Listeria monocytogenes: survival and adaptation in the gastrointestinal tract
- 1Alimentary Pharmabiotic Centre, University College Cork, Cork, Ireland
- 2School of Microbiology, University College Cork, Cork, Ireland
- 3School of Pharmacy, University College Cork, Cork, Ireland
The foodborne pathogen Listeria monocytogenes has the capacity to survive and grow in a diverse range of natural environments. The transition from a food environment to the gastrointestinal tract begins a process of adaptation that may culminate in invasive systemic disease. Here we describe recent advances in our understanding of how L. monocytogenes adapts to the gastrointestinal environment prior to initiating systemic infection. We will discuss mechanisms used by the pathogen to survive encounters with acidic environments (which include the glutamate decarboxylase and arginine deiminase systems), and those which enable the organism to cope with bile acids (including bile salt hydrolase) and competition with the resident microbiota. An increased understanding of how the pathogen survives in this environment is likely to inform the future design of novel prophylactic approaches that exploit specific pharmabiotics; including probiotics, prebiotics, or phages.
Introduction
Listeria monocytogenes is a Gram positive foodborne pathogen capable of causing infection of the fetus in pregnant women and meningitis, meningoencephalitis or febrile gastroenteritis in non-pregnant individuals. The pathogen continues to cause large common-source outbreaks from ready-to-eat food sources and represents a significant cause of food-related mortality (Jackson et al., 2011; Mccollum et al., 2013). The pathogen exists in the environment as a saprophyte and can access the human food chain either directly or through infection or carriage in farm animals (zoonotic disease). Listeria possesses a number of molecular mechanisms with which to adapt to the different stages of the pathogenic lifecycle (reviewed previously in Gahan and Hill, 2005; Gray et al., 2006; Sleator et al., 2009; Camejo et al., 2011; Cossart, 2011). Here we focus upon recent advances in our understanding of how L. monocytogenes adapts to the environment of the gastrointestinal (GI) tract and makes the transition from saprophyte to pathogen.
Potential for Faecal or Gall Bladder Carriage
L. monocytogenes faecal carriage in farm animals has been reported as 21.3% in cattle and 1.5% in sheep (Esteban et al., 2009), while it can be detected in the faeces of 2.1% of asymptomatic individuals in human population studies (Cobb et al., 1996). An interesting study followed the appearance of L. monocytogenes in the faeces of three human volunteers (one male, two females) over approximately 1 year (Grif et al., 2003). L. monocytogenes was isolated in 31 of the 868 (3.57%) stool samples analyzed, which suggests between 5 and 9 annual exposures to L. monocytogenes per person. In the majority of cases L. monocytogenes carriage was transient, lasting between 1 and 4 days (Grif et al., 2003).
Active systemic infection in mice following intravenous inoculation results in faecal shedding of L. monocytogenes that can last for up to 9 days post-infection (Nichterlein et al., 1994). It is significant that L. monocytogenes can colonize the murine gall bladder following oral or intravenous administration (Hardy et al., 2004; Bron et al., 2006). It can also be isolated from the gall bladder in infected guinea pigs (Jensen et al., 2008) and turkeys (Huff et al., 2005), but not in infected sheep (Zundel and Bernard, 2006). There is also evidence suggesting that L. monocytogenes may be a rare cause of human cholecystitis (infection of the gall bladder) (Allerberger et al., 1989; Descy et al., 2012; Bruminhent et al., 2013). In mice efficient growth of L. monocytogenes in the gall bladder results in rapid shedding into the GI tract and faeces (Hardy et al., 2006). Growth in this bile-rich environment may therefore provide a significant source of faecal shedding during infection. We have recently demonstrated that gall bladder bile actually serves as an efficient growth environment for a number of bacterial species (including non-pathogens), suggesting that L. monocytogenes is not unusual in its ability to grow in this environment (Dowd et al., 2011). The ability of the pathogen to cross epithelial barriers may make this a potential site for bacterial replication, a phenomenon which is perhaps further influenced by specific host conditions (Dowd et al., 2011; Bruminhent et al., 2013). Furthermore, to our knowledge the possibility of ascending bile duct infection by L. monocytogenes has not been investigated.
Insights into Stress Adaptation in the GI Tract
Given the evidence of transient faecal carriage of L. monocytogenes (Grif et al., 2003), it is likely that the pathogen is a common allochthonous passenger in the human gut rather than an autochthonous commensal. It is clear that the alternative sigma factor, Sigma B, plays a significant role in adaptation to the gastrointestinal environment (Sleator et al., 2009; Toledo-Arana et al., 2009). Mutation of sigB results in decreased virulence of L. monocytogenes when administered orally to guinea pigs or mice (Nadon et al., 2002; Garner et al., 2006). However, systemic infection is not affected. Transcriptomic analyses have identified the members of the Sigma B regulon, the exact components of which may differ marginally between L. monocytogenes isolates (Hain et al., 2008; Sleator et al., 2009), and which contain genes encoding systems for bile, acid, and salt adaptation (Hain et al., 2008; Oliver et al., 2009; Chaturongakul et al., 2011; Mujahid et al., 2013).
