Identification of potential genetic components involved in the deviant quorum-sensing signaling pathways of Burkholderia glumae through a functional genomics approach
- Department of Plant Pathology and Crop Physiology, Louisiana State University Agricultural Center, Baton Rouge, LA, USA
Burkholderia glumae is the chief causal agent for bacterial panicle blight of rice. The acyl-homoserine lactone (AHL)-mediated quorum-sensing (QS) system dependent on a pair of luxI and luxR homologs, tofI and tofR, is the primary cell-to-cell signaling mechanism determining the virulence of this bacterium. Production of toxoflavin, a major virulence factor of B. glumae, is known to be dependent on the tofI/tofR QS system. In our previous study, however, it was observed that B. glumae mutants defective in tofI or tofR produced toxoflavin if they grew on the surface of a solid medium, suggesting that alternative signaling pathways independent of tofI or tofR are activated in that growth condition for the production of toxoflavin. In this study, potential genetic components involved in the tofI- and tofR-independent signaling pathways for toxoflavin production were sought through screening random mini-Tn5 mutants of B. glumae to better understand the intercellular signaling pathways of this pathogen. Fifteen and three genes were initially identified as the potential genetic elements of the tofI- and tofR-independent pathways, respectively. Especially, the ORF (bglu_2g06320) divergently transcribed from toxJ, which encodes an orphan LuxR protein and controls toxoflavin biosynthesis, was newly identified in this study as a gene required for the tofR-independent toxoflavin production and named as toxK. Among those genes, flhD, dgcB, and wzyB were further studied to validate their functions in the tofI-independent toxoflavin production, and similar studies were also conducted with qsmR and toxK for their functions in the tofR-independent toxoflavin production. This work provides a foundation for future comprehensive studies of the intercellular signaling systems of B. glumae and other related pathogenic bacteria.
Introduction
Burkholderia glumae causes bacterial panicle blight of rice and produces major virulence factors, such as toxoflavin and lipase, under the control of the quorum sensing (QS) system mediated by the luxI homolog, tofI, and the luxR homolog, tofR (Kim et al., 2004, 2007; Devescovi et al., 2007). Kim et al. (2004) used biosensors and thin layer chromatography to determine the acyl-homoserine lactone (AHL)-type autoinducers of B. glumae and found that N-octanoyl homoserine lactone (C8-HSL) and N-hexanoyl homoserine lactone (C6-HSL) are produced by the LuxI homolog, TofI. C8-HSL is considered as the functional autoinducer binding to TofR for promoting the virulence-related phenotypes including toxoflavin production and flagellum-mediated motility, while the role of C6-HSL is still vague (Kim et al., 2004, 2007). Several important genes for the virulence of B. glumae regulated by C8-HSL and its cognate receptor TofR include: the tox gene clusters (operons) for toxoflavin biosynthesis (toxABCDE) and transport (toxFGHI) (Kim et al., 2004; Shingu and Yoneyama, 2004; Suzuki et al., 2004), lipA encoding the LipA lipase (Devescovi et al., 2007), genes for flagellar biogenesis and qsmR encoding an IclR-type transcriptional regulator (Kim et al., 2007), and katG encoding a protective catalase (Chun et al., 2009). In addition, genes for the type III secretion system (T3SS) was demonstrated to be a part of the regulon of the tofI/tofR QS system (Kang et al., 2008). As these genes under the control of the tofI/tofR QS is important for the survival, colonization and pathogenesis of B. glumae, it will be beneficial to expand the knowledge upon the intercellular signaling network involving tofI and tofR for gaining a better understanding of the pathogenic behaviors of this pathogen.
In our previous study, a series of deletion mutants of B. glumae for tofI and tofR were generated for comprehensive characterization of the tofI/tofR QS system, using a Louisiana strain of B. glumae, 336gr-1 (Chen et al., 2012). Consistent with the previous studies with mutant derivatives of the B. glumae strain BGR1, ΔtofI or ΔtofR derivatives of B. glumae 336gr-1 did not produce toxoflavin in Luria-Bertani (LB) broth (Chen et al., 2012). However, these mutants produced high levels of toxoflavin when they were grown on solid media, including Luria broth (LB) agar and King's B (KB) agar (Chen et al., 2012). These results indicate the presence of previously unknown signaling/regulatory pathways for the production of toxoflavin that are activated in certain growth conditions (e.g., solid media) in tofI- and tofR-independent manners (Chen et al., 2012). In this study, ΔtofI and ΔtofR derivatives of B. glumae 336gr-1 were randomly mutagenized with mini-Tn5 Cm and the mutants showing lost ability of toxoflavin production on LB agar were screened in an attempt to identify and characterize the tofI- and tofR-independent signaling/regulatory pathways for toxoflavin production. For more sensitive visual detection of altered toxoflavin production by each transposon mutant, a DNA construct that harbors the promoterless gusA reporter gene fusion to the promoter region of the toxoflavin biosynthesis operon toxABCDE, PtoxABCDE::gusA, was introduced into the ΔtofI and ΔtofR mutants, and the expression of the fused gusA reporter gene in each mutant was monitored based on the blue coloration of each mutant on LB agar containing the substrate of β-glucuronidase, 5-bromo-4-chloro-1H-indol-3-yl β-D-glucopyranosiduronic acid (X-gluc). Mutated genes of the primarily screened mutants, which did not show blue coloration on LB agar with X-gluc, were identified by sequencing the flanking regions of mini-Tn5 Cm inserted in the genome and the functions of selected genes were further studied in terms of their regulatory roles in toxoflavin production and virulence.
Materials and Methods
Growth Conditions of Bacterial Strains
The bacterial strains and plasmid constructs used in this study are listed in Table 1. Media used for routine cultures of bacterial strains were LB broth or LB agar (Sambrook, 2001) with appropriate amendments of antibiotics. Liquid cultures were incubated in a shaking incubator at 200 rpm at 37°C. The antibiotics and their working concentrations used in this study were: ampicillin (Amp), 100 μg/ml; kanamycin (Km), 50 μg/ml; nitrofurantoin (Nit), 100 μg/ml; and gentamycin (Gm), 20 μg/ml. The substrate of β-glucuronidase, X-gluc), was applied at a working concentration of 2 mM.
