Haemophilus haemolyticus Interaction with Host Cells Is Different to Nontypeable Haemophilus influenzae and Prevents NTHi Association with Epithelial Cells
- 1Wesfarmers Centre of Vaccines and Infectious Diseases, Telethon Kids Institute, The University of Western Australia, Perth, WA, Australia
- 2School of Paediatrics and Child Health, The University of Western Australia, Perth, WA, Australia
- 3Department of Paediatrics, Princess Margaret Hospital for Children, Perth, WA, Australia
- 4School of Public Health, Curtin University, Perth, WA, Australia
Nontypeable Haemophilus influenzae (NTHi) is an opportunistic pathogen that resides in the upper respiratory tract and contributes to a significant burden of respiratory related diseases in children and adults. Haemophilus haemolyticus is a respiratory tract commensal that can be misidentified as NTHi due to high levels of genetic relatedness. There are reports of invasive disease from H. haemolyticus, which further blurs the species boundary with NTHi. To investigate differences in pathogenicity between these species, we optimized an in vitro epithelial cell model to compare the interaction of 10 H. haemolyticus strains with 4 NTHi and 4 H. influenzae-like haemophili. There was inter- and intra-species variability but overall, H. haemolyticus had reduced capacity to attach to and invade nasopharyngeal and bronchoalveolar epithelial cell lines (D562 and A549) within 3 h when compared with NTHi. H. haemolyticus was cytotoxic to both cell lines at 24 h, whereas NTHi was not. Nasopharyngeal epithelium challenged with some H. haemolyticus strains released high levels of inflammatory mediators IL-6 and IL-8, whereas NTHi did not elicit an inflammatory response despite higher levels of cell association and invasion. Furthermore, peripheral blood mononuclear cells stimulated with H. haemolyticus or NTHi released similar and high levels of IL-6, IL-8, IL-10, IL-1β, and TNFα when compared with unstimulated cells but only NTHi elicited an IFNγ response. Due to the relatedness of H. haemolyticus and NTHi, we hypothesized that H. haemolyticus may compete with NTHi for colonization of the respiratory tract. We observed that in vitro pre-treatment of epithelial cells with H. haemolyticus significantly reduced NTHi attachment, suggesting interference or competition between the two species is possible and warrants further investigation. In conclusion, H. haemolyticus interacts differently with host cells compared to NTHi, with different immunostimulatory and cytotoxic properties. This study provides an in vitro model for further investigation into the pathogenesis of Haemophilus species and the foundation for exploring whether H. haemolyticus can be used to prevent NTHi disease.
Introduction
Haemophilus haemolyticus is a respiratory tract commensal that is closely related to the opportunistic pathogen nontypeable Haemophilus influenzae (NTHi). NTHi is a major cause of otitis media (OM) in children (Murphy et al., 2009b; Wiertsema et al., 2011) and exacerbations of chronic obstructive pulmonary disease (COPD) in adults (Thanavala and Lugade, 2011; Alikhan and Lee, 2014). Additionally, NTHi causes sinusitis, conjunctivitis, pneumonia, bacteraemia and meningitis (Shann et al., 1984; Dworkin et al., 2007; Cripps, 2010; Laupland et al., 2011; van Wessel et al., 2011). The burden of invasive disease due to NTHi is steadily increasing, particularly in infants and the elderly (Laupland et al., 2011). There is added concern due to the emergence of antibiotic resistance within the species (Van Eldere et al., 2014). Although H. haemolyticus is generally considered a commensal, there are occasional reports of isolation of this bacterium from normally sterile sites (Anderson et al., 2012; Morton et al., 2012; Hariadi et al., 2015). The distinction of H. haemolyticus as a true commensal is complicated by the fact that H. haemolyticus can be misidentified as NTHi using routine microbiological tests (reviewed in Pickering et al., 2014b). Whilst in-depth investigations into the genetics of H. haemolyticus and NTHi have been conducted in order to develop discriminatory tests to distinguish these closely related species (McCrea et al., 2008, 2010a; Sandstedt et al., 2008; Norskov-Lauritsen, 2011; Binks et al., 2012; Pickering et al., 2014b), the interaction of H. haemolyticus with host cells has not been previously investigated.
Colonization of the human upper respiratory tract with NTHi precedes infection and studies have shown an association between a high density of NTHi carriage and subsequent development of OM (Smith-Vaughan et al., 2006, 2013). Although the progression from NTHi colonization to disease is not entirely understood, in vitro and in vivo studies have revealed that NTHi can persist in biofilms and/or intracellularly within the respiratory tract (Murphy et al., 2009a; Clementi and Murphy, 2011; Novotny et al., 2013; Jalalvand and Riesbeck, 2014). This makes NTHi respiratory infections such as OM difficult to treat with antibiotics (Dagan, 2000). Current preventative strategies against development of NTHi disease include the licensed pneumococcal conjugate vaccine (PHID-CV) that incorporates the NTHi lipoprotein Protein D (Prymula and Schuerman, 2009). PHiD-CV does not prevent NTHi colonization (van den Bergh et al., 2013; Hammitt et al., 2014; Feazel et al., 2015) and the impact of this vaccine on NTHi OM has been variable (Prymula et al., 2011; van den Bergh et al., 2013; Tregnaghi et al., 2014; Leach et al., 2015). Additionally, clinical Protein D-negative NTHi strains have been identified (Smith-Vaughan et al., 2014) highlighting the potential for PHID-CV evasion by NTHi. Indeed, sub-unit vaccines against NTHi are limited given the considerable antigenic variation within NTHi (Cripps et al., 2002; Price et al., 2015). A Cochrane review of six clinical trials with an oral whole-cell killed NTHi vaccine developed to reduce acute exacerbations in COPD patients revealed mixed results and studies that did show a benefit to COPD patients had too small numbers to warrant widespread vaccination (Teo et al., 2014).
An alternative approach to vaccination is the use of probiotic therapies with the potential to modulate the host microbiome and prevent infection. Data from trials of probiotics to prevent OM are conflicting with some studies showing protection (Skovbjerg et al., 2009; Lehtoranta et al., 2012; Di Pierro et al., 2014) and others demonstrating a lesser or no impact (Taipale et al., 2011; Cohen et al., 2013). To our knowledge, no studies have directly addressed the prevention of NTHi colonization with a commensal or probiotic species. We propose that H. haemolyticus may interfere with NTHi colonization of the respiratory tract and could therefore be exploited as a probiotic to reduce the burden of NTHi disease. This is not only due to the fact that both species colonize the respiratory tract and are highly related at a genomic level (Price et al., 2015) but also that H. haemolyticus is less often isolated from populations with high rates of NTHi carriage and disease (Fenger et al., 2012; Pickering et al., 2014c; Aho et al., unpublished). In order to explore the potential therapeutic use of H. haemolyticus, investigations into the interaction of H. haemolyticus with the host were required.
In this study, we used an established in vitro model of NTHi infection (Swords et al., 2000) to characterize H. haemolyticus association and invasion of human respiratory epithelial cells. As H. haemolyticus is generally considered to be a commensal, we hypothesized that nasopharyngeal H. haemolyticus isolates would be able to colonize host epithelial cells but not necessarily invade them like NTHi. To take into consideration the high level of strain variation, we compared host cell association and invasion of 10 H. haemolyticus isolates (including four naturally occurring Protein D-negative isolates and an invasive H. haemolyticus isolate) with four NTHi strains. We also assessed four representative isolates from a clade of “fuzzy” haemophili that have recently been differentiated from NTHi and H. haemolyticus by whole genome sequencing and are referred to as “H. influenzae-like haemophili” (Price et al., 2015). Inflammatory responses from epithelial cells challenged with these strains were evaluated and further assessed using peripheral blood mononuclear cells (PBMCs). Finally, we investigated whether pre-treatment of epithelial cells with H. haemolyticus could block NTHi attachment to cells to determine whether H. haemolyticus can inhibit NTHi colonization. This is the first study to assess in vitro interactions of H. haemolyticus with host cells. This study provides in vitro models to further investigate the pathogenesis of haemophili, the biological role of H. haemolyticus, and to determine whether H. haemolyticus may be used to prevent NTHi colonization and therefore disease.
Materials and Methods
Microbial Strains and Growth Conditions
The isolates assessed in this study were first described in four previous studies (Bakaletz et al., 1988; Nizet et al., 1996; Wiertsema et al., 2011; Morton et al., 2012) and are detailed in Table 1. All isolates were X and V factor dependent and were identified as either NTHi, H. haemolyticus or H. influenzae-like haemophili by hpd#3 PCR (Wang et al., 2011), HRM hpd PCR (Pickering et al., 2014a), fucP PCR, or whole genome sequencing (Price et al., 2015).