A comprehensive transcriptomic analysis of listerial growth both in vivo and in vitro was carried out using tiling arrays in order to map the expression of genes and small RNAs (sRNAs) during the passage from saprophytism to virulence (Toledo-Arana et al., 2009). The study identified 50 sRNAs as well as antisense RNAs that point to an added layer of regulatory complexity. The work definitively established that Sigma B is the predominant regulator of virulence genes and survival factors in the GI tract, with PrfA acting as the main regulator of virulence genes in blood (Toledo-Arana et al., 2009). Adaptation to the murine GI tract resulted in significant reshaping of the transcriptional profile, with the upregulation of 437 genes and the downregulation of 769 genes, many of which are regulated by Sigma B (including inlA, inlB, inlH, and bsh) (Toledo-Arana et al., 2009).
Further studies of L. monocytogenes gene expression profiles in the GI tract of germ-free mice or mice mono-colonized by Lactobacillus species have provided insights into how the pathogen adapts to the physical and chemical environment of the gut (Archambaud et al., 2012). In an elegant experimental setup transgenic mice optimized for listerial infection (E-cadhum mice) were mono-colonized with either Lb. paracasei CNCM I-3689 or Lb. casei BL23 prior to oral infection with L. monocytogenes. Passage from broth to the GI tract resulted in the upregulation of 520 protein coding genes and seven non-coding sRNAs, whereas 523 genes and three sRNAs were downregulated. The presence of Lactobacillus spp. resulted in enhanced effects upon numerous gene systems suggesting a major influence of the microbiota upon gene expression in L. monocytogenes. Many of the listerial genes that were positively influenced by the microbiota are potentially involved in adaptation to the gut environment and include genes encoding pathways involved in ethanolamine and propanediol metabolism that have been implicated in the gastrointestinal survival of Listeria (Cummins et al., 2013) and other bacteria (Sriramulu et al., 2008; Archambaud et al., 2012).
Functional genetic approaches have been employed to determine the key systems required for survival in the GI tract (Sleator et al., 2009). We recently employed a mariner transposon-based signature-tagged mutagenesis approach to identify loci likely to be play a role during the gastrointestinal phase of L. monocytogenes infection (Cummins et al., 2013). The study determined the importance of a gene encoding a Sigma B-regulated internalin-like protein (equivalent to lmo0610) (Mcgann et al., 2008) for pathogenesis via the gastrointestinal route of infection. A mutant in an enzyme putatively involved in propanediol utilization was also unable to infect mice efficiently via the oral route, providing further evidence that ethanolamine/propanediol pathways are important for listerial growth in the GI tract (Archambaud et al., 2012). We also identified a number of putative regulators, an iron uptake system (Jin et al., 2006) and metabolic systems required for colonization of the GI tract and subsequent infection using our approach (Cummins et al., 2013).
Adaptation to Low pH
During passage through the GI tract L. monocytogenes encounters low pH environments in both stomach and duodenum. In humans the low pH of the stomach provides a significant barrier to L. monocytogenes infection and patients taking medications which reduce gastric acid (including proton pump inhibitors) are at increased risk of infection (Bavishi and Dupont, 2011). Listeria possesses a number of systems that regulate intracellular pH during exposure to acidic environments (reviewed in Smith et al., 2013 and Ryan et al., 2008).
The glutamate decarboxylase (GAD) system is particularly important for mediating pH homeostasis. The GAD system in L. monocytogenes is complex and comprises two glutamate/GABA antiporters (GadT1 and GadT2) and three glutamate decarboxylase enzymes (GadD1, GadD2, and GadD3) (Figure 1). A recent study demonstrated that the GadD2/T2 and GadD3 systems are present in all strains analyzed but the GadD1/T1 system is present in only 36.6% of strains analyzed and is absent from serotype 4b and lineage IV strains (Chen et al., 2012). Indeed the gadD1 and gadT1 genes are present on a five-gene stress survival islet (SSI-1) (Ryan et al., 2010) that also contains a gene encoding a putative penicillin V acylase that is required for maximum bile tolerance (Begley et al., 2005b; Ryan et al., 2010). Deletion of SSI-1 from L. monocytogenes strain LO28 reduced the acid and salt tolerance of the strain and significantly affected ability to grow in foods, suggesting the importance of SSI-1 for survival during the infectious cycle (Ryan et al., 2010).
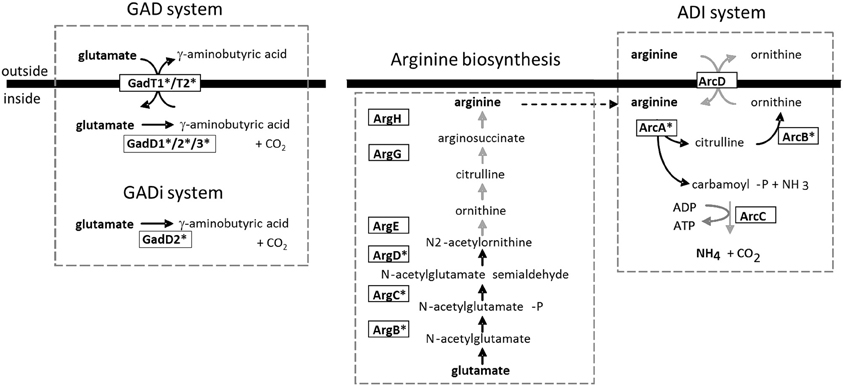
Figure 1. Two major acid tolerance mechanisms in L. monocytogenes. The GAD system is proposed to function through the import of glutamate into the cell (via GadT1 or GadT2) which is converted to GABA via Gad enzymes (GadD1, D2, or D3) in the cytoplasm. The GABA is then thought to be exported from the cell by means of the antiport systems (GadT1/T2). The consumption of protons during the reaction is contributes to the reduction in intracellular hydrogen ion concentration (Cotter et al., 2001). Karatzas et al. (2012) have recently proposed the existence of an intracellular GADi system that operates in the absence of added glutamate. The ADI system has also been proposed to operate via utilization of intracellular arginine as well as via arginine import (Ryan et al., 2009). The arginine biosynthetic pathway is shown. Black arrows indicate genes that are upregulated during growth of Listeria in the GI tract of germ-free mice and further upregulated through in situ exposure to Lactobacillus spp. (Archambaud et al., 2012). In such cases the enzyme is highlighted with an asterisk.
