that BteA can still identify its target when expressed at high levels. It remains to be seen if deletion of the LRT will abolish the toxicity of BteA or the other LRT domain-containing effectors. Thus, although much is known about the BteA-family LRT domains there are many unanswered questions, including the mechanism of the LRT-PM association.
Several BteA-like LRT MLDs are present in MARTX toxins produced by Vibrio splendidus and the insect pathogens P. luminescens and P. asymbiotica (French et al., 2009). Interestingly, the putative effectors attached to these MLDs show no similarity with the catalytic portion of BteA or other known effectors, and are yet to have any ascribed function (French et al., 2009; Satchell, 2011). Despite the lack of knowledge of their associated effectors, the localization of the isolated LRT domain from one of the P. luminescence MARTX toxin effectors was similar to that of BteA. Therefore, this family of LRT domains is likely functionally conserved across toxin types and the effectors attached to the BteA-like LRT domains likely assert their toxicity through direct contact with PM lipid rafts or raft-associated proteins (French et al., 2009).
These differently sized, mostly unrelated bacterial motifs found attached to distinct types of enzymes represent assorted domains that are each able to independently identify the eukaryotic PM. Despite these differences, all of these MLDs likely serve to enhance the natural affinity of their cognate catalytic domains for their membrane-bound targets in order to promote bacterial infection and disease.
Additional Means of Regulating Intracellular Targeting
Apart from the host cell modifications and dedicated MLDs responsible for intracellular targeting, additional structural motifs that affect localization have been identified within several effectors. While these motifs have not necessarily been shown to independently localize to the PM, they have been shown to affect the localization of their associated effectors.
Coiled-coils Assist PM Targeting
In general, coiled-coil motifs are thought to mediate protein–protein binding through interactions between the amphipathic a-helices that make up these domains (Burkhard et al., 2001). In silico analysis performed using two independent prediction programs found that of the 39 Salmonella T3SS1 and T3SS2 effectors analyzed, 19 are predicted to contain putative coiled-coil domains. Of these coiled-coil containing effectors, 12 had previously been shown to be membrane-associated, including SopB (see Knodler et al., 2011 for specific proteins and references). Mutations designed to interrupt the structure of the coiled-coils completely abolished the targeting and membrane localization of four different Salmonella effectors, highlighting the importance of these domains for proper targeting. Despite this, GFP-fusions to the isolated coiled-coil were strictly cytosolic, indicating that they were necessary but not sufficient for membrane association. Coiled-coil domain swapping experiments between the effectors were unsuccessful, showing that each effector relies upon its cognate coiled-coil motif and that they are not functionally interchangeable. Interestingly, these motifs do not overlap with any of the characterized effector-targeting/chaperone-binding regions (see below), suggesting that coiled-coil domains either represent an additional level of spatial regulation or that they are simply employed by several PM-associated effectors for binding their target (Figure 2).
Chaperone-Binding and Membrane Localization
In addition to possessing signal sequences, sub-cellular targeting motifs, and catalytic domains, specific binding sites for individual chaperone proteins have also been found within several T3SS effectors. Although no specific function has been confirmed, chaperone-binding is thought to play a variety of roles in the delivery of their associated effectors into the host cell including; identification of the T3SS apparatus within the bacterial cell (Birtalan et al., 2002), structural maintenance of the effectors prior to export (Luo et al., 2001; Page et al., 2002), facilitating transport through the T3SS needle (Zheng et al., 2012), and influencing the order in which the effectors are delivered (Boyd et al., 2000). Interestingly, the chaperone-binding site (CBS) often overlaps with the MLD within effectors (Boyd et al., 2000; Birtalan et al., 2002; Letzelter et al., 2006). The MLD from YopO/YpkA, a Rho-targeting effector produced by Yersinia, completely coincides with the CBS. The YopO/YpkA MLD spans residues 20–90 and is rich in I/L and K/R residues (Table 2), whereas residues 20–77 are essential for chaperone-binding (Figure 2; Letzelter et al., 2006). The CBS is also found within the BteA LRT domain (French et al., 2009), suggesting that coordination of chaperone-binding and intracellular targeting is a conserved strategy utilized by different classes of effectors. In support of the predicted roles for chaperones in T3SS, deletion of the YopO/YpkA shared MLD/CBS motif prevents the export and translocation of their associated effectors (Boyd et al., 2000), although the MLD/CBS are not required for proper delivery as some effectors are still translocated when their MLD/CBS are deleted (Letzelter et al., 2006). This overlap between the MLD and CBS suggests that, due to its highly hydrophobic/charged nature, the MLD is likely a favorable spot for chaperone-binding along with membrane association. However, further biochemical and biophysical studies are warranted to determine the role of chaperone-binding to effector intracellular targeting within the host.