Bacteria were cultured from glycerol stocks, or resuspended lyophilised pellets, onto chocolate agar plates and incubated overnight at 37°C in 5% CO2. Inoculum for all subsequent experiments was harvested from agar plates as previously described for NTHi (Swords et al., 2000). Bacterial viability was measured as previously described (Kirkham et al., 2013).
Epithelial Cell Culture and Viability
All cell culture reagents were from Gibco, Life Technologies, Tullamarine, Victoria, Australia, unless otherwise stated. Cell cultures were maintained in 125 cm3 flasks (BD Biosciences, Macquarie Park, NSW, Australia) at 37°C, 5% CO2.Immortalized Detroit 562 human pharyngeal carcinoma epithelial cells (ATCC® CCL-138) were cultured in Minimal Essential Media (MEM) with Earle's salts and supplemented with 10% heat inactivated fetal calf serum (Sigma-Aldrich), 1 mM sodium pyruvate, 2 mM L-glutamine, 1 × non-essential amino acids, and 100 U Penicillin/Streptomycin. Cells were weaned from antibiotics 24 h prior to suspension in 24-well tissue culture plates (Corning, Life Technologies). Immortalized A549 human lung carcinoma epithelial cells (ATCC® CCL-185) were cultured in RPMI 1640 (Invitrogen, Life Technologies) supplemented with 10% heat inactivated fetal calf serum. At confluence, flasks of each cell line were disrupted with 0.05% trypsin/EDTA, washed in 1 × PBS and resuspended in their respective culture media. Cells were counted with a haemocytometer and viability was determined by trypan blue staining. Then cells were seeded into 24-well plates at 1 × 105/mL and grown to confluence for bacterial challenge.
Bacterial Association and Invasion of Respiratory Epithelial Cell Lines
A standard gentamicin invasion assay was used to determine the ability of H. haemolyticus and NTHi to attach to and invade epithelial cells (Swords et al., 2000), with the following modifications. Monolayers of cells were challenged at a multiplicity of infection (MOI) of 10:1 bacteria:cells for 1, 3, or 24 h. At the 1 h gentamicin treatment step for evaluation of intracellular bacteria, media only was added to the cells being evaluated for total bacterial association for 1 h. 100 μg/mL of gentamicin was used instead of 50 μg/mL. All isolates were checked to be sensitive to 100 μg/mL gentamicin and resistant to 2% saponin as determined by viable count. Assays were conducted in triplicate and on at least three separate occasions. Supernatants from each triplicate well were pooled after bacterial challenge, filter-sterilized and stored at −80°C for measurement of inflammatory mediators.
PBMC Stimulation with Bacteria
Adult PBMCs were collected from five healthy donors and processed and prepared as previously described (Kirkham et al., 2013). PBMCs were stimulated with thawed preparations of either NTHi86-028NP or H. haemolyticus33390 at 10:1 bacteria:cells. Cells in control wells were treated with either PBS (cells only; unstimulated), 1 ng/mL lipopolysaccharide (LPS, from Escherichia coli R515; Alexis Biochemicals, Sapphire Biosciences, NSW, Australia) or 1 μg/mL Staphylococcal enterotoxin B (SEB, from Staphylococcus aureus; Sigma Aldrich). At 24 h post-treatment, plates were centrifuged for 5 min at 200 g and supernatants were harvested. Triplicate wells were combined and stored at -80°C for measurement of inflammatory mediators.
Cytokine and Chemokine Analysis
IL-6, IL-10, IFNγ, and TNFα levels (pg/mL) were measured in filter-sterilized supernatants from epithelial and PBMC cultures using the Bioplex 200 system (Bio-Rad Laboratories Inc., Hercules, CA, USA) and an in-house multiplex bead-based assay as previously described (Blyth et al., 2011). Measurement of IL-1β and IL-8 was added into this Bioplex assay using commercially available antibody pairs (Bioscientific, Kirrawee, NSW, Australia) and following validation to ensure that there was no cross reactivity with any of the other cytokines.
Staining and Flow Cytometry
As NTHi is difficult to distinguish from H. haemolyticus by standard microbiological culture, we used the fluorescent dye Cell Trace Violet (CTV, excitation = 390 nm, emission = 445 nm; Invitrogen) to label NTHi86-028NP for experiments involving bacterial co-culture. Overnight growth of bacteria was harvested from chocolate agar plates and resuspended in cell culture media to an OD600nm 0.2 (which is equivalent to ~108 CFU/mL). Bacterial suspensions were incubated with 20 μM CTV for 5 min before pelleting at maximum speed in a benchtop centrifuge for 7 min. Harvested bacteria were resuspended in 10 mL of pre-warmed epithelial cell culture media, protected from light and incubated at 37°C for 10 min before use. The CTV stain did not affect the ability of NTHi86-028NP to attach to or invade A549 cells (data not shown). Flow cytometry was conducted on a Flow and cell cytometer Canto II device (BD Biosciences) that was calibrated using Cytometer Setup and Tracking beads (BD Biosciences). Forward scatter and side scatter were set to capture bacterial populations. Flow cytometry data was acquired using FACSDiva software and then analyzed using Flow Jo v7 software (FloJo LLC, Oregon, USA).
Competitive Colonization Assay with Detroit 562 Cells
The optimal challenge dose of ~1 × 107 CFU/mL for the association and invasion experiments was not possible for the competitive colonization experiment due to the high background fluorescence of unstained bacteria. We therefore used a higher challenge dose of 1 × 109 CFU/mL CTV-NTHi (MOI 1000:1). NTHi86-028NP was prepared and stained as above however the bacterial suspension was concentrated to 1 × 109 CFU/mL by harvesting 100 mL and resuspending in 10 mL prior to addition to epithelial cells. Overnight growth of H. haemolyticus33390 was harvested from chocolate agar plates and resuspended in cell culture media to an OD600 nm 0.2 (which is equivalent to ~108 CFU/mL). D562 monolayers were treated with either 10-fold increasing doses (1 × 103 to 1 × 107 CFU/mL) of H. haemolyticus33390 or media alone for 1 h at 37°C, 5% CO2. Cells were then washed three times with PBS to remove unbound H. haemolyticus and challenged with 1 × 109 CFU/mL CTV-labeled NTHi86-028NP in culture media for 1 h. Two control wells were treated with 1 × 109 CFU/mL unstained NTHi to give background fluorescence levels. All wells were washed three times with 200 μL PBS to remove unbound NTHi and then 200 μL of PBS was aliquoted into each well. Each plate was then read immediately with an Enspire plate reader (Perkin Elmer, Massachusetts, USA).
Transwell Assay with A549 Cells
High density 3 μM pore Transwell inserts (BD Biosciences) were seeded with 1 × 104 A549 cells and cultured until confluent. As the high density inserts are opaque, it was not possible to gauge confluence of the cells on these inserts with a microscope. Therefore, two transparent low density 3 μM pore inserts were used to gauge confluence of A549 monolayers on the high density inserts. Transepithelial electrical resistance (TER) measurements were variable with A549 cells, therefore fluorescently conjugated dextrans RITC (Rhodamine B Isothiocyanate conjugated to 70 kDa dextran) and FITC (Fluoroscein Isothiocyanate conjugated to 3 kDa dextran; both Sigma) were used to determine the permeability of confluent epithelial cells seeded onto Transwells, as previously described (Kowapradit et al., 2010). The concentration of each fluorescent tracer was measured by the Enspire multi plate reader (FITC excitation = 485 nm, emission = 544 nm, RITC excitation = 520 nm, emission = 590 nm) using standard curves of known concentrations of RITC and FITC. Epithelial monolayers that were deemed confluent by microscopy were confirmed to be exclusive of passive bacterial transfer by the presence of 3 kDa but not 70 kDa fluorescent tracers in the lower compartment of the Transwell cultures.
When the low density inserts were deemed confluent with A549 cells, cells were counted from these two inserts and the average cell count was used to determine the bacterial challenge dose required for an MOI of 10:1. Media was removed from the inner chamber of the Transwell inserts and replaced with either media alone or media containing H. haemolyticus (MOI 10:1), prepared as described above for the bacterial association experiments. Treated cells were incubated at 37°C, 5% CO2 for 1 h. Inserts were then washed thoroughly with PBS, and CTV-stained NTHi (MOI of 10:1) was added to the inner chambers. Following a 3 h incubation at 37°C, 5% CO2, 100 μL of media from above and 100 μL of media from below the Transwells was removed for FACs analysis.
