Defining the metabolic requirements for the growth and colonization capacity of Campylobacter jejuni
- Hannover Medical School, Institute for Medical Microbiology and Hospital Epidemiology, Hannover, Germany
During the last decade Campylobacter jejuni has been recognized as the leading cause of bacterial gastroenteritis worldwide. This facultative intracellular pathogen is a member of the Epsilonproteobacteria and requires microaerobic atmosphere and nutrient rich media for efficient proliferation in vitro. Its catabolic capacity is highly restricted in contrast to Salmonella Typhimurium and other enteropathogenic bacteria because several common pathways for carbohydrate utilization are either missing or incomplete. Despite these metabolic limitations, C. jejuni efficiently colonizes various animal hosts as a commensal intestinal inhabitant. Moreover, C. jejuni is tremendously successful in competing with the human intestinal microbiota; an infectious dose of few hundreds bacteria is sufficient to overcome the colonization resistance of humans and can lead to campylobacteriosis. Besides the importance and clear clinical manifestation of this disease, the pathogenesis mechanisms of C. jejuni infections are still poorly understood. In recent years comparative genome sequence, transcriptome and metabolome analyses as well as mutagenesis studies combined with animal infection models have provided a new understanding of how the specific metabolic capacity of C. jejuni drives its persistence in the intestinal habitat of various hosts. Furthermore, new insights into the metabolic requirements that support the intracellular survival of C. jejuni were obtained. Because C. jejuni harbors distinct properties in establishing an infection in comparison to pathogenic Enterobacteriaceae, it represents an excellent organism for elucidating new aspects of the dynamic interaction and metabolic cross talk between a bacterial pathogen, the microbiota and the host.
Introduction
Campylobacteriosis has emerged as major bacterial food-borne disease in industrialized countries in recent years (Epps et al., 2013). Mainly C. jejuni is associated with acute Campylobacter enteritis in humans causing more than 80% of the registered Campylobacter infections. Predominant sources of infections are contaminated meat (especially chicken), raw milk and water. The clinical manifestations of Campylobacter enteritis are indistinguishable from Salmonellosis and range from mild watery to severe, inflammatory and bloody diarrhea accompanied with abdominal pain and fever (Allos, 2001). Such variations in the disease outcome might correlate with the well documented different virulence potential of individual C. jejuni isolates and are possibly linked to dissimilarities in motility and surface structures involved in the direct interaction with the host. These variable structures include the lipooligosaccharide, the capsule and the glycosylation pattern of the flagellin (Wilson et al., 2010). Interestingly, not only variable surface structures but also metabolic traits are highly variable between C. jejuni isolates: Genes supporting the oxygen-independent respiration and the catabolism of amino acids and peptides are particular over-represented in robust-colonizing strains compared to poor-colonizing strains (Ahmed et al., 2002; Hofreuter et al., 2006; Hepworth et al., 2007; Seal et al., 2007; Hiett et al., 2008). Such observations suggest that the physiological properties of C. jejuni play a crucial role for its pathogenesis. While some aspects of C. jejuni pathogenesis have been described in detail previously (Young et al., 2007; Janssen et al., 2008; van Putten et al., 2009; Gilbreath et al., 2011; Szymanski and Gaynor, 2012), this review specifically summarizes our increasing knowledge about the in vitro and in vivo metabolism of C. jejuni and its impact on the virulence and colonization process of this important pathogen.
General Physiological Properties of C. jejuni
Though C. jejuni shows fastidious growth characteristics in vitro and easily looses viability as well as culturability, it resides as a commensal in a wide range of diverse animal hosts (e.g., chicken, cattle, sheep, goat, dog, duck and pig) and can be isolated from various environmental sources or refrigerated foods. This life style implies that C. jejuni is able to resist varying temperatures, oxygen concentrations, pH values, osmotic environments and nutrient availabilities as reviewed comprehensively elsewhere (Park, 2002; Murphy et al., 2006). C. jejuni belongs to the group of thermophilic Campylobacter species that grow preferentially between 42°C and 37°C, but do not proliferate below 30°C (Penner, 1988). It was suggested that the absence of cold-shock proteins might be responsible for the inability of this pathogen to grow at lower temperatures (Hazeleger et al., 1998). Nonetheless, C. jejuni shows respiration and ATP generation at temperatures as low as 4°C, maintaining its metabolic activities at low temperatures for an extended time period (Hazeleger et al., 1998). C. jejuni survives at 4°C even better than at 25°C (Blaser et al., 1980), which leads to the common problem that refrigerated meat contaminated with C. jejuni during the slaughter process represents a particular frequent source of C. jejuni infections (Bhaduri and Cottrell, 2004). While C. jejuni is able to withstand low temperature, atmospheric oxygen concentration affects its viability dramatically. The microaerophilic nature of C. jejuni requires an atmosphere with reduced oxygen and elevated carbon dioxide concentrations for its efficient cultivation in vitro: Variable oxygen tolerance has been described for C. jejuni isolates, but gas mixtures of 5% oxygen, 10% carbon dioxide and 85% nitrogen provide optimal cultivation conditions for most C. jejuni isolates (Bolton and Coates, 1983). C. jejuni has with about 800 bacteria a lower infective dose than Salmonella Typhimurium or pathogenic Escherichia coli strains (Black et al., 1988; Kothary and Babu, 2001), indicating that C. jejuni is well adapted to survive the harsh, acidic environment of the human stomach. Moreover, it demonstrates the proficiency with which C. jejuni is able to multiply in the gastrointestinal tract of humans and to consume the nutritional sources present in the host intestine in order to overcome the microbiota-mediated colonization resistance. Therefore, characterizing the metabolic properties of C. jejuni that allows this successful human pathogen and widespread animal commensal to thrive in its diverse hosts has gained increasing attention in recent years.
Finding a Nutritional Niche: The Low-Carb, High-Protein Diet of C. jejuni
The Non-glycolytic Nature of C. jejuni
C. jejuni is a chemoheterotrophic bacterium with metabolic properties that clearly distinguish it from other enteropathogenic bacteria. Most striking is its restricted carbohydrate catabolism: Early physiological studies examining the substrate utilization of Campylobacter identified its incapability to use glucose and other carbohydrates as growth substrates, and since then Campylobacter is generally considered to be a non-saccharolytic bacterium. These observations were supported by genome sequence analysis (Parkhill et al., 2000; Velayudhan and Kelly, 2002; Gundogdu et al., 2007) and recent growth-independent BIOLOG phenotype microarray analyses based on the tetrazolium redox dye chemistry that allows monitoring the respiratory activity of metabolically active cells (Bochner, 2009). This approach confirmed that pentoses and hexoses like glucose, fructose, galactose, rhamnose and the disaccharides lactose, maltose, trehalose and sucrose do not enhance the respiratory activity of C. jejuni (Line et al., 2010; Gripp et al., 2011; Muraoka and Zhang, 2011). However, these studies did reveal that certain strains like C. jejuni NCTC 11168 are capable to catabolize fucose in contrast to other isolates. The observed metabolic diversity is explained by the occurrence of a 9 kb genomic island, comprised in C. jejuni NCTC 11168 of the open reading frames Cj0480 to Cj0490, which are absent in C. jejuni 81–176 (Hofreuter et al., 2006; Muraoka and Zhang, 2011; Stahl et al., 2011). This gene region encodes for a putative fucose permease FucP (Cj0486) with homology to major facilitator superfamily (MFS) transporters. It was demonstrated that FucP enhanced the growth of C. jejuni NCTC 11168 when cultivated in chemically defined media containing 25 or 50 mM fucose as an additional carbon and energy source (Muraoka and Zhang, 2011; Stahl et al., 2011). The catabolic pathway of fucose in fucP-positive C. jejuni strains is not clear yet. Preliminary data suggest that the fucose utilization does not occur as observed for other intestinal bacteria like E. coli or Bacteroides species but as described in Xanthomonas campestris generating pyruvate and lactate (Stahl et al., 2011). Both end products are utilized by C. jejuni and efficiently promote its in vitro growth (Mendz et al., 1997; Velayudhan and Kelly, 2002; Thomas et al., 2011). Interestingly, no secreted fucosidase enzymes that cleave fucose residues from glycosylated host proteins have been described for the fucose-catabolizing C. jejuni strains. This suggests that fucP positive C. jejuni isolates might rely on the fucose released from intestinal mucins and host glycans by commensal bacteria like Bacteroides thetaiotaomicron as demonstrated for enterohaemorrhagic Escherichia coli (Pacheco et al., 2012). Alternatively, the activity of an induced host fucosidase could make free fucose available for C. jejuni similar as described for the human fucosidase FUCA2 of cultured human gastric and pancreatic adenocarcinoma cells upon infection with Helicobacter pylori (Liu et al., 2009).
The incapability to catabolize glucose distinguishes C. jejuni from many other gastrointestinal pathogens like Salmonella Typhimurium, enteropathogenic E. coli or Listeria monocytogenes (Dandekar et al., 2012; Fuchs et al., 2012) but also from its close relative H. pylori (Mendz et al., 1993). C. jejuni and H. pylori have an interrupted Embden–Meyerhof–Parnas (EMP) pathway because they lack the glycolytic enzyme phosphofructokinase, which catalyzes the phosphorylation of fructose-6-phosphate to fructose-1,6-bisphosphate. However, H. pylori possesses a complete pentose phosphate pathway and an Entner–Doudoroff pathway (Doig et al., 1999) that enable the catabolism of glucose to pyruvate (Mendz et al., 1994), while C. jejuni does not encode for the glucose-6-phosphate dehydrogenase (EC 1.1.1.49) and the 6-phosphogluconolactonase (EC 3.1.1.31) of the oxidative pentose phosphate pathway branch but harbors all enzymes of the reductive branch of the pentose phosphate pathway (Parkhill et al., 2000; Fouts et al., 2005). Consequently, gluconeogenesis seems to be required for the biosynthesis of glucose and its derivatives that are precursor substrates for the LPS and capsule biosynthesis as well as the N- and O-glycosylation of various secreted proteins (Karlyshev et al., 2005). The genome of C. jejuni harbors all enzymes necessary for gluconeogenic synthesis of glucose from phosphoenolpyruvate (PEP) (Parkhill et al., 2000), but gluconeogenesis has not been experimentally proven yet. Only few studies have characterized the intermediary metabolism of C. jejuni and the anaplerotic reactions fueling gluconeogenesis, but it was demonstrated that the PEP carboxykinase PckA (Cj0932c), the pyruvate kinase Pyk (Cj0392c) and the pyruvate carboxylase (PycA, Cj1037c; PycB, Cj0933c) comprise a crucial metabolic junction between catabolism and anabolism in C. jejuni (Velayudhan and Kelly, 2002). These enzymes are required for the oxaloacetate—PEP—pyruvate conversion that comprises a metabolic triangle, which plays the central role in controlling the carbon flow by connecting the tricarboxylic acid (TCA) cycle with the lower portion of the EMP pathway (Sauer and Eikmanns, 2005). Interestingly, C. jejuni does not encode for a PEP carboxylase catalyzing the generation of oxaloacetate from PEP and is lacking a PEP synthase to mediate the first ATP-consuming gluconeogenic reaction that synthesizes PEP from pyruvate. Consequently, the PEP carboxykinase PckA, catalyzing the synthesis of PEP by the decarboxylation of oxaloacetate, plays an important role in the intermediary metabolism of C. jejuni (Velayudhan and Kelly, 2002). Further studies are needed to identify the preferred substrates fueling the lower portion of the EMP pathway and to investigate how the intermediary metabolism is regulated and fine-tuned in C. jejuni in order to establish a balance between anabolism and catabolism.
Organic Acids and Amino Acids Fuel the Growth of C. jejuni
Because the gluconeogenesis seems to play a crucial role in the physiology of C. jejuni, it raises the question of which substrates are efficiently utilized by this pathogen to fuel its intermediary metabolism and cope with its necessities for carbohydrate, lipid and protein biosynthesis. Various studies using the API test system (Elharrif and Megraud, 1986), the detection of CO2 release upon incubation of C. jejuni with 14C-labeled substrates (Westfall et al., 1986), substrate oxidation experiments with an oxygen electrode system (Mohammed et al., 2004) and in vitro growth experiments (Velayudhan and Kelly, 2002; Hinton, 2006; Guccione et al., 2008; Wright et al., 2009) revealed that C. jejuni catabolizes organic acids like lactate, pyruvate, acetate and intermediates of the TCA cycle as well as a restricted number of amino acids. The importance of organic acids for the proliferation of C. jejuni was further demonstrated by studies that identified pyruvate, 2-oxoglutarate, fumarate, succinate, malate and lactate as chemoattractants of C. jejuni (Hugdahl et al., 1988; Vegge et al., 2009).
The L- and D-lactate catabolism of C. jejuni has been described in detail: L-lactate is taken up by C. jejuni NCTC 11168 with the symport of protons by the transporter protein Cj0076c that belongs to the lactate permease LctP family (TC 2.A.14). In addition, other not yet identified transporter proteins are probably involved in the uptake of L- and D-lactate (Thomas et al., 2011). The imported L-lactate is subsequently oxidized to pyruvate, which itself is a growth substrate of C. jejuni (Mendz et al., 1997; Velayudhan et al., 2004) though no pyruvate carrier has been described yet. C. jejuni does not harbor a pyruvate dehydrogenase found in other enteropathogenic bacteria but catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA through a pyruvate:acceptor oxidoreductase (POR; Cj1476c) similar as described for H. pylori (St Maurice et al., 2007). However, the POR enzyme of H. pylori is composed of the four subunits PorABCD (Hughes et al., 1998), whereas the pyruvate-flavodoxin oxidoreductase of C. jejuni is comprised of one polypeptide with four functional domains. It is conserved in other Campylobacter species and has homologs in host-associated bacteria like Fusobacterium nucleatum, Fusobacterium necrophorum, Cetobacterium somerae or Sebaldella termitidis, as well as environmental bacteria such as Psychrilyobacter atlanticus and Orenia marismortui. Interestingly, the pyruvate-flavodoxin oxidoreductase of C. jejuni is responsible for its sensitivity against nitazoxanide, an antiprotozoal drug that inhibits PORs but not pyruvate dehydrogenases (Hoffman et al., 2007). The POR catalyzed generation of acetyl-CoA both fuels the TCA cycle of C. jejuni and is used for fatty acid biosynthesis (Leach et al., 1997; Gundogdu et al., 2007; Kirkpatrick et al., 2009). Furthermore, acetyl-CoA can be converted to acetyl-P and subsequently to acetate by the consecutive activities of the phosphate acetyltransferase Pta (Cj0688) and the acetate kinase AckA (Cj0689). The generated acetate is secreted by a yet unknown transporter during the logarithmic growth phase (Wright et al., 2009). In the late logarithmic growth phase an “acetate switch,” which is well characterized for E. coli (Wolfe, 2005), could be observed for C. jejuni: The previously secreted acetate is taken up and can be used as a growth substrate through the conversion to acetyl-CoA by the acetyl-CoA synthetase ACS (Cj1537c) (Wright et al., 2009).
It is well established that the utilization of amino acids plays an important role in fueling the central metabolism of C. jejuni. Strikingly, however, only few glucogenic amino acids are degraded by this pathogen and support its proliferation, and some amino acids such as arginine and lysine have been described as chemorepellents (Rahman et al., 2014). For most C. jejuni isolates the growth-promoting amino acids are aspartate, glutamate, proline and serine (Leach et al., 1997; Guccione et al., 2008; Hofreuter et al., 2008). This finding is in agreement with the metabolic profiling of supernatants from C. jejuni liquid cultures demonstrating a significant depletion of these four amino acids from nutrient-rich brain heart infusion (BHI) or Mueller-Hinton (MH) medium (Guccione et al., 2008; Wright et al., 2009). The utilization of the growth-promoting amino acids in liquid cultures occurs in sequential phases: Aspartate and serine are first catabolized and facilitate the rapid growth of C. jejuni followed by the usage of glutamate. Proline seems to be a less-preferred growth substrate of C. jejuni because its consumption from the culture medium occurred less rapidly in comparison to the depletion of aspartate, serine and glutamate (Leach et al., 1997; Weingarten et al., 2009; Wright et al., 2009). Accordingly, L-aspartate, L-glutmate and L-serine but not L-proline are effective chemoattractants for C. jejuni (Hugdahl et al., 1988; Vegge et al., 2009).
