Membrane vesicle production by Chlamydia trachomatis as an adaptive response
- 1Department of Microbiology, Immunology, and Parasitology, Louisiana State University Health Sciences Center, New Orleans, LA, USA
- 2Department of Neonatology, Ministry of Education Key Laboratory of Child Development and Disorder, The Children's Hospital, Chongqing Medical University, Chongqing, China
- 3Department of Chemistry, University of Georgia, Athens, GA, USA
Bacteria have evolved specific adaptive responses to cope with changing environments. These adaptations include stress response phenotypes with dynamic modifications of the bacterial cell envelope and generation of membrane vesicles (MVs). The obligate intracellular bacterium, Chlamydia trachomatis, typically has a biphasic lifestyle, but can enter into an altered growth state typified by morphologically aberrant chlamydial forms, termed persistent growth forms, when induced by stress in vitro. How C. trachomatis can adapt to a persistent growth state in host epithelial cells in vivo is not well understood, but is an important question, since it extends the host-bacterial relationship in vitro and has thus been indicated as a survival mechanism in chronic chlamydial infections. Here, we review recent findings on the mechanistic aspects of bacterial adaptation to stress with a focus on how C. trachomatis remodels its envelope, produces MVs, and the potential important consequences of MV production with respect to host-pathogen interactions. Emerging data suggest that the generation of MVs may be an important mechanism for C. trachomatis intracellular survival of stress, and thus may aid in the establishment of a chronic infection in human genital epithelial cells.
Introduction
Chlamydia trachomatis serovars D-K account for the most prevalent bacterial sexually transmitted infections (STIs) worldwide. Despite aggressive control efforts, C. trachomatis infections have continued to constitute a serious public health risk (World Health Organization, 2011; Rekart et al., 2013). Infection may result in cervicitis, and in some women, C. trachomatis may ascend into the endometrium and Fallopian tubes, where it can establish a chronic infection leading to diseases such as pelvic inflammatory disease (PID), ectopic pregnancy, and infertility. C. trachomatis infections of women also pose a risk to infants, as infants born from mothers with C. trachomatis infections can develop conjunctivitis and/or pneumonia. Finally, epidemiological evidence indicates that C. trachomatis infection of the reproductive tract also may increase the risk of HIV transmission, making the study and understanding of the pathogenicity of this bacterium imperative.
In adapting to an intracellular niche, C. trachomatis has evolved a notably reduced genome of ~1 million base pairs that supports a unique developmental cycle (Stephens et al., 1998). This cycle typically involves two forms: infectious elementary bodies (EBs) and dividing, metabolically active reticulate bodies (RBs) (Moulder, 1991). Under stressful conditions in vitro, however, C. trachomatis can enter into an alternative viable but non-dividing growth form, termed a persistent growth form (Beatty et al., 1994b). This persistent growth state extends the host-bacterial relationship in vitro and has thus been proposed to be linked to chronic infection and adverse outcomes, including elicitation of tissue-damaging host responses, in vivo (Hogan et al., 2004; Darville and Hiltke, 2010). Persistent forms are also less responsive to antimicrobial therapy in vitro (Wyrick and Knight, 2004; Reveneau et al., 2005) and in vivo (Byrne, 2001; Phillips-Campbell et al., 2014). Several excellent reviews provide extensively insightful descriptions of the chlamydial persistent growth state and the potential to establish a chronic relationship with the host (Beatty et al., 1994b; Hogan et al., 2004; McClarty et al., 2007; Wyrick, 2010; Schoborg, 2011; Lo et al., 2012).
C. trachomatis growth exclusively takes place within an inclusion, a membrane-bound vacuole. Despite the presence of this barrier, C. trachomatis actively communicates with the host cells. One method of interaction is the coordination of trafficking specific subsets of host vesicles to and from the inclusion, enabling delivery of the inclusion components and nutrients required for infection (Fields and Hackstadt, 2002). C. trachomatis also secretes numerous effectors with host cell-modulating activities across the complex membranes of the bacterium and the inclusion or host cytoplasm to host cell compartments. Although diverse secretory pathways, including the type II (Sec), type III, and type V (autotransporter) secretion systems, have been shown to play a role in the translocation of protein effectors (Crane et al., 2006; Valdivia, 2008; Chen et al., 2010a; Mueller et al., 2014), a mechanism for robust delivery of complex bacterial components to host cells is likely to be mediated by membrane vesicles (MVs) that emerge from the envelope of growing bacteria (Giles et al., 2006; Giles and Wyrick, 2008; Wang et al., 2011a; Frohlich et al., 2012). As part of bacterial growth and/or envelope stress responses, the formation of MVs is a universal feature found in all Gram-negative bacteria (Beveridge, 1999; Kulp and Kuehn, 2010), Mycobacterium spp. (Prados-Rosales et al., 2011), and Gram-positive bacteria such as Bacillus spp. (Dubey and Ben-Yehuda, 2011). Herein, we review recent findings on the mechanistic aspects of bacterial adaptation to stress with a focus on how C. trachomatis remodels its envelope, produces MVs, and the potential important consequences of MV production with respect to host-pathogen interactions.
Molecular Architecture of the Cell Envelope During the Chlamydial Developmental Cycle
One of the critical developmental events common to Chlamydia spp. is the ability of the organisms to adapt their envelopes for the purpose of interacting with the host cell. Like other Gram-negative bacteria, the C. trachomatis envelope consists of an outer membrane (OM), an inner membrane (IM), and a periplasm (Figure 1A). Recent elegant studies by several groups have revealed that chlamydiae possess functional peptidoglycan necessary for bacterial division, despite the lack of the cytoskeletal protein FtsZ (McCoy and Maurelli, 2006; Ouellette et al., 2012; Pilhofer et al., 2013; Liechti et al., 2014). Nevertheless, a unique feature of the chlamydial envelope, distinguishing them from other Gram-negative bacteria, is the presence of a disulfide bond cross-linked major outer membrane protein (MOMP) with the periplasmic localized OmcB and the lipoprotein OmcA only in EBs (Hackstadt et al., 1985; Hatch, 1996). Both OmcB and OmcA contain abundant cysteines. This cross-linkage is believed to contribute to cell wall rigidity and osmotic stability of the EBs.
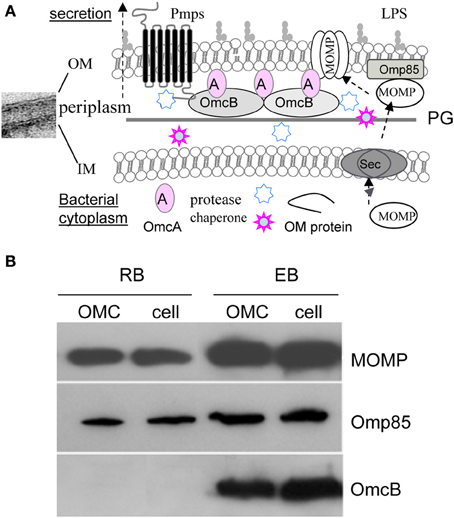
Figure 1. (A) A schematic diagram of the envelope structure of C. trachomatis. Showing are the representative OM associated components [Lipopolysaccharide (LPS), peptidoglycan (PG), MOMP, Pmps, OmcA, OmcB, and Omp85], and putative chaperone and protease in the periplasm in EBs. The potential process of MOMP or other OM protein transport across the inner membrane via the Sec pathway, and insertion and assembly into the OM is indicated. (B) Immunoblot of C. trachomatis serovar F Omp85. Similar to MOMP, Omp85 is present in both EBs and RBs as a component of the OM complex (OMC), however, OmcB exists only in EBs. The OMCs were prepared from the insoluble fraction of isolated EBs or RBs in phosphate buffered saline containing 2% Sarkysol as described previously (Caldwell et al., 1981). The lysates of EBs and RBs, and their OMCs were dissolved in Laemmli sample buffer (1:1, v:v) supplemented with 5% 2-mercaptoethanol, 10 mM dithiothreitol and heated for 10 min at 100°C, isolated on 10% SDS-PAGE, followed by immunoblotting using polyclonal antibodies to Omp85 or OmcB, and a monoclonal antibody to C. trachomatis serovar F MOMP.
Proteins of the OM are important for various purposes including envelope architecture, virulence, transport, cell division, induction of inflammatory cytokine production, and immune evasion (Hatch, 1996; Stephens and Lammel, 2001; Abdelsamed et al., 2013). All of these tasks are presumably related to the generation and function of MVs that will be discussed in the later sections of this review. MOMP is the most abundant surface-exposed protein of both RBs and EBs (Caldwell et al., 1981; Hatch, 1996) and it functions as an adherin and a porin with its β-barrel structure (Wang et al., 2006; Sun et al., 2007). MOMP also has alternative conformations that may adapt to specific chlamydial growth stages (Feher et al., 2013) and may impart different levels of immunogenicity. Also located in the OM is a family of polymorphic membrane proteins (Pmps) or autotransporters unique to Chlamydia spp. (Stephens et al., 1998). C. trachomatis encodes nine Pmps (PmpA-I) that are either temporally or constitutively expressed (Tanzer and Hatch, 2001; Tan et al., 2010). Such temporal expression may promote antigenic variation, tissue tropism, and differential disease severity (Gomes et al., 2006; Tan et al., 2009; Taylor et al., 2011; Abdelsamed et al., 2013). Caldwell's group has reported that PmpD is a species-common, pan-neutralizing target (Crane et al., 2006). Other OM proteins studied include plasmid encoded Pgp3 (Chen et al., 2010b), PorB (Kubo and Stephens, 2000), HSP70 (DnaK) (Raulston, 1995), and Omp85 (YaeT) (Stephens and Lammel, 2001) that exists in both EBs and RBs (Figure 1B) (Liu et al., 2010). In bacteria, Omp85 facilitates the insertion of assembly intermediates from the periplasm to the OM (Ricci and Silhavy, 2012).