Recombinant DNA Techniques
General DNA manipulations were conducted following standard methods (Sambrook, 2001). A Strata Clone™ PCR cloning kit (Agilent Technologies, Santa Clara, CA, USA) was used for cloning PCR products into a pSC-A-amp/kan vector. Cloned PCR products in pSC-A-amp/kan were sequenced for confirmation at GeneLab in the LSU School of Veterinary Medicine or at Macrogen USA (http://www.macrogenusa.com) using M13 forward and reverse primers. A GenePulser unit (BioRad Laboratories, Hercules, CA, USA) was used to perform electroporation for transforming E. coli competent cells under 1.5 kV, with 1 μl of ligated DNA and 25 μl of competent cells. Triparental mating was used for the transformation of B. glumae with DNA constructs (Figurski and Helinski, 1979). Digested DNA fragments were extracted from the agarose gel using GenElute™ Gel extraction kits (Sigma-Aldrich, St. Louis, MO, USA). The concentrations of the purified DNA and RNA were measured using a NanoDrop DN-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). All the restriction enzymes used in this study were purchased from New England Biolabs (Beverly, MA, USA). The genomic library used in this study was constructed previously in our laboratory (Karki et al., 2012a).
Construction of pKGpToxA-GUS
To generate the DNA construct for a gusA fusion to the promoter region of toxABCDE (PtoxABCDE::gusA), the DNA fragment containing 562 bp upstream and 108 bp continuous coding sequences of toxA was amplified with the primer set, toxA PF/toxA PR (Table 2). The resultant PCR product was initially cloned into pSC-A-amp/kan, generating pSC-A-ptoxA. The cloned region was then introduced into pBB2GUS, a broad host vector containing a promoterless gusA, using the unique HindIII site to obtain pBB2GUS-ptoxA. The region containing the PtoxABCDE::gusA fusion was cut with KpnI and SpeI and ligated to the suicide vector, pKNOCK-Gm (Alexeyev, 1999), to generate pKGpToxA-GUS. This DNA construct was then introduced into the ΔtofI strain, LSUPB145, and the ΔtofR strain, LSUPB289, through tri-parental mating, generating LSUPB178 and LSUPB324, respectively.
Random Mutation of B. glumae and Screening of Random B. glumae Mutants
Mini-Tn5 Cm, a mini-Tn5 transposon carrying a Cm resistant gene (De Lorenzo et al., 1990), was introduced to LSUPB178 (a ΔtofI derivative carrying the PtoxABCDE::gusA fusion in the genome) and LSUPB324 (a ΔtofR derivative carrying the PtoxABCDE::gusA fusion in the genome) by tri-parental mating for random mutagenesis. After the tri-parental mating, mini-Tn5 Cm mutants that did not show blue coloration (indicating no β-glucuronidase gene activity) on LBGm/Cm/Nir/X−gluc plates after 2 days of incubation at 37°C were collected for identifying the genes disrupted by mini-Tn5 Cm.
Identification of Genes Disrupted by Mini-Tn5 Cm
The mutated genes of the screened mutants were identified following a previously developed method (Kwon and Ricke, 2000) with some modifications. Briefly, genomic DNA of mutant strains were extracted using a GenElute™ Bacterial Genomic DNA kit (Sigma-Aldrich, St. Louis, MO, USA). Genomic DNA (ca. 4 mg) was digested with two restriction enzymes, PstI and NlaIII, and the digested DNA fragments were ligated to the Y-shaped linker. Then, Y Linker Primer, CTGCTCGAATTCAAGCTTCT (specific to Y linker sequence), and Tn5 primer, GGCCAGATCTGATCAAGAGA (specific to transposon), were used to amplify the DNA sequences flanking where the transposon was inserted. The amplified PCR products were sequenced and the sequence data were BLAST-searched against the National Center for Biotechnology Information (NCBI) database for identification of the sequenced region.
Directional Mutation for Validation of Gene Functions
A 300—400 bp internal DNA sequence of a gene of interest was amplified using a primer set indicated in Table 2 and cloned into a pKNOCK suicide vector, pKNOCK-Km or pKNOCK-Gm (Alexeyev, 1999). Specifically, PCR products for 355 bp of qsmR, 304 bp of flhD, 402 bp of toxK, 376 bp of dgcB and 370 bp of wzyB were cloned into PSC-A-amp/kan to generate PSC-qsmR, PSC-flhD, PSC-toxK, PSC-dgcB and PSC-wzyB, respectively.KpnI and SacII sites were used to transfer the qsmR region in PSC-qsmR to pKNOCK-Km to generate pKKmqsmR. EcoRI site was used to move the flhD and toxK regions in PSC-flhD and PSC-toxK to pKNOCK-Km vector, generating pKKmflhD and pKKmtoxK, respectively. SpeI and HindIII were used to transfer the dgcB region from pSC-dgcB to pKNOCK-Gm, generating pKGmdgcB. SpeI and KpnI were used to transfer the wzyB region from pSC-wzyB to pKNOCK-Gm, generating pKGmwzyB. Each final DNA construct in a pKNOCK suicide vector was introduced into B. glumae strains by tri-parental mating. Diagnostic PCR was conducted to confirm if the internal sequence of each gene was successfully integrated to the bacterial genome through homologous recombination. Primer pairs listed in Table 2, FLHD-C1/FLHD-C2, toxK-C1/toxK-C2, qsmR-C1/qsmR-C2, DGCB-C1/DGCB-C2, and Oap-C1/Oap-C2, were used for the diagnostic PCRs to confirm the targeted mutations of flhD, toxK, qsmR, dgcB, and wzyB, respectively.
Quantification of Toxoflavin Production by B. glumae Strains Grown on LB Agar
Toxoflavin produced by B. glumae strains grown on LB agar was quantified following a previous method (Chen et al., 2012) with miscellaneous modifications. Briefly, overnight bacterial cultures were concentrated to OD600 = 100 (~1011 cfu/ml) and one loopful of each inoculum was streaked on the entire area of a LB agar plate. After 24 h incubation at 37°C, the bacterial culture was scraped off from the surface of the LB agar. Five grams of the LB agar where bacteria were grown on was then cut into small pieces with a razor blade. The chopped agar pieces were mixed with 5 ml chloroform and left to sit at room temperature for 30 min. The chloroform fraction was then transferred to a new tube and air-dried under a fume hood. The remaining materials in the dried tube were dissolved in 1 ml of 80% methanol. The OD393nm values were measured to determine the quantity of toxoflavin, using a spectrophotometer. The sample from uncultured LB agar was used to set the zero of OD393nm.
Virulence Assay with Onion Bulb Scales
Virulence tests for the mutant strains of B. glumae were conducted using onion bulbs as a surrogate host, following a previously described method (Karki et al., 2012b).