Serine is imported by the high-capacity, low affinity serine transport protein SdaC (Velayudhan et al., 2004), a serine/H+ symporter of the hydroxy/aromatic amino acid permease (HAAAP) family TC 2.A.42.2. The sdaC gene (cj1625c) is organized in an operon with sdaA (cj1624c) encoding for a serine dehydratase that catalyzes the deamination of serine to pyruvate. SdaA harbors, like various respiratory enzymes of C. jejuni, an oxygen-labile [4Fe–4S] cluster that might contribute to the oxygen sensitivity of this pathogen (Velayudhan et al., 2004). Serine utilization seems to be a variable catabolic trait of C. jejuni because not all tested isolates were able to grow with this amino acid a sole carbon source (Hofreuter et al., 2008). The molecular basis for this metabolic diversity is not known yet, as C. jejuni strains unable to utilize serine have no mutations in the sdaA and sdaC genes though the serine dehydratase activity was fairly reduced (Hofreuter et al., 2012).
In contrast to serine catabolism, proline utilization seems to be a more conserved metabolic trait of C. jejuni as also shown for the usage of aspartate and glutamate (Hofreuter et al., 2008). A directed mutagenesis approach demonstrated that the growth of C. jejuni 81–176 with proline is mediated by the PutP transporter and the enzyme PutA (Hofreuter et al., 2012). PutP (CJJ81176_1494) belongs to the Na+/solute symporter family TC 2A.21, which is widespread in Gram-positive and Gram-negative bacteria (Jung et al., 2012). It is highly conserved in C. jejuni and shows about 80% amino acid identity to respective transporter proteins of C. coli, C. lari, C. upsaliensis and C. fetus, whereas no homologs are present in other Campylobacter species. In addition, the proline symporter protein (PutPCj) of C. jejuni shows 75% identity to the PutP transporter (PutPHp) of the closely related H. pylori (Hofreuter et al., 2012). So the PutPCj-mediated proline import might have comparable properties as the PutPHp-catalyzed uptake of proline, which depends entirely on Na+ as a coupling ion and binds specifically to L-proline with high affinity (Rivera-Ordaz et al., 2013). The PutA (CJJ81176_1495) enzyme of C. jejuni is predicted to possess both proline dehydrogenase and delta-1-pyrroline-5-carboxylate dehydrogenase activities and to use FAD and NADH as cofactors, respectively. It catalyzes the oxidation of the imported proline to glutamate. The putA and putP genes of C. jejuni and other Epsilonproteobacteria compromise an operon structure, whereas the adjacent located putA and putP genes of Enterobacteria have an inverse orientation (Hofreuter et al., 2012). Furthermore, the PutA protein of C. jejuni does not contain the N-terminal DNA-binding domain, which is involved in the repression of putAP gene cluster in Salmonella Typhimurium during the absence of proline (Ostrovsky de Spicer and Maloy, 1993).
C. jejuni takes up glutamtate through an ABC transporter system encoded by the peb locus (cj0919c-cj0922c) harboring genes for two permeases, one ATP-binding protein and one periplasmic substrate-binding protein Peb1A (Cj0921c). The latter has been first described as surface-bound antigen of C. jejuni with homology to amino acid-binding proteins like GlnH and HisJ (Pei and Blaser, 1993). Further studies characterized Peb1A as a glutamate- and aspartate-binding protein (Leon-Kempis Mdel et al., 2006; Müller et al., 2007), and an isogenic C. jejuni peb1A mutant showed impaired growth with aspartate or glutamate as sole carbon sources (Leon-Kempis Mdel et al., 2006). This phenotype correlated with an abolished glutamate uptake. In addition to the Peb ABC transporter, mutations of the permease PaqP (Cj0467) and the ATPase PaqQ (Cj0469) of the Paq (pathogenesis-associated glutamine) ABC transporter system negatively affected the uptake of glutamate in C. jejuni 81–176 (Lin et al., 2009). Imported glutamate is unlikely to be converted directly to 2-oxoglutarate through deamination since C. jejuni lacks a glutamate dehydrogenase (EC 1.4.1.2, EC 1.4.1.3, EC 1.4.1.4), which is present in other representatives of the Campylobacteriaceae. Instead, glutamate might either be converted to glutamine by the type I glutamine synthetase GlnA (Cj0699c) or is substrate of the aspartate:glutamate transaminase AspB (Cj0762c) catalyzing the generation of aspartate and 2-oxoglutarate from oxaloacetate and glutamate (Guccione et al., 2008). AspB plays an important role in the intermediary metabolism of C. jejuni and its efficient proliferation, since the inactivation of aspB results in an severe growth defect of the respective mutant in nutrient rich MH or BHI media as well as in defined medium with 20 mM glutamate or serine as sole energy sources (Guccione et al., 2008; Novik et al., 2010).
Besides serine, proline and glutamate catabolism, the utilization of aspartate is crucial for the metabolic fitness of C. jejuni. This is reflected by the identification of two chemoreceptors, Tlp1 (Hartley-Tassell et al., 2010) and CcmL (Tlp3) (Rahman et al., 2014), that interact with aspartate. The uptake of aspartate is convoluted and involves various transporters: While the periplasmic binding protein Peb1A interacts with aspartate and participates in its uptake, the peb1A mutant still possesses capacity to import aspartate although with a 20-fold reduction (Leon-Kempis Mdel et al., 2006). The direct involvement of the other Peb ABC transporter components like the two Peb permeases and the Peb ATP-binding protein has not been experimentally proven yet. In addition to the Peb1A-mediated aspartate uptake, the C4-dicarboxylate antiporters DcuA (Cj0088) and DcuB (Cj0671) have been shown to transport aspartate into C. jejuni under oxygen-limited conditions. Their activities were redundant so only a dcuA/dcuB double mutant showed reduced growth with aspartate (Guccione et al., 2008). It was suggested that aspartate uptake through the C4-dicarboxylate transport protein DctA (Cj1192) was responsible for the remaining growth of the dcuA/dcuB double mutant (Guccione et al., 2008). Interestingly, also a paqP mutant but not a paqQ mutant of the Paq ABC transporter showed a reduced level of aspartate uptake (Lin et al., 2009). The fate of imported aspartate is multifaceted involving central anabolic as well as catabolic pathways of C. jejuni: Aspartate represents an efficient carbon and energy source for C. jejuni as it directly fuels the TCA cycle by the aspartate ammonia lyse AspA (Cj0087) catalyzing the deamination to fumarate (Guccione et al., 2008; Novik et al., 2010). This AspA-catalyzed reaction of aspartate to fumarate also plays a role in the response of C. jejuni 81–176 to high pressure and its recovery from cell injury (Bieche et al., 2012). Fumarate might be converted to oxaloacetate, which can be used as substrate for the gluconeogenesis and synthesis of essential carbohydrates. Alternatively, fumarate represents an important electron acceptor for the oxygen-independent respiration of C. jejuni (Sellars et al., 2002), generating a significant amount of succinate, which is secreted during the growth by C. jejuni (Guccione et al., 2008). In addition, aspartate is the precursor for the biosynthesis of several proteinogenic amino acids (lysine, methionine, threonine, isoleucine) as well as β-alanine. The latter is generated from aspartate by the activity of an aspartate alpha-decarboxylase PanD (Cj0296c) and serves as precursor for the synthesis of pantothenate and coenzyme A.
Genome sequence analysis showed that C. jejuni encodes for an asparaginase (AnsB) facilitating the conversion of asparagine to aspartate and for a glutamate synthase (GltBD) catalyzing the generation of glutamate from glutamine and 2-oxoglutarate (Gundogdu et al., 2007). These enzymes seem functional since asparaginase activity was detected in cell extracts of C. jejuni (Guccione et al., 2008) and 14C-labeled glutamine was catabolized under CO2 production (Westfall et al., 1986). These studies indicated that C. jejuni is able to catabolize asparagine as well as glutamine and both amino acids are chemoattractants for C. jejuni (Vegge et al., 2009). Although glutamine uptake in C. jejuni is facilitated by the conserved Paq transporter system (Lin et al., 2009), growth experiments revealed that just a subset of the tested C. jejuni isolates could use glutamine as sole carbon and energy source (Hofreuter et al., 2008). These results imply that the direct uptake of glutamine by the Paq and any additional transporter systems is not sufficient to promote pronounced proliferation of C. jejuni under the tested in vitro conditions. Only C. jejuni strains encoding for a secreted γ-glutamyltranspeptidase (GGT) were able to utilize glutamine efficiently for growth (Hofreuter et al., 2008). This periplasmic GGT enzyme facilitates the hydrolysis of glutamine to glutamate and ammonia similar to what has been described for H. pylori (Shibayama et al., 2007). Whereas the occurrence of GGT seems to be a conserved genetic trait in H. pylori (Leduc et al., 2010), not more than a third of the analyzed C. jejuni isolates harbored the GGT gene: Significant variations in the ggt frequency (between 8 and 30%) resulted from differences in the sequence type (ST) / clonal complex (CC) composition of the analyzed C. jejuni collections as determined by multilocus sequence typing (MLST): The ggt gene was strongly associated with ST-45 CC and ST-22 strains (Revez et al., 2011; de Haan et al., 2012) but typically absent in the common ST-21 isolates (Gripp et al., 2011; Zautner et al., 2011; de Haan et al., 2012). Surprisingly, GGT-activity provides C. jejuni strains not only an expanded amino catabolism but also a survival advantage in the presence of bactericidal isothiocyanates as shown for C. jejuni 81–176 (Dufour et al., 2012).
Most of the analyzed C. jejuni strains, including the reference strain NCTC 11168, were not able to grow with asparagine as the primary source of carbon and energy although all possess an L-asparaginase gene that encodes for a cytoplasmic AnsB enzyme (Hofreuter et al., 2008). This suggests that efficient asparagine import does not occur in C. jejuni. In contrast to C. jejuni NCTC 11168, certain isolates like C. jejuni 81–176 harbor an ansB allele variation that encodes for an additional sec-dependent secretion signal. Such a modified AnsB enzyme (AnsBs) is translocated to the periplasm where it catalyzes the deamination of asparagine to aspartate. Consequently, the growth of ansBs- and ggt-positive C. jejuni isolates with asparagine and glutamine relies on the conversion of both amino acids to aspartate and glutamate in the periplasm and the subsequent uptake of the deaminated amino acids (Hofreuter et al., 2008) through the DcuAB, DctA and Peb transporters (Leon-Kempis Mdel et al., 2006; Guccione et al., 2008). Interestingly, many ggt-positive C. jejuni strains encode also for a secreted asparaginase but lack the fucP-gene cluster, while C. jejuni isolates without the GGT- and AnsBs-mediated expanded amino acid catabolism harbor the genes required for fucose utilization (Gripp et al., 2011; Zautner et al., 2011; de Haan et al., 2012). This correlation may reflect the selection pressure to compensate the restricted amino acid catabolism with enhanced carbohydrate utilization. Future studies are required to elucidate how these differences in substrate utilization might correlate with a variable fine-tuning of the intermediary metabolism of GGT/AnsBs-positive and -negative C. jejuni strains and how these metabolic variations could reflect the physiological adaptations of certain C. jejuni strains to different hosts.
Besides the transport systems for the growth-promoting amino acids serine, proline, aspartate and glutamate, C. jejuni harbors homologs of the LIV (leucine, isoleucine, valine) branched-chain amino acid ABC transporter system of E. coli (Ribardo and Hendrixson, 2011). Though none of the branched-chain amino acids directly fuels the growth of C. jejuni (Guccione et al., 2008; Hofreuter et al., 2008), isoleucine interacts with the transducer-like protein Tlp3 of C. jejuni NCTC 11168 and induces a positive chemotactic response (Rahman et al., 2014). The LIV-locus in C. jejuni is comprised of six genes encoding for the periplasmic binding proteins LivJ and LivK, the two permeases LivH and LivM, and the cytoplasmic ATPases LivG and LivF. Targeted mutagenesis confirmed that the individual LIV ABC transporter components are responsible for the uptake of the branched amino acids, particularly of leucine (Ribardo and Hendrixson, 2011). Expression of the liv genes in C. jejuni is not repressed in the presence of leucine (Ribardo and Hendrixson, 2011) as shown for E. coli (Quay and Oxender, 1976), further exemplifying the metabolic specificity of C. jejuni.
Several additional putative amino acid transporters are present within the sequenced genomes of C. jejuni. For example, Cj0903c with homology to members of the sodium:alanine symporter family and SstT (Cj1097), the putative sodium/serine-threonine symporter, are conserved in C. jejuni and future studies are needed to elucidate their substrate specificity. Besides the aspartate/glutamate-binding protein Peb1A, C. jejuni harbors CjaA, CjaC and GlnH, three additional periplasmic proteins that belong to the family 3 of solute-binding proteins, which might also facilitate the uptake of amino acids. CjaA (Cj0982c) is an N-glycosylated 30 kDa lipoprotein attached to the inner membrane (Pawelec et al., 1997; Wyszynska et al., 2008) that binds cysteine (Müller et al., 2005). Its direct involvement in the import of cysteine has not been experimentally proven so far, and growth experiments suggested that CjaA might not be solely involved in the uptake of cysteine (Vorwerk et al., 2014). CjaC (Cj0734c) is a N-glycosylated 28 kDa protein with unknown substrate specificity that is anchored in the cytoplasmic membrane of C. jejuni and shows best homologies to various periplasmic binding proteins of ABC-type amino acid transporters (Pawelec et al., 1998; Wyszynska et al., 2007). Interestingly, a hisJ mutant of S. Typhimurium could be complemented by heterologous expression of the C. jejuni cjaC gene indicating that CjaC participates in the uptake of histidine in C. jejuni as well (Garvis et al., 1996). The third family 3 solute-binding protein, GlnH (Cj0817), remains uncharacterized. Taken together, though much progress has been made in recent years to characterize the amino acid catabolism of C. jejuni, future work is required to identify the transport systems involved in the uptake of amino acids that do not directly promote the growth of C. jejuni as energy or carbon sources.
Whereas the importance of amino acid uptake and utilization for the growth of C. jejuni is well documented, only few studies have investigated the de novo biosynthesis of amino acids in this pathogen, like e.g., the involvement of ilvE and cysM in the synthesis of leucine and cysteine, respectively (Ribardo and Hendrixson, 2011; Vorwerk et al., 2014). Other studies have examined the amino acid biosynthesis capability of C. jejuni by heterologous expression of certain genes of arginine (Hani and Chan, 1994; Hani et al., 1999), cysteine (Garvis et al., 1997), leucine (Labigne et al., 1992) and aromatic amino acid (Wösten et al., 1996) biosynthesis pathways in respective auxotrophic Escherichia coli mutants. Therefore, information about the complete biosynthesis pathways of amino acids are primarily obtained through the in silico analysis of available genome sequences of C. jejuni isolates: In contrast to H. pylori, which has limited capacity for the synthesis of amino acids and requires arginine, histidine, isoleucine, leucine, methionine, phenylalanine and valine for growth (Doig et al., 1999), C. jejuni NCTC 11168 encodes for enzymes enabling the synthesis of all amino acids (Gundogdu et al., 2007). Future studies are required to clarify how conserved the functionality of the predicted amino acid biosynthesis pathways are among C. jejuni strains as several amino acid auxotrophies, e.g., for methionine, proline or the branched amino acids isoleucine, leucine and valine, have been previously described in various C. jejuni isolates (Tenover et al., 1985; Blaser et al., 1986; Tenover and Patton, 1987).
Peptidases and Peptide Catabolism of C. jejuni
The central role of amino acid catabolism for the proliferation of C. jejuni suggests that peptides may also be important growth-promoting substrates for this pathogen, especially, since the digestion of proteins in the gastrointestinal tract of its hosts generates a variety of peptides besides free amino acids (Adibi and Mercer, 1973). Several putative peptidases and proteases are encoded by the C. jejuni genome (Hofreuter et al., 2006; Gundogdu et al., 2007), and some, like ClpP (Cj0192c), HtrA (Cj1228c), CJJ81176_1086 (Cj1068), CJJ81176_1228 (Cj1215), Cj0511 or Pgp1 (Cj1345c; CJJ81176_1344), have been associated with the virulence of C. jejuni (Brondsted et al., 2005; Cohn et al., 2007; Novik et al., 2010; Boehm et al., 2012; Frirdich et al., 2012; Karlyshev et al., 2014).