Topics for further Consideration and Concluding Remarks
The wide array of PM targeting mechanisms and dedicated MLDs that bacterial effectors utilize to identify the host cell PM and/or their PM-associated targets highlight the importance of spatial recognition to effector function. This is superbly illustrated in the findings that several bacterial effectors are no longer toxic to cells when their targeting motifs are removed, despite not affecting their catalytic abilities. While all effectors possess a certain affinity for their substrates, directing their localization to these targets circumvents the crowded environment of a cell and can enhance catalysis. Still, several membrane-associated effectors are able to identify their substrate by possessing a strong affinity for their target or having a dedicated binding site associated with the catalytic site. Determining the contribution of these affinities to intracellular targeting will further clarify the roles of dedicated targeting motifs.
The presence of so many effectors that utilize different host cell enzyme modifications for mediating their intracellular targeting shows how pathogens have evolved their virulence repertoire to effectively modulate host functions. Conservation of these lipidation motifs sites also suggests that these sites have been selected for or acquired during frequent contact with their hosts. The fact that these effectors are produced by both extra- and intracellular pathogens of animals, plants, and insects is intriguing and provides additional evidence of pathogens evolving to mediate their survival and occupy specific environmental/host niches. Furthermore, the varied types of effectors that utilize these short sequences for modifications illustrates the effectiveness of this strategy for identifying substrates and shows that effector-targeting can be streamlined to avoid integration of complex mechanisms of targeting. In depth genetic analyses are needed to identify the origins of these motifs and to determine how they have become so widely distributed amongst so many diverse bacterial species and toxins.
Similarly, the assortment of different dedicated MLDs, the range of sizes, and mechanisms of intracellular effector-targeting is somewhat surprising given that they all seem to be directed to similar locations. This can be explained by the presence of additional targeting components apart from what is sufficient to identify the PM, such as inclusion of hydrophobic/charged motifs, coiled-coil domains, and/or ubiquitination motifs. These additional mechanisms likely provide information necessary for site-specific targeting, following their initial association with the PM. Catalytic domains themselves can also influence spatial specificity because they have distinct binding partners and substrates. This is evident in the variety of toxic catalytic domains that are delivered by shared MLDs, such as those attached to the 4HBMs. Although they are presumably directed to similar intracellular locations by the same mechanism, their catalytic domains have completely different PM-associated targets (Just et al., 1995; Sheahan and Satchell, 2007; Orth et al., 2009; Geissler et al., 2010). Despite the lack of information regarding their activities or intracellular targets, this is also likely the case with the BteA-like LRTs and their associated catalytic domains, because sequence comparisons show limited homology downstream of the LRTs (French et al., 2009). For these two families of MLDs, multiple recombination events have likely occurred between several different bacteria and types of toxins in order to generate the diversity of catalytic domains associated with these conserved MLDs.
Although several functionally interchangeable effector-targeting mechanisms have been identified, many motifs are effector-specific. It remains to be seen how these strategies are capable of delivering their cognate toxic cargoes to specific sites within the cell, how these mechanisms differ from other MLDs, and why the specific sites are targeted. In addition, given the importance of modulating host cell membranes for bacterial infection/persistence, as more effectors involved in virulence are identified, additional novel and universal PM targeting motifs are also likely to be identified.
With the structural and biochemical data that has been gathered as well as the computational and biophysical tools available, it appears that further characterization of bacterial effector-membrane interactions at the molecular level is imminent. Determination of the binding mechanisms, origins of these domains, and the transfer/recombination events that led to their association with different catalytic domains will be required to resolve how assorted toxins produced by phylogenetically diverse bacteria employ similar targeting domains. Understanding these interactions will provide further insight into the pathogenesis of numerous medically and agriculturally relevant pathogens.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I wish to thank Karla Satchell for helpful discussions, guidance, and support and Gregory Tyson for critically reviewing the manuscript. In addition, I apologize to any colleagues whose work was not included herein due to spatial constraints.
References
Agbor, T. A., and McCormick, B. A. (2011). Salmonella effectors: important players modulating host cell function during infection. Cell. Microbiol. 13, 1858–1869.