Results
Fifteen Protein-Coding Genes Were Identified as Potential Genetic Components for the Tofi-Independent Production of Toxoflavin
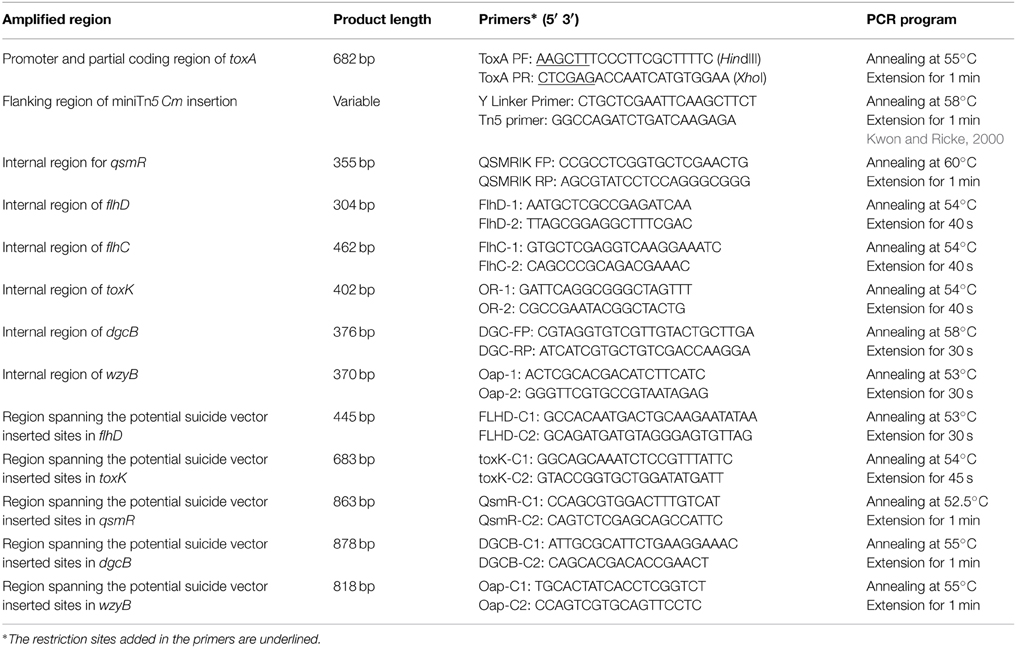
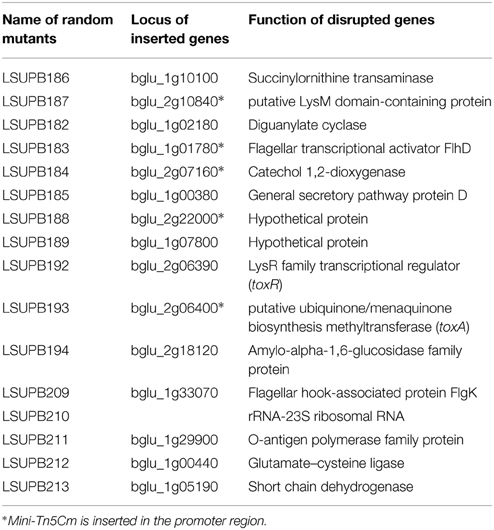
Both LSUPB178 (a ΔtofI derivative carrying the PtoxA::gusA fusion in the genome) and LSUPB324 (a ΔtofR derivative carrying the PtoxA::gusA fusion in the genome) showed blue coloration on LB agar containing X-gluc, indicating that the introduced gusA fusion to the toxA promoter is functional (data not shown). After the random mutagenesis of LSUPB178 with mini-Tn5 Cm, the mutant colonies that did not exhibit blue color were picked from the selection plates (LB agar amended with Gm, Cm, Nit, and X-gluc). It was considered that mini-Tn5 Cm is inserted in a potential genetic component of the tofI-independent toxoflavin production pathway in the mutants screened. Among the 4400 random mini-Tn5 Cm derivatives of LSUPB178 observed, 16 mutant derivatives were initially screened and their mutated genes were identified through BLAST searches of the flanking sequences (Table 3). All the 16 mutant strains screened were confirmed to be toxoflavin-deficient on LB agar at 37°C (data not shown). Fifteen out of the 16 mutants screened turned out to have mini-Tn5 Cm insertion in protein-coding regions and one mutant had the insertion in the middle of a 23S ribosomal RNA sequence. The 15 protein-coding genes identified include toxR and toxA, which are known to be required for toxoflavin production (Herrmann and Weaver, 1999), indicating the validity of this experiment (Table 3). Among the 15 genes identified, three genes (flhD encoding a transcriptional activator for flagellar biogenesis, dgcB encoding a putative diaguanylate cyclase, and wzyB encoding a putative O-antigen polymerase family protein) were selected for the extended functional studies described below.
Three Protein-Coding Genes Were Identified as Potential Genetic Components for the tofR-Independent Production of Toxoflavin
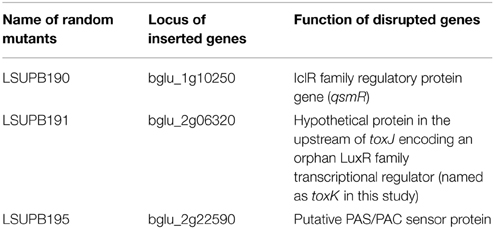
Similar to LSUPB178, LSUPB324 (ΔtofR:: PtoxABCDE−gusA) exhibited blue color on LB agar X-gluc (data not shown). Three mini-Tn5 Cm derivatives of LSUPB324 that did not produce blue color were characterized for the mutated genes (Table 4), and loss of the function in toxoflavin production was observed with all the three mutants (data not shown). Particularly, one toxoflavin-deficient mini-Tn5 Cm mutant, LSUPB191, turned out to be disrupted in the ORF, bglu_2g06320 (Table 4). This ORF is located upstream of and divergently transcribed from toxJ, a known regulatory gene for toxoflavin biosynthesis encoding an orphan LuxR protein. Due to its apparent function in toxoflavin production, the ORF was newly named as toxK in this study. Among the three identified genes, qsmR and toxK were chosen for the further functional studies described below.
Targeted Mutation of flhD, dgcB, wzyB, qsmR, or toxK Resulted in Impaired Toxoflavin Production
To confirm the initially observed functions of flhD,dgcB, and wzyB in tofI-independent toxoflavin production, independent mutants of these genes were generated via homologous recombination. For this, internal regions of flhD,dgcB, and wzyB were cloned in a suicide vector (pKNOCK-Km or pKNOCK-Gm) and introduced into the ΔtofI background, LSUPB145, generating the corresponding mutants, LSUPB503, LSUPB462, and LSUPB515, respectively (see the Materials and Methods section for detail). The same strategy was applied for the targeted mutation of qsmR and toxK in the ΔtofR background, LSUPB169, generating the corresponding mutants LSUPB277 and LSUPB445, respectively. These independent mutants of the five genes were then examined for their phenotypes in toxoflavin production in comparison with their parental strains.
flhD
Compared to LSUPB145, its flhD− derivative LSUPB503 showed a substantial reduction in toxoflavin production (Figure 1). There was little difference between LSUPB145 and LSUPB503 during the first 24 h of incubation period, but during the additional 24 h incubation period, reduction of toxoflavin amount was observed with LSUPB503, while more than 5 times increase of toxoflavin was observed with LSUPB145 during the same incubation period (Figure 1). Null mutation of flhC, which is located downstream of flhD, in the ΔtofI background through homologous recombination of an internal coding region also caused loss of toxoflavin production (data not shown).