The role of peptidases in the catabolism and nutrient acquisition of C. jejuni has not been characterized in detail so far. BIOLOG phenotype microarray analysis suggested that dipeptides like glycyl-glutamine and glycyl-proline enhance the respiratory activity of C. jejuni and can be used as carbon sources though strain specific differences exist (Gripp et al., 2011; Muraoka and Zhang, 2011). This variability in peptide catabolism might be the consequence of C. jejuni isolates having variable number of peptidases: GGT, the putative S15 family dipeptidyl-peptidase CJJ81176_1680 or the putative subtilisin-like serine peptidases Cj1365 / CJJ81176_1367 and CJJ81176_1371 occur in a subset of C. jejuni strains (Champion et al., 2005; Hofreuter et al., 2006; Hepworth et al., 2007; Gonzalez et al., 2009; Zautner et al., 2011). Only ggt-positive C. jejuni strains can efficiently use the tripeptide glutathione as carbon/energy (Hofreuter et al., 2008) and cysteine source (Vorwerk et al., 2014). A recent study using the BIOLOG phenotype microarray technology suggested that the C. jejuni NCTC 11168 transporter protein Cj0917c, which has homology to the carbon starvation protein A (CstA) of E. coli, is involved in the catabolism of several tri- and dipeptides (Rasmussen et al., 2013). Additional experiments are necessary to demonstrate the growth-promoting effect of respective peptides. Moreover, the Campylobacter peptide transporter A (CptA; CJJ81176_0236), a member of the Proton-dependent Oligopeptide Transporter (POT) family, has been described to promote the growth of C. jejuni 81–176 with the dipeptides Cys-Gly, Arg-Trp and Arg-Ile (Vorwerk et al., 2014).
Taken together, C. jejuni shows an intriguing metabolic diversity. The diverse growth properties of C. jejuni isolates result from the variable presence or absence of metabolic genes involved in the strain-specific utilization of particular substrates such as fucose, asparagine or glutamine and peptides. In addition, C. jejuni isolates are equipped with different sets of group A chemoreceptor tlp genes that response to a variety of potential nutrients (Day et al., 2012; Rahman et al., 2014). Such a variable presence of chemosensory receptor genes in C. jejuni suggests that different strains may not respond equivalently to certain nutrients and consequently cannot utilize and benefit from the same growth substrates.
Finding the Optimal Oxygen Level: How Does a Microaerophilic Pathogen Persists in the Anaerobic Environment of the Intestine?
In contrast to facultative anaerobe pathogens like S. Typhimurium or enteropathogenic E. coli, C. jejuni faces the challenging situation to proliferate as obligate microaerophilic bacterium in the intestine of its hosts where lower levels than the preferred 5% oxygen exist. It has been suggested that C. jejuni is adapted to this disadvantageous circumstance by its colonization pattern and distinct respiratory capability: (i) C. jejuni colonizes preferentially the mucus layer and the intestinal crypt close to the epithelium (Lee et al., 1986; Beery et al., 1988) where the oxygen tension is higher than in the intestinal lumen. In addition, the region between the mid small intestine and the mid colon, which is the preferred colonization site of C. jejuni, harbors higher oxygen tensions than the distal colon and rectum (He et al., 1999). (ii) Early biochemical characterizations of C. jejuni demonstrated the presence of a surprisingly complex and highly branched respiratory chain, allowing this pathogen to use a variety of electron donors and several other electron acceptors besides oxygen (Carlone and Lascelles, 1982; Hoffman and Goodman, 1982; Hitchcock et al., 2010). Menaquinone-6 and methyl-substituted menaquinone-6 mediate in C. jejuni the electron transfer along the respiration chain between electron donors and receptors (Carlone and Anet, 1983; Moss et al., 1984). The menBCDEF menaquinone biosynthesis genes found in enterobacteria to catalyze the transformation of chorismate to menaquinone are absent in C. jejuni. Instead C. jejuni harbors enzymes of an alternative menaquinone biosynthesis pathway similar to the futalosine pathway described for Streptomyces coelicolor (Hiratsuka et al., 2008; Li et al., 2011). This modified futalosine pathway of C. jejuni is also employed by H. pylori (Arakawa et al., 2011) and uses 6-amino-6-deoxyfutalosine instead of futalosine as an intermediate for the synthesis of menaquinone. Consequently, orthologs to enzymes MqnA and MqnB of S. coelicolor are not required by C. jejuni and replaced by the menaquinone biosynthetic enzyme A2 (MqnA2; Cj1285c) and the 5′-methylthioadenosine nucleosidase MTAN (Cj0117) (Li et al., 2011). MTAN might be a promising new drug target for C. jejuni as demonstrated for H. pylori (Wang et al., 2012).
Genome sequence analysis of C. jejuni (Parkhill et al., 2000; Sellars et al., 2002) revealed that the respiratory electron chain of C. jejuni is comprised of two terminal, membrane-bound oxidases. Both enzymes mediate the oxygen-dependent respiration of C. jejuni but harbor strikingly different oxygen affinities: the cyanide-insensitive oxidase CioAB (initially named CydAB; Cj0081, Cj0082) exhibits low affinity for oxygen, whereas the cyanide-sensitive, cb-type cytochrome c oxidase CcoNOQP (Cj1490c-Cj1487c) shows high affinity to oxygen and might be crucial for respiration under oxygen-limited conditions (Jackson et al., 2007). The CioAB oxidase receives electrons directly from the oxidized menaquinone pool, whereas oxygen reduction by the CcoNOQP oxidase involves electron transfer from the oxidized menaquinone pool via the proton-translocating cytochrome bc complex (PetABC: Cj1186c-Cj1184c) and a periplasmic c-type cytochrome (Cj1153).
Though C. jejuni is equipped with various enzymes facilitating oxygen-independent respiration, no growth can be observed under strictly anaerobic conditions (Veron et al., 1981). It was suggested that the class I ribonucleotide reductase (NrdAB-type RNR) of C. jejuni is responsible for its inability to grow anaerobically because this enzyme requires low amounts of oxygen for the DNA synthesis (Sellars et al., 2002). This prerequisite for oxygen brings the disadvantage that non-specific electron transfer from the respiratory chain to oxygen occurs (Cabiscol et al., 2000), leading to the generation of toxic reactive oxygen species (ROS) like hydroxyl (·OH) and superoxide (O−2) radicals as well as hydrogen peroxide (H2O2). C. jejuni harbors a variety of ROS-detoxifying enzymes including the superoxide dismutase SodB (Pesci et al., 1994), the alkyl hydroxide reductase AhpC (Baillon et al., 1999), the catalase KatA (Cj1385) (Day et al., 2000) as well as the thiolperoxidases Tpx and Bcp (Atack et al., 2008). Surprisingly, these enzymes are unable to provide sufficient oxygen tolerance in aerobic conditions for C. jejuni. A recent study linked the oxygen-labile, iron-sulfur (4Fe-4S)-containing metabolic enzymes pyruvate:acceptor oxidoreductase POR and the oorDABC (cj0535-cj0538) encoded 2-oxoglutarate:acceptor oxidoreductase (OOR) to the oxygen sensitivity of C. jejuni (Kendall et al., 2014). The enzymatic activity of POR and OOR are partially protected by the hemerythrin proteins HerA (Cj0241c) and HerB (Cj1224), but C. jejuni lacks sufficient mechanisms to repair POR and OOR once damaged through exposure to atmospheric oxygen concentrations (Kendall et al., 2014).
A Variety of Electron Donors Fuel the Respiratory Activity of C. jejuni
The reduction equivalents nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH) serve in many bacteria as major electron sources for the respiratory electron transport chain, which generates through the proton-translocating NADH:quinone oxidoreductase (Nuo/NDH-1) complex a proton gradient that drives the oxidative phosphorylation (Haddock and Jones, 1977). Strikingly, NADH is a poor respiratory electron donor in C. jejuni in contrast to FADH (Hoffman and Goodman, 1982) as consequence of the specific NDH-1 complex property encoded by the nuo gene cluster cj1566c to cj1579c in C. jejuni NCTC 11168: The genes nuoE and nuoF encoding for the NADH dehydrogenase subunits of the NDH-1 complex are replaced in C. jejuni NCTC 11168 by the genes cj1575c and cj1574c (Smith et al., 2000). It was shown that Cj1574c, probably in conjunction with Cj1575c, mediates the electron transfer from the reduced, flavin monoculeotide containing flavodoxin FldA (Cj1382c) to the NDH-1 complex (Weerakoon and Olson, 2008). Consequently, the NDH-1 complex of C. jejuni seems to participate rather in the oxidation of flavin mononucleotides than of NADH. It was further demonstrated that reduced FldA is generated by the oxidation of 2-oxoglutarate to succinyl-CoA catalyzed by the OOR, whereas the putative ferredoxins FdxA (Cj0333c) and Cj0369c are no electron acceptors for OOR (Weerakoon and Olson, 2008).
Several other electron donors besides FADH have been described to fuel the respiration chain of C. jejuni: some of them, like hydrogen, formate, lactate or succinate, are generated through the catabolic activity of the host gut microbiota (Bernalier-Donadille, 2010), which suggests that C. jejuni might benefit from metabolic cross-feeding. Hydrogen can be used as an electron donor by C. jejuni through the activity of the membrane-bound NiFe-type hydrogenase HydABCD (Cj1267c-Cj1264c) (Hoffman and Goodman, 1982; Weerakoon et al., 2009). The accessory factors encoded by the hypFBCDEA operon (cj0622-cj0627) are required for assembling of the hydrogenase enzyme complex and insertion of the nickel cofactor. An ABC-transporter system (Cj1584c-Cj1580c) of C. jejuni NCTC 11168 has recently been identified as a high-affinity nickel uptake system and was named NikZYXWV (Howlett et al., 2012). Under low nickel concentrations the inactivation of nikZ (cj1584c), encoding for a periplasmic binding protein, led to an abolished hydrogenase activity of the mutant strain. This result demonstrated the importance of the Nik-transporter system for the acquisition of nickel as a cofactor for the enzyme. Yet hydrogenase activity of the nikZ mutant was observed in the presence of high nickel concentrations, indicating the presence of additional nickel transporters (Howlett et al., 2012). In contrast to studies with E. coli, showing the importance of the nickel chaperone SlyD for the hydrogenase activity, a mutation in the slyD ortholog gene cj0115 of C. jejuni NCTC 11168 did not abolish the mutants nickel uptake capacity and hydrogenase activity (Howlett et al., 2012).
Formate is mainly generated by the mixed-acid fermentation of the intestinal microbiota and is sensed as chemoattractant by C. jejuni through the Tlp7 chemoreceptor (Vegge et al., 2009; Tareen et al., 2010). It can be oxidized by C. jejuni to CO2, protons and electrons through a membrane-bound formate dehydrogenase (FDH) complex comprised of the selenocysteine-containing subunit FdhH (Cj1511c), the iron-sulfur subunit FdhB (Cj1510c) and the formate dehydrogenase cytochrome-b subunit FdhC (Cj1509c), which requires FdhD (Cj1508c) for its activity. The released electrons are directly transferred from the FDH complex to the menaquinone pool (Weerakoon et al., 2009). The FDH activity of C. jejuni is controlled by the accessory proteins FdhT (Cj1500), FdhU (Cj1501) and a high-affinity TupABC-like tungstate transporter (Cj1538-Cj1540) indicating that tungstate might be incorporated into the FDH complex (Smart et al., 2009; Pryjma et al., 2012; Shaw et al., 2012).
Lactate is oxidized by C. jejuni to pyruvate by the membrane-associated NAD-independent respiratory lactate dehydrogenase complex (L-iLDH; Cj0075c, Cj0074c; Cj0073c) though inactivation of these genes in C. jejuni NCTC 11168 did not abolish the growth of respective mutants with lactate (Thomas et al., 2011). However, a second L-iLDH, the oxidoreductase Cj1585c, was identified and demonstrated to be responsible for the observed redundancy in the catabolism of L-lactate. Only a double mutation inactivating cj0075c and cj1585c abolished the growth of C. jejuni NCTC 11168 with 20 mM L-lactate as a carbon source but it did not affect in the utilization of D-lactate. The gene locus of cj1585c is not conserved in C. jejuni (Hofreuter et al., 2006) and is replaced by a dimethyl sulfoxide reductase (dmsABC) gene cluster encoding for an anaerobic dimethyl sulfoxide reductase complex in strains like C. jejuni 81–176 (Hofreuter et al., 2006), 81116 (Pearson et al., 2007), M1 (Friis et al., 2010) or 327 (Takamiya et al., 2011). This finding is verified by the observation that a mutant in the Cj0075c homolog of wild-type strain C. jejuni 81116, which naturally lacks the L-iLDH Cj1585c, was unable to grow with L-lactate (Thomas et al., 2011).
The TCA cycle intermediate succinate is not only a carbon and energy source for C. jejuni but serves also as an electron donor. Consequently, succinate is oxidized to fumarate accompanied with the generation of FADH2 and the subsequent electron transfer to the menaquinone pool. One predicted succinate dehydrogenase (succinate:quinone oxidoreductase) SdhABC (Cj0437-Cj0439) of C. jejuni was misannotated and not involved in the conversion of succinate to fumarate (Weingarten et al., 2009). Instead, the putative FrdABC fumarate reductase complex comprised of a membrane-associated diheme cytochrome B (FrdC, Cj0408), the flavoprotein FrdA (Cj0409) and the Fe-S protein FrdB (Cj0410) showed properties of a succinate:quinone reductase and was solely responsible for the oxidation of succinate to fumarate (Weingarten et al., 2009).
C. jejuni is unable to catabolize gluconate due to the absence of an Entner-Doudoroff pathway, but it can use gluconate as electron donor through a temperature-regulated flavin-containing gluconate dehydrogenase (Pajaniappan et al., 2008). Proteomic analysis demonstrated that the gluconate dehydrogenase (GADH) expression increased in C. jejuni upon a temperature shift from 37 to 42°C correlating with an elevated GADH activity. The co-transcribed genes cj0414 and cj0415 encode for the two components of the gluconate-oxidizing oxidoreductase, which is predicted to be localized in the periplasm peripherally associated with the cytoplasmic membrane and to transfer electrons to the periplasmic cytochrome c (Pajaniappan et al., 2008). Interestingly, both GADH subunits are conserved in C. jejuni but absent from other Campylobacter species.
Another example of the unique respiratory capacity of C. jejuni is its sulfite respiration system, which uses sulfite and metabisulfite as electron donors. This sulfite:cytochrome c oxidoreductase (SOR) system is conserved in C. jejuni and can also be found in C. lari but not in any other examined Campylobacter and Helicobacter species. The periplasmic proteins SorA (Cj0005c) and SorB (Cj0004c) with properties of a molybdopterin oxidoreductase and a monoheme cytochrome c552, respectively, catalyze the oxidation of sulfite to sulfate accompanied by an electron transfer to cytochrome c (Myers and Kelly, 2005).
In summary, the highly branched electron transport chain of C. jejuni enables respiration with a variety of electron donors that are secreted catabolic end products of the surrounding microbiota of the intestinal habitat. This cross-feeding might provide an important advantage for C. jejuni to overcome the colonization resistance of its host and to establish its nutritional niche.
The Versatility of Oxygen-independent Respiration in C. jejuni
C. jejuni is unable to grow under anaerobic conditions but is well adapted to the oxygen-limited conditions of its intestinal habitat by harboring a variety of respiration systems using alternative electron acceptors to oxygen, like nitrate, nitrite, S- and N-oxides or fumarate (Sellars et al., 2002).
Under oxygen-limited cultivation conditions of C. jejuni, fumarate is reduced to succinate by the bifunctional FrdABC complex and the methlymenaquinol:fumarate reductase complex comprised of MrfA (Cj0437), MrfB (Cj0438) and MrfE (Cj0439), initially described as SdhABC complex (Weingarten et al., 2009; Guccione et al., 2010). While the fumarate reductase FrdABC complex acts in the cytoplasm of C. jejuni, the MfrABE-mediated fumarate reduction occurs in the periplasm where MfrA is exported by the twin arginine translocase (TAT) system (Hitchcock et al., 2010). The periplasmic MrfABE complex of C. jejuni also participates in the reduction of the C4-dicarboxylate mesaconate and the C4-monocarboxylate crotonate (Guccione et al., 2010). Interestingly, both substances are fumarate analogs, which are produced as metabolic intermediates by anaerobic bacteria of the intestinal microbiota, suggesting a metabolic interaction between C. jejuni and species of Clostridium and Fusobacterium (Guccione et al., 2010).
