Low occurrence of Pseudomonas aeruginosa in agricultural soils with and without organic amendment
- 1CNRS, Ecole Nationale Vétérinaire de Lyon, and Université Lyon 1, UMR 5557 Ecologie Microbienne, Université de Lyon, Villeurbanne, France
- 2UMR Ecosol, IRD-Université de Ouagadougou, UFR/SVT, Ouagadougou, Burkina Faso
- 3Plateforme GenoSol, INRA-Université Bourgogne, AgroSup, UMR1347 Agroécologie, Dijon, France
- 4INRA-Université Bourgogne, AgroSup, UMR1347 Agroécologie, Dijon, France
- 5INRA, US 1106 InfoSol, Orléans, France
- 6INRA, UMR 1091 Environnement et Grandes Cultures, Thiverval-Grignon, France
The occurrence of Pseudomonas aeruginosa was monitored at a broad spatial scale in French agricultural soils, from various soil types and under various land uses to evaluate the ability of soil to be a natural habitat for that species. To appreciate the impact of agricultural practices on the potential dispersion of P. aeruginosa, we further investigated the impact of organic amendment at experimental sites in France and Burkina Faso. A real-time quantitative PCR (qPCR) approach was used to analyze a set of 380 samples selected within the French RMQS (“Réseau de Mesures de la Qualité des Sols”) soil library. In parallel, a culture-dependent approach was tested on a subset of samples. The results showed that P. aeruginosa was very rarely detected suggesting a sporadic presence of this bacterium in soils from France and Burkina Faso, whatever the structural and physico-chemical characteristics or climate. When we analyzed the impact of organic amendment on the prevalence of P. aeruginosa, we found that even if it was detectable in various manures (at levels from 103 to 105 CFU or DNA targets (g drywt)−1 of sample), it was hardly ever detected in the corresponding soils, which raises questions about its survival. The only case reports were from a vineyard soil amended with a compost of mushroom manure in Burgundy, and a few samples from two fields amended with raw urban wastes in the sub-urban area of Ouagadougou, Burkina Faso. In these soils the levels of culturable cells were below 10 CFU (g drywt)−1.
Introduction
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacillus and an opportunistic pathogen considered as one of the major agents of nosocomial infections. It can cause infections among patients with weakened defense barriers, such as severely burned or artificially ventilated patients (Richard et al., 1994), and is the main pathogen associated with respiratory tract infection in cystic fibrosis patients (Saiman and Siegel, 2004). Infections in healthy individuals can occur as keratitis (Song et al., 2000), otitis (Heslop and Ovesen, 2006), and others. P. aeruginosa can also be detected in human and animal fecal samples (Mushin and Ziv, 1973; Lavenir et al., 2008). It has been identified as an animal pathogen responsible for various infections (Daly et al., 1999; Ledbetter et al., 2007). The global increase of the worldwide population, combined with the increasing numbers of patients at risk might then favor the incidence of infection by this opportunistic pathogen. Knowing more about its ecology and natural reservoirs is therefore important to avoid infections and outbreaks.
This species is intrinsically resistant to a wide range of antimicrobials, has wide metabolic versatility and can be found in wide variety of ecological environments. It is described as preferentially living in aquatic habitats and colonizing moist niches. Within hospital settings, it was isolated from sinks and tubs (Römling et al., 1994), tap water outlets (Reuter et al., 2002), or water pipes (Lavenir et al., 2008). In outdoor environments it was isolated from a wide range of water sources including rivers (Pirnay et al., 2005; Selezska et al., 2012), open ocean (Khan et al., 2007), and can be recovered in high numbers from recreative waters (Barben et al., 2005) or wastewater (Lavenir et al., 2007; Lee et al., 2008).
Considering terrestrial environments, its isolation from plants (Green et al., 1974; Cho et al., 1975) or vegetables (Wright et al., 1976) as well as its detection in agricultural soils (Green et al., 1974; Marques et al., 1979) have been reported. However, whether these environments act as a reservoir or a transient recipient of P. aeruginosa is still under debate and data on the factors driving its survival and dissemination are scarce. The oldest report on its occurrence in soils was from Ringen and Drake (1952). Later, Green et al. (1974) recovered it from Californian soils and concluded about agricultural soil as a natural habitat for the bacterium since the soils sampled had no known organic fertilizer or animal pasturing background, and irrigation water was free of the bacterium. Further studies showed that P. aeruginosa is often present within polluted soils from various geographical regions and participates to the degradation of the hydrocarbons (Garcia-Junco et al., 2001; Norman et al., 2002; Kaszab et al., 2010). To our knowledge, no recent studies have been performed to better appreciate its relative abundance within the global soil bacterial communities, and to assess the influence of soil characteristics and anthropogenic constraints on its distribution. For instance the presence of P. aeruginosa in various water sources and in fecal material raises questions about the potential dispersion in soil through common agricultural practices i.e., irrigation and organic amendment, and then on the role of human activity for such dispersion. The presence of P. aeruginosa has been previously reported in both farmyard manure (Lavenir et al., 2007; Colinon et al., 2013) and composted industrial wastes (Kaszab et al., 2011). Human activities could also act as indirect selective pressures through the addition of chemicals i.e., pesticides, antibiotic, hydrocarbons known to be metabolized by P. aeruginosa and that could favor and enrich indigenous P. aeruginosa populations. This study is then dedicated to filling this knowledge gap.
Here we present a study investigating the prevalence of P. aeruginosa in agricultural soils as influenced by soil properties and the potential impact of agricultural practices on this prevalence. Culture-dependent and culture-independent (i.e., real-time quantitative PCR, qPCR) approaches were performed to quantify the occurrence of P. aeruginosa. More precisely we combined a previously developed SYBR Green qPCR assay targeting the ecfX gene (Colinon et al., 2013) and culturing of bacteria on a semi-selective media (nalidixic-supplemented cetrimide agar base) coupled to ecfX gene amplification to confirm isolate identity. ecfX encodes an ECF (extracytoplasmic function) sigma factor which is restricted to P. aeruginosa and was previously validated to improve both sensitivity and specificity of P. aeruginosa detection in environmental samples (Lavenir et al., 2007). Presented data are from a study conducted on a broad spatial scale using various soil types, under various land uses, and from various geographical origins, partly using the collection belonging to the French national soil survey (Arrouays et al., 2002). In addition we monitored the impact of various organic amendments, including cow and horse manure, sewage sludge, and composted or raw urban wastes, spread onto experimental fields in France and in Burkina Faso.