Many chlamydial envelope components are ligands recognized by host pattern-recognition receptors that induce inflammatory cytokine production and generate adaptive immune responses (Wang et al., 2010; Taylor et al., 2011; Abdelsamed et al., 2013). Some envelope components may also contribute to immune evasion. To this end, several chlamydial envelope components have been considered vaccine candidates (Crane et al., 2006; Schautteet et al., 2011; Hafner et al., 2014). Chlamydia spp. may quantitatively or qualitatively change their envelope structures as a result of adaptation to developmental signals or environmental cues (Beatty et al., 1994b; Hatch, 1996; Carrasco et al., 2011). The plasticity of the envelope has important implications for the design of control strategies against bacterial infection because of its importance in determining susceptibility to host defenses and antibiotics (Tan et al., 2009; Hurdle et al., 2011).
Stress Induces Dramatic Changes in Chlamydia Spp. In Vitro
Matsumoto and Manire described the first ultrastructure of an aberrant RB form induced by penicillin in C. psittaci-infected L929 cells as viewed by transmission electron microscopy (TEM) (Matsumoto and Manire, 1970). They found that, in the presence of penicillin, RBs no longer divided, the RB to EB transition was disrupted, and aberrant multinucleated RBs accumulated. Concurrently, abundant vesicles were pinched off the aberrant RBs. Removal of penicillin allowed for the chlamydiae to reenter a normal growth state. Many elegant studies since have revealed that the viable but nondividing altered persistent form occurs when Chlamydia spp. are exposed to a variety of stress conditions. These include IFNγ (Beatty et al., 1994a), sub-inhibitory concentrations of antibiotics (Engel, 1992; Wyrick and Knight, 2004), nutrient deprivation (Beatty et al., 1994a; Raulston, 1997; Harper et al., 2000), co-infection with either herpes simplex virus (Vanover et al., 2008; Prusty et al., 2012) or Toxoplasma gondii (Romano et al., 2013) in vitro, and infection with Chlamydia-phage (Hsia et al., 2000). These stressors are often encountered by pathogens during infection in vivo (Wyrick, 2010). IFNγ, one of the stressors commonly studied, is a key component of immunity to intracellular pathogens. IFNγ induces expression of indoleamine-2, 3dioxygenase (IDO) that catalyzes the initial step in the degradation of L-tryptophan to N-formylkynurenine and kynurenine in eukaryotic cells (Beatty et al., 1994a). Such tryptophan depletion profoundly interferes with chlamydial growth, which, depending on the IFNγ concentration and exposure time, induces the bacteria to enter into a persistent growth form or results in bacterial eradication. Exposure to β-lactam antibiotics is another well-studied stressor that causes chlamydiae to enter into a persistent growth state and is used in simulating an inadequate antimicrobial treatment of chlamydiae infection (Matsumoto and Manire, 1970; Gerard et al., 2001; Giles and Wyrick, 2008; Carrasco et al., 2011; Wang et al., 2011a; Ouellette et al., 2012; Phillips-Campbell et al., 2014). Chlamydia spp. have a wide range of responses to stress. A clear shift of gene expression profiles specific to each stressor has been consistently found in many host cell types (Beatty et al., 1993, 1994b; Gerard et al., 2001, 2013; Molestina et al., 2002; Belland et al., 2003a,b; Ouellette et al., 2006; Lo et al., 2012). Changes in gene expression induced by stress at the levels of transcription and translation are consistent with the nature of a persistent growth form that undergoes DNA replication, but not division by binary fission. Interestingly, IFNγ exposure resulted in a global transcriptional upregulation lacking increased translation in chlamydiae (Ouellette et al., 2006). In contrast, penicillin exposure induced an alteration in transcription coupled to a change in translation (Ouellette et al., 2006). These data support that the impact of each stressor on bacteria is mediated by different mechanisms, and the bacterial response to each stress may be distinct.
To explore the mechanisms by which C. trachomatis survives under persistence inducing conditions, we utilized human primary endocervical epithelial cells, the main site of C. trachomatis infection in vivo. We found that normal C. trachomatis forms could develop in cultured primary cells similarly to HeLa cells (Wang et al., 2011a). Figures 2A–C show that exposure to ampicillin or IFNγ led to aberrant RB phenotypes and an accumulation of abundant MVs generated by C. trachomatis in cultured cells. Additionally, confocal microscopy and cell fractionation analyses demonstrated that the secreted chlamydial T3S effectors, CopN and Tarp, and the secreted protease, CPAF (Zhong et al., 2001) were decreased in ampicillin-induced persistent forms (Wang et al., 2011a), the latter of which is consistent with previous observations using IFNγ exposure or iron-deprivation persistence models (Shaw et al., 2002; Heuer et al., 2003). Changes in the spectrum of host cytosolic chlamydial proteins may underlie the host-pathogen relationship because of their importance in modulating host cell signaling (Valdivia, 2008; Zhong, 2011; Mueller et al., 2014).
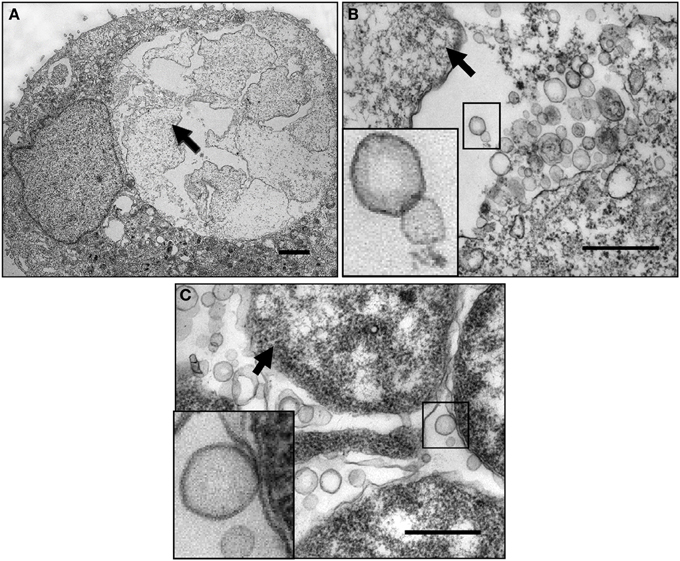
Figure 2. Micrograph of chlamydial MVs in cultured human cervical epithelial cells. (A) Ampicillin-induced persistent growth forms in infected human primary endocervical epithelial cells. (B) A high magnification image showing ampicillin-induced C. trachomatis MVs. The boxed MVs are enlarged in the insert. (C) IFNγ-induced C. trachomatis MVs in infected HeLa cells. The boxed bacterial membranes and the MVs are enlarged in the insert. Note: the vesicles appear to be single-membrane structures different from the double membrane structures of intact chlamydial organisms. They are associated with the bacterial surface, clustered or scattered within the inclusion lumen. Arrows indicate persistent forms. The primary cell cultures were established from endocervical tissue explants as previously described (Herbst-Kralovetz et al., 2008). Cells were infected with serovar F EBs resulting in a 30% infection rate. Ampicillin (10 μg/ml) was added to the culture at 16 h post-infection. For the IFNγ exposure model, HeLa cells were exposed to RPMI 1640 medium containing 50 units/ml of IFNγ for 24 h prior to infection. Fresh IFNγ-containing medium was added after infection. Cells were harvested at 36 h post-infection, fixed and processed for TEM as described previously (Belland et al., 2003a). Scale bars: (A) 500 nm. (B,C) 200 nm.
