Metabolic aspects of bacterial persisters
- Department of Microbial Genetics, Faculty of Science, Interfaculty Institute of Microbiology and Infection Medicine Tübingen (IMIT), University of Tübingen, Tübingen, Germany
Persister cells form a multi-drug tolerant subpopulation within an isogenic culture of bacteria that are genetically susceptible to antibiotics. Studies with different Gram negative and Gram positive bacteria have identified a large number of genes associated with the persister state. In contrast, the revelation of persister metabolism has only been addressed recently. We here summarize metabolic aspects of persisters, which includes an overview about the bifunctional role of selected carbohydrates as both triggers for the exit from the drug tolerant state and metabolites which persisters feed on. Also alarmones as indicators for starvation have been shown to influence persister levels via different signaling cascades involving the activation of toxin-antitoxin systems and other regulatory factors. Finally, recent data obtained by 13C-isotopolog profiling demonstrated an active amino acid anabolism in Staphylococcus aureus cultures challenged with high drug concentrations. Understanding the metabolism of persister cells poses challenges but also paves the way for the development of anti-persister compounds.
Introduction
The treatment of recurrent bacterial infections is often a tedious trial due to antibiotic recalcitrance. This is not solely caused by resistance but also implies persister cells which are (multi-) drug-tolerant (Lewis, 2010). Persisters were first described in 1944 when killing of S. aureus with penicillin was found to leave a few survivor cells behind (Bigger, 1944). Notably the antibiotic tolerance of persisters is not genetically manifested, as progenies of persisters are as susceptible as the parent strains (Keren et al., 2004a). Consistent with a number of studies, persisters among an isogenic bacterial culture temporarily reside in a slow- or non-growing state and arise both stochastically and in response to environmental cues. For instance, biofilms accommodate a high level of persisters (Lewis, 2005) and their number within a culture depends greatly on the growth stage, with stationary cultures exhibiting much higher persister levels compared to the exponential phase (Keren et al., 2004a; Lechner et al., 2012). This correlation was confirmed by further studies that established a strong influence between inoculum age and persister frequency (Luidalepp et al., 2011). Retarded protein synthesis as well as protein aggregate accumulation were found to affect the persister levels of a culture (Kwan et al., 2013; Leszczynska et al., 2013), as does bacterial compound signaling (Keller and Surette, 2006). Molecules such as indole, 2′ Amino-acetophenone, or CSP pheromone, some of which are quorum sensing (QS) messengers, can induce drug tolerance and the persister state in different bacteria (Leung and Levesque, 2012; Vega et al., 2012, 2013; Que et al., 2013). Akin to QS, some bacteria have been shown to produce so called resuscitation-promoting factors, converting dormant cells back to a more active state. Among several examples apparently based upon similar mechanisms, the addition of spent culture medium to dormant S. aureus cells led to accelerated awakening (Mukamolova et al., 1999; Pinto et al., 2013; Pascoe et al., 2014). The retention of a viable state over longer periods of time and particularly the reversion from dormancy to a growing state requires metabolic activity. One major question is how persisters maintain a critical degree of metabolism over extended periods of time without being killed during hostile conditions. Here, we sum up recent findings on the involvement of metabolism in the persister state (Figure 1) and illustrate the experimental difficulties and challenges accessing this topic. The reader is also referred to a recent review article by Amato et al. (2014).