Materials and Methods
Sources of French Soil Samples
A set of 380 soil samples was selected within the French RMQS (“Réseau de Mesures de la Qualité des Sols” = French soil quality monitoring network) for the qPCR detection of P. aeruginosa (Figure 1A). The RMQS is a soil library which contains 2200 soils sampled with a 16 km × 16 km systematic grid over the whole French territory, and representative of the different soil types, land cover, land management type and climatic conditions occurring in France [for more details, see (Arrouays et al., 2002)]. The 380 RMQS samples were chosen from 5 different geographical regions (Brittany, South-East, North, Center, Bassin Parisien). These regions were chosen in order to get samples with various geographic, pedo-climatic, and land use characteristics, as well as exposure to various organic amendments. Sixty-three out of the 380 soils received repeated organic amendment over several years from various sources before sampling: bovine manure (n = 49), liquid swine manure (n = 10), poultry dropping (n = 1), or sewage sludge (n = 3) (Figure 1B). Ninety-seven out of 380 soils had no known history of amendment. This information was not available for the remaining soils (220 out of 380). Twenty-six locations from Burgundy within the Center region were selected among the 380 samples to search for the presence of P. aeruginosa using the culture-dependent approach. Additional soil samples were collected from various agricultural areas in France including Limousin, Burgundy, Rhône-Alpes, North, and Bassin Parisien regions (Figure 1A) and from 2 experimental sites from the INRA research center in Chinon (Center region) and Mâcon (Burgundy region). These agricultural soils were planted with maize, wheat or grapevine. During the year of collection, some soils received organic amendments mainly farmyard manure, and/or were exposed to animal grazing, others did not (Table 1). The samples, i.e., ten samplings per field composing one sample, were collected from the upper layer (0–5, 0–10 or 0–20 cm), sifted through 2 mm-mesh sieves and stored at room temperature for no longer than 1 week. They were collected during various campaigns between 2006 and 2011. Soil physico-chemical characteristics for some sites tested with qPCR- and culture-based approaches are presented in Table S1 to show the extent diversity of the soils studied.
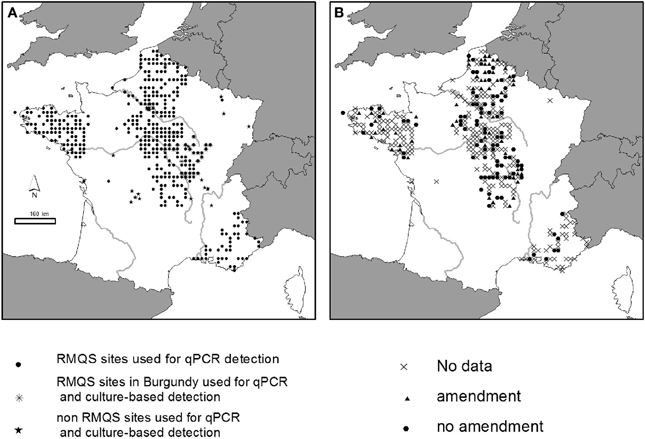
Figure 1. Sites studied in France. (A) Soils used for qPCR and culture-based detection of P. aeruginosa. (B) Soils that received or not organic amendment.
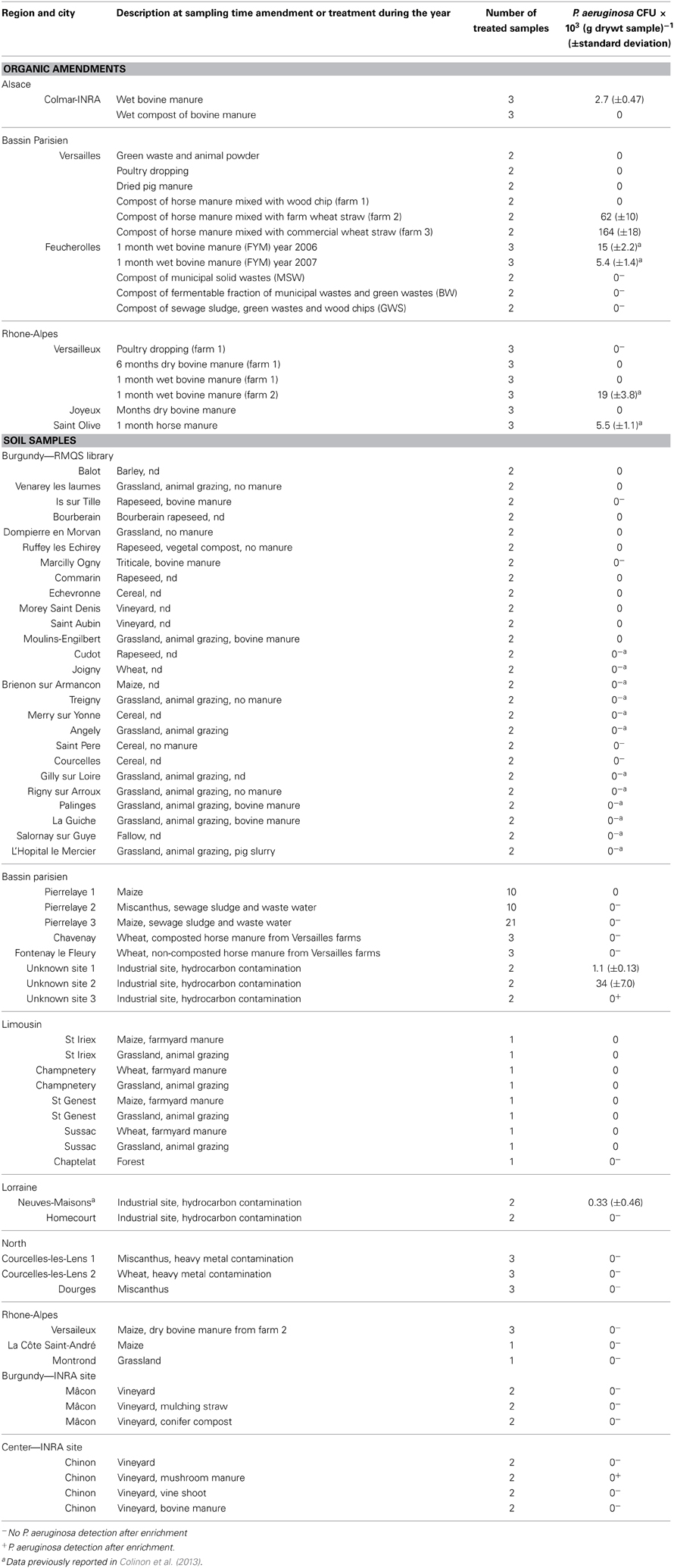
Table 1. Number of CFU (colony forming units) of P. aeruginosa in organic amendments and agricultural soils amended or not with organic wastes.
We also included soils from industrial sites contaminated with hydrocarbons in Bassin Parisien (soils kindly provided by Biogénie Europe SAS, Echarcon, France) and in the Lorraine region (soils kindly provided by Dr. C. Leyval, UMR CNRS 7137, Nancy, France).
Sources of Organic Amendments
We included poultry dropping, 1 month or 6 months-old bovine and horse manures obtained from 5 farms in the Dombes area (Rhône-Alpes), 1 month-old bovine manure from Colmar (Alsace) as well as various organic amendments i.e., bovine manure, compost of horse manure, poultry dropping, dehydrated pig manure and various municipal composted wastes, that are used on the Feucherolles experimental site (Bassin Parisien; see below) (Table 2), or on various fields around Versailles (Bassin Parisien). Part of these amendments was provided by INRA of Grignon.
