Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows
- Dipartimento di Scienze della Vita e Biologia dei sistemi – Life Sciences and Systems Biology, University of Torino, Torino, Italy
Lactic Acid Bacteria (LAB) are ancient organisms that cannot biosynthesize functional cytochromes, and cannot get ATP from respiration. Besides sugar fermentation, they evolved electrogenic decarboxylations and ATP-forming deiminations. The right balance between sugar fermentation and decarboxylation/deimination ensures buffered environments thus enabling LAB to survive in human gastric trait and colonize gut. A complex molecular cross-talk between LAB and host exists. LAB moonlight proteins are made in response to gut stimuli and promote bacterial adhesion to mucosa and stimulate immune cells. Similarly, when LAB are present, human enterocytes activate specific gene expression of specific genes only. Furthermore, LAB antagonistic relationships with other microorganisms constitute the basis for their anti-infective role. Histamine and tyramine are LAB bioactive catabolites that act on the CNS, causing hypertension and allergies. Nevertheless, some LAB biosynthesize both gamma-amino-butyrate (GABA), that has relaxing effect on gut smooth muscles, and beta-phenylethylamine, that controls satiety and mood. Since LAB have reduced amino acid biosynthetic abilities, they developed a sophisticated proteolytic system, that is also involved in antihypertensive and opiod peptide generation from milk proteins. Short-chain fatty acids are glycolytic and phosphoketolase end-products, regulating epithelial cell proliferation and differentiation. Nevertheless, they constitute a supplementary energy source for the host, causing weight gain. Human metabolism can also be affected by anabolic LAB products such as conjugated linoleic acids (CLA). Some CLA isomers reduce cancer cell viability and ameliorate insulin resistance, while others lower the HDL/LDL ratio and modify eicosanoid production, with detrimental health effects. A further appreciated LAB feature is the ability to fix selenium into seleno-cysteine. Thus, opening interesting perspectives for their utilization as antioxidant nutraceutical vectors.
Short History of Lactic Acid Bacteria
Lactic acid bacteria (LAB) include a large number of bacterial genera among which the best known are lactobacilli, lactococci, enterococci, streptococci, leuconostoc, and pediococci. These genera differ for morphology, pH and salt tolerance, temperature optimum, habitats, and pathogenic potential. At present, it is very difficult to establish a clear demarcation line between beneficial and virulent species, being some problematic traits more linked to strain than to species. However, Lactobacilli and Lactococci are considered GRAS (generally regarded as safe).
LAB were among the first living organisms on the earth: they appeared about three billions years ago, in the transition period from anaerobiosis to aerobiosis. Apparently, they seem to be well adapted to both anaerobic and aerobic life conditions, since they bear all the necessary proteins for respiration and several enzymes involved in fermentative pathways. Nevertheless, during evolution they never acquired the ability to biosynthesize heme, an essential component of cytochromes, so they can shift to respiratory metabolism only with exogenous hemine supplementation. In the case of pathogenic streptococci, they can subtract heme from host hemoglobin by means of hemolysin, a protein considered a virulence factor. Therefore, most LAB species are compelled to live on fermentation metabolism with lower energy yields (Carr et al., 2002).
Furthermore, LAB inability to synthesize heme molecules implies that they cannot get functional heme enzymes such as peroxidases and catalases that are the most efficient proteins involved in hydroxyl radical scavanging. This results in an impaired resistance to O2 toxicity. In spite of this, LAB can tolerate oxygen and are defined microaerophylic organisms (Brioukhanov and Netrusov, 2007).
Some strategies have been set up by these bacteria to replace heme enzyme anti reactive oxygen species (ROS) efficacy, for instance the use of metals as radical scavengers: zinc constitutes an ion trap for oxygen radicals, (Salvatore et al., 2007) manganese can act as a superoxide dismutase-like system (De Angelis and Gobbetti, 1999), while selenium acts in the selenocysteine-harboring proteins (Calomme et al., 1995). It is known that LAB species are able to accumulate high intracellular amounts of manganese (up to 25 mM), selenium, and zinc.
During evolution, due to the low energy gain by fermentation and to the harsh life condition in an “oxidant” world (like the one in which we now live), LAB have been compelled to specialize their metabolism rather toward stress defence than to acquire strategic biosynthetic abilities. Therefore, they developed symbiont/parasite relationships with plants and animals which can supply vitamins, proteins, and amino acids. It is worth noting that LAB had acquired the ability to recognize several sugars, such as for instance xylose, cellobiose, ribose, arabinose, glucose, and fructose, before they developed the ability to ferment lactose to lactate, which was made possible only after mammal's expansion on the Earth. Therefore, they firstly colonized fruits and vegetable ecological niches, and later cheese, wine, and especially milk, which constitute their election habitat being rich in lactose (Carr et al., 2002).
It is possible that, in early life, sugar catabolism was not as predominant as in current LAB metabolism and that parallel pathways, like acid and amino acid decarboxylations and arginine deimination (ADI), could simultaneously be activated to get energy on poor media such as cell lysis products (ribose, fatty acids, amino acids) (Konings, 2006).
However, commensal LAB living in both gut and other mucosal ecological niches, although fed with abundant nutrients, still have a stressful life and often are compelled to cope with antagonistic host factors as well as with yeast or bacteria sharing the same habitat. These harsh conditions allowed the evolution of interesting metabolic and cross-talk features.
LAB Biochemistry
Most LAB biochemical pathways have been fully elucidated so far, due to the strategic importance of these bacteria in the food industry both as starters and as biocontrol agents for food deterioration. Recent interest on LAB as ascertained probiotic agents has completed the information about LAB metabolism and its role in the context of the gut ecosystem.
LAB Energy Metabolism
LAB energy metabolism is chiefly based upon lactic fermentation, ADI, acid, and amino acid decarboxylation. While sugar fermentation has a true energetic role, the other pathways can solve different and more complex functions. Lactic fermentation is an appreciated feature in both gut and industrial LAB since its end-product, lactate, by causing acidification, ensures a control over less friendly bacteria.
Sugar fermentations
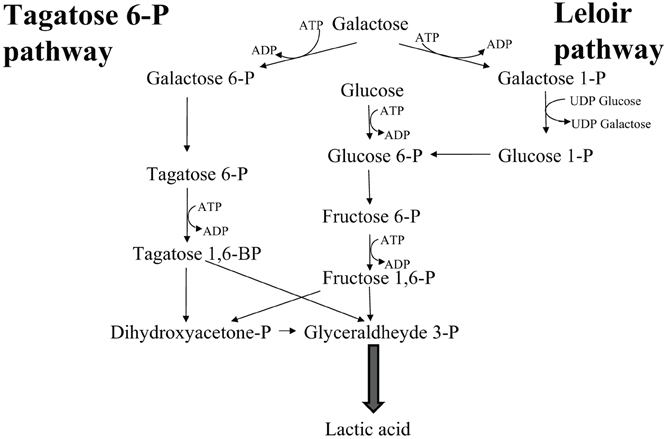
Homolactic fermentation. Homolactic fermentation always results from glycolysis. Obligate homofermentative LAB (pediococci, lactococci, streptococci, and some lactobacilli) produce 100% lactic acid through the Embden-Mayeroff route from different hexoses, which are internalized by means of specific membrane transporters such as permeases and symporters, and then isomerized to glucose or fructose. Galactose constitutes an exception since it can be either isomerized to glucose by the Leloir reaction mediated by high energy compounds (UDPGlu-UDPGal) or drifted to glycolysis through the D-tagatose 6-phosphate route, generating diidroxiacetonphosphate, hence glyceraldehyde phosphate (Figure 1).
When a disaccharide, such as lactose or saccharose, is present it can be hydrolyzed into the two monosaccharides in the external environment or rather be uptaken as disaccharide and then hydrolyzed inside the cell. From each exose, two moles of lactic acid are produced at the end of the glycolytic process as the result of pyruvate reduction to lactate, through NADH re-oxidation to NAD+. Lactic acid can be either in the D or in the L optical form, depending on each species' genetic determinants encoding either D-lactate (D-LDH) or L-lactate (L-LDH) dehydrogenase, respectively. Some species can produce both D and L lactate as the result of racemase activity, or due to the presence of genetic determinants for both LDH isoforms. In this case the second genetic determinant can derive by horizontal gene transfer. The most common catabolic pathway, i.e., the conversion of the disaccharide lactose into lactate, generates therefore four moles of lactic acid and four moles of ATP. No gas is produced in the process. This low energy gain can sometimes be improved by proton-substrate symport, i.e., lactic acid excretion, generating a proton gradient: since this system is electrogenic it can increase the energy yield of LAB.
Facultative homofermenters can direct part of the pyruvic acid that is generated by glycolysis toward the production of formate, acetate, and ethanol. Pyruvate-formate-lyase can convert pyruvate (C3) into formate (C1) and acetylCoA (C2). The latter can undergo transferase reaction into acetyl phosphate and then conversion into acetate leading to ATP synthesis. Acetate can be either accumulated in the growth medium or alternatively reduced to ethanol via acetaldehyde with NADH consumption, depending on the pH, and reduced pyridine coenzymes availability. This route allows one additional ATP mole gain, but less lactic acid is produced. Since formate can be decarboxylated/oxidized, an additional CO2 mole can be produced by this pathway (gas producing bacteria).
Heterolactic fermentation. Heterolactic fermentation concerns LAB which lack the glycolytic enzyme fructose 1,6 bisphosphate aldolase (Leuconostoc, Oenococcus, and some Lactobacillus species) so they cannot metabolize hexoses through the Embden-Mayeroff pathway. Therefore, they utilize the pentose-phosphate route with the transketolase reaction joining the glycolysis with the three carbon metabolite glyceraldehyde 3-phosphate. The remaining C2 unit, acetyl phosphate, is then converted into ethanol or into acetate (the latter reaction resulting in an additional ATP mole gain), depending on NAD+/NADH ratio, as previously described for the facultative homofermenters. Nevertheless, acetate production is not so frequent due to the necessity of NADH re-oxidation. The energetic yield of the transketolase pathway is lower than the homolactic fermentation giving rise only to 1 ATP mole, 0.5 moles of lactate, and 0.5 moles of ethanol or acetate, per mole of consumed hexose, plus one CO2 mole deriving from the hexose/pentose conversion (by decarboxylation). So, the heterofermentative route is gas producing.
All heterofermentative LAB can also degrade pentoses, while not all homofermenters are also pentose degraders.
Arginine deimination
Besides sugar level phosphorylation, LAB can get energy from ADI to citrulline and citrulline cleavage to ornithine plus carbamoyl phosphate. The latter is then converted into ATP and carbon dioxide, or rather used for pyrimidine biosynthesis. This route also generates two moles of ammonia (one from the arginine-citrulline transition, the second from carbamoyl phosphate hydrolysis) useful to compensate the acidity generated by sugar catabolism to lactic acid, acetic acid, and formic acid, in both homo- and hetero-fermenting conditions (De Angelis et al., 2002).