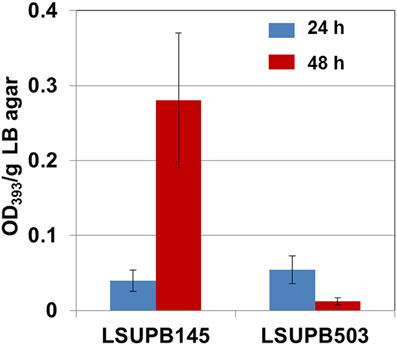
Figure 1. Toxoflavin production of LSUPB503 (flhD−/Δ tofI) compared to its parental strain, LSUPB145 (Δ tofI), grown on LB agar after 24 and 48 h of incubation at 37°C. The amounts of toxoflavin are indicated with the OD393/g LB agar values.
dgcB
Both of the two independent dgcB null mutants, LSUPB462 (a dgcB::pKGmdgcB derivative of the ΔtofI strain LSUPB145) and LSUPB182 (a dgcB::mini-Tn5 Cm derivative of LSUPB145), were toxoflavin-deficient on LB agar medium in common (data not shown). This indicates the important role of the DgcB diguanylate cyclase in the tofI-independent pathway for toxoflavin production. To know if dgcB is also critical for toxoflavin production when the tofI/tofR QS system is intact, LSUPB460 (a dgcB− derivative of the wild type strain 336gr-1) was generated and examined for toxoflavin production. As shown in Figure 2, LSUPB460 produced a similar level of toxoflavin production compared to 336gr-1. This suggests that dgcB plays a positive role in toxoflavin production but this function is dispensable in the presence of the intact tofI/tofR QS system. To explore the relationship between dgcB and other QS elements tofR and tofM (Chen et al., 2012), pKGmdgcB was introduced into LSUPB169 (a ΔtofR mutant) and LSUPB286 (a ΔtofM mutant) for generating dgcB− derivatives of these QS mutant strains. But dgcB− derivatives could be obtained from only LSUPB286 but not LSUPB169, and the dgcB−/ΔtofM strain, LSUPB464, also showed a toxoflavin-deficient phenotype like its parent, LSUPB286 (Figure 2).
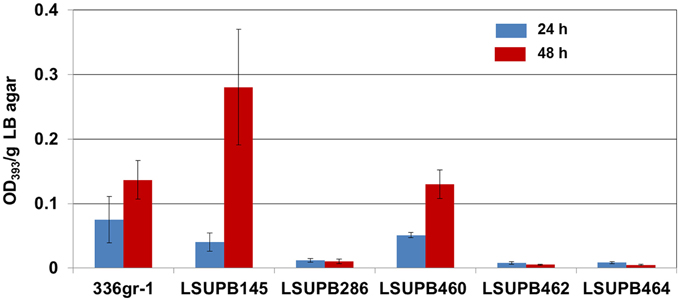
Figure 2. Toxoflavin production of the dgcB mutants, LSUPB460 (dgcB−), LSUPB462 (dgcB−/ΔtofI) and LSUPB464 (dgcB−/ΔtofM), compared to their parental strains, 336gr-1 (wild type), LSUPB145 (ΔtofI) and LSUPB286 (ΔtofM), grown on LB agar after 24 and 48 h of incubation at 37°C. The amounts of toxoflavin are indicated with the OD393/g LB agar values.
wzyB
LSUPB515, the wzyB disruptive mutant in LSUPB145 background grew very slowly on both LB agar (data not shown) and LB broth (Figure 3A). Because of the extremely slow growth of the bacterial cells, it was difficult to conduct reliable toxoflavin quantification for LSUPB515. To determine if wzyB is also required for the tofR-independent toxofalvin production, a disruptive mutant in LSUPB169 background, LSUPB516, was generated and tested for its ability to produce toxoflavin. LSUPB516 showed normal cell growth unlike LSUPB515 (Figure 3A) and a toxoflavin-deficient phenotype (Figure 3B), indicating that wzyB is required for both tofI- and tofR-independent toxoflavin production. Generation of a wzyB null mutant derivative of 336gr-1 to determine the role of this gene in the wild type background was not successful.
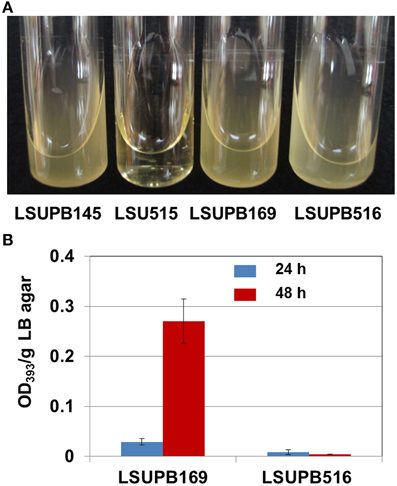
Figure 3. Bacterial growth and toxoflavin production of wzyB mutants. (A) Bacterial growth in LB broth at 37°C: LSUPB145 (ΔtofI), LSUPB515 (wzyB−/ΔtofI), LSUPB169 (ΔtofR) and LSUPB516 (wzyB−/ΔtofR). Photo was taken 48 h after inoculation. (B) Toxoflavin production of the wzyB mutant, LSUPB516 (wzyB−/ΔtofR), compared to its parental strains, LSUPB169 (ΔtofR), grown on LB agar after 24 and 48 h of incubation at 37°C. The amounts of toxoflavin are indicated with the OD393/g LB agar values.
qsmR
LSUPB277, qsmR mutated in LSUPB169 background, had the same toxoflavin loss phenotype as the random mutant LSUPB190, which confirmed the important role of qsmR in tofR-independent toxoflavin production (Figure 4). To determine the role of qsmR in toxoflavin production in the presence and absence of the intact QS system, LSUPB273 (a qsmR mutant in the wild type background 336gr-1) and LSUPB275 (a qsmR mutant in the ΔtofI background LSUPB145) were also generated and tested for their phenotypes in toxoflavin production. As shown in Figure 4, all the qsmR knock out mutants tested were deficient in toxoflavin production regardless of the genetic background of their parental strains, indicating the essential role of qsmR in the production of toxoflavin.