MV Produced During Bacterial Infection are Present in C. Trachomatis Infected Cells In Vivo in Humans
We have been particularly intrigued by the observed vesicular structures that are derived from both pathogen and host cells during C. trachomatis infection in culture, as they may serve as a vital element for host-pathogen interactions. C. trachomatis is likely to encounter numerous known and not-yet-identified conditions in vivo. Of these, varying levels of IFNγ in the endocervix during chlamydial infection in vivo are likely crucial as demonstrated by studies in vitro, in animal models, and in observational studies in humans (Arno et al., 1990; Beatty et al., 1994a; Byrne, 2001; Aiyar et al., 2014; Lewis et al., 2014). The direct involvement of persistent growth forms in pathogenesis in vivo in humans is challenging to prove, but several TEM studies have visualized atypical pleomorphic RBs and aberrant C. trachomatis forms in individuals with chronic infections, in Fallopian tube tissues, and in the synovium of reactive arthritis patients (Patton et al., 1994; Nanagara et al., 1995; Mazzoli et al., 2000; Bragina et al., 2001). Very recently, the Quayle laboratory developed methodology to sample endocervical cells and components of the endocervical environment in C. trachomatis infected women. By using a cytobrush to retrieve cells followed by immediate placement of these brushes in modified TEM fixative, the ultrastructure of “in vivo” chlamydial growth forms in endocervical epithelial cells could later be visualized by TEM (Lewis et al., 2014). These TEM analyses revealed that chlamydial infection in the human endocervix could result in a variety of inclusion types, containing normal forms, a mixture of relatively “normal” and aberrant forms, or inclusions with highly aberrant forms, and this varies between patients (Lewis et al., 2014). Since MVs are so small in size and lack unique biological markers for identification by standard immunostaining techniques, we have begun to take advantage of these samples to examine MV production in vivo. High magnification TEM images were obtained from cross sections of several inclusions and indicated the in vivo presence of C. trachomatis MVs in inclusions sampled from two different patients (Figures 3A–D). MVs appeared to be single membrane structures and apparently associated with the OM of chlamydial organisms similar to those observed in vitro (Figure 2). The double membrane structures of C. trachomatis forms are intact, which is an indication of faithful sample preservation. This preliminary but novel observation provides an important link to those observations made in vitro, indicating that MVs are a component of C. trachomatis infection in vivo in humans. Further examination of samples using this methodology is in progress to determine the relationship between MV formation and the bacterial growth state in natural human infections.
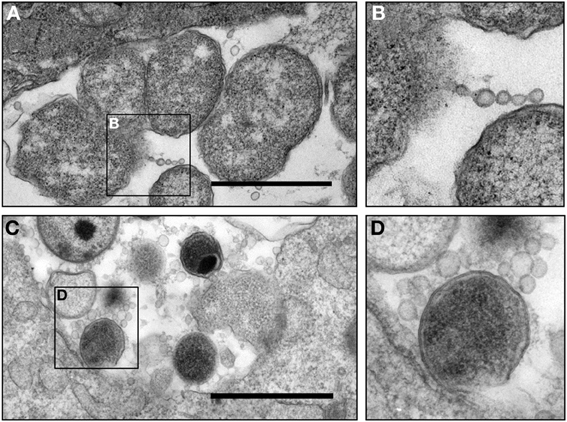
Figure 3. Micrograph of chlamydial MVs in cervical epithelial cells from C. trachomatis infected patients. (A,B) Presence of C. trachomatis vesicles associated with an aberrant C. trachomatis form in an inclusion. Vesicles appeared in the form of a chain or single round vesicular structures. This sample was obtained from a patient (patient 2) presented in Lewis et al. in this journal (Lewis et al., 2014); (C,D) presence of C. trachomatis MVs associated with a normal C. trachomatis form in an inclusion obtained from an epithelial cell from a second C. trachomatis infected patient. C. trachomatis infected human epithelial cells were collected by cytobrush from patients with an NAAT and culture positive C. trachomatis infection, and were immediately placed in a modified TEM fixative (Lewis et al., 2014) (currently under review for this Journal) and processed as previously described (Belland et al., 2003a). Scale bar = 500 nm.
The Importance of MVs During C. Trachomatis Infection
The best-studied mechanism that is related to OM changes in Gram-negative bacteria is the formation of MVs during both physiological adaptation and responses to envelope stress. Bacterial MVs are not only compositionally similar to the OM, containing LPS, phospholipids, and OM proteins, but also contain selective periplasmic or cytoplasmic components, such as toxins, DNAs, and RNAs, depending on the strain (Kadurugamuwa and Beveridge, 1999; Beveridge, 1999; Kuehn and Kesty, 2005). Research demonstrates the potential of MVs as vehicles of pathogenicity, as they can deliver complex bacterial molecules to target cells (Kuehn and Kesty, 2005; Bomberger et al., 2009; Amano et al., 2010; Elmi et al., 2012). The MVs also involve immune activation or suppression, stress responses, and attachment and internalization of the bacteria. Because of the cargo carrier nature and potent built-in adjuvanticity of most MVs studied to date, they are being utilized as vaccines (Collins, 2011; Unal et al., 2011). Engineered MVs in E. coli have exhibited a promising robust and tunable platform for the development of recombinant multivalent vaccines (Chen et al., 2010c; Bartolini et al., 2013).
Although chlamydial MVs were first observed over four decades ago (Matsumoto and Manire, 1970; Stirling and Richmond, 1980), the implications of these multi-functional MVs are only beginning to be elucidated (Giles et al., 2006; Giles and Wyrick, 2008; Wang et al., 2011a; Frohlich et al., 2012). A puzzle remains with regard to what role chlamydial MVs play during infection. Research data, including our own, suggest that the generation of chlamydial protein containing MVs occurs during productive infection and is enhanced by stress (Figures 2, 3) (Matsumoto and Manire, 1970; Giles et al., 2006; Giles and Wyrick, 2008; Wang et al., 2011a; Frohlich et al., 2012). The presence of MVs, not only during the β-lactam induced aberrant state, but also during both tryptophan starvation via IFNγ exposure and in vivo infection demonstrate a universal mechanism that appears to be responsive to stress rather than merely a by-product of dysfunctional membrane biogenesis during β-lactam exposure.
Protein Composition of Chlamydial MVs
A challenge to studying chlamydial MVs is the obligate intracellular lifestyle of C. trachomatis and the complexity of isolating vesicle populations derived from the pathogen as opposed to the host cells. To more comprehensively understand the properties of chlamydial MVs, we developed a method that permits efficient isolation and enrichment of intact chlamydial MVs for biochemical analysis (Frohlich et al., 2012). This approach uses a combination of nitrogen cavitation, magnetic bead immunological capture, and isopycnic centrifugation of C. trachomatis infected L929 cells. This method not only allows for the study of MVs, but also isolation of chlamydial forms, host organelles, and host cytoplasm. Therefore, a “snapshot” of the total host and chlamydial environments can be obtained from a single experiment.
The integrity of chlamydial MVs isolated by this approach was confirmed by TEM analysis (Figure 4) (Frohlich et al., 2012). MOMP and several important C. trachomatis cytotoxic and/or secreted proteins (CPAF, Pgp3, CT159, and CT166) are found to be associated with MVs induced by ampicillin as determined by immunoblotting analyses (Figure 4) (Frohlich et al., 2012). These data support that MVs are generated by C. trachomatis as a means of carrying and delivering chlamydial proteins or antigens. Since those factors identified as being associated with chlamydial MVs often also stimulate traditional secretion pathways, it is possible that MV delivery provides an alternative and specific mode of protein delivery. It has been proposed that translocation of chlamydial proteases, CPAF and HtrA, from the bacterial cytoplasm to the periplasm is Sec-dependent (Wu et al., 2011; Zhong, 2011), but secretion of these proteins to an extrabacterial location or the host cytosol is likely mediated by MVs. One could envision a stress or developmental cycle-specific alternative protein delivery system employed to further circumvent host processes designed to limit chlamydial growth. Interestingly, evidence supports the association of chlamydial protein-containing MVs with the endoplasmic reticulum (Giles and Wyrick, 2008; Frohlich et al., 2012). Considering the widespread strategy of chlamydial exploitation of host cellular machinery, it is likely that the delivery of chlamydial proteins to the host by MVs, at least in some cases, hijacks the host cells' own protein delivery systems to provide a targeted and specific localization of chlamydial proteins. Further experimentation is needed to confirm and understand these processes.
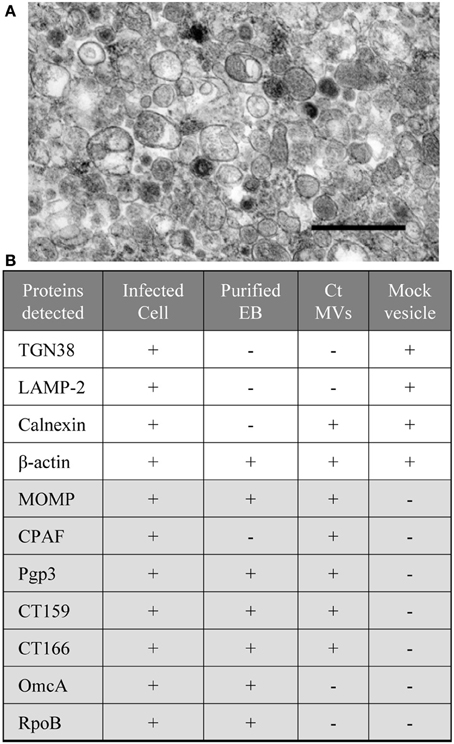
Figure 4. Compositional analysis of C. trachomatis MV fractions. (A) Micrographs of isolated MV fractions from C. trachomatis infected L929 cells by method described previously (Frohlich et al., 2012). At the ultrastructural level, the MVs show heterogeneity in their size and morphology. Scale bar = 500 nm; (B) C. trachomatis (Ct) or host cell proteins detected by immunoblotting with the lysates of infected cells, EBs, chlamydial MVs, and the control vesicles from mock infected cells. Note: coexistence of host ER marker (calnexin) with selective chamydial proteins (MOMP, CPAF, CT159, and CT160). Proteins were separated by 12% SDS-PAGE, followed by immunoblotting with antibodies to the protein of interest as described previously (Frohlich et al., 2012).
