Rapid detection of predation of Escherichia coli O157:H7 and sorting of bacterivorous Tetrahymena by flow cytometry
- 1Foodborne Toxin Detection and Prevention Research Unit, Western Regional Research Center, US Department of Agriculture, Agricultural Research Service, Albany, CA, USA
- 2Produce Safety and Microbiology Research Unit, Western Regional Research Center, US Department of Agriculture, Agricultural Research Service, Albany, CA, USA
Protozoa are known to harbor bacterial pathogens, alter their survival in the environment and make them hypervirulent. Rapid non-culture based detection methods are required to determine the environmental survival and transport of enteric pathogens from point sources such as dairies and feedlots to food crops grown in proximity. Grazing studies were performed on a soil isolate of Tetrahymena fed green fluorescent protein (GFP) expressing Escherichia coli O157:H7 to determine the suitability of the use of such fluorescent prey bacteria to locate and sort bacterivorous protozoa by flow cytometry. In order to overcome autofluorescence of the target organism and to clearly discern Tetrahymena with ingested prey vs. those without, a ratio of prey to host of at least 100:1 was determined to be preferable. Under these conditions, we successfully sorted the two populations using short 5–45 min exposures of the prey and verified the internalization of E. coli O157:H7 cells in protozoa by confocal microscopy. This technique can be easily adopted for environmental monitoring of rates of enteric pathogen destruction vs. protection in protozoa.
Introduction
Protozoa in the environment have been implicated as both potential hosts harboring pathogens (Barker and Brown, 1994) and as agents enhancing pathogen survival and pathogenicity (Rasmussen et al., 2005; Bichai et al., 2008). Presence of the shiga toxin-encoding prophage in Escherichia coli O157:H7 (EcO157) is reported to enhance their survival in the food vacuoles of grazing Tetrahymena pyriformis (Steinberg and Levin, 2007). The passage of Salmonella enterica through and excretion from a soilborne Tetrahymena species is reported to convey increased survival of the organism (Brandl et al., 2005). However, protozoan predation was linked to decreases in EcO157 populations in dairy wastewater (Ravva et al., 2010, 2013) but only three ciliate protozoa were isolated in pure culture (Ravva et al., 2010). For these reasons and difficulties in culturing environmental protozoa, it is important to be able to identify protists in the environment which are actively ingesting bacteria and providing a safe-haven for environmental persistence and transport of EcO157 and other human enteric pathogens.
The study of bacterivory by protozoans by flow cytometry presents several challenges, not the least of which is the dynamic range demand on the instrument. In order to discern bacteria from protozoa, they must bear some separately identifiable characteristic. Size is an obvious choice and the different populations can be observed according to their different locations on a side scatter vs. forward scatter plot. Logarithmic scaling is required to overcome the wide variation in size between the bacteria and protozoa. Creative adjustment of signal threshold and detector voltage levels is required when the ratio of protozoa to bacteria is low (Rifa et al., 2002).
However, this gives us no information about protozoa that have ingested bacteria. Generally, this means that the bacteria are labeled with some fluorescent or otherwise identifiable compound. Examples have included chemical staining of the prey bacteria (Gonzalez, 1999; First et al., 2012), in vivo expression of fluorescent proteins (Fu et al., 2003) and even bioluminescence (Nelson et al., 2003).
Nonetheless, it is commonly observed that a variety of living cells also exhibit a degree of fluorescence, i.e., auto fluorescence, in the absence of any added fluorescent label. Protozoa typically being much larger than bacteria, they can be weakly auto fluorescent and yet on a cellular basis produce a signal of the same magnitude or greater than a single fluorescently labeled bacterium (First et al., 2012).
These problems are further compounded when studying organisms in environmental samples where the presence of other matter, living and otherwise, makes the identification and sorting of target populations even more difficult. In such cases, the use of nucleic acid staining has proven useful in locating the target organisms in the detritus (Lindström et al., 2002).
In this paper we examine some of these issues using a ciliate protozoan Tetrahymena species and pathogenic EcO157 transformed to express green or red fluorescent proteins (GFP or RFP).