Dimethyl sulfoxide (DMSO) and trimethylamine N-oxide (TMAO) are additional electron acceptors of C. jejuni in vitro (Sellars et al., 2002): The gene cj0264c of C. jejuni NCTC 11168 is conserved in C. jejuni and encodes for a molybdopterin-containing oxidoreductase, which is responsible for its DMSO and TMAO reductase activity. The TMAO- and DMSO-dependent respiration improved the in vitro growth of C. jejuni NCTC 11168 under oxygen-limited but not under microaerobic conditions. Future studies have to clarify if the putative cytochrome C-type heme-binding periplasmic protein, encoded by cj0265c, participates in the elector transfer to DMSO, TMAO and other N- or S- oxides (Sellars et al., 2002). Beside Cj0264c, C. jejuni 81–176 harbors a putative oxidoreductase with homology to the anaerobic dimethylsulfoxide reductase DmsA (Hofreuter et al., 2006). This oxidoreductase has 28% protein sequence identity with Cj0264c and may catalyze the reduction of DMSO / TMAO as well. The dmsA gene (cju34 / cjj81176_1570) of C. jejuni 81–176 is organized in a gene cluster together with three other gene genes encoding for the iron-sulfur containing DMSO reductase subunit DmsB (CJJ81176_1571), the DMSO reductase anchor subunit DmsC (CJJ81176_1572) and chaperon protein TorD (CJJ81176_1572) that are predicted to participate with DmsA at the electron transfer to DMSO / TMAO or other S- and N-oxides (Hofreuter et al., 2006).
Several human pathogens harbor nitrate and nitrite respiration systems (Sparacino-Watkins et al., 2014). C. jejuni encodes for the periplasmic located Nap-type nitrate reductase (Cj0780-Cj0785) catalyzing the reduction of nitrate to nitrite (Pittman and Kelly, 2005). Nitrite can subsequently be used as electron acceptor and reduced to ammonia by C. jejuni through the concerted activity of the pentaheme nitrite reductase NrfA (Cj1357c) and the electron donor protein NrfH (Cj1358c) of the tetraheme NapC/NirT cytochrome C family (Pittman et al., 2007). Exposure to nitrite leads to a nitrosative stress response in C. jejuni and the nitrite reductase NrfA catalyzes not only the reduction of nitrite to ammonia but participates with the single domain globin Cgb (Cj1586) in the detoxification of nitric oxide (Pittman et al., 2007).
S. Typhimurium uses the TtrRSBCA system for the oxygen-independent respiration with the alternative electron acceptor tetrathionate (Hensel et al., 1999). The recently described thiosulfate/tetrathionate-dependent respiration of C. jejuni is mediated by the bifunctional tetrathionate reductase/thiosulfate dehydrogenase TsdA (Liu et al., 2013) and seems to be a variable property in C. jejuni that promotes growth only of certain isolates in oxygen-limited conditions. Some C. jejuni strains like 81116 (Pearson et al., 2007) or M1 (Friis et al., 2010) harbor an active TsdA (C8j_0815) enzyme that catalyzes the reduction of tetrathionate to thiosulfate in the presence of additional electron donors like formate (Liu et al., 2013). In addition, these strains can use thiosulfate as an electron donor that is converted by TsdA to tetrathionate under microaerobic conditions. The C. jejuni isolates NCTC 11168 (Parkhill et al., 2000), RM1221 (Fouts et al., 2005) or 81–176 (Hofreuter et al., 2006) harbor truncated alleles of tsdA (c8j_0815) but possess like C. jejuni 81116 a less-active TsdA homolog (C8j_0040), which catalyzes the conversions between thiosulfate and tetrathionate at low levels (Liu et al., 2013). Future studies are needed to examine if the minimal thiosulfate oxidation and tetrathionate reduction driven by C8j_0040 can contribute to the growth of the C. jejuni strains harboring truncated tsdA genes. Furthermore, animal infection studies are required to clarify if tetrathionate respiration in C. jejuni provides a similar benefit for the colonization process as described for S. Typhimurium (Winter et al., 2010).
Physiological Factors Promoting the Colonization Process of C. jejuni
Transposon mutagenesis projects and in silico analysis of metabolic pathways led to the identification of genes essential for the proliferation of C. jejuni in vitro (Metris et al., 2011; Stahl and Stintzi, 2011). In addition, recent studies provided comprehensive data of genes expressed during the in vitro growth of C. jejuni when cultivated in nutrient rich media like BHI or Brucella broth by high-throughput deep sequencing of messenger RNA transcripts (Chaudhuri et al., 2011; Dugar et al., 2013; Porcelli et al., 2013). However, the knowledge of the nutritional requirements and metabolic traits that facilitate the growth of C. jejuni during the colonization of various hosts is limited. Still, our understanding about the metabolic necessities of C. jejuni required for a successful host infection has significantly improved in recent years by in vivo transcriptome analyses and screening of isogenic C. jejuni mutants in colonization models using 1 day old chickens, neonatal piglets and immunodeficient, antibiotic treated or germfree mice.
Taveirne et al. (2013) applied the RNAseq approach to study the in vivo transcriptome of C. jejuni after infection of 1-day-old chicks. Comparing the gene expression profile of C. jejuni isolated from the caecum of the chicks with the transcriptome during in vitro cultivation identified several gene groups that were more highly expressed in vivo including, for example, genes involved in the oxidative stress response (katA), iron (chuABCD; cjj81176_1601-cjj81176_1604) and phosphate (pstSCAB; cjj81176_0642-cjj81176_0645) transport. Elevated expression of the single-domain globin gene cbg (Elvers et al., 2004) indicated that C. jejuni was also exposed to nitrosative stress during colonization of the chicken caecum (Taveirne et al., 2013). One of the most increased transcripts in vivo was katA (Taveirne et al., 2013), the product of which is essential to counteract the damaging effect of H2O2(Grant and Park, 1995). Such oxidative stress might occur through the production of H2O2 by the intestinal epithelial cells at the colonization site of C. jejuni as indicated by ex vivo experiments with biopsy samples and infection of cultured HCT-8 cells (Corcionivoschi et al., 2012). In addition, the production of reactive oxygen species like superoxide anions and H2O2 are central for the innate immune defense (Rada and Leto, 2008). The KatA-mediated detoxification of H2O2 is crucial for C. jejuni in vivo, as the inactivation of katA reduced the colonization of the chicken caecum by the respective C. jejuni mutant (Bingham-Ramos and Hendrixson, 2008; Palyada et al., 2009). The severity of the observed colonization defects differed between these two studies, and this variability is likely due to dissimilar experimental settings or different C. jejuni and chicken strains used in the experiments. The ankyrin repeat-containing protein Cj1386 of C. jejuni NCTC 11168 is required for its full catalase activity by facilitating the correct heme trafficking to KatA (Flint et al., 2012). Similar to the katA mutant, a cj1386 mutant exhibited a reduced capability to colonize the caecum of chicken and to compete with the wild-type strain in the intestinal colonization of neonate piglets (Flint et al., 2012), further illustrating the importance of H2O2 detoxification for the pathogenesis of C. jejuni (Figure 1). The periplasmic c-type cytochrome Cj1153 has been suggested to participate in the defense against H2O2through the reduction and detoxification of exogenous H2O2to water and oxygen via electron transfer to the potential cytC peroxidases DocA (Cj0020c) and Cj0358 (Sellars et al., 2002; Hendrixson and DiRita, 2004). But in contrast to KatA, both peroxidases play no role in the resistance of C. jejuni against H2O2; nonetheless docA and cj0358 mutants were defective in the colonization of chicken (Hendrixson and DiRita, 2004; Bingham-Ramos and Hendrixson, 2008). The up-regulation of katA and additional oxidative stress response genes like sodB, ahpC and tpx was also observed in a microarray study analyzing the gene expression profile of C. jejuni in a rabbit ileal loop infection model (Stintzi et al., 2005). This observation hinted once more to the importance to counteract oxidative stress for a successful colonization process of C. jejuni and was confirmed by infection experiments demonstrating that AhpC and SodB activities promote the colonization of C. jejuni in chickens and mice (Palyada et al., 2009; Novik et al., 2010).
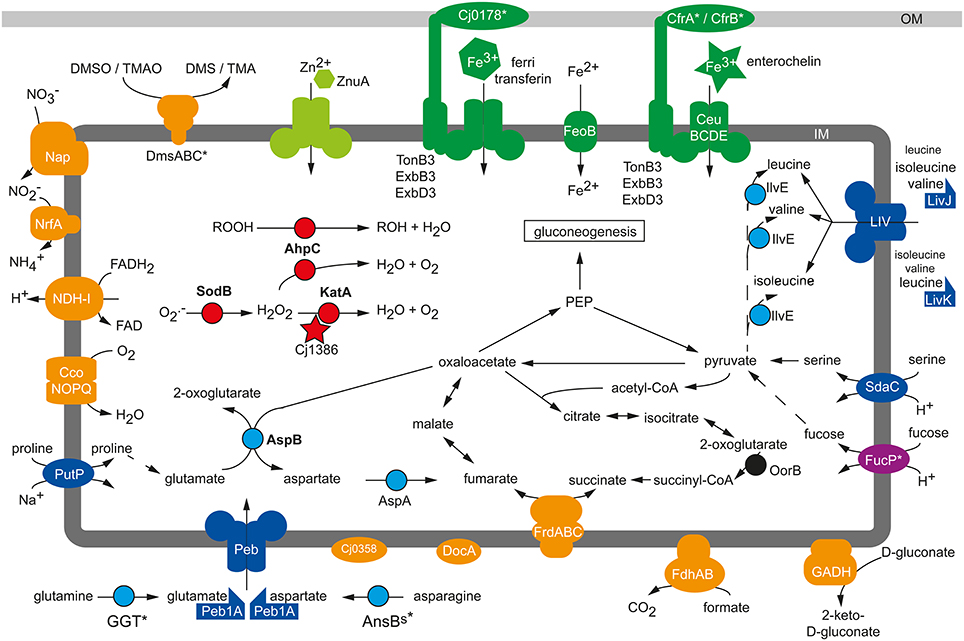
Figure 1. Physiological colonization factors of C. jejuni. Schematic presentation of metabolic factors being required for the efficient colonization process of C. jejuni in chicken, murine and porcine infection experiments. Blue, amino acid metabolism; red, reactive oxygen defense; orange, respiration; green, micronutrient utilization; IM, inner membrane; OM, outer membrane, *, Strain-specific gene; not conserved in all C. jejuni isolates.
Microarray-based gene expression analysis of C. jejuni isolated from the caecum of infected chickens suggested that an optimal respiratory adaptation of C. jejuni to its intestinal habitat is central for the colonization process (Woodall et al., 2005). This is reflected by the upregulation of the petABC and ccoNOQP operons in vivo, both of which facilitate the enhanced respiration of C. jejuni with the cb-type cytochrome c oxidase, which harbors high affinity to oxygen. Mutation of ccoNOQP abolished the ability of C. jejuni to colonize chicken, illustrating the crucial role of the terminal oxidoreductase for this microaerophilic pathogen to maintain its oxygen-dependent respiratory activity in the avian gastrointestinal tract (Weingarten et al., 2008). In contrast, inactivation of the terminal cyanide-insensitive oxidase CioA did not affect the colonization process (Weingarten et al., 2008), though the cioA expression was significantly higher in vivo than in vitro (Stintzi et al., 2005). Infection experiments with isogenic mutants of the highly branched respiratory chain revealed that especially the usage of alternative electron acceptors is crucial for the persistence of C. jejuni in the oxygen-limited environment of the intestine (Figure 1): The nap and nrf genes, facilitating the oxygen-independent respiration with nitrate and nitrite, were up-regulated in vivo (Woodall et al., 2005) and the NapA-mediated nitrate respiration of C. jejuni supported colonization in chickens (Weingarten et al., 2008). Interestingly, an increased intestinal nitrate level produced through an inflammatory host response also promoted the anaerobic respiration and luminal growth of E. coli in a murine infection model (Winter et al., 2013). Though the nitrite reductase activity of C. jejuni participates in nitrite respiration and protects against nitrosative stress in vitro, a nrfA mutant showed no general chicken colonization defect of the respective mutant in two independent studies (Pittman et al., 2007; Weingarten et al., 2008). Such discrepancy might be the result of different C. jejuni and chicken strains used in the studies as well as dissimilar infection doses and protocols. DMSO and TMAO have been proposed as electron acceptors that could be used by C. jejuni in aquatic environments since neither substances is found in significant amounts in the gastrointestinal tract of animals (Sellars et al., 2002). However, TMAO might eventually be present in the gastrointestinal tract under certain circumstances, as its precursor TMA is commonly produced by the human microbiota through the catabolism of carnitine and oxidized to TMAO in the host liver (Koeth et al., 2013; Zhu et al., 2014). This could explain why the inactivation of the dmsA gene in C. jejuni 81–176 resulted in a mutant with diminished persistence capability in immunodeficient mice when co-infected with the wild-type strain (Hofreuter et al., 2006). In contrast, the TMAO/DMSO reductase Cj0264c of C. jejuni NCTC 11168 was in single-infection experiments negligible for the colonization of chicken (Weingarten et al., 2008). Further work is needed to clarify if these different observations are the consequences of co- vs. single-infections, distinctive functions of DmsA and Cj0264c, or result from the dissimilarities in the C. jejuni strains and animal infection models used. Additional studies are required to identify the definite electron acceptors for the DmsA and Cj0264c oxidoreductases during the persistence of C. jejuni in the host intestine.
Increased in vivo gene expression levels were found for the C4-dicarboxylate transporter genes ducA and dcuB, the aspartase gene aspA, the fumarate reductase frdABC genes and the methylmenaquinol:fumarate reductase mfr genes (Woodall et al., 2005) participating in the fumarate respiration of C. jejuni under microaerobic and oxygen-limited conditions (Guccione et al., 2008, 2010). Higher in vitro expression of these genes was also observed under microaerobic and oxygen-limited growth conditions for the robust colonizer C. jejuni NCTC11168-O in comparison to its poor-colonizing variant C. jejuni NCTC11168-GS, likewise hinting toward an important role of oxygen-independent respiration for the persistence of C. jejuni in its host (Gaynor et al., 2004). Unexpectedly, the MrfA activity was dispensable for the C. jejuni chicken colonization (Weingarten et al., 2008), besides the impaired fumarate respiration under oxygen-limited conditions (Guccione et al., 2010). In contrast, the bifunctional fumarate reductase FrdA was required for optimal colonization (Weingarten et al., 2009). It is not yet clear to what extent the abolished utilization of growth substrates like the TCA cycle intermediates succinate or 2-oxoglutarate is responsible for the colonization defect of the frdA mutant in comparison to its inability to perform fumarate respiration. The importance of a functional TCA cycle for a C. jejuni infection was shown by the inactivation of the 2-oxoglutarate:acceptor oxidoreductase (oorB) resulting in a mutant with significantly reduced colonization capability in chicken compared to the wild-type strain as well (Weerakoon et al., 2009).
Intestinal pathogens have to overcome the gut microbiota-mediated colonization resistance of the host in order to establish a stable infection (Stecher et al., 2013). This might involve the successful competition with the intestinal microflora for common nutrients or, more often, the occupation of a specific metabolic niche in order to circumvent a direct competition with the commensal gut bacteria (Kamada et al., 2013). Murine infection models demonstrated that microbiota-derived molecular hydrogen promotes the growth of H. pylori (Olson and Maier, 2002) and S. Typhimurium (Maier et al., 2013) in a hydrogenase dependent manner, but hydrogenase activity of C. jejuni was not required for the persistence in chicken (Weerakoon et al., 2009). Other defects in systems fueling the respiration chain with electrons, like mutation in the formate dehydrogenase Fdh and the NDH-I complex, reduced the colonization capability of C. jejuni in chicken (Weerakoon et al., 2009). The gluconate dehydrogenase GADH, mainly active at 42°C, provided a host-specific benefit and was required by C. jejuni to persist in the chicken but not in the murine intestine (Pajaniappan et al., 2008), probably reflecting the different body temperatures of the avian and murine host. Whereas C. jejuni has to compete with commensal E. coli for gluconate (Chang et al., 2004), it generally does not catabolize carbohydrates that are utilized by commensal E. coli. C. jejuni strains harboring the FucP transporter are able to catabolize fucose (Muraoka and Zhang, 2011; Stahl et al., 2011), which is one of the less preferred growth substrates used by commensal and pathogenic E. coli in vivo (Fabich et al., 2008). The utilization of fucose is not essential for C. jejuni in vivo, as fucP mutants could be recovered from mono-infected chickens in similar amounts as the wild-type and had no disadvantage in chicken co-infection experiments with high infectious doses (Muraoka and Zhang, 2011). In low dose co-infection experiments with chicken the wild-type strain either outcompeted the fucP mutant (Muraoka and Zhang, 2011) or had no advantage (Stahl et al., 2011). In latter study, the additional administration of fucose allowed the wild-type strain to outcompete the mutant (Stahl et al., 2011), suggesting that sufficient amount of free fucose is not always available for C. jejuni under normal circumstances. Still, fucose catabolism seems to support the colonization of fucP-positive C. jejuni isolates in certain settings since a C. jejuni fucP mutant was recovered in lower numbers than the wild-type strain from co-infection experiments with neonatal piglets (Stahl et al., 2011).