On the other hand, ornithine is exchanged with extracellular arginine in an ornithine/arginine antiport system ensuring the continuity of the reactions. In wine, LAB possessing this pathway have been selected, since the wine environment is still rich in arginine after yeast alcoholic fermentation. Although the ADI route is somehow similar to an inverted urea cycle, from an evolutionary view point these pathways are not related at all (Liu et al., 1996).
Acid and amino acid decarboxylations
Apart from its secondary involvement in pH buffering, the arginine deiminase route is chiefly an ATP generating pathway useful to complement substrate level phosphorylation. On the contrary, acid and amino acid decarboxylations solve more complex roles and their evolutionary history is worth to know. Decarboxylations are coupled with electrogenic antiport systems generating a proton gradient across the cytoplasm membrane and the overall system supports a double role: acidity control and proton motive force (PMF) generation, i.e., supplementary energy in organisms unable of respiration. In the case of acid decarboxylations, the reaction concerns dicarboxylic and tricarboxylic acids, like malate and citrate, and the reaction products are not informational molecules.
It has been hypothesized that, early in evolution, acid decarboxylase activities were mainly directed toward acidification control. Later, free energy conservation mechanisms have been acquired by coupling decarboxylation with electrogenic antiport systems, which allowed more sophisticated cellular responses, like generation of metabolic energy and a better overall acid stress resistance. These pathways are believed to having evolved through horizontal gene transfer allowing antiporter proteins genes to be located in the same operon as decarboxylase genes, thus undergoing the same transcriptional control (Makarova and Koonin, 2007) On the other hand, LAB genomes are known to encode a highly variable number of genes that suggests that during evolution gene acquisition/loss has played a crucial role in determining the present situation. LAB genome size ranges from the 1.8 Mb of L. gasseri to 3.3 Mb of L. plantarum and also the G+C content vary significantly (from 34 to 46%) (Siezen et al., 2004), suggesting frequent recombination events. Furthermore, the presence of soluble acid decarboxylases and membrane bound precursor/product exchangers in LAB (Lucas et al., 2005), constitutes an intermediate event between the simple decarboxylation occurring in the cytoplasm of strictly anaerobes (like Clostridia and Fusobacterium), where no free energy is conserved, and the membrane bound complexes (including decarboxylases and transporters) found in gamma proteobacteria in which active sodium extrusion occurs (Makarova et al., 2006).
For what concerns amino acid decarboxylases, their physiological roles are similar to those of acid decarboxylases, (Molenaar et al., 1993). Nonetheless, some additional functions like bacteria–bacteria interactions and host-symbiont communication were found, being some products informational molecules. Glutamate can be decarboxylated to gamma-amino-butyrate (GABA), histidine to histamine, phenylalanine to β-phenylethylamine, tyrosine to tyramine, triptophane to triptamine, ornithine to putrescine, lysine to cadaverine (Konings, 2006).
Amino acid decarboxylations are catalyzed by PLP or pyruvoil dependent enzymes that can be either soluble (Pessione et al., 2005) or membrane-located. In Enterococcus a membrane bound tyrosine and phenylalanine decarboxylase has been described (Pessione et al., 2009): this cellular location can allow a better interaction between decarboxylase enzyme and transporter protein. This may constitute a further evolutive step toward a more efficient system to get energy and to counteract acidic stress in a less acid-tolerant genus than other LAB (Figure 2).
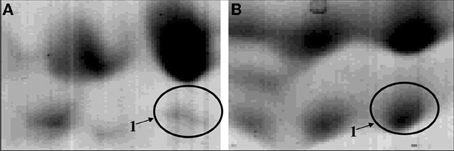
Figure 2. Zoom of the proteomic maps of the membrane-enriched protein fraction of a tyramine and beta-phenylethylamine producer Enterococcus faecalis grown in absence (A) or presence (B) of tyrosine. The circle indicated the enzyme TDC (tyrosine/phenylalanine decarboxylase).
Some produced amines are bio-active molecules (histamine, tyramine, triptamine, beta-phenylethyl amine, GABA), acting at vascular or central nervous system level of human host (Moreno-Arribas et al., 2003), while others are informational messengers for microbial cross-talk and for stress control (at least for putrescine in some bacterial species). All of them can undergo further bio-transformations by monoamino- and diamino-oxidases. These routes can thus be considered as “escape-pathways” during sugar depletion or excessive acidification (i.e., stationary phase stress) but also as means to interact with the external environment. It has to be considered that most LAB genomes harbor genes for both the ADI pathway enzymes and amino acid decarboxylases: often these two routes are reciprocally regulated (Lactobacillus, Lactococcus) (Lamberti et al., 2011a,b) but sometimes they do not compete and are parallely used (Enterococcus), underlining their similar but different physiological function. Proteomic studies can add useful information about the physiological and environmental modulations acting on these foundmental pathways. From a human point of view, while acid decarboxylations are appreciated because they reduce acidity (i.e., in wine), amino acid decarboxylations, and ADI are both regarded as problematic since they can give rise to ammonia and undesired bio-active amines such as histamine, that is an allergy mediator, or tyramine which is involved in hypertension and more severe syndromes such as cerebral hemorrhage (Millichap and Yee, 2003). Conversely GABA and beta-phenylethylamine, are appreciated molecules since they can control appetite, mood, and smooth muscle activity, as it will be better elucidated in the next paragraphs (Inoue et al., 2003).
LAB proteolytic system
The choice to use amino acids not only as a nitrogen source but also for energy metabolism, requires high amino acid availability, and, for this reason, LAB have evolved complex proteolytic and peptidolytic systems. These include three components: (1) extracellular or membrane bound proteases (mainly PrtP and CEP), hydrolyzing proteins in oligopeptides; (2) membrane transport proteins (Opp, Dtp, and Dpp ATP-binding cassette transporters), catalyzing intracellular uptake of oligopeptides, di/tripeptides, and dipeptides, respectively; (3) intracellular peptidases, which cleave peptides into free amino acids (Konings, 2002).
In Lactococci, proteases are prevalently cell-envelope anchored (CEP proteases), except in Lactococcus lactis subsp. cremoris which is able to secrete proteinases. In Lactobacilli some of them have an extracellular location (Lb. lactis and Lb. helveticus) while others are cell-wall associated (Lb. delbruekii ssp bulgaricus). Most LAB proteolytic enzymes are synthesized as pre-pro-enzymes, whose signal peptide is cleaved upon membrane translocation: the resulting membrane bound pro-proteinase undergoes autocatalytic cleavage to obtain its mature, active, conformation. These proteases are serine proteinases, with high molecular weight and 5.5–6.5 pH optima, which is compatible with the environment in which LAB live.
However, other systems could be present to allow free amino acid availability at the external side of the cytoplasmic membrane. Actually, some peptidases have been detected on the inner face of the cytoplasmic membrane by antibody recognition (PepX in Lactococci) or have been supposed to be associated to an anionic specific permease, although they had been cytosolic-predicted on the basis of genome sequences data. For instance, PepA does not bear a signal peptide or a true hydrophobic domain that could justify its membrane location. Nevertheless, its high substrate specificity in releasing N-terminal acidic residues from peptides could account for its role in supplying free glutamate, for the glutamate-GABA pathway antiporter, at membrane level. In E. faecalis a similar function can be performed by ClpP: this protease is able to hydrolyze tetra- and tri-peptides containing the recognition sites tyrosine-tryptophan or tyrosine-leucine (Katayama et al., 1988). This may result in free tyrosine availability for the tyrosine-tyramine decarboxylation route.
An important role that is performed by both released and cell-wall associated LAB proteinases in the human gut are the digestion of not fully hydrolyzed proteins. They may shorten long and medium-sized peptides into smaller peptides. This role can also be played by intracellular peptidases that are released by LAB cell autolysis. Since the LAB proteolytic system has evolved in the milk ecological niche, is particularly suitable for diary protein hydrolysis. Apart from their digestive-assimilative function, these enzymes have received great attention because they are able to generate bio-active peptides from both caseins and milk whey proteins (Law and Haandrikman, 1997). Diary proteins are digested in the human stomach and tenuous intestine by endogenous proteolytic enzymes. This digestion is completed by proteases supplied by the gut microflora. Casein is made up of four main proteins: alpha s1 casein, alpha s2 casein, beta-casein, and k-casein, differing in amino acid sequence, phosphorylation, and glycosylation degree, hydropaticity index. The hydrolysis of all these components, whose ratio is about 38:11:38:13, can give rise to bioactive molecules acting at different biological levels. Similarly, hydrolytic cleavage of milk whey proteins (alpha lacto albumine, beta lactoglobulin, lactoferrin, and immunoglobulins) can also generate bioactive peptides. The proteolytic enzymes that are released by LAB have been analyzed by 2DE-MS and proved to be very different in the different LAB species and strains, giving rise to a different pool of bioactive peptides. However, their activity, although very high at the normal intestinal pH (6.5–7.0), proved to be different toward alphas1 and beta casein (Hébert et al., 1999). The produced peptides are generally stable, although more investigations are needed to assess their real life span in vivo. The most interesting bioactive peptides resulting by LAB proteolytic activity are involved in immune system, cardiovascular system and central nervous system regulation, nutritional supplementation (such as metals assumption), and antimicrobial functions (antibiotic-like compounds). Sometimes each single peptide can accomplish two or more different physiological functions (Clare and Swaisgood, 2000).
LAB Biosynthetic Capabilities
In spite of their limited biosynthetic abilities (especially for amino acid and vitamin synthesis), LAB can produce molecules of interest among which the most interesting are exopolysaccharides (EPS) and fructooligosaccharides (FOS), short chain fatty acids (SCFA), conjugated linoleic acids (CLA), and selenoproteins (Figure 3).
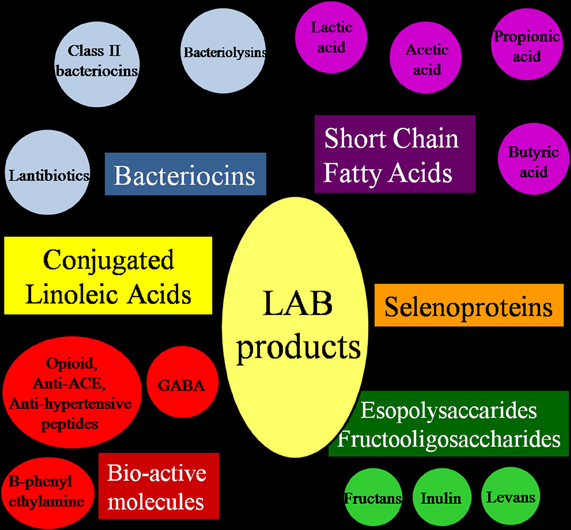
Figure 3. Molecules contributing to the nutraceutical and probiotic potential of Lactic Acid Bacteria.
