Isolation and characterization of a CO2-tolerant Lactobacillus strain from Crystal Geyser, Utah, U.S.A.
- 1Department of Geosciences, Jackson School of Geosciences, The University of Texas at Austin, Austin, TX, USA
- 2Center for Petroleum and Geosystems Engineering, Cockrell School of Engineering, The University of Texas at Austin, Austin, TX, USA
When CO2 is sequestered into the deep subsurface, changes to the subsurface microbial community will occur. Capnophiles, microorganisms that grow in CO2-rich environments, are some organisms that may be selected for under the new environmental conditions. To determine whether capnophiles comprise an important part of CO2-rich environments, an isolate from Crystal Geyser, Utah, U.S.A., a CO2- rich spring considered a carbon sequestration analog, was characterized. The isolate was cultured under varying CO2, pH, salinity, and temperature, as well as different carbon substrates and terminal electron acceptors (TEAs) to elucidate growth conditions and metabolic activity. Designated CG-1, the isolate is related (99%) to Lactobacillus casei in 16S rRNA gene identity, growing at PCO2 between 0 and 1.0 MPa. Growth is inhibited at 2.5 MPa, but stationary phase cultures exposed to this pressure survive beyond 5 days. At 5.0 MPa, survival is at least 24 h. CG-1 grows in neutral pH, 0.25 M NaCl, and between 25° and 45°C and consumes glucose, lactose, sucrose, or crude oil, likely performing lactic acid fermentation. Fatty acid profiles between 0.1 and 1.0 MPa suggests decreases in cell size and increases in membrane rigidity. Transmission electron microscopy reveals rod shaped bacteria at 0.1 MPa. At 1.0 MPa, cells are smaller, amorphous, and produce abundant capsular material. Its ability to grow in environments regardless of the presence of CO2 suggests we have isolated an organism that is more capnotolerant than capnophilic. Results also show that microorganisms are capable of surviving the stressful conditions created by the introduction of CO2 for sequestration. Furthermore, our ability to culture an environmental isolate indicates that organisms found in CO2 environments from previous genomic and metagenomics studies are viable, metabolizing, and potentially affecting the surrounding environment.
Introduction
The sequestering of anthropogenic CO2 via pumping into the formation waters of deep saline aquifers will create new environmental subsurface conditions (Kharaka et al., 2006; Kaszuba and Janecky, 2009; Little and Jackson, 2010). CO2 will reside in the reservoir as a plume of supercritical fluid and will gradually dissolve into the water. Given that high aqueous CO2 concentrations are generally toxic to most microorganisms as the molecule interferes with intracellular functions (Hong and Pyun, 1999; Spilimbergo and Bertucco, 2003; Watanabe et al., 2003; Damar and Balaban, 2006; Mitchell et al., 2008; Santillan et al., 2013), this change will initially stress the native microbial population. However, over longer time scales, these new conditions may create a niche for CO2-tolerant bacteria that will select for a new community of microorganisms capable of resisting CO2 toxicity.
One particular set of organisms that may be selected for in this new environment are capnophiles: organisms that rely on CO2 for growth and survival. They are phylogenetically diverse, perform a variety of metabolic activities, and share very little evolutionary relationship to each other (Song et al., 2007; Bringel et al., 2008; Arioli et al., 2009). Capnophiles have been isolated from a variety of habitats including plants, mammalian cavities, wastewater, and animal rumens (Ballestra et al., 1996; Bringel et al., 2008; Ueda et al., 2008; Arioli et al., 2009). While a large degree of attention has been given to these bacteria for food and disease studies, little has been given to their presence and environmental significance in the deep subsurface. Capnophiles may, for example, use CO2 for metabolic processes such as in acetyl-CoA production, amino acid synthesis, or fermentation (e.g., Wynne and Foster, 1948; Maren, 1967; Smith and Ferry, 2000; Song et al., 2007; Arioli et al., 2009; Hertzberger et al., 2013).