Statistical Analyses
Statistical analyses were conducted using GraphPad Prism version 5.02 (GraphPad Inc., California, USA) and p < 0.05 was considered statistically significant. Student's t-tests were used to compare log transformed bacterial viability and Mann-Whitney U-tests were used to compare epithelial viability and bacterial association and invasion of individual isolates with reference strains. Kruskal-Wallis test with Dunn's post-test analysis was used to compare intra-species association and invasion of epithelial cells, epithelial cytokine release and PBMC cytokine release. Student's t-tests were used to determine differences in bacterial fluorescence following different pre-treatments in competitive colonization assays.
Results
H. haemolyticus Viability in Cell Culture Media Diminished Significantly More than NTHi
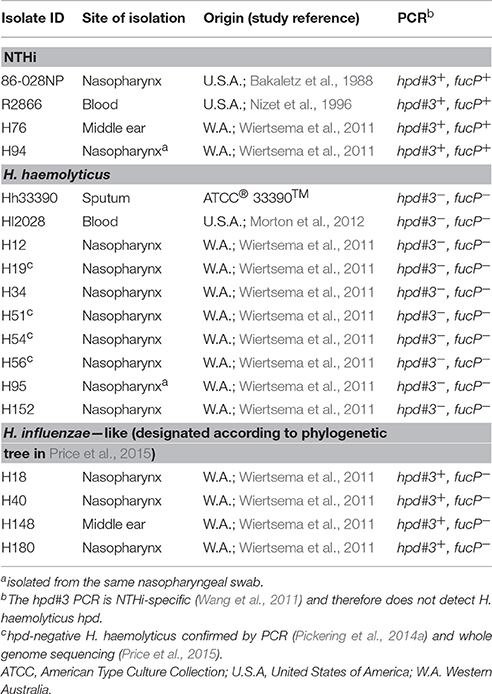
The viability of reference strains H. haemolyticus33390 and NTHi86-028NP remained relatively stable in D562 and A549 media for the first 3 h of incubation. However, by 24 h the viability of H. haemolyticus33390 dropped significantly in D562 media (Figure 1A, p < 0.001) and A549 media (Figure 1B, p < 0.05) when compared to NTHi viability at 24 h.
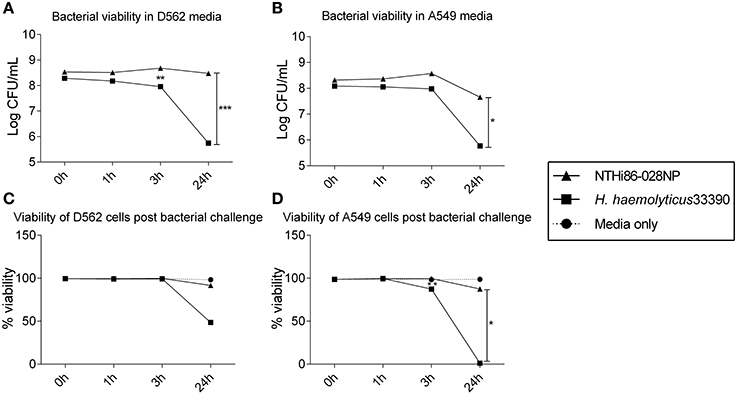
Figure 1. Viability of respiratory epithelial cells co-cultured with H. haemolyticus and NTHi. Viability of NTHi86-028NP and H. haemolyticus33390 in D562 (A) and A549 (B) cell culture media (no epithelial cells) over 24 h as determined by viable count.Viability of D562 (C) and A549 (D) respiratory epithelial cells incubated with media only (circles), H. haemolyticus33390 (squares), or NTHi86-028NP (triangles) over 24 h as determined by trypan blue counts. Student's t-tests were used to compare log transformed bacterial viability and Mann Whitney U-tests were used to compare post bacterial challenge epithelial cell viability, where *p < 0.05, **p < 0.01, ***p < 0.001.
H. haemolyticus was Cytotoxic, Particularly to A549 Epithelial Cells
H. haemolyticus33390 challenge had no impact on D562 cell viability at 3 h, however the epithelial viability reduced to 50% by 24 h (Figure 1C). This cell death was even more pronounced with A549 cells, where viability following H. haemolyticus33390 challenge decreased from ~85% at 3 h to complete cell death at 24 h (Figure 1D). In contrast, NTHi had minimal impact on the viability of both cell lines over the same time period. Several H. haemolyticus isolates were also cytotoxic at 24 h (data not shown). In consideration of the viability results, a 3 h time point and D562 cells were chosen for further analysis of H. haemolyticus host cell association, invasion, and inflammatory properties.
H. haemolyticus Associated with Epithelial Cells but Not As Well As NTHi
There was significant intra-species variability in the capacity of each haemophilus group to attach to and invade D562 cells (Figures 2A,B; p < 0.01). However, overall the NTHi isolates displayed a greater ability to associate with and invade D562 epithelia (range: association 76–122%, invasion 44–79%) compared with H. haemolyticus isolates (range: association 48–93%, invasion 0–70%) and H. influenzae-like isolates (range: association 49–101%, invasion 0–62%) (Figures 2A,B). The invasive H. haemolyticus isolate HI2028 and invasive H. influenzae-like middle ear isolate H148 had high levels of attachment (96 ± 1% and 99 ± 1%, respectively, Figure 2A) and invasion (58 ± 1% and 58 ± 1%, respectively, Figure 2B) that were comparable to the NTHi strains and significantly higher than the H. haemolyticus33390 reference strain (p < 0.01). However, this was not specific to the invasive H. haemolyticus strain as there were also carriage isolates of H. haemolyticus (H12, H54, H95) that had attachment and invasion properties similar to NTHi.
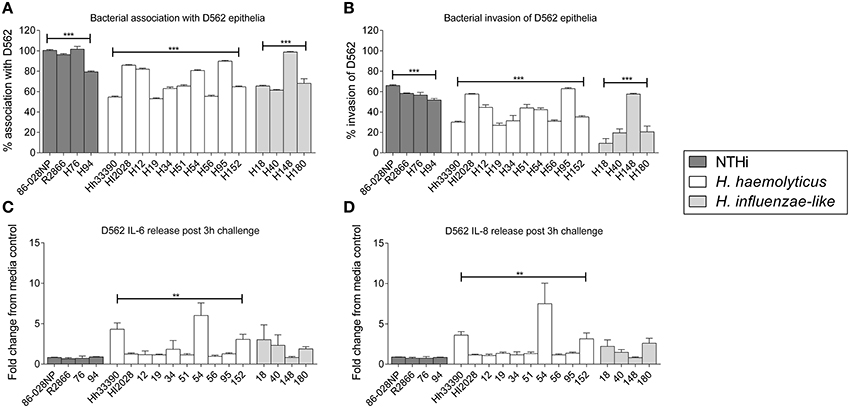
Figure 2. Interactions of clinical strains of haemophili with respiratory epithelia. Association (A) and invasion (B) of D562 epithelial monolayers with haemophilus reference strains and clinical isolates at 3 h. Level of IL-6 (C) and IL-8 (D) present in cell culture supernatant at 3 h post-bacterial challenge and presented as the fold change of cytokine in the supernatant from cells incubated with media only. Results are represented as the mean ± SEM of three separate experiments, each conducted in triplicate excluding R2866 which was conducted once in triplicate. Kruskal-Wallis and Dunn's post-test was used to compare intra-species variability where **p < 0.01; ***p < 0.001.
Protein D is Not Essential for H. haemolyticus Association with Host Cells
The absence of the protein D (hpd) gene in four clinical H. haemolyticus isolates (H19, H51, H54, and H56) was confirmed by whole genome sequencing (Price et al., 2015). Based on previous NTHi data, we hypothesized that the H. haemolyticus hpd-negative isolates would have significantly reduced association with host cells. This was not observed, with the four strains displaying significant variability in attachment (p < 0.01) and invasion (p < 0.01), Figures 2A,B. Compared with the H. haemolyticus33390 reference strain, two of the hpd-negative H. haemolyticus isolates (H19, H56) had similar association properties (p = 0.75 and p = 0.44, respectively) and 2 (H51, H54) had significantly higher association properties (p < 0.01 and p < 0.01, respectively).