EPS and FOS
Like most bacteria, LAB can synthesize cell-wall structural polysaccharides (PS) such as peptidoglycan and lipoteichoic acids and exocellular polymers. The latter include both capsular PS where the PS is covalently bound to the cell surface and the EPS which form a loosely bound slime layer that can also be secreted into the environment.
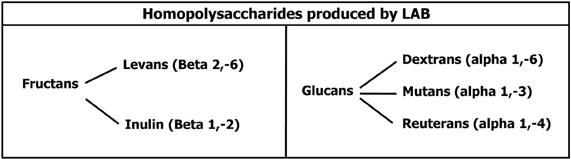
EPS can be divided in homopolysaccharides (Homo-EPS) and heteropolysaccharides (Hetero-EPS). Homo-EPS consist of either D-glucose (glucans) or D-fructose (fructans) residues, with different types of linkage and branching degree. Hetero-EPS are constructed from multiple copies of an oligosaccharide and show little structural similarity to one another: glucose, galactose, xylose, mannose, arabinose, and rhamnose are the most represented sugars but also amino-sugars and polyols can be occasionally present as well as glucuronic acid. They are often highly branched with different binding types. From a biochemical point of view the synthesis of Hetero-EPS is an energy-intensive process that requires four separate reactions: (1) sugar internalization; (2) synthesis of sugar nucleotide precursors, providing the energy for the polymerization reaction; (3) assembly of the monosaccharide repeating unit by the combined action of different types of glycosyltransferases located at the cytoplasmic membrane level; (4) export of the EPS. Glucose-6-phosphate is the key metabolite that can be diverted from the glycolytic catabolic pathway to be addressed to the synthesis of UDP-glucose and dTDP-glucose, but also the Leloir route, when present, can be used to convert galactose to UDPGal and UDPGlu. In mesophylic LAB, the enzymes required for sugar uptake and for the synthesis of sugar nucleotides are chromosomally encoded, while the genetic determinants for EPS specific enzymes are plasmid located (Laws et al., 2001). Conversely, homo-EPS synthesis requires only one extracellular glycosyltransferase and the sucrose is used as substrate instead of activated sugars. This enzyme can thus synthesize alpha-glucans (dextran, reuteran, mutan) or beta-fructans (levans and inuline). The low-molecular mass oligosaccharides are known as FOS and glucooligosaccharides (Figure 4).
Based on the identification of sucrose binding boxes, several putative encoding genes for both glucansucrases and fructansucrases have been identified in the genome sequences of LAB (van Hijum et al., 2006).
Generally, LAB divert a small percentage of their sugar substrates toward the biosynthesis of EPS, whose physiological role is probably diverse and complex: due to their high water-binding properties, EPS constitute a protection factor against dessication and osmotic stress (Kumar et al., 2007). Their role in protecting the bacterial cell from phage attack has also been suggested (Moineau et al., 1996), but more recent experiments failed to reveal a significant phage resistant phenotype in EPS producer strains (Deveau et al., 2002). The same is true for the firstly hypothesized role as extracellular energy/carbon reserve, since most producers species lack the onset of genes necessary for their own EPS degradation (Badel et al., 2011). Therefore, this reserve would constitute a benefit only in the case of sintrophyc/symbiotic life with other bacteria. Actually EPS are biosynthesized by most LAB species under quorum-sensing control and they are related with biofilm formation and adhesion to solid surfaces. In the biofilm EPS also play a crucial role in sequestering essential cations (Kumar et al., 2007). In this context, EPS role as carbon reserve makes sense considering the overall population. However, their main biological function in vivo is to protect bacterial cells from toxic compounds (ethanol, sulphur dioxide, and toxic metal ions), from antibiotics and from host immune system, especially phagocytosis. A second important role is connected with adhesion to eukaryotic cells (plants, tooth surface), cellular recognition, via binding to lectins (Cerning, 1990), and immune system modulation. Krinos and co-workers (2001) demonstrated that, like capsular PS, also EPS can affect the surface antigenicity of the different strains (different EPS giving rise to different combinations) ultimately resulting in maintenance or elimination of specific strains in the gut ecological niche.
In the past decade fructo-oligosaccharides (FOS = inulin and levans), have found growing interest as prebiotics, i.e., compounds supporting growth of probiotic organisms. Prebiotics are molecules that are not metabolized by humans but favor both adhesion and persistence of probiotic bacteria in the gut environment. Inulin production has been observed in Lactobacillus reuteri and all inulosucrases share a common C-terminal motif (LPXTG) for cell-wall anchoring. Surface display of these enzymes results in enhanced adhesion of bacteria to human tissue cells, contributing to the maintenance of a healthy urogenital mucosa. Therefore, the presence of fructansucrase enzymes in commensal LAB can solve two distinct problems: from one side helping tissue colonization and persistence and from the other side supply with “self made” prebiotics (e.g., fructans) the commensal flora.
SCFA
Although SCFA are not produced by anabolic reaction but rather as the final catabolites of the energy metabolism, their importance in host-bacteria interactions is of such growing interest that will be treated in this paragraph. The most studied SCFA are butyrate, propionate, and acetate. Lactobacilli can produce SCFA by the fermentation of pyruvate, which is generated during the glycolytic pathway, but also by the phosphoketolase route in heterofermenting conditions. Acetate can contribute to ennvironment acidification and this is an appreciated feature since LAB can compete with other less acidophylic organisms (most pathogenic bacterial species) present in the same ecosystem. In vivo acetate enters the peripheral circulation to be metabolized by muscles and other tissues, while propionate is taken up by the liver. Both molecules can modulate sugar metabolism after a standard meal, by lowering glycaemia and improving insulin sensitivity. Butyrate has been claimed to have detrimental effects on body weight and obesity, since it represents an additional, easy available, energy source which is not directly introduced with the diet, but present as the result of commensal bacteria metabolism (non-digestible PS fermentation in the colon) (Turnbaugh et al., 2006). Once this SCFA is absorbed in the large intestine, it is not only converted to fat within the liver but it also positively regulates host genes promoting lipogenesis and fat storage into adipocytes (DiBaise et al., 2008). However, butyrate is readily absorbed by colonocytes where it exerts a control over uncontrolled proliferation and also stimulates differentiation, thus promoting the switch from neoplastic to normal phenotypes. It is known that butyrate differently affects normal colonocytes and tumor colon cells. On the former it exerts a trophic action inducing growth and proliferation, on the latter it rather induces cell-cycle arrest, differentiation, and apoptosis (Iacomino et al., 2001). Butyrate selectivity on the two different cell populations is considered a good weapon to treat cancer. The reason of the different effects of butyrate is probably linked to its different concentrations within the two type of cells: normal colonocytes rapidly utilize (through mitochondrial beta-oxidation) butyrate as the major carbon/energy source thus supporting ATP synthesis and proliferation. The remaining low levels cannot suppress cell cycle progression or induce apoptosis (Boosalis et al., 2001). On the contrary, colorectal tumor cells, due to the switch toward anaerobic metabolism, display an impaired capability to oxidatively catabolize butyrate. Therefore, in this type of neoplastic cells, its intracellular concentration is higher. This high level of butyrate may constitute a critical threshold responsible for the activation of signals that induce cell cycle arrest, differentiation, and apoptosis (Lupton, 2004). The effect on cell cycle arrest is partly due to inhibition on specific histone deacetylase (Davie, 2003), and this action can also be obtained by the use of propionate, but not acetate. Some authors also hypothesized that butyrate and propionate act in a synergistic way (Minucci and Pelicci, 2006). Furthermore, comparative transcriptomic and proteomic studies on butyrate-treated and control colonocytes, underlined a butyrate effect on extracellular matrix components which are important in cell–cell interactions, and on the angiogenetic process controlling the expression level of VEGF (vascular endothelial growth factor) and HIF-1 (hypoxia-inducible-factor-1) (Pellizzaro et al., 2002). Finally, butyrate can control oxidative and metabolic stress at molecular level, by enhancing repair responses (Sauer et al., 2007) and it can inhibit the synthesis of pro-inflammatory (IL12, TNF, gamma IF) cytokines, also contributing to tumor development control (Inan et al., 2000).
It is reasonable to conclude that butyrate can contribute to the host health when a neoplastic risk is present, while it is a problematic metabolite in case of obesity and metabolic disorders.
CLA
LAB can also biosynthesize CLA that are positional and geometric isomers of the cis9, cis12 octa-deca-dienoic acid (linoleic acid). So far, the best producing strains belong to the genera Lactobacillus and Streptococcus. The most frequently found isomers of linoleic acid have the unsaturation site at positions 9, 11 or 10, 12, namely, Δtrans 9, cis11 octa decadienoic acid, and, in lower amounts, Δcis10, trans 12 linoleic acid.
CLA can have positive effects on inflammation, cancer (apoptosis induction), metabolic disorders (insulin resistance, body weight control), and cardio-vascular diseases. Nevertheless, some detrimental influences on mammalian health have been observed, like a tumor-stimulating action or deleterious effects on glucose metabolism and lipid profiles. There is some evidence suggesting that different CLA isomers can exert differential effects, the more dangerous being the trans 10, cis 12 isomer. Also CLA concentrations play a role in the balance toward beneficial or detrimental effects (Rose, 1997): anti-carcinogenic effects are observable at CLA dosages of 0.5–1% (w:w) of the total diet and studies showed that humans generally excrete 20 mg of linoleic acid per day. This substrate can then be available for CLA producing bacteria in the intestine. Higher dietary intake of linoleic acid may imply risks (Ewaschuk et al., 2006). All these considerations suggest that further studies are needed to better characterize CLA producing LAB and their metabolic products.
LAB selenoproteins biosynthesis
The capability to produce metal-fixing enzymes is a further appreciated feature in LAB, that render them interesting commensal organisms. Actually, several Lactobacillus species are able to intracellularly fix sodium selenite (Figure 5) into selenocysteins and selenomethionines, thus providing a more bio-available form of this metal, which is generally poorly adsorbed by human cells in its inorganic form (Calomme et al., 1995).
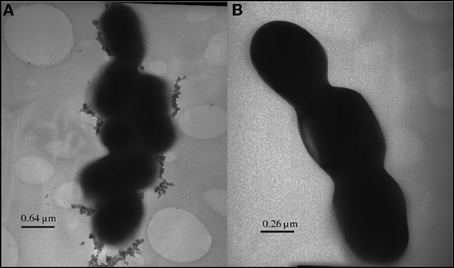
Figure 5. Inorganic selenium granules displayed by TEM on the surface of a selenium-fixing Lactobacillus reuteri strain.