Clearly the GAD system plays a significant role in pH homeostasis in L. monocytogenes as mutants in gadT2/D2 (Cotter et al., 2001) are significantly compromised in acid tolerance. Evidence suggests that GadD1/T1 is important for growth in mildly acidic environments rather than under extremes of acid stress (Cotter et al., 2005). The ability of individual strains to grow at mildly acidic pH correlates with the presence of the GadD1/T1 present on SSI-1 (Cotter et al., 2005). Recently it has been demonstrated that GABA may accumulate in L. monocytogenes under minimal growth conditions, providing evidence for an intracellular GAD system (GADi) that operates separately from the canonical extracellular GAD (GADe) system (Karatzas et al., 2010, 2012). Interestingly all elements of the GAD system are transcriptionally upregulated in Listeria during colonization of the GI tract (Archambaud et al., 2012).
L. monocytogenes strains also possess an arginine deiminase (ADI) pathway and an agmantine (AgDI) deiminase system that contribute to pH homeostasis (Ryan et al., 2008; Smith et al., 2013). The ADI system is not present in the non-pathogenic species L. innocua. The ADI system operates through the catabolism of arginine to produce ornithine and ammonium ions (NH+4) which protect the cells from low pH (Figure 1). In a typical bacterial ADI system extracellular arginine feeds the ADI system via an arginine/ornithine antiporter (Cotter and Hill, 2003). However, we noted that the L. monocytogenes ADI contributes to pH homeostasis in the absence of extracellular arginine and we have evidence to suggest that the system also utilizes arginine that is synthesized via the cellular arginine biosynthetic pathway (encoded by the arg genes). In L. monocytogenes LO28 the arginine catabolic genes of the ADI system (including arcA and arcC) and the arginine biosynthetic genes (including argG) are induced by low pH and anaerobicity and are under the influence of the transcriptional regulator ArgR (Ryan et al., 2009). The data suggest that L. monocytogenes may synthesize arginine under anaerobic conditions to regulate cellular homeostasis through the ADI system (Ryan et al., 2009). It is interesting to note that the ADI system is also regulated by Sigma B (Hain et al., 2008) and the genes encoding both the arginine deiminase (arcA) and enzymes of the biosynthetic pathway (including argB, argC, and argD) are significantly induced in the GI tract of germ-free mice and are further induced by the presence of lactobacilli (Archambaud et al., 2012).
Adaptation to Bile Acids
Bile acids are produced from cholesterol in the liver, stored in the gall bladder and are released into the duodenum postprandially (Begley et al., 2005a). Bile acids can disrupt bacterial membrane structure, cause dissociation of membrane proteins, induce DNA damage and trigger oxidative stress in bacterial cells (Begley et al., 2005a). These cellular insults are reflected in a recent analysis of the proteome in L. monocytogenes cells exposed to bile acids under anaerobic conditions (Payne et al., 2013). The study found significant alterations in proteins associated with DNA repair, chaperone activity and oxidative stress responses including elevated levels of excinuclease ABC proteins (UvrABC), DNA mismatch repair proteins and DnaK (Payne et al., 2013).
Similarly a recent analysis of the transcriptomic response of L. monocytogenes to mammalian bile has provided insights into how bile may act as a specific signal during gastrointestinal transit (Quillin et al., 2011). The study demonstrated that bile exposure regulates many virulence factors in L. monocytogenes. In particular the work identified a TetR-type regulator [renamed bile-regulated transcription factor A (BrtA)] that senses bile (in particular the bile acid cholic acid) and regulates expression of two multidrug resistance (MDR) efflux pumps (MdrM and MdrT) that mediate bile tolerance and liver/gall bladder colonization (Quillin et al., 2011). This finding may be particularly relevant given the broader role of MdrM/T in mediating secretion of cyclic-di-AMP, a signaling molecule that triggers STING-dependent production of interferon-beta and promotes in vivo survival of the pathogen (Crimmins et al., 2008; Woodward et al., 2010; Schwartz et al., 2012; Burdette and Vance, 2013).
L. monocytogenes strains express a bile salt hydrolase (BSH) enzyme that has the potential to detoxify individual conjugated bile acids and contributes to listerial survival in the GI tract (Dussurget et al., 2002; Begley et al., 2005b). BSH is an enzyme that is specific to commensal bacteria that inhabit the GI tract and a limited number of pathogens that infect via this route (Jones et al., 2008). The importance of the enzyme for gastrointestinal survival is evidenced by the wide distribution of this activity amongst commensals (Jones et al., 2008). BSH expression in L. monocytogenes is under the influence of Sigma B and is strongly enhanced during growth in the intestine and following interaction with commensal lactobacilli (Archambaud et al., 2012). Recent work suggests that BSH and Sigma B may not be essential for growth of L. monocytogenes in the murine gall bladder (Dowd et al., 2011) where the pathogen instead relies upon expression of pathways required for nutrient acquisition or synthesis of monomeric biomolecules (Dowd et al., 2011; Faith et al., 2012). In contrast, when the pH of bile is reduced (as occurs in the duodenum) the toxicity of bile acids is increased (Begley et al., 2005a) and the pathogen requires BSH, another bile resistance protein called bile exclusion protein (BilE) (Sleator et al., 2005) and other potentially Sigma B-regulated detoxification mechanisms (Dowd et al., 2011).