Materials and Methods
Strains and Culture
Tetrahymena strain SSU was a soil isolate kindly provided by MT Brandl (Brandl et al., 2005) of Produce Safety and Microbiology Research Unit, USDA, ARS, Albany, CA. The protozoa were enriched by the addition of E. coli strain DH5-α cells to aqueous soil suspensions, purified and characterized by 18S rRNA gene sequencing (Brandl et al., 2005). The stock culture was grown and maintained in Neff's medium (Neff et al., 1964). Protozoa concentrations were determined (Ravva et al., 2010) using the most-probable number (MPN) method (Blodgett, 2010).
The prey organisms in the grazing experiments were EcO157 strains MM123 and MM127 derived from the Odwalla apple juice outbreak strain RM1484 (Cooley et al., 2006; Ravva et al., 2006, 2010). MM123 and MM127 are spontaneous rifampicin-resistant (100 μg mL−1) mutants of RM2315 and RM2318 containing plasmids expressing genes for green fluorescent protein GFP (pWM1029) and red fluorescent protein DsRed (pWM1032, RFP), respectively. The EcO157 strains expressing fluorescent proteins were transformed by WG Miller (Produce Safety and Microbiology Unit) using the plasmids (pWM1029 and pWM1032) he constructed (Miller, Unpublished data). The strains were grown at 37°C on Luria-Bertani (LB) broth supplemented with 50 μg mL−1 kanamycin. Cultures were resuspended in 0.01M PBS (pH 7.2) to an optical density of 0.3 at 600 nm to establish a working suspension of 108 cfu mL−1 and diluted accordingly as needed. Actual concentrations were monitored by serial dilution and plating on LB agar supplemented with 50 μg mL−1 kanamycin. The fluorescent colonies were counted on a UV Transilluminator (Model 3-3000, Fotodyne, Hartland, WI).
Grazing Studies
Grazing experiments were conducted using Tetrahymena SSU at a concentration of about 104 protozoan cells mL−1 and live prey concentrations ranging from 104 to 108 EcO157 cells mL−1 giving prey-predator ratios of 1 to 10,000:1. Tetrahymena SSU grown in Neff's medium was amended with 0.01M PBS containing the appropriate concentration of EcO157 cells and incubated at 25 ± 2°C for various intervals during a 4-h period. One-milliliter samples were taken at various intervals for fixation and flow cytometry. Since working with live pathogens require additional safety measures, the uptake of live vs. heat-killed cells at a prey concentration of 106 EcO157 cells mL−1 was compared initially at various intervals during a 90-min period. Samples for flow cytometry were fixed with 1% formalin for 10 min at room temperature prior to sorting.
Flow Cytometry and Sorting
The instrument used was a FACSVantage SE flow cytometer (BD Biosciences, San Jose, CA) with an Enterprise II, water-cooled Argon laser (Coherent, Santa Clara, CA). The green fluorescence of GFP was quantified using a 530/30 nm bandpass filter and a 630/22 nm bandpass filter was used for the red fluorescence of RFP. Sheath fluid was either FACSFlow (BD Biosciences) or preservative free BioSure flow cytometry sheath solution (BioSure, Grass Valley, CA). The minimum useable nozzle diameter was found to be 70 μm with these organisms. Nonetheless, formalin fixation was required to obtain the Tetrahymena intact after sorting. EcO157 cells were inactivated during the fixation process.
Confocal Imaging
Internalization of GFP-EcO157 (MM123) in Tetrahymena was monitored after sorting using a Leica TCS-NT confocal microscope equipped HC PL FLUOTAR 20×/0.50 or 40×/0.70 or 63×/1.2 W PL APO objectives, with argon (488 nm), krypton (568 nm), and He/Ne (633 nm) lasers (Leica Microsystems, Wetzlar, Germany) and using Leica TCS NT Software (v. 2.5) for image analysis and preparation. The BP520/50 emission filter set with 488nm laser excitation was used for visualizing the GFP fluorescence. The GFP fluorescence was assigned a green color in the compiled images.
Results
We compared both GFP and RFP EcO157 for the grazing studies and found that there was no significant advantage of one over the other. Both strains gave good signal strength but the Tetrahymena was observed to autofluoresce with approximately equal intensity to the average single bacterial fluorescence signal in either case (data not shown). Because of this autofluorescence, the uptake of bacteria is preferably several-fold in order to significantly resolve the bacterivorous protists from the non-feeding population.