The catabolism of free amino acids, derived through proteolytic protein degradation by the host and its intestinal microbiota, plays a central role for the colonization process of C. jejuni (Figure 1). Microarray experiments showed the upregulation of the serine dehydratase sdaA gene in C. jejuni colonizing the chicken caecum (Woodall et al., 2005), and an sdaA mutant with abolished serine utilization exhibited a severe colonization defect in chicken (Velayudhan et al., 2004) and mice (Hofreuter et al., 2012). Similarly, the GGT-dependent utilization of glutamine/glutathione contributed to the colonization of C. jejuni in the murine and avian intestine (Hofreuter et al., 2006; Barnes et al., 2007). Furthermore, a C. jejuni peb1A mutant, abolished in the growth with glutamate and aspartate in vitro, showed a colonization defect in mice, as did a putP mutant unable to catabolize proline (Pei et al., 1998; Hofreuter et al., 2012). With the exception of serine, the usage of amino acids provided no general but rather a tissue-specific benefit for C. jejuni during infection experiments with immune deficient Myd88−/− mice: Glutamine / glutathione and proline catabolism were required for the efficient colonization of the murine intestine, but had no supporting effect during the persistence of C. jejuni in the murine liver after intraperitoneal infection (Hofreuter et al., 2006, 2008, 2012). In contrast, asparagine utilization through the secreted asparaginase AnsBs did not promote the intestinal colonization of C. jejuni 81–176 but rather enhanced its colonization in the liver of mice (Hofreuter et al., 2008). Future studies have to clarify if the observed colonization defect of the C. jejuni ansBs mutant in the murine liver is solely the consequence of a growth disadvantage or if the secreted asparaginase of C. jejuni has an additional immune modulatory effect as described for S. Typhimurium (Kullas et al., 2012). The asparaginase-catalyzed deamination of asparagine leads to aspartate, which represents a growth substrate but also a precursor of the alternative electron acceptor fumarate for C. jejuni, as describe above. Thus, the colonization defect in chicken and mice of an aspA mutant (Guccione et al., 2008; Novik et al., 2010) might be the combined effect of a reduced carbon and energy source availability as well as a diminished capability to perform fumarate respiration in the oxygen-limited intestinal environment. Moreover, the C. jejuni aspartate aminotransferase aspB mutant was defective in mouse colonization (Novik et al., 2010). Amino acids that cannot be used as energy sources by C. jejuni promote its infection process as well. This observation was made in chicken infection experiments with a C. jejuni transposon mutant library that identified the putative amino acid transporter Cj0903c and the periplasmic binding protein LivJ of the LIV branched-chain amino acid ABC transporter as colonization factors of C. jejuni (Hendrixson and DiRita, 2004). Additionally, inactivation of the periplasmic binding proteins LivK, which facilitates primarily the high level uptake of leucine, showed a reduced colonization capacity of C. jejuni in chicken when a low inoculum dose was used (Ribardo and Hendrixson, 2011). The mutation of the branched-chain amino acid aminotransferase ilvE gene resulted in a similar colonization phenotype as observed for the livJ and livK mutants. Unexpectedly, inactivation of other components of the LIV ABC transporter system did not affect the colonization levels of C. jejuni in chicken, indicating that other permeases might be involved in the uptake of the branched amino acids as well (Ribardo and Hendrixson, 2011).
C. jejuni requires not only energy sources but also micronutrients like iron, nickel, molybdate, tungsten, cobalt and zinc (Stahl et al., 2012) for the successful competition with the host microbiota. The growth-supporting effect of zinc has been demonstrated in vivo (Figure 1): A C. jejuni mutant with an inactivated periplasmic binding protein ZnuA (Cj0143c) of a high-affinity zinc ABC transporter system (Cj0143c-Cj0141c) showed a caecum colonization defect in chickens with normal microbiota (Davis et al., 2009) but not in chickens harboring a limited intestinal flora (Gielda and DiRita, 2012). Interestingly, an increased amount of zinc, magnesium and iron was measured in the caecum of chickens with limited flora whereas the concentrations of copper and manganese were reduced (Gielda and DiRita, 2012).
In addition to zinc, the availability of iron plays a central role in the pathogenesis of pathogens (Braun, 2001). Iron occurs in the host organism generally in its oxidized form as ferric iron (Fe3+). To reduce the availability of Fe3+ for pathogens, specific iron-binding proteins like lactoferrin and transferrin sequester ferric iron in host organisms. To overcome this iron limitation commensal and pathogenic bacteria secret siderophores, specific iron chelators with higher affinity to Fe3+ than the iron-binding proteins of the host (Skaar, 2010). Several studies have demonstrated that the acquisition of iron is important for its infection process. However, C. jejuni does not produce and encode Fe3+-binding siderophores (Baig et al., 1986; Gundogdu et al., 2007). Instead, it benefits from the host and its microbiota by encoding for several iron uptake systems that bind and import the iron-chelating siderophores of other bacteria such as ferric-enterobactin and salmochelin, the fungal ferrichrome or the host-derived iron-binding substances transferrin/lactoferrin, hemin and hemoglobin (Miller et al., 2009; Naikare et al., 2013). The outer membrane ferric-enterobactin receptor CfrA (Cj0755) of C. jejuni NCTC 11168 is expressed under iron-restricted growth conditions and required for colonization of chicken (Palyada et al., 2004). Interestingly, the robust colonizer C. jejuni 81–176 lacks the cfrA gene (Hofreuter et al., 2006), as do various other C. jejuni strains (Zeng et al., 2013a). These strains harbor the alternative ferric-enterobactin receptor cfrB (cjj81176_0471), which occurs in C. jejuni NCTC 11168 and other cfrA-positive strains as a pseudogene (Xu et al., 2010). Inactivation of cfrB in the bovine C. jejuni isolate JL11, which lacks the cfrA gene, clearly demonstrated that CfrB is required by this strain for the colonization of chicken (Xu et al., 2010). The CeuBCDE (Cj1352-Cj1355) ABC transporter system mediates, in cooperation with CfrA, the uptake of enterobactin/enterochelin into C. jejuni, and a C. jejuni NCTC 11168 ceuE mutant showed a similar chicken colonization defect as the cfrA mutant (Palyada et al., 2004). A further putative siderophore receptor is encoded by cj0178 in C. jejuni NCTC 11168 and RM1221 but is absent in the isolate C. jejuni 81–176 (Hofreuter et al., 2006). The outer membrane protein Cj0178 (CtuA) was characterized as a receptor for the iron-binding host glycoproteins ferri-transferrin, ferri-lactoferrin and ferri-ovotransferrin (Miller et al., 2008). A cj0178 mutant strain of C. jejuni NCTC 11168 showed a severe colonization defect in chicken infection experiments (Palyada et al., 2004) and a slightly attenuated phenotype in the rabbit ileal loop model (Stintzi et al., 2005). The chuABCDZ (cj1613c-cj1617) gene cluster is widespread in C. jejuni and encodes for an iron uptake system that facilitates the utilization of the host compounds like hemoglobin and hemin as iron sources (Pickett et al., 1992; Ridley et al., 2006). Microarray and RNAseq analysis revealed the upregulation of chu genes in C. jejuni NCTC 11168 and 81–176 isolated from the caecum of infected chicken (Woodall et al., 2005; Taveirne et al., 2013), suggesting that the utilization of the host-derived heme is required for the colonization process of C. jejuni. However, a C. jejuni NCTC 11168 chuA mutant showed the same ability to colonize the chicken intestine as the wild-type strain (Naikare et al., 2013).
The uptake of bacterial or host-derived siderophores by C. jejuni requires, in addition to specific receptor and ABC transporter proteins, a periplasma-bridging TonB-ExbB-ExbD system, which energizes the translocation of the siderophores across the outer membrane (Braun, 2001). C. jejuni isolates are equipped with different numbers of TonB-ExbBD systems: While C. jejuni NCTC 11168 encodes for three TonB-ExbB-ExbD systems, C. jejuni 81–176 harbors only the TonB2-ExbB2-ExbD2 system, which seems to be conserved in C. jejuni (Hofreuter et al., 2006; Zeng et al., 2013b). It was recently demonstrated that TonB3 of C. jejuni NCTC 11168 mediates the uptake of a wide range of siderophores from bacteria and vertebrates, including enterobactin and salmochelin, and a tonB3 mutant showed an abolished colonization capability in chicken infection experiments (Naikare et al., 2013). Moreover, a C. jejuni NCTC 11168 tonB1 mutant exhibited like the tonB3 mutant, an attenuated colonization phenotype in chicken, whereas the inactivation of tonB2 in C. jejuni NCTC 11168 did not dramatically affect the enterobactin utilization as well as colonization capability (Naikare et al., 2013). C. jejuni relies not only on the utilization of siderophore-bound ferric iron but can also use ferrous (Fe2+) iron. Some C. jejuni strains take up Fe2+ through the FeoB transport protein (Naikare et al., 2006). Though a feoB mutant of C. jejuni NCTC 11168 had no growth disadvantage compared the wild-type strain during in vitro co-cultivation experiments, it showed a significant colonization defect in chickens and was outcompeted by the parental strain during a co-infection experiment with newborn piglets (Naikare et al., 2006). Future studies have to clarify how the puzzling diversity in the iron uptake systems affects the virulence and colonization fitness as well as the oxygen detoxification of different C. jejuni strains.
A Discreet Tenant: The Persistence of C. jejuni Inside Invaded Epithelial Cells
C. jejuni colonizes efficiently the gut of its diverse hosts where the majority of the population proliferates in the extracellular space of the mucus-filled intestinal crypts (Lee et al., 1986; Beery et al., 1988). Only a small fraction of the C. jejuni population can also be found inside intestinal epithelial cells when biopsies of infected humans (van Spreeuwel et al., 1985) or piglets (Babakhani et al., 1993) are analyzed. Similar to other facultative intracellular pathogens, like e.g., S. Typhimurium or Legionella pneumophila, C. jejuni resides during its intracellular stage inside a specific membrane-bound compartment, the so-called Campylobacter-containing-vacuole (CCV) (Watson and Galan, 2008). Though significant progress has been made in characterizing the metabolism of various facultative intracellular pathogens and in identifying bacterial factors facilitating efficient growth inside the host cells (Eisenreich et al., 2010), our knowledge about the metabolic properties that allow C. jejuni to persist in its intracellular compartment is still limited. In contrast to S. Typhimurium and L. pneumophila, C. jejuni does not multiply intracellularly (Watson and Galan, 2008). The absence of intracellular growth indicates that reduced metabolic activity might be sufficient for C. jejuni to maintain its membrane potential and viability inside its intracellular compartment. Accordingly, a metabolic downshift was recently described by quantitative proteome analysis of C. jejuni cells isolated from the CCVs of infected T84 colonic epithelial cells (Liu et al., 2012). This study showed that 20 h after infection 225 of the over 1400 detected C. jejuni proteins exhibit different levels compared to the proteome of C. jejuni cells isolated 2 h post-infection. Interestingly, 211 of the 225 proteins were found at lower levels, whereas only 14 proteins showed a 2- to 11-fold higher level, most prominently the acetyl-CoA synthetase ACS. The levels of the catalase KatA and the thiol peroxidase Tpx showed slightly but significant higher levels at 20 h compared to 2 h after infection. Surprisingly, KatA activity was not required by C. jejuni to survive within Hep-2 cells (Day et al., 2000), whereas the superoxide dismutase sodB mutant of C. jejuni showed abolished intracellular survival (Pesci et al., 1994; Novik et al., 2010). Interestingly, other proteins involved in oxidative stress response like AhpC, SodB, Trx and TrxB showed unaltered or slightly decreased levels, similar to proteins involved in the protection of C. jejuni against environmental stress (GroEL, DnaK, HtrA, GroES, GrpE, ClpB, DnaJ) (Liu et al., 2012).
The quiescent status of this pathogen inside the CCV is further enforced by the observation that many C. jejuni proteins with lower levels during the extended intracellular persistence are required for protein synthesis (Rps and Rpl proteins), the biosynthesis of amino acids (HisA, DapA, LysC, PheA), purines (PurH, PurL, PurM) and fatty acids (FabH, FabZ). In addition, several protein- and peptide-degrading proteins (ClpB, HslU, M16 and M24/M37 peptidase) and transporters involved in the uptake of amino acids (LivFJK, SdaC), phosphate (PstS) and metals like iron (Cj0175c), tungsten (TupA) or nickel (NikZ) showed lower levels 20 h post-infection (Liu et al., 2012). The metabolic downshift observed for intracellular C. jejuni is further reflected by decreased levels of proteins involved in the intermediary metabolism and gluconeogenesis like Tal, TpiA, GapA, FbaA and Pyk (Liu et al., 2012).
Most strikingly, C. jejuni undergoes a remarkable respiratory reprogramming when residing in its intracellular compartment for an extended time period: Proteins involved in the aerobic respiration like the cbb3-type cytochrome c oxidase subunits CcoO and CcoP and the ubiquinol-cytochrome c reductase (PetA, PetC) exhibited decreased protein levels after 20 h inside cultured epithelial cells. Such a decreased level was also seen for the formate dehydrogenase (FdhAB), the nitrate (NapAB) and nitrite (NrfA) reductases. C. jejuni seems to adapt its respiration mode to the oxygen-restricted conditions inside the CCV (Watson and Galan, 2008; Liu et al., 2012; Pryjma et al., 2012) by fumarate respiration: The fumarate reductase frdA mutant of C. jejuni 81–176 showed a significant intracellular survival defect inside COS-1 cells (Liu et al., 2012) and the importance of fumarate respiration was further supported by the reduced intracellular survival rate of an aspartate ammonia-lyase aspA mutant (Novik et al., 2010). In contrast to the frdA mutant, an mrfA mutant showed no decreased viability in the CCV after 20 h (Liu et al., 2012). C. jejuni 81–176 mutants with abolished formate (fdhB; cjj81176_1502), nitrate (napG; cjj81176_0802) and TMAO or other N- and S-oxide (torA; cjj81176_0291) respiration showed also no intracellular survival defect (Liu et al., 2012). Moreover, the cyanide-insensitive oxidase subunit CioA (CJJ81176_0118) with low affinity to oxygen is not required for the intracellular survival of C. jejuni 81–176 (Liu et al., 2012).
Additional insights into the intracellular lifestyle of C. jejuni provided microarray experiments analyzing the expression of C. jejuni 81–176 genes during the adhesion to and invasion into the human INT 407 cell line. This approach identified the stringent response regulator protein SpoT (Cj1272c) as being upregulated during the first 6 h post-infection of the cultured epithelial cells (Gaynor et al., 2005). SpoT modulates the guanosine tetraphosphate (ppGpp) metabolism in C. jejuni and inactivation of spoT in C. jejuni 81–176 resulted in a decreased synthesis of ppGpp similar as described for other bacterial pathogens (Dalebroux et al., 2010). The spoT mutant of C. jejuni 81–176 showed only a slight defect in adherence to and invasion into INT 407 cells, but a more pronounced defect in its intracellular survival over 20 h in comparison to the wild-type strain (Gaynor et al., 2005). Moreover, inactivation of spoT resulted in a decreased level of poly-phosphate (poly-P) in C. jejuni 81–176 comparable to a mutant strain lacking the polyphosphate kinase 1 (ppk1) mutant. PPK1 (Cj1359; CJJ81176_1361) uses ATP for the generation of poly-P, which participates in the stress response of C. jejuni and enables the survival during osmotic shock and low-nutrient stress. It was demonstrated that PPK1 is required for extended survival in infected INT407 cells (Candon et al., 2007), but as the ppk1 mutation results, similar to the mutation of spoT, in pleiotropic effects, it does not allow the identification of mechanisms solely required for the intracellular survival. Poly-P might not only be important as a metabolic regulator and protector against various stresses like hydrogen peroxide and osmotic stress, it might also serve as an energy and phosphate source (Kornberg, 1995) that enhances the viability of C. jejuni while residing in the nutrient restricted environment of the CCV.
Though progress has been made in recent years to describe the nutrient requirements that allow C. jejuni to thrive in the gastrointestinal lumen of its hosts, how C. jejuni maintains its viability inside invaded epithelial cells and energizes its intracellular persistence remains ill-defined.