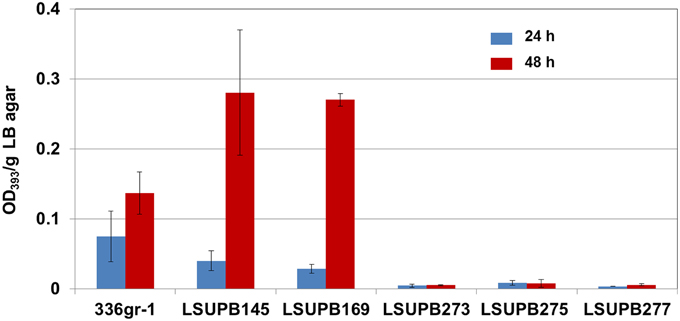
Figure 4. Toxoflavin production of the qsmR mutants, LSUPB273 (qsmR−), LSUPB275 (qsmR−/ΔtofI) and LSUPB277 (qsmR−/ΔtofR), compared to their parental strains, 336gr-1 (wild type), LSUPB145 (ΔtofI) and LSUPB169 (ΔtofR), grown on LB agar after 24 and 48 h of incubation at 37°C. The amounts of toxoflavin are indicated with the OD393/g LB agar values.
toxK
As shown in Figure 5, toxoflavin production was almost abolished in the toxK mutant in LSUPB169 background, LSUPB445, indicating that this putative gene in front of toxJ encoding an orphan LuxR protein is essential for toxoflavin production of B. glumae in the absence of tofR. The function of toxK in the wild type and ΔtofI backgrounds has not been determined due to the failure of generation of toxK− and toxK−/ΔtofI strains through homologous recombination.
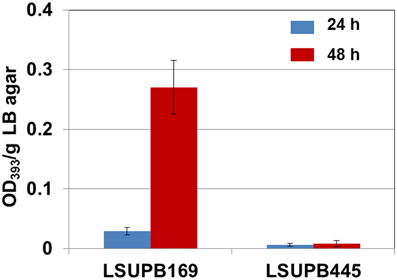
Figure 5. Toxoflavin production of the toxK mutant LSUPB445 (toxK−/ΔtofR) compared to its parental strain, LSUPB169 (ΔtofR), grown on LB agar after 24 and 48 h of incubation at 37°C. The amounts of toxoflavin are indicated with the OD393/g LB agar values.
Onion Maceration Caused by Mutants
The virulence functions of the five selected genes were determined with the virulence assay system using onion bulb scales as a surrogate host (Karki et al., 2012a,b). The strains included in the virulence assay were; 336gr-1 (wild type), LSUPB145 (ΔtofI), LSUPB169 (ΔtofR), LSUPB286 (ΔtofM), LSUPB503 (flhD− in ΔtofI), LSUPB445 (toxK− in ΔtofR), LSUPB273 (qsmR−), LSUPB275 (qsmR− in ΔtofI), LSUPB277 (qsmR− in ΔtofR), LSUPB460 (dgcB−), LSUPB462 (dgcB− in ΔtofI), LSUPB464 (dgcB− in ΔtofM), and LSUPB516 (wzyB− in ΔtofR). As shown in Figure 6, all the mutants tested exhibited reduced virulence compared to their parental strains even though none of them were completely avirulent, indicating the positive roles of the five genes in bacterial pathogenesis.
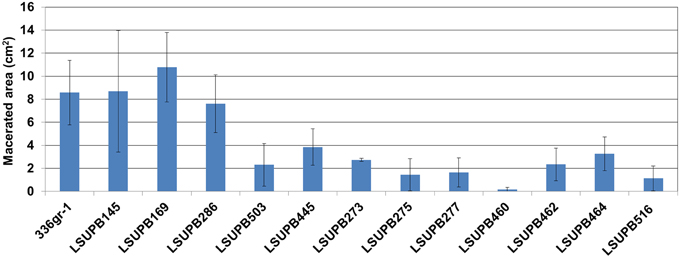
Figure 6. Virulence of Burkholderia glumae 336gr-1 and its various mutant derivatives determined by the maceration areas they produced on onion bulb scales. The two diameters of each maceration area (a and b) were measured to calculate the size of the area, using the formula: Area (cm2) = πab. Each error bar indicates the standard deviation from four replications.
Discussion
Genome-Wide Screening by Random Transposon Mutagensis Revealed Novel Regulators of Toxoflavin in B. glumae
The toxA promoter and gusA transcriptional fusion was introduced to ΔtofI and ΔtofR mutants and random mutagenesis with mini-Tn5 Cm was implemented to search for the genes that could affect the expression of the toxABCDE operon, thus the toxoflavin biosynthesis, in tofI- or tofR-independent ways. Through this approach, fifteen and three protein-coding genes were identified as potential regulators for the tofI- and tofR-independent toxoflavin biosynthesis of B. glumae 336gr-1, respectively. Identification of toxA and toxR, which are known genes required for toxoflavin biosynthesis, verifies the reliability of this experiment. The reason for the abolishment of the PtoxA::gusAexpression by disruption of toxA may be due to the fact that toxR, which is an essential regulatory factor for toxoflavin biosynthesis, requires a residual amount of toxoflavin as a co-inducer for its functionality (Kim, 2004). Among the potential genetic elements for tofI- or tofR-independent toxoflavin production identified from screening of random mini-Tn5 Cm mutants, five genes (flhD encoding a flagella transcriptional activator, dgcB encoding a diguanylate cyclase, wzyB encoding an O-antigen polymerase family protein, qsmR encoding an IclR-type transcriptional regulator, and toxK encoding a hypothetical protein and divergently transcribed from toxJ encoding an orphan LuxR homolog essential for toxoflavin production) were further characterized in this study for their functions in toxoflavin production in the presence or absence of the intact TofI/TofR QS system.
Based on the annotated genome B. glumae BGR1 (Lim et al., 2009), it is unlikely that possible polar effects from the transposon or directional mutations can cause misinterpretation of the observed mutant phenotypes for the five genes investigated in this study. flhD (bglu_1g01780) and flhC (bglu_1g01790) comprise an operon, but their products are known to work together as one functional unit (Kim et al., 2007). In this study, both flhD− and flhC− mutants showed the toxoflavin-deficient phenotype in the ΔtofI background. In case of the other genes, dgcB (bglu_1g02180), qsmR (bglu_1g10250), and toxK (bglu_2g06320) are apparently monocistronic, while wzyB (bglu_1g29900) is located at the end of a putative operon structure containing three genes. Thus, polarity may not be a major concern with the five genes investigated in this study.