The plot in Figure 1 shows a typical time course of fluorescence uptake by Tetrahymena feeding on GFP-EcO157 with an initial concentration of 106 mL−1. Neither heat nor formalin fixation had an adverse effect on GFP fluorescence of the individual E. coli bacteria. Therefore, either method could be used to inactivate the pathogenic bacteria for safety concern when sorting but we chose to use formalin fixation subsequent to feeding so that the bacteria could be fed to Tetrahymena live. In any case, the plot shows a rapid uptake of EcO157 whether live or heat killed, the latter yielding somewhat more peak fluorescence internalization.
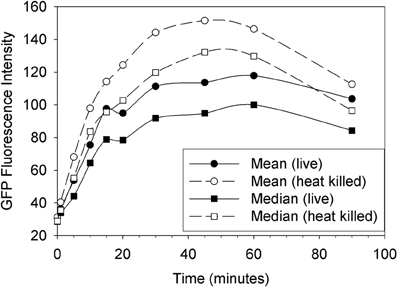
Figure 1. Uptake of live vs. heat killed cells of GFP-EcO157 by Tetrahymena monitored over time as the mean and median green fluorescence intensity. Open symbols indicate the case where the E. coli were heat killed prior to feeding. Initial EcO157 concentration was 106 cells mL−1. Predator-prey ratio was 1:100.
As Tetrahymena feeds on EcO157 cells, there is a broadening of the green fluorescence of the population and a shift to higher average fluorescence (Figure 2). Because neither Tetrahymena nor the GFP-EcO157 cells have a narrow fluorescence distribution (fluorescence due to autofluorescence in the former case and GFP in the latter) we do not see discrete increases in fluorescence, i.e. it is not possible from the histograms to discern separate populations according to the numbers of bacteria ingested per cell.
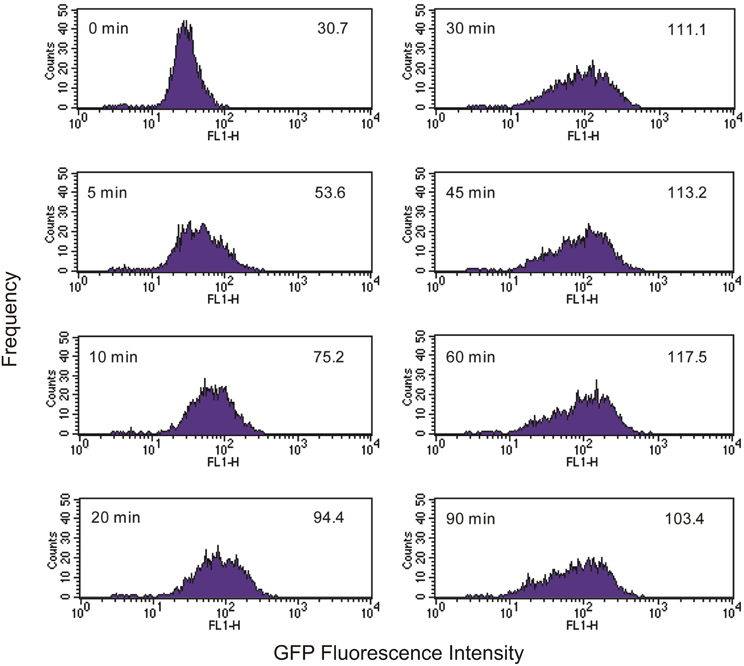
Figure 2. Time course of green fluorescence uptake of Tetrahymena (104 cells mL−1) in the presence of an initial concentration of 106 mL−1 live GFP-EcO157. Time and mean fluorescence intensities are indicated in each histogram. Predator-prey ratio was 1:100. See Figure 1 for comparative uptake of heat killed cells.
The overall fluorescence of a protozoan cell at any given time is determined by the rate at which it is ingesting fluorescent bacteria, the rate at which it is excreting or digesting fluorescent bacteria, the rate at which the GFP is inactivated inside the Tetrahymena as well as the cell's autofluorescence. The ingestion rate is expected to be a function of bacterial concentration and at low enough concentration, the rates of digestion and/or excretion may predominate such that significant fluorescence accumulation is not possible.