Some H. haemolyticus Isolates Induced High Pro-Inflammatory Responses from Epithelial Cells whereas NTHi Did Not
There was significant variation in epithelial release of IL6 (p < 0.01, Figure 2C) and IL-8 (p < 0.01, Figure 2D) in response to challenge with the 10 H. haemolyticus isolates investigated. In contrast, there was negligible variability in the IL-6 and IL-8 cellular responses to the different NTHi strains (p = 0.35 and p = 0.77, respectively). The D562 IL-6 and IL-8 responses to NTHi challenge were lower than those produced by unstimulated D562 cells. The invasive H. haemolyticus HI2028 isolate behaved like NTHi in terms of eliciting low levels of IL-6 and IL-8 compared with other H. haemolyticus strains but this was not unique to the invasive strain. Similarly, the H. influenzae-like middle ear isolate H148 lacked inflammatory properties compared to the other H. influenzae-like strains, which matched the increased association and invasion profile for the H148 strain. Strain H54, an hpd-negative isolate, was the most immunostimulatory of all strains assessed, but this high IL-6 and IL-8 response was not observed for the other hpd-negative isolates (H19, H51, and H56). IFNγ, TNFα, IL-10, and IL-1β measurements were below the limit of detection in culture supernatant at the 3 h time-point (data not shown).
NTHi-Treatment of PBMCs Elicited IFNγ Production whereas H. haemolyticus-Treatment Did Not
Extracellular levels of IL-6, IL-10, TNFα, and IL-1β were significantly increased following 24 h stimulation of PBMCs with either NTHi86-028NP (p < 0.01) or H. haemolyticus33390 (p < 0.05) when compared to unstimulated cells (Figure 3). H. haemolyticus and LPS induced significant release of IL-8 from PBMCs compared with unstimulated cells (p < 0.01 and p < 0.05, respectively). IL-8 production from PBMCs was also elevated in response to NTHi but this was not significant. In contrast, NTHi and SEB induced significant release of IFNγ from PBMCs (p < 0.01 and p < 0.05, respectively) whereas H. haemolyticus did not. There was minimal IFNγ released from PBMCs treated with H. haemolyticus (mean 28 ± 10 pg/mL) compared with NTHi (mean 1718 ± 560 pg/mL).
Figure 3. Inflammatory mediator release (pg/mL) from PBMCs at 24 h post-treatment with NTHi or H. haemolyticus. Concentration of inflammatory mediators IL-6 (A), IL-8 (B), IL-10 (C), TNFα (D), INFγ (E), and IL-1β (F) following treatment of adult PBMCs with either media alone (unstim), lipopolysaccharide (LPS), Staphylococcal enterotoxin B (SEB), NTHi86-028NP, or H. haemolyticus33390. The horizontal bars depict the median analyte level for each treatment group (n = 5). *p < 0.05, **p < 0.01; ***p < 0.001 when compared with unstimulated cells using a Kruskal-Wallis and Dunn's post-test.
Pre-Treatment of Epithelial Cells with H. haemolyticus Inhibited NTHi Association and Transcytosis
Incubation of D562 cells for 1 h with increasing doses of H. haemolyticus33390 resulted in a significant reduction in the subsequent association of CTV-labeled NTHi86-028NP (Figure 4A). The greatest reduction of NTHi association was observed when D562 cells were pre-incubated with the lowest H. haemolyticus dose (MOI 1:1, p < 0.0001).
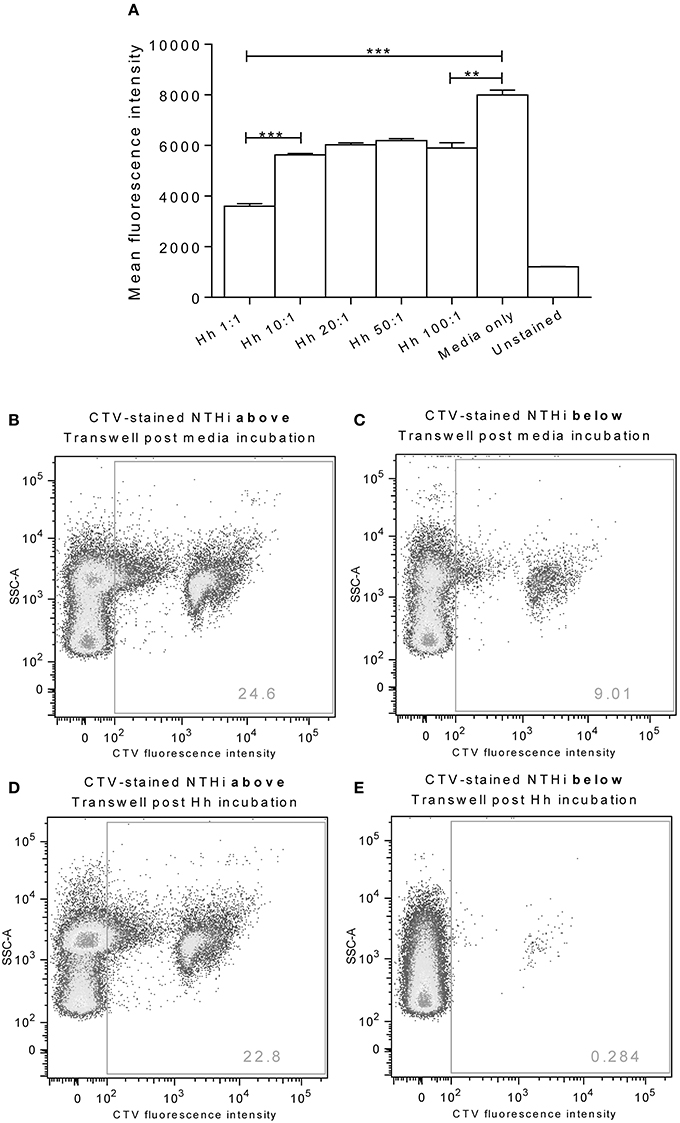
Figure 4. Pre-colonization of epithelia with H. haemolyticus impacts subsequent NTHi interactions. Monolayers of D562 cells were challenged with increasing doses of H. haemolyticus33390 or media for 1 h, washed thoroughly and then challenged with 1000:1 CTV-stained NTHi86-028NP (A). At 1 h post NTHi-infection, wells were thoroughly washed, and fluorescence was measured to identify attached NTHi. Results are represented as mean ± SEM of three technical repeats. Student's unpaired t-test was used to compare between treatments where **p < 0.01, ***p < 0.001. A549 monolayers grown on Transwells were incubated with media (B,C) or 10:1 H. haemolyticus33390 (D,E) for 1 h. Monolayers were then washed with media, challenged with CTV-stained NTHi (MOI 10:1) and incubated at 37°C 5% CO2 for a further 3 h. Pre-colonization with H. haemolyticus (MOI 10:1) reduced the proportion of CTV-stained NTHi detected in the lower compartment of the Transwell (E, 0.3%) compared with pre-incubation with media alone (C, 9.0%).
A Transwell chamber assay was used to examine the ability of H. haemolyticus to block NTHi transcytosis through a monolayer of A549 epithelial cells (Figures 4B–E). We observed that pre-treatment of Transwell cultured A549 cells with H. haemolyticus33390 (MOI 10:1, ~1 × 107 CFU/mL) reduced NTHi transcytosis by 30-fold (Figure 4E) when compared with cells treated with NTHi alone (Figure 4C); 0.3 vs. 9% fluorescence.
Discussion
To our knowledge, this is the first study to assess interactions of H. haemolyticus with host cells. We have developed in vitro models that can be used to further investigate the interaction of H. haemolyticus with the host. The loss in H. haemolyticus viability in cell culture media over 24 h indicates bacterial death but the mechanisms behind this (i.e., autolysis or allolysis) have not been determined. We observed that, in contrast to NTHi, H. haemolyticus is cytotoxic to immortalized nasopharyngeal and bronchoalveolar epithelial cell lines over a 24 h incubation period. This was surprising given the predominantly commensal nature of H. haemolyticus. As H. haemolyticus does not grow well in liquid media, it is possible that bacterial death results in release of cytotoxic components over 24 h. Indeed, filtered cell culture media from some H. haemolyticus strains incubated for 24 h with D562 cells was cytotoxic to the immortalized epithelial cell lines (data not shown), suggesting that a soluble cytotoxic component is released into the cell culture media. It is possible that some H. haemolyticus strains express a toxin that is not expressed by NTHi. Indeed, H. ducreyi is known to secrete cytolethal distending toxin, which arrests the epithelial cell cycle (Gargi et al., 2013). Comparative genomics has revealed putative H. haemolyticus toxins that are under further investigation in our laboratory. It is also possible that our method for preparing inoculum (harvesting bacteria from agar plates rather than liquid culture) results in a high proportion of dead bacteria that are not accounted for by viability counting. These dead bacteria may lead to an increase in toxic load to the epithelial cells, possibly from lipooligosaccharide (LOS) or other toxins. However, the H. haemolyticus and NTHi inocula were prepared in the same manner and the NTHi preparation was not cytotoxic. Differences in the phosphorylcholine (ChoP) decoration of LOS between H. haemolyticus and NTHi have been identified and have been suggested to provide an explanation for NTHi pathogenesis through promotion of bacterial adherence and invasion of host cells (McCrea et al., 2010b; Post et al., 2016). Detailed comparison of LOS structures from nine H. haemolyticus and six NTHi strains found that only 4/9 (44%) of the H. haemolyticus strains tested expressed ChoP decorated LOS in comparison with 5/6 of NTHi strains (Post et al., 2016). Interestingly, we found that a similar proportion of the H. haemolyticus strains in our study (4/10) had better attachment to epithelial cells that was similar to the NTHi strains (>80% association of challenge inoculum; Figure 1). Whether ChoP decoration of LOS facilitates H. haemolyticus association with epithelial cells remains to be determined. It is important to understand the basis of the observed cytotoxicity and intra-species variation if H. haemolyticus is to be further assessed as a potential probiotic. Identification of virulence genes and recombination hotspots in H. haemolyticus will highlight regions that will require attenuation or natural selection.