Flhd is Required for the Toxoflavin Production by B. glumae 336gr-1 in the ΔtofI Background
The flhDC operon encoding the FlhDC complex was characterized as a flagellar transcriptional activator, which was controlled by QS in B. glumae BGR1 (Kim et al., 2007). It was reported that a flhD deficient mutant of BGR1 still produced toxoflavin but lost pathogenicity as well as flagellum-mediated motility (Kim et al., 2007). In this study with another strain of B. glumae, 336gr-1, it was observed that flhD as well as flhC were required for the production of toxoflavin in the absence of tofI. At this point, it has not been determined if flhD− or flhC− derivatives of 336gr-1 still produce toxoflavin like BGR1. Mutation of these genes in the wild type as well as in theΔtofR backgrounds is currently under way. Due to the significant difference between BGR1 and 336gr-1 in the QS-mediated regulation of toxoflavin, it is possible that flhDC is not involved in toxoflavin production in BGR1. Nevertheless, this study indicates that flhDC has a regulatory function in toxoflavin production in addition to flagellum-mediated motilities in at least some strains of B. glumae.
dgcB is Required for Toxoflavin Production in the Absence tofI but Not in the Wild Type Background Having the Intact tofI/tofR QS system
The results in this study indicate that dgcB is required for toxoflavin production in the absence of tofI but not in the presence of the intact TofI/TofR QS system. dgcB codes for a diguanylate cyclase, which synthesizes cyclic di-guanosine monophosphate (c-di-GMP) from two guanosine triphosphate (GTP) molecules. c-di-GMP is a small diffusible signal molecule that influences a wide range of cellular functions including bacterial virulence (Romling, 2012). The phenotype of the dgcB mutant, LSUPB462, observed in this study strongly implies that the c-di-GMP signaling plays a pivotal role in the regulation of virulence factors in B. glumae. Involvement of c-di-GMP signaling pathways for the pathogenesis by plant pathogenic bacteria has been studied mainly with Xanthomonas spp. Ham (2013). Regulatory functions of c-di-GMP for the expression of EPS and biofilm were recently reported with B. cenocepacia (Fazli et al., 2011, 2013; Deng et al., 2012). Even though this study adds to our knowledge about the function of c-di-GMP signaling in plant pathogenic bacteria, more studies including identification and functional characterization of other c-di-GMP signaling components, such as phosphodiesterases and c-di-GMP-binding protein, should be followed to clearly understand the role of this signaling system for the bacterial pathogenesis by B. glumae.
wzyB Encoding an O-Antigen Polymerase May Play an Important Role in the tofI-Independent Intercellular Signaling for Toxoflavin Production
wzyB was initially identified as a genetic element for the tofI-independent toxoflavin production through screening of random mini-Tn5 Cm mutant derivatives of the ΔtofI strain, LSUPB145. LSUPB515, an independent wzyB mutant derivative of LSUPB145 generated through homologous recombination, grew slowly due to an unknown reason and produced no observable toxoflavin. LSUPB516, a wzyB mutant derivative of the ΔtofR strain LSUPB169 through homologous recombination, also lost the ability to produce toxoflavin on a solid medium. These observations indicate that wzyB is required for both tofI- and tofR-independent toxoflavin production. However, it remains to be determined whether or not wzyB is also required for the toxoflavin production in the presence of the intact TofI/TofR QS system because wzyB mutation in the wild type background has been failed due to an unknown reason.
O-antigen is one of the three major components (the other two are lipid A and a core oligosaccharide) of lipopolysaccharides (LPS) in the outer membrane of Gram negative bacteria; and is structurally very diverse and highly immunogenic (Raetz and Whitfield, 2002; Stone et al., 2012). Study of O-antigen and its synthesis pathway can provide important antibiotic targets for potential clinical use. O-antigen polymerase family proteins (encoded by wzy homologs) are responsible for polymerizing the O-antigen units to O-antigen chain (Daniels et al., 1998; Chin et al., 2010). It is also well known that LPS is a common virulence determinant (Thomsen et al., 2003; Ellis and Kuehn, 2010). As an important synthesis enzyme for O-antigen and LPS, O-antigen polymerase should be important for maintaining the virulence of pathogenic bacteria. Even though not much study has been focused on wzy homologs, it was recently reported that non-polar mutation of the wzy gene in Salmonella enterica serovar Typhimurium caused attenuated virulence in mice (Kong et al., 2011). It would be an appealing hypothesis that the outer membrane-bound O-antigen produced by wzy genes mediates signal transduction dependent on “physical contact” of bacterial cells populated on a solid surface.
Furthermore, it would be worthy to make more investigation on the relationship between motility genes, especially flhDC, and wzyB as well as other LPS synthesis genes. There have been reports about how the subunits of and enzymes involved in assembling LPS influence the motility function of bacteria. The mutation of an O-antigen gene in Salmonella enterica serovar Typhimurium resulted in defective bacterial swarming and, in the same study, it was also suggested that the role of O-antigen is to improve the wettability of the bacterial colony (Toguchi et al., 2000). The mutation of wzxE gene, which is responsible for transporting O-antigen units across the inner membrane to the LPS assembling site (Chin et al., 2010), caused E. coli to lose both swimming and swarming motility (Girgis et al., 2007). The mutation of waaL, an O-antigen ligase gene, in Vibrio fischeri resulted in loss of motility and ability to survive with the wild type in the co-colony assay (Buttner, 2012). In another study, mutation of waaL blocked the expression of flhDC genes and aborted the swarming motility of human pathogen Proteus mirabilis in soft agar, but overexpression of flhDC genes in trans overcame this defect (Morgenstein et al., 2010). Therefore, motility assays are under way in our laboratory to determine the functional relationship between wzyB and the QS-regulated motility system in B. glumae.
qsmR is an Essential Regulatory Gene for the Toxoflavin Production of B. glumae 336gr-1
In this study, null mutation of qsmR resulted in lost function of toxoflavin production in the wild type (336gr-1), ΔtofI (LSUPB145) and ΔtofR (LSUPB169) backgrounds, indicating that qsmR is another essential regulatory factor for toxoflavin production in B. glumae 336gr-1. qsmR was previously reported as a positive regulator for flhDC, which was activated by C8-HSL and TofR complex in another strain of B. glumae, BGR1 (Kim et al., 2007). In the same study, disruption of qsmR resulted in loss of motility and pathogenicity in rice, but no observable difference in toxoflavin production in the solid medium condition (Kim et al., 2007). However, the qsmR− derivative of BGR1 showed decreased production and degradation of toxoflavin in the liquid medium condition from 12 h of incubation (Kim et al., 2007). In this study with B. glumae 336gr-1, the qsmR mutant (LSUPB273) did not produce observable toxoflavin on LB agar and still retained partial virulence on onion, which was somewhat different from the results of Kim et al. with B. glumae BGR1. These results from this and previous studies suggest that toxoflavin biosynthesis is regulated by qsmR more tightly in 336gr-1 than in BGR1. qsmR was also shown to regulate the katG catalase gene, which is required for the full virulence of B. glumae BGR1(Chun et al., 2009).