The selenocysteine is defined as the 21st amino acid, is encoded by the UGA codon, and possess its own tRNA, which uses serine as an intermediate (SerSec t-RNA). Selenomethionine is non-specifically incorporated into proteins at the place of methionine and it is also important for trans-selenation reactions. Several bacterial enzymes have been demonstrated to contain selenocysteines in their active site and all belong to the oxidoreductase class: among these glutathione peroxidase, an enzyme of crucial importance to control oxidative stress and all related diseases in both bacteria and eukaryotic cells. Lactobacillus reuteri, for instance, express a selenocysteine-lyase (Lamberti et al., 2011a,b), a PLP-dependent enzyme having a key role for new seleno-proteins biosynthesis, supplying the optimal substrate (H2Se) to selenophosphate synthetase, which catalyze the production of the “activated” selenium form useful to selenocysteine sinthetase to produce L-selenocysteinil-tRNA (Lacourciere and Stadtman, 1998). LAB can also function as chelators for other nutraceutical important metals at the colon level. Investigations carried out in different prokaryotes leaded to the characterization of many mechanism for “channeling” zinc (Znll) ions, including zinc-binding, zinc-importing proteins, and zinc export systems (Blencowe and Morby, 2003). Zinc-bearing and zinc-extruding LAB can thus function as immunomodulators useful to control viral gastroenteritis (Salvatore et al., 2007).
LAB Ecology
The contribution of LAB to the overall intestinal ecosystem has been only recently elucidated. Traditional culture methods failed to shed light on the actual microbial populations resident in the gut of adult humans, but the application of culture-independent techniques such as PCR combined with DGGE (Denaturing gradient gel electrophoresis) resulted in a more detailed knowledge of what really happens at gut level. First of all it is important to differentiate resident and transient bacteria: Vaughan and co-workers (2002) reported a list of Lactobacillus species permanently colonizing the intestine: L. acidophilus, L. brevis, L. casei, L. crispatus, L. delbrueckii, L. fermentum, L. fructivorans, L. gasseri, L. paracasei, L. plantarum, L. rhamnosus, L. ruminis, L. sakei, L. salivarius, L. vaginalis. It is worth mention that LAB represent the 0.01–1.8% of the total bacterial community living in the human gut (Louis et al., 2007).
LAB–Gut Microbiota Interactions and LAB Antibacterial Potential
Both antagonism and cooperation contribute to the reciprocal relationships between LAB and other intestinal microorganisms. Biofilm formation, phage- and pheromones-mediated genetic exchange and synthrophies are well established strategies for cooperating with other gut microorganisms. Nonetheless, the antagonistic behavior is prevalent.
Nutritional, enzymatic, and metabolic competition
Cell wall targeted enzymes such as N-acetylmuramidase and N-acetylmuramoyl-L-alanine amidase are proteins involved in peptidoglycan renewal, acting as general cell-wall lysis factors (Salazar and Asenjo, 2007). They are often produced to control bacterial populations sharing the same ecological niche. Similarly, chitinase, a protein hydrolyzing the prominent component of yeast and fungi cell walls, i.e., chitin, has been found in the exoproteome of LAB (Genovese, unpublished results).
However, nutrient competition is the best known aspecific interference, common to all microbial populations, set up by LAB. Since LAB are predominantly saccharolytic, but also amino acid utilizers, they can subtract nutrients to both saccharolytic and proteolytic species living in the gut environment. Furthermore, their peculiar fermentative metabolism, generating acids, constitutes a specific metabolic competition for coping with other endogenous or exogenous microbial species together with carbon dioxide, ethanol, hydrogen peroxide (produced and fastly eliminated by the NADH oxidase activity). The direct antimicrobial effects of organic acids (lactic, acetic, and propionic acid) is due to their interference with bacterial cytoplasm membrane potential and inhibition of active transport, while carbon dioxide (formed during LAB heterolactic fermentation) and hydrogen peroxide prevent the growth of some bacteria by creating an unfavorable environment.
It is worth noting that the mentioned LAB protelytic activity toward caseins can also give rise to antimicrobial peptides such as k-casecidin, a pentapeptide (Phe-Phe-Ser-Asp-Lys) derived by k-casein hydrolysis, and isracidin, an oligo peptide originated from alpha s1 casein. Both molecules proved to be very active against Staphylococus aureus, and the latter also stimulates phagocytosis. Artificial removal of the Lys C-terminal and Phe N-terminal from k-casecidin strongly reduces the original activity of the natural pentapeptide (Matin and Otani, 2002). L.acidophylus, L.plantarum, L.helveticus, L.rhamnosus, and Lactococcus lactis produce other alpha s1 casein-derived peptides also, which have a completely different amino acid sequence. The latter proved to be active against Enterobacter sakazakii, a gram negative species causing infections in newborns (Hayes et al., 2006).
Bacteriocin production
A promising feature of LAB is the production of interference molecules, the bacteriocins (Montalbán-López et al., 2011). Bacteriocins are proteinaceous molecules that are synthesized at ribosomal level (and not as secondary metabolites) which can interfere with the growth of most bacteria. They have bactericidal action and are selective for prokaryotes. Bacteriocins are produced at the end of the exponential growth-phase and their spectrum of action can vary, depending on the producing species. Producer strains are immune to their own bacteriocins because they possess genes that encode immunity mechanisms, which enable a distinction between “self” and “non-self.” Protection can be provided by a dedicated immunity protein and/or by a specialized ABC-transporter system that pumps the bacteriocin outside the producer membrane (Draper et al., 2009).
LAB are particularly prolific in bacteriocins production and can biosynthesize different types of antagonistic molecules. The lantibiotics, so named because they contain post-translationally modified aminoacids such as lanthionine (two alanines linked by a sulphur), β-metyl-lanthionine, dehydroalanine, and dehydrobutyrine, are small peptides (19–38 amino acids in length) mainly active against Gram positive bacteria (Figure 6).
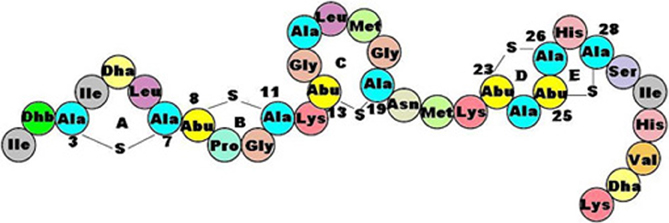
Figure 6. General structure of a lantibiotic: covalently bound alanines and post-translationally modified amino acids are peculiar features of these anti-microbial peptides.
They can be divided into two subclasses on the basis of their structure and mode of action: the elongated amphiphilic cationic lantibiotics (Nisin, Lactococcin, and Pediocin) are active through the formation of pores, leading to the dissipation of membrane potential and the efflux of small metabolites from target cells. By contrast, the globular lantibiotics (for example, Mersadicin) act through enzyme inhibition, interfering with cell-wall biosynthesis. The class II bacteriocins are small (<10 kDa) heat-stable peptides, not undergoing extensive post-translational modifications, although they may contain D-amino acids. The best known class II bacteriocin, the so-called pediocin-like, has a narrow but very specific activity against the food pathogen Listeria monocytogenes (Kazazic et al., 2002). Finally, bacteriolysins are large, heat-labile antimicrobial proteins causing the lysis of sensitive cells by catalyzing cell-wall hydrolysis. Their structure contains a catalytic N-terminal domain that shows homologies to endopeptidases, and a C-terminus that represents the target recognition site (Lai et al., 2002).
All these natural antimicrobial compounds are the most interesting antagonist metabolic end-products synthesized by LAB. Last years' research has been focused on the study of their isolation, structure, and mechanism of action (Millette et al., 2007).
LAB-Human Host Interactions
LAB colonization and diet
LAB, especially Lactobacilli, colonize the new-born as soon as the infant is breast-fed. On the contrary, artificially-fed new-borns generally host different bacterial species such as Clostridia and Bacteroides. The presence of LAB, that are prevalently saccharolytic, acidogenic and bacteriocin producers, acts as a selective agent, protecting the gut environment from putrefactive and sulphate-reducing bacteria known to be involved in the production of gas, thiols, phenols, indole, histolesive proteases, and carcinogens (Holzapfel et al., 1998). While sugar fermentations chiefly occur in the proximal colon (pH 5–6), proteins and peptides putrefaction is typical of the distal colon (pH 7). LAB presence is maximal in the tenue and proves to be absolutely beneficial for the health and the well being of the host, but, to be maintained is necessary a specific diet.
Actually, it is well established that diet plays a crucial role in selecting microbial populations in the complex and dynamic ecosystem of human gut bacteria. Food containing LAB, such as yogurt and cheese, can enrich gut LAB populations since these bacteria can easily survive to the gastric transit. Actually, LAB are naturally acidophilic organisms (growth pH optimum 3.5–6.5) and they can further improve their acidic pH resistance by setting up strategies that produce alkaline metabolites (the referred ADI pathway and amino acid decarboxylations routes). Furthermore, as human or artificial milk can select LAB or Clostridia in the new-born, similarly, a “meat-fat” rich diet contributes to select and expand Clostridia in the adult while a non-metabolizable sugar diet (especially fructose-, inuline-, and cellulose-rich nutrients that escape regular digestion in the human body) favors LAB. The epidemiological link between the “meat-fat” rich diet and colon cancer (Hill, 1975) can partly be explained by the capability of Clostridia to metabolize bile acids to carcinogenic molecules responsible for neoplastic transformation of the gut mucosa (Murray et al., 1980). Thus, the role of LAB is not only confined to preventing colonization by exogenous bacteria but also to cope against toxins and mutagenic molecules of endogenous origin.
LAB host cross-talk and stress chaperones
It has recently been established that “symbiosis” can trigger adaptive responses in both bacteria and host, which are detectable by transcriptomic and proteomic studies (host-microbiota interactomics), and concern specific metabolic pathways activation, immune system stimulation/depression, and stress responses.
From the host side, activation of important genes involved in cell signaling pathways and immunomodulation has been observed in an in vitro experiment using CaCo-2 epithelial cells exposed to Lactobacillus acidophilus, a natural inhabitant of the human small intestine. The use of other food-derived LAB cannot trigger the same biological response, thus demonstrating that the long symbiosis has selected peculiar genetic characters for host-microbe interaction (O'Flaherty and Klaenhammer, 2011). Furthermore, the importance of LAB cell wall glycome in cytokine induction in intestinal epithelial cells has been proved with the use of mutants knock-out for protein glycosylation ability. These mutants not only failed to stimulate cytokines, but they were also unable to adhere to CaCo2 cells and showed a significant impaired persistence in the mice gut environment (Tytgat et al., 2011).
From the bacterial side, adaptation responses, promoting persistence in the host have been described. In LAB, genetic loci specifically induced by gut transit are those involved in nutritional adaptation (hydrolases, ABC transporters, and PTS systems) but also in anti-stress mechanisms such as membrane modification, surface glycosylation and anti-oxidative defences (Lebeer et al., 2011). Bacterial stress protein production (enhanced synthesis of chaperones GroEL, GroES, DnaK, Hrc, Clp, Cts) in response to host factors has also been established (Henderson et al., 2006). It is now well demonstrated that, besides helping correct protein folding, chaperones can perform multiple functions such as preventing denaturation, rearranging proteins after oxidative and pH stress and performing targeted proteolysis on the irreversible denaturated proteins. Among these functions is the adhesion to extracellular matrix proteins of the host tissues, and the stimulation of the host immune system. To perform these additional functions chaperones should be surface or extracellularly located.