Al-Quadan, T., and Kwaik, Y. A. (2011). Molecular characterization of exploitation of the polyubiquitination and farnesylation machineries of Dictyostelium discoideum by the AnkB F-box effector of Legionella pneumophila. Front. Microbiol. 2:23. doi:10.3389/fmicb.2011.00023
Al-Quadan, T., Price, C. T., London, N., Schueler-Furman, O., and Abukwaik, Y. (2011). Anchoring of bacterial effectors to host membranes through host-mediated lipidation by prenylation: a common paradigm. Trends Microbiol. 19, 573–579.
Arbuzova, A., Wang, L., Wang, J., Hangyas-Mihalyne, G., Murray, D., Honig, B., and Mclaughlin, S. (2000). Membrane binding of peptides containing both basic and aromatic residues. Experimental studies with peptides corresponding to the scaffolding region of caveolin and the effector region of MARCKS. Biochemistry 39, 10330–10339.
Axtell, M. J., and Staskawicz, B. J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377.
Backert, S., and Meyer, T. F. (2006). Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9, 207–217.
Bakowski, M. A., Braun, V., Lam, G. Y., Yeung, T., Heo, W. D., Meyer, T., Finlay, B. B., Grinstein, S., and Brumell, J. H. (2010). The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella-containing vacuole. Cell Host Microbe 7, 453–462.
Birtalan, S. C., Phillips, R. M., and Ghosh, P. (2002). Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9, 971–980.
Black, D. S., and Bliska, J. B. (2000). The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol. Microbiol. 37, 515–527.
Block, A., and Alfano, J. R. (2011). Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14, 39–46.
Bosis, E., Salomon, D., and Sessa, G. (2011). A simple yeast-based strategy to identify host cellular processes targeted by bacterial effector proteins. PLoS ONE 6, e27698. doi:10.1371/journal.pone.0027698
Boucrot, E., Beuzon, C. R., Holden, D. W., Gorvel, J. P., and Meresse, S. (2003). Salmonella typhimurium SifA effector protein requires its membrane-anchoring C-terminal hexapeptide for its biological function. J. Biol. Chem. 278, 14196–14202.
Boyd, A. P., Lambermont, I., and Cornelis, G. R. (2000). Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J. Bacteriol. 182, 4811–4821.
Burkhard, P., Stetefeld, J., and Strelkov, S. V. (2001). Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88.
Cascales, E., and Christie, P. J. (2003). The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1, 137–149.
Dowen, R. H., Engel, J. L., Shao, F., Ecker, J. R., and Dixon, J. E. (2009). A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J. Biol. Chem. 284, 15867–15879.
Egerer, M., and Satchell, K. J. (2010). Inositol hexakisphosphate-induced autoprocessing of large bacterial protein toxins. PLoS Pathog. 6, e1000942. doi:10.1371/journal.ppat.1000942
Ensminger, A. W., and Isberg, R. R. (2009). Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12, 67–73.
Fotiadis, C. T., Dimou, M., Georgakopoulos, D. G., Katinakis, P., and Tampakaki, A. P. (2012). Functional characterization of NopT1 and NopT2, two type III effectors of Bradyrhizobium japonicum. FEMS Microbiol. Lett. 327, 66–77.
French, C. T., Panina, E. M., Yeh, S. H., Griffith, N., Arambula, D. G., and Miller, J. F. (2009). The Bordetella type III secretion system effector BteA contains a conserved N-terminal motif that guides bacterial virulence factors to lipid rafts. Cell. Microbiol. 11, 1735–1749.
Fronzes, R., Christie, P. J., and Waksman, G. (2009). The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7, 703–714.
Galan, J. E. (2009). Common themes in the design and function of bacterial effectors. Cell. Host Microbe 5, 571–579.
Gao, J., Liao, J., and Yang, G. Y. (2009). CAAX-box protein, prenylation process and carcinogenesis. Am. J. Transl. Res. 1, 312–325.
Geissler, B., Ahrens, S., and Satchell, K. J. (2012). Plasma membrane association of three classes of bacterial toxins is mediated by a basic-hydrophobic motif. Cell. Microbiol. 14, 286–298.
Geissler, B., Tungekar, R., and Satchell, K. J. (2010). Identification of a conserved membrane localization domain within numerous large bacterial protein toxins. Proc. Natl. Acad. Sci. U.S.A. 107, 5581–5586.
Hernandez, L. D., Hueffer, K., Wenk, M. R., and Galan, J. E. (2004). Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science 304, 1805–1807.