Potential Functions of Chlamydial MVs in Infected Host Cells—Cargo Delivery
Although the exact destination of MVs is difficult to determine, TEM in combination with biochemical and immunodetection analyses suggest that MVs can connect with bacterial cells, accumulate in the inclusion lumen, associate with the inclusion membrane, evert from but still associate with the inclusion, and be found beyond the confines of the inclusion (Giles et al., 2006; Giles and Wyrick, 2008). Previous studies have suggested that a subset of these vesicles are antigen-containing structures that emerge from the inclusion membrane to release antigens from the inclusion without the need for inclusion disruption (Richmond and Stirling, 1981; Giles et al., 2006). These vesicles appeared to contain inclusion membrane proteins (Incs) co-localized with C. trachomatis antigens, including MOMP, LPS, GroEL2, and GroEL3, but not GroEL1 (Giles et al., 2006). We found that the isolated MV fraction was rich in CPAF, Pgp3, CT159, and CT166 (Frohlich et al., 2012). The functions of these proteins relating to chlamydial virulence or modulating host cellular functions have been studied (Belland et al., 2001; Zhong et al., 2001; Chen et al., 2010b; Thalmann et al., 2010). It is possible that distinct subpopulations are trafficked through cellular machinery alongside host vesicles. Presumably, the release of bioactive contents mediated by MVs influences the fate of host cells. How does MV translocation across the inclusion membrane and cytosol and/or surface of the host cell occur? How do these chlamydial antigen-containing vesicles influence host cell signaling and antigen presentation? Is the generation of MVs eliciting a variety of specific and highly regulated adaptive responses to protect C. trachomatis from the offending stress, or do they modulate innate and/or adaptive immunity? Do MVs produced by C. trachomatis relate to exosome formation by cervical epithelial cells with C. trachomatis infection? Certainly, any one of these events alone or in concert would confer a selective advantage in adaptation to and survival in the ever-changing intracellular niche.
The Potential Role of MVs in the Innate Immune Response
The MVs derived from many Gram-negative bacteria are heavily laden with complexes of pathogen-associated molecular patterns, such as LPS, flagellin, CpG DNA, and virulence factors. These MVs are strongly recognized by the host immune system, resulting in the upregulation of pro-inflammatory cytokine secretion (Amano et al., 2010; Kulp and Kuehn, 2010). To determine if chlamydial MVs, induced by ampicillin, elicit an innate immune response, we investigated the impact of isolated MVs on the secretion of cytokines by epithelial cells. An immortalized human endocervical epithelial cell line (A2EN) infected with C. trachomatis was used as our model of infection. A2EN cells retain site appropriate expression of hormone receptors, responsiveness to exogenous hormone stimulation, expression of TLRs, and responsiveness to TLR agonists by secreting cytokines, chemokines, and anti-microbial peptides (Buckner et al., 2013).MVs isolated from C. trachomatis infected cell cultures exposed to ampicillin were added to monolayers of A2EN cells. Purified host cell vesicles without C. trachomatis infection, heat-killed Pseudomonas aeroginosa PAO1, and A2EN cells productively infected with chlamydial organisms were used as controls. Culture supernatants were collected at 24 and 48 h. Chlamydial MVs induced a modest, albeit statistically significant (p = 0.0230 based on performing a One Way ANOVA with a Bonferroni post-test), 2-fold increase in CXCL-8 at 24 h compared to the mock MV control, although, no significant increase of IL-6, TNF-α, or GM-CSF was observed (Figure 5). At 48 h, however, the differences observed were not significant as both chlamydial MVs and the mock vesicle control induced a similar increase in CXCL-8 and GM-CSF levels, perhaps due to the species differences between the mouse L929 derived vesicles and the human A2EN cells. As previously published, C. trachomatis infection in A2EN cells failed to upregulate secretion of CXCL8 (Buckner et al., 2013). Productive infection with C. trachomatis conducted at the same time as the MV experiments recapitulated these published observations and provided an additional control lending to the consistent response of A2EN cells across different laboratories (data not shown). These data suggest that MVs have more capacity to elicit an innate immune response than productive chlamydial infection, and that the molecules on the surface or contained within these vesicles may be structurally distinct compared to those present on or within whole chlamydial particles. The significance of such an early (at 24 h) but weak stimulatory effect on endocervical cells by chlamydial MVs remains to be determined. These results are clearly different from previously reported observations with MVs from other Gram-negative bacteria, as MVs derived from these other bacteria induce a robust and significant increase in pro-inflammatory cytokines and chemokines and induce antibody production (Amano et al., 2010; Nakao et al., 2011; Sharpe et al., 2011; Zhao et al., 2013). However, given the obligate intracellular nature of C. trachomatis, a robust immune response would not likely be favorable to continued or long-term infection. These results support other observations of immune-evasion tactics used by chlamydiae, even in the delivery of MVs.
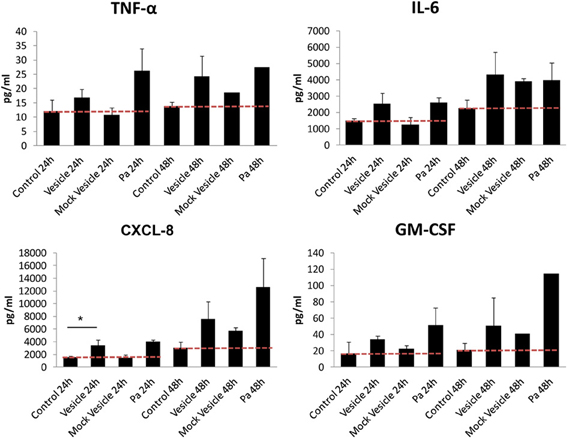
Figure 5. Cytokine response of A2EN cells to vesicles isolated from a C. trachomatis infection of L929 cells. MVs isolated from C. trachomatis infected cell cultures exposed to ampicillin (10 μg/ml) were added to a monolayer of A2EN cells. Purified host cell vesicles from a mock infection, heat-killed Pseudomonas aeroginosa PAO1 (Pa), and A2EN cells productively infected with chlamydial organisms were used as controls. Cytokine measurements were made using multiplex analyses as previously described (Buckner et al., 2013). *p = 0.0230. The red dashed line represents baseline control values.
The Potential Role of MVs in Exchange of Genetic Materials
It has been demonstrated that bacterial MV production provides a general advantage for survival and social networks in many bacteria species (Kuehn and Kesty, 2005; Amano et al., 2010; Bielig et al., 2011; Unal et al., 2011). Of great interest to the study of MVs is their participation in inter- and intra-species exchange of bacterial material in addition to their immuomodulatory functions (Kadurugamuwa and Beveridge, 1999; Renelli et al., 2004; Chiura et al., 2011; Barteneva et al., 2013). MVs may bind to bacterial or host cells, fuse with the target cell membrane, and deliver their cargo to the cytosol of the target cells. Recent evidence that protein and DNA transfer between distantly related species raises the prospect of a widely distributed mechanism of bacterial communication (Dubey and Ben-Yehuda, 2011). This poses an interesting potential for C. trachomatis given the vast sampling of both normal and pathogenic microorganisms potentially inhabiting the same niche in the genital tract and could potentially provide a significant adaptation and survival advantage.
While there is no evidence that chlamydial MVs deliver products to other species, it is possible that MVs could be used to deliver not only proteins but also genetic materials among chlamydiae. Recently, it has become evident that Chlamydia spp. have all the necessary recombination machinery despite an inclusion-confined lifestyle (Zhang et al., 1995; Demars et al., 2007; Joseph et al., 2011; Harris et al., 2012). Recombination has been documented between strains with different tissues tropisms, the lymphogranuloma venereum with the urogenital biovars, and within different genital strains in vitro and in vivo (Jeffrey et al., 2010, 2013; Harris et al., 2012). Whole genome sequencing analyses with a collection of diverse clinical isolates and laboratory strains have revealed that the recombination or mutation events often occur in several regions of the chromosome encoding surface-exposed proteins, such as MOMP, Pmps and the T3S system effector Tarp (Gomes et al., 2004; Brunelle and Sensabaugh, 2006; Gomes et al., 2006; Joseph et al., 2011; Harris et al., 2012). Different C. trachomatis strains can recombine following mixed infection for a relatively short time period in vitro (Jeffrey et al., 2013). Their potential capabilities for DNA exchange might explain, at least in part, the acquisition of virulence genes and fitness traits, as well as variations in these surface exposed proteins. Whether or not MVs directly contribute to the horizontal gene transfer between chlamydial organisms, as is observed in other bacteria, is unknown, but it begs the question: what impact does MV-mediated cell-to-cell communication have on pathogenesis and bacterial survival within the host? How might these MVs directly or indirectly affect the genital tract microbiome? Further addressing these questions will help in understanding the occurrence of antigen variation and diversity in C. trachomatis during infection and promote the development of novel genetic tools to study genetics and pathogenesis of C. trachomatis.