This phenomenon is known as moonlighting and several moonlighting proteins have been described so far, both in eukarya and in bacteria, including LAB. They include not only stress chaperones (GroEL, DNAk) but also glycolytic and TCA cycle enzymes. Moonlighting proteins are extracellularly transferred without any export signal and without specific cell-wall or membrane anchoring motif: thus the surface or extracellular localization can be related to the environmental pH. Antikainen and co-workers (Antikainen et al., 2007a) demonstrated that higher extracellular pH can cause release of the weakly linked proteins in Lactobacillus crispatus but cell-wall renewal, that is frequent during the exponential growth phase, can also be involved in the detachment of such proteins (Sánchez et al., 2008). Generally surface-exposed and released moonlighting proteins exert different biological functions, namely adhesion (cell-wall bound) and immune system modulation (extracellular chaperones).
LAB adhesion to host tissues and immunomodulation
Adhesion. The bacteria-human mucosa interaction is a dynamic equilibrium and several factors can alterate this balance. Intestinal movements and bacterial chemotaxis cause a weak association between bacteria and the surface of the gut epithelium where electrostatic and hydrophobic bonds are prevalent. In a following step, a more specific interaction between molecular determinants on bacterial surface and specific gut mucosa receptors occurs. Specific targets on human tissues are collagen type I and IV, fibronectin, and laminin. In LAB, good binding-effectors are PS, lipoteichoic acids as well as proteins such as lectins and adhesines. The adhesion potential of a certain LAB strain is therefore crucial for survival and persistence in the gut environment.
Several surface located moonlighting proteins can act as adhesines. Both the chaperone GroEL and an elongation factor, EF-Tu, display adhesive capabilities, toward human tissues in Lactobacillus johnsonii. EF-Tu is a guanosine nucleotide binding protein playing a central role in protein synthesis (when expressed inside the cell) but able to bind fibronectin or mucine and epithelial cells when exposed on the cell surface (Granato et al., 2004). Trigger factor is also connected with adhesion: it has been described to be exposed on the surface of Lactobacillus plantarum and, in L. reuteri NCIB11951, a collagen I binding protein, shares high sequence homology with E. coli trigger factor (Aleljung et al., 1994). Apart from these proteins, also glycolytic enzymes can have adhesive properties: GAPDH is able to bind fibronectin, plasmin, and mucine in several bacterial species (Alvarez et al., 2003) and a similar role may be performed by PGK (phosphoglycerate kinase) that is referred to be extracellularly located in Lactobacillus rhamnosus GG (Sánchez et al., 2008). Some of these proteins will be treated in a separated chapter for their peculiar attitude to behave also as plasminogen receptors.
Immune system modulation. About 70% of the immune system is localized in the gastro-intestinal tract as GALT (gut-associated lymphoid tissue). A reciprocal relationship exists between LAB and immune system. From one side the immune system selects the LAB species to be accepted from the other side LAB can modulate immunological functions. The most evident LAB's effects concern the enhancement of the ratio between anti-inflammatory (IL-10, β-TGF) and pro- inflammatory (IL-1 beta, IL-3, IL-4) cytokines (Pessi et al., 2000) and the selection of T-lymphocyte populations (Karimi et al., 2009). Different LAB components, such as teichoic and lipoteichoich acids, lipoproteins, and EPS may be the inducers of the immune response (Weidenmaier and Peschel, 2008). Some of the referred adhesive proteins also exhibit immunomodulatory properties when they are secreted. This is particularly true for the chaperone GroEL which has been demonstrated to interact with macrophages and stimulate cytokines secretion. In L. johnsonii GroEL also induces a strong aggregation of the pathogen Helicobacter pylori, contributing to decrease of the bacterial load and thus facilitating clearance of the aggregated pathogens by the mucus (Granato et al., 2004). In L. reuteri a strict link between adhesion and regulatory T-cell induction has been demonstrated as well (Smits et al., 2005). Last but not least, an indirect immune stimulating activity can be triggered by proteolytic intestinal LAB species through the production of immunomodulating peptides from casein. These molecules primarly enhance lymphocyte proliferation and macrophage phagocytosis but they are probably important in regulating the development of the immune system in new borns and might contribute to attenuate allergic reaction and tumor development as well (Korhonen and Pihlanto, 2006).
LAB interaction with the plasminogen-plasmin system
Lactobacillus crispatus, Lactobacillus acidophilus, L. amylovorus, L. gallinarum, L. gasseri, and Lactobacillus johnsonii display the ability to bind plasminogen on their cell wall (Hurmalainen et al., 2007). Several adhesion-involved glycolytic enzymes (enolase, GAPDH, phosphoglycerate kinase, and phosphoglycerate mutase), bile salts hydrolase and the stress protein DnaK, have been demonstrated to behave as plasminogen (Pg) receptors. They all share a common feature, a C-terminus lysine. Some reports suggest that plasminogen binding capability in LAB can constitute a risk because plasminogen can be activated to plasmin thus triggering further proteolytic cascades, resulting in the degradation of the gut extracellular matrix. Nevertheless, LAB lack the intrinsic potential for plasminogen activation typical of pathogenic species. Actually, pathogenic bacteria like Staphylococcus aureus and Streptococcus pneumoniae can convert plasminogen into plasmin by specific virulence factors (staphylokinases and streptokinases, respectively) (Bergmann et al., 2003) that, so far, were never found in the genome of LAB. According to some authors (Antikainen et al., 2007b) LAB can exploit the endogenous system of the host (urokinase and tissue plasminogen activators) to trigger proteolysis, according to others (Sánchez et al., 2008). LAB can quench plasminogen, subtracting it from pathogenic bacteria. At present, this is still a controversial question concerning LAB and more research is needed to fully shed light on this bacteria-host interaction aspect.
LAB Proved Beneficial Actions on Human Health
The most important beneficial interactions between gut LAB and humans include metabolism regulation, infection control, and inflammation/allergy modulation. The last three effects are in some way connected with the immune system regulatory action exterted by LAB.
Metabolism
Energy/mineral recovery
LAB can enhance energy recovery from nutrients by degrading non-metabolizable sugars and supply the host with their own β-galactosidase thus by-passing enzymatic lactose intolerance. Some casein phosphopeptides (CPP) show the capacity of maintaining in solution calcium ions, even in alkaline pH. This results in increased absorption of calcium useful in osteoporosis, but also in enhanced absorption of iron, zinc, and manganese, useful as enzyme co-factors or prostetic groups. To perform this function is necessary that LAB digest casein, especially alpha s1 and alpha s2 caseins, releasing CPP phosphorylated on Ser and Thr residues. Since beta and k-caseins are poor in hydroxylated amino acids, they are less suitable for phosporylation, and the peptides derived from them are less active in metal binding (Meisel, 1998).
Cholesterol-lowering
When released in the gut by LAB, SCFA can cause a decrease in the hepatic cholesterol synthesis and a redistribution of cholesterol from blood to the liver (Pereira and Gibson, 2002). Also deconjugation of bile acids by LAB can play a similar role in plasma cholesterol-lowering. Deconjugated bile acids are not well absorbed by the gut mucosa and therefore excreted. As a consequence, new cholesterol is driven from blood to liver, for de novo bile acid synthesis (St-Onge et al., 2000). In Lactobacillus reuteri a specific choloylglycine hydrolase, catalyzing the initial gateway reaction for bile acids deconjugation, is biosynthesized (Martoni et al., 2008). Cholesterol-lowering capability has also been demonstrated by LAB-produced EPS (Pigeon et al., 2002). Benefits on cardiovascular pathologies and hearth disease are thus expected.
Infection and Immunomodulation
LAB have longly been known as being able to control infections because of their direct antibacterial action (mediated by bacteriocins and acid production) and to their indirect (immunologically mediated) action against viruses. The role of Lactobacilli in preventing traveller's diarrhea, growth of Helicobacter pylori and toxin producing E. coli has been demonstrated in both in vitro and in vivo studies. Lactobacillus rhamnosus GG has also been successfully employed for preventing C. difficile colitis, as well as to treat atopic eczema by means of modulation over Interleukin-10 (Pessi et al., 2000), while Lactobacillus reuteri can control IgE-mediated allergies by acting at regulatory T cells level (Karimi et al., 2009). Antiinflammatory (Kitazawa et al., 1998) and antitumoral activities have also been reported to be due to LAB-produced EPS. Rhamnose Hetero-EPS proved to be effective against gastric ulcer (Badel et al., 2011) and a stimulation of TNF has been demonstated (DeVuyst et al., 2007). Experiments using cocktails of Lactobacillus and Bifidobacterium also revealed the ability to cause apoptosis of colon carcinomas.
Informational Molecules and Oxidative Stress
A very cutting-edge evidence is the possibility that LAB can control both the human antioxidant defences and the production of informational molecules acting on mood, blood pressure, and more generally on the gut-brain axis.
Lactobacillus helveticus, Lactobacillus delbrueckii subsp. bulgaricus SS1, and Lactococcus lactis subsp. cremoris FT4 can all modulate blood pressure by producing angiotensin 1-converting enzyme inhibitory peptides (ACE inhibitors) from milk proteins. The most active molecules are tripeptides made up of Val-Pro-Pro and Ile-Pro-Pro, but other, casein-derived, antihyperthensive peptides like alpha s1 casokinin-5 (Phe-Phe-Val-Ala-Pro), alpha s1 casokinin-6 (Thre-Thre-Met-Pro-Leu-Trp), betacasokinin (Lys-Val-Leu-Pro-Val), and a dipeptide Tyr-Pro, proved to be effective in blood pressure control as well (Yamamoto et al., 1999). Since their production is the result of the proteolytic activity of some LAB strains over casein, it is dependent upon casein availability in the intestine and from the proteolytic potential of each single strain. Generally LAB peptidases, by shortening the poly/oligopeptide chain, contribute to enhance the anti ACE potential. Interestingly, the final active short peptides are resistant to both pH variations and human digestive enzymes (Sipola et al., 2002; Gobbetti et al., 2004). Antithrombotic peptides derived from human and bovine k-caseinglycopeptides have also been found in five-days-old newborns after breast and formula feeding, respectively (Chabance et al., 1995). Even if these molecules can be produced as the result of human proteolysis on food, LAB proteolytic activity on casein can for sure contribute to their release in vivo. These molecules, named casoplatelin and thrombin inhibitory peptide, are 6–11 amino acids long oligopeptides, preventing the aggregation of ADP-activated platelets and the binding of human fibrinogen (lambda chain) on the platelet surface receptor. A shorter molecule, casopiastrin, also displayed fibrinogen binding inhibiting activity (Jollès et al., 1986).