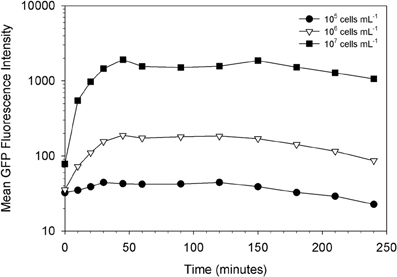
Indeed, we found that the initial concentration of prey is a vital parameter in determining the maximum uptake of fluorescence. This is clearly shown in Figures 3, 4. At the lower initial concentrations of 104 and 105 bacteria mL−1, the feeding Tetrahymena did not develop enough fluorescence to be clearly discerned from the non-feeding population. On the other hand, as Figure 4 shows, there were very few cells which had no increased fluorescence when the initial bacterial concentrations were 107 and 108 bacteria mL−1. The mean fluorescence of the individual E. coli and unfed Tetrahymena in this experiment were 18 and 16, respectively, on the scale shown.
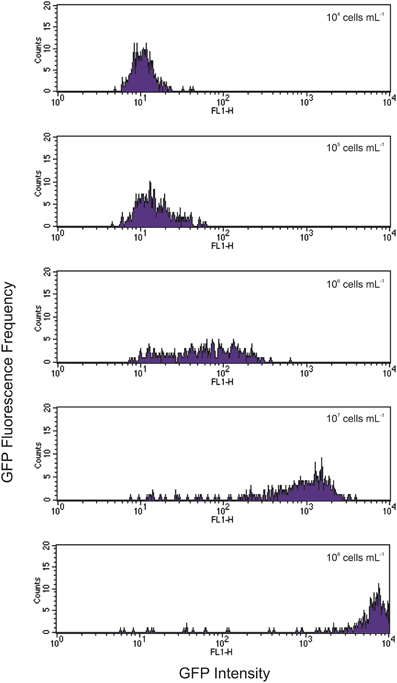
Figure 4. Histograms showing increasing levels of green fluorescence in Tetrahymena after 1 h of feeding with initial concentrations of GFP-EcO157 from 104 cells mL−1 (top) to 108 cells mL−1 (bottom). Each histogram represents a 10-fold increase in initial concentration.
We sorted Tetrahymena with high and low levels of green fluorescence. Figure 5 shows confocal images of example cells sorted from these regions verifying the presence and absence of ingested GFP-EcO157 in the two sorted groups. In this example, the main confounding parameters were the high concentration of background bacteria which were 1000-fold more prevalent (107 EcO157 cells: 104 protozoa mL−1) than the Tetrahymena beside the approximately equal fluorescence intensity of the empty Tetrahymena and the single GFP-EcO157 cell. However, even when we added additional debris the feeding Tetrahymena could be identified from the background.
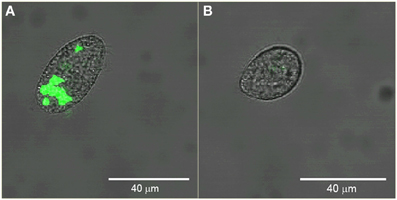
Figure 5. Confocal images of example Tetrahymena sorted from high (A) and low (B) green fluorescence gate regions. Initial EcO157 concentration was 107 mL−1. Cells were fixed after 30 min of grazing. Bars in images are 40 μm in length.
Discussion
The feeding of fluorescent protein labeled prey bacteria is a useful tool to identify and sort bacterivorous protozoa, at least in the case of the ciliate Tetrahymena. The ultimate level of fluorescence incorporation is determined by the rates of uptake, excretion and loss due to digestion. It is reported that GFP is unstable and loses fluorescence in the acidic food vacuoles of protozoa even when the bacteria are not completely digested (Parry et al., 2001). We found the fluorescence to be sufficiently persistent in fixed cells to be analyzed and sorted by FACS as well as to be visualized under confocal microscopy. On the other hand, if GFP was persistent in undigested, intact bacteria excreted by Tetrahymena (Brandl et al., 2005; Steinberg and Levin, 2007), we should have been able to identify fluorescent vesicles but this was not the case during a 4 h incubation in this study. However, we did observe vesicles filled with GFP-EcO157 (the same strain) expelled from environmental protozoa 2 days after ingestion (Ravva et al., 2010) and internalized GFP-EcO157 cells were detected in protozoa even after 14 days.