Potential Effects of the Food Matrix and Pre-Adaptation
Relatively few studies have analyzed the influence of the food matrix upon subsequent infectious potential in L. monocytogenes. Previous physiological studies have implied that extracellular levels of glutamate, which serves as a substrate of the GAD system (Cotter et al., 2001), or carnitine, which is taken up principally by the OpuC transporter (Sleator et al., 2001; Wemekamp-Kamphuis et al., 2002), could have the potential to influence infection of the GI tract. Furthermore, acid or salt adaptation during growth in food may influence stress-hardening of the pathogen, thereby enhancing subsequent survival in the gut (Sleator et al., 2009). Both transcriptomic and proteomic approaches are being used in order to understand the molecular mechanisms that underpin cross-adaptation to a variety of stresses (Bergholz et al., 2012; Melo et al., 2013) and the interplay between different regulatory networks (Chaturongakul et al., 2011).
A number of studies have investigated the effects of acid shock upon expression of in vivo-relevant genes regulated by PrfA and Sigma B (including bsh, inlA, and inlB) (Phan-Thanh and Mahouin, 1999; Abram et al., 2008; Xayarath et al., 2009; Neuhaus et al., 2013). Recent work demonstrated that acid shock significantly induced these systems and enhanced the invasive capacity for epithelial cells and the ability to infect Caenorhabditis elegans (an alternative host model system) (Neuhaus et al., 2013). Other work has demonstrated that oxygen restriction in the growth environment significantly enhances subsequent infectivity of L. monocytogenes in guinea pigs (Bo Andersen et al., 2007). Indeed hypoxia has been shown to be a potent inducer of genes required for gastrointestinal survival and invasion (including inlA and inlB) (Toledo-Arana et al., 2009).
A better understanding of how environmental conditions influence the Sigma B regulon has the potential to provide added insights into adaptation and infection by pathogen. Interesting recent work has determined that light (in the form of both blue or red light) is a potent inducer of the Sigma B regulon in L. monocytogenes (Ondrusch and Kreft, 2011). The signal from blue light is sensed by a blue light detector (encoded by lmo0799) (Ondrusch and Kreft, 2011; Tiensuu et al., 2013) and a mutation in lmo0799 enhances the red-light effect (Ondrusch and Kreft, 2011). Exposure of L. monocytogenes to blue light induced inlA and inlB expression and leads to enhanced invasion of epithelial cells (Ondrusch and Kreft, 2011). However, the relevance of this finding for in vivo infection is not yet clear. Furthermore, a recent study indicates that oscillating day and night cycles co-ordinate differentiation of L. monocytogenes colonies by inducing alternating opaque and translucent rings on soft agar plates. This differentiation was mediated by Lmo0799 through induction of Sigma B, though both PrfA and ActA were also required for ring formation (Tiensuu et al., 2013). Significantly, bacteria forming opaque rings demonstrated enhanced levels of exopolysaccharide (EPS) production and elevated stress resistance indicating a significant physiological adaptation which may be significant in different phases of the infectious cycle (Tiensuu et al., 2013).
Role of the Microbiota in Resistance to Listeria Colonization
L. monocytogenes is a non-fastidious and tractable target for demonstrating the barrier effect of probiotic commensals upon colonization or infection. In vitro cell culture studies indicate that cell invasion by L. monocytogenes can be inhibited by a variety of commensal bacteria (Corr et al., 2007a; Gomes et al., 2012; Nakamura et al., 2012). Experiments using germ-free mice or rats also clearly demonstrate that the gastrointestinal microbiota plays an important role as a barrier to L. monocytogenes infection (Czuprynski and Balish, 1981; Bambirra et al., 2007; Archambaud et al., 2012).
The mechanisms by which probiotic commensals may protect against gastrointestinal pathogens include direct antagonism, immunomodulation, enhancement of epithelial barrier function and bacterial signaling events (including quorum sensing) (Corr et al., 2009). A number of these potential mechanisms have been examined in Listeria oral infection models. We have shown that bacteriocin production by Lb. salivarius UCC118 is a dominant mechanism by which this potentially probiotic commensal mediates a level of protection against oral infection in conventional mice (Corr et al., 2007b). The work indicates that bacteriocin production in situ may provide a mechanistic basis by which the microbiota protects against foodborne bacteria. It also represents a potential proof-of-concept for the development of bacteriocin-producing commensal strains to protect against a number of foodborne agents (Rea et al., 2010, 2011).
The study by Archambaud and coworkers which utilized germ-free and mono-colonized E-cadhum mice demonstrated the significant crosstalk that occurs between probiotic commensals and both the host and the pathogen (Archambaud et al., 2012). In particular the study revealed that probiotic commensal strains modulate the host interferon response against L. monocytogenes and reduce dissemination of the pathogen in the host. The study demonstrates that crosstalk between the microbiota and the host provides a mechanism for modulating defences against the pathogen in vivo (Archambaud et al., 2012).