Experimental Sites with Organic Amendments in France and Burkina Faso
The site of Feucherolles was used for 8 years to assess the effects of urban composts on soil fertility and their environmental impact (Houot et al., 2002). The field experiment was cropped with a wheat-maize succession for the time the experiment lasted. Organic amendments included (1) a municipal solid waste compost (MSW) made from residual municipal wastes after selective collection of clean, dry packaging, (2) a biowaste compost (BW) made from selectively collected fermentable fractions of municipal wastes co-composted with green wastes, (3) a compost produced from the co-composting of sewage sludge, green wastes and wood chips (GWS), and (4) a farmyard manure (FYM) issued from cow breeding. These four organic treatments were compared to an organic input-free control. The experiment was a randomized complete-block designed with four replicates. Samples were collected from the upper layer (0–10 cm) in September (before amendments were added), October (1 month after amendments were added) and December (3 months after amendments were added) for two successive years (2006 and 2007). Each treatment was performed on four plot replicates and samples were made of a composite sampling (10 samplings per plot replicate). The culture approach was tested on all replicates, i.e., a total of 20 soil samples (4 replicates × 5 treatments) per sampling date.
Four sahelian sites (Tabtenga, Zagtouli, Yagma and Toubwéogo) located in the sub-urban area of Ouagadougou, Burkina Faso, were added to our study in order to include soil types from regions of different climate conditions from those observed in France. This also enabled us to assess the impact of the local farmers' practices, which differed from the French farmers' and involved the use of untreated domestic and urban wastes on agricultural fields. The sites in Burkina Faso were planted with sorghum and impacted by untreated wastes (UW) due to long-term practices (between 6 and 18 years) by local farmers. The amendments induced an increase in organic carbon content, and in pH values (Table S1). The samplings were performed during three campaigns conducted in March 2007, June 2008 and February 2011. At each site sampled in 2007 (Tabtenga, Toubwéogo, and Yagma) two transects per field were analyzed. In 2008, three transects per field were made at Tabtenga, Toubwéogo, and Zagtouli. At Tabtenga in 2011 one transect was made in the control plot and 6 samples were analyzed, while 3 transects were made in the amended field and 4 samples per transect were taken. At each site in 2007, 2008, and 2011 soils were sampled from the upper layer (0–5 cm) and the samples were made of a composite sampling (10 samplings over a 20 m transect). During the 2008 campaign additional soil samples from a bellowed horizon (5–20 cm) were also collected.
Enumeration and Identification of Culturable Pseudomonas aeruginosa
P. aeruginosa enumeration was performed using Cetrimide Agar Base (CAB) medium (Oxoïd, Cambridge, UK) supplemented with nalidixic acid (15 mg l−1) and cycloheximide (200 mg l−1). Soil bacterial cells were extracted by blending 5 g of soil with 50 ml of a saline solution (NaCl 0.8%) for 90 s in a Warring Blender (Eberbach Corporation, Ann Arbor, MI, USA). The homogenized soil suspensions were serially diluted in sterile saline solution, and aliquots of the appropriate dilutions were spread onto agar plates. In all cases, three plates were inoculated per dilution and incubated at 28°C for up to 3 days. To ensure that 28°C would be the appropriate temperature to recover P. aeruginosa from environmental samples we performed preliminary tests and compared plating at 28 and 37°C for all soil samples from Burkina Faso and for some French soils including the industrial ones and some manure amendments. In parallel we also checked on more than 50 clinical and environmental strains from our own collection and from international collections whether strains grow differently at 28 and 37°C. For both tests results were similar whatever the temperature of incubation. We then decided to do further screening at 28°C. One hundred μl were usually spread per 90-mm diameter plate. To improve P. aeruginosa detection sensitivity, 1 ml of soil suspension were used for the soil samples from Burkina Faso as these soils contained high quantities of sand, low quantities of organic matter, and therefore, yielded clear soil suspensions. In addition, enrichment assays were performed by transferring 2 g of soil into 20 ml of a salt solution supplemented with acetamide as described previously (Green et al., 1974). Inoculated enrichment broths were incubated for 3 days at 28°C with shaking at 180 rpm. Serial dilutions were performed and plated on CAB agar medium.
All the greenish and clearly yellowish colonies were collected from CAB plates and confirmed as P. aeruginosa by PCR screening with the ecfX gene encoding for an ECF (extracytoplasmic function) sigma factor, as previously described (Colinon et al., 2013) and by oxidase assay. All ecfX- and oxidase-positive isolates were submitted to partial 16S rDNA sequencing to confirm their identification. The detection limit using cultivation-based measurements varied from 10 to 100 CFU (g drywt soil)−1 (Colinon et al., 2013).
As our study evaluated the impact of amendment but that samplings were mostly performed long after amendment i.e., at least a month, we set up a laboratory experiment to evaluate how long cells could survive and be detected in soils using the culture approach. An agricultural soil from a field planted with maize (La Côte Saint André, Rhône-Alpes region; Table 1) was chosen to conduct that experiment (Table 1). A clinical strain PAO1 and an environmental strain ATCC 31479 were added at a concentration of 1.2 × 106 and 1.8 × 106 CFU (g drywt)−1 of soil, respectively. Microcosms of 10 g of sieved (2 mm) soil were inoculated with cell-water suspensions. Microcosms were incubated at 20°C for 38 days. They were set up in triplicates and were sacrified at each time point (day 3, 6, 16, 24, 38). The same experiment was conducted on sterilized soil from La Côte Saint André (gamma irradiation sterilization; Ionisos, Dagneux, France). Microcosms were inoculated with PAO1 and ATCC 31479 at levels of 1.09 x 108 and 1.42 x 108 CFU (g drywt)−1 of soil, respectively. Microcosms were set up in triplicates and were analyzed at each time point (2, 6, 17, 44, and 58).
DNA Extraction and Purification
The isolated strain genomic DNA was extracted by gentle alkaline lysis (using sarkosyl and NaCl) and purified according to Johnson (1994), and DNA concentration was estimated using a Nanodrop® ND-1000 spectrophotometer (Labtech International, Paris, France) at a 260 nm wavelength. DNA from soils of the RMQS grid were extracted by the GenoSol platform at INRA Dijon according to a single procedure optimized by Ranjard et al. (2003). DNA from soils sampled in Pierrelaye, Feucherolles and Burkina Faso and amendment samples (0.5 g) were extracted in triplicates with the FastDNA® SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA) and purified on S-400-HR mini-columns (Pharmacia, St Quentin Yvelines, France) following the manufacturer's instructions. The DNA extracts were resolved by electrophoresis in 0.8% agarose gels, stained with ethidium bromide and photographed using a Gel Doc 1000 camera (Bio-Rad, Ivry sur Seine, France). DNA concentrations in the crude and purified soil extracts were determined by electrophoretic comparison with a standard curve as previously described (Ranjard et al., 2003).