Outlook
Campylobacteriosis of humans in industrialized countries is assumed to be an accidental and transitory event in the life style of C. jejuni. Such short-term interactions prevent co-evolution and potential fine-tuning of Campylobacter virulence factors. Consequently, the evolutionary forces that shape the metabolic and pathogenic properties of C. jejuni occur during the interactions with the microbiota of its primary hosts rather than within the human gastrointestinal ecosystem. Still, the metabolic framework of C. jejuni is sufficient to overcome the colonization resistance in the human gastrointestinal tract and to efficiently promote its proliferation, leading consequently to the development of diarrhea. These findings illustrate the necessity for future comprehensive studies to investigate the extent to which metabolic factors mediate the long-term persistence of C. jejuni in its natural hosts like chicken and cattle. Specifically targeting such growth-promoting metabolic pathways for the development of drugs might not only lower the colonization burden in animal hosts with C. jejuni but will consequently lower the exposure doses for humans. Moreover, the restricted carbohydrate catabolism of C. jejuni in comparison with other enteropathogenic pathogens like S. Typhimurium (Becker et al., 2006; Steeb et al., 2013) should facilitate the discovery of “metabolic Achilles heels” for the successful design of drugs against Campylobacter.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to thank Alyssa Ingmundson, Juliane Mohr and Hanne Vorwerk for critical review of the manuscript. The research is supported by the Deutsche Forschungsgemeinschaft with the DFG grants HO 4553/1-1 and HO 4553/2-1 as part of the priority program 1316 “Host-Adapted Metabolism of Bacterial Pathogens” as well as the FBI-Zoo grant from the German Ministry for Education and Research (BMBF).
References
Adibi, S. A., and Mercer, D. W. (1973). Protein digestion in human intestine as reflected in luminal, mucosal, and plasma amino acid concentrations after meals. J. Clin. Invest. 52, 1586–1594. doi: 10.1172/JCI107335
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ahmed, I. H., Manning, G., Wassenaar, T. M., Cawthraw, S., and Newell, D. G. (2002). Identification of genetic differences between two Campylobacter jejuni strains with different colonization potentials. Microbiology 148, 1203–1212.
Allos, B. M. (2001). Campylobacter jejuni Infections: update on emerging issues and trends. Clin. Infect. Dis. 32, 1201–1206. doi: 10.1086/319760
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arakawa, C., Kuratsu, M., Furihata, K., Hiratsuka, T., Itoh, N., Seto, H., et al. (2011). Diversity of the early step of the futalosine pathway. Antimicrob. Agents Chemother. 55, 913–916. doi: 10.1128/AAC.01362-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Atack, J. M., Harvey, P., Jones, M. A., and Kelly, D. J. (2008). The Campylobacter jejuni thiol peroxidases Tpx and Bcp both contribute to aerotolerance and peroxide-mediated stress resistance but have distinct substrate specificities. J. Bacteriol. 190, 5279–5290. doi: 10.1128/JB.00100-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Babakhani, F. K., Bradley, G. A., and Joens, L. A. (1993). Newborn piglet model for campylobacteriosis. Infect. Immun. 61, 3466–3475.
Baig, B. H., Wachsmuth, I. K., and Morris, G. K. (1986). Utilization of exogenous siderophores by Campylobacter species. J. Clin. Microbiol. 23, 431–433.
Baillon, M. L., van Vliet, A. H., Ketley, J. M., Constantinidou, C., and Penn, C. W. (1999). An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181, 4798–4804.
Barnes, I. H., Bagnall, M. C., Browning, D. D., Thompson, S. A., Manning, G., and Newell, D. G. (2007). Gamma-glutamyl transpeptidase has a role in the persistent colonization of the avian gut by Campylobacter jejuni. Microb. Pathog. 43, 198–207. doi: 10.1016/j.micpath.2007.05.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Becker, D., Selbach, M., Rollenhagen, C., Ballmaier, M., Meyer, T. F., Mann, M., et al. (2006). Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307. doi: 10.1038/nature04616
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beery, J. T., Hugdahl, M. B., and Doyle, M. P. (1988). Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 54, 2365–2370.
Bernalier-Donadille, A. (2010). Fermentative metabolism by the human gut microbiota. Gastroenterol. Clin. Biol. 34(Suppl. 1), S16–S22. doi: 10.1016/S0399-8320(10)70016-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhaduri, S., and Cottrell, B. (2004). Survival of cold-stressed Campylobacter jejuni on ground chicken and chicken skin during frozen storage. Appl. Environ. Microbiol. 70, 7103–7109. doi: 10.1128/AEM.70.12.7103-7109.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bieche, C., de Lamballerie, M., Chevret, D., Federighi, M., and Tresse, O. (2012). Dynamic proteome changes in Campylobacter jejuni 81-176 after high pressure shock and subsequent recovery. J. Proteomics 75, 1144–1156. doi: 10.1016/j.jprot.2011.10.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bingham-Ramos, L. K., and Hendrixson, D. R. (2008). Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect. Immun. 76, 1105–1114. doi: 10.1128/IAI.01430-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Black, R. E., Levine, M. M., Clements, M. L., Hughes, T. P., and Blaser, M. J. (1988). Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157, 472–479. doi: 10.1093/infdis/157.3.472
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Blaser, M. J., Hardesty, H. L., Powers, B., and Wang, W. L. (1980). Survival of Campylobacter fetus subsp. jejuni in biological milieus. J. Clin. Microbiol. 11, 309–313.
Blaser, M. J., Perez, G. P., Smith, P. F., Patton, C., Tenover, F. C., Lastovica, A. J., et al. (1986). Extraintestinal Campylobacter jejuni and Campylobacter coli infections: host factors and strain characteristics. J. Infect. Dis. 153, 552–559. doi: 10.1093/infdis/153.3.552
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bochner, B. R. (2009). Global phenotypic characterization of bacteria. FEMS Microbiol. Rev. 33, 191–205. doi: 10.1111/j.1574-6976.2008.00149.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boehm, M., Hoy, B., Rohde, M., Tegtmeyer, N., Baek, K. T., Oyarzabal, O. A., et al. (2012). Rapid paracellular transmigration of Campylobacter jejuni across polarized epithelial cells without affecting TER: role of proteolytic-active HtrA cleaving E-cadherin but not fibronectin. Gut Pathog. 4:3. doi: 10.1186/1757-4749-4-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bolton, F. J., and Coates, D. (1983). A study of the oxygen and carbon dioxide requirements of thermophilic Campylobacters. J. Clin. Pathol. 36, 829–834. doi: 10.1136/jcp.36.7.829
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Braun, V. (2001). Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 291, 67–79. doi: 10.1078/1438-4221-00103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brondsted, L., Andersen, M. T., Parker, M., Jorgensen, K., and Ingmer, H. (2005). The HtrA protease of Campylobacter jejuni is required for heat and oxygen tolerance and for optimal interaction with human epithelial cells. Appl. Environ. Microbiol. 71, 3205–3212. doi: 10.1128/AEM.71.6.3205-3212.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cabiscol, E., Tamarit, J., and Ros, J. (2000). Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3, 3–8.
Candon, H. L., Allan, B. J., Fraley, C. D., and Gaynor, E. C. (2007). Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J. Bacteriol. 189, 8099–8108. doi: 10.1128/JB.01037-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carlone, G. M., and Anet, F. A. (1983). Detection of menaquinone-6 and a novel methyl-substituted menaquinone-6 in Campylobacter jejuni and Campylobacter fetus subsp. fetus. J. Gen. Microbiol. 129, 3385–3393.
Carlone, G. M., and Lascelles, J. (1982). Aerobic and anaerobic respiratory systems in Campylobacter fetus subsp. jejuni grown in atmospheres containing hydrogen. J. Bacteriol. 152, 306–314.
Champion, O. L., Gaunt, M. W., Gundogdu, O., Elmi, A., Witney, A. A., Hinds, J., et al. (2005). Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. U.S.A. 102, 16043–16048. doi: 10.1073/pnas.0503252102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, D. E., Smalley, D. J., Tucker, D. L., Leatham, M. P., Norris, W. E., Stevenson, S. J., et al. (2004). Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U.S.A. 101, 7427–7432. doi: 10.1073/pnas.0307888101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chaudhuri, R. R., Yu, L., Kanji, A., Perkins, T. T., Gardner, P. P., Choudhary, J., et al. (2011). Quantitative RNA-seq analysis of the Campylobacter jejuni transcriptome. Microbiology 157, 2922–2932. doi: 10.1099/mic.0.050278-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cohn, M. T., Ingmer, H., Mulholland, F., Jorgensen, K., Wells, J. M., and Brondsted, L. (2007). Contribution of conserved ATP-dependent proteases of Campylobacter jejuni to stress tolerance and virulence. Appl. Environ. Microbiol. 73, 7803–7813. doi: 10.1128/AEM.00698-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corcionivoschi, N., Alvarez, L. A., Sharp, T. H., Strengert, M., Alemka, A., Mantell, J., et al. (2012). Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe 12, 47–59. doi: 10.1016/j.chom.2012.05.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dalebroux, Z. D., Svensson, S. L., Gaynor, E. C., and Swanson, M. S. (2010). ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171–199. doi: 10.1128/MMBR.00046-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dandekar, T., Astrid, F., Jasmin, P., and Hensel, M. (2012). Salmonella enterica: a surprisingly well-adapted intracellular lifestyle. Front. Microbiol. 3:164. doi: 10.3389/fmicb.2012.00164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Davis, L. M., Kakuda, T., and DiRita, V. J. (2009). A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. J. Bacteriol. 191, 1631–1640. doi: 10.1128/JB.01394-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Day, C. J., Hartley-Tassell, L. E., Shewell, L. K., King, R. M., Tram, G., Day, S. K., et al. (2012). Variation of chemosensory receptor content of Campylobacter jejuni strains and modulation of receptor gene expression under different in vivo and in vitro growth conditions. BMC Microbiol. 12:128. doi: 10.1186/1471-2180-12-128
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Day, W. A. Jr., Sajecki, J. L., Pitts, T. M., and Joens, L. A. (2000). Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68, 6337–6345. doi: 10.1128/IAI.68.11.6337-6345.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
de Haan, C. P., Llarena, A. K., Revez, J., and Hanninen, M. L. (2012). Association of Campylobacter jejuni metabolic traits with multilocus sequence types. Appl. Environ. Microbiol. 78, 5550–5554. doi: 10.1128/AEM.01023-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Doig, P., de Jonge, B. L., Alm, R. A., Brown, E. D., Uria-Nickelsen, M., Noonan, B., et al. (1999). Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol. Mol. Biol. Rev. 63, 675–707.
Dufour, V., Alazzam, B., Ermel, G., Thepaut, M., Rossero, A., Tresse, O., et al. (2012). Antimicrobial activities of isothiocyanates against Campylobacter jejuni isolates. Front. Cell. Infect. Microbiol. 2:53. doi: 10.3389/fcimb.2012.00053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dugar, G., Herbig, A., Forstner, K. U., Heidrich, N., Reinhardt, R., Nieselt, K., et al. (2013). High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 9:e1003495. doi: 10.1371/journal.pgen.1003495
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eisenreich, W., Dandekar, T., Heesemann, J., and Goebel, W. (2010). Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412. doi: 10.1038/nrmicro2351
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Elharrif, Z., and Megraud, F. (1986). Characterization of thermophilic Campylobacter. 1. carbon-substrate utilization tests. Curr. Microbiol. 13, 117–122. doi: 10.1007/BF01568505
Elvers, K. T., Wu, G., Gilberthorpe, N. J., Poole, R. K., and Park, S. F. (2004). Role of an inducible single-domain hemoglobin in mediating resistance to nitric oxide and nitrosative stress in Campylobacter jejuni and Campylobacter coli. J. Bacteriol. 186, 5332–5341. doi: 10.1128/JB.186.16.5332-5341.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Epps, S. V., Harvey, R. B., Hume, M. E., Phillips, T. D., Anderson, R. C., and Nisbet, D. J. (2013). Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int. J. Environ. Res. Public Health 10, 6292–6304. doi: 10.3390/ijerph10126292
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fabich, A. J., Jones, S. A., Chowdhury, F. Z., Cernosek, A., Anderson, A., Smalley, D., et al. (2008). Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 76, 1143–1152. doi: 10.1128/IAI.01386-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Flint, A., Sun, Y. Q., and Stintzi, A. (2012). Cj1386 is an ankyrin-containing protein involved in heme trafficking to catalase in Campylobacter jejuni. J. Bacteriol. 194, 334–345. doi: 10.1128/JB.05740-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fouts, D. E., Mongodin, E. F., Mandrell, R. E., Miller, W. G., Rasko, D. A., Ravel, J., et al. (2005). Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. doi: 10.1371/journal.pbio.0030015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Friis, C., Wassenaar, T. M., Javed, M. A., Snipen, L., Lagesen, K., Hallin, P. F., et al. (2010). Genomic characterization of Campylobacter jejuni strain M1. PLoS ONE 5:e12253. doi: 10.1371/journal.pone.0012253
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frirdich, E., Biboy, J., Adams, C., Lee, J., Ellermeier, J., Gielda, L. D., et al. (2012). Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog. 8:e1002602. doi: 10.1371/journal.ppat.1002602
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fuchs, T. M., Eisenreich, W., Heesemann, J., and Goebel, W. (2012). Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra- and intracellular habitats. FEMS Microbiol. Rev. 36, 435–462. doi: 10.1111/j.1574-6976.2011.00301.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garvis, S. G., Puzon, G. J., and Konkel, M. E. (1996). Molecular characterization of a Campylobacter jejuni 29-kilodalton periplasmic binding protein. Infect. Immun. 64, 3537–3543.
Garvis, S. G., Tipton, S. L., and Konkel, M. E. (1997). Identification of a functional homolog of the Escherichia coli and Salmonella typhimurium cysM gene encoding O-acetylserine sulfhydrylase B in Campylobacter jejuni. Gene 185, 63–67. doi: 10.1016/S0378-1119(96)00631-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaynor, E. C., Cawthraw, S., Manning, G., MacKichan, J. K., Falkow, S., and Newell, D. G. (2004). The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186, 503–517. doi: 10.1128/JB.186.2.503-517.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaynor, E. C., Wells, D. H., MacKichan, J. K., and Falkow, S. (2005). The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 56, 8–27. doi: 10.1111/j.1365-2958.2005.04525.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gielda, L. M., and DiRita, V. J. (2012). Zinc competition among the intestinal microbiota. MBio 3, e00171–e00112. doi: 10.1128/mBio.00171-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gilbreath, J. J., Cody, W. L., Merrell, D. S., and Hendrixson, D. R. (2011). Change is good: variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter. Microbiol. Mol. Biol. Rev. 75, 84–132. doi: 10.1128/MMBR.00035-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gonzalez, M., Hakkinen, M., Rautelin, H., and Hanninen, M. L. (2009). Bovine Campylobacter jejuni strains differ from human and chicken strains in an analysis of certain molecular genetic markers. Appl. Environ. Microbiol. 75, 1208–1210. doi: 10.1128/AEM.01879-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grant, K. A., and Park, S. F. (1995). Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology 141, 1369–1376. doi: 10.1099/13500872-141-6-1369
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gripp, E., Hlahla, D., Didelot, X., Kops, F., Maurischat, S., Tedin, K., et al. (2011). Closely related Campylobacter jejuni strains from different sources reveal a generalist rather than a specialist lifestyle. BMC Genomics 12:584. doi: 10.1186/1471-2164-12-584
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guccione, E., Hitchcock, A., Hall, S. J., Mulholland, F., Shearer, N., van Vliet, A. H., et al. (2010). Reduction of fumarate, mesaconate and crotonate by Mfr, a novel oxygen-regulated periplasmic reductase in Campylobacter jejuni. Environ. Microbiol. 12, 576–591. doi: 10.1111/j.1462-2920.2009.02096.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guccione, E., Leon-Kempis Mdel, R., Pearson, B. M., Hitchin, E., Mulholland, F., van Diemen, P. M., et al. (2008). Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69, 77–93. doi: 10.1111/j.1365-2958.2008.06263.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gundogdu, O., Bentley, S. D., Holden, M. T., Parkhill, J., Dorrell, N., and Wren, B. W. (2007). Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics 8:162. doi: 10.1186/1471-2164-8-162
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hani, E. K., and Chan, V. L. (1994). Cloning, characterization, and nucleotide sequence analysis of the argH gene from Campylobacter jejuni TGH9011 encoding argininosuccinate lyase. J. Bacteriol. 176, 1865–1871.
Hani, E. K., Ng, D., and Chan, V. L. (1999). Arginine biosynthesis in Campylobacter jejuni TGH9011: determination of the argCOBD cluster. Can. J. Microbiol. 45, 959–969. doi: 10.1139/w99-095
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hartley-Tassell, L. E., Shewell, L. K., Day, C. J., Wilson, J. C., Sandhu, R., Ketley, J. M., et al. (2010). Identification and characterization of the aspartate chemosensory receptor of Campylobacter jejuni. Mol. Microbiol. 75, 710–730. doi: 10.1111/j.1365-2958.2009.07010.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hazeleger, W. C., Wouters, J. A., Rombouts, F. M., and Abee, T. (1998). Physiological activity of Campylobacter jejuni far below the minimal growth temperature. Appl. Environ. Microbiol. 64, 3917–3922.