Conclusions
Increased understanding of the mechanisms by which L. monocytogenes adapts to the host GI tract has the potential to inform the development of novel treatment or prevention strategies. For instance a recent small molecule screening approach indicated that fluoro-phenyl-styrene-sulfonamide (FPSS) can inhibit Sigma B activity, thereby downregulating the expression of key virulence related loci (inlA, inlB, bsh) (Palmer et al., 2011). There is significant evidence that Listeria-specific bacteriophages or phage lysins can control bacterial growth in the GI tract (Coffey et al., 2010; Mai et al., 2010). Significantly, the US FDA has recently approved the use of a bacteriophage-based additive for the control of listerial growth in foods (Coffey et al., 2010). Finally, the continued investigation of potential probiotic (Corr et al., 2007b; Archambaud et al., 2012) and prebiotic (Ebersbach et al., 2010) candidates for their ability to control L. monocytogenes in the GI tract should inform the future development of functional foods with the potential to reduce disease incidence.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors' work has been supported in part by grants from Science Foundation Ireland in the form of a center grant (Alimentary Pharmabiotic Centre; Grant Numbers SFI/12/RC/2273 and 12/RC/2273).
References
Abram, F., Starr, E., Karatzas, K. A., Matlawska-Wasowska, K., Boyd, A., Wiedmann, M., et al. (2008). Identification of components of the Sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74, 6848–6858. doi: 10.1128/AEM.00442-08
Allerberger, F., Langer, B., Hirsch, O., Dierich, M. P., and Seeliger, H. P. (1989). Listeria monocytogenes cholecystitis. Z Gastroenterol. 27, 145–147.
Archambaud, C., Nahori, M. A., Soubigou, G., Becavin, C., Laval, L., Lechat, P., et al. (2012). Impact of lactobacilli on orally acquired listeriosis. Proc. Natl. Acad. Sci. U.S.A. 109, 16684–16689. doi: 10.1073/pnas.1212809109
Bambirra, F. H., Lima, K. G., Franco, B. D., Cara, D. C., Nardi, R. M., Barbosa, F. H., et al. (2007). Protective effect of Lactobacillus sakei 2a against experimental challenge with Listeria monocytogenes in gnotobiotic mice. Lett. Appl. Microbiol. 45, 663–667. doi: 10.1111/j.1472-765X.2007.02250.x
Bavishi, C., and Dupont, H. L. (2011). Systematic review: the use of proton pump inhibitors and increased susceptibility to enteric infection. Aliment. Pharmacol. Ther. 34, 1269–1281. doi: 10.1111/j.1365-2036.2011.04874.x
Begley, M., Gahan, C. G., and Hill, C. (2005a). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Begley, M., Sleator, R. D., Gahan, C. G., and Hill, C. (2005b). Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73, 894–904. doi: 10.1128/IAI.73.2.894-904.2005
Bergholz, T. M., Bowen, B., Wiedmann, M., and Boor, K. J. (2012). Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl. Environ. Microbiol. 78, 2602–2612. doi: 10.1128/AEM.07658-11
Bo Andersen, J., Roldgaard, B. B., Christensen, B. B., and Licht, T. R. (2007). Oxygen restriction increases the infective potential of Listeria monocytogenes in vitro in Caco-2 cells and in vivo in guinea pigs. BMC Microbiol. 7:55. doi: 10.1186/1471-2180-7-55
Bron, P. A., Monk, I. R., Corr, S. C., Hill, C., and Gahan, C. G. (2006). Novel luciferase reporter system for in vitro and organ-specific monitoring of differential gene expression in Listeria monocytogenes. Appl. Environ. Microbiol. 72, 2876–2884. doi: 10.1128/AEM.72.4.2876-2884.2006
Bruminhent, J., Lynch, T. K., Gefen, J., and Santoro, J. (2013). Listeria monocytogenes cholecystitis: a possible new syndrome. Am. J. Med. Sci. 345, 414–417. doi: 10.1097/MAJ.0b013e3182761cda
Burdette, D. L., and Vance, R. E. (2013). STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 14, 19–26. doi: 10.1038/ni.2491
Camejo, A., Carvalho, F., Reis, O., Leitao, E., Sousa, S., and Cabanes, D. (2011). The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2, 379–394. doi: 10.4161/viru.2.5.17703
Chaturongakul, S., Raengpradub, S., Palmer, M. E., Bergholz, T. M., Orsi, R. H., Hu, Y., et al. (2011). Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors SigmaB, SigmaC, SigmaH, and SigmaL in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 187–200. doi: 10.1128/AEM.00952-10
Chen, J., Fang, C., Zheng, T., Zhu, N., Bei, Y., and Fang, W. (2012). Genomic presence of gadD1 glutamate decarboxylase correlates with the organization of ascB-dapE internalin cluster in Listeria monocytogenes. Foodborne Pathog. Dis. 9, 175–178. doi: 10.1089/fpd.2011.1022
Cobb, C. A., Curtis, G. D., Bansi, D. S., Slade, E., Mehal, W., Mitchell, R. G., et al. (1996). Increased prevalence of Listeria monocytogenes in the faeces of patients receiving long-term H2-antagonists. Eur. J. Gastroenterol. Hepatol. 8, 1071–1074. doi: 10.1097/00042737-199611000-00008
Coffey, B., Mills, S., Coffey, A., Mcauliffe, O., and Ross, R. P. (2010). Phage and their lysins as biocontrol agents for food safety applications. Annu. Rev. Food Sci. Technol. 1, 449–468. doi: 10.1146/annurev.food.102308.124046
Corr, S. C., Gahan, C. G., and Hill, C. (2007a). Impact of selected Lactobacillus and Bifidobacterium species on Listeria monocytogenes infection and the mucosal immune response. FEMS Immunol. Med. Microbiol. 50, 380–388. doi: 10.1111/j.1574-695X.2007.00264.x