Pseudomonas aeruginosa-Specific qPCR for Soil Samples
Pseudomonas aeruginosa-specific qPCR was set up using the ECF5 (5′-AAGCGTTCGTCCTGCACAA-3′) and ECF2 (5′-TCATCCTTCGCCTCCCTG-3′) primers. These primers amplify a 146-bp long fragment of the ecfX gene (Colinon et al., 2013). The Eva Green SMX 1000R (Bio-Rad) was used, according to the manufacturer's instructions. The reactions were carried out in a 20-μl reaction mixture containing 500 nmol l−1 of each primer. Non-template controls, including the reaction mixture with sterile water instead of DNA template, were added to each run. Quantitative PCR was performed in a LightCycler 480 system (Roche Diagnostics, Meylan, France). The following PCR protocol was applied: initial denaturation at 95°C for 5 min, followed by 45 cycles with denaturation at 98°C for 10 s, annealing and elongation at 63°C for 20 s. Subsequently a melting curve was recorded by increasing the temperature from 65 to 98°C (+1°C every 10 s). For reproducibility, all qPCRs were triplicated on separate plates. P. aeruginosa strain UCBPP-PA14 genomic DNA was used to generate a standard curve. Quantification was performed by comparison with a 2- to 5-fold diluted standard (1 fg to 100 pg, 5 μl per reaction). Data analysis was performed using the LightCycler® 480 software (Roche Diagnostics). PCR product specificity was checked by melting curve analysis and agarose gel electrophoresis. Based on the results of previous optimization tests (Colinon et al., 2013), 5 ng of soil DNA extracts (5 μl each per reaction) were screened in triplicates using 25 ng μl−1 of T4 Gene 32 protein. qPCR results were interpreted as undoubtedly positive only when the fluorescence signal obtained corresponded to at least 50 fg of P. aeruginosa cells (7 genome equivalent) at the appropriate corresponding Tm.
Sensitivity of the qPCR approach was determined to be around 5 × 104 cells (g drywt soil)−1 (Colinon et al., 2013).
Control Tests for DNA Quality and P. aeruginosa DNA Persistance
The quality of genomic DNA is critical for success of the qPCR. Before being screened for the presence of P. aeruginosa we then checked for the presence of inhibitors in DNA extracts and their impact on our ability to amplify added DNA target or intrinsic DNA target. Two tests were then performed. In the first one we added 106 copies of the circularized pGEM-T Easy plasmid DNA (2.5 μl per 20 μl of PCR reaction) to 5 ng of each DNA. The plasmid DNA was quantified afterwards by comparison with a 10-fold diluted standard of pGEM-T Easy plasmid DNA (101 to 107 copies, 2.5 μl each per reaction). qPCR reactions and cycling conditions were as stated above, except for the primers we used which were SP6 and T7 universal primers, the annealing temperature which was set at 55°C, and the PCR kit which was the LightCycler§ 480 SYBR Green I Master kit (Roche Diagnostics). A 10-fold reduction of the copy number amplified by qPCR with respect to the copy number introduced in the reaction mixture was considered as 10% amplification efficiency, or, conversely, as 90% inhibition.
In the second test we performed a 16S rRNA primer-based assay as described in previous studies (Lopez-Gutierrez et al., 2004). Samples were screened once using 2 ng of template DNA. The assay was only conducted on RMQS DNA extracts.
Results
Detection of P. aeruginosa in Organic Amendments
The culture dependent approach was performed on various organic materials including poultry dropping, animal manures of various ages and humidity content, composted or not, and various composts of municipal wastes (Table 1). No P. aeruginosa were isolated from poultry dropping, dehydrated pig manure and the mix of green waste and animal powder samples. Analysis of manures collected from farms in Bassin Parisien, Rhône-Alpes and Alsace showed the presence of P. aeruginosa at levels of 5.5 (±1.1) × 103 to 164 (±18) × 103 CFU (g drywt)−1 for horse manure and at level of 2.7 (±0.47) × 103 to 19 (±3.8) × 103 CFU (g drywt)−1 for bovine manure. It has to be noted that P. aeruginosa was not isolated from 6 months-old bovine manure and was still detected after a compost treatment of horse manure. qPCR enabled quantification of P. aeruginosa at level of 23 (±3.5) × 103 and 76 (±15) × 103 cells (g drywt)−1 in the manure samples used at the Feucherolles site in 2006 and 2007, respectively, as reported in our previous study (Colinon et al., 2013). Here we performed qPCR on DNA extracts from manure samples collected in Rhône-Alpes and Alsace. In these samples P. aeruginosa was detected at a level of 28 (±7.5) × 103 and 43 (±9.7) × 103 cells (g drywt)−1 in the horse manure samples from St-Olive and the bovine manure from Versailleux, respectively. Positive results were confirmed by sequencing amplified products (data not shown). qPCR did not allow detection of P. aeruginosa in the other samples.
Analysis of the 3 composts derived from municipal wastes or sewage sludge used on the plots of the experimental site of Feucherolles did not indicate the presence of P. aeruginosa. Neither the culture approach nor the qPCR enabled detection of P. aeruginosa in these samples.
Screening of Soil DNA for the Presence of P. aeruginosa Using qPCR
Five percent of the 380 RMQS soil DNAs showed inhibition on qPCR above 10%. None of the DNA samples from Feucherolles, Pierrelaye and Burkina Faso soil samples showed significant inhibition (<1% inhibition). The ability to amplify indigenous DNA target was also tested using universal primers to detect 16rDNA sequences among the RMQS DNA extracts. The 16S rRNA gene copy numbers of bacteria ranged from 6.31 × 105 to 1.02 × 109 copies (g drywt)−1 in 33 out of 380 DNA samples and from 1.02 × 109 to 1.06 × 1011 copies (g drywt)−1 of soil in the remaining DNAs.
All DNA samples were analyzed whether or not inhibition and differences in 16S rDNA copies were observed.
The survey of P. aeruginosa distribution based on the qPCR detection showed that none of the 380 DNA samples of the RMQS soil set emitted a noteworthy positive fluorescence signal. Similarly none of the samples from the Feucherolles, the Pierrelaye and the Burkina Faso sites gave a positive signal. We previously estimated the detection limit to be 5 × 104 cells (g drywt)−1 of soil (Colinon et al., 2013). Then the absence of amplification from soil DNA extracts indicated that either P. aeruginosa is not present within these indigenous soil bacterial communities or is present at a low level, below the detection threshold of this assay.
Detection of P. aeruginosa in Soil based on Culture Dependent Approach
To confirm or not the absence of P. aeruginosa in the RMQS soil samples we screened a subset of the RMQS library including 26 soils from the Burgundy region, using the culture-dependent approach (Table 1). To avoid changes in bacterial community composition or in physiological cell properties, all these samples were processed directly after being sampled and had not been dried and stored as were the samples from the RMQS library used for DNA extraction. The results showed an absence of P. aeruginosa even after an enrichment step. Similarly, detection with the culture dependent method was mostly unsuccessful with the agricultural soils sampled in other French regions including Bassin Parisien, Limousin, Lorraine, North, and Rhône-Alpes (Table 1). As observed in the control soil, direct isolation of P. aeruginosa from the organic-amended soils of the 2 INRA sites at Mâcon and Chinon was unsuccessful whatever the amendments. However the enrichment procedure enabled us to isolate P. aeruginosa from the vineyard soil amended with mushroom manure in Chinon (Table 1).