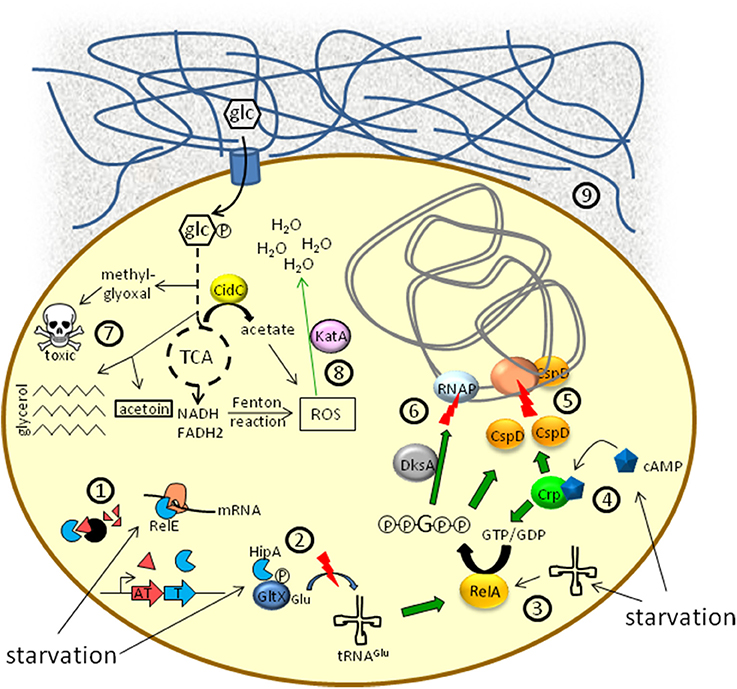
Figure 1. Schematic overview of metabolic aspects associated with the persister state. ①, ② Toxin/antitoxin-systems: In a number of bacteria, Lon or Clp proteases, activated in response to starvation, degrade antitoxin proteins ①. Liberated toxins (e.g., RelE) can cleave mRNA or employ a ppGpp-dependent signal transduction ② to induce growth arrest. ③ Uncharged tRNAs due to amino acid starvation lead to the synthesis of ppGpp via RelA. ④ Nutrient limitation favors the synthesis of the second messenger cAMP by adenylate cyclase. cAMP binds to the cAMP receptor protein (Crp) and the cAMP/Crp complex activates the expression of both relA and cspD. ⑤ Inhibition of DNA replication by CspD. ⑥ Modulation of RNA polymerase (RNAP) activity by the DksA/ppGpp complex. ⑦ Metabolic flux alterations result in a decreased TCA cycle activity and increased persistence. Synthesis of methylglyoxal leads to growth inhibition. Acetoin and triglyceride synthesis represent alternative pathways for the deprivation of pyruvate and acetyl-CoA from the TCA cycle. ⑧ Different branches of metabolism can produce reactive oxygen species (ROS) as hazardous side products, impairing persister formation. Enzymes counteracting ROS activity (e.g., KatA) are upregulated in persister cells. ⑨ Biofilms containing protein- and/or aminosugar-polymer structures (blue meshwork) may represent environments of low supply of nutrients, such as glucose (glc), which favors persister formation.
Genes Linking Metabolism and the Persister State
The number of identified genes associated with the persister state is steadily increasing and toxin-antitoxin (TA) systems act as key regulators in this regard (Lewis, 2010; Schuster and Bertram, 2013). These systems usually comprise a toxin that blocks or corrupts essential cellular functions and an antitoxin abrogating the toxin's activity. TA systems participate in multiple processes in bacteria, ranging from stress response to regulation of metabolism and survival inside host cells (McKenzie et al., 2012; Helaine et al., 2014). The issue of how TA systems are controlled and how this leads to persister formation has been illuminated in a number of cases. For example, glucose starvation and shortage of amino acids activate RelE-toxin homologs in E. coli (Christensen-Dalsgaard et al., 2010). Proteome analysis of starving Mycobacterium tuberculosis cells revealed an increased abundance of TA system proteins under nutrient limited conditions (Albrethsen et al., 2013). The alarmone ppGpp is part of the stringent response signaling pathway, which is switched on in response to amino acid depletion. ppGpp abundance and TA system activity appear to be tightly intertwined to control the metabolic state of bacterial cells (Traxler et al., 2008; Bokinsky et al., 2013; Germain et al., 2013; Maisonneuve et al., 2013). For example, the TA toxin HipA phosphorylates the glutamyl-tRNA synthetase GltX which inhibits the loading of tRNAGlu and consequently mimics nutrient limitation resulting in ppGpp synthesis. Pseudomonas aeruginosa actively responds to nutrient limitation via a ppGpp-dependent mechanism directing cells to a state of increased antibiotic tolerance (Nguyen et al., 2011). Metabolic stress can also lead to a different scenario, in which ppGpp and the cognate hydrolase SpoT influence persister formation (Amato et al., 2013). Low levels of SpoT thus increase ppGpp abundance that is also associated with DNA gyrase inhibition and reduction of RNA polymerase activity. In S. aureus a similar role for ppGpp was demonstrated, as its permanent synthesis leads to growth inhibition and impaired virulence, facilitating persistent infections (Gao et al., 2010). Also the E. coli cold shock protein CspD that is expressed during stationary phase and is induced by glucose starvation is influenced by ppGpp (Yamanaka and Inouye, 1997). The lack of nutrients leads to a CspD-dependent inhibition of DNA replication, resulting in increased persister formation (Kim and Wood, 2010). Interestingly, also another second messenger, cyclic AMP (cAMP), is part of the regulatory network of CspD. cAMP, whose physiological level is associated with nutrient availability, increases cspD transcription in complex with its receptor protein Crp (Uppal et al., 2014). Moreover, the cAMP-Crp complex also activates the expression of relA, resulting in a further increase of the intracellular ppGpp level (Nakagawa et al., 2006). This example illustrates how the metabolic state of a cell can be coupled to persister formation via different pathways to achieve a subtle and precise regulation. In the light of these results, ppGpp seems to be an important mediator between metabolism and persister formation.