Our hypothesis was that H. haemolyticus would not associate with and/or invade epithelial cells in the same capacity as NTHi. All 10 H. haemolyticus strains investigated were able to infect cells within 3 h (with most association and invasion occurring in the first hour). Although H. haemolyticus was hypothesized to have reduced cell association and invasion overall compared with NTHi, there was considerable intra-species variation. Such strain to strain variation was expected, given how closely related H. haemolyticus is to the genetically heterologous NTHi (Erwin and Smith, 2007; Price et al., 2015) and the ability for recombination between these species (Sondergaard et al., 2014). The greatest difference between H. haemolyticus and NTHi attachment and invasion was evident between the reference strains, highlighting the necessity for assessing multiple strains within a species to gain an understanding of strain diversity.
Protein D, the NTHi vaccine antigen in PHiD-CV, has been demonstrated to be important for adherence of NTHi to airway epithelial cells (Johnson et al., 2011). In contrast, Protein D does not appear to be required for H. haemolyticus association with host cells, with the four different Protein D-negative strains exhibiting higher or equal attachment to and invasion of cells than Protein D-positive strains. A protein D knockout in H. haemolyticus would need to be constructed and compared with the parent strain to confirm that Protein D is not essential for H. haemolyticus adherence to epithelial cells. The identification of naturally-occurring Protein D-negative NTHi carriage isolates (Smith-Vaughan et al., 2014) also suggests that this vaccine antigen is not required for NTHi colonization of the host.
The invasive H. haemolyticus isolate HI2028 (Morton et al., 2012) interacted with the host cells in a similar manner to the NTHi isolates, with comparable cell association, invasion, and low inflammatory responses. However, this was not exclusive to HI2028 and other non-invasive H. haemolyticus carriage isolates displayed similar host interactions. Comparative genomics of the virulome for HI2028 vs. other haemophili may provide insight into whether there are specific genes (possibly acquired from NTHi) that are required for H. haemolyticus to become invasive.
We also assessed the host-cell interactions of four strains that belong to a recently described clade of “H. influenzae-like” haemophili (Price et al., 2015). These strains are identified as NTHi by the #hpd3 PCR (Wang et al., 2011) but are lacking the fucose operon that is used to identify NTHi with the multi-locus sequence typing scheme (http://haemophilus.mlst.net/; de Gier et al., 2015; Price et al., 2015). Overall, the H. influenzae-like haemophili were more similar in their host cell interactions to the H. haemolyticus than the NTHi strains assessed in this study, with lower attachment and invasion of epithelial cells and higher immunostimulatory properties. Interestingly, the one H. influenzae-like strain that displayed NTHi-like properties (H148) was isolated from the middle ear effusion of a child with recurrent acute OM whereas the other H. influenzae-like strains were nasopharyngeal isolates. The intra- and inter-species variability in host interactions that were observed in this study highlights the complexity of species definition in the genomic era. Awareness of the H. influenzae-like cluster and further analysis of the phenotypic traits of such strains is required to assist in resolving Haemophilus speciation.
H. haemolyticus was overall more immunostimulatory to epithelial cells than NTHi, with negligible release of IL-8 and IL-6 at 3 h post-NTHi stimulation. This suggests that NTHi is able to down-regulate or evade the initial immune responses in this model. The majority of the literature on host epithelial responses to NTHi are either with bacterial lysates or purified bacterial components rather than live NTHi. In support of our findings, assessment of A549 epithelial cell inflammatory responses to NTHi found that live NTHi did not stimulate activation of the IL-8 promoter, whereas treatment of the A549 cells with a soluble cytoplasmic fraction of NTHi induced robust IL-8 promoter activation within 4 h (Wang et al., 2003). This is further supported in a study describing NTHi isolates that did not stimulate mRNA expression of any cytokine or chemokine in human middle ear epithelial cells (Tong et al., 2001). The mechanisms for NTHi evasion of the host epithelial inflammatory response are not fully understood but it appears that some strains of H. haemolyticus also possess this property. Interestingly, gut commensals have been shown to either attenuate host production of inflammatory cytokines (Neish et al., 2000) or elicit a local inflammatory response (Chervonsky, 2012) to regulate colonization. It is possible that H. haemolyticus may use similar methods for regulating colonization of the respiratory tract. The host response to polymicrobial challenge has not been taken into consideration in our study and the synergistic effect of NTHi and H. haemolyticus, as well as other respiratory flora, warrants investigation. For example, there is an increase in IL-8 production from A549 and D562 cells when they are simultaneously challenged with NTHi and Streptococcus pneumoniae, compared with single species challenge (Ratner et al., 2005). Understanding regulation of microbial colonization at the epithelial surface may lead to the development of strategies to promote healthy colonization of the human respiratory tract and ultimately to the prevention of respiratory tract infections.
Further differences in cell-mediated responses to NTHi and H. haemolyticus were observed with PBMCs, where NTHi86-028NP challenge released high levels of IFNγ but H. haemolyticus33390 did not. IFNγ is critical in clearing intracellular bacterial infections such as Listeria monocytogenes and Salmonella typhimurium (Suzue et al., 2003). Intracellular survival provides a reservoir of bacteria for reinfection and is thought to contribute to the recurrent nature of diseases such as OM and exacerbations in bronchiectasis patients (Coates et al., 2008; King et al., 2008). We have shown that NTHi can persist intracellularly in the middle ear of children with recurrent OM (Thornton et al., 2011). Although a specific role for IFNγ in clearing NTHi infection has not been identified, PBMCs from children with chronic suppurative lung disease (Pizzutto et al., 2015) and adults with bronchiectasis (King et al., 2008) have been shown to have impaired IFNγ production in response to NTHi challenge. The lack of IFNγ response from H. haemolyticus treated PBMCs may suggest that H. haemolyticus cannot survive inside PBMCs or it may just be that the H. haemolyticus has died in cell culture media (as observed with the epithelial cell culture media by 24 h). Indeed, we have previously shown that live preparations of NTHi (instead of heat-killed or ethanol-killed) are required for IFNγ production from PBMCs (Kirkham et al., 2013). A limitation of this study is that bacterial viability was not assessed due to the difficulty in distinguishing a loss in bacterial viability from cell-mediated killing. There was significantly more IL-8 production from H. haemolyticus treated PBMCs compared with NTHi-challenge, whether this is due to release of a soluble factor from live or dead H. haemolyticus remains to be determined.
Pre-treatment of D562 monolayers with all doses of H. haemolyticus reduced the proportion of NTHi that were subsequently able to associate with the epithelial cells. The lowest dose of H. haemolyticus (MOI 1:1) resulted in the greatest reduction of NTHi association, suggesting that the inhibitory effect of H. haemolyticus on NTHi attachment may be more than just competition for binding sites. These data, together with the immunostimulatory capacity of H. haemolyticus, suggest that immune activation may have a role in prevention of NTHi attachment and subsequent invasion. It is important to note that NTHi attachment was inhibited by H. haemolyticus, despite needing to use a high NTHi challenge dose (1 × 109 CFU/mL) to detect fluorescence above background levels for unstained NTHi. In attempts to improve sensitivity for assessment of whether H. haemolyticus could block NTHi association and infection, a Transwell assay was investigated. We observed that pre-colonization of A549 monolayers with H. haemolyticus reduced the proportion of NTHi that could translocate through the epithelial cells. Whilst this result was also promising, the Transwell model we employed has limitations and others have suggested A549 cells do not express functional tight junction proteins when grown as a monolayer (Balis et al., 1984; Winton et al., 1998). This is likely to have an impact on the integrity of the epithelial barrier, and suggests that more complex models are required to investigate the potential of H. haemolyticus to prevent NTHi association and invasion.