Hicks, S. W., Charron, G., Hang, H. C., and Galan, J. E. (2011). Subcellular targeting of Salmonella virulence proteins by host-mediated S-palmitoylation. Cell Host Microbe 10, 9–20.
Isaksson, E. L., Aili, M., Fahlgren, A., Carlsson, S. E., Rosqvist, R., and Wolf-Watz, H. (2009). The membrane localization domain is required for intracellular localization and autoregulation of YopE in Yersinia pseudotuberculosis. Infect. Immun. 77, 4740–4749.
Isberg, R. R., O’Connor, T. J., and Heidtman, M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24.
Ivanov, S. S., Charron, G., Hang, H. C., and Roy, C. R. (2010). Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J. Biol. Chem. 285, 34686–34698.
Jank, T., Bohmer, K. E., Tzivelekidis, T., Schwan, C., Belyi, Y., and Aktories, K. (2012). Domain organization of Legionella effector SetA. Cell. Microbiol. 14, 852–868.
Just, I., Selzer, J., Wilm, M., Von Eichel-Streiber, C., Mann, M., and Aktories, K. (1995). Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375, 500–503.
Kamitani, S., Kitadokoro, K., Miyazawa, M., Toshima, H., Fukui, A., Abe, H., Miyake, M., and Horiguchi, Y. (2010). Characterization of the membrane-targeting C1 domain in Pasteurella multocida toxin. J. Biol. Chem. 285, 25467–25475.
Kazmierczak, B. I., and Engel, J. N. (2002). Pseudomonas aeruginosa ExoT acts in vivo as a GTPase-activating protein for RhoA, Rac1, and Cdc42. Infect. Immun. 70, 2198–2205.
Kenny, B., Devinney, R., Stein, M., Reinscheid, D. J., Frey, E. A., and Finlay, B. B. (1997). Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520.
Kitadokoro, K., Kamitani, S., Miyazawa, M., Hanajima-Ozawa, M., Fukui, A., Miyake, M., and Horiguchi, Y. (2007). Crystal structures reveal a thiol protease-like catalytic triad in the C-terminal region of Pasteurella multocida toxin. Proc. Natl. Acad. Sci. U.S.A. 104, 5139–5144.
Knodler, L. A., Ibarra, J. A., Perez-Rueda, E., Yip, C. K., and Steele-Mortimer, O. (2011). Coiled-coil domains enhance the membrane association of Salmonella type III effectors. Cell. Microbiol. 13, 1497–1517.
Krall, R., Sun, J., Pederson, K. J., and Barbieri, J. T. (2002). In vivo rho GTPase-activating protein activity of Pseudomonas aeruginosa cytotoxin ExoS. Infect. Immun. 70, 360–367.
Krall, R., Zhang, Y., and Barbieri, J. T. (2004). Intracellular membrane localization of Pseudomonas ExoS and Yersinia YopE in mammalian cells. J. Biol. Chem. 279, 2747–2753.
Letzelter, M., Sorg, I., Mota, L. J., Meyer, S., Stalder, J., Feldman, M., Kuhn, M., Callebaut, I., and Cornelis, G. R. (2006). The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 25, 3223–3233.
London, N., Lamphear, C. L., Hougland, J. L., Fierke, C. A., and Schueler-Furman, O. (2011). Identification of a novel class of farnesylation targets by structure-based modeling of binding specificity. PLoS Comput. Biol. 7, e1002170. doi:10.1371/journal.pcbi.1002170
Luo, Y., Bertero, M. G., Frey, E. A., Pfuetzner, R. A., Wenk, M. R., Creagh, L., Marcus, S. L., Lim, D., Sicheri, F., Kay, C., Haynes, C., Finlay, B. B., and Strynadka, N. C. (2001). Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Biol. 8, 1031–1036.
Mallo, G. V., Espina, M., Smith, A. C., Terebiznik, M. R., Aleman, A., Finlay, B. B., Rameh, L. E., Grinstein, S., and Brumell, J. H. (2008). SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34. J. Cell Biol. 182, 741–752.
Martin, D. D., Beauchamp, E., and Berthiaume, L. G. (2011). Post-translational myristoylation: fat matters in cellular life and death. Biochimie 93, 18–31.
Mason, D., Mallo, G. V., Terebiznik, M. R., Payrastre, B., Finlay, B. B., Brumell, J. H., Rameh, L., and Grinstein, S. (2007). Alteration of epithelial structure and function associated with PtdIns(4,5)P2 degradation by a bacterial phosphatase. J. Gen. Physiol. 129, 267–283.