Corr, S. C., Li, Y., Riedel, C. U., O'Toole, P. W., Hill, C., and Gahan, C. G. (2007b). Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U.S.A. 104, 7617–7621. doi: 10.1073/pnas.0700440104
Corr, S. C., Hill, C., and Gahan, C. G. (2009). Understanding the mechanisms by which probiotics inhibit gastrointestinal pathogens. Adv. Food Nutr. Res. 56, 1–15. doi: 10.1016/S1043-4526(08)00601-3
Cossart, P. (2011). Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. U.S.A. 108, 19484–19491. doi: 10.1073/pnas.1112371108
Cotter, P. D., Gahan, C. G., and Hill, C. (2001). A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40, 465–475. doi: 10.1046/j.1365-2958.2001.02398.x
Cotter, P. D., and Hill, C. (2003). Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67, 429–453. doi: 10.1128/MMBR.67.3.429-453.2003
Cotter, P. D., Ryan, S., Gahan, C. G., and Hill, C. (2005). Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71, 2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005
Crimmins, G. T., Herskovits, A. A., Rehder, K., Sivick, K. E., Lauer, P., Dubensky, T. W., et al. (2008). Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc. Natl. Acad. Sci. U.S.A. 105, 10191–10196. doi: 10.1073/pnas.0804170105
Cummins, J., Casey, P. G., Joyce, S. A., and Gahan, C. G. (2013). A mariner transposon-based signature-tagged mutagenesis system for the analysis of oral infection by Listeria monocytogenes. PLoS ONE 8:e75437. doi: 10.1371/journal.pone.0075437
Czuprynski, C. J., and Balish, E. (1981). Pathogenesis of Listeria monocytogenes for gnotobiotic rats. Infect. Immun. 32, 323–331.
Descy, J., de Mol, P., Hayette, M. P., Huynen, P., Meex, C., and Melin, P. (2012). Acute cholecystitis with Listeria monocytogenes. Acta Clin. Belg. 67, 295–297. doi: 10.1179/ACB.67.4.2062676
Dowd, G. C., Joyce, S. A., Hill, C., and Gahan, C. G. (2011). Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile. Infect. Immun. 79, 369–379. doi: 10.1128/IAI.00330-10
Dussurget, O., Cabanes, D., Dehoux, P., Lecuit, M., Buchrieser, C., Glaser, P., et al. (2002). Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45, 1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x
Ebersbach, T., Jorgensen, J. B., Heegaard, P. M., Lahtinen, S. J., Ouwehand, A. C., Poulsen, M., et al. (2010). Certain dietary carbohydrates promote Listeria infection in a guinea pig model, while others prevent it. Int. J. Food Microbiol. 140, 218–224. doi: 10.1016/j.ijfoodmicro.2010.03.030
Esteban, J. I., Oporto, B., Aduriz, G., Juste, R. A., and Hurtado, A. (2009). Faecal shedding and strain diversity of Listeria monocytogenes in healthy ruminants and swine in Northern Spain. BMC Vet. Res. 5:2. doi: 10.1186/1746-6148-5-2
Faith, N. G., Kim, J. W., Azizoglu, R., Kathariou, S., and Czuprynski, C. (2012). Purine biosynthesis mutants (purA and purB) of serotype 4b Listeria monocytogenes are severely attenuated for systemic infection in intragastrically inoculated A/J Mice. Foodborne Pathog. Dis. 9, 480–486. doi: 10.1089/fpd.2011.1013
Gahan, C. G., and Hill, C. (2005). Gastrointestinal phase of Listeria monocytogenes infection. J. Appl. Microbiol. 98, 1345–1353. doi: 10.1111/j.1365-2672.2005.02559.x
Garner, M. R., Njaa, B. L., Wiedmann, M., and Boor, K. J. (2006). Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74, 876–886. doi: 10.1128/IAI.74.2.876-886.2006
Gomes, B. C., Rodrigues, M. R., Winkelstroter, L. K., Nomizo, A., and de Martinis, E. C. (2012). In vitro evaluation of the probiotic potential of bacteriocin producer Lactobacillus sakei 1. J. Food Prot. 75, 1083–1089. doi: 10.4315/0362-028X.JFP-11-523
Gray, M. J., Freitag, N. E., and Boor, K. J. (2006). How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect. Immun. 74, 2505–2512. doi: 10.1128/IAI.74.5.2505-2512.2006
Grif, K., Patscheider, G., Dierich, M. P., and Allerberger, F. (2003). Incidence of fecal carriage of Listeria monocytogenes in three healthy volunteers: a one-year prospective stool survey. Eur. J. Clin. Microbiol. Infect. Dis. 22, 16–20. doi: 10.1007/s10096-002-0835-9
Hain, T., Hossain, H., Chatterjee, S. S., Machata, S., Volk, U., Wagner, S., et al. (2008). Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigmaB regulon. BMC Microbiol. 8:20. doi: 10.1186/1471-2180-8-20
Hardy, J., Francis, K. P., Deboer, M., Chu, P., Gibbs, K., and Contag, C. H. (2004). Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303, 851–853. doi: 10.1126/science.1092712
Hardy, J., Margolis, J. J., and Contag, C. H. (2006). Induced biliary excretion of Listeria monocytogenes. Infect. Immun. 74, 1819–1827. doi: 10.1128/IAI.74.3.1819-1827.2006
Huff, G. R., Huff, W. E., Beasley, J. N., Rath, N. C., Johnson, M. G., and Nannapaneni, R. (2005). Respiratory infection of turkeys with Listeria monocytogenes Scott A. Avian Dis. 49, 551–557. doi: 10.1637/7375-05040R.1
Jackson, K. A., Biggerstaff, M., Tobin-D'Angelo, M., Sweat, D., Klos, R., Nosari, J., et al. (2011). Multistate outbreak of Listeria monocytogenes associated with Mexican-style cheese made from pasteurized milk among pregnant, Hispanic women. J. Food Prot. 74, 949–953. doi: 10.4315/0362-028X.JFP-10-536
Jensen, A., Williams, D., Irvin, E. A., Gram, L., and Smith, M. A. (2008). A processing plant persistent strain of Listeria monocytogenes crosses the fetoplacental barrier in a pregnant guinea pig model. J. Food Prot. 71, 1028–1034.