Despite the presence of P. aeruginosa in the studied horse manure-containing amendments (composts of horse manure from farms in Versailles) no P. aeruginosa was recovered from the soils that had previously received amendments originating from these farms [i.e., soils from Fontenay le Fleury and Chavenay (Bassin Parisien)] (Table 1). Detection was also unsuccessful with samples from soils that received either bovine manure or pig slurry, or that were exposed to animal grazing in Limousin and Burgundy.
It has to be noticed that negative results were also obtained with soil samples from agricultural fields either planted with maize and miscanthus which received sewage sludge and were irrigated for more than 100 years with untreated and further treated urban wastewater (soils from Pierrelaye, Bassin Parisien). We expected these soils to contain P. aeruginosa as that species is frequently detected in wastewaters (Lavenir et al., 2007; Lee et al., 2008). As a comparison and because of the literature reports about the presence of P. aeruginosa in environments contaminated with hydrocarbons, we included some soil samples collected from hydrocarbon-polluted industrial sites. Two out of the 3 soils from Bassin Parisien and one located in Lorraine (soil from Neuves-Maisons) led to the direct isolation of P. aeruginosa on selective medium. Statistically different counts were observed as the numbers of CFU were 1.1 (±0.13) × 103 (p value = 0.0013) 34 (±7.0) × 103 (p value = 0.00022), and 0.33 (±0.46) × 103 (p value = 0.0015) (g drywt)−1 in the three soils, respectively. The enrichment step with acetamide enabled us to isolate P. aeruginosa in the third sample from Bassin Parisien but not in the sample from the Homécourt soil in Lorraine.
Temporal Impact of Organic Amendments on P. aeruginosa Dissemination at Experimental Sites
The experiment conducted to evaluate whether P. aeruginosa can still be detected by plate-counting after addition in soil showed that both strains were still detectable up to day 58 (Table 2) in sterilized microcosms. Their level was still high reaching 2.9 (±2.3) × 107 and 1.3 (±0.67) × 107 for PA01 and ATCC 31479, respectively. On the opposite the level of both strains declined in the non-sterile microcosm. PAO1 and ATCC 31479 were not detectable after 24 and 38 days, respectively (supplementary data; Table S2).
At the experimental site of Feucherolles, samples collected before amendment, 1 month and 3 months after amendment, did not led to the detection of P. aeruginosa whatever the amendment used and despite the presence of P. aeruginosa in the farmyard manure (Table 2) during year 2006. Detection was unsuccessful with or without adding the enrichment step in the isolation procedure. As the experiment was conducted again in 2007, soils were analyzed following the same sampling time scale. Similarly no P. aeruginosa was isolated.
The presence of P. aeruginosa was also investigated in soils amended with untreated urban wastes from 4 agricultural sites in the vicinity of Ouagadougou, Burkina Faso (Table 2). No P. aeruginosa was detected in the unamended soils whatever the site and the sampling year. None of the samples from the upper soil layer, except 1 from the site of Tabtenga from the 2011 sampling campaign, led to a direct isolation of P. aeruginosa. The positive sample enabled us to obtain only 1 colony. This sample was analyzed 3 times and either no or 1 colony was obtained. Then the estimation of P. aeruginosa abundance was below 10 CFU g−1 dry soil. Our enrichment assays led to the detection of P. aeruginosa in 1 soil sample from the Yagma site, and in 4 out of 12 samples collected in the Tabtenga amended field during the 2011 campaign. None of the samples from the 5–20 cm layer led to the detection of P. aeruginosa.
A genetic typing of some isolates obtained from the direct isolation and the enrichment procedure was performed using Multiple Locus Variable Number of Tandem Repeat (VNTR) Analysis (MLVA). Data showed that those isolates had different MLVA types and differed from other environmental and clinical isolates (Youenou et al., 2014) suggesting that the detection of P. aeruginosa in the soil samples from Burkina Faso was not due to a laboratory contamination.
Discussion
In this study we conducted a survey of the prevalence of P. aeruginosa in French agricultural soils using both a culture-based approach and a culture-independent one to evaluate the capacity of soil environment to constitute a reservoir or a transient recipient of P. aeruginosa. This study intended to gain new insights on the distribution of P. aeruginosa in non-aquatic environments. Ringen and Drake (1952) addressed this point and observed that P. aeruginosa occurred in only 3 out of 100 soil samples. Recently through a synthesis of available literature Selezska et al. (2012) concluded that “P. aeruginosa is obviously an aquatic rather than soil organism” and as stated by Ringen and Drake (1952) the “isolations of P. aeruginosa from […] soil seem to be one of chance.” Despite of their different abiotic and biotic properties, land use and geographical origin, none among the wide range of the agricultural soils we selected in the RMQS soil library enabled the detection of P. aeruginosa based on the qPCR approach suggesting that none of these environmental conditions sustain this species growth. However P. aeruginosa could be present at a low abundance below the detection limit of our qPCR approach. Based on the culture dependent approach none of the RMQS sites and other French agricultural fields led to the isolation of P. aeruginosa with or without a previous enrichment procedure. These observations are consistent with the conclusions of Selezska et al. (2012), but contrast with previous reports from Green et al. (1974) who recovered it from 24% of the Californian soils they tested. However it has to be noted that they used an enrichment step with acetamide and could not isolate P. aeruginosa without it. A recent study on Hungarian compost-amended soils also mentioned the presence of P. aeruginosa but at a low level with counts between 100 and 102 MPN (Most Probable Number) per g of soil (Kaszab et al., 2011). In our work, we failed to recover isolates from French agricultural samples, suggesting that if soil is a natural habitat for P. aeruginosa then it is a minor population or that it colonizes specific soil niches (rhizosphere, macrofauna) not investigated in the present study.
The widespread presence of P. aeruginosa in various water sources (Khan et al., 2007; Lavenir et al., 2007; Lee et al., 2008), its carriage by human and animals (Lavenir et al., 2008; Szmolka et al., 2012) and its presence in compost manure (Edrington et al., 2009) raised the question of the role of anthropogenic activities on P. aeruginosa dissemination. We looked for the presence of P. aeruginosa in various organic amendments derived from animal farms as spreading bovine manure, pig slurry or poultry dropping are common French farmer practices to enrich soil in organic matter. We could not isolate P. aeruginosa directly from fresh horse and bovine feces on 6 tested samples from Versailleux and St Olive (data not shown) but confirmed its presence in horse and bovine manures collected from geographically distant farms in France (Bassin Parisien and Rhône-Alpes). Similarly Kaszab et al. (2011), previously reported the lack of detection of P. aeruginosa in various raw wastes except wheat straw whereas they were able to detect it after composting treatments. Interestingly we could not isolate this species from 6 months-old manure. As these samples were relatively dry (5% water content) compared to the 1 month-old manure (at least 50% water content) this observation suggest that dehydration could eliminate or lower P. aeruginosa abundance. This observation is in agreement with the hypothesis that P. aeruginosa preferentially inhabit wet environments. Our data also showed that compost of horse manure still contained P. aeruginosa cells whereas composting is a treatment used to lower the amount of pathogens in organic wastes. This observation suggests that the heat treatment used in our composting process (50°C for 4 weeks) does not eliminate P. aeruginosa and that P. aeruginosa tolerates changes in temperature. Increase in temperature might enable it to replicate during composting and might not affect its culturability, as previously reported (Kaszab et al., 2011).