Reports about the genetic alterations in the energy metabolism of bacterial cells provide a rather inconsistent picture. E. coli mutants lacking ubiF or sucB, encoding for enzymes involved in ubiquinone biosynthesis, or the TCA cycle, respectively, showed decreased persister levels compared to the wild-type strain (Ma et al., 2010). Both enzymes contribute to the generation of the intracellular ATP pool. However, the inhibition of ATP synthesis by carbonyl cyanide m-chlorophenylhydrazone (CCCP) led to an increased persister formation in another study (Kwan et al., 2013). The same effect was observed for the membrane binding protein TisB of the tisAB TA-system. TisB expression decreases the proton motive force (PMF) and impedes energy production causing an elevated persister level (Unoson and Wagner, 2008; Dörr et al., 2010).
Experimental Approaches and Metabolic Peculiarities of Persister Cells
Major drawbacks in the analysis of persister cells' metabolism are both the natural heterogeneity of the bacterial population and the fact that antibiotics used to isolate persisters destroy their naïve state. Furthermore, the persister state is of temporary nature only and these cells usually merely represent a small subpopulation within a culture. It is therefore of utmost importance to distinguish between results stemming from persister and non-persister cells, which requires efficient means to separate them. Lytic antibiotics or unstable GFP-variants have been used before to address this issue (Keren et al., 2004b; Shah et al., 2006). Obtained transcriptome patterns of suchlike differentiated M. tuberculosis or E. coli persisters indicated a downregulation of metabolic genes, and therefore a decreased metabolism in persisters (Shah et al., 2006; Keren et al., 2011). Different approaches have been taken to examine the metabolism of persisters more directly. The group of Brynildsen used phenotype microarrays and a fluorescent dye to assay the activity of bacterial reductases as a proxy for metabolic activity (Orman and Brynildsen, 2013a,b). Based on these results, a less active metabolism is apparently not a requirement, but it increases the chance for a cell to enter the persister state. Another powerful technique, termed isotopolog profiling, is based upon feeding of 13C-isotope labeled carbohydrates to cultures and subsequent analysis of labeled intermediates. This allows deducing relative activities of metabolic pathways or even networks in a time-resolved manner by comparing ratios of labeled and unlabeled compounds (Eisenreich et al., 2010). Isotopolog profiling provides information of relative metabolic fluxes but not on the quantities or absolute concentrations of metabolites. Measuring the decrease of energy substrates in the medium over time can be theoretically used to determine the metabolic level. We used isotopolog profiling to investigate, which metabolic pathways are active in stationary growth phase S. aureus cells that had been challenged with daptomycin (Lechner et al., 2014). De novo biosynthesis of amino acids was observed, and their labeling patterns suggested an active glycolysis, TCA cycle and pentose phosphate pathway. Of note, analysis of 13C-labeling pattern of Asp and Glu indicated an increased activity of the TCA cycle.