Jin, B., Newton, S. M., Shao, Y., Jiang, X., Charbit, A., and Klebba, P. E. (2006). Iron acquisition systems for ferric hydroxamates, haemin and haemoglobin in Listeria monocytogenes. Mol. Microbiol. 59, 1185–1198. doi: 10.1111/j.1365-2958.2005.05015.x
Jones, B. V., Begley, M., Hill, C., Gahan, C. G., and Marchesi, J. R. (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 13580–13585. doi: 10.1073/pnas.0804437105
Karatzas, K. A., Brennan, O., Heavin, S., Morrissey, J., and O'Byrne, C. P. (2010). Intracellular accumulation of high levels of gamma-aminobutyrate by Listeria monocytogenes 10403S in response to low pH: uncoupling of gamma-aminobutyrate synthesis from efflux in a chemically defined medium. Appl. Environ. Microbiol. 76, 3529–3537. doi: 10.1128/AEM.03063-09
Karatzas, K. A., Suur, L., and O'Byrne, C. P. (2012). Characterization of the intracellular glutamate decarboxylase system: analysis of its function, transcription, and role in the acid resistance of various strains of Listeria monocytogenes. Appl. Environ. Microbiol. 78, 3571–3579. doi: 10.1128/AEM.00227-12
Mai, V., Ukhanova, M., Visone, L., Abuladze, T., and Sulakvelidze, A. (2010). Bacteriophage administration reduces the concentration of Listeria monocytogenes in the gastrointestinal tract and its translocation to spleen and liver in experimentally infected mice. Int. J. Microbiol. 2010, 624234. doi: 10.1155/2010/624234
Mccollum, J. T., Cronquist, A. B., Silk, B. J., Jackson, K. A., O'Connor, K. A., Cosgrove, S., et al. (2013). Multistate outbreak of listeriosis associated with cantaloupe. N.Engl. J. Med. 369, 944–953. doi: 10.1056/NEJMoa1215837
Mcgann, P., Raengpradub, S., Ivanek, R., Wiedmann, M., and Boor, K. J. (2008). Differential regulation of Listeria monocytogenes internalin and internalin-like genes by SigmaB and PrfA as revealed by subgenomic microarray analyses. Foodborne Pathog. Dis. 5, 417–435. doi: 10.1089/fpd.2008.0085
Melo, J., Schrama, D., Hussey, S., Andrew, P. W., and Faleiro, M. L. (2013). Listeria monocytogenes dairy isolates show a different proteome response to sequential exposure to gastric and intestinal fluids. Int. J. Food Microbiol. 163, 51–63. doi: 10.1016/j.ijfoodmicro.2013.03.001
Mujahid, S., Orsi, R. H., Vangay, P., Boor, K. J., and Wiedmann, M. (2013). Refinement of the Listeria monocytogenes SigmaB regulon through quantitative proteomic analysis. Microbiology 159, 1109–1119. doi: 10.1099/mic.0.066001-0
Nadon, C. A., Bowen, B. M., Wiedmann, M., and Boor, K. J. (2002). Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70, 3948–3952. doi: 10.1128/IAI.70.7.3948-3952.2002
Nakamura, S., Kuda, T., An, C., Kanno, T., Takahashi, H., and Kimura, B. (2012). Inhibitory effects of Leuconostoc mesenteroides 1RM3 isolated from narezushi, a fermented fish with rice, on Listeria monocytogenes infection to Caco-2 cells and A/J mice. Anaerobe 18, 19–24. doi: 10.1016/j.anaerobe.2011.11.006
Neuhaus, K., Satorhelyi, P., Schauer, K., Scherer, S., and Fuchs, T. M. (2013). Acid shock of Listeria monocytogenes at low environmental temperatures induces prfA, epithelial cell invasion, and lethality towards Caenorhabditis elegans. BMC Genomics 14:285. doi: 10.1186/1471-2164-14-285
Nichterlein, T., Kretschmar, M., and Hof, H. (1994). Fecal shedding of Listeria monocytogenes during murine listeriosis after intravenous infection. Zentralbl. Bakteriol. 281, 192–195. doi: 10.1016/S0934-8840(11)80569-4
Oliver, H. F., Orsi, R. H., Ponnala, L., Keich, U., Wang, W., Sun, Q., et al. (2009). Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and Sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. doi: 10.1186/1471-2164-10-641
Ondrusch, N., and Kreft, J. (2011). Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PLoS ONE 6:e16151. doi: 10.1371/journal.pone.0016151
Palmer, M. E., Chaturongakul, S., Wiedmann, M., and Boor, K. J. (2011). The Listeria monocytogenes SigmaB regulon and its virulence-associated functions are inhibited by a small molecule. MBio 2: pii: e00241-11. doi: 10.1128/mBio.00241-11
Payne, A., Schmidt, T. B., Nanduri, B., Pendarvis, K., Pittman, J. R., Thornton, J. A., et al. (2013). Proteomic analysis of the response of Listeria monocytogenes to bile salts under anaerobic conditions. J. Med. Microbiol. 62, 25–35. doi: 10.1099/jmm.0.049742-0