Maurer-Stroh, S., and Eisenhaber, F. (2005). Refinement and prediction of protein prenylation motifs. Genome Biol. 6, R55.
Mesmin, B., Robbe, K., Geny, B., Luton, F., Brandolin, G., Popoff, M. R., and Antonny, B. (2004). A phosphatidylserine-binding site in the cytosolic fragment of Clostridium sordellii lethal toxin facilitates glucosylation of membrane-bound Rac and is required for cytotoxicity. J. Biol. Chem. 279, 49876–49882.
Mulgrew-Nesbitt, A., Diraviyam, K., Wang, J., Singh, S., Murray, P., Li, Z., Rogers, L., Mirkovic, N., and Murray, D. (2006). The role of electrostatics in protein-membrane interactions. Biochim. Biophys. Acta 1761, 812–826.
Nagai, H., and Roy, C. R. (2003). Show me the substrates: modulation of host cell function by type IV secretion systems. Cell. Microbiol. 5, 373–383.
Nimchuk, Z., Marois, E., Kjemtrup, S., Leister, R. T., Katagiri, F., and Dangl, J. L. (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101, 353–363.
Orth, J. H., Preuss, I., Fester, I., Schlosser, A., Wilson, B. A., and Aktories, K. (2009). Pasteurella multocida toxin activation of heterotrimeric G proteins by deamidation. Proc. Natl. Acad. Sci. U.S.A. 106, 7179–7184.
Page, A. L., Sansonetti, P., and Parsot, C. (2002). Spa15 of Shigella flexneri, a third type of chaperone in the type III secretion pathway. Mol. Microbiol. 43, 1533–1542.
Panina, E. M., Mattoo, S., Griffith, N., Kozak, N. A., Yuk, M. H., and Miller, J. F. (2005). A genome-wide screen identifies a Bordetella type III secretion effector and candidate effectors in other species. Mol. Microbiol. 58, 267–279.
Patel, J. C., Hueffer, K., Lam, T. T., and Galan, J. E. (2009). Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137, 283–294.
Phillips, R. M., Six, D. A., Dennis, E. A., and Ghosh, P. (2003). In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J. Biol. Chem. 278, 41326–41332.
Price, C. T., Al-Khodor, S., Al-Quadan, T., Santic, M., Habyarimana, F., Kalia, A., and Kwaik, Y. A. (2009). Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 5, e1000704. doi:10.1371/journal.ppat.1000704
Price, C. T., Al-Quadan, T., Santic, M., Jones, S. C., and Abu Kwaik, Y. (2010a). Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J. Exp. Med. 207, 1713–1726.
Price, C. T., Jones, S. C., Amundson, K. E., and Kwaik, Y. A. (2010b). Host-mediated post-translational prenylation of novel dot/icm-translocated effectors of Legionella pneumophila. Front. Microbiol. 1:131. doi:10.3389/fmicb.2010.00131
Pruitt, R. N., Chumbler, N. M., Rutherford, S. A., Farrow, M. A., Friedman, D. B., Spiller, B., and Lacy, D. B. (2012). Structural determinants of the Clostridium difficile toxin A glucosyltransferase activity. J. Biol. Chem. 287, 8013–8020.
Rabin, S. D., Veesenmeyer, J. L., Bieging, K. T., and Hauser, A. R. (2006). A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect. Immun. 74, 2552–2561.
Reinert, D. J., Jank, T., Aktories, K., and Schulz, G. E. (2005). Structural basis for the function of Clostrium difficile toxin B. J. Mol. Biol. 351, 973–981.
Reinicke, A. T., Hutchinson, J. L., Magee, A. I., Mastroeni, P., Trowsdale, J., and Kelly, A. P. (2005). A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J. Biol. Chem. 280, 14620–14627.
Resh, M. D. (1994). Myristylation and palmitylation of Src family members: the fats of the matter. Cell 76, 411–413.
Robert-Seilaniantz, A., Shan, L., Zhou, J. M., and Tang, X. (2006). The Pseudomonas syringae pv. tomato DC3000 type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol. Plant Microbe Interact. 19, 130–138.
Roy, M. O., Leventis, R., and Silvius, J. R. (2000). Mutational and biochemical analysis of plasma membrane targeting mediated by the farnesylated, polybasic carboxy terminus of K-ras4B. Biochemistry 39, 8298–8307.
Satchell, K. J. (2011). Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu. Rev. Microbiol. 65, 71–90.