Based on the observed abundance in the various manure samples tested in this study (i.e., about 104 CFU g−1 dry soil) and on the mean amount of added manure to soil about 1 to 20 ton (ha year)−1 as indicated by farmers in the RMQS inquiry) we estimated the amount of P. aeruginosa introduced into soil to be about 1 × 1010 to 20 × 1010 cells (ha year)−1. These amounts are added in the first 20–30 cm once a year or every 2 years as usually done by French farmers. The RMQS library gave us the opportunity to screen 63 soils that were known to receive various amount and sources of organic amendments. None of the 49 and 10 soils that received bovine manure or pig slurry, respectively, enabled detection of P. aeruginosa. Similarly, most of the non-RMQS soil samples that received manure or that were used for animal grazing led to the detection of P. aeruginosa. The only samples in which P. aeruginosa was detected were collected from a mushroom manure-amended vineyard site (Chinon) in France and from urban waste-amended fields (Yagma and Tabtenga sites) in Burkina Faso. However the abundance of P. aeruginosa was very low: few isolates were successfully obtained from the Chinon and the Burkina Faso sites. Furthermore these isolates were mainly obtained after an enrichment procedure. The soil in Chinon presented the distinctive feature of being amended with mushroom manure. Unfortunately, at the time of sampling we could not obtain mushroom manure samples to check whether manure was the source of P. aeruginosa in the soil or whether P. aeruginosa was an autochtonous member of the soil microflora. P. aeruginosa was not isolated from the control soil. We then hypothesized that P. aeruginosa was present in the amendment because mushroom manure contains horse manure and P. aeruginosa was shown from this study and from a previous report (Lavenir et al., 2007) to be present in horse manure. The capacity of P. aeruginosa to persist in manure and then in amended soils has not been reported yet, but such persistence had already been demonstrated for other pathogenic bacteria (Pell, 1997; Gerba and Smith, 2005). In our study, the experimental site of Feucherolles gave us the opportunity to investigate the survival of P. aeruginosa in soils after various organic amendments were applied, including farmyard manure and compost of municipal wastes. Despite the presence of P. aeruginosa in the manure added in 2006 and 2007, we failed to recover it in the soil samples from the manure-amended plots 1 and 3 months after amendments were spread. These results were obtained for 2 years, consecutively. Our observations then suggested that P. aeruginosa was not indigenous to the bacterial community of that soil, (no P. aeruginosa was detected in the control soil), and that it may not survive easily in soils [no P. aeruginosa was detected in the manure-amended field, probably due to antagonistic interactions with the indigenous microflora (predation, substrate competition) or unfavorable physico-chemical soil properties]. Our very preliminary data on P. aeruginosa survival in soil suggested that exogenous cells added to soil can adapt to soil abiotic conditions but its persistence is affected by indigenous microflora. Identification of the soil parameters influencing P. aeruginosa survival should be investigated further also taking into account strain origin (human or animal) and intrinsic characteristics (i.e., antibiotic resistance properties) as well as the nature of P. aeruginosa sources (waste water, organic amendment. However we cannot rule out that P. aeruginosa survived whatever the conditions but became non-culturable, as in aquatic environments (Khan et al., 2010).
The presence of P. aeruginosa was also investigated in soils amended with untreated urban wastes in Burkina Faso. Despite long-term amendment practices (from 6 to 20 years), P. aeruginosa was only found in 2007 in an amended field in Yagma and in 2011 in an amended field in Tabtenga. It should be noted that few samples were positive per site and the level of P. aeruginosa was found lower than 10 cells per g of soil. We expected to detect it given that untreated wastes can contain potential pathogenic bacteria such as Salmonella or E. coli in a higher density than treated amendments, and fecal coliforms and streptococci, which indicate fecal contamination, frequently reach a density of 107–108 CFU g−1 dry soil (around 103 in treated composts) (Deportes et al., 1998; Hassen et al., 2001). Moreover, P. aeruginosa has already been found in a Nigerian soil amended with urban waste (Achudume and Olawale, 2009). The fact that P. aeruginosa was not detected in the unamended fields whatever the site suggests that the amendment was at the origin of the presence of P. aeruginosa. Amendments can be a direct source of P. aeruginosa when wastes contain pathogens and do not undergo any treatment to eliminate them. They can also act indirectly: we observed that the use of organic amendments in fields increased total biomass, as well as the total number of heterotrophic bacteria (data not shown). Tracking P. aeruginosa into these soils is made difficult due to the complex quality and the combined origins of these untreated wastes that are made of domestic, animal and hospital wastes. However, due to the potential pathogenicity of P. aeruginosa and its innate resistance to antimicrobial agents, these wastes would need to be treated before being applied to soils in order to avoid human infection. The treatment has to be suitable for this pathogen as our study and previous reports (Kaszab et al., 2011) showed that P. aeruginosa is not eliminated and could be enriched during composting.
An intriguing observation is the frequent isolation of P. aeruginosa from hydrocarbon contaminated sites both aquatic (Bartha, 1977; Bhattacharya et al., 2000) and terrestrial (Norman et al., 2002; Kaszab et al., 2010) hydrocarbon-impacted environments. Despite the low number of samples tested our detection was very successful since P. aeruginosa was detected in 4 out of the 5 samples. This observation raises the question of its origin in these soils: indigenous soil population or exogenous population added with the hydrocarbons. Similarly we can wonder the role of P. aeruginosa intrinsic properties in its ability to opportunistically take advantages of contamination. In such environments P. aeruginosa can degrade hydrocarbons and/or act as a helper population that facilitates accessibility to hydrocarbons due to the synthesis of biosurfactants (i.e., rhamnolipids) (Garcia-Junco et al., 2001). Its ability to synthesize pyocyanin an inhibitor of other bacterial species, may also increase its competitiveness and survival in oil-contaminated sites (Norman et al., 2002).