Recent studies provided first insights into the metabolic state of persisters associated with biofilms that provide a protective niche for bacteria against antibiotics and other harmful conditions (Mah and O'Toole, 2001; Donlan and Costerton, 2002; Davenport et al., 2014). This is due, in part, to metabolic downshifts in biofilm dwelling cells. Impaired nutrient penetration and consumption by peripheral cells result in decreased nutrient supply in this environment. Genes involved in TCA cycle and energy production were downregulated in tobramycin challenged and biofilm embedded Burkholderia cenocepacia persisters (Van Acker et al., 2013). Metabolic activity can lead to H2O2 generation by the reduction of molecular oxygen caused by the respiratory chain (Gonzalez-Flecha and Demple, 1995). H2O2 can thereby accidentally drive the Fe2+-dependent Fenton reaction leading to the formation of reactive oxygen species (ROS), which attack essential cellular functions (Imlay et al., 1988). Therefore, long-term survival of a bacterial cell could benefit from an impaired metabolism. In addition, a reduced energy level simultaneously prevents the PMF-dependent uptake of the aminoglycoside tobramycin, as detailed below. In M. tuberculosis, redirections in the carbon flux were correlated to growth arrest and antibiotic tolerance (Baek et al., 2011). Acyl-CoA is thereby converted to triglycerides, draining the fuel for the TCA cycle. A further example for increased persistence due to the change of metabolic fluxes is the synthesis of methylglyoxal, which impedes growth of E. coli (Girgis et al., 2012). Single deletions of the genes encoding the two metabolic enzymes glycerol-3-phosphate dehydrogenase (glpD) or transketolase A (tktA) lead to accumulation of dihydroxyacetone phosphate (DHAP) which is finally converted to methylglyoxal. Interestingly, the glyoxylate shunt is upregulated in B. cenocepacia persister cells, bypassing NADH production and possible ROS formation via the TCA cycle, illustrating an additional protective mechanism. Another link between persister level and ROS formation in biofilm was established in P. aeruginosa, where mutants defective in the stringent response were more susceptible toward antibiotic treatment (Nguyen et al., 2011). Starvation apparently leads to increased antioxidant countermeasures by an upregulation of catalase activity and the restriction of the synthesis of pro-oxidant substances. Furthermore, the metabolic regulator catabolite repression control (Crc) protein decreases the metabolic activity of P. aeruginosa in biofilms conferring increased tolerance toward ciprofloxacin (Zhang et al., 2012). These results indicate that a metabolic adaptation process especially in regard to the TCA cycle is involved in the maintenance of the persister state. Besides biofilm cells, the importance of a metabolic downshift was also confirmed by long-term survival assays of planktonic S. aureus cells, in which mutants lacking the TCA cycle enzymes aconitase or succinate dehydrogenase showed an enhanced stationary-phase survival level (Somerville et al., 2002; Gaupp et al., 2010). Retarded metabolic flux through or disruption of the TCA cycle was found in clinical S. epidermidis isolates with enhanced survival during β-lactam treatment (Thomas et al., 2013). Reduced ROS formation was determined as one critical feature in this regard. In line, ROS activity seems to be involved in programmed cell death, as shown in S. aureus (Thomas et al., 2014). ROS formation is apparently linked to acetate production which is again tightly regulated by the two antagonistic factors CidC and AlsSD. CidC is an oxidase converting pyruvate to acetate and is activated by the CidR regulator during the presence of glucose. The CidR regulon also comprises the alsSD operon encoding for an α-acetolactate synthase/decarboxylase leading to acetoin synthesis from pyruvate, thereby reducing the amount of acetate by CidC. These data illustrate the strong connection of TCA cycle dependent ROS formation and its negative influence on the long-term survival of cells thereby requiring alternative metabolic pathways to avoid their production.