Additionally, recent studies with B. glumae BGR1 demonstrated that qsmR is a key player for both bacterial pathogenesis and survival in stressful conditions. From a proteomics search, the type II secretion system of B. glumae BGR1 encoded by gsp genes was found to be regulated by QS, and qsmR was shown to be a major positive regulator for gsp genes (Goo et al., 2010). In a study about how QS systems help representative Burkholderia species survive stationary phases, the sub-QS regulator qsmR was determined to be critical for activating the production of oxalate and maintaining normal pH levels of bacterial culture media (Goo et al., 2012). Without functional qsmR, there was a survival defect in B. glumae and B. thailandensis populations, which correlated with the high pH and lack of oxalate (Goo et al., 2012). Taken this and the previous studies on qsmR together, it is thought that qsmR plays an important role in general in the expression of various virulence factors in B. glumae despite some functional variations among different strains.
Discovery of the toxK Gene Adds More Complexity to the Known Regulatory Cascade for Toxoflavin Biosynthesis Involving the Orphan LuxR Protein, ToxJ
The ORF (gene ID of the reference sequence: bglu_2g06320) located upstream of and divergently transcribed from toxJ, the QS-dependent toxoflavin regulator encoding an orphan LuxR protein, was identified as a new genetic element required for the tofR-independent toxoflavin production of B. glumae 336gr-1 and named as toxK. There is one lux box like sequence (tof box) between toxJ and toxK, and it was previously proven that TofR and C8-HSL complex can bind on that site and activate the expression of toxJ (Kim et al., 2004). It may be the case that the tof box between toxJ and toxK promotes the expression of both genes, even though this notion should be proved experimentally. The deduced protein encoded by toxK contains 144 residues with 15.5 kDa of estimated molecular mass without an obvious homolog. Instead, the deduced protein (ID: ACR31068) has a conserved domain found in heterogeneous ribonucleoproteins (hnRNPs), implying that binding to target RNAs is its major functional mechanism.
It is interesting that the orphan lux homolog, toxJ, is next to an RNA-binding protein-coding gene instead of a luxI homolog. Regarding its location, size and predicted RNA-binding activity, toxK may be considered as a “chaperone” that modulates the expression and function of toxJ. More information about the function of toxK will be obtained when toxK mutants in the wild type and ΔtofR backgrounds are available and more phenotypic assays with toxK mutants in various genetic backgrounds are performed. In addition, future molecular studies to determine if mRNA of toxJ is a target of toxK and how toxK affect the expression of toxJ as well as other potential target genes will provide valuable insights into the functional basis of toxJ and, further, into a type of functional mechanism adopted by other LuxR solos.
In conclusion, potential genetic components involved in the deviant quorum-sensing signaling pathways of B. glumae for the production of the major virulence factor, toxoflavin, were identified in this study. In particular, the genes known for other functions, flhDC, dgcB, wzyB, and qsmR, as well as a previously unknown gene, toxK, were newly found to function in the tofI- and tofR-independent toxofalvin production via directional mutation of each gene. The information obtained from this study provides not only great insights into the complex cell-to-cell signaling network of this bacterium but a good foundation of future studies for comprehensive functional analyses of the individual genes identified in this study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the USDA NIFA (Hatch Project #: LAB93918 and LAB94203), the Louisiana State University Agricultural Center, and the Louisiana Board of Regents (the Research and Development Program: LEQSF(2008-11)-RD-A-02). RC was supported by the Economic Development Graduate Assistantship and the Flagship Graduate Assistantship from Louisiana State University Graduate School.
References
Alexeyev, M. F. (1999). The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26, 824–828.
Buttner, D. (2012). Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. doi: 10.1128/MMBR.05017-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, R., Barphagha, I. K., Karki, H. S., and Ham, J. H. (2012). Dissection of quorum-sensing genes in Burkholderia glumae reveals non-canonical regulation and the new regulatory gene tofM for toxoflavin production. PLoS ONE 7:e52150. doi: 10.1371/journal.pone.0052150
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chin, K. H., Lee, Y. C., Tu, Z. L., Chen, C. H., Tseng, Y. H., Yang, J. M., et al. (2010). The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J. Mol. Biol. 396, 646–662. doi: 10.1016/j.jmb.2009.11.076
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chun, H., Choi, O., Goo, E., Kim, N., Kim, H., Kang, Y., et al. (2009) The quorum sensing-dependent gene katG of Burkholderia glumae is important for protection from visible light. J. Bacteriol. 191, 4152–4157. doi: 10.1128/JB.00227-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Daniels, C., Vindurampulle, C., and Morona, R. (1998). Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 28, 1211–1222. doi: 10.1046/j.1365-2958.1998.00884.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
De Lorenzo, V., Herrero, M., Jakubzik, U., and Timmis, K. N. (1990). Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172, 6568–6572.
Deng, Y., Schmid, N., Wang, C., Wang, J., Pessi, G., Wu, D., et al. (2012). Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence thorugh cyclic dimeric quanosne monophosphate turnover. Proc. Natl. Acad. Sci. U.S.A. 109, 15479–15484. doi: 10.1073/pnas.1205037109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Devescovi, G., Bigirimana, J., Degrassi, G., Cabrio, L., Lipuma, J. J., Kim, J., et al. (2007). Involvement of a quorum-sensing-regulated lipase secreted by a clinical isolate of Burkholderia glumae in severe disease symptoms in rice. Appl. Environ. Microbiol. 73, 4950–4958. doi: 10.1128/AEM.00105-07
Ditta, G., Stanfield, S., Corbin, D., and Helinski, D. R. (1980). Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 77, 7347–7351. doi: 10.1073/pnas.77.12.7347
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ellis, T. N., and Kuehn, M. J. (2010). Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74, 81–94. doi: 10.1128/MMBR.00031-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fazli, M., McCarthy, Y., Givskov, M., Ryan, R. P., and Tolker-Nielsen, T. (2013). The exopolysaccharide gene cluster Bcam1330-Bcam1341 is involved in Burkholderia cenocepacia biofilm formation, and its expression is regulated by c-di-GMP and Bcam1349. Microbiol. Open 2, 105–122. doi: 10.1002/mbo3.61
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fazli, M., O'connell, A., Nilsson, M., Niehaus, K., Dow, J. M., Givskov, M., et al. (2011). The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 82, 327–341. doi: 10.1111/j.1365-2958.2011.07814.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Girgis, H. S., Liu, Y. C., Ryu, W. S., and Tavazoie, S. (2007). A comprehensive genetic characterization of bacterial motility. PLoS Genet. 3:154. doi: 10.1371/journal.pgen.0030154