The most interesting molecules produced with the contribution of LAB protease system on dairy food are the opioid peptides (exorphins) and opioid antagonist peptides (casoxins). In mammals' central and peripheral nervous system there are receptors (K, delta, and mu) for endogenously produced opioid peptides (endorphins) which have inhibitory activity on adenylate cyclase enzyme. Protein fragments with opioid-like activity have been found as the result of both human digestive pH/proteases and microbial proteases over casein molecules. These peptides are made-up of 4–10 aminoacids and can trigger a myorelaxant effect by acting on the mu receptors (beta-casomorphins) or on the delta receptor (alpha s1 casein-derived peptide). The N-terminal residue, which is crucial for triggering biological activity, is Tyr in beta-casomorphins and Arg in alpha s1 casein-derived peptide (Loukas et al., 1983). Nevertheless, the two molecules display different half-life due to their different structure and the presence of several proline residues (six) in beta-casomorphins seem to be the factor enhancing peptide resistance to enzymatic digestion in human digestive tract. Here they can affect intestinal transit time, water balance, and aminoacid uptake. Once absorbed into blood, they reach the brain and peripheral receptors where they exert their relaxing action inducing sleeping and calmness (Chabance et al., 1998). Opposite to this action is the anti-oppioid activity displayed by peptides derived from k-casein hydrolysis. These molecules called casoxins function as oppioid-antagonists over both the mu and k-type receptors. Their possible physiological action on human is counteracting the life-threatening depression of the CNS and respiratory system (Chiba and Yoshikawa, 1986). Finally, some peptides derived from casein by LAB proteolys can exert mixed functions such as morphine-like and immunostimulating (Kayser and Meisel, 1996).
Some Lactobacillus (Cho et al., 2007) and Lactococcus species (Mazzoli et al., 2010) are also able to produce, by glutamate decarboxylation (see paragraph on energy metabolism), GABA, the most widely distributed neurotransmitter in the vertebrate central nervous system, that also acts as modulatory effector at the gut level. Although GABA cannot by-pass the blood-brain barrier, it can act as a relaxing molecule over the gut smooth muscles, can lower the blood pressure in mild hypertensive patients (Inoue et al., 2003) and also plays regulatory and trophic roles on pancreas (Erlander et al., 1991). Even if not all the cited LAB are stable inhabitants of human gut, they can be acquired by food and can persist in suitable conditions.
For what concerns the antioxidant potential, as referred in a previous paragraph, some LAB strains are able to fix inorganic selenite into seleno amino acids thus opening the possibility to biosynthesize in vivo selenoproteins, essential factors for oxidative stress control. Selenium has been demonstrated to be a nutritionally essential trace element since 1957 (Schwarz and Foltz, 1957). However, dietary selenium intake vary widely among different countries, from 7 mcg per day in China up to 100–200 mcg of Canada, USA, and Venezuela (Rayman, 2008). The reason of such differences lies in the different soils in which vegetables are grown and animal are raised. It is now evident that the selenium content of food is strictly linked to soil selenium availability and where this availability is scarse severe endemic selenium deficiencies occur (Zhao et al., 2007). Furthermore, serum selenium concentrations reflect food selenium content, thus demonstrating that no alternative endogenous way to get this metal is possible (Combs, 2001).
Even if it is widely accepted that selenium deficiency (exception made for very specific and rare syndroms) does not cause illness by itself, nevertheless, this metal appears to be a key nutrient for countracting bacterial and viral infections (Beck et al., 2003), for thyroid hormone balance (Tinggi, 2008), for limiting oxidized LDL associated atherogenesis and consequent heart disease (Furman et al., 2004), and for controlling age-related disorders such as oxidative status, immunodeficiency, general inflammation (Méplan, 2011), and prostate cancer (Klein et al., 2003). Many evidences suggest a cancer-protective effect of selenium supplementation since this metal proves to be an effective tool for controlling DNA damage risk associated with neoplastic degeneration. An inverse correlation between selenium blood levels and cancer mortality has also been observed.
The recommended dietary intake of selenium for humans is 55 mcg per day (Alzate et al., 2008). The Lactobacillus biosynthesized selenium proteins not only represent a more bioavailable dietary source but there is also a correlation between this organic form and the concentration of this metal in the external environment, opening interesting perspectives on metal release in the gut environment by LAB, with consequent oxidative stress control.
LAB Lights and Shadows
LAB, like every organism on this earth, are not perfect. The described metabolic and biochemical features underline the risk that they can produce toxic metabolites suh as ammonia and bio-active undesired amines like histamine and tyramine. Their ability to produce butyrate can be severely evaluated when a metabolic syndrome is present since this metabolite, although having a protective effect against neoplastic degeneration, can increase the human calories gain and also inhibit lypolytic processes. Similarly, some CLA isomers produced by LAB could be deleterious, so a careful typing of gut LAB single strains and their metabolites can help to avoid undesiderd effects. For what concerns the ability to activate plasmin-mediated proteolytic cascades, by using endogenous factors, this risk is counterbalanced by the possible quenching effect of released palsminogen receptors on the plasminogen itself.
A separate comment is needed for the Genus Enterococcus. While some species like E. faecium are currently considered safe and even employed as starters in the food industry and as probiotics in both food and pharmaceutical preparations, other species like E. faecalis can cause problems due to different reasons. It should be remembered that enterococci are more tolerant to salt, less acidophilic, and less acydogenic than other LAB, thus constituting a group per se. First of all they are naturally resistant to many antimicrobial agents, and also bear transmissible antibiotic resistance factors. Furthermore, their pheromone production is very high thus enhancing the frequency of recombination. A second risk is linked to the fact that most pathogenicity characters expressed by enterococci are not species but rather strain related. Among these are worth mentioning gelatinase, serine proteases, endocarditis antigen, and the aggregation factor. A further problem is connected with the oxidative catabolism of steroid molecules. During their enterohepatic circulation, bile acids are bio-transformed by the intestinal microbiota into a variety of metabolites some of which are carcinogenics, especially the C7 dehydroxy- and the C7 keto-derivatives (Murray et al., 1980). Although dehydrogenation, epimerization, and dehydroxylation are mainly peformed by Clostridia, also some Enterococcus faecalis strains have been reported to have C7 dehydroxylating activity. These reactions are directed not only toward bile acids or cholesterol but also to different steroid molecules present in the gut (Groh et al., 1993).
It is always difficult to establish once and for all when the benefits are higher than the risks, especially when dealing with bacteria, whose frequency of mutation, genetic recombination, and evolution can change the scenario in few years. Only some decades ago E. coli was considered safe and Clostridia highly pathogenic. Nowadays, some E. coli strains have been proved to cause death and Clostridia have even been proposed as probiotics (Cartman, 2011). For sure the “omics” approach can help us, now and in the near future, to shed light on some controversial aspects still concerning LAB biology, and on how environmental condition can affect their biology. Nevertheless, considering the absence of the main bacterial pathogenicity characters (toxins, invasion potential) and all the beneficial traits supplied by LAB in the context of the gut ecosystem (acidification, prebiotic FOS biosynthesis, lactose recovery, bacteriocins release, positive bio-active molecules production acting on mood and appetite, immune system modulation, and selenium organication) we can conclude that this group of bacteria, is not only well adapted to live as commensal with humans, but can also help the human host to maintan his/her healty status and, may be, to enhance his/her performances and longevity.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
A particular thank to Dr. Roberto Mazzoli for helpful discussions and critical reading of the manuscript and to Dr. Alessandro Pessione for figure managing.
References
Aleljung, P., Shen, W., Rozalska, B., Hellman, U., Ljungh, A., and Wadström, T. (1994). Purification of collagen-binding proteins of Lactobacillus reuteri NCIB 1. Curr. Microbiol. 28, 231–236.
Alvarez, R. A., Blaylock, M. W., and Baseman, J. B. (2003). Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol. Microbiol. 48, 1417–1425.
Alzate, A., Fernández-Fernández, A., Pérez-Conde, M. C., Gutiérrez, A. M., and Cámara, C. (2008). Comparison of biotransformation of inorganic selenium by Lactobacillus and Saccharomyces in lactic fermentation process of yogurt and kefir. J. Agric. Food Chem. 56, 8728–8736.
Antikainen, J., Kuparinen, V., Lähteenmäki, K., and Korhonen, T. K. (2007a). pH-dependent association of enolase and glyceraldehyde-3-phosphate dehydrogenase of Lactobacillus crispatus with the cell wall and lipoteichoic acids. J. Bacteriol. 189, 4539–4543.
Antikainen, J., Kuparinen, V., Lahteenmaki, K., and Korhonen, T. K. (2007b). Enolases from gram-positive bacterial pathogens and commensal lactobacilli share functional similarity in virulence-associated traits. FEMS Immunol. Med. Microbiol. 51, 526–534.
Badel, S., Bernardi, T., and Michaud, P. (2011). New perspectives for Lactobacilli exopolysaccharides. Biotechnol. Advances 29, 54–66.
Beck, M. A., Levander, O., and Handy, J. (2003). Selenium deficiency and viral infection. J. Nutr. 133, 1463S–1467S.
Bergmann, S., Wild, D., Diekmann, O., Frank, R., Bracht, D., Chhatwal, G. S., and Hammerschmidt, S. (2003). Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49, 411–423.
Blencowe, D. K., and Morby, A. P. (2003). Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 27, 291–311.
Boosalis, M. S., Bandyopadhyay, R., Bresnick, E. H., Pace, B. S., Van DeMark, K., Zhang, B., Faller, D. V., and Perrine, S. P. (2001). Short-chain fatty acid derivatives stimulate cell proliferation and induce STAT-5 activation. Blood 97, 3259–3267.
Brioukhanov, A. L., and Netrusov, A. I. (2007). Aerotolerance of strictly anaerobic microorganisms and factors of defense against oxidative stress: a review. Appl. Biochem. Microbiol. 43, 567–582.
Calomme, M., Hu, J., Van Den Branden, K., and Vanden Berghe, D. (1995). Seleno-lactobacillus – an organic selenium source. Biol. Trace Elem. Res. 47, 379–383.
Carr, F. J., Chill, D., and Maida, N. (2002). The lactic acid bacteria: a literature survey. Crit. Rev. Microbiol. 28, 281–370.
Cerning, J. (1990). Exocellular polysaccharides produced by lactic acid bacteria. FEMS Microbiol. Rev. 87, 113–130.
Chabance, B., Jollès, P., Izquierdo, C., Mazoyer, E., Francoual, C., Drouet, L., and Fiat, A. M. (1995). Characterization of an antithrombotic peptide from kappa-casein in newborn plasma after milk ingestion. Br. J. Nutr. 73, 583–590.