One of the hurdles in utilizing this technique is the incorporation of sufficient fluorescence in the protozoa to obtain a separately identifiable population from within environmental samples. This can be overcome by using a sufficient concentration of labeled prey that gives signal significantly higher than the autofluorescence of the target protozoa and other biological and non-biological components. In the environment, the concentration of host and prey would almost certainly be lower than the levels used here. But there seems to be a concentration of prey below which bacterivory is disfavored or at least below which the ingestion rate is too slow relative to excretion and digestion to allow sufficient accumulation of fluorescence to discern. This observation is consistent with the report of Steinberg and Levin that Tetrahymena grazing on EcO157 ceases below concentrations of about 105 bacteria mL−1 (Steinberg and Levin, 2007) which they attribute to minimum consumption requirement for growth (Watson et al., 1981).
Ingestion rates have been measured by using uniformly fluorescent microspheres (Lavin et al., 1990). Because of the high uniformity of such particles, populations with discrete numbers of ingested particles can clearly be discerned on fluorescence histograms of the protozoa. Although it is not possible with GFP-EcO157 which are not so uniform in size and fluorescence, this method may be used to determine within 5–45 min if environmental protozoa are responsible for significant decreases (Ravva et al., 2010, 2013) of enteric pathogenic bacteria in agricultural environments. Since, isolation and characterization of protozoa in the environment is extremely difficult for use in direct tests to measure the uptake of pathogenic bacteria, flow cytometry appears to be an ideal method in sorting and culturing protozoa based on their relative uptake of fluorescence-labeled organisms. We used flow cytometry earlier to characterize bacteria from urban aerosols (Hernlem and Ravva, 2007). Feasibility studies with environmental samples are needed to determine if protozoa preferentially harbor and transport enteric pathogens from point sources (dairies and feed lots) to fruit and vegetable crops grown in proximity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Reference to a company or product named by the USDA is only for purposes of information and does not imply approval or recommendation of the product to the exclusion of others that may also be suitable. The work was funded by the U.S. Department of Agriculture, Agricultural Research Service under CRIS projects 5325-42000-046 and 5325-42000-048. We thank Anne Bates for confocal microscopy.
References
Barker, J., and Brown, M. R. (1994). Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140, 1253–1259. doi: 10.1099/00221287-140-6-1253
Bichai, F., Payment, P., and Barbeau, B. (2008). Protection of waterborne pathogens by higher organisms in drinking water: a review. Can. J. Microbiol. 54, 509–524. doi: 10.1139/w08-039
Blodgett, R. (2010).Bacteriological Analytical Manual Online, eds G. J. Jackson, R. I. Merker and R. Bandler Available online at: http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/ucm109656.htm
Brandl, M. T., Rosenthal, B. M., Haxo, A. F., and Berk, S. G. (2005). Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Appl. Environ. Microbiol. 71, 1562–1569. doi: 10.1128/AEM.71.3.1562-1569.2005
Cooley, M. B., Chao, D., and Mandrell, R. E. (2006). Escherichia coli O157:H7 survival and growth on lettuce is altered by the presence of epiphytic bacteria. J. Food. Protect. 69, 2329–2335.
First, M. R., Park, N. Y., Berrang, M. E., Meinersmann, R. J., Bernhard, J. M., Gast, R. J., et al. (2012). Ciliate ingestion and digestion: flow cytometric measurements and regrowth of a digestion-resistant Campylobacter jejuni. J. Eukaryot. Microbiol. 59, 12–19. doi: 10.1111/j.1550-7408.2011.00589.x
Fu, Y., O'Kelly, C., Sieracki, M., and Distel, D. L. (2003). Protistan grazing analysis by flow cytometry using prey labeled by in vivo expression of fluorescent proteins. Appl. Environ. Microbiol. 69, 6848–6855. doi: 10.1128/AEM.69.11.6848-6855.2003
Gonzalez, J. M. (1999). Bacterivory rate estimates and fraction of active bacterivores in natural protist assemblages from aquatic systems. Appl. Environ. Microbiol. 65, 1463–1469.