Anti-Persister Strategies and the Interplay between Carbohydrate Supply and Persister Killing
The importance of persisters in bacterial infections is more and more corroborated (Fauvart et al., 2011). Recent studies indicate that physiology and metabolism could be an Achilles heel for the development of new anti-persister strategies (Allison et al., 2011). A number of compounds counteract the persister state by targeting indispensable cellular processes or by activating resuscitation. These drugs include the acyldepsipeptide ADEP4 which permanently activates Clp proteases or a biphenyl-derivative termed C10 that reverts cells to an antibiotic susceptible state (Kim et al., 2011; Conlon et al., 2013). Manipulating bacterial signaling via artificial QS inhibitors is another approach (Pan and Ren, 2013) and also ppGpp was identified as a potential anti-persister/-biofilm target. A dodecamer peptide termed 1018 was reported to label the alarmone for degradation, thereby inhibiting formation or dispersal of biofilms as sources for recurrent and persistent infections. This peptide was active against at least seven Gram positive or Gram negative bacterial species (de la Fuente-Nunez et al., 2014). In 2011 the group of James J. Collins described that the addition of selected carbohydrates enhanced the killing of persisters by aminoglycoside antibiotics. They established a relationship between the metabolism of selected sugars, the generation of PMF and the enhanced uptake of the drug (Allison et al., 2011). The rate of increased killing is thereby mainly determined by the rate of substrate utilization. Particularly fructose was an effective compound in combination with gentamicin to eradicate E. coli as well as S. aureus persisters. Subsequent to this finding on metabolite enabled killing, the non-susceptibility of persisters toward aminoglycosides treatment aided in the identification of the most utilizable substrates for such cells (Orman and Brynildsen, 2013b). Another successful approach of metabolite induced killing was demonstrated by combating P. aeruginosa biofilms with a combination of mannitol and tobramycin (Barraud et al., 2013) and also arginine and nitrate were described as useful additives in this regard (Borriello et al., 2006). By contrast, an excess of glucose in S. epidermidis led to a higher level of dormant cells in a biofilm, presumably due to the accumulation of acidic degradation products resulting from glucose metabolism (Cerca et al., 2011).
Conclusion
Recent studies highlight the importance of investigating the two interconnected fields of persister state and metabolic activity in bacteria in more detail. Adaptation of the metabolism is a key prerequisite for persisters to cope with hostile conditions. In particular, the modulation of TCA cycle activity appears as a hallmark in persister metabolism. This regulation must be precisely controlled to avoid ROS formation with potentially destructive implications for persister cells. Multiple lines of evidence suggest that the metabolism of persisters can be tuned to alter their susceptibility toward antibiotics or to trigger programmed cell-death-like processes (Rice and Bayles, 2003). In a number of cases, this is achieved simply by supplementing selected carbohydrates. Based upon these findings, new effective anti-persister therapies could be developed to reduce the risk of relapsing or chronic infections. This could result in the development of concerted combination therapies, exploiting the natural metabolic activity of persister cells.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Friedrich Götz for support. This work was supported by grant BE4038/2 within the priority programme 1316 “host adapted metabolism of bacterial pathogens” of the Deutsche Forschungsgemeinschaft. Funding for the open access charge was provided by the Open Access Publishing Fund of the University of Tübingen.
References
Albrethsen, J., Agner, J., Piersma, S. R., Hojrup, P., Pham, T. V., Weldingh, K., et al. (2013). Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol. Cell. Proteomics 12, 1180–1191. doi: 10.1074/mcp.M112.018846
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Allison, K. R., Brynildsen, M. P., and Collins, J. J. (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220. doi: 10.1038/nature10069
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Amato, S. M., Fazen, C. H., Henry, T. C., Mok, W. W., Orman, M. A., Sandvik, E. L., et al. (2014). The role of metabolism in bacterial persistence. Front. Microbiol. 5:70. doi: 10.3389/fmicb.2014.00070
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Amato, S. M., Orman, M. A., and Brynildsen, M. P. (2013). Metabolic control of persister formation in Escherichia coli. Mol. Cell 50, 475–487. doi: 10.1016/j.molcel.2013.04.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baek, S. H., Li, A. H., and Sassetti, C. M. (2011). Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 9:e1001065. doi: 10.1371/journal.pbio.1001065