Chabance, B., Marteau, P., Rambaud, J. C., Migliore-Samour, D., Boynard, M., Perrotin, P., Guillet, R., Jollès, P., and Fiat, A. M. (1998). Casein peptide release and passage to the blood in humans during digestion of milk or yogurt. Biochimie 80, 155–165.
Chiba, H., and Yoshikawa, M. (1986). “Biologically functional peptides from food proteins,” in Protein Tailoring for Food and Medical Use, eds R. E. Feeney and J. R. Whitaker (New York, NY: Marcel Dekker), 123–153.
Cho, Y. R., Chang, J. Y., and Chang, H. C. (2007). Production of gamma-aminobutyric acid (GABA) by Lactobacillus buchneri isolated from kimchi and its neuroprotective effect on neuronal cells. J. Microbiol. Biotechnol. 17, 104–109.
Clare, D. A., and Swaisgood, H. E. (2000). Bioactive milk peptides: a prospectus. J. Dairy Sci. 83, 1187–1195.
Davie, J. R. (2003). Inhibition of histone deacetylase activity by butyrate. J. Nutr. 133(Suppl. 7), 2485S–2493S.
De Angelis, M., and Gobbetti, M. (1999). Lactobacillus sanfranciscensis CB1, manganese, oxygen, superoxide dismutase and metabolism. Appl. Microbiol. Biotechnol. 51, 358–363.
De Angelis, M., Mariotti, L., Rossi, J., Servili, M., Fox, P. F., Rollán, G., and Gobbetti, M. (2002). Arginine catabolism by sourdough lactic acid bacteria: purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl. Environ. Microbiol. 68, 6193–6201.
Deveau, H., Van Calsteren, M. R., and Moineau, S. (2002). Effect of exopolysaccharides on phage-host interactions in Lactococcus lactis. Appl. Environ. Microbiol. 68, 4364–4369.
DeVuyst, L., De Vin, F., and Kamerling, J. P. (2007). “Exopolysacchides from lactic acid bacteria,” in Comprehensive Glycoscience, Vol. 2, ed J. P. Kamerling (Oxford, Elsevier), 477–518.
DiBaise, J. K., Zhang, H., Crowell, M. D., Krajmalnik-Brown, R., Decker, G. A., and Rittmann, B. E. (2008). Gut microbiota and its possible relationship with obesity. Mayo Clin. Proc. 83, 460–469.
Draper, L. A., Grainger, K., Deegan, L. H., Cotter, P. D., Hill, C., and Ross, R. P. (2009). Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin (3147). Mol. Microbiol. 71, 1043–1054.
Erlander, M. G., Tillakaratne, N. J., Feldblum, S., Patel, N., and Tobin, A. J. (1991). Two genes encode distinct glutamate decarboxylases. Neuron 7, 91–100.
Ewaschuk, J. B., Walker, J. W., Diaz, H., and Madsen, K. L. (2006). Bioproduction of conjugated linoleic acid by probiotic bacteria occurs in vitro and in vivo in mice. J. Nutr. 136, 1483–1487.
Furman, C., Rundlöf, A. K., Larigauderie, G., Jaye, M., Bricca, G., Copin, C., Kandoussi, A. M., Fruchart, J. C., Arnér, E. S., and Rouis, M. (2004). Thioredoxin reductase 1 is upregulated in atherosclerotic plaques: specific induction of the promoter in human macrophages by oxidized low-density lipoproteins. Free Radic. Biol. Med. 37, 71–85.
Gobbetti, M., Minervini, F., and Rizzello, C. G. (2004). Angiotensin I converting enzyme-inhibitory and antimicrobial bioactive peptides. Int. J. Dairy Technol. 57, 173–188.
Granato, D., Bergonzelli, G. E., Pridmore, R. D., Marvin, L., Rouvet, M., and Corthésy-Theulaz, I. E. (2004). Cell surface-associated elongation factor Tu mediates the attachmentof Lactobacillus jonhsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 72, 2160–2169.
Groh, H., Schade, K., and Hörhold-Schubert, C. (1993). Steroid metabolism with intestinal microorganisms. J. Basic Microbiol. 33, 59–72.
Hayes, M., Ross, R. P., Fitzgerald, G. F., Hill, C., and Stanton, C. (2006). Casein-derived antimicrobial peptides generated by Lactobacillus acidophilus DPC(6026). Appl. Environ. Microbiol. 72, 2260–2264.
Hébert, E. M., Raya, R. R., and de Giori, G. S. (1999). Characterisation of a cell-envelope proteinase from Lactobacillus helveticus. Biotechnol. Lett. 21, 831–834.
Henderson, B., Allan, E., and Coates, A. R. M. (2006). Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect. Immun. 74, 3693–3706.
Hill, M. J. (1975). The role of colon anaerobes in the metabolism of bile acids and steroids, and its relation to colon cancer. Cancer 36(Suppl. 6), 2387–2400.
Holzapfel, W. H., Haberer, P., Snel, J., Schillinger, U., and Huis in't Veld, J. H. (1998). Overview of gut flora and probiotics. Int. J. Food Microbiol. 41, 85–101.
Hurmalainen, V., Edelman, S., Antikainen, J., Baumann, M., Lahteenmaki, K., and Korhonen, T. K. (2007). Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology 153, 1112–1122.
Iacomino, G., Tecce, M. F., Grimaldi, C., Tosto, M., and Russo, G. L. (2001). Transcriptional response of a human colon adenocarcinoma cell line to sodium butyrate. Biochem. Biophys. Res. Commun. 285, 1280–1289.
Inan, M. S., Rasoulpour, R. J., Yin, L., Hubbard, A. K., Rosenberg, D. W., and Giardina, C. (2000). The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line. Gastroenterology 118, 724–734.
Inoue, K., Shirai, T., Ochiai, H., Kasao, M., Hayakawa, K., Kimura, M., and Sansawa, H. (2003). Blood-pressure lowering effect of a novel fermented milk containing gammaaminobutyric acid (GABA) in mild hypertensives. Eur. J. Clin. Nutr. 57, 490–495.
Jollès, P., Lévy-Toledano, S., Fiat, A. M., Soria, C., Gillessen, D., Thomaidis, A., Dunn, F. W., and Caen, J. P. (1986). Analogy between fibrinogen and casein. Effect of an undecapeptide isolated from kappa-casein on platelet function. Eur. J. Biochem. 158, 379–382.
Karimi, K., Inman, M. D., Bienenstock, J., and Forsythe, P. (2009). Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 179, 186–193.
Katayama, Y., Gottesman, S., Pumphrey, J., Rudikoff, S., Clark, W. P., and Maurizi, M. R. (1988). The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning, and mutational analysis of the ATP-binding component. J. Biol. Chem. 263, 15226–15236.
Kayser, H., and Meisel, H. (1996). Stimulation of human peripheral blood lymphocytes by bioactive peptides derived from bovine milk proteins. FEBS Lett. 383, 18–20.
Kazazic, M., Nissen-Meyer, J., and Fimland, G. (2002). Mutational analysis of the role of charged residues in target-cell binding, potency and specificity of the pediocin-like bacteriocin sakacin P. Microbiology 148, 2019–2027.
Kitazawa, H., Harata, T., Uemura, J., Saito, T., Kaneko, T., and Itoh, T. (1998). Phosphate group requirement for mitogenic activation of lymphocytes by an extracellular phosphopolysaccharide from Lactobacillus delbrueckii ssp. bulgaricus. Int. J. Food Microbiol. 40, 169–175.
Klein, E. A., Thompson, I. M., Lippman, S. M., Goodman, P. J., Albanes, D., Taylor, P. R., and Coltman, C. (2003). SELECT: the selenium and vitamin E cancer prevention trial. Urol. Oncol. 21, 59–65.
Konings, W. N. (2002). The cell membrane and the struggle for life of lactic acid bacteria. Antonie Van Leeuwenhoek 82, 3–27.
Konings, W. N. (2006). Microbial transport: adaptations to natural environments. Antonie Van Leeuwenhoek 90, 325–342.
Korhonen, H., and Pihlanto, A. (2006). Bioactive peptides: production and functionality. Int. Dairy J. 16, 945–960.
Krinos, C. M., Coyne, M. J., Weinacht, K. G., Tzianabos, A. O., Kasper, D. L., and Comstock, L. E. (2001). Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414, 555–558.
Kumar, A. S., Mody, K., and Jha, B. (2007). Bacterial exopolysaccharides–a perception. J. Basic Microbiol. 47, 103–117.
Lacourciere, G. M., and Stadtman, T. C. (1998). The NIFS protein can function as a selenide delivery protein in the biosynthesis of selenophosphate. J. Biol. Chem. 273, 30921–30926.
Lai, A. C., Tran, S., and Simmonds, R. S. (2002). Functional characterization of domains found within a lytic enzyme produced by Streptococcus equi subsp. zooepidemicus. FEMS Microbiol. Lett. 215, 133–138.
Lamberti, C., Mangiapane, E., Pessione, A., Mazzoli, R., Giunta, C., and Pessione, E. (2011a). Proteomic characterization of a selenium-metabolizing probiotic Lactobacillus reuteri Lb2 BM for nutraceutical applications. Proteomics 11, 2212–2221.
Lamberti, C., Purrotti, M., Mazzoli, R., Fattori, P., Barello, C., Coïsson, J. D., Giunta, C., and Pessione, E. (2011b). ADI pathway and histidine decarboxylation are reciprocally regulated in Lactobacillus hilgardii ISE 5211, proteomic evidence. Amino Acids 41, 517–527.
Law, J., and Haandrikman, A. (1997). Proteolytic enzymes of lactic acid bacteria. Int. Dairy J. 7, 1–11.
Laws, A., Gu, Y., and Marshall, V. (2001). Biosynthesis, characterization and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol. Adv. 19, 597–625.
Lebeer, S., Claes, J. J., Vandenplas, S., Verhoeven, T. L. A., Tytgat, H., Schoofs, G., De Keersmaecker, S. C. J., and Vanderleyden, J. (2011). “In vivo espression technology reveals new insights in the intestinal adaptation of Lactobacillus rhamnosus GG,” in Abstract book in 10th Symposium on Lactic Acid Bacteria, Egmond aan Zee, The Netherlands, August 28–September 1 2011, poster D002.
Liu, S., Pritchard, G. G., Hardman, M. J., and Pilone, G. J. (1996). Arginine catabolism in wine lactic acid bacteria: is it via the arginine deiminase pathway or the arginase-urease pathway? J. Appl. Bacteriol. 81, 486–492.
Louis, P., Scott, K. P., Duncans, S. H., and Flint, H. J. (2007). Understanding the effects of diet on bacterial metabolism in the large intestine J. Appl. Microbiol. 102, 1197–1208.
Loukas, S., Varoucha, D., Zioudrou, C., Streaty, R. A., and Klee, W. A. (1983). Opioid activities and structures of alpha-casein-derived exorphins. Biochemistry 22, 4567–4573.