He, G., Shankar, R. A., Chzhan, M., Samouilov, A., Kuppusamy, P., and Zweier, J. L. (1999). Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. U.S.A. 96, 4586–4591. doi: 10.1073/pnas.96.8.4586
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hendrixson, D. R., and DiRita, V. J. (2004). Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52, 471–484. doi: 10.1111/j.1365-2958.2004.03988.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hensel, M., Hinsley, A. P., Nikolaus, T., Sawers, G., and Berks, B. C. (1999). The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32, 275–287. doi: 10.1046/j.1365-2958.1999.01345.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hepworth, P. J., Leatherbarrow, H., Hart, C. A., and Winstanley, C. (2007). Use of suppression subtractive hybridisation to extend our knowledge of genome diversity in Campylobacter jejuni. BMC Genomics 8:110. doi: 10.1186/1471-2164-8-110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hiett, K. L., Stintzi, A., Andacht, T. M., Kuntz, R. L., and Seal, B. S. (2008). Genomic differences between Campylobacter jejuni isolates identify surface membrane and flagellar function gene products potentially important for colonizing the chicken intestine. Funct. Integr. Genomics 8, 407–420. doi: 10.1007/s10142-008-0087-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hinton, A. Jr. (2006). Growth of Campylobacter in media supplemented with organic acids. J. Food Prot. 69, 34–38.
Hiratsuka, T., Furihata, K., Ishikawa, J., Yamashita, H., Itoh, N., Seto, H., et al. (2008). An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321, 1670–1673. doi: 10.1126/science.1160446
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hitchcock, A., Hall, S. J., Myers, J. D., Mulholland, F., Jones, M. A., and Kelly, D. J. (2010). Roles of the twin-arginine translocase and associated chaperones in the biogenesis of the electron transport chains of the human pathogen Campylobacter jejuni. Microbiology 156, 2994–3010. doi: 10.1099/mic.0.042788-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hoffman, P. S., and Goodman, T. G. (1982). Respiratory physiology and energy conservation efficiency of Campylobacter jejuni. J. Bacteriol. 150, 319–326.
Hoffman, P. S., Sisson, G., Croxen, M. A., Welch, K., Harman, W. D., Cremades, N., et al. (2007). Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni. Antimicrob. Agents Chemother. 51, 868–876. doi: 10.1128/AAC.01159-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hofreuter, D., Mohr, J., Wensel, O., Rademacher, S., Schreiber, K., Schomburg, D., et al. (2012). Contribution of amino acid catabolism to the tissue specific persistence of Campylobacter jejuni in a murine colonization model. PLoS ONE 7:e50699. doi: 10.1371/journal.pone.0050699
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hofreuter, D., Novik, V., and Galan, J. E. (2008). Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4, 425–433. doi: 10.1016/j.chom.2008.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hofreuter, D., Tsai, J., Watson, R. O., Novik, V., Altman, B., Benitez, M., et al. (2006). Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74, 4694–4707. doi: 10.1128/IAI.00210-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Howlett, R. M., Hughes, B. M., Hitchcock, A., and Kelly, D. J. (2012). Hydrogenase activity in the foodborne pathogen Campylobacter jejuni depends upon a novel ABC-type nickel transporter (NikZYXWV) and is SlyD-independent. Microbiology 158, 1645–1655. doi: 10.1099/mic.0.054130-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hugdahl, M. B., Beery, J. T., and Doyle, M. P. (1988). Chemotactic behavior of Campylobacter jejuni. Infect. Immun. 56, 1560–1566.
Hughes, N. J., Clayton, C. L., Chalk, P. A., and Kelly, D. J. (1998). Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180, 1119–1128.
Jackson, R. J., Elvers, K. T., Lee, L. J., Gidley, M. D., Wainwright, L. M., Lightfoot, J., et al. (2007). Oxygen reactivity of both respiratory oxidases in Campylobacter jejuni: the cydAB genes encode a cyanide-resistant, low-affinity oxidase that is not of the cytochrome bd type. J. Bacteriol. 189, 1604–1615. doi: 10.1128/JB.00897-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Janssen, R., Krogfelt, K. A., Cawthraw, S. A., van Pelt, W., Wagenaar, J. A., and Owen, R. J. (2008). Host-pathogen interactions in Campylobacter infections: the host perspective. Clin. Microbiol. Rev. 21, 505–518. doi: 10.1128/CMR.00055-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jung, H., Hilger, D., and Raba, M. (2012). The Na(+)/L-proline transporter PutP. Front Biosci (Landmark Ed) 17, 745–759. doi: 10.2741/3955
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kamada, N., Chen, G. Y., Inohara, N., and Nunez, G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karlyshev, A. V., Ketley, J. M., and Wren, B. W. (2005). The Campylobacter jejuni glycome. FEMS Microbiol. Rev. 29, 377–390.
Karlyshev, A. V., Thacker, G., Jones, M. A., Clements, M. O., and Wren, B. W. (2014). Campylobacter jejuni gene cj0511 encodes a serine peptidase essential for colonisation. FEBS Open Bio 4, 468–472. doi: 10.1016/j.fob.2014.04.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kendall, J. J., Barrero-Tobon, A. M., Hendrixson, D. R., and Kelly, D. J. (2014). Hemerythrins in the microaerophilic bacterium Campylobacter jejuni help protect key iron-sulphur cluster enzymes from oxidative damage. Environ. Microbiol. 16, 1105–1121. doi: 10.1111/1462-2920.12341
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kirkpatrick, A. S., Yokoyama, T., Choi, K. J., and Yeo, H. J. (2009). Campylobacter jejuni fatty acid synthase II: structural and functional analysis of beta-hydroxyacyl-ACP dehydratase (FabZ). Biochem. Biophys. Res. Commun. 380, 407–412. doi: 10.1016/j.bbrc.2009.01.115
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koeth, R. A., Wang, Z., Levison, B. S., Buffa, J. A., Org, E., Sheehy, B. T., et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585. doi: 10.1038/nm.3145
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kornberg, A. (1995). Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J. Bacteriol. 177, 491–496.
Kothary, M. H., and Babu, U. S. (2001). Infective dose of foodborne pathogens in volunteers: a review. J. Food Saf. 21, 49–73. doi: 10.1111/j.1745-4565.2001.tb00307.x
Kullas, A. L., McClelland, M., Yang, H. J., Tam, J. W., Torres, A., Porwollik, S., et al. (2012). L-asparaginase II produced by Salmonella typhimurium inhibits T cell responses and mediates virulence. Cell Host Microbe 12, 791–798. doi: 10.1016/j.chom.2012.10.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Labigne, A., Courcoux, P., and Tompkins, L. (1992). Cloning of Campylobacter jejuni genes required for leucine biosynthesis, and construction of Leu-negative mutant of C. jejuni by shuttle transposon mutagenesis. Res. Microbiol. 143, 15–26. doi: 10.1016/0923-2508(92)90030-R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leach, S., Harvey, P., and Wali, R. (1997). Changes with growth rate in the membrane lipid composition of and amino acid utilization by continuous cultures of Campylobacter jejuni. J. Appl. Microbiol. 82, 631–640. doi: 10.1111/j.1365-2672.1997.tb02873.x
Leduc, D., Gallaud, J., Stingl, K., and de Reuse, H. (2010). Coupled amino acid deamidase-transport systems essential for Helicobacter pylori colonization. Infect. Immun. 78, 2782–2792. doi: 10.1128/IAI.00149-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, A., O'Rourke, J. L., Barrington, P. J., and Trust, T. J. (1986). Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacter jejuni: a mouse cecal model. Infect. Immun. 51, 536–546.
Leon-Kempis Mdel, R., Guccione, E., Mulholland, F., Williamson, M. P., and Kelly, D. J. (2006). The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 60, 1262–1275. doi: 10.1111/j.1365-2958.2006.05168.x
Li, X., Apel, D., Gaynor, E. C., and Tanner, M. E. (2011). 5′-methylthioadenosine nucleosidase is implicated in playing a key role in a modified futalosine pathway for menaquinone biosynthesis in Campylobacter jejuni. J. Biol. Chem. 286, 19392–19398. doi: 10.1074/jbc.M111.229781
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lin, A. E., Krastel, K., Hobb, R. I., Thompson, S. A., Cvitkovitch, D. G., and Gaynor, E. C. (2009). Atypical roles for Campylobacter jejuni amino acid ATP binding cassette transporter components PaqP and PaqQ in bacterial stress tolerance and pathogen-host cell dynamics. Infect. Immun. 77, 4912–4924. doi: 10.1128/IAI.00571-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Line, J. E., Hiett, K. L., Guard-Bouldin, J., and Seal, B. S. (2010). Differential carbon source utilization by Campylobacter jejuni 11168 in response to growth temperature variation. J. Microbiol. Methods 80, 198–202. doi: 10.1016/j.mimet.2009.12.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, T. W., Ho, C. W., Huang, H. H., Chang, S. M., Popat, S. D., Wang, Y. T., et al. (2009). Role for alpha-L-fucosidase in the control of Helicobacter pylori-infected gastric cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 14581–14586. doi: 10.1073/pnas.0903286106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, X., Gao, B., Novik, V., and Galan, J. E. (2012). Quantitative Proteomics of Intracellular Campylobacter jejuni Reveals Metabolic Reprogramming. PLoS Pathog. 8:e1002562. doi: 10.1371/journal.ppat.1002562
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, Y. W., Denkmann, K., Kosciow, K., Dahl, C., and Kelly, D. J. (2013). Tetrathionate stimulated growth of Campylobacter jejuni identifies a new type of bi-functional tetrathionate reductase (TsdA) that is widely distributed in bacteria. Mol. Microbiol. 88, 173–188. doi: 10.1111/mmi.12176
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maier, L., Vyas, R., Cordova, C. D., Lindsay, H., Schmidt, T. S., Brugiroux, S., et al. (2013). Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe 14, 641–651. doi: 10.1016/j.chom.2013.11.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mendz, G. L., Ball, G. E., and Meek, D. J. (1997). Pyruvate metabolism in Campylobacter spp. Biochim. Biophys. Acta 1334, 291–302. doi: 10.1016/S0304-4165(96)00107-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mendz, G. L., Hazell, S. L., and Burns, B. P. (1993). Glucose utilization and lactate production by Helicobacter pylori. J. Gen. Microbiol. 139, 3023–3028. doi: 10.1099/00221287-139-12-3023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mendz, G. L., Hazell, S. L., and Burns, B. P. (1994). The Entner-Doudoroff pathway in Helicobacter pylori. Arch. Biochem. Biophys. 312, 349–356. doi: 10.1006/abbi.1994.1319
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Metris, A., Reuter, M., Gaskin, D. J., Baranyi, J., and van Vliet, A. H. (2011). In vivo and in silico determination of essential genes of Campylobacter jejuni. BMC Genomics 12:535. doi: 10.1186/1471-2164-12-535
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, C. E., Rock, J. D., Ridley, K. A., Williams, P. H., and Ketley, J. M. (2008). Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni. J. Bacteriol. 190, 1900–1911. doi: 10.1128/JB.01761-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, C. E., Williams, P. H., and Ketley, J. M. (2009). Pumping iron: mechanisms for iron uptake by Campylobacter. Microbiology 155, 3157–3165. doi: 10.1099/mic.0.032425-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mohammed, K. A., Miles, R. J., and Halablab, M. A. (2004). The pattern and kinetics of substrate metabolism of Campylobacter jejuni and Campylobacter coli. Lett. Appl. Microbiol. 39, 261–266. doi: 10.1111/j.1472-765X.2004.01574.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moss, C. W., Kai, A., Lambert, M. A., and Patton, C. (1984). Isoprenoid quinone content and cellular fatty acid composition of Campylobacter species. J. Clin. Microbiol. 19, 772–776.
Müller, A., Leon-Kempis Mdel, R., Dodson, E., Wilson, K. S., Wilkinson, A. J., and Kelly, D. J. (2007). A bacterial virulence factor with a dual role as an adhesin and a solute-binding protein: the crystal structure at 1.5 A resolution of the PEB1a protein from the food-borne human pathogen Campylobacter jejuni. J. Mol. Biol. 372, 160–171. doi: 10.1016/j.jmb.2007.06.041
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Müller, A., Thomas, G. H., Horler, R., Brannigan, J. A., Blagova, E., Levdikov, V. M., et al. (2005). An ATP-binding cassette-type cysteine transporter in Campylobacter jejuni inferred from the structure of an extracytoplasmic solute receptor protein. Mol. Microbiol. 57, 143–155. doi: 10.1111/j.1365-2958.2005.04691.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muraoka, W. T., and Zhang, Q. (2011). Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J. Bacteriol. 193, 1065–1075. doi: 10.1128/JB.01252-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murphy, C., Carroll, C., and Jordan, K. N. (2006). Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J. Appl. Microbiol. 100, 623–632. doi: 10.1111/j.1365-2672.2006.02903.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Myers, J. D., and Kelly, D. J. (2005). A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology 151, 233–242. doi: 10.1099/mic.0.27573-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naikare, H., Butcher, J., Flint, A., Xu, J., Raymond, K. N., and Stintzi, A. (2013). Campylobacter jejuni ferric-enterobactin receptor CfrA is TonB3 dependent and mediates iron acquisition from structurally different catechol siderophores. Metallomics 5, 988–996. doi: 10.1039/c3mt20254b
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naikare, H., Palyada, K., Panciera, R., Marlow, D., and Stintzi, A. (2006). Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74, 5433–5444. doi: 10.1128/IAI.00052-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Novik, V., Hofreuter, D., and Galan, J. E. (2010). Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect. Immun. 78, 3540–3553. doi: 10.1128/IAI.00109-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Olson, J. W., and Maier, R. J. (2002). Molecular hydrogen as an energy source for Helicobacter pylori. Science 298, 1788–1790. doi: 10.1126/science.1077123
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ostrovsky de Spicer, P., and Maloy, S. (1993). PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc. Natl. Acad. Sci. U.S.A. 90, 4295–4298. doi: 10.1073/pnas.90.9.4295
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pacheco, A. R., Curtis, M. M., Ritchie, J. M., Munera, D., Waldor, M. K., Moreira, C. G., et al. (2012). Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117. doi: 10.1038/nature11623
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pajaniappan, M., Hall, J. E., Cawthraw, S. A., Newell, D. G., Gaynor, E. C., Fields, J. A., et al. (2008). A temperature-regulated Campylobacter jejuni gluconate dehydrogenase is involved in respiration-dependent energy conservation and chicken colonization. Mol. Microbiol. 68, 474–491. doi: 10.1111/j.1365-2958.2008.06161.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palyada, K., Sun, Y. Q., Flint, A., Butcher, J., Naikare, H., and Stintzi, A. (2009). Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. doi: 10.1186/1471-2164-10-481
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palyada, K., Threadgill, D., and Stintzi, A. (2004). Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186, 4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, S. F. (2002). The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int. J. Food Microbiol. 74, 177–188. doi: 10.1016/S0168-1605(01)00678-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pawelec, D., Jakubowska-Mroz, J., and Jagusztyn-Krynicka, E. K. (1998). Campylobacter jejuni 72Dz/92 cjaC gene coding 28 kDa immunopositive protein, a homologue of the solute-binding components of the ABC transport system. Lett. Appl. Microbiol. 26, 69–76. doi: 10.1046/j.1472-765X.1998.00276.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pawelec, D., Rozynek, E., Popowski, J., and Jagusztyn-Krynicka, E. K. (1997). Cloning and characterization of a Campylobacter jejuni 72Dz/92 gene encoding a 30 kDa immunopositive protein, component of the ABC transport system; expression of the gene in avirulent Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 19, 137–150. doi: 10.1111/j.1574-695X.1997.tb01083.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pearson, B. M., Gaskin, D. J., Segers, R. P., Wells, J. M., Nuijten, P. J., and van Vliet, A. H. (2007). The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828). J. Bacteriol. 189, 8402–8403. doi: 10.1128/JB.01404-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pei, Z., and Blaser, M. J. (1993). PEB1, the major cell-binding factor of Campylobacter jejuni, is a homolog of the binding component in gram-negative nutrient transport systems. J. Biol. Chem. 268, 18717–18725.
Pei, Z., Burucoa, C., Grignon, B., Baqar, S., Huang, X. Z., Kopecko, D. J., et al. (1998). Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66, 938–943.