We evidenced that P. aeruginosa is very rarely present in soils from agricultural lands and that recovery is more likely from environments exposed to intense anthropogenic activities, i.e., soils amended with untreated wastes in the tropical context or exposed to hydrocarbon contamination. We then concluded that soils are not reservoir for this species and that the encountered biotic and abiotic conditions may not favor its growth and spread. In some circumstances, i.e., the use of organic amendments, it was isolated from a few soil samples but always at a very low level suggesting that the risk for community-acquired infections among humans might be low. However the duration of its persistence and the environmental conditions and/or the alternative soil niches (plant roots, soil macrofauna) that could favor its persistence need further investigations.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
RMQS soil sampling and physico-chemical analyses were supported by a French Scientific Group of Interest on soils: the “GIS Sol,” which involves the French Ministries in charge of Environment and Agriculture, the French Agency for Energy and Environment (ADEME), the National Institute for Agronomic Research (INRA), the French Institute for Research and Development (IRD) and the National Institute of the Geographic and Forest Information (IGN). We thank all the soil surveyors and technical assistants involved in sampling the sites. We thank Mélanie Lelièvre (Plateforme Genosol, Dijon, France), and the technical staff of the French soil library (Unité INFOSOL, INRA, Orléans, France) and of the platform DTAMB (IFR 41—Université de Lyon, France) for their dedicated technical assistance in DNA extraction, qPCR optimization, and soil preparation. We thank the technical assistant and the students of professor Edmond Hien (University of Ouagadougou) for their help in sampling the sites in Burkina Faso. We thank Isabelle Lamy and Christian Dron for sampling in Pierrelaye and Courcelles. This work was funded by the “Agence Nationale de la Recherche” (ANR) (programs 07 SEST project 018-01, 05 SEST project 009-01, 08 CES project 012-05). Céline Colinon was funded with a postdoctoral grant from the ANR 07 SEST project 018-01 and Amélie Deredjian with a PhD grant from the National Center for Scientific Research (CNRS). We thank the CNRS and the Ministry of Foreign Affairs (CORUS program) for supporting part of this work financially.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fcimb.2014.00053/abstract
References
Achudume, A. C., and Olawale, J. T. (2009). Occurrence of antibiotic resistant bacteria in waste site of Ede south west Nigeria. J. Environ. Biol. 30, 187–189.
Arrouays, D., Jolivet, C., Boulonne, L., Bodineau, G., Saby, N., and Grolleau, E. (2002). A new initiative in France: a multi-institutional soil quality monitoring network. C. R. Acad. Agric. Fr. 88, 93–105.
Barben, J., Hafen, G., and Schmid, J. (2005). Pseudomonas aeruginosa in public swimming pools and bathroom water of patients with cystic fibrosis. J. Cyst. Fibros. 4, 227–231. doi: 10.1016/j.jcf.2005.06.003
Bartha, R. (1977). The microbiology of aquatic oil spills. Adv. Appl. Microbiol. 22, 225–266. doi: 10.1016/S0065-2164(08)70164-3
Bhattacharya, M., Roy, S. S., Biswas, D., and Kumar, R. (2000). Effect of Mg(2+) ion in protein secretion by magnesium-resistant strains of Pseudomonas aeruginosa and Vibrio parahaemolyticus isolated from the coastal water of Haldia port. FEMS Microbiol. Lett. 185, 151–156. doi: 10.1111/j.1574-6968.2000.tb09054.x
Cho, J. J., Schroth, M. N., Kominos, S. K., and Green, S. K. (1975). Ornamental plants as carriers of Pseudomonas aeruginosa. Phytopathology 65, 425–431. doi: 10.1094/Phyto-65-425
Colinon, C., Deredjian, A., Hien, E., Brothier, E., Bouziri, L., Cournoyer, B., et al. (2013). Detection and enumeration of Pseudomonas aeruginosa in soil and manure assessed by an ecfX qPCR assay. J. Appl. Microbiol. 114, 1734–1749. doi: 10.1111/jam.12189
Daly, M., Power, E., Bjorkroth, J., Sheehan, P., O'Connell, A., Colgan, M., et al. (1999). Molecular analysis of Pseudomonas aeruginosa: epidemiological investigation of mastitis outbreaks in Irish dairy herds. Appl. Environ. Microbiol. 65, 2723–2729.
Deportes, I., Benoit-Guyod, J. L., Zmirou, D., and Bouvier, M. C. (1998). Microbial disinfection capacity of municipal solid waste (MSW) composting. J. Appl. Microbiol. 85, 238–246. doi: 10.1046/j.1365-2672.1998.00484.x
Edrington, T. S., Fox, W. E., Callaway, T. R., Anderson, R. C., Hoffman, D.W., and Nisbet, D. J. (2009). Pathogen prevalence and influence of composted dairy manure application on antimicrobial resistance profiles of commensal soil bacteria. Foodborne Pathog. Dis. 6, 217–224. doi: 10.1089/fpd.2008.0184
Garcia-Junco, M., De Olmedo, E., and Ortega-Calvo, J. J. (2001). Bioavailability of solid and non-aqueous phase liquid (NAPL)-dissolved phenanthrene to the biosurfactant-producing bacterium Pseudomonas aeruginosa 19SJ. Environ. Microbiol. 3, 561–569. doi: 10.1046/j.1462-2920.2001.00223.x
Gerba, C. P., and Smith, J. E. Jr. (2005). Sources of pathogenic microorganisms and their fate during land application of wastes. J. Environ. Qual. 34, 42–48.
Green, S. K., Schroth, M. N., Cho, J. J., Kominos, S. K., and Vitanza-Jack, V. B. (1974). Agricultural plants and soil as a reservoir for Pseudomonas aeruginosa. Appl. Microbiol. 28, 987–991.
Hassen, A., Belguith, K., Jedidi, N., Cherif, A., Cherif, M., and Boudabous, A. (2001). Microbial characterization during composting of municipal solid waste. Bioresour. Technol. 80, 217–225. doi: 10.1016/S0960-8524(01)00065-7
Heslop, A., and Ovesen, T. (2006). Severe acute middle ear infections: microbiology and treatment. Int. J. Pediatr. Otorhinolaryngol. 70, 1811–1816. doi: 10.1016/j.ijporl.2006.06.009
Houot, S., Clergeot, D., Michelin, J., Francou, C., Bourgeois, S., Caria, G., et al. (2002). “Agronomic value and environmental impacts of urban composts used in agriculture,” in Microbiology of Composting, ed H. Insam (New York, NY: Springer), 458–472.
Johnson, J. L. (1994). “Similarity analysis of DNAs,” in Methods for General and Molecular Bacteriology, eds P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (Washington, DC: ASM press), 655–682.