In summary, we have demonstrated that the interaction of H. haemolyticus with the host is fundamentally different to NTHi, despite their genetic similarity and ability to recombine. We have indicated that the H. influenzae-like cluster of NTHi strains interact with the host in a more similar manner to H. haemolyticus than NTHi, in terms of epithelial association and cytokine responses. Our data suggest that H. haemolyticus may block NTHi colonization, providing the foundation for further assessment of H. haemolyticus as a bacterial therapy to prevent NTHi diseases.
Authors Contributions
LK and PR conceived the project. LK, JP, and RT designed the experiments. JP, AP, KC, and CG executed the experiments and all authors contributed to data interpretation. JP and LK prepared the manuscript and all authors provided critical review.
Funding
This work was funded by the Australian National Health and Medical Research Council (NHMRC; project grants 1011172 and 1086580) and the Western Australian Department of Health and Channel 7 Telethon Trust.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
JP is a recipient of NHMRC, UWA, and Princess Margaret Hospital Foundation postgraduate scholarships. RT is recipient of a BrightSpark postdoctoral fellowship and LK is recipient of an NHMRC Career Development Fellowship 1061428. We would like to thank Dr Daniel Morton at Oklahoma University, USA for provision of H. haemolyticus HI2028 and Dr Jeroen Langereis at Radboud University, The Netherlands for provision of NTHi strain R2866.
Abbreviations
A549, human lung carcinoma epithelial cells; CFU, colony forming units; ChoP, phosphorylcholine; CTV, cell trace violet; D562, Detroit 562 human pharyngeal carcinoma epithelial cells; IFN, interferon; IL, interleukin; LOS, lipooligosaccharide; LPS, lipopolysaccharide; NTHi, nontypeable Haemophilus influenzae; OM, otitis media; PBMCs, peripheral blood mononuclear cells; RT, room temperature; SEB, Staphylococcus enterotoxin B; SEM, standard error of the mean; TNF, tumor necrosis factor.
References
Alikhan, M. M., and Lee, F. E. (2014). Understanding nontypeable Haemophilus influenzae and chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 20, 159–164. doi: 10.1097/MCP.0000000000000023
Anderson, R., Wang, X., Briere, E. C., Katz, L. S., Cohn, A. C., Clark, T. A., et al. (2012). Haemophilus haemolyticus isolates causing clinical disease. J. Clin. Microbiol. 50, 2462–2465. doi: 10.1128/JCM.06575-11
Bakaletz, L. O., Tallan, B. M., Hoepf, T., DeMaria, T. F., Birck, H. G., and Lim, D. J. (1988). Frequency of fimbriation of nontypable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect. Immun. 56, 331–335.
Balis, J. U., Bumgarner, S. D., Paciga, J. E., Paterson, J. F., and Shelley, S. A. (1984). Synthesis of lung surfactant-associated glycoproteins by A549 cells: description of an in vitro model for human type II cell dysfunction. Exp. Lung Res. 6, 197–213. doi: 10.3109/01902148409109248
Binks, M. J., Temple, B., Kirkham, L. A., Wiertsema, S. P., Dunne, E. M., Richmond, P. C., et al. (2012). Molecular surveillance of true nontypeable Haemophilus influenzae: an evaluation of PCR screening assays. PLoS ONE 7:e34083. doi: 10.1371/journal.pone.0034083
Blyth, C. C., Currie, A. J., Wiertsema, S. P., Conway, N., Kirkham, L. A., Fuery, A., et al. (2011). Trivalent influenza vaccine and febrile adverse events in Australia, 2010: clinical features and potential mechanisms. Vaccine 29, 5107–5113. doi: 10.1016/j.vaccine.2011.05.054
Chervonsky, A. V. (2012). Intestinal commensals: influence on immune system and tolerance to pathogens. Curr. Opin. Immunol. 24, 255–260. doi: 10.1016/j.coi.2012.03.002
Clementi, C. F., and Murphy, T. F. (2011). Non-typeable Haemophilus influenzae invasion and persistence in the human respiratory tract. Front. Cell. Infect. Microbiol. 1:1. doi: 10.3389/fcimb.2011.00001
Coates, H., Thornton, R., Langlands, J., Filion, P., Keil, A. D., Vijayasekaran, S., et al. (2008). The role of chronic infection in children with otitis media with effusion: evidence for intracellular persistence of bacteria. Otolaryngol. Head Neck Surg. 138, 778–781. doi: 10.1016/j.otohns.2007.02.009
Cohen, R., Martin, E., de La Rocque, F., Thollot, F., Pecquet, S., Werner, A., et al. (2013). Probiotics and prebiotics in preventing episodes of acute otitis media in high-risk children: a randomized, double-blind, placebo-controlled study. Pediatr. Infect. Dis. J. 32, 810–814. doi: 10.1097/INF.0b013e31828df4f3
Cripps, A. W. (2010). Nontypeable Haemophilus influenzae and childhood pneumonia. P. N. G. Med. J. 53, 147–150. Avaliable online at: http://pngimr.org.pg/png_med_journal/sept-dec%202010%20nontypeable%20haemophilus%20influenzae.pdf
Cripps, A. W., Foxwell, R., and Kyd, J. (2002). Challenges for the development of vaccines against Haemophilus influenzae and Neisseria meningitidis. Curr. Opin. Immunol. 14, 553–557. doi: 10.1016/S0952-7915(02)00373-4
Dagan, R. (2000). Treatment of acute otitis media - challenges in the era of antibiotic resistance. Vaccine 19, S9–S16. doi: 10.1016/S0264-410X(00)00272-3
de Gier, C., Kirkham, L. A., and Norskov-Lauritsen, N. (2015). Complete deletion of the fucose operon in Haemophilus influenzae is associated with a cluster in multilocus sequence analysis-based phylogenetic Group II related to Haemophilus haemolyticus: implications for identification and typing. J. Clin. Microbiol. 53, 3773–3778. doi: 10.1128/JCM.01969-15
Di Pierro, F., Colombo, M., Zanvit, A., Risso, P., and Rottoli, A. S. (2014). Use of Streptococcus salivarius K12 in the prevention of streptococcal and viral pharyngotonsillitis in children. Drug Healthc. Patient Saf. 6, 15–20. doi: 10.2147/DHPS.S59665
Dworkin, M. S., Park, L., and Borchardt, S. M. (2007). The changing epidemiology of invasive Haemophilus influenzae disease, especially in persons > or = 65 years old. Clin. Infect. Dis. 44, 810–816. doi: 10.1016/j.jiac.2016.01.009
Erwin, A. L., and Smith, A. L. (2007). Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol. 15, 355–362. doi: 10.1016/j.tim.2007.06.004
Feazel, L. M., Santorico, S. A., Robertson, C. E., Bashraheil, M., Scott, J. A., Frank, D. N., et al. (2015). Effects of vaccination with 10-valent Pneumococcal Non-typeable Haemophilus influenzae Protein D Conjugate Vaccine (PHiD-CV) on the nasopharyngeal microbiome of kenyan toddlers. PLoS ONE 10:e0128064. doi: 10.1371/journal.pone.0128064
Fenger, M. G., Ridderberg, W., Olesen, H. V., and Nørskov-Lauritsen, N. (2012). Low occurrence of 'non-haemolytic Haemophilus haemolyticus' misidentified as Haemophilus influenzae in cystic fibrosis respiratory specimens, and frequent recurrence of persistent H. influenzae clones despite antimicrobial treatment. Int. J. Med. Microbiol. 302, 315–319. doi: 10.1016/j.ijmm.2012.10.001
Gargi, A., Tamilselvam, B., Powers, B., Prouty, M. G., Lincecum, T., Eshraghi, A., et al. (2013). Cellular interactions of the cytolethal distending toxins from Escherichia coli and Haemophilus ducreyi. J. Biol. Chem. 288, 7492–7505. doi: 10.1074/jbc.M112.448118
Hammitt, L. L., Ojal, J., Bashraheil, M., Morpeth, S. C., Karani, A., Habib, A., et al. (2014). Immunogenicity, impact on carriage and reactogenicity of 10-valent pneumococcal non-typeable Haemophilus influenzae protein D conjugate vaccine in Kenyan children aged 1-4 years: a randomized controlled trial. PLoS ONE 9:e85459. doi: 10.1371/journal.pone.0085459
Hariadi, N. I., Zhang, L., Patel, M., Sandstedt, S. A., Davis, G. S., Marrs, C. F., et al. (2015). Comparative profile of heme acquisition genes in disease-causing and colonizing nontypeable Haemophilus influenzae and Haemophilus haemolyticus. J. Clin. Microbiol. 53, 2132–2137. doi: 10.1128/JCM.00345-15
Jalalvand, F., and Riesbeck, K. (2014). Haemophilus influenzae: recent advances in the understanding of molecular pathogenesis and polymicrobial infections. Curr. Opin. Infect. Dis. 27, 268–274. doi: 10.1097/QCO.0000000000000056
Johnson, R. W., McGillivary, G., Denoel, P., Poolman, J., and Bakaletz, L. O. (2011). Abrogation of nontypeable Haemophilus influenzae protein D function reduces phosphorylcholine decoration, adherence to airway epithelial cells, and fitness in a chinchilla model of otitis media. Vaccine 29, 1211–1221. doi: 10.1016/j.vaccine.2010.12.003