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Goo, E., Kang, Y., Kim, H., and Hwang, I. (2010). Proteomic analysis of quorum sensing-dependent proteins in Burkholderia glumae. J. Proteome. Res. 9, 3184–3199. doi: 10.1021/pr100045n
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Goo, E., Majerczyk, C. D., An, J. H., Chandler, J. R., Seo, Y. S., Ham, H., et al. (2012). Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proc. Natl. Acad. Sci. U.S.A. 109, 19775–19780. doi: 10.1073/pnas.1218092109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Grant, S. G., Jessee, J., Bloom, F. R., and Hanahan, D. (1990). Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U.S.A. 87, 4645–4649. doi: 10.1073/pnas.87.12.4645
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ham, J. H. (2013) Intercellular and intracellular signalling systems that globally control the expression of virulence genes in plant pathogenic bacteria. Mol. Plant Pathol. 14, 308–322. doi: 10.1111/mpp.12005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Herrmann, K. M., and Weaver, L. M. (1999) The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 473–503. doi: 10.1146/annurev.arplant.50.1.473
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kang, Y., Kim, J., Kim, S., Kim, H., Lim, J. Y., Kim, M., et al. (2008). Proteomic analysis of the proteins regulated by HrpB from the plant pathogenic bacterium Burkholderia glumae. Proteomics 8, 106–121. doi: 10.1002/pmic.200700244
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Karki, H. S., Barphagha, I. K., and Ham, J. H. (2012a). A conserved two-component regulatory system, PidS/PidR, globally regulates pigmentation and virulence-related phenotypes of Burkholderia glumae. Mol. Plant Pathol. 13, 785–794. doi: 10.1111/j.1364-3703.2012.00787.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Karki, H. S., Shrestha, B. K., Han, J. W., Groth, D. E., Barphagha, I. K., Rush, M. C., et al. (2012b). Diversities in virulence, antifungal activity, pigmentation and DNA fingerprint among strains of Burkholderia glumae. PLoS ONE 7:e45376. doi: 10.1371/journal.pone.0045376
Kim, J., Kang, Y., Choi, O., Jeong, Y., Jeong, J. E., Lim, J. Y., et al. (2007). Regulation of polar flagellum genes is mediated by quorum sensing and FlhDC in Burkholderia glumae. Mol. Microbiol. 64, 165–179. doi: 10.1111/j.1365-2958.2007.05646.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kim, J., Kim, J. G., Kang, Y., Jang, J. Y., Jog, G. J., Lim, J. Y., et al. (2004). Quorum sensing and the LysR-type transcriptional activator toxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 54, 921–934. doi: 10.1111/j.1365-2958.2004.04338.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kong, Q. K., Yang, J. S., Liu, Q., Alamuri, P., Roland, K. L., and Curtiss, R. (2011). Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar Typhimurium. Infec. Immun. 79, 4227–4239. doi: 10.1128/IAI.05398-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M. II, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kwon, Y. M., and Ricke, S. C. (2000). Efficient amplification of multiple transposon-flanking sequences. J. Microbiol. Method 41, 195–199. doi: 10.1016/S0167-7012(00)00159-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lim, J., Lee, T. H., Nahm, B. H., Choi, Y. D., Kim, M., and Hwang, I. (2009). Complete genome sequence of Burkholderia glumae BGR1. J. Bacteriol. 191, 3758–3759. doi: 10.1128/JB.00349-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McClean, K. H., Winson, M. K., Fish, L., Taylor, A., Chhabra, S. R., Camara, M., et al. (1997). Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology, 143(Pt 12), 3703–3711. doi: 10.1099/00221287-143-12-3703
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Morgenstein, R. M., Clemmer, K. M., and Rather, P. N. (2010). Loss of the WaaL O-antigen ligase prevents surface activation of the flagellar gene cascade in Proteus mirabilis. J. Bacteriol. 192, 3213–3221. doi: 10.1128/JB.00196-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Raetz, C. R. H., and Whitfield, C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. doi: 10.1146/annurev.biochem.71.110601.135414
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Romling, U. (2012). Cyclic di-GMP, an established secondary messenger still speeding up. Environ. Microbiol. 14, 1817–1829. doi: 10.1111/j.1462-2920.2011.02617.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sambrook, J. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Press: New York, NY.
Shingu, Y., and Yoneyama, K. (2004). Essential regulator gene toxR for toxoflavin biosynthesis of Burkholderia glumae. J. Gen. Plant Pathol. 70, 108–114. doi: 10.1007/s10327-003-0101-8
Simon, R., Priefer, U., and Puhler, A. (1983). A broad host range mobilization systemfor in vitro genetic engineering: transposition mutagenesis in gram negative bacteria. Biotechnology 1, 784–791. doi: 10.1038/nbt1183-784
Stone, J. K., Mayo, M., Grasso, S. A., Ginther, J. L., Warrington, S. D., Allender, C. J., et al. (2012). Detection of Burkholderia pseudomallei O-antigen serotypes in near-neighbor species. BMC Microbiol. 12:250. doi: 10.1186/1471-2180-12-250
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Suzuki, F., Sawada, H. A., Zegami, K., and Tsuchiya, K. (2004). Molecular characterization of the tox operon involved in toxoflavin biosynthesis of Burkholderia glumae. J. Gen. Plant Pathol. 70, 97–107. doi: 10.1007/s10327-003-0096-1
Thomsen, L. E., Chadfield, M. S., Bispham, J., Wallis, T. S., Olsen, J. E., and Ingmer, H. (2003). Reduced amounts of LPS affect both stress tolerance and virulence of Salmonella enterica serovar Dublin. FEMS Microbiol. Lett. 228, 225–231. doi: 10.1016/S0378-1097(03)00762-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Toguchi, A., Siano, M., Burkart, M., and Harshey, R. M. (2000). Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182, 6308–6321. doi: 10.1128/JB.182.22.6308-6321.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: Burkholderia glumae, bacterial panicle blight of rice, bacterial grain rot of rice, quorum-sensing, toxoflavin
Citation: Chen R, Barphagha IK and Ham JH (2015) Identification of potential genetic components involved in the deviant quorum-sensing signaling pathways of Burkholderia glumae through a functional genomics approach. Front. Cell. Infect. Microbiol. 5:22. doi: 10.3389/fcimb.2015.00022
Received: 29 November 2014; Accepted: 22 February 2015;
Published: 10 March 2015.
Edited by:
Vittorio Venturi, International Centre for Genetic Engineering and Biotechnology, ItalyReviewed by:
Leo Eberl, University of Zürich, SwitzerlandShien Lu, Mississippi State University, USA
Nian Wang, University of Florida, USA
Copyright © 2015 Chen, Barphagha and Ham. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jong Hyun Ham, Department of Plant Pathology and Crop Physiology, Louisiana State University Agricultural Center, 302 Life Sciences Building, Baton Rouge, LA 70803, USA jham@agcenter.lsu.edu
 Ruoxi Chen
Ruoxi Chen Inderjit K. Barphagha
Inderjit K. Barphagha  Jong Hyun Ham
Jong Hyun Ham