Molecular Mechanisms Controlling MV Formation in C. Trachomatis
Despite tremendous efforts, mechanisms by which bacteria produce MVs while sustaining their viability remain unclear. Based on studies in model organisms, including P. aeruginosa and E. coli, several mechanisms of MV formation have been proposed. First, the loss of OM-PG cross-links may allow excess lipid formation to induce MV formation. Second, accumulated periplasmic proteins simply push out the OM, trapping large amounts of protein inside the resulting vesicle. Third, the involvement of integral membrane proteins or signal molecules [such as Pseudomonas quorum sensing (PQS) molecules] induces membrane curvature and vesiculation (Schertzer and Whiteley, 2012). Fourth, the presence of rough LPS in bacteria contributes directly and indirectly to the formation of MVs (Kadurugamuwa and Beveridge, 1995; Sabra et al., 2003). Finally, genetic mechanisms may exist to regulate MV formation (McBroom et al., 2006; Schwechheimer et al., 2013). These MV inducing factors that have been proposed may not be mutually exclusive, and multiple dynamic surface components may contribute to MV production in bacteria (Kulp and Kuehn, 2010; Schwechheimer et al., 2013).
We know surprisingly little about OM biogenesis, and even less is known about the related vesicle formation in C. trachomatis. It is likely that after synthesis in the bacterial cytoplasm, chlamydial protein translocation to the OM involves complex steps conserved in Gram-negative bacteria (Hagan et al., 2010; Silhavy et al., 2010). These include: (i) translocation across the IM via the Sec pathway; (ii) folding facilitated by periplasmic chaperones/proteases, and (iii) assembly and insertion into the lipid phase of the OM, a process facilitated by a β-barrel assembly machine (BAM) complex that consists of Omp85 and its interacting lipoproteins (Ricci and Silhavy, 2012). Also necessary for maintenance of EB envelope integrity is the disulfide bond cross-linking of MOMP with OmcA and OmcB (Hatch, 1996).
We hypothesize that both bacterial and host factors contribute to the generation and release of MVs from C. trachomatis. We envision different mechanistic scenarios involved in MV formation during productive infection or the persistent growth state induced by ampicillin. In the case of productive infection, MV generation may reflect local OM deformation and/or a physiological turnover of envelope components because of envelope remodeling, a critical process in chlamydial development. In fact, RB multiplication and division require a rapid surface expansion or alteration achieved by the synthesis, assembly, and insertion of lipid or non-lipid OM components, while transition from a large RB to a small EB undergoes the opposite. In contrast, ampicillin induced hyper-vesiculation phenotypes may be the result of an envelope stress response. Exposure to ampicillin blocks RB division, although DNA replication remains. These can, in turn, result in attenuation of OM protein expression, accumulation of misfolded proteins in the periplasm, activation of proteolytic processes, and interference with the correct assembly and insertion of proteins in the OM. All of these are likely to induce heightened vesiculation. MV formation may offer a means to remove stress caused by the accumulated “toxic” waste or unfolded proteins. Chlamydial HtrA, a key player in envelope stress response (Huston et al., 2007, 2008; Zhong, 2011), is likely to play a role in controlling the formation of MVs, as is observed in other bacteria (McBroom et al., 2006). Given the strong capacity of chlamydial exploitation of host cellular machinery, MV cargo delivery may partially depend on a yet to be defined host trafficking pathway. Whether or not a potential host factor contributes to the formation of chlamydial MVs remains to be determined. Nevertheless, studying molecular mechanisms underlying envelope adaptation to development or stress offers a powerful tool and new route to understanding how C. trachomatis adapts to and survives in nutrient rich but hostile intracellular niches.
Summary and Perspective
The production of MVs in bacteria is a universal mechanism whereby virulence factors, signaling molecules, and genetic materials can be packaged and effectively transported to target cells. Major challenges in the field of vesicle research with obligate intracellular bacteria include (i) developing new comprehensive approaches for vesicle isolation and characterization, (ii) isolating pure populations from bacteria or host cells, and (iii) monitoring vesicle dynamics under physiologic relevant conditions. Nevertheless, the information already available indicates that MVs produced during chlamydial infection are present in cultured cell lines, primary human endocervical epithelial cells infected with C. trachomatis, and more importantly, in clinical specimens from C. trachomatis infected patients. These data provide preliminary evidence that the MV is a component of chlamydial infection in vivo. We hypothesize that C. trachomatis varies OM organization and produces MVs during the developmental cycle and in response to stress. Many mysteries remain with regard to how MVs contribute to host-pathogen interactions, the relationship of developmental envelope biogenesis within the context of MV formation and regulation, and the modes of MV cellular targeting and delivery. Improved understanding of the mechanisms of MV mediated mass material exchanges and the effects on C. trachomatis survival may have relevance to several important aspects of chlamydial biology, including the C. trachomatis persistent growth state in vivo. Recent progresses in chlamydial genetics may have great potential to inspire significant advance in MV studies in C. trachomatis (Wang et al., 2011b; Gérard et al., 2013; Gong et al., 2013; Song et al., 2013; Bauler and Hackstadt, 2014). Since global control of C. trachomatis infection will best be achieved with a vaccine, developing a greater understanding of the functional role and the mechanisms of envelope modification and MV production may open new therapeutic avenues against this medically important intracellular bacterium.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from NIH AI093565, LSU School of Medicine's Dean's Research Bridge Funding, and the Louisiana Vaccine Center and the South Louisiana Institute for Infectious Disease Research sponsored by the Louisiana Board of Regents. We thank Drs., Wandy Beatty for conducting all TEM experiments, Yanguang Cong for assistance in protein analysis, Guangming Zhong, Ted Hackstadt, You-xun Zhang, and Tom Hatch for valuable reagents, and Pris Wyrick and Kenneth Johnston for kind advice and helpful discussions. We apologize to those whose work we could not cover due to space limitations.
References
Abdelsamed, H., Peters, J., and Byrne, G. I. (2013). Genetic variation in Chlamydia trachomatis and their hosts: impact on disease severity and tissue tropism. Future Microbiol. 8, 1129–1146. doi: 10.2217/fmb.13.80
Aiyar, A., Quayle, A. J., Buckner, L. B., Sherchand, S. P., Chang, T. L., Zea, A. H., et al. (2014). Influence of the tryptophan-indole-IFNγ axis on human genital Chlamydia trachomatis infection: role of vaginal co-infections. Front. Cell. Infect. Microbiol. 4:72. doi: 10.3389/fcimb.2014.00072
Amano, A., Takeuchi, H., and Furuta, N. (2010). Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes Infect. 12, 791–798. doi: 10.1016/j.micinf.2010.05.008
Arno, J. N., Ricker, V. A., Batteiger, B. E., Katz, B. P., Caine, V. A., and Jones, R. B. (1990). Interferon-gamma in endocervical secretions of women infected with Chlamydia trachomatis. J. Infect. Dis. 162, 1385–1389.
Barteneva, N. S., Maltsev, N., and Vorobjev, I. A. (2013). Microvesicles and intercellular communication in the context of parasitism. Front. Cell. Infect. Microbiol. 3:49. doi: 10.3389/fcimb.2013.00049
Bartolini, E., Ianni, E., Frigimelica, E., Petracca, R., Galli, G., Berlanda Scorza, F., et al. (2013). Recombinant outer membrane vesicles carrying Chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J. Extracell. Vesicles 2:20181. doi: 10.3402/jev.v2i0.20181
Bauler, L. D., and Hackstadt, T. (2014). Expression and targeting of secreted proteins from Chlamydia trachomatis. J. Bacteriol. 196, 1325–1334. doi: 10.1128/JB.01290-13
Beatty, W., Byrne, G., and Morrison, R. (1993). Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. U.S.A. 90, 3998–4002. doi: 10.1073/pnas.90.9.3998
Beatty, W. L., Belanger, T. A., Desai, A. A., Morrison, R. P., and Byrne, G. I. (1994a). Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 62, 3705–3711.
Beatty, W. L., Morrison, R. P., and Byrne, G. I. (1994b). Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58, 686–699.
Belland, R. J., Nelson, D. E., Virok, D., Crane, D. D., Hogan, D., Sturdevant, D., et al. (2003a). Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. U.S.A. 100, 15971–15976. doi: 10.1073/pnas.2535394100
Belland, R. J., Scidmore, M. A., Crane, D. D., Hogan, D. M., Whitmire, W., Mcclarty, G., et al. (2001). Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. U.S.A. 98, 13984–13989. doi: 10.1073/pnas.241377698
Belland, R. J., Zhong, G., Crane, D. D., Hogan, D., Sturdevant, D., Sharma, J., et al. (2003b). Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U.S.A. 100, 8478–8483. doi: 10.1073/pnas.1331135100
Beveridge, T. J. (1999). Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733.