Phan-Thanh, L., and Mahouin, F. (1999). A proteomic approach to study the acid response in Listeria monocytogenes. Electrophoresis 20, 2214–2224. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2214::AID-ELPS2214>3.0.CO;2-G
Quillin, S. J., Schwartz, K. T., and Leber, J. H. (2011). The novel Listeria monocytogenes bile sensor BrtA controls expression of the cholic acid efflux pump MdrT. Mol. Microbiol. 81, 129–142. doi: 10.1111/j.1365-2958.2011.07683.x
Rea, M. C., Dobson, A., O'Sullivan, O., Crispie, F., Fouhy, F., Cotter, P. D., et al. (2011). Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4639–4644. doi: 10.1073/pnas.1001224107
Rea, M. C., Sit, C. S., Clayton, E., O'Connor, P. M., Whittal, R. M., Zheng, J., et al. (2010). Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U.S.A. 107, 9352–9357. doi: 10.1073/pnas.0913554107
Ryan, S., Begley, M., Gahan, C. G., and Hill, C. (2009). Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11, 432–445. doi: 10.1111/j.1462-2920.2008.01782.x
Ryan, S., Begley, M., Hill, C., and Gahan, C. G. (2010). A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 109, 984–995. doi: 10.1111/j.1365-2672.2010.04726.x
Ryan, S., Hill, C., and Gahan, C. G. (2008). Acid stress responses in Listeria monocytogenes. Adv. Appl. Microbiol. 65, 67–91. doi: 10.1016/S0065-2164(08)00603-5
Schwartz, K. T., Carleton, J. D., Quillin, S. J., Rollins, S. D., Portnoy, D. A., and Leber, J. H. (2012). Hyperinduction of host beta interferon by a Listeria monocytogenes strain naturally overexpressing the multidrug efflux pump MdrT. Infect. Immun. 80, 1537–1545. doi: 10.1128/IAI.06286-11
Sleator, R. D., Watson, D., Hill, C., and Gahan, C. G. (2009). The interaction between Listeria monocytogenes and the host gastrointestinal tract. Microbiology 155, 2463–2475. doi: 10.1099/mic.0.030205-0
Sleator, R. D., Wemekamp-Kamphuis, H. H., Gahan, C. G., Abee, T., and Hill, C. (2005). A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 55, 1183–1195. doi: 10.1111/j.1365-2958.2004.04454.x
Sleator, R. D., Wouters, J., Gahan, C. G., Abee, T., and Hill, C. (2001). Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl. Environ. Microbiol. 67, 2692–2698. doi: 10.1128/AEM.67.6.2692-2698.2001
Smith, J. L., Liu, Y., and Paoli, G. C. (2013). How does Listeria monocytogenes combat acid conditions? Can. J. Microbiol. 59, 141–152. doi: 10.1139/cjm-2012-0392
Sriramulu, D. D., Liang, M., Hernandez-Romero, D., Raux-Deery, E., Lunsdorf, H., Parsons, J. B., et al. (2008). Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J. Bacteriol. 190, 4559–4567. doi: 10.1128/JB.01535-07
Tiensuu, T., Andersson, C., Ryden, P., and Johansson, J. (2013). Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol. Microbiol. 87, 909–924. doi: 10.1111/mmi.12140
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
Wemekamp-Kamphuis, H. H., Wouters, J. A., Sleator, R. D., Gahan, C. G., Hill, C., and Abee, T. (2002). Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl. Environ. Microbiol. 68, 4710–4716. doi: 10.1128/AEM.68.10.4710-4716.2002
Woodward, J. J., Iavarone, A. T., and Portnoy, D. A. (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. doi: 10.1126/science.1189801
Xayarath, B., Marquis, H., Port, G. C., and Freitag, N. E. (2009). Listeria monocytogenes CtaP is a multifunctional cysteine transport-associated protein required for bacterial pathogenesis. Mol. Microbiol. 74, 956–973. doi: 10.1111/j.1365-2958.2009.06910.x
Keywords: Listeria, stress, acid, bile, gastrointestinal, virulence, pathogenesis, infection
Citation: Gahan CGM and Hill C (2014) Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front. Cell. Infect. Microbiol. 4:9. doi: 10.3389/fcimb.2014.00009
Received: 02 December 2013; Accepted: 18 January 2014;
Published online: 05 February 2014.
Edited by:
Stephanie M. Seveau, The Ohio State University, USAReviewed by:
Olivier Dussurget, Université Paris Diderot, FranceKathryn Boor, Cornell University, USA
Copyright © 2014 Gahan and Hill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cormac G. M. Gahan, Alimentary Pharmabiotic Centre, School of Microbiology and School of Pharmacy, University College Cork, Cork, Ireland e-mail: c.gahan@ucc.ie
 Cormac G. M. Gahan
Cormac G. M. Gahan Colin Hill
Colin Hill