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barraud, N., Buson, A., Jarolimek, W., and Rice, S. A. (2013). Mannitol enhances antibiotic sensitivity of persister bacteria in Pseudomonas aeruginosa biofilms. PLoS ONE 8:e84220. doi: 10.1371/journal.pone.0084220
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bigger, J. W. (1944). The bactericidal action of penicillin on Staphylococcus pyogenes. Ir. J. Med. Sci. 19, 16. doi: 10.1007/BF02948462
Bokinsky, G., Baidoo, E. E., Akella, S., Burd, H., Weaver, D., Alonso-Gutierrez, J., et al. (2013). HipA-triggered growth arrest and beta-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J. Bacteriol. 195, 3173–3182. doi: 10.1128/JB.02210-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Borriello, G., Richards, L., Ehrlich, G. D., and Stewart, P. S. (2006). Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 50, 382–384. doi: 10.1128/AAC.50.1.382-384.2006
Cerca, F., Andrade, F., Franca, A., Andrade, E. B., Ribeiro, A., Almeida, A. A., et al. (2011). Staphylococcus epidermidis biofilms with higher proportions of dormant bacteria induce a lower activation of murine macrophages. J. Med. Microbiol. 60, 1717–1724. doi: 10.1099/jmm.0.031922-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Christensen-Dalsgaard, M., Jorgensen, M. G., and Gerdes, K. (2010). Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75, 333–348. doi: 10.1111/j.1365-2958.2009.06969.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conlon, B. P., Nakayasu, E. S., Fleck, L. E., Lafleur, M. D., Isabella, V. M., Coleman, K., et al. (2013). Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370. doi: 10.1038/nature12790
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Davenport, E. K., Call, D. R., and Beyenal, H. (2014). Differential protection from tobramycin by extracellular polymeric substances from Acinetobacter baumannii and Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 58, 4755–4761. doi: 10.1128/AAC.03071-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
de la Fuente-Nunez, C., Reffuveille, F., Haney, E. F., Straus, S. K., and Hancock, R. E. (2014). Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10:e1004152. doi: 10.1371/journal.ppat.1004152
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dörr, T., Vulić, M., and Lewis, K. (2010). Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317. doi: 10.1371/journal.pbio.1000317
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eisenreich, W., Dandekar, T., Heesemann, J., and Goebel, W. (2010). Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412. doi: 10.1038/nrmicro2351
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fauvart, M., De Groote, V. N., and Michiels, J. (2011). Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J. Med. Microbiol. 60, 699–709. doi: 10.1099/jmm.0.030932-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, W., Chua, K., Davies, J. K., Newton, H. J., Seemann, T., Harrison, P. F., et al. (2010). Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 6:e1000944. doi: 10.1371/journal.ppat.1000944
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaupp, R., Schlag, S., Liebeke, M., Lalk, M., and Gotz, F. (2010). Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms. J. Bacteriol. 192, 2385–2394. doi: 10.1128/JB.01472-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Germain, E., Castro-Roa, D., Zenkin, N., and Gerdes, K. (2013). Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254. doi: 10.1016/j.molcel.2013.08.045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Girgis, H. S., Harris, K., and Tavazoie, S. (2012). Large mutational target size for rapid emergence of bacterial persistence. Proc. Natl. Acad. Sci. U.S.A. 109, 12740–12745. doi: 10.1073/pnas.1205124109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gonzalez-Flecha, B., and Demple, B. (1995). Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J. Biol. Chem. 270, 13681–13687. doi: 10.1074/jbc.270.23.13681
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Helaine, S., Cheverton, A. M., Watson, K. G., Faure, L. M., Matthews, S. A., and Holden, D. W. (2014). Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208. doi: 10.1126/science.1244705
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Imlay, J. A., Chin, S. M., and Linn, S. (1988). Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240, 640–642. doi: 10.1126/science.2834821
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keller, L., and Surette, M. G. (2006). Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4, 249–258. doi: 10.1038/nrmicro1383
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keren, I., Kaldalu, N., Spoering, A., Wang, Y., and Lewis, K. (2004a). Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18. doi: 10.1016/S0378-1097(03)00856-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keren, I., Minami, S., Rubin, E., and Lewis, K. (2011). Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio 2, e00100–e00111. doi: 10.1128/mBio.00100-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keren, I., Shah, D., Spoering, A., Kaldalu, N., and Lewis, K. (2004b). Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J. S., Heo, P., Yang, T. J., Lee, K. S., Cho, D. H., Kim, B. T., et al. (2011). Selective killing of bacterial persisters by a single chemical compound without affecting normal antibiotic-sensitive cells. Antimicrob. Agents Chemother. 55, 5380–5383. doi: 10.1128/AAC.00708-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, Y., and Wood, T. K. (2010). Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391, 209–213. doi: 10.1016/j.bbrc.2009.11.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kwan, B. W., Valenta, J. A., Benedik, M. J., and Wood, T. K. (2013). Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 57, 1468–1473. doi: 10.1128/AAC.02135-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lechner, S., Lewis, K., and Bertram, R. (2012). Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol. 22, 235–244. doi: 10.1159/000342449
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lechner, S. P. M., Lange, B., Huber, C., Eisenreich, W., Herbig, A., Nieselt, K., et al. (2014). Metabolic and transcriptional activities of Staphylococcus aureus challenged with high-doses of daptomycin. Int. J. Med. Microbiol. doi: 10.1016/j.ijmm.2014.05.008. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leszczynska, D., Matuszewska, E., Kuczynska-Wisnik, D., Furmanek-Blaszk, B., and Laskowska, E. (2013). The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS ONE 8:e54737. doi: 10.1371/journal.pone.0054737
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leung, V., and Levesque, C. M. (2012). A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J. Bacteriol. 194, 2265–2274. doi: 10.1128/JB.06707-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lewis, K. (2005). Persister cells and the riddle of biofilm survival. Biochemistry (Mosc.) 70, 267–274. doi: 10.1007/s10541-005-0111-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lewis, K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. doi: 10.1146/annurev.micro.112408.134306
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luidalepp, H., Joers, A., Kaldalu, N., and Tenson, T. (2011). Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J. Bacteriol. 193, 3598–3605. doi: 10.1128/JB.00085-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, C., Sim, S., Shi, W., Du, L., Xing, D., and Zhang, Y. (2010). Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol. Lett. 303, 33–40. doi: 10.1111/j.1574-6968.2009.01857.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mah, T. F., and O'Toole, G. A. (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39. doi: 10.1016/S0966-842X(00)01913-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maisonneuve, E., Castro-Camargo, M., and Gerdes, K. (2013). (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150. doi: 10.1016/j.cell.2013.07.048
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McKenzie, J. L., Robson, J., Berney, M., Smith, T. C., Ruthe, A., Gardner, P. P., et al. (2012). A VapBC toxin-antitoxin module is a posttranscriptional regulator of metabolic flux in mycobacteria. J. Bacteriol. 194, 2189–2204. doi: 10.1128/JB.06790-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mukamolova, G. V., Kormer, S. S., Kell, D. B., and Kaprelyants, A. S. (1999). Stimulation of the multiplication of Micrococcus luteus by an autocrine growth factor. Arch. Microbiol. 172, 9–14. doi: 10.1007/s002030050733
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nakagawa, A., Oshima, T., and Mori, H. (2006). Identification and characterization of a second, inducible promoter of relA in Escherichia coli. Genes Genet. Syst. 81, 299–310. doi: 10.1266/ggs.81.299
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nguyen, D., Joshi-Datar, A., Lepine, F., Bauerle, E., Olakanmi, O., Beer, K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986. doi: 10.1126/science.1211037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orman, M. A., and Brynildsen, M. P. (2013a). Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob. Agents Chemother. 57, 3230–3239. doi: 10.1128/AAC.00243-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orman, M. A., and Brynildsen, M. P. (2013b). Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob. Agents Chemother. 57, 4398–4409. doi: 10.1128/AAC.00372-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pan, J., and Ren, D. (2013). Structural effects on persister control by brominated furanones. Bioorg. Med. Chem. Lett. 23, 6559–6562. doi: 10.1016/j.bmcl.2013.10.070
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pascoe, B., Dams, L., Wilkinson, T. S., Harris, L. G., Bodger, O., Mack, D., et al. (2014). Dormant cells of Staphylococcus aureus are resuscitated by spent culture supernatant. PLoS ONE 9:e85998. doi: 10.1371/journal.pone.0085998
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pinto, D., Sao-Jose, C., Santos, M. A., and Chambel, L. (2013). Characterization of two resuscitation promoting factors of Listeria monocytogenes. Microbiology 159, 1390–1401. doi: 10.1099/mic.0.067850-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Que, Y. A., Hazan, R., Strobel, B., Maura, D., He, J., Kesarwani, M., et al. (2013). A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS ONE 8:e80140. doi: 10.1371/journal.pone.0080140
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rice, K. C., and Bayles, K. W. (2003). Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 50, 729–738. doi: 10.1046/j.1365-2958.2003.t01-1-03720.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schuster, C. F., and Bertram, R. (2013). Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340, 73–85. doi: 10.1111/1574-6968.12074
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shah, D., Zhang, Z., Khodursky, A., Kaldalu, N., Kurg, K., and Lewis, K. (2006). Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. doi: 10.1186/1471-2180-6-53
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Somerville, G. A., Chaussee, M. S., Morgan, C. I., Fitzgerald, J. R., Dorward, D. W., Reitzer, L. J., et al. (2002). Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70, 6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thomas, V. C., Kinkead, L. C., Janssen, A., Schaeffer, C. R., Woods, K. M., Lindgren, J. K., et al. (2013). A dysfunctional tricarboxylic acid cycle enhances fitness of Staphylococcus epidermidis during beta-lactam stress. MBio 4:e00437-13. doi: 10.1128/mBio.00437-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thomas, V. C., Sadykov, M. R., Chaudhari, S. S., Jones, J., Endres, J. L., Widhelm, T. J., et al. (2014). A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog. 10:e1004205. doi: 10.1371/journal.ppat.1004205
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Traxler, M. F., Summers, S. M., Nguyen, H. T., Zacharia, V. M., Hightower, G. A., Smith, J. T., et al. (2008). The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Unoson, C., and Wagner, E. G. (2008). A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 70, 258–270. doi: 10.1111/j.1365-2958.2008.06416.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Uppal, S., Shetty, D. M., and Jawali, N. (2014). Cyclic AMP receptor protein regulates cspD, a bacterial toxin gene, in Escherichia coli. J. Bacteriol. 196, 1569–1577. doi: 10.1128/JB.01476-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Acker, H., Sass, A., Bazzini, S., De Roy, K., Udine, C., Messiaen, T., et al. (2013). Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS ONE 8:e58943. doi: 10.1371/journal.pone.0058943
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vega, N. M., Allison, K. R., Khalil, A. S., and Collins, J. J. (2012). Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 8, 431–433. doi: 10.1038/nchembio.915
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vega, N. M., Allison, K. R., Samuels, A. N., Klempner, M. S., and Collins, J. J. (2013). Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. U.S.A. 110, 14420–14425. doi: 10.1073/pnas.1308085110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamanaka, K., and Inouye, M. (1997). Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 179, 5126–5130.
Zhang, L., Chiang, W. C., Gao, Q., Givskov, M., Tolker-Nielsen, T., Yang, L., et al. (2012). The catabolite repression control protein Crc plays a role in the development of antimicrobial-tolerant subpopulations in Pseudomonas aeruginosa biofilms. Microbiology 158, 3014–3019. doi: 10.1099/mic.0.061192-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Staphylococcus aureus, persisters, metabolism, toxin-antitoxin system, ppGpp, biofilm
Citation: Prax M and Bertram R (2014) Metabolic aspects of bacterial persisters. Front. Cell. Infect. Microbiol. 4:148. doi: 10.3389/fcimb.2014.00148
Received: 08 July 2014; Paper pending published: 02 September 2014;
Accepted: 05 October 2014; Published online: 22 October 2014.
Edited by:
Thomas Dandekar, University of Wuerzburg, GermanyReviewed by:
Alain Charbit, University Paris Descartes, FrancePetra Dersch, Helmholtz Center for Infection Research, Germany
Copyright © 2014 Prax and Bertram. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralph Bertram, Department of Microbial Genetics, Faculty of Science, Interfaculty Institute of Microbiology and Infection Medicine Tübingen (IMIT), University of Tübingen, Waldhäuser Str. 70/8, 72076 Tübingen, Germany e-mail: ralph.bertram@uni-tuebingen.de
 Marcel Prax
Marcel Prax  Ralph Bertram
Ralph Bertram