Schmid, M. C., Scheidegger, F., Dehio, M., Balmelle-Devaux, N., Schulein, R., Guye, P., Chennakesava, C. S., Biedermann, B., and Dehio, C. (2006). A translocated bacterial protein protects vascular endothelial cells from apoptosis. PLoS Pathog. 2, e115. doi:10.1371/journal.ppat.0020115
Segal, G., Purcell, M., and Shuman, H. A. (1998). Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A. 95, 1669–1674.
Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J. E., and Innes, R. W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301, 1230–1233.
Sheahan, K. L., and Satchell, K. J. (2007). Inactivation of small Rho GTPases by the multifunctional RTX toxin from Vibrio cholerae. Cell. Microbiol. 9, 1324–1335.
Thieme, F., Szczesny, R., Urban, A., Kirchner, O., Hause, G., and Bonas, U. (2007). New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N-myristoylation motif. Mol. Plant Microbe Interact. 20, 1250–1261.
Veesenmeyer, J. L., Howell, H., Halavaty, A. S., Ahrens, S., Anderson, W. F., and Hauser, A. R. (2010). Role of the membrane localization domain of the Pseudomonas aeruginosa effector protein ExoU in cytotoxicity. Infect. Immun. 78, 3346–3357.
Vogel, J. P., Andrews, H. L., Wong, S. K., and Isberg, R. R. (1998). Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876.
Wu, S., Lu, D., Kabbage, M., Wei, H. L., Swingle, B., Records, A. R., Dickman, M., He, P., and Shan, L. (2011). Bacterial effector HopF2 suppresses Arabidopsis innate immunity at the plasma membrane. Mol. Plant Microbe Interact. 24, 585–593.
Yeung, T., Heit, B., Dubuisson, J. F., Fairn, G. D., Chiu, B., Inman, R., Kapus, A., Swanson, M., and Grinstein, S. (2009). Contribution of phosphatidylserine to membrane surface charge and protein targeting during phagosome maturation. J. Cell Biol. 185, 917–928.
Zhang, W., Crocker, E., Mclaughlin, S., and Smith, S. O. (2003). Binding of peptides with basic and aromatic residues to bilayer membranes: phenylalanine in the myristoylated alanine-rich C kinase substrate effector domain penetrates into the hydrophobic core of the bilayer. J. Biol. Chem. 278, 21459–21466.
Zhang, Y., and Barbieri, J. T. (2005). A leucine-rich motif targets Pseudomonas aeruginosa ExoS within mammalian cells. Infect. Immun. 73, 7938–7945.
Zhang, Y., Deng, Q., Porath, J. A., Williams, C. L., Pederson-Gulrud, K. J., and Barbieri, J. T. (2007). Plasma membrane localization affects the RhoGAP specificity of Pseudomonas ExoS. Cell. Microbiol. 9, 2192–2201.
Zhao, G., and London, E. (2005). Behavior of diphtheria toxin T domain containing substitutions that block normal membrane insertion at Pro345 and Leu307: control of deep membrane insertion and coupling between deep insertion of hydrophobic subdomains. Biochemistry 44, 4488–4498.
Zheng, Z., Ma, D., Yahr, T. L., and Chen, L. (2012). The transiently ordered regions in intrinsically disordered ExsE are correlated with structural elements involved in chaperone binding. Biochem. Biophys. Res. Commun. 417, 129–134.
Zhu, W., Banga, S., Tan, Y., Zheng, C., Stephenson, R., Gately, J., and Luo, Z. Q. (2011). Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE 6, e17638. doi:10.1371/journal.pone.0017638
Keywords: plasma membrane, bacterial effectors, toxins, intracellular targeting
Citation: Geissler B (2012) Bacterial toxin effector-membrane targeting: outside in, then back again. Front. Cell. Inf. Microbio. 2:75. doi: 10.3389/fcimb.2012.00075
Received: 14 February 2012; Accepted: 14 May 2012;
Published online: 31 May 2012.
Edited by:
Ken Bradley, University of California Los Angeles, USAReviewed by:
Cammie Lesser, Harvard Medical School, USARebekah DeVinney, University of Calgary, Canada
Copyright: © 2012 Geissler. This is an openaccess article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Brett Geissler, Department of Microbiology-Immunology, Feinberg School of Medicine, Northwestern University, Searle 3-520, 745 North Fairbanks Court, Chicago, IL 60611, USA. e-mail: b-geissler@northwestern.edu