Bielig, H., Dongre, M., Zurek, B., Wai, S. N., and Kufer, T. A. (2011). A role for quorum sensing in regulating innate immune responses mediated by Vibrio cholerae outer membrane vesicles (OMVs). Gut Microbes 2, 274–279. doi: 10.4161/gmic.2.5.18091
Bomberger, J. M., Maceachran, D. P., Coutermarsh, B. A., Ye, S., O'toole, G. A., and Stanton, B. A. (2009). Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5:e1000382. doi: 10.1371/journal.ppat.1000382
Bragina, E. Y., Gomberg, M. A., and Dmitriev, G. A. (2001). Electron microscopic evidence of persistent chlamydial infection following treatment. J. Eur. Acad. Dermatol. Venereol. 15, 405–409. doi: 10.1046/j.1468-3083.2001.00342.x
Brunelle, B. W., and Sensabaugh, G. F. (2006). The ompA gene in Chlamydia trachomatis differs in phylogeny and rate of evolution from other regions of the genome. Infect. Immun. 74, 578–585. doi: 10.1128/IAI.74.1.578-585.2006
Buckner, L. R., Lewis, M. E., Greene, S. J., Foster, T. P., and Quayle, A. J. (2013). Chlamydia trachomatis infection results in a modest pro-inflammatory cytokine response and a decrease in T cell chemokine secretion in human polarized endocervical epithelial cells. Cytokine 63, 151–165. doi: 10.1016/j.cyto.2013.04.022
Byrne, G. I. (2001). Chlamydial treatment failures: a persistent problem? J. Eur. Acad. Dermatol. Venereol. 15, 381–381. doi: 10.1046/j.1468-3083.2001.00343.x
Caldwell, H. D., Kromhout, J., and Schachter, J. (1981). Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31, 1161–1176.
Carrasco, J. A., Tan, C., Rank, R. G., Hsia, R. C., and Bavoil, P. M. (2011). Altered developmental expression of polymorphic membrane proteins in penicillin-stressed Chlamydia trachomatis. Cell. Microbiol. 13, 1014–1025. doi: 10.1111/j.1462-5822.2011.01598.x
Chen, D., Lei, L., Lu, C., Flores, R., Delisa, M. P., Roberts, T. C., et al. (2010a). Secretion of the chlamydial virulence factor CPAF requires the Sec-dependent pathway. Microbiology 156, 3031–3040. doi: 10.1099/mic.0.040527-0
Chen, D., Lei, L., Lu, C., Galaleldeen, A., Hart, P. J., and Zhong, G. (2010b). Characterization of Pgp3, a Chlamydia trachomatis plasmid-encoded immunodominant antigen. J. Bacteriol. 192, 6017–6024. doi: 10.1128/JB.00847-10
Chen, D. J., Osterrieder, N., Metzger, S. M., Buckles, E., Doody, A. M., Delisa, M. P., et al. (2010c). Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc. Natl. Acad. Sci. U.S.A. 107, 3099–3104. doi: 10.1073/pnas.0805532107
Chiura, H. X., Kogure, K., Hagemann, S., Ellinger, A., and Velimirov, B. (2011). Evidence for particle-induced horizontal gene transfer and serial transduction between bacteria. FEMS Microbiol. Ecol. 76, 576–591. doi: 10.1111/j.1574-6941.2011.01077.x
Collins, B. S. (2011). Gram-negative outer membrane vesicles in vaccine development. Discov. Med. 12, 7–15.
Crane, D. D., Carlson, J. H., Fischer, E. R., Bavoil, P., Hsia, R. C., Tan, C., et al. (2006). Chlamydia trachomatis polymorphic membrane protein D is a species-common pan-neutralizing antigen. Proc. Natl. Acad. Sci. U.S.A. 103, 1894–1899. doi: 10.1073/pnas.0508983103
Darville, T., and Hiltke, T. (2010). Pathogenesis of genital tract disease due to Chlamydia trachomatis. J. Infect. Dis. S114–S125. doi: 10.1086/652397
Demars, R., Weinfurter, J., Guex, E., Lin, J., and Potucek, Y. (2007). Lateral gene transfer in vitro in the intracellular pathogen Chlamydia trachomatis. J. Bacteriol. 189, 991–1003. doi: 10.1128/JB.00845-06
Dubey, G. P., and Ben-Yehuda, S. (2011). Intercellular nanotubes mediate bacterial communication. Cell 144, 590–600. doi: 10.1016/j.cell.2011.01.015
Elmi, A., Watson, E., Sandu, P., Gundogdu, O., Mills, D. C., Inglis, N. F., et al. (2012). Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect. Immun. 80, 4089–4098. doi: 10.1128/IAI.00161-12
Engel, J. N. (1992). Azithromycin-induced block of elementary body formation in Chlamydia trachomatis. Antimicrob. Agents Chemother. 36, 2304–2309. doi: 10.1128/AAC.36.10.2304
Feher, V. A., Randall, A., Baldi, P., Bush, R. M., De La Maza, L. M., and Amaro, R. E. (2013). A 3-dimensional trimeric beta-barrel model for Chlamydia MOMP contains conserved and novel elements of Gram-negative bacterial porins. PLoS ONE 8:e68934. doi: 10.1371/journal.pone.0068934
Fields, K. A., and Hackstadt, T. (2002). The chlamydial inclusion: escape from the endocytic pathway1. Annu. Rev. Cell Dev. Biol. 18, 221–245. doi: 10.1146/annurev.cellbio.18.012502.105845
Frohlich, K., Hua, Z., Wang, J., and Shen, L. (2012). Isolation of Chlamydia trachomatis and membrane vesicles derived from host and bacteria. J. Microbiol. Methods 91, 222–230. doi: 10.1016/j.mimet.2012.08.012
Gerard, H. C., Carter, J. D., and Hudson, A. P. (2013). Chlamydia trachomatis is present and metabolically active during the remitting phase in synovial tissues from patients with chronic Chlamydia-induced reactive arthritis. Am. J. Med. Sci. 346, 22–25. doi: 10.1097/MAJ.0b013e3182648740
Gerard, H. C., Krausse-Opatz, B., Wang, Z., Rudy, D., Rao, J. P., Zeidler, H., et al. (2001). Expression of Chlamydia trachomatis genes encoding products required for DNA synthesis and cell division during active versus persistent infection. Mol. Microbiol. 41, 731–741. doi: 10.1046/j.1365-2958.2001.02550.x
Gérard, H. C., Mishra, M. K., Mao, G., Wang, S., Hali, M., Whittum-Hudson, J. A., et al. (2013). Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomedicine 9, 996–1008. doi: 10.1016/j.nano.2013.04.004
Giles, D. K., Whittimore, J. D., Larue, R. W., Raulston, J. E., and Wyrick, P. B. (2006). Ultrastructural analysis of chlamydial antigen-containing vesicles everting from the Chlamydia trachomatis inclusion. Microbes Infect. 8, 1579–1591. doi: 10.1016/j.micinf.2006.01.018
Giles, D. K., and Wyrick, P. B. (2008). Trafficking of chlamydial antigens to the endoplasmic reticulum of infected epithelial cells. Microbes Infect. 10, 1494–1503. doi: 10.1016/j.micinf.2008.09.001
Gomes, J. P., Bruno, W. J., Borrego, M. J., and Dean, D. (2004). Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 186, 4295–4306. doi: 10.1128/JB.186.13.4295-4306.2004
Gomes, J. P., Nunes, A., Bruno, W. J., Borrego, M. J., Florindo, C., and Dean, D. (2006). Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J. Bacteriol. 188, 275–286. doi: 10.1128/JB.188.1.275-286.2006
Gong, S., Yang, Z., Lei, L., Shen, L., and Zhong, G. (2013). Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J. Bacteriol. 195, 3819–3826. doi: 10.1128/JB.00511-13
Hackstadt, T., Todd, W. J., and Caldwell, H. D. (1985). Disulfide-mediated interactions of the chlamydial major outer membrane protein: role in the differentiation of chlamydiae? J. Bacteriol. 161, 25–31.
Hafner, L. M., Wilson, D. P., and Timms, P. (2014). Development status and future prospects for a vaccine against Chlamydia trachomatis infection. Vaccine 32, 1563–1571. doi: 10.1016/j.vaccine.2013.08.020
Hagan, C. L., Kim, S., and Kahne, D. (2010). Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892. doi: 10.1126/science.1188919
Harper, A., Pogson, C. I., Jones, M. L., and Pearce, J. H. (2000). Chlamydial development is adversely affected by minor changes in amino acid supply, blood plasma amino acid levels, and glucose deprivation. Infect. Immun. 68, 1457–1464. doi: 10.1128/IAI.68.3.1457-1464.2000
Harris, S. R., Clarke, I. N., Seth-Smith, H. M., Solomon, A. W., Cutcliffe, L. T., Marsh, P., et al. (2012). Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat. Genet. 44, S1. doi: 10.1038/ng.2214
Hatch, T. P. (1996). Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 178, 1–5.