Hernlem, B. J., and Ravva, S. V. (2007). Application of flow cytometry and cell sorting to the bacterial analysis of environmental aerosol samples. J. Environ. Monit. 9, 1317–1322. doi: 10.1039/b710512f
Lavin, D. P., Fredrickson, A. G., and Srienc, F. (1990). Flow cytometric measurement of rates of particle uptake from dilute suspensions by a ciliated protozoan. Cytometry 11, 875–882. doi: 10.1002/cyto.990110804
Lindström, E. S., Weisse, T., and Stadler, P. (2002). Enumeration of small ciliates in culture by flow cytometry and nucleic staining. J. Microbiol. Meth. 49, 173–182. doi: 10.1016/S0167-70120100366-9
Neff, R. J., Ray, S. A., Benton, W. F., and Wilborn, M. (1964). “Induction of synchronous encystment (differentiation),” in Acanthamoeba spp. Methods in Cell Physiology, ed D. M. Prescott (Washington, DC: Academic Press), 55–83. doi: 10.1016/S0091-679X(08)62086-5
Nelson, S. M., Cooper, A. A. A., Taylor, E. L., and Salisbury, V. C. (2003). Use of bioluminescent Escherichia coli O157:H7 to study intra-protozoan survival of bacteria within Tetrahymena pyriformis. FEMS Microbiol. Lett. 223, 95–99. doi: 10.1016/S0378-1097(03)00349-5
Parry, J. D., Heaton, K., Drinkall, J., and Jones, H. L. J. (2001). Feasibility of using GFP-expressing Escherichia coli, coupled with fluorimetry, to determine protozoan ingestion rates. FEMS Microbiol. Ecol. 35, 11–17. doi: 10.1111/j.1574-6941.2001.tb00783.x
Rasmussen, M. A., Carlson, S. A., Franklin, S. K., McCuddin, Z. P., Wu, M. T., and Sharma, V. K. (2005). Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect. Immun. 73, 4668–4675. doi: 10.1128/IAI.73.8.4668-4675.2005
Ravva, S. V., Sarreal, C. Z., Duffy, B., and Stanker, L. H. (2006). Survival of Escherichia coli O157:H7 in wastewater from dairy lagoons. J. Appl. Microbiol. 101, 891–902. doi: 10.1111/j.1365-2672.2006.02956.x
Ravva, S. V., Sarreal, C. Z., and Mandrell, R. E. (2010). Identification of protozoa in dairy lagoon wastewater that consume Escherichia coli O157:H7 preferentially. PLoS ONE 5:e15671. doi: 10.1371/journal.pone.0015671
Ravva, S. V., Sarreal, C. Z., and Mandrell, R. E. (2013). Altered protozoan and bacterial communities and survival of Escherichia coli O157:H7 in monensin-treated wastewater from a dairy lagoon. PLoS ONE 8:e54782. doi: 10.1371/journal.pone.0054782
Rifa, T. G., Latatu, A., Ayo, B., Iriberri, J., and Comas-Riu, J. (2002). Flow cytometric detection and quantification of heterotrophic nanoflagellates in enriched seawater and cultures. Syst. Appl. Microbiol. 25, 100–108. doi: 10.1078/0723-2020-000085
Steinberg, K. M., and Levin, B. R. (2007). Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc. Soc. Biol. 274, 1921–1929. doi: 10.1098/rspb.2007.0245
Keywords: enteropathogenic E. coli, foodborne pathogen, GFP, flow cytometry, protozoa, environmental transport, Tetrahymena, E. coli O157:H7
Citation: Hernlem BJ, Ravva SV and Sarreal CZ (2014) Rapid detection of predation of Escherichia coli O157:H7 and sorting of bacterivorous Tetrahymena by flow cytometry. Front. Cell. Infect. Microbiol. 4:57. doi: 10.3389/fcimb.2014.00057
Received: 05 February 2014; Accepted: 15 April 2014;
Published online: 05 May 2014.
Edited by:
Alain Hartmann, Institut National de la Recherche Agronomique, FranceReviewed by:
Philip R. Hardwidge, Kansas State University, USABeatriz Arellano Reynoso, The Universidad Nacional Autónoma de México, Mexico
Copyright © 2014 Hernlem, Ravva and Sarreal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subbarao V. Ravva, Produce Safety and Microbiology Research Unit, Western Regional Research Center, US Department of Agriculture, Agricultural Research Service, 800 Buchanan St, Albany, CA 94710, USA e-mail: subbarao.ravva@ars.usda.gov
 Bradley J. Hernlem
Bradley J. Hernlem Subbarao V. Ravva
Subbarao V. Ravva Chester Z. Sarreal
Chester Z. Sarreal