King, P. T., Ngui, J., Farmer, M. W., Hutchinson, P., Holmes, P. W., and Holdsworth, S. R. (2008). Cytotoxic T lymphocyte and natural killer cell responses to non-typeable Haemophilus influenzae. Clin. Exp. Immunol. 152, 542–551. doi: 10.1111/j.1365-2249.2008.03667.x
Kirkham, L. A., Corscadden, K. J., Wiertsema, S. P., Currie, A. J., and Richmond, P. C. (2013). A practical method for preparation of pneumococcal and nontypeable Haemophilus influenzae inocula that preserves viability and immunostimulatory activity. BMC Res. Notes 6:522. doi: 10.1186/1756-0500-6-522
Kowapradit, J., Opanasopit, P., Ngawhirunpat, T., Apirakaramwong, A., Rojanarata, T., Ruktanonchai, U., et al. (2010). In vitro permeability enhancement in intestinal epithelial cells (Caco-2) monolayer of water soluble quaternary ammonium chitosan derivatives. AAPS Pharm. Sci. Tech. 11, 497–508. doi: 10.1208/s12249-010-9399-7
Laupland, K. B., Schonheyder, H. C., Ostergaard, C., Knudsen, J. D., Valiquette, L., Galbraith, J., et al. (2011). Epidemiology of Haemophilus influenzae bacteremia: a multi-national population-based assessment. J. Infect. 62, 142–148. doi: 10.1016/j.jinf.2010.11.009
Leach, A. J., Wigger, C., Hare, K., Hampton, V., Beissbarth, J., Andrews, R., et al. (2015). Reduced middle ear infection with non-typeable Haemophilus influenzae, but not Streptococcus pneumoniae, after transition to 10-valent pneumococcal non-typeable H. influenzae protein D conjugate vaccine. BMC Pediatr. 15:162. doi: 10.1186/s12887-015-0483-8
Lehtoranta, L., Soderlund-Venermo, M., Nokso-Koivisto, J., Toivola, H., Blomgren, K., Hatakka, K., et al. (2012). Human bocavirus in the nasopharynx of otitis-prone children. Int. J. Pediatr. Otorhinolaryngol. 76, 206–211. doi: 10.1016/j.ijporl.2011.10.025
McCrea, K. W., Wang, M. L., Xie, J., Sandstedt, S. A., Davis, G. S., Lee, J. H., et al. (2010a). Prevalence of the sodC gene in nontypeable Haemophilus influenzae and Haemophilus haemolyticus by microarray-based hybridization. J. Clin. Microbiol. 48, 714–719. doi: 10.1128/JCM.01416-09
McCrea, K. W., Xie, J., LaCross, N., Patel, M., Mukundan, D., Murphy, T. F., et al. (2008). Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. J. Clin. Microbiol. 46, 406–416. doi: 10.1128/JCM.01832-07
McCrea, K. W., Xie, J., Marrs, C. F., and Gilsdorf, J. R. (2010b). Prevalence of genetic differences in phosphorylcholine expression between nontypeable Haemophilus influenzae and Haemophilus haemolyticus. BMC Microbiol. 10, 286. doi: 10.1186/1471-2180-10-286
Morton, D. J., Hempel, R. J., Whitby, P. W., Seale, T. W., and Stull, T. L. (2012). An invasive Haemophilus haemolyticus isolate. J. Clin. Microbiol. 50, 1502–1503. doi: 10.1128/JCM.06688-11
Murphy, T., Bakaletz, L., and Smeesters, P. (2009a). Microbial interactions in the respiratory tract. Pediatr. Infect. Dis. J. 28(10 Suppl.), S121–S126. doi: 10.1097/INF.0b013e3181b6d7ec
Murphy, T. F., Faden, H., Bakaletz, L. O., Kyd, J. M., Forsgren, A., Campos, J., et al. (2009b). Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr. Infect. Dis. J. 28, 43–48. doi: 10.1097/INF.0b013e318184dba2
Neish, A. S., Gewirtz, A. T., Zeng, H., Young, A. N., Hobert, M. E., Karmali, V., et al. (2000). Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science 289, 1560–1563. doi: 10.1126/science.289.5484.1560
Nizet, V., Colina, K. F., Almquist, J. R., Rubens, C. E., and Smith, A. L. (1996). A virulent nonencapsulated Haemophilus influenzae. J. Infect. Dis. 173, 180–186. doi: 10.1093/infdis/173.1.180
Norskov-Lauritsen, N. (2011). Increased level of intragenomic 16S rRNA gene heterogeneity in commensal strains closely related to Haemophilus influenzae. Microbiology 157(Pt 4), 1050–1055. doi: 10.1099/mic.0.047233-0
Novotny, L. A., Clements, J. D., and Bakaletz, L. O. (2013). Kinetic analysis and evaluation of the mechanisms involved in the resolution of experimental nontypeable Haemophilus influenzae-induced otitis media after transcutaneous immunization. Vaccine 31, 3417–3426. doi: 10.1016/j.vaccine.2012.10.033
Pickering, J., Binks, M. J., Beissbarth, J., Hare, K. M., Kirkham, L. A., and Smith-Vaughan, H. (2014a). A PCR-high-resolution melt assay for rapid differentiation of nontypeable Haemophilus influenzae and Haemophilus haemolyticus. J. Clin. Microbiol. 52, 663–667. doi: 10.1128/JCM.02191-13
Pickering, J., Richmond, P. C., and Kirkham, L.-A. S. (2014b). Molecular tools for differentiation of nontypeable Haemophilus influenzae from Haemophilus haemolyticus. Front. Microbiol. 5:664. doi: 10.3389/fmicb.2014.00664
Pickering, J., Smith-Vaughan, H., Beissbarth, J., Bowman, J. M., Wiertsema, S., Riley, T. V., et al. (2014c). Diversity of nontypeable Haemophilus influenzae strains colonizing australian aboriginal and non-aboriginal children. J. Clin. Microbiol. 52, 1352–1357. doi: 10.1128/JCM.03448-13
Pizzutto, S. J., Upham, J. W., Yerkovich, S. T., and Chang, A. B. (2015). High pulmonary levels of IL-6 and IL-1beta in children with chronic suppurative lung disease are associated with low systemic IFN-gamma production in response to non-typeable Haemophilus influenzae. PLoS ONE 10:e0129517. doi: 10.1371/journal.pone.0129517
Post, D. M., Ketterer, M. R., Coffin, J. E., Reinders, L. M., Munson, R. S. Jr. Bair, T., et al. (2016). Comparative analyses of the lipooligosaccharides from nontypeable Haemophilus influenzae and Haemophilus haemolyticus show differences in sialic acid and phosphorylcholine modifications. Infect. Immun. 84, 765–774. doi: 10.1128/IAI.01185-15
Price, E. P., Sarovich, D. S., Nosworthy, E., Beissbarth, J., Marsh, R. L., Pickering, J., et al. (2015). Haemophilus influenzae: using comparative genomics to accurately identify a highly recombinogenic human pathogen. BMC Genomics 16:641. doi: 10.1186/s12864-015-1857-x
Prymula, R., Hanovcova, I., Splino, M., Kriz, P., Motlova, J., Lebedova, V., et al. (2011). Impact of the 10-valent pneumococcal non-typeable Haemophilus influenzae Protein D conjugate vaccine (PHiD-CV) on bacterial nasopharyngeal carriage. Vaccine 29, 1959–1967. doi: 10.1016/j.vaccine.2010.12.086
Prymula, R., and Schuerman, L. (2009). 10-valent pneumococcal nontypeable Haemophilus influenzae PD conjugate vaccine: synflorix. Expert Rev. Vaccines 8, 1479–1500. doi: 10.1586/erv.09.113
Ratner, A. J., Lysenko, E. S., Paul, M. N., and Weiser, J. N. (2005). Synergistic proinflammatory responses induced by polymicrobial colonization of epithelial surfaces. Proc. Natl. Acad. Sci. U.S.A. 102, 3429–3434. doi: 10.1073/pnas.0500599102
Sandstedt, S. A., Zhang, L., Patel, M., McCrea, K. W., Qin, Z., Marrs, C. F., et al. (2008). Comparison of laboratory-based and phylogenetic methods to distinguish between Haemophilus influenzae and H. haemolyticus. J. Microbiol. Methods 75, 369–371. doi: 10.1016/j.mimet.2008.06.023
Shann, F., Gratten, M., Germer, S., Linnemann, V., Hazlett, D., and Payne, R. (1984). Aetiology of pneumonia in children in Goroka Hospital, Papua New Guinea. Lancet 2, 537–541. doi: 10.1016/S0140-6736(84)90764-5
Skovbjerg, S., Roos, K., Holm, S. E., Grahn Hakansson, E., Nowrouzian, F., Ivarsson, M., et al. (2009). Spray bacteriotherapy decreases middle ear fluid in children with secretory otitis media. Arch. Dis. Child. 94, 92–98. doi: 10.1136/adc.2008.137414
Smith-Vaughan, H., Byun, R., Nadkarni, M., Jacques, N. A., Hunter, N., Halpin, S., et al. (2006). Measuring nasal bacterial load and its association with otitis media. BMC Ear Nose Throat Disord. 6:10 doi: 10.1186/1472-6815-6-10
Smith-Vaughan, H. C., Binks, M. J., Marsh, R. L., Kaestli, M., Ward, L., Hare, K. M., et al. (2013). Dominance of Haemophilus influenzae in ear discharge from Indigenous Australian children with acute otitis media with tympanic membrane perforation. BMC Ear Nose Throat Disord. 13:12. doi: 10.1186/1472-6815-13-12
Smith-Vaughan, H. C., Chang, A. B., Sarovich, D. S., Marsh, R. L., Grimwood, K., Leach, A. J., et al. (2014). Absence of an important vaccine and diagnostic target in carriage- and disease-related nontypeable Haemophilus influenzae. Clin. Vaccine Immunol. 21, 250–252. doi: 10.1128/CVI.00632-13
Sondergaard, A., Witherden, E. A., Norskov-Lauritsen, L., and Tristram, S. (2014). Horizontal transfer of chromosomally-encoded resistance to beta-lactam antibiotics between Haemophilus influenzae and Haemophilus haemolyticus, in International Pasteurellaccae Conference 2014 (Prato).