Herbst-Kralovetz, M. M., Quayle, A. J., Ficarra, M., Greene, S., Rose, W. A., Chesson, R., et al. (2008). Quantification and comparison of toll-like receptor expression and responsiveness in primary and immortalized human female lower genital tract epithelia. Am. J. Reprod. Immunol. 59, 212–224. doi: 10.1111/j.1600-0897.2007.00566.x
Heuer, D., Brinkmann, V., Meyer, T. F., and Szczepek, A. J. (2003). Expression and translocation of chlamydial protease during acute and persistent infection of the epithelial HEp-2 cells with Chlamydophila (Chlamydia) pneumoniae. Cell. Microbiol. 5, 315–322. doi: 10.1046/j.1462-5822.2003.00278.x
Hogan, R. J., Mathews, S. A., Mukhopadhyay, S., Summersgill, J. T., and Timms, P. (2004). Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 72, 1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004
Hsia, R., Ohayon, H., Gounon, P., Dautry-Varsat, A., and Bavoil, P. M. (2000). Phage infection of the obligate intracellular bacterium, Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect. 2, 761–772. doi: 10.1016/S1286-4579(00)90356-3
Hurdle, J. G., O'neill, A. J., Chopra, I., and Lee, R. E. (2011). Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 9, 62–75. doi: 10.1038/nrmicro2474
Huston, W., Swedberg, J., Harris, J., Walsh, T., Mathews, S., and Timms, P. (2007). The temperature activated HtrA protease from pathogen Chlamydia trachomatis acts as both a chaperone and protease at 37degreesC. FEBS Lett. 581, 3382–3386. doi: 10.1016/j.febslet.2007.06.039
Huston, W., Theodoropoulos, C., Mathews, S., and Timms, P. (2008). Chlamydia trachomatis responds to heat shock, penicillin induced persistence, and IFN-gamma persistence by altering levels of the extracytoplasmic stress response protease HtrA. BMC Microbiol. 8:190. doi: 10.1186/1471-2180-8-190
Jeffrey, B. M., Suchland, R. J., Eriksen, S. G., Sandoz, K. M., and Rockey, D. D. (2013). Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol. 13:142. doi: 10.1186/1471-2180-13-142
Jeffrey, B. M., Suchland, R. J., Quinn, K. L., Davidson, J. R., Stamm, W. E., and Rockey, D. D. (2010). Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect. Immun. 78, 2544–2553. doi: 10.1128/IAI.01324-09
Joseph, S. J., Didelot, X., Gandhi, K., Dean, D., and Read, T. D. (2011). Interplay of recombination and selection in the genomes of Chlamydia trachomatis. Biol. Direct 6, 28. doi: 10.1186/1745-6150-6-28
Kadurugamuwa, J. L., and Beveridge, T. J. (1995). Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177, 3998–4008.
Kadurugamuwa, J. L., and Beveridge, T. J. (1999). Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other Gram-negative bacteria. Microbiology 145, 2051–2060. doi: 10.1099/13500872-145-8-2051
Kubo, A., and Stephens, R. S. (2000). Characterization and functional analysis of PorB, a Chlamydia porin and neutralizing target. Mol. Microbiol. 38, 772–780. doi: 10.1046/j.1365-2958.2000.02167.x
Kuehn, M. J., and Kesty, N. C. (2005). Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19, 2645–2655. doi: 10.1101/gad.1299905
Kulp, A., and Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184. doi: 10.1146/annurev.micro.091208.073413
Lewis, M. E., Belland, R. J., Abdelrahman, Y. M., Beatty, W., Aiyar, A. A., Zea, A. H., et al. (2014). Morphologic and molecular evaluation of Chlamydia trachomatis growth in the human endocervix reveals distinct growth patterns. Front. Cell. Infect. Microbiol. 4:71. doi: 10.3389/fcimb.2014.00071
Liechti, G. W., Kuru, E., Hall, E., Kalinda, A., Brun, Y. V., Vannieuwenhze, M., et al. (2014). A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510. doi: 10.1038/nature12892
Liu, X., Afrane, M., Clemmer, D. E., Zhong, G., and Nelson, D. E. (2010). Identification of Chlamydia trachomatis outer membrane complex proteins by differential proteomics. J. Bacteriol. 192, 2852–2860. doi: 10.1128/JB.01628-09
Lo, C.-C., Xie, G., Bonner, C. A., and Jensen, R. A. (2012). The alternativetranslational profile that underlies the immune-evasive state of persistence in chlamydiaceae exploits differential tryptophan contents of the protein repertoire. Microbiol. Mol. Biol. Rev. 76, 405–443. doi: 10.1128/MMBR.05013-11
Matsumoto, A., and Manire, G. P. (1970). Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J. Bacteriol. 101, 278–285.
Mazzoli, S., Bani, D., Salvi, A., Ramacciotti, I., Romeo, C, and Bani, T. (2000). “In vivo evidence of Chlamydia trachomatis miniature reticulary bodies (MRB) as persistence markers in patients with chronic chlamydial prostatitis,” in Proceedings- European Society For Chlamydia Research, 40.
McBroom, A. J., Johnson, A. P., Vemulapalli, S., and Kuehn, M. J. (2006). Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188, 5385–5392. doi: 10.1128/JB.00498-06
McClarty, G., Caldwell, H. D., and Nelson, D. E. (2007). Chlamydial interferon gamma immune evasion influences infection tropism. Curr. Opin. Microbiol. 10, 47–51. doi: 10.1016/j.mib.2006.12.003
McCoy, A. J., and Maurelli, A. T. (2006). Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol. 14, 70–77. doi: 10.1016/j.tim.2005.12.004
Molestina, R. E., Klein, J. B., Miller, R. D., Pierce, W. H., Ramirez, J. A., and Summersgill, J. T. (2002). Proteomic analysis of differentially expressed Chlamydia pneumoniae genes during persistent infection of HEp-2 cells. Infect. Immun. 70, 2976–2981. doi: 10.1128/IAI.70.6.2976-2981.2002
Moulder, J. W. (1991). Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55, 143–190.
Mueller, K. E., Plano, G. V., and Fields, K. A. (2014). New frontiers in type iii secretion biology: the Chlamydia perspective. Infect. Immun. 82, 2–9. doi: 10.1128/IAI.00917-13
Nakao, R., Hasegawa, H., Ochiai, K., Takashiba, S., Ainai, A., Ohnishi, M., et al. (2011). Outer membrane vesicles of Porphyromonas gingivalis elicit a mucosal immune response. PLoS ONE 6:e26163. doi: 10.1371/journal.pone.0026163
Nanagara, R., Li, F., Beutler, A., Hudson, A., and Schumacher, H. R. Jr. (1995). Alteration of Chlamydia trachomatis biologic behavior in synovial membranes. Suppression of surface antigen production in reactive arthritis and Reiter's syndrome. Arthritis Rheum. 38, 1410–1417. doi: 10.1002/art.1780381008
Ouellette, S. P., Hatch, T. P., Abdelrahman, Y. M., Rose, L. A., Belland, R. J., and Byrne, G. I. (2006). Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol. Microbiol. 62, 1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x
Ouellette, S. P., Karimova, G., Subtil, A., and Ladant, D. (2012). Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol. Microbiol. 85, 164–178. doi: 10.1111/j.1365-2958.2012.08100.x
Patton, D., Askienazy-Elbhar, M., Henry-Suchet, J., Campbell, L. A., Cappuccio, A., Tannous, W., et al. (1994). Detection of Chlamydia trachomatis in fallopian tube tissue in women with postinfectious tubal infertility. Am. J. Obstet. Gynecol. 171, 95–101. doi: 10.1016/S0002-9378(94)70084-2
Phillips-Campbell, R., Kintner, J., and Schoborg, R. V. (2014). Induction of the Chlamydia muridarum stress/persistence response increases azithromycin treatment failure in a murine model of infection. Antimicrob. Agents Chemother. 58, 1782–1784. doi: 10.1128/AAC.02097-13
Pilhofer, M., Aistleitner, K., Biboy, J., Gray, J., Kuru, E., Hall, E., et al. (2013). Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat. Commun. 4:2856. doi: 10.1038/ncomms3856
Prados-Rosales, R., Baena, A., Martinez, L. R., Luque-Garcia, J., Kalscheuer, R., Veeraraghavan, U., et al. (2011). Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J. Clin. Invest. 121, 1471–1483. doi: 10.1172/JCI44261
Prusty, B. K., Böhme, L., Bergmann, B., Siegl, C., Krause, E., Mehlitz, A., et al. (2012). Imbalanced oxidative stress causes chlamydial persistence during non-productive human herpes virus co-infection. PLoS ONE 7:e47427. doi: 10.1371/journal.pone.0047427
Raulston, J. E. (1995). Chlamydial envelope components and pathogen-host cell interactions. Mol. Microbiol. 15, 607–616. doi: 10.1111/j.1365-2958.1995.tb02370.x
Raulston, J. E. (1997). Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect. Immun. 65, 4539–4547.
Rekart, M. L., Gilbert, M., Meza, R., Kim, P. H., Chang, M., Money, D. M., et al. (2013). Chlamydia public health programs and the epidemiology of pelvic inflammatory disease and ectopic pregnancy. J. Infect. Dis. 207, 30–38. doi: 10.1093/infdis/jis644
Renelli, M., Matias, V., Lo, R. Y., and Beveridge, T. J. (2004). DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150, 2161–2169. doi: 10.1099/mic.0.26841-0
Reveneau, N., Crane, D. D., Fischer, E., and Caldwell, H. D. (2005). Bactericidal activity of first-choice antibiotics against gamma interferon-induced persistent infection of human epithelial cells by Chlamydia trachomatis. Antimicrob. Agents Chemother. 49, 1787–1793. doi: 10.1128/AAC.49.5.1787-1793.2005
Ricci, D. P., and Silhavy, T. J. (2012). The bam machine: a molecular cooper. Biochim. Biophys. Acta 1818, 1067–1084. doi: 10.1016/j.bbamem.2011.08.020
Richmond, S. J., and Stirling, P. (1981). Localization of chlamydial group Antigen in McCoy cell monolayers infected with Chlamydia trachomatis or Chlamydia psittaci. Infect. Immun. 34, 561–570.