Penner, J. L. (1988). The genus Campylobacter: a decade of progress. Clin. Microbiol. Rev. 1, 157–172.
Pesci, E. C., Cottle, D. L., and Pickett, C. L. (1994). Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62, 2687–2694.
Pickett, C. L., Auffenberg, T., Pesci, E. C., Sheen, V. L., and Jusuf, S. S. (1992). Iron acquisition and hemolysin production by Campylobacter jejuni. Infect. Immun. 60, 3872–3877.
Pittman, M. S., Elvers, K. T., Lee, L., Jones, M. A., Poole, R. K., Park, S. F., et al. (2007). Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol. Microbiol. 63, 575–590. doi: 10.1111/j.1365-2958.2006.05532.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pittman, M. S., and Kelly, D. J. (2005). Electron transport through nitrate and nitrite reductases in Campylobacter jejuni. Biochem. Soc. Trans. 33, 190–192. doi: 10.1042/BST0330190
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Porcelli, I., Reuter, M., Pearson, B. M., Wilhelm, T., and van Vliet, A. H. (2013). Parallel evolution of genome structure and transcriptional landscape in the Epsilonproteobacteria. BMC Genomics 14:616. doi: 10.1186/1471-2164-14-616
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pryjma, M., Apel, D., Huynh, S., Parker, C. T., and Gaynor, E. C. (2012). FdhTU-modulated formate dehydrogenase expression and electron donor availability enhance recovery of Campylobacter jejuni following host cell infection. J. Bacteriol. 194, 3803–3813. doi: 10.1128/JB.06665-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Quay, S. C., and Oxender, D. L. (1976). Regulation of branched-chain amino acid transport in Escherichia coli. J. Bacteriol. 127, 1225–1238.
Rada, B., and Leto, T. L. (2008). Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib. Microbiol. 15, 164–187. doi: 10.1159/000136357
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rahman, H., King, R. M., Shewell, L. K., Semchenko, E. A., Hartley-Tassell, L. E., Wilson, J. C., et al. (2014). Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PLoS Pathog. 10:e1003822. doi: 10.1371/journal.ppat.1003822
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rasmussen, J. J., Vegge, C. S., Frokiaer, H., Howlett, R. M., Krogfelt, K. A., Kelly, D. J., et al. (2013). Campylobacter jejuni carbon starvation protein A (CstA) is involved in peptide utilization, motility and agglutination, and has a role in stimulation of dendritic cells. J. Med. Microbiol. 62, 1135–1143. doi: 10.1099/jmm.0.059345-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Revez, J., Rossi, M., Ellstrom, P., de Haan, C., Rautelin, H., and Hanninen, M. L. (2011). Finnish Campylobacter jejuni strains of multilocus sequence type ST-22 complex have two lineages with different characteristics. PLoS ONE 6:e26880. doi: 10.1371/journal.pone.0026880
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ribardo, D. A., and Hendrixson, D. R. (2011). Analysis of the LIV system of Campylobacter jejuni reveals alternative roles for LivJ and LivK in commensalism beyond branched-chain amino acid transport. J. Bacteriol. 193, 6233–6243. doi: 10.1128/JB.05473-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ridley, K. A., Rock, J. D., Li, Y., and Ketley, J. M. (2006). Heme utilization in Campylobacter jejuni. J. Bacteriol. 188, 7862–7875. doi: 10.1128/JB.00994-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rivera-Ordaz, A., Bracher, S., Sarrach, S., Li, Z., Shi, L., Quick, M., et al. (2013). The sodium/proline transporter PutP of Helicobacter pylori. PLoS ONE 8:e83576. doi: 10.1371/journal.pone.0083576
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sauer, U., and Eikmanns, B. J. (2005). The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29, 765–794. doi: 10.1016/j.femsre.2004.11.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Seal, B. S., Hiett, K. L., Kuntz, R. L., Woolsey, R., Schegg, K. M., Ard, M., et al. (2007). Proteomic analyses of a robust versus a poor chicken gastrointestinal colonizing isolate of Campylobacter jejuni. J. Proteome Res. 6, 4582–4591. doi: 10.1021/pr070356a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sellars, M. J., Hall, S. J., and Kelly, D. J. (2002). Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184, 4187–4196. doi: 10.1128/JB.184.15.4187-4196.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shaw, F. L., Mulholland, F., Le Gall, G., Porcelli, I., Hart, D. J., Pearson, B. M., et al. (2012). Selenium-dependent biogenesis of formate dehydrogenase in Campylobacter jejuni is controlled by the fdhTU accessory genes. J. Bacteriol. 194, 3814–3823. doi: 10.1128/JB.06586-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shibayama, K., Wachino, J., Arakawa, Y., Saidijam, M., Rutherford, N. G., and Henderson, P. J. (2007). Metabolism of glutamine and glutathione via gamma-glutamyltranspeptidase and glutamate transport in Helicobacter pylori: possible significance in the pathophysiology of the organism. Mol. Microbiol. 64, 396–406. doi: 10.1111/j.1365-2958.2007.05661.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Skaar, E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6:e1000949. doi: 10.1371/journal.ppat.1000949
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smart, J. P., Cliff, M. J., and Kelly, D. J. (2009). A role for tungsten in the biology of Campylobacter jejuni: tungstate stimulates formate dehydrogenase activity and is transported via an ultra-high affinity ABC system distinct from the molybdate transporter. Mol. Microbiol. 74, 742–757. doi: 10.1111/j.1365-2958.2009.06902.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, M. A., Finel, M., Korolik, V., and Mendz, G. L. (2000). Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch. Microbiol. 174, 1–10. doi: 10.1007/s002030000174
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sparacino-Watkins, C., Stolz, J. F., and Basu, P. (2014). Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 43, 676–706. doi: 10.1039/c3cs60249d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stahl, M., Butcher, J., and Stintzi, A. (2012). Nutrient acquisition and metabolism by Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2:5. doi: 10.3389/fcimb.2012.00005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stahl, M., Friis, L. M., Nothaft, H., Liu, X., Li, J., Szymanski, C. M., et al. (2011). L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. U.S.A. 108, 7194–7199. doi: 10.1073/pnas.1014125108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stahl, M., and Stintzi, A. (2011). Identification of essential genes in Campylobacter jejuni genome highlights hyper-variable plasticity regions. Funct. Integr. Genomics 11, 241–257. doi: 10.1007/s10142-011-0214-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stecher, B., Berry, D., and Loy, A. (2013). Colonization resistance and microbial ecophysiology: using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol. Rev. 37, 793–829. doi: 10.1111/1574-6976.12024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Steeb, B., Claudi, B., Burton, N. A., Tienz, P., Schmidt, A., Farhan, H., et al. (2013). Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog. 9:e1003301. doi: 10.1371/journal.ppat.1003301
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stintzi, A., Marlow, D., Palyada, K., Naikare, H., Panciera, R., Whitworth, L., et al. (2005). Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 73, 1797–1810. doi: 10.1128/IAI.73.3.1797-1810.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
St Maurice, M., Cremades, N., Croxen, M. A., Sisson, G., Sancho, J., and Hoffman, P. S. (2007). Flavodoxin:quinone reductase (FqrB): a redox partner of pyruvate:ferredoxin oxidoreductase that reversibly couples pyruvate oxidation to NADPH production in Helicobacter pylori and Campylobacter jejuni. J. Bacteriol. 189, 4764–4773. doi: 10.1128/JB.00287-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Szymanski, C. M., and Gaynor, E. C. (2012). How a sugary bug gets through the day: recent developments in understanding fundamental processes impacting Campylobacter jejuni pathogenesis. Gut Microbes 3, 135–144. doi: 10.4161/gmic.19488
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takamiya, M., Ozen, A., Rasmussen, M., Alter, T., Gilbert, T., Ussery, D. W., et al. (2011). Genome Sequence of Campylobacter jejuni strain 327, a strain isolated from a turkey slaughterhouse. Stand. Genomic Sci. 4, 113–122. doi: 10.4056/sigs.1313504
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tareen, A. M., Dasti, J. I., Zautner, A. E., Gross, U., and Lugert, R. (2010). Campylobacter jejuni proteins Cj0952c and Cj0951c affect chemotactic behaviour towards formic acid and are important for invasion of host cells. Microbiology 156, 3123–3135. doi: 10.1099/mic.0.039438-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Taveirne, M. E., Theriot, C. M., Livny, J., and DiRita, V. J. (2013). The complete Campylobacter jejuni transcriptome during colonization of a natural host determined by RNAseq. PLoS ONE 8:e73586. doi: 10.1371/journal.pone.0073586
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tenover, F. C., Knapp, J. S., Patton, C., and Plorde, J. J. (1985). Use of auxotyping for epidemiological studies of Campylobacter jejuni and Campylobacter coli infections. Infect. Immun. 48, 384–388.
Tenover, F. C., and Patton, C. M. (1987). Naturally occurring auxotrophs of Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 25, 1659–1661.
Thomas, M. T., Shepherd, M., Poole, R. K., van Vliet, A. H., Kelly, D. J., and Pearson, B. M. (2011). Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on L-lactate. Environ. Microbiol. 13, 48–61. doi: 10.1111/j.1462-2920.2010.02307.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Putten, J. P., van Alphen, L. B., Wosten, M. M., and de Zoete, M. R. (2009). Molecular mechanisms of Campylobacter infection. Curr. Top. Microbiol. Immunol. 337, 197–229. doi: 10.1007/978-3-642-01846-6_7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Spreeuwel, J. P., Duursma, G. C., Meijer, C. J., Bax, R., Rosekrans, P. C., and Lindeman, J. (1985). Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut 26, 945–951. doi: 10.1136/gut.26.9.945
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vegge, C. S., Brondsted, L., Li, Y. P., Bang, D. D., and Ingmer, H. (2009). Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl. Environ. Microbiol. 75, 5308–5314. doi: 10.1128/AEM.00287-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Velayudhan, J., Jones, M. A., Barrow, P. A., and Kelly, D. J. (2004). L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immun. 72, 260–268. doi: 10.1128/IAI.72.1.260-268.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Velayudhan, J., and Kelly, D. J. (2002). Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: an essential role for phosphoenolpyruvate carboxykinase. Microbiology 148, 685–694.
Veron, M., Lenvoisefuret, A., and Beaune, P. (1981). Anaerobic Respiration of Fumarate as a Differential Test between Campylobacter fetus and Campylobacter jejuni. Curr. Microbiol. 6, 349–354. doi: 10.1007/BF01567010
Vorwerk, H., Mohr, J., Huber, C., Wensel, O., Schmidt-Hohagen, K., Gripp, E., et al. (2014). Utilization of host-derived cysteine-containing peptides overcomes the restricted sulfur metabolism of Campylobacter jejuni. Mol. Microbiol. 93, 1224–1245. doi: 10.1111/mmi.12732
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, S., Haapalainen, A. M., Yan, F., Du, Q., Tyler, P. C., Evans, G. B., et al. (2012). A picomolar transition state analogue inhibitor of MTAN as a specific antibiotic for Helicobacter pylori. Biochemistry 51, 6892–6894. doi: 10.1021/bi3009664
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Watson, R. O., and Galan, J. E. (2008). Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 4:e14. doi: 10.1371/journal.ppat.0040014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weerakoon, D. R., Borden, N. J., Goodson, C. M., Grimes, J., and Olson, J. W. (2009). The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb. Pathog. 47, 8–15. doi: 10.1016/j.micpath.2009.04.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weerakoon, D. R., and Olson, J. W. (2008). The Campylobacter jejuni NADH:ubiquinone oxidoreductase (complex I) utilizes flavodoxin rather than NADH. J. Bacteriol. 190, 915–925. doi: 10.1128/JB.01647-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weingarten, R. A., Grimes, J. L., and Olson, J. W. (2008). Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl. Environ. Microbiol. 74, 1367–1375. doi: 10.1128/AEM.02261-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weingarten, R. A., Taveirne, M. E., and Olson, J. W. (2009). The dual-functioning fumarate reductase is the sole succinate:quinone reductase in Campylobacter jejuni and is required for full host colonization. J. Bacteriol. 191, 5293–5300. doi: 10.1128/JB.00166-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Westfall, H. N., Rollins, D. M., and Weiss, E. (1986). Substrate utilization by Campylobacter jejuni and Campylobacter coli. Appl. Environ. Microbiol. 52, 700–705.
Wilson, D. L., Rathinam, V. A., Qi, W., Wick, L. M., Landgraf, J., Bell, J. A., et al. (2010). Genetic diversity in Campylobacter jejuni is associated with differential colonization of broiler chickens and C57BL/6J IL10-deficient mice. Microbiology 156, 2046–2057. doi: 10.1099/mic.0.035717-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Winter, S. E., Winter, M. G., Xavier, M. N., Thiennimitr, P., Poon, V., Keestra, A. M., et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711. doi: 10.1126/science.1232467
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wolfe, A. J. (2005). The acetate switch. Microbiol. Mol. Biol. Rev. 69, 12–50. doi: 10.1128/MMBR.69.1.12-50.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Woodall, C. A., Jones, M. A., Barrow, P. A., Hinds, J., Marsden, G. L., Kelly, D. J., et al. (2005). Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect. Immun. 73, 5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wösten, M. M., Dubbink, H. J., and van der Zeijst, B. A. (1996). The aroA gene of Campylobacter jejuni. Gene 181, 109–112. doi: 10.1016/S0378-1119(96)00482-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wright, J. A., Grant, A. J., Hurd, D., Harrison, M., Guccione, E. J., Kelly, D. J., et al. (2009). Metabolite and transcriptome analysis of Campylobacter jejuni in vitro growth reveals a stationary-phase physiological switch. Microbiology 155, 80–94. doi: 10.1099/mic.0.021790-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wyszynska, A., Tomczyk, K., and Jagusztyn-Krynicka, E. K. (2007). Comparison of the localization and post-translational modification of Campylobacter coli CjaC and its homolog from Campylobacter jejuni, Cj0734c/HisJ. Acta Biochim. Pol. 54, 143–150.
Wyszynska, A., Zycka, J., Godlewska, R., and Jagusztyn-Krynicka, E. K. (2008). The Campylobacter jejuni/coli cjaA (cj0982c) gene encodes an N-glycosylated lipoprotein localized in the inner membrane. Curr. Microbiol. 57, 181–188. doi: 10.1007/s00284-008-9171-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, F., Zeng, X., Haigh, R. D., Ketley, J. M., and Lin, J. (2010). Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J. Bacteriol. 192, 4425–4435. doi: 10.1128/JB.00478-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Young, K. T., Davis, L. M., and Dirita, V. J. (2007). Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5, 665–679. doi: 10.1038/nrmicro1718
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zautner, A. E., Herrmann, S., Corso, J., Tareen, A. M., Alter, T., and Gross, U. (2011). Epidemiological association of different Campylobacter jejuni groups with metabolism-associated genetic markers. Appl. Environ. Microbiol. 77, 2359–2365. doi: 10.1128/AEM.02403-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zeng, X., Mo, Y., Xu, F., and Lin, J. (2013a). Identification and characterization of a periplasmic trilactone esterase, Cee, revealed unique features of ferric enterobactin acquisition in Campylobacter. Mol. Microbiol. 87, 594–608. doi: 10.1111/mmi.12118
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zeng, X., Xu, F., and Lin, J. (2013b). Specific TonB-ExbB-ExbD energy transduction systems required for ferric enterobactin acquisition in Campylobacter. FEMS Microbiol. Lett. 347, 83–91. doi: 10.1111/1574-6968.12221
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, Y., Jameson, E., Crosatti, M., Schafer, H., Rajakumar, K., Bugg, T. D., et al. (2014). Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl. Acad. Sci. U.S.A. 111, 4268–4273. doi: 10.1073/pnas.1316569111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Campylobacter jejuni, intermediary metabolism, amino acid catabolism, peptide catabolism, respiration, colonization, intracellular survival
Citation: Hofreuter D (2014) Defining the metabolic requirements for the growth and colonization capacity of Campylobacter jejuni. Front. Cell. Infect. Microbiol. 4:137. doi: 10.3389/fcimb.2014.00137
Received: 15 July 2014; Accepted: 11 September 2014;
Published online: 29 September 2014.
Edited by:
Wolfgang Eisenreich, Technische Universität München, GermanyReviewed by:
Odile Tresse, French National Institute for Agricultural Research, FranceDavid Kelly, The University of Sheffield, UK
Copyright © 2014 Hofreuter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dirk Hofreuter, Hannover Medical School, Institute for Medical Microbiology and Hospital Epidemiology, Carl-Neuberg-Str. 1, D-30625 Hannover, Germany e-mail: hofreuter.dirk@mh-hannover.de
 Dirk Hofreuter
Dirk Hofreuter