Kaszab, E., Kriszt, B., Atzel, B., Szabo, G., Szabo, I., Harkai, P., et al. (2010). The occurrence of multidrug-resistant Pseudomonas aeruginosa on hydrocarbon-contaminated sites. Microb. Ecol. 59, 37–45. doi: 10.1007/s00248-009-9551-7
Kaszab, E., Szoboszlay, S., Dobolyi, C., Háhn, J., Pék, N., and Kriszt, B. (2011). Antibiotic resistance profiles and virulence markers of Pseudomonas aeruginosa strains isolated from composts. Bioresour. Technol. 102, 1543–1548. doi: 10.1016/j.biortech.2010.08.027
Khan, N. H., Ahsan, M., Taylor, W. D., and Kogure, K. (2010). Culturability and survival of marine, freshwater and clinical Pseudomonas aeruginosa. Microbes Environ. 25, 266–274. doi: 10.1264/jsme2.ME09178
Khan, N. H., Ishii, Y., Kimata-Kino, N., Esaki, H., Nishino, T., Nishimura, M., et al. (2007). Isolation of Pseudomonas aeruginosa from open ocean and comparison with freshwater, clinical, and animal isolates. Microb. Ecol. 53, 173–186. doi: 10.1007/s00248-006-9059-3
Lavenir, R., Jocktane, D., Laurent, F., Nazaret, S., and Cournoyer, B. (2007). Improved reliability of Pseudomonas aeruginosa PCR detection by the use of the species-specific ecfX gene target. J. Microbiol. Methods 70, 20–29. doi: 10.1016/j.mimet.2007.03.008
Lavenir, R., Sanroma, M., Gibert, S., Crouzet, O., Laurent, F., Kravtsoff, J., et al. (2008). Spatio-temporal analysis of infra-specific genetic variations among a Pseudomonas aeruginosa water network hospital population: invasion and selection of clonal complexes. J. Appl. Microbiol. 105, 1491–1501. doi: 10.1111/j.1365-2672.2008.03907.x
Ledbetter, E. C., Hendricks, L. M., Riis, R. C., and Scarlett, J. M. (2007). In vitro fluoroquinolone susceptibility of Pseudomonas aeruginosa isolates from dogs with ulcerative keratitis. Am. J. Vet. Res. 68, 638–642. doi: 10.2460/ajvr.68.6.638
Lee, D. Y., Lauder, H., Cruwys, H., Falletta, P., and Beaudette, L. A. (2008). Development and application of an oligonucleotide microarray and real-time quantitative PCR for detection of wastewater bacterial pathogens. Sci. Total Environ. 398, 203–211. doi: 10.1016/j.scitotenv.2008.03.004
Lopez-Gutierrez, J. C., Henry, S., Hallet, S., Martin-Laurent, F., Catroux, G., and Philippot, L. (2004). Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57, 399–407. doi: 10.1016/j.mimet.2004.02.009
Marques, A. M., Congregado, F., and Simon-Pujol, D. M. (1979). Antibiotic and heavy metal resistance of Pseudomonas aeruginosa isolated from soils. J. Appl. Bacteriol. 47, 347–350. doi: 10.1111/j.1365-2672.1979.tb01765.x
Mushin, R., and Ziv, G. (1973). An epidemiological study of Pseudomonas aeruginosa in cattle and other animals by pyocine typing. J. Hyg. (Lond). 71, 113–122. doi: 10.1017/S0022172400046271
Norman, R. S., Frontera-Suau, R., and Morris, P. J. (2002). Variability in Pseudomonas aeruginosa lipopolysaccharide expression during crude oil degradation. Appl. Environ. Microbiol. 68, 5096–5103. doi: 10.1128/AEM.68.10.5096-5103.2002
Pell, A. N. (1997). Manure and microbes: public and animal health problem? J. Dairy Sci. 80, 2673–2681. doi: 10.3168/jds.S0022-0302(97)76227-1
Pirnay, J. P., Matthijs, S., Colak, H., Chablain, P., Bilocq, F., Van Eldere, J., et al. (2005). Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ. Microbiol. 7, 969–980. doi: 10.1111/j.1462-2920.2005.00776.x
Ranjard, L., Lejon, D. P., Mougel, C., Schehrer, L., Merdinoglu, D., and Chaussod, R. (2003). Sampling strategy in molecular microbial ecology: influence of soil sample size on DNA fingerprinting analysis of fungal and bacterial communities. Environ. Microbiol. 5, 1111–1120. doi: 10.1046/j.1462-2920.2003.00521.x
Reuter, S., Sigge, A., Wiedeck, H., and Trautmann, M. (2002). Analysis of transmission pathways of Pseudomonas aeruginosa between patients and tap water outlets. Crit. Care Med. 30, 2222–2228. doi: 10.1097/00003246-200210000-00008
Richard, P., Le Floch, R., Chamoux, C., Pannier, M., Espaze, E., and Richet, H. (1994). Pseudomonas aeruginosa outbreak in a burn unit: role of antimicrobials in the emergence of multiply resistant strains. J. Infect. Dis. 170, 380–383. doi: 10.1093/infdis/170.2.377
Ringen, L. M., and Drake, C. H. (1952). A study of the incidence of Pseudomonas aeruginosa from various natural sources. J. Bacteriol. 64, 841–845.
Römling, U., Wingender, J., Müller, H., and Tümmler, B. (1994). A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60, 1734–1738.
Saiman, L., and Siegel, J. (2004). Infection control in cystic fibrosis. Clin. Microbiol. Rev. 17, 57–71. doi: 10.1128/CMR.17.1.57-71.2004
Selezska, K., Kazmierczak, M., Müsken, M., Garbe, J., Schobert, M., Häussler, S., et al. (2012). Pseudomonas aeruginosa population structure revisited under environmental focus: impact of water quality and phage pressure. Environ. Microbiol. 14, 1952–1967. doi: 10.1111/j.1462-2920.2012.02719.x
Song, K. P., Chan, T. K., Ji, Z. L., and Wong, S. W. (2000). Rapid identification of Pseudomonas aeruginosa from ocular isolates by PCR using exotoxin A-specific primers. Mol. Cell. Probes 14, 199–204. doi: 10.1006/mcpr.2000.0306
Szmolka, A., Cramer, N., and Nagy, B. (2012). Comparative genomic analysis of bovine, environmental, and human strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 335, 113–122. doi: 10.1111/j.1574-6968.2012.02642.x
Wright, C., Kominos, S. D., and Yee, R. B. (1976). Enterobacteriaceae and Pseudomonas aeruginosa recovered from vegetable salads. Appl. Environ. Microbiol. 31, 453–454.
Keywords: Pseudomonas aeruginosa, qPCR, cultivation, soil, organic amendment
Citation: Deredjian A, Colinon C, Hien E, Brothier E, Youenou B, Cournoyer B, Dequiedt S, Hartmann A, Jolivet C, Houot S, Ranjard L, Saby NPA, and Nazaret S (2014) Low occurrence of Pseudomonas aeruginosa in agricultural soils with and without organic amendment. Front. Cell. Infect. Microbiol. 4:53. doi: 10.3389/fcimb.2014.00053
Received: 30 December 2013; Accepted: 09 April 2014;
Published online: 29 April 2014.
Edited by:
Ed Topp, Agriculture and Agri-Food Canada, CanadaReviewed by:
Huw Williams, Imperial College London, UKMatthew J. Wargo, University of Vermont College of Medicine, USA
Copyright © 2014 Deredjian, Colinon, Hien, Brothier, Youenou, Cournoyer, Dequiedt, Hartmann, Jolivet, Houot, Ranjard, Saby, and Nazaret. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sylvie Nazaret, CNRS, UMR 5557 Ecole Nationale Vétérinaire de Lyon, and Université Lyon 1, Université de Lyon, Mendel Bldg., 5th Floor, F-69622 Villeurbanne Cedex, France e-mail: sylvie.nazaret@univ-lyon1.fr
†These authors have contributed equally to this work
 Amélie Deredjian
Amélie Deredjian Céline Colinon1†
Céline Colinon1†  Elisabeth Brothier
Elisabeth Brothier Benjamin Youenou
Benjamin Youenou Samuel Dequiedt
Samuel Dequiedt Alain Hartmann
Alain Hartmann Claudy Jolivet
Claudy Jolivet Nicolas P. A. Saby
Nicolas P. A. Saby Sylvie Nazaret
Sylvie Nazaret