Suzue, K., Asai, T., Takeuchi, T., and Koyasu, S. (2003). In vivo role of IFN-gamma produced by antigen-presenting cells in early host defense against intracellular pathogens. Eur. J. Immunol. 33, 2666–2675. doi: 10.1002/eji.200323292
Swords, W. E., Buscher, B. A., Ver Steeg Ii, K., Preston, A., Nichols, W. A., Weiser, J. N., et al. (2000). Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37, 13–27. doi: 10.1046/j.1365-2958.2000.01952.x
Taipale, T., Pienihakkinen, K., Isolauri, E., Larsen, C., Brockmann, E., Alanen, P., et al. (2011). Bifidobacterium animalis subsp. lactis BB-12 in reducing the risk of infections in infancy. Br. J. Nutr. 105, 409–416. doi: 10.1017/S0007114510003685
Teo, E., House, H., Lockhart, K., Purchuri, S. N., Pushparajah, J., Cripps, A. W., et al. (2014). Haemophilus influenzae oral vaccination for preventing acute exacerbations of chronic bronchitis and chronic obstructive pulmonary disease. Cochrane Database Syst Rev. 9:CD010010. doi: 10.1002/14651858.CD010010.pub2
Thanavala, Y., and Lugade, A. A. (2011). Role of nontypeable Haemophilus influenzae in otitis media and chronic obstructive pulmonary disease. Adv. Otorhinolaryngol. 72, 170–175. doi: 10.1159/000324785
Thornton, R. B., Rigby, P. J., Wiertsema, S. P., Filion, P., Langlands, J., Coates, H. L., et al. (2011). Multi-species bacterial biofilm and intracellular infection in otitis media. BMC Pediatr. 11:94. doi: 10.1186/1471-2431-11-94
Tong, H. H., Chen, Y., James, M., Van Deusen, J., Welling, D. B., and DeMaria, T. F. (2001). Expression of cytokine and chemokine genes by human middle ear epithelial cells induced by formalin-killed Haemophilus influenzae or its lipooligosaccharide htrB and rfaD mutants. Infect. Immun. 69, 3678–3684. doi: 10.1128/IAI.69.6.3678-3684.2001
Tregnaghi, M. W., Saez-Llorens, X., Lopez, P., Abate, H., Smith, E., Posleman, A., et al. (2014). Efficacy of Pneumococcal Nontypable Haemophilus influenzae Protein D Conjugate Vaccine (PHiD-CV) in young latin american children: a double-blind randomized controlled trial. PLoS Med. 11:e1001657. doi: 10.1371/journal.pmed.1001657
van den Bergh, M. R., Spijkerman, J., Swinnen, K. M., Francois, N. A., Pascal, T. G., Borys, D., et al. (2013). Effects of the 10-valent pneumococcal nontypeable Haemophilus influenzae protein D-conjugate vaccine on nasopharyngeal bacterial colonization in young children: a randomized controlled trial. Clin. Infect. Dis. 56, e30–e39. doi: 10.1093/cid/cis922
Van Eldere, J., Slack, M. P., Ladhani, S., and Cripps, A. W. (2014). Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect. Dis. 14, 1281–1292. doi: 10.1016/S1473-3099(14)70734-0
van Wessel, K., Rodenburg, G. D., Veenhoven, R. H., Spanjaard, L., van der Ende, A., and Sanders, E. A. (2011). Nontypeable Haemophilus influenzae invasive disease in The Netherlands: a retrospective surveillance study 2001-2008. Clin. Infect. Dis. 53, e1–e7. doi: 10.1093/cid/cir268
Wang, B., Cleary, P. P., Xu, H., and Li, J. D. (2003). Up-regulation of interleukin-8 by novel small cytoplasmic molecules of nontypeable Haemophilus influenzae via p38 and extracellular signal-regulated kinase pathways. Infect. Immun. 71, 5523–5530. doi: 10.1128/IAI.71.10.5523-5530.2003
Wang, X., Mair, R., Hatcher, C., Theodore, M. J., Edmond, K., Wu, H. M., et al. (2011). Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. Int. J. Med. Microbiol. 301, 303–309. doi: 10.1016/j.ijmm.2010.11.004
Wiertsema, S. P., Kirkham, L. A., Corscadden, K. J., Mowe, E. N., Bowman, J. M., Jacoby, P., et al. (2011). Predominance of nontypeable Haemophilus influenzae in children with otitis media following introduction of a 3+0 pneumococcal conjugate vaccine schedule. Vaccine 29, 5163–5170. doi: 10.1016/j.vaccine.2011.05.035
Winton, H. L., Wan, H., Cannell, M. B., Gruenert, D. C., Thompson, P. J., Garrod, D. R., et al. (1998). Cell lines of pulmonary and non-pulmonary origin as tools to study the effects of house dust mite proteinases on the regulation of epithelial permeability. Clin. Exp. Allergy 28, 1273–1285. doi: 10.1046/j.1365-2222.1998.00354.x
Keywords: colonization, Haemophilus haemolyticus, host epithelium, inflammatory mediators, in vitro, NTHi, PBMCs
Citation: Pickering JL, Prosser A, Corscadden KJ, de Gier C, Richmond PC, Zhang G, Thornton RB and Kirkham L-AS (2016) Haemophilus haemolyticus Interaction with Host Cells Is Different to Nontypeable Haemophilus influenzae and Prevents NTHi Association with Epithelial Cells. Front. Cell. Infect. Microbiol. 6:50. doi: 10.3389/fcimb.2016.00050
Received: 12 February 2016; Accepted: 18 April 2016;
Published: 09 May 2016.
Edited by:
W. Edward Swords, Wake Forest University Health Sciences, USAReviewed by:
Kevin Mason, The Ohio State University, USAJohn Wayne Rumsey, SSC and Seminole County Public Schools, USA
Copyright © 2016 Pickering, Prosser, Corscadden, de Gier, Richmond, Zhang, Thornton and Kirkham. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lea-Ann S. Kirkham, lea-ann.kirkham@uwa.edu.au
 Janessa L. Pickering
Janessa L. Pickering Amy Prosser2
Amy Prosser2  Camilla de Gier
Camilla de Gier Peter C. Richmond
Peter C. Richmond Ruth B. Thornton
Ruth B. Thornton Lea-Ann S. Kirkham
Lea-Ann S. Kirkham