Lucas, P. M., Wolken, W. A., Claisse, O., Lolkema, J. S., and Lonvaud-Funel, A. (2005). Histamine-producing pathway encoded on an unstable plasmid in Lactobacillus hilgardii (0006). Appl. Environ. Microbiol. 71, 1417–1424.
Lupton, J. R. (2004). Microbial degradation products influence colon cancer risk: the butyrate controversy. J. Nutr. 134, 479–482.
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., Pavlov, A., Pavlova, N., Karamychev, V., Polouchine, N., Shakhova, V., Grigoriev, I., Lou, Y., Rohksar, D., Lucas, S., Huang, K., Goodstein, D. M., Hawkins, T., Plengvidhya, V., Welker, D., Hughes, J., Goh, Y., Benson, A., Baldwin, K., Lee, J. H., Díaz-Muñiz, I., Dosti, B., Smeianov, V., Wechter, W., Barabote, R., Lorca, G., Altermann, E., Barrangou, R., Ganesan, B., Xie, Y., Rawsthorne, H., Tamir, D., Parker, C., Breidt, F., Broadbent, J., Hutkins, R., O'Sullivan, D., Steele, J., Unlu, G., Saier, M., Klaenhammer, T., Richardson, P., Kozyavkin, S., Weimer, B., and Mills, D. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 15611–15616.
Makarova, K. S., and Koonin, E. V. (2007). Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 189, 1199–1208.
Martoni, C., Bhatena, J., Urbanska, A. M., and Prakash, S. (2008). Microencapsulated bile salt hydrolase producing Lactobacillus reuteri for oral targeted delivery in the gastrointestinal tract. Appl. Microbiol. Biotechnol. 81, 225–233.
Matin, M. A., and Otani, H. (2002). Cytotoxic and antibacterial activities of chemically synthesized kappa-casecidin and its partial peptide fragments. J. Dairy Res. 69, 329–334.
Mazzoli, R., Pessione, E., Dufour, M., Laroute, V., Giuffrida, M. G., Giunta, C., Cocaign-Bousquet, M., and Loubière, P. (2010). Glutamate-induced metabolic changes in Lactococcus lactis NCDO 2118 during GABA production: combined transcriptomic and proteomic analysis. Amino Acids 39, 727–737.
Méplan, C. (2011). Trace elements and ageing, a genomic perspective using selenium as an example. J. Trace Elem. Med. Biol. S11–S16.
Millette, M., Dupont, C., Archambault, D., and Lacroix, M. (2007). Partial characterization of bacteriocins produced by human Lactococcus lactis and Pediococccus acidilactici isolates. J. Appl. Microbiol. 102, 274–282.
Millichap, J. G., and Yee, M. M. (2003). The diet factor in pediatric and adolescent migraine. Pediatr. Neurol. 28, 9–15.
Minucci, S., and Pelicci, P. G. (2006). Histone deacetylase inhibitors and the promise af epigenetic (and more) treatments for cancer. Nat. Rev. Cancer 6, 38–51.
Moineau, S., Borkaev, M., Holler, B. J., Walker, S. A., Kondo, J. K., and Vedamuthu, E. R. (1996). Isolation and characterization of lactococcal bacteriophages from cultured buttermilk plants in the United States. J. Dairy Sci. 79, 2104–2111.
Molenaar, D., Bosscher, J. S., ten Brink, B., Driessen, A. J., and Konings, W. N. (1993). Generation of a proton motive force by histidine decarboxylation and electrogenic histidine/histamine antiport in Lactobacillus buchneri. J. Bacteriol. 175, 2864–2870.
Montalbán-López, M., Sánchez-Hidalgo, M., Valdivia, E., Martínez-Bueno, M., and Maqueda, M. (2011). Are bacteriocins underexploited? Novel applications for old antimicrobials. Curr. Pharm. Biotechnol. 12, 1205–1220.
Moreno-Arribas, M. V., Polo, M. C., Jorganes, F., and Muñoz, R. (2003). Screening of biogenic amine production by lactic acid bacteria isolated from grape must and wine. Int. J. Food Microbiol. 84, 117–123.
Murray, W. R., Backwood, A., Trotter, J. M., Calman, K. C., and MacKay, C. (1980). Faecal bile acids and clostridia in the aetiology of colorectal cancer. Br. J. Cancer 41, 923–928.
O'Flaherty, S., and Klaenhammer, T. (2011). Gene expression analysis of human intestinal epithelial cells exposed to lactic acid bacteria. Abstract book in 10th Symposium on Lactic Acid Bacteria, Egmond aan Zee, The Netherlands, August 28–September 1 2011, poster C076.
Pellizzaro, C., Coradini, D., and Daidone, M. G. (2002). Modulation of angiogenesis-related proteins synthesis by sodium butyrate in colon cancer cell line HT29. Carcinogenesis 23, 735–740.
Pereira, D. I., and Gibson, G. R. (2002). Effects of consumption of probiotics and prebiotics on serum lipid levels in humans. Crit. Rev. Biochem. Mol. Biol. 37, 259–281.
Pessi, T., Sütas, Y., Hurme, M., and Isolauri, E. (2000). Interleukin-10 generation in atopic children following oral Lactobacillus rhamnosus GG. Clin. Exp. Allergy 30, 1804–1808.
Pessione, E., Mazzoli, R., Giuffrida, M. G., Lamberti, C., Garcia-Moruno, E., Barello, C., Conti, A., and Giunta, C. (2005). A proteomic approach to studying biogenic amine producing lactic acid bacteria. Proteomics 5, 687–698.
Pessione, E., Pessione, A., Lamberti, C., Coïsson, D. J., Riedel, K., Mazzoli, R., Bonetta, S., Eberl, L., and Giunta, C. (2009). First evidence of a membrane-bound, tyramine and beta-phenylethylamine producing, tyrosine decarboxylase in Enterococcus faecalis: a two-dimensional electrophoresis proteomic study. Proteomics 9, 2695–2710.
Pigeon, R. M., Cuesta, E. P., and Gililliand, S. E. (2002). Binding of free bile acids by cells of yogurt starter culture bacteria. J. Dairy Sci. 85, 2705–2710.
Rayman, M. P. (2008). Food-chain selenium and human health: emphasis on intake. Br. J. Nutr. 100, 254–268.
Rose, D. P. (1997). Effects of dietary fatty acids on breast and prostate cancers: evidence from in vitro experiments and animal studies. Am. J. Clin. Nutr. 66(Suppl. 6), 1513S–1522S.
Salazar, O., and Asenjo, J. A. (2007). Enzymatic lysis of microbial cells. Biotechnol. Lett. 29, 985–994.
Salvatore, S., Hauser, B., Devreker, T., Vieira, M. C., Luini, C., Arrigo, S., Nespoli, L., and Vandenplas, Y. (2007). Probiotics and zinc in acute infectious gastroenteritis in children: are they effective? Nutrition 23, 498–506.
Sánchez, B., Bressollier, P., and Urdaci, M. C. (2008). Exported proteins in probiotic bacteria: adhesion to intestinal surfaces, host immunomodulation and molecular cross-talking with the host. FEMS Immunol. Med. Microbiol. 54, 1–17.
Sauer, J., Richter, K. K., and Pool-Zobel, B. L. (2007). Physiological concentrations of butyrate favorably modulate genes of oxidative and metabolic stress in primary human colon cells. J. Nutr. Biochem. 18, 736–745.
Schwarz, K., and Foltz, C. M. (1957). Selenium as an integral part of factor3 against dietary liver necrosis. J. Am. Chem. Soc. 79, 3292–3293.
Siezen, R. J., van Enckevort, F. H., Kleerebezem, M., and Teusink, B. (2004). Genome data mining of lactic acid bacteria: the impact of bioinformatics. Curr. Opin. Biotechnol. 15, 105–115.
Sipola, M., Finckenberg, P., Vapaatalo, H., Pihlanto-Leppälä, A., Korhonen, H., Korpela, R., and Nurminen, M. L. (2002). Alpha-lactorphin and beta-lactorphin improve arterial function in spontaneously hypertensive rats. Life Sci. 71, 1245–1253.
Smits, H. H., Engering, A., van der Kleij, D., de Jong, E. C., Schipper, K., van Capel, T. M., Zaat, B. A., Yazdanbakhsh, M., Wierenga, E. A., van Kooyk, Y., and Kapsenberg, M. L. (2005). Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 115, 1260–1267.
St-Onge, M. P., Farnworth, E. R., and Jones, P. J. (2000). Consumption of fermented and nonfermented dairy products: effects on cholesterol concentrations and metabolism. Am. J. Clin. Nutr. 71, 674–681.
Tinggi, U. (2008). Selenium: its role as antioxidant in human health. Environ. Health Prev. Med. 13, 102–108.
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031.
Tytgat, H. L. P., Lebeer, S., Claes, I., Schoofs, G., Verhoeven, T. L. A., De Keersmaecker, S. C. J., Marchal, K., and Vanderleyden, J. (2011). “Glycobiology of LGG,” in Abstract book in 10th Symposium on Lactic Acid Bacteria, Egmond aan Zee, The Netherlands, August 28–September 1 2011, SL 15.
van Hijum, S. A., Kralj, S., Ozimek, L. K., Dijkhuizen, L., and van Geel-Schutten, I. G. H. (2006). Structure-function relationship of glucansucrase and fructansucrase enzymes from Lactic acid bacteria. Microbiol. Mol. Biol. Rev. 70, 157–176.
Vaughan, E. E., de Vries, M. C., Zoetendal, E. G., Kaouther Ben Amor Akkerman, A. D. L., and De Vos, W. M. (2002). The intestinal LABs. Antonie Van Leeuwenhoek 82, 341–352.
Weidenmaier, C., and Peschel, A. (2008). Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 4, 276–287.
Yamamoto, N., Maeno, M., and Takano, T. (1999). Purification and characterization of an antihypertensive peptide from a yogurt-like product fermented by Lactobacillus helveticus CPN4. J. Dairy Sci. 82, 1388–1393.
Keywords: selenium, bioactive molecules, proteolysis, exopolysaccharides, CLA, short chain fatty acids, bacteriocins, diet
Citation: Pessione E (2012) Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front. Cell. Inf. Microbio. 2:86. doi: 10.3389/fcimb.2012.00086
Received: 23 March 2012; Paper pending published: 09 April 2012;
Accepted: 01 June 2012; Published online: 22 June 2012.
Edited by:
Lorenza Putignani, Children's Hospital and Research Institute Bambino Gesù Hospital, ItalyReviewed by:
Subramanian Dhandayuthapani, University of Texas Health Science Center, USAHua Xie, Meharry Medical College, USA
Copyright: © 2012 Pessione. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Enrica Pessione, Dipartimento di Scienze della Vita e Biologia dei sistemi – Life Sciences and Systems Biology, University of Torino, Via Accademia Albertina 13, 10123 Torino, Italy. e-mail: enrica.pessione@unito.it