Romano, J. D., De Beaumont, C., Carrasco, J. A., Ehrenman, K., Bavoil, P. M., and Coppens, I. (2013). Fierce competition between Toxoplasma and Chlamydia for host cell structures in dually infected cells. Eukaryotic Cell 12, 265–277. doi: 10.1128/EC.00313-12
Sabra, W., Lünsdorf, H., and Zeng, A.-P. (2003). Alterations in the formation of lipopolysaccharide and membrane vesicles on the surface of Pseudomonas aeruginosa PAO1 under oxygen stress conditions. Microbiology 149, 2789–2795. doi: 10.1099/mic.0.26443-0
Schautteet, K., De Clercq, E., and Vanrompay, D. (2011). Chlamydia trachomatis vaccine research through the years. Infect. Dis. Obstet. Gynecol. 2011:963513. doi: 10.1155/2011/963513
Schertzer, J. W., and Whiteley, M. (2012). A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 3. doi: 10.1128/mBio.00297-11
Schoborg, R. V. (2011). Chlamydia persistence—a tool to dissect chlamydia–host interactions. Microbes Infect. 13, 649–662. doi: 10.1016/j.micinf.2011.03.004
Schwechheimer, C., Sullivan, C. J., and Kuehn, M. J. (2013). Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry 52, 3031–3040. doi: 10.1021/bi400164t
Sharpe, S. W., Kuehn, M. J., and Mason, K. M. (2011). Elicitation of epithelial cell-derived immune effectors by outer membrane vesicles of nontypeable Haemophilus influenzae. Infect. Immun. 79, 4361–4369. doi: 10.1128/IAI.05332-11
Shaw, A. C., Vandahl, B. B., Larsen, M. R., Roepstorff, P., Gevaert, K., Vandekerckhove, J., et al. (2002). Characterization of a secreted Chlamydia protease. Cell. Microbiol. 4, 411–424. doi: 10.1046/j.1462-5822.2002.00200.x
Silhavy, T. J., Kahne, D., and Walker, S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. doi: 10.1101/cshperspect.a000414
Song, L., Carlson, J. H., Whitmire, W. M., Kari, L., Virtaneva, K., Sturdevant, D. E., et al. (2013). Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect. Immun. 81, 636–644. doi: 10.1128/IAI.01305-12
Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., et al. (1998). Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759. doi: 10.1126/science.282.5389.754
Stephens, R. S., and Lammel, C. J. (2001). Chlamydia outer membrane protein discovery using genomics. Curr. Opin. Microbiol. 4, 16–20. doi: 10.1016/S1369-5274(00)00158-2
Stirling, P., and Richmond, S. J. (1980). Production of outer membrane blebs during chlamydial replication. FEMS Microbiol. Lett. 9, 1574–6968. doi: 10.1111/j.1574-6968.1980.tb05616.x
Sun, G., Pal, S., Sarcon, A. K., Kim, S., Sugawara, E., Nikaido, H., et al. (2007). Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis. J. Bacteriol. 189, 6222–6235. doi: 10.1128/JB.00552-07
Tan, C., Hsia, R.-C., Shou, H., Carrasco, J. A., Rank, R. G., and Bavoil, P. M. (2010). Variable expression of surface-exposed polymorphic membrane proteins in in vitro-grown Chlamydia trachomatis. Cell. Microbiol. 12, 174–187. doi: 10.1111/j.1462-5822.2009.01389.x
Tan, C., Hsia, R. C., Shou, H., Haggerty, C. L., Ness, R. B., Gaydos, C. A., et al. (2009). Chlamydia trachomatis-infected patients display variable antibody profiles against the nine-member polymorphic membrane protein family. Infect. Immun. 77, 3218–3226. doi: 10.1128/IAI.01566-08
Tanzer, R. J., and Hatch, T. P. (2001). Characterization of outer membrane proteins in Chlamydia trachomatis LGV serovar L2. J. Bacteriol. 183, 2686–2690. doi: 10.1128/JB.183.8.2686-2690.2001
Taylor, B. D., Darville, T., Tan, C., Bavoil, P. M., Ness, R. B., and Haggerty, C. L. (2011). The role of Chlamydia trachomatis polymorphic membrane proteins in inflammation and sequelae among women with pelvic inflammatory disease. Infect. Dis. Obstet. Gynecol. 2011:989762. doi: 10.1155/2011/989762
Thalmann, J., Janik, K., May, M., Sommer, K., Ebeling, J., Hofmann, F., et al. (2010). Actin re-organization induced by Chlamydia trachomatis serovar D—evidence for a critical role of the effector protein CT166 targeting Rac. PLoS ONE 5:e9887. doi: 10.1371/journal.pone.0009887
Unal, C. M., Schaar, V., and Riesbeck, K. (2011). Bacterial outer membrane vesicles in disease and preventive medicine. Semin. Immunopathol. 33, 395–408. doi: 10.1007/s00281-010-0231-y
Valdivia, R. H. (2008). Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11, 53–59. doi: 10.1016/j.mib.2008.01.003
Vanover, J., Sun, J., Deka, S., Kintner, J., Duffourc, M. M., and Schoborg, R. V. (2008). Herpes simplex virus co-infection-induced Chlamydia trachomatis persistence is not mediated by any known persistence inducer or anti-chlamydial pathway. Microbiology 154, 971–978. doi: 10.1099/mic.0.2007/012161-0
Wang, J., Frohlich, K. M., Buckner, L., Quayle, A. J., Luo, M., Feng, X., et al. (2011a). Altered protein secretion of Chlamydia trachomatis in persistently infected human endocervical epithelial cells. Microbiology 157, 2759–2771. doi: 10.1099/mic.0.044917-0
Wang, J., Zhang, Y., Lu, C., Lei, L., Yu, P., and Zhong, G. (2010). A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J. Immunol. 185, 1670–1680. doi: 10.4049/jimmunol.1001240
Wang, Y., Berg, E. A., Feng, X., Shen, L., Smith, T., Costello, C. E., et al. (2006). Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci. 15, 122–134. doi: 10.1110/ps.051616206
Wang, Y., Kahane, S., Cutcliffe, L. T., Skilton, R. J., Lambden, P. R., and Clarke, I. N. (2011b). Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 7:e1002258. doi: 10.1371/journal.ppat.1002258
World Health Organization. D.O.R.H.A.R. (2011). Prevalence and Incidence of Selected Sexually Transmitted Infections. Geneva: World Health Organization.
Wu, X., Lei, L., Gong, S., Chen, D., Flores, R., and Zhong, G. (2011). The chlamydial periplasmic stress response serine protease cHtrA is secreted into host cell cytosol. BMC Microbiol. 11:87. doi: 10.1186/1471-2180-11-87
Wyrick, P. B. (2010). Chlamydia trachomatis persistence in vitro: an overview. J. Infect. Dis. 201, S88–S95. doi: 10.1086/652394
Wyrick, P. B., and Knight, S. T. (2004). Pre-exposure of infected human endometrial epithelial cells to penicillin in vitro renders Chlamydia trachomatis refractory to azithromycin. J. Antimicrob. Chemother. 54, 79–85. doi: 10.1093/jac/dkh283
Zhang, D. J., Fan, H., Mcclarty, G., and Brunham, R. C. (1995). Identification of the Chlamydia trachomatis RecA-encoding gene. Infect. Immun. 63, 676–680.
Zhao, K., Deng, X., He, C., Yue, B., and Wu, M. (2013). Pseudomonas aeruginosa outer membrane vesicles modulate host immune responses by targeting the toll-like receptor 4 signaling pathway. Infect. Immun. 81, 4509–4518. doi: 10.1128/IAI.01008-13
Zhong, G. (2011). Chlamydia trachomatis secretion of proteases for manipulating host signaling pathways. Front. Microbiol. 2:14. doi: 10.3389/fmicb.2011.00014
Keywords: Chlamydia trachomatis, membrane vesicles, adaptive response, persistent growth state, stress
Citation: Frohlich KM, Hua Z, Quayle AJ, Wang J, Lewis ME, Chou C-W, Luo M, Buckner LR and Shen L (2014) Membrane vesicle production by Chlamydia trachomatis as an adaptive response. Front. Cell. Infect. Microbiol. 4:73. doi: 10.3389/fcimb.2014.00073
Received: 21 February 2014; Accepted: 19 May 2014;
Published online: 10 June 2014.
Edited by:
Nicole Borel, University of Zurich, SwitzerlandReviewed by:
Priscilla B. Wyrick, East Tennessee State University College of Medicine, USAJennifer Hall, East Tennessee State University, USA
Copyright © 2014 Frohlich, Hua, Quayle, Wang, Lewis, Chou, Luo, Buckner and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Shen, Department of Microbiology Immunology and Parasitology, Louisiana State University Health Sciences Center, New Orleans, LA 70112, USA e-mail: lshen@lsuhsc.edu
†Present address: Kyla M. Frohlich, Department of Biological Sciences, The RNA Institute, University at Albany, SUNY, Albany, USA
 Kyla M. Frohlich1†
Kyla M. Frohlich1†  Alison J. Quayle
Alison J. Quayle Jin Wang
Jin Wang Maria E. Lewis
Maria E. Lewis Miao Luo
Miao Luo Lyndsey R. Buckner
Lyndsey R. Buckner Li Shen
Li Shen