LuxR solos in Photorhabdus species
- 1Bereich Mikrobiologie, Biozentrum, Ludwig-Maximilians-Universität München, München, Germany
- 2Fachbereich Biowissenschaften, Merck Stiftungsprofessur für Molekulare Biotechnologie, Goethe-Universität Frankfurt, Frankfurt am Main, Germany
Bacteria communicate via small diffusible molecules to mediate group-coordinated behavior, a process designated as quorum sensing. The basic molecular quorum sensing system of Gram-negative bacteria consists of a LuxI-type autoinducer synthase producing acyl-homoserine lactones (AHLs) as signaling molecules, and a LuxR-type receptor detecting the AHLs to control expression of specific genes. However, many proteobacteria possess one or more unpaired LuxR-type receptors that lack a cognate LuxI-like synthase, referred to as LuxR solos. The enteric and insect pathogenic bacteria of the genus Photorhabdus harbor an extraordinarily high number of LuxR solos, more than any other known bacteria, and all lack a LuxI-like synthase. Here, we focus on the presence and the different types of LuxR solos in the three known Photorhabdus species using bioinformatics analyses. Generally, the N-terminal signal-binding domain (SBD) of LuxR-type receptors sensing AHLs have a motif of six conserved amino acids that is important for binding and specificity of the signaling molecule. However, this motif is altered in the majority of the Photorhabdus-specific LuxR solos, suggesting the use of other signaling molecules than AHLs. Furthermore, all Photorhabdus species contain at least one LuxR solo with an intact AHL-binding motif, which might allow the ability to sense AHLs of other bacteria. Moreover, all three species have high AHL-degrading activity caused by the presence of different AHL-lactonases and AHL-acylases, revealing a high quorum quenching activity against other bacteria. However, the majority of the other LuxR solos in Photorhabdus have a N-terminal so-called PAS4-domain instead of an AHL-binding domain, containing different amino acid motifs than the AHL-sensors, which potentially allows the recognition of a highly variable range of signaling molecules that can be sensed apart from AHLs. These PAS4-LuxR solos are proposed to be involved in host sensing, and therefore in inter-kingdom signaling. Overall, Photorhabdus species are perfect model organisms to study bacterial communication via LuxR solos and their role for a symbiotic and pathogenic life style.
Introduction
Photorhabdus spec. are pathogenic enteric bacteria that maintain a mutualistic interaction with heterorhabditid nematodes and can infect a wide variety of insect species. To date, three different Photorhabdus species are known: P. luminescens, P. temperata, and P. asymbiotica (Fischer-Le Saux et al., 1999). Whereas the first two species are highly pathogenic toward insects, P. asymbiotica additionally infects men (Gerrard et al., 2003). P. asymbiotica is associated with severe soft-tissue and systemic infections in humans, and is considered as an emerging threat (Gerrard et al., 2004). Commonly, all Photorhabdus bacteria colonize the gut of the infective juvenile stage of Heterorhabditis spec. nematodes. Upon entering insect larvae, the nematodes directly inject the bacteria by regurgitation into the insect's hemocoel. Once inside the insect, the bacteria rapidly replicate and quickly establish a lethal septicemia in the host by production of a broad range of different toxins that kill the insect within 48 h. At this stage the dying insect glows, due to bacterial luciferase production. Bioconversion of the insect's body by a huge set of bacterial exoenzymes produces a rich food source both for the bacteria and the nematodes. Furthermore, the bacteria support nematode reproduction probably by providing essential nutrients that are required for efficient nematode proliferation (Han and Ehlers, 2000). Moreover, the bacteria produce several antibiotics to defend the insect cadaver from invasion of other bacteria. When the insect cadaver is depleted, the nematodes and bacteria re-associate and emerge from the carcass in search of a new insect host (see Clarke, 2008; Waterfield et al., 2009, for review). During this complex life cycle the bacteria constantly have to monitor their host environment, and to communicate with each other as well as with their hosts to perfectly adapt to the respective conditions.
Bacterial communication via small molecules to mediate group-coordinated behavior, referred to as quorum sensing (QS), is well recognized. Typically, Gram-negative bacteria use small diffusible molecules, e.g., acyl-homoserine lactones (AHLs) derived from fatty acids, whereas Gram-positive bacteria use peptide derivatives for communication. However, recently the production of AHLs by a Gram-positive bacterium belonging to the Exiguobacterium genera isolated from marine water was discovered (Biswa and Doble, 2013). The prototypical quorum sensing system of Gram-negative bacteria consists of a LuxI-like autoinducer synthase that produces AHLs as signals and a LuxR-type receptor that detects the AHLs to control expression of specific genes (Waters and Bassler, 2005). The AHLs are constantly synthesized by LuxI, and are sensed by the cognate LuxR-like receptor when exceeding a threshold concentration. Upon AHL-binding, LuxR binds to the promoter/operator regions of the target genes/operons causing altered gene expression in response to the cell count. Thus, bacteria respond to AHLs and adapt bacterial group-behavior by regulation of gene expression when the cell density reaches a certain size (quorum). Signaling via AHLs has been linked to diverse phenotypes like the production of virulence factors, motility, antibiotic production, sporulation, bioluminescence or biofilm formation (Waters and Bassler, 2005). LuxI/LuxR based quorum sensing systems have been intensively studied. The first system was described in V. fischeri showing that AHLs are used to regulate light production dependent on the cell density (Nealson and Hastings, 1979). Furthermore, LuxR-based cell-cell communication is medically relevant as many pathogenic bacteria use these quorum sensing systems for an effective infection process (Rutherford and Bassler, 2012).
Moreover, many bacterial genomes encode additional LuxR homologs lacking a cognate LuxI synthase. These LuxR homologs are designated as LuxR orphans (Patankar and González, 2009) or LuxR solos (Subramoni and Venturi, 2009). Genome sequencing of 265 proteobacterial genomes revealed that 68 of these encode at least one LuxI and one LuxR homolog (Case et al., 2008). Furthermore, 45 of these 68 genomes encode more LuxR-type proteins than LuxI homologs like Pseudomonas aeruginosa, which harbors beside the two classical QS systems, LasI/LasR and RhlI/RhlR, the LuxR solo QscR (Oinuma and Greenberg, 2011). Further 45 genomes do not harbor a luxI-like AHL synthase encoding gene, but encode at least one LuxR homolog. The three Photorhabdus species all lack a LuxI homolog, but have an extraordinary high number of genes that encode potential LuxR solos and are therefore assumed to have a huge capacity for cell-cell and/or inter-kingdom communication (Heermann and Fuchs, 2008). Therefore, these bacteria are perfect model organisms to study bacterial cell-cell communication via LuxR solos. Here we focus on the function of the multiple LuxR solos of the three known Photorhabdus species for their life cycle, their role in cell-cell communication and inter-kingdom signaling, as well as the specificity of signal perception.
Materials and Methods
Bacterial Strains
The strains Photorhabdus luminescens TT01 (Fischer-Le Saux et al., 1999), Photorhabdus temperata NC19 (Tailliez et al., 2010), Photorhabdus asymbiotica ATCC43949 (Wilkonson et al., 2009), and Vibrio harveyi BB120 (Bassler et al., 1997) were used in this study.
Quorum Quenching Bioassay
Bioluminescence of Vibrio harveyi wild-type (BB120) was used as read-out to analyze degradative activity of AHLs in Photorhabdus species supernatants. Therefore, V. harveyi BB120 strain was grown at 30°C in LM medium [10% (w/v) peptone, 5% (w/v) yeast extract, 20% (w/v) NaCl] supplemented with carbenicillin (100 μg/ml). An overnight culture of V. harveyi BB120 was inoculated at Optical Density (OD) at 600 nm = 0.2 in LM and a volume of 80 μl was pipetted into each well of a 96-microtiter plate. Cells were then grown for 4.5 h aerobically at 30°C until the mid-exponential growth phase, the growth phase where bioluminescence naturally occurs (Anetzberger et al., 2012). After 4.5 h of growth a volume of 80 μl of the cell-free supernatant of P. luminescens, P. temperata, or P. asymbiotica, respectively, was added. Thus, P. luminescens and P. temperata cultures were grown at 30°C, and P. asymbiotica culture was grown at 37°C in CASO Medium. After 4 d and 7 d, cells were adjusted to OD 600 nm = 1 before the supernatant was harvested via centrifugation. As control the same volume of LM medium and supernatant of a V. harveyi BB120 culture, harvested after 8 h and 1 day of growth, was added. OD and luminescence were monitored every hour with a Sunrise plate reader (Tecan, Crailsheim) and a Centro luminometer (Berthold Technologies, Bad Wildbad), respectively.
Bioinformatics Studies
For bioinformatics identification of LuxR solos the genome assembly and annotation reports of Photorhabdus luminescens subsp. laumondii TT01 (NCBI reference sequence NC_005126.1), of Photorhabdus temperata subsp. khanii NC19 (BioSample SAMN02597464, 19 contigs, GenBank Assembly ID: GCA_000517265.1), of Photorhabdus asymbiotica subsp. asymbiotica ATCC43949 (NCBI reference sequence NC_012962.1), and of the P. asymbiotica plasmid pPAU1 (NCBI reference sequence NC_012961.1) were used. LuxR solos in the three above-mentioned Photorhabdus species were first identified based on the presence of the C-terminal “HTH LUXR” motif (SMART00421) using SMART 7 software (Simple Modular Architecture Research Tool) (Letunic et al., 2012) and BLAST software (Altschul et al., 1990). Protein domains were identified using SMART 7 software and are identified with a maximal p-value of 2e-07 for the “PAS_4”-domain (PFAM08448), of 2e-06 for the “HTH LUXR” motif (SMART00421) and of 1.80e-24 for the “Autoind_bind”-domain (PFAM03472). Furthermore, homologous proteins in P. asymbiotica were identified using STRING 9.1 database (Franceschini et al., 2013) based on the LuxR solos of P. luminescens (Heermann and Fuchs, 2008).
To elucidate the relationship between the LuxR solos of Photorhabdus species, the protein sequences of the 100 LuxR solos of the three Photorhabdus species, of LuxR from Vibrio fischeri, of TraR from Agrobacterium tumefaciens, of SdiA from Escherichia coli and of QscR and LasR from Pseudomonas aeruginosa were aligned and a phylogenetic tree was generated based on the alignment using CLC Mainworkbench 7 (CLC Bio Qiagen, Hilden, Germany). In the next step, the amino acid residues at the positions of the WYDPWG-motif in the signal-binding domain (SBD) of AHL-sensors were added as metadata layers.
AHL-lactonases and AHL-acylases in the three Photorhabdus species were identified using SMART 7 software (Letunic et al., 2012), BLAST software (Altschul et al., 1990) and STRING 9.1 database (Franceschini et al., 2013) based on the presence of known AHL-lactonases and AHL-acylases from Bacillus cereus, Agrobacterium tumefaciens, Arthrobacter spec., Rhodococcus erythropolis, Pseudomonas aeruginosa, Streptomyces spec., Dictyoglomus thermophilum, and Anabaena spec. Furthermore, the genomes of the three Photorhabdus species were analyzed for the presence of the “Beta-lactamase” motif (PF00144).
Results and Discussion
Photorhabdus Species Harbor Three Different Types of LuxR Solos
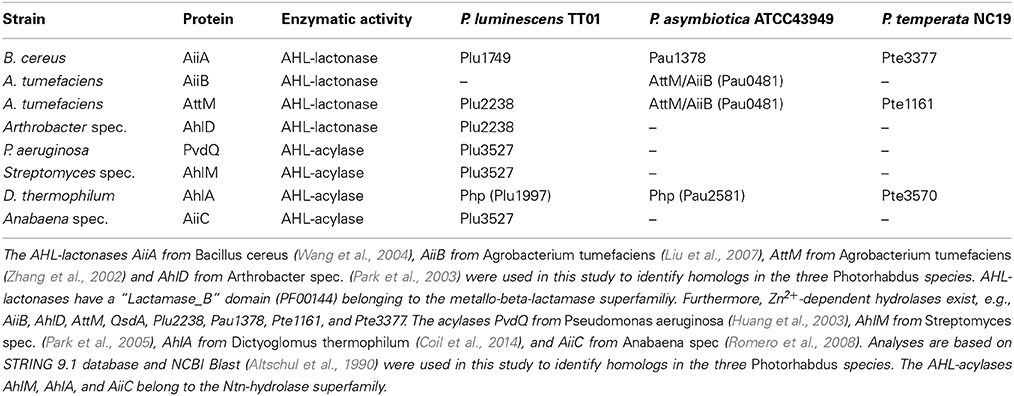
We analyzed the genome of each one representative strain of the three known Photorhabdus species P. luminescens, P. temperata, and P. asymbiotica for the presence of genes that encode potential LuxR solos. We found that all three Photorhabdus species harbor an exceptionally high number of LuxR solos, with 40, 22, and 38 LuxR solos in P. luminescens, P. asymbiotica, and P. temperata, respectively (Figure 1). A previous study revealed that P. luminescens has 39 LuxR solos (Heermann and Fuchs, 2008), but SMART 7 software (Letunic et al., 2012) actually identified Plu4288 as LuxR solo as well, which increased the number of LuxR solos to 40 in this organism. These LuxR solos all contain a C-terminal DNA-binding domain (DBD) with a helix-turn-helix motif, the “HTH LUXR” motif, which is typical for LuxR-type proteins and that was used to identify LuxR solos in Photorhabdus species. However, LuxR-type receptors are composed of two functional domains, the C-terminal DBD and the N-terminal SBD that are connected with a short linker (Nasser and Reverchon, 2006). Furthermore, the size of the protein rather than the degree of similarity is crucial whether a protein with such a domain organization meets the criteria to be a LuxR homolog, which is about 250 amino acids in length (Fuqua et al., 1994; Subramoni and Venturi, 2009). All the LuxR homologs identified in the three Photorhabdus species meet these criteria, however, diverse domains can make up the SBD, which is the domain that is important for signal-sensing specificity of the receptor. The entire 100 LuxR solos of the three Photorhabdus species differ in their N-terminal SBD, for which reason they are grouped into three types: LuxR solos with a PAS4-domain, LuxR solos with an AHL-domain (Autoind_bind) and LuxR solos with yet undefined SBD domains (Figure 1). Presumably, these variable SBD-domains enable the bacteria to sense diverse signals, like exogenous AHLs, exogenous or endogenous non-AHLs, or eukaryotic signals, and thereby influence different cellular processes (Subramoni and Venturi, 2009). The overall number of the LuxR solos might therefore reflect the diversity of invertebrate or vertebrate hosts that Photorhabdus species can infect or colonize.
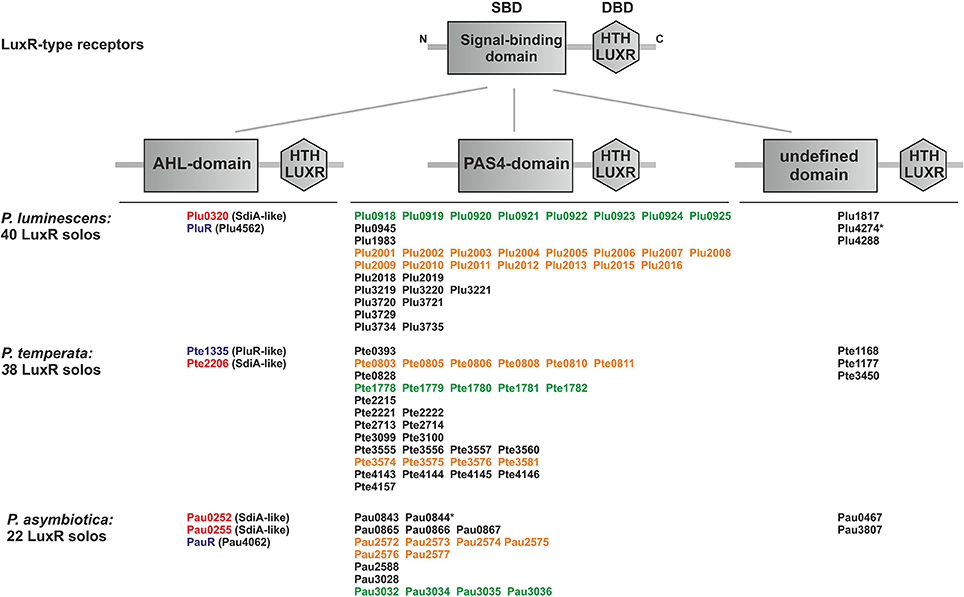
Figure 1. Presence and domain structure of the LuxR solos in P. luminescens, P. asymbiotica, and P. temperata. LuxR-type receptors share a modular domain structure, with a N-terminal signal-binding domain (SBD) (gray box) and a C-terminal DNA-binding domain (DBD) with the conserved “HTH LUXR” motif (gray hexagon), which is illustrated in the upper part. The N-terminus is marked with an “N” and the C-terminus with a “C.” The LuxR solos of Photorhabdus species, P. luminescens TT01, P. temperata NC19, and P. asymbiotica ATCC43949, are grouped into three types based on the different N-terminal domain. The “HTH LUXR” motif (SMART00421) is indicated by a hexagon, the AHL-domain (PFAM03472: “Autoind_bind”-domain) and the PAS4-domain (PFAM08448: “PAS_4”-domain) by boxes. LuxR solos marked with an asterisk additionally have an N-terminal transmembrane domain (not illustrated). Homologous proteins or cluster are marked with the colors red, purple, orange and green. LuxR solos were identified using SMART 7 software (Letunic et al., 2012) and BLAST software (Altschul et al., 1990).
LuxR solos containing a PAS4-domain
The majority of the LuxR solos in Photorhabdus spec. contain a N-terminal PAS4-domain, which are in sum 80 of the 100 identified LuxR solos. However, the signals sensed by these LuxR solos are yet unidentified. Commonly, PAS-domains are ubiquitous, they appear in archaea, eubacteria and eukarya and are involved in binding of a diverse set of small regulatory molecules either covalently or non-covalently (Hefti et al., 2004). In the fruit fly Drosophila melanogaster PAS3 domains have been proved as insect juvenile hormone receptors (Dubrovsky, 2005). The homologous PAS4-domains in Photorhabdus are assumed to bind hormone-like molecules and are therefore proposed to be major players in inter-kingdom signaling via the detection of hormone-specific signals from the eukaryotic hosts (Heermann and Fuchs, 2008). As this is most likely, it cannot be excluded that also other bacteria might be sensed via one or more of the PAS4-LuxR receptors. Moreover, the majority of the PAS4-domain-containing LuxR solos in Photorhabdus spec. are organized in large gene clusters, which are plu0915-plu0925 and plu2001-plu2016 in P. luminescens, pau2572-pau2577, and pau3032-pau3036 in P. asymbiotica and the operons pte0803-pte0811, pte1178-pte1779, and pte3555-pte3581 in P. temperata. The function of this genetic clustering as well as the high redundancy of all these PAS4-LuxR solos is unknown to date. Presumably, the high redundancy of PAS4-LuxR solos might be a co-evolutional result by adaptation of the bacteria to the variety of insect hosts they can infect. This idea is underlined by the finding that several LuxR solos from plant-associated bacteria are known to respond to plant signaling molecules and are therefore assumed to have undergone co-evolution with the related host plant (Covaceuszach et al., 2013). Putative signals sensed by the PAS4-LuxR solos might be diffusible substances like hormones. The phytopathogens belonging to the genus Xanthomonas use a fatty acid signal belonging to the Diffusible Signal Factor (DSF) family for cell-cell signaling to control the virulence of Xanthomonas campestris pv. campestris (Xcc) to plants. Recently, a second sensor for DSF was identified, which is a complex sensor kinase having a N-terminal PAS4-domain essential for sensing of DSF (An et al., 2014). This supports the idea that the PAS4-LuxR solos of Photorhabdus spec. are adapted to distinct signals from the invertebrate hosts (insects and nematodes) or in case of P. asymbiotica additionally to vertebrate hosts, especially humans, to expand or fine-tune their regulatory network.
LuxR solos containing an AHL-domain
Besides the high number of PAS4-LuxR solos, all three Photorhabdus species contain at least one LuxR solo that is homologous to the LuxR solo SdiA. P. luminescens and P. temperata each have one SdiA homolog, Plu0320 and Pte2206, respectively, whereas the human pathogenic P. asymbiotica has two SdiA homologs, Pau0252 and Pau0255. The LuxR solos SdiA of Escherichia coli and Salmonella enterica are known to detect exogenously produced AHLs and therefore these organisms are able to sense mixed microbial communities (Wang et al., 1991; Michael et al., 2001). All four SdiA homologs in Photorhabdus spec. have a N-terminal “Autoind_bind” domain (PFAM03472) (Figure 1), which is typical for AHL-sensors and responsible for AHL-sensing. Similar to SdiA, the four LuxR solos Plu0320, Pau0252, Pau0255, and Pte2206, respectively, can be assumed to detect AHLs as well. Since no luxI gene is present in any of the Photorhabdus genomes, it is most likely that these LuxR solos can respond to exogenously produced AHLs, produced by other bacteria, probably of the insect gut, which are then sensed by Photorhabdus spec. during the infection process. Moreover, AHLs have never been detected using different analytical chemistry methods (HPLC/MS, GC/MS) in any Photorhabdus strain, despite the analysis of >100 strains (Darko Kresovic and Helge B. Bode, data not shown). As the human pathogen P. asymbiotica has two putative AHL-sensors (Figure 1), it is most likely that AHL-sensing plays an even more important role for vertebrate than invertebrate host infection. Interestingly, also the three homologous LuxR solos PluR (Plu4562) of P. luminescens, PauR (Pau4062) of P. asymbiotica, and PluR (Pte1335) of P. temperata contain a predicted “Autoind_bind” domain like the AHL sensors. However, the LuxR solo PluR senses α-pyrones named photopyrones (PPYs) instead of AHLs (Brachmann et al., 2013), which shows that the presence of an “Autoind_bind” domain is not automatically going along with AHL-sensing. PluR was the first example of a LuxR solo that senses an endogenous signal and is therefore part of a bacterial cell-cell communication system in P. luminescens regulating expression of the pcfABCDEF operon leading to cell clumping (Brachmann et al., 2013). P. luminescens produces eight different PPYs, out of which three (PPYA, PPYB, PPYD) are mainly present in the culture fluid. Among these, PluR senses PPYD with the highest sensitivity in a concentration as low as 3.5 nM. Furthermore, the ketosynthase-like protein PpyS was identified as the PPY synthase and therefore as producer of the signaling molecules. All proteins are also present with high homology in P. temperata, which indicates a similar strategy for cell-cell communication in this genus (Brachmann et al., 2013). The insect and human pathogen P. asymbiotica harbors a pcf regulon that is highly homologous to that of P. luminescens. It consists of the pcfABCDEF (pau4068-pau4063) operon and an adjacent gene encoding a LuxR solo that we named PauR (Pau4062). However, P. asymbiotica contains neither a ppyS homolog nor a luxI-like synthase and does not produce any PPYs or AHLs. Therefore, it is assumed that P. asymbiotica uses yet unidentified signaling molecules for cell-cell communication, which are sensed by PauR. As we will focus on below, this is rather due to the presence of a conserved amino acid motif within the SBD, which is intact in the SdiA homologs, but different in both PluR proteins and PauR. When PluR was originally detected it met the criteria to be defined as LuxR solo. Since PluR has a cognate synthase, but which is different from LuxI, it is thus part of an entire cell-cell communication system. Therefore, it is disputable whether the designation LuxR solo is actually appropriate at least for PluR.
LuxR solos containing a non-defined signal-binding domain
Besides LuxR solos with defined SBDs, all three Photorhabdus species harbor LuxR solos with yet unidentified SBDs, namely Plu1817, Plu4274, Plu4288, Pau0467, Pau3804, Pte1168, Pte1177, and Pte3450. In contrast to many other LuxR solos, which are often hydrophobic and membrane associated (Welch et al., 2000), Pau0844 and Plu4274 are predicted to contain a N-terminal transmembrane domain (TM). Although the exact mechanism of DNA-binding by a membrane-integrated protein is not yet fully understood, other examples of those membrane-integrated DNA-binding receptors are known, e.g., the pH-sensor CadC of Escherichia coli (Haneburger et al., 2011). The presence of an intact TM within the SBD could give indication for sensing an extremely hydrophobic and non-water-soluble signaling molecule. The LuxR solo Pau0844 is the only LuxR solo that harbors a TM in combination with a PAS4-domain, which might therefore sense an extremely hydrophobic eukaryotic signal. Overall, the LuxR solos with yet undefined SBDs might expand the range of signals that are sensed and therefore the communication network. However, their roles in cell-cell communication and/or inter-kingdom signaling as well as the related signaling molecules are unknown to date.
Conserved Amino Acid Motifs in the Signal-binding Domain of LuxR Solos
Comparison of all LuxR solos of Photorhabdus spec. indicates that signal-specificity relies on the SBD, given that the major differences are located in the SBD and the similarities in the DBD. In general, proteins of the LuxR family are less conserved in their amino acid sequence, which only makes up between 18 and 25% identity (Subramoni and Venturi, 2009). However, LuxR-type proteins sensing AHLs harbor six highly conserved amino acid in the N-terminal SBD that are important for signal-binding, signal molecule specificity and shaping the signal-binding pocket. These six conserved amino acids are W57, Y61, D70, P71, W85, and G113 (with respect to TraR), which are either hydrophobic or aromatic and displaying a conserved motif for AHL-sensors (Patankar and González, 2009). Bioinformatics analyses of the amino acids of the Photorhabdus-specific LuxR solos at these positions reveals that the conserved WYDPWG-motif of AHL-sensors is only completely present in the SdiA homologs Plu0320 of P. luminescens, Pau0252 and Pau0255 of P. asymbiotica, and Pte2206 of P. temperata, supporting the idea that these LuxR solos sense exogenous AHLs. All other LuxR solos in Photorhabdus spec. have at least two substitutions at different positions in the WYDPWG-motif. Remarkably, amino acid Y61 (with respect to TraR) is present in 97% of the LuxR solos of the three Photorhabdus species. This amino acid is known to be involved in binding of the acyl chain of the signaling molecule via hydrophobic interactions, e.g., in TraR (Churchill and Chen, 2011) or LuxR (Nasser and Reverchon, 2006). Therefore, there seems to be a common mechanism to arrange distinct signaling molecules in the signal-binding pocket, independent from the exact chemical nature of the signaling molecule. The position D70 (with respect to TraR) is known to be important for binding the amide group of the cognate AHL within AHL-binding LuxR receptors (Churchill and Chen, 2011). This position is also conserved among 80% of the LuxR solos in the three Photorhabdus species, whereas the remaining LuxR solos (19%), mainly from P. asymbiotica, have the substitution D70N at this position, which might be important to sense eukaryotic signals during vertebrate infection (Figure 2). Although not sensing AHLs (Brachmann et al., 2013), the three LuxR solos, PauR and both PluR homologs of P. asymbiotica, P. luminescens, and P. temperata, respectively, have a predicted “Autoind_bind” domain. However, only the two conserved amino acids Y61 and D70 (with respect to TraR) are present in these LuxR solos. Moreover, both PluR proteins share a TYDQCS-motif, which is therefore probably specific for PPY-sensing (Figure 2). Individual replacement of Y66 and D75 with alanine within PluR prevented PPY-binding, revealing that these amino acids are important for PPY-binding and/or specificity (Brachmann et al., 2013). In contrast, PauR has a slightly different SBD-motif, which is TYDQYI, indicating that this LuxR solo, although highly homologous to both PluR proteins, senses a signal that is different from PPYs as well as AHLs. Overall, variations in the WYDPWG-motif that is typical for AHL-sensing probably expand the diversity of signals that can be sensed by LuxR-type proteins having an “Autoind_bind” domain. Additionally, the 80 LuxR solos of Photorhabdus spec. with a PAS4-domain share a more variable amino acid motif in the SBD, reflecting a higher variety of signals they might sense (Figure 2). However, the precise signals that are sensed by PAS4-LuxR solos are yet unknown. The huge diversity of the SBD-motifs of PAS4-LuxR solos in Photorhabdus and the variations in the conserved amino acid motifs probably gives the bacteria the capacity to respond to a broad range of signals that occur in the different environments or hosts.
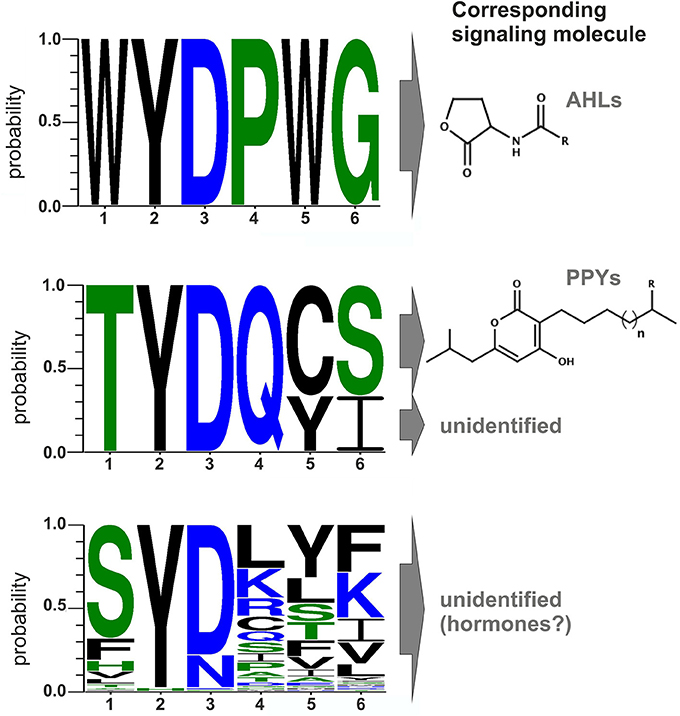
Figure 2. Conserved amino acid motifs of LuxR-type proteins with different domains and their corresponding signaling molecules. Upper part: Motif of the six conserved amino acid positions in typical AHL-sensors. Protein sequences of LuxR from Vibrio fischeri, TraR from Agrobacterium tumefaciens, SdiA from Escherichia coli, QscR and LasR from Pseudomonas aeruginosa were used to generate the alignment. Middle part: Motif of the six conserved amino acids positions of PluR (Plu4562) from P. luminescens, PluR (Pte1335) from P. temperata and PauR (Pau4062) from P. asymbiotica. PluR from P. luminescens and from P. temperata are sensing photopyrones as signaling molecule, however this is yet unidentified for PauR. Lower part: Motif of the six conserved amino acid positions of the overall 80 PAS4-LuxR solos in all three Photorhabdus species, whereas the corresponding signal molecules are yet unknown but possible are eukaryotic hormones. All alignments were generated with CLC Mainworkbench 7 software (CLC Bio Qiagen, Hilden, Germany). The sequence logo was made with WebLogo3 (Crooks et al., 2004).
A closer look at the protein sequence and phylogeny of the Photorhabdus-specific LuxR solos reveals that related LuxR solos, independent of their host organism, share the similar amino acid motif in the SBD (Figure 3). More precisely, LuxR solos encoded by one operon tend to have a similar amino acid motif at the six conserved positions of the SBD. For example the plu0919-plu0925 operon and the homologous pte4143-pte4146 operon encoding the respective PAS4-LuxR solos both show replacement of amino acid W85 (with respect to TraR) to L and of G113 (with respect to TraR) either to V or L in the WYDPWG-motif. This in total applies for 14 of 80 LuxR solos that have a PAS4-domain. However, over 30 LuxR solos show a substitution of W85 to Y containing a PAS4-domain. These homologous changes may originate via gene duplications within one species and might give these LuxR solos the ability to sense similar signals. On the other hand, closely related LuxR solos also incline to the same kind of substitution of the amino acid motif in the SBD (Figure 3). The homologous PluR LuxR solos, Plu4562 from P. luminescens and Pte1335 from P. temperata, share the identical substitutions of four conserved amino acids, forming a TYDQCS-motif allowing both to sense PPYs. The closely related LuxR solo PauR from P. asymbiotica harbors the slightly different TYDQYI-motif at the similar positions, suggesting binding of a distinct signaling molecule. This could be due to the fact that cell-cell-communication via PPY might be inappropriate when infecting vertebrates. Comparably, the LuxR solos of the novel subfamily of plant-associated bacteria have substitutions in the WYDPWG-motif, which are W57M and Y61W (with respect to TraR) that might allow the binding of plant signal molecules rather than AHLs (Patankar and González, 2009; González and Venturi, 2013). The P. luminescens specific PAS4-LuxR solos Plu2018, Plu3221, Plu3720, Plu3729, and Plu3735 are closely related to each other and all have an identical amino acid motif within the SBD, which is SYDLYK. This shows that phylogeny of the LuxR solos correlates with the protein sequence as well as the six conserved amino acid positions within the SBD.
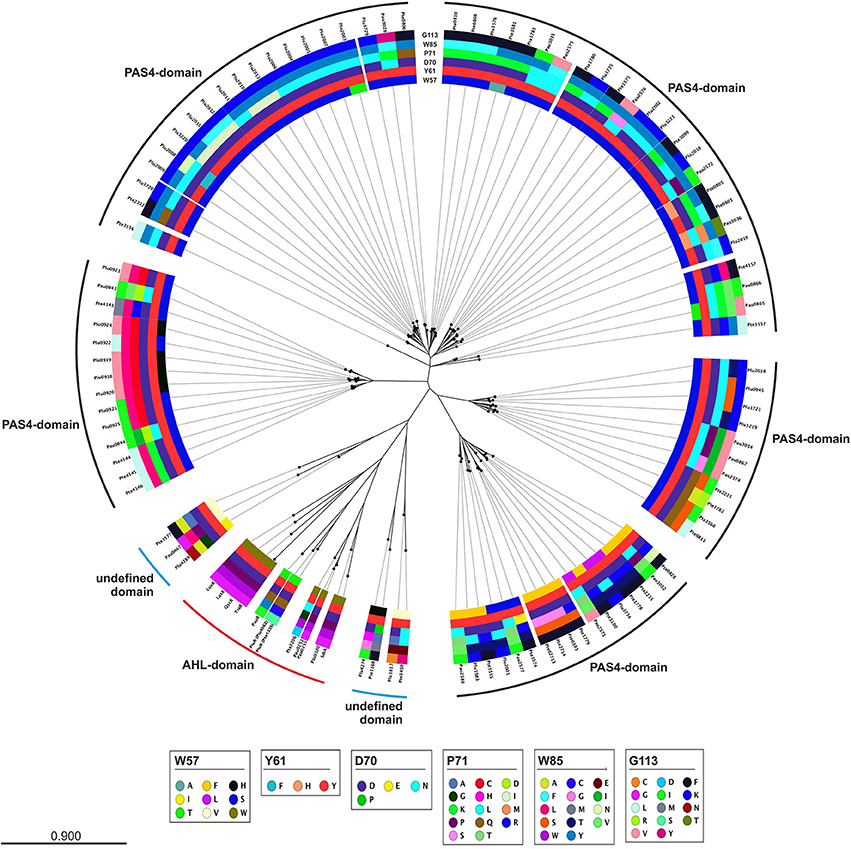
Figure 3. Phylogenetic tree of the LuxR solos present in P. luminescens, P. temperata, and P. asymbiotica. Protein sequences of the overall LuxR solos of the three Photorhabdus species, of LuxR from Vibrio fischeri, of TraR from Agrobacterium tumefaciens, of SdiA from Escherichia coli as well as of QscR and LasR from Pseudomonas aeruginosa were aligned and a phylogenetic tree was generated. Based on this alignment the different amino acid motifs at the six conserved positions were identified by deviation from the WYDPWG-motif in the SBD of AHL-sensors. A special focus on the amino acids at positions of the WYDPWG-motif is shown from the inner to the outer circle. The amino acid W57, with respect to TraR, is marked in brown, Y61 in red, D70 in purple, P71 in dark red, W85 in pink and G113 in light pink, however substitutions within this positions are marked in different colors. LuxR-type proteins with an AHL-binding domain are marked with a red dash, LuxR-type proteins with a PAS4-domain are marked with a black dash and LuxR-type proteins with a N-terminal yet undefined domain are marked with a blue dash. Alignment and radial phylogenetic tree was generated with the CLC Mainworkbench 7 (CLC Bio Qiagen, Hilden, Germany). The scale bar indicates the length of the branches.
Furthermore, variations of the amino acids at the conserved positions to motifs different from WYDPWG offers the possibility to sense different signals or altering the affinity toward a specific signal, as the latter is true for EsaR from Pantoea stewartii (Shong et al., 2013). However, LuxR solos with similar amino acid motifs in the SBD of different species might in turn lead to the binding of the same signaling molecules.
Quorum Quenching via AHL-lactonases and AHL-acylases in Photorhabdus Species
As identified above, each Photorhabdus species harbor a SdiA-homolog, which are the LuxR solos Plu0320, Pte2206, Pau0252, and Pau0255 of P. luminescens, P. temperata, and P. asymbiotica, respectively, containing an AHL-binding domain. Although the functions of these LuxR solos are yet unknown, it might be the sensing of external AHLs. One reason for sensing external AHLs could be to interfere with LuxR quorum sensing systems of their environment by degradation of the QS signaling molecule of neighboring bacteria. This process is designated as quorum quenching (QQ), interferes with the communication of a bacterial population, and can be used to influence their activity, e.g., virulence. For this purpose, many bacteria produce lactonases and acylases, which hydrolyze the lactone ring and cleave the amide bond, respectively, of AHLs. These enzymes are found in both Gram-negative AHL producers and non-AHL producers as well as in Gram-positive bacteria (Williams, 2007). The virulence of the plant pathogens Pseudomonas solanacearum, diverse Erwinia species and the human pathogen Pseudomonas aeruginosa is influenced via QQ enzymes (Dong et al., 2000). We analyzed the genome of all three Photorhabdus species for the presence of genes that encode putative AHL-lactonases and AHL-acylases. We identified several homologs of known AHL-lactonase and AHL-acylases in all three Photorhabdus species (Table 1). Particularly, homologs for the AHL-lactonase AiiA from Bacillus spec., which inactivates the QS signal and decreases the virulence of Erwinia carotovora (Park et al., 2008), are found in all three Photorhabdus species, which are Plu1749, Pau1378, and Pte3377. In addition, AiiA showed a strong enzymatic activity toward distinct AHLs, which vary in length and the substitution at the C3 position of the acyl chain (Wang et al., 2004). Park et al. (2003) demonstrated that the AHL-lactonase AhlD of Arthrobacter spp. blocks the communication of other bacteria and yet P. luminescens could use the same strategy via the homolog Plu2238. The AHL-lactonase AttM of A. tumefaciens, encoded on the At plasmid, is important for the attachment to plant cells (Liu et al., 2007) and a homolog is found in each Photorhabdus species, which are Plu2238, Pau0481, and Pte1161. However, the plasmid pPAU1 from P. asymbiotica harbors no known AHL-lactonase or AHL-acylase or homologs revealing that QQ by Photorhabdus is not especially important for vertebrate infection, or that the similar QQ enzymes for vertebrate as well as for invertebrate infection are used. However, we not only detected the presence of these putative AHL-lactonases and AHL-acylases in Photorhabdus species. Moreover, we analyzed the potential QQ ability of Photorhabdus species culture fluids. Therefore, the culture fluids of all three Photorhabdus spec. at different growth phases were examined using the marine bacterium Vibrio harveyi and its regulation of bioluminescence via accumulation of the AHLs as reporter. Thereby, all three culture fluids emerged to have high QQ activity, putatively by degradation of the AHL HAI-1 of V. harveyi (Figure 4). Especially, the culture supernatant of P. luminescens had the highest AHL-degrading activity against HAI-1 of V. harveyi compared to those of P. temperata as well as P. asymbiotica (Figure 4). However, the number of putative AHL-lactonases and AHL-acylases homologs predicted in all three Photorhabdus species is similar (Table 1). These differences in QQ could therefore only be explained with the theory that the respective genes might be differently regulated in the three Photorhabdus species and that therefore not all enzymes are fully produced or activated under the in vitro experimental conditions used. Anyway, our findings indicate that the three Photorhabdus species can interfere with the AHL-based quorum sensing systems of other bacteria. This could be an important step in the infection process, probably to shut down virulence of opportunistic pathogens or food competitors that are present in the host gut or in the soil to prevent them from pitching into their prey or to stop them from antibiotic production. The expression of the different AHL-lactonases and/or AHL-acylases encoding genes could be activated via the SdiA-homologous LuxR solos Plu0320, Pte2206, Pau0252, and Pau0255 of P. luminescens, P. temperata, and P. asymbiotica, respectively, after sensing the external AHLs. However, whether the AHL-lactonases and/or AHL-acylases encoding genes are under control of the SdiA-homologous LuxR solos in Photorhabdus spec. is indeed possible, but has to be investigated.
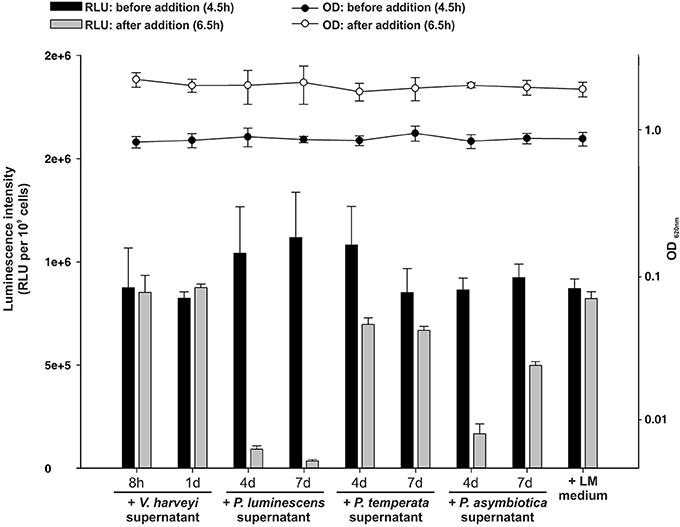
Figure 4. Quorum quenching by Photorhabdus species culture fluids. Bioluminescence of Vibrio harveyi wild-type BB120 was used as read-out to analyze degradative activity of Photorhabdus spec. supernatants. The supernatants of P. luminescens, P. temperata, and P. asymbiotica were harvested after 4 and 7 days of growth and added to the BB120 reporter strain in the mid-exponential growth phase, when bioluminescence occurs. As controls V. harveyi supernatant and LM medium, harvested after 8 h and 1 day of growth, was added. Luminescence (RLU) was measured before (black bars) and after (gray bars) the addition of the Photorhabdus spec. supernatant and optical density (OD) was measured before (black circles) and after (white circles) the addition of the Photorhabdus spec. supernatant. Error bars represent standard deviation of at least three independently performed experiments. RLU, relative light units.
Overall, the defense of the dead insect larvae, which is a rich food source, is crucial for the survival of Photorhabdus as well as the nematode partner. Thus, disturbing of the ambient mixed microbial community and its communication might be an important step for a successful infection process and reproduction.
Conclusions
LuxR solos emerge to be more and more important players in cell-cell communication or inter-kingdom-signaling as they offer possibilities using alternative communication molecules to AHLs. In all three Photorhabdus species an extraordinary high number of LuxR solos were identified, making them to optimal model organisms to study the function of LuxR solos in bacteria. Thereby, regulation via LuxR solos can be proposed to be important at different steps of the Photorhabdus life cycle and infection process (Figure 5). However, the majority of the LuxR solos in all three Photorhabdus species contain a PAS4-domain, which lends support for the theory that host sensing is highly important at different steps in the Photorhabdus life cycle. In contrast, sensing other bacteria by detecting their AHLs as well as quorum sensing seems to be only important for the pathogenic steps of the life cycle rather than for the symbiosis parts. We have seen that the LuxR solos contain different SBDs, which include diverse amino acid motifs at conserved positions. The diversity of these motifs gives rise to the speculation that signal-binding of all these LuxR solos goes far beyond AHL-signaling, as we have recently demonstrated for PluR for the first time (Brachmann et al., 2013). One can only guess the variety of signals perceived by all these LuxR solos and their function in cell-cell communication and inter-kingdom-signaling. It will be the goal of the near future research to unravel the various signaling molecules and to correlate them to the specific LuxR solos or amino acid motifs in the SBD of these proteins. The presence of all those different types of LuxR solos gains first insight into the complexity of the communication network between bacteria among each other as well as with their hosts. As most of the LuxR solos that have been investigated so far are involved in regulation of insect or plant pathogenicity, the homologous receptors or the related signaling molecules might be promising specific drug targets in human pathogens as well.
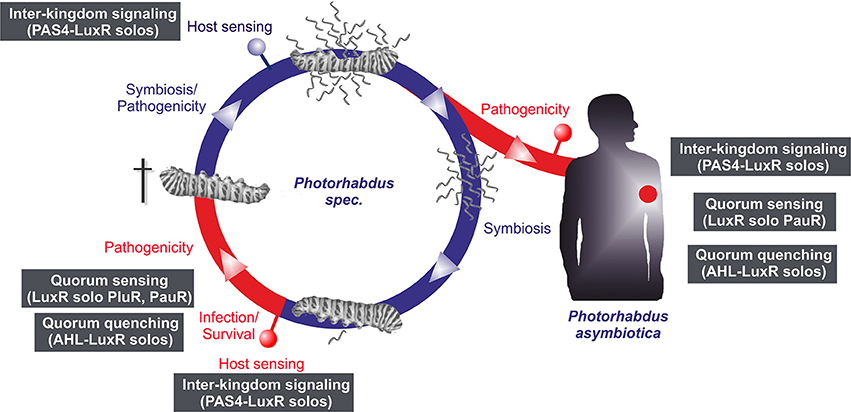
Figure 5. Life and infection cycle of Photorhabdus spec. and involvement of the different LuxR solos. Life cycle of P. luminescens, P. temperata, and P. asymbiotica (left panel). The bacteria colonize the upper gut of heterorhabditid nematodes that invade insect larvae. After release into the insect's hemolymph the bacteria produce several toxins that rapidly kill the prey. After death of the insect host, the bacteria degrade the cadaver, and additionally support nematode development. When the cadaver is depleted from nutrients, bacteria and nematode re-associate and leave the carcass in search for a new victim. The pathogenic part of the cycle is drawn in red, the symbiotic part in blue. P. asymbiotica can additionally infect humans by inducing systemic and soft tissue infections (right panel). Putative involvement of the different LuxR solos at steps of the infection process or host sensing is indicated by the gray boxes (see text for details).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work in the Heermann lab was financially supported by the Deutsche Forschungsgemeinschaft (HE 5247/4-1 and SPP 1617). Work in the Bode lab was supported by the DFG (SPP 1617). We thank Dr. Kirsten Jung and Dr. Laure Plener for providing the Vibrio harveyi strain. We are grateful to Dr. Heinrich Jung for critical reading of the manuscript.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
An, S.-Q., Allan, J. H., McCarthy, Y., Febrer, M., Dow, J. M., and Ryan, R. P. (2014). The PAS domain-containing histidine kinase RpfS is a second sensor for the diffusible signal factor of Xanthomonas campestris. Mol. Microbiol. 92, 586–597. doi: 10.1111/mmi.12577
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anetzberger, C., Reiger, M., Fekete, A., Schell, U., Stambrau, N., Plener, L., et al. (2012). Autoinducers act as biological timers in Vibrio harveyi. PLoS ONE 7:e48310. doi: 10.1371/journal.pone.0048310
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bassler, B. L., Greenberg, E. P., and Stevens, A. M. (1997). Cross-species induction of luminescence in the Quorum-Sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045.
Biswa, P., and Doble, M. (2013). Production of acylated homoserine lactone by Gram-positive bacteria isolated from marine water. FEMS Microbiol. Lett. 343, 34–41. doi: 10.1111/1574-6968.12123
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brachmann, A. O., Brameyer, S., Kresovic, D., Hitkova, I., Kopp, Y., Manske, C., et al. (2013). Pyrones as bacterial signaling molecules. Nat. Chem. Biol. 9, 573–578. doi: 10.1038/nchembio.1295
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Case, R. J., Labbate, M., and Kjelleberg, S. (2008). AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2, 345–349. doi: 10.1038/ismej.2008.13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Churchill, M. E. A., and Chen, L. (2011). Structural basis of acyl-homoserine lactone-dependent signaling. Chem. Rev. 111, 68–85. doi: 10.1021/cr1000817
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clarke, D. J. (2008). Photorhabdus: a model for the analysis of pathogenicity and mutualism. Cell. Microbiol. 10, 2159–2167. doi: 10.1111/j.1462-5822.2008.01209.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coil, D. A., Badger, J. H., Forberger, H. C., Riggs, F., Madupu, R., Fedorova, N., et al. (2014). Complete genome sequence of the extreme thermophile Dictyoglomus thermophilum H-6-12. Genome Announc. 2, 1–2. doi: 10.1128/genomeA.00109-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Covaceuszach, S., Degrassi, G., Venturi, V., and Lamba, D. (2013). Structural insights into a novel interkingdom signaling circuit by cartography of the ligand-binding sites of the homologous quorum sensing LuxR-Family. Int. J. Mol. Sci. 14, 20578–20596. doi: 10.3390/ijms141020578
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dong, Y. H., Xu, J. L., Li, X. Z., and Zhang, L. H. (2000). AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. U.S.A. 97, 3526–3531. doi: 10.1073/pnas.97.7.3526
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dubrovsky, E. B. (2005). Hormonal cross talk in insect development. Trends Endocrinol. Metab. 16, 6–11. doi: 10.1016/j.tem.2004.11.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fischer-Le Saux, M., Viallard, V., Brunel, B., Normand, P., and Boemare, N. E. (1999). Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. Iuminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov. and P. asymbiotica sp. nov. Int. J. Syst. Evol. Microbiol. 49, 1645–1656.
Franceschini, A., Szklarczyk, D., Frankild, S., Kuhn, M., Simonovic, M., Roth, A., et al. (2013). STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, 808–815. doi: 10.1093/nar/gks1094
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275.
Gerrard, J. G., McNevin, S., Alfredson, D., Forgan-Smith, R., and Fraser, N. (2003). Photorhabdus species: bioluminescent bacteria as emerging human pathogens? Emerging Infect. Dis. 9, 251–254. doi: 10.3201/eid0902.020222
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gerrard, J., Waterfield, N., Vohra, R., and ffrench-Constant, R. (2004). Human infection with Photorhabdus asymbiotica: an emerging bacterial pathogen. Microbes Infect. 6, 229–237. doi: 10.1016/j.micinf.2003.10.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
González, J. F., and Venturi, V. (2013). A novel widespread interkingdom signaling circuit. Trends Plant Sci. 18, 167–174. doi: 10.1016/j.tplants.2012.09.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haneburger, I., Eichinger, A., Skerra, A., and Jung, K. (2011). New insights into the signaling mechanism of the pH-responsive, membrane-integrated transcriptional activator CadC of Escherichia coli. J. Biol. Chem. 286, 10681–10689. doi: 10.1074/jbc.M110.196923
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Han, R. R., and Ehlers, R. U. R. (2000). Pathogenicity, development, and reproduction of Heterorhabditis bacteriophora and Steinernema carpocapsae under axenic in vivo conditions. J. Invertebr. Pathol. 75, 55–58. doi: 10.1006/jipa.1999.4900
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heermann, R., and Fuchs, T. M. (2008). Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics 9:40. doi: 10.1186/1471-2164-9-40
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hefti, M. H., Françoijs, K.-J., De Vries, S. C., Dixon, R., and Vervoort, J. (2004). The PAS fold: a redefinition of the PAS domain based upon structural prediction. Europ. J. Biochem. 271, 1198–1208. doi: 10.1111/j.1432-1033.2004.04023.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, J. J., Han, J.-I., Zhang, L.-H., and Leadbetter, J. R. (2003). Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 69, 5941–5949. doi: 10.1128/AEM.69.10.5941-5949.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, D., Thomas, P. W., Momb, J., Hoang, Q. Q., Petsko, G. A., Ringe, D., et al. (2007). Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens. Biochemistry 46, 11789–11799. doi: 10.1021/bi7012849
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Michael, B., Smith, J. N., Swift, S., Heffron, F., and Ahmer, B. M. (2001). SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183, 5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nasser, W., and Reverchon, S. (2006). New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Anal. Bioanal. Chem. 387, 381–390. doi: 10.1007/s00216-006-0702-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nealson, K. H., and Hastings, J. W. (1979). Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43, 496–518.
Oinuma, K.-I., and Greenberg, E. P. (2011). Acyl-homoserine lactone binding to and stability of the orphan Pseudomonas aeruginosa quorum-sensing signal receptor QscR. J. Bacteriol. 193, 421–428. doi: 10.1128/JB.01041-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, S. J., Park, S. Y., Ryu, C.-M., Park, S.-H., and Lee, J. K. (2008). The role of AiiA a quorum-quenching enzyme from Bacillus thuringiensis on the rhizosphere competence. J. Microbiol. Biotechnol. 18, 1518–1521.
Park, S.-Y., Kang, H.-O., Jang, H.-S., Lee, J.-K., Koo, B.-T., and Yum, D.-Y. (2005). Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 71, 2632–2641. doi: 10.1128/AEM.71.5.2632-2641.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, S.-Y., Lee, S. J., Oh, T.-K., Oh, J.-W., Koo, B.-T., Yum, D.-Y., et al. (2003). AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149, 1541–1550. doi: 10.1099/mic.0.26269-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Patankar, A. V., and González, J. E. (2009). Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 33, 739–756. doi: 10.1111/j.1574-6976.2009.00163.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Romero, M., Diggle, S. P., Heeb, S., CÃ mara, M., and Otero, A. (2008). Quorum quenching activity in Anabaenasp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol. Lett. 280, 73–80. doi: 10.1111/j.1574-6968.2007.01046.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rutherford, S. T., and Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shong, J., Huang, Y.-M., Bystroff, C., and Collins, C. H. (2013). Directed evolution of the quorum-sensing regulator EsaR for increased signal sensitivity. ACS Chem. Biol. 8, 789–795. doi: 10.1021/cb3006402
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Subramoni, S., and Venturi, V. (2009). LuxR-family “solos”: bachelor sensors/regulators of signalling molecules. Microbiology 155, 1377–1385. doi: 10.1099/mic.0.026849-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tailliez, P., Laroui, C., Ginibre, N., Paule, A., Pages, S., and Boemare, N. (2010). Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa: X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int. J. Syst. Evol. Microbiol. 60, 1921–1937. doi: 10.1099/ijs.0.014308-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, L.-H., Weng, L.-X., Dong, Y.-H., and Zhang, L.-H. (2004). Specificity and enzyme kinetics of the quorum-quenching N-Acyl homoserine lactone lactonase (AHL-lactonase). J. Biol. Chem. 279, 13645–13651. doi: 10.1074/jbc.M311194200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X., de Boer, P. A. J., and Rothfield, L. I. (1991). A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 10, 3363–3372.
Waterfield, N. R., Ciche, T., and Clarke, D. (2009). Photorhabdus and a Host of Hosts. Annu. Rev. Microbiol. 63, 557–574. doi: 10.1146/annurev.micro.091208.073507
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Microbiol. 21, 319–349. doi: 10.1146/annurev.cellbio.21.012704.131001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Welch, M., Todd, D. E., Whitehead, N. A., McGowan, S. J., Bycroft, B. W., and Salmond, P. C. (2000). N-acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19, 631–641. doi: 10.1093/emboj/19.4.631
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wilkonson, P., Waterfield, N. R., Crossman, L., Corton, C., Sanchez-Contreras, M., Vlisidou, I., et al. (2009). Comparative genomics of the emerging human pathogen Photorhabdus asymbiotica with the insect pathogen Photorhabdus luminescens. BMC Genomics 10:302. doi: 10.1186/1471-2164-10-302
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Williams, P. (2007). Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153, 3923–3938. doi: 10.1099/mic.0.2007/012856-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, H. B., Wang, L. H., and Zhang, L.-H. (2002). Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 99, 4638–4643. doi: 10.1073/pnas.022056699
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: LuxR solos, quorum sensing, cell-cell communication, quorum quenching, entomopathogenic bacteria
Citation: Brameyer S, Kresovic D, Bode HB and Heermann R (2014) LuxR solos in Photorhabdus species. Front. Cell. Infect. Microbiol. 4:166. doi: 10.3389/fcimb.2014.00166
Received: 09 September 2014; Accepted: 24 October 2014;
Published online: 18 November 2014.
Edited by:
Vittorio Venturi, International Centre for Genetic Engineering and Biotechnology, ItalyReviewed by:
Brian Ahmer, The Ohio State University, USAEric Déziel, Institut National de la Recherche Scientifique, Canada
Copyright © 2014 Brameyer, Kresovic, Bode and Heermann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Helge B. Bode, Fachbereich Biowissenschaften, Merck Stiftungsprofessur für Molekulare Biotechnologie, Goethe-Universität Frankfurt, Max-von-Laue-Str. 9, 60438 Frankfurt am Main, Germany e-mail: h.bode@bio.uni-frankfurt.de;
Ralf Heermann, Bereich Mikrobiologie, Biozentrum, Ludwig-Maximilians-Universität München, Großhaderner Str. 2-4, 82152 Martinsried/München, Germany e-mail: heermann@lmu.de
 Sophie Brameyer
Sophie Brameyer Darko Kresovic2
Darko Kresovic2  Helge B. Bode
Helge B. Bode Ralf Heermann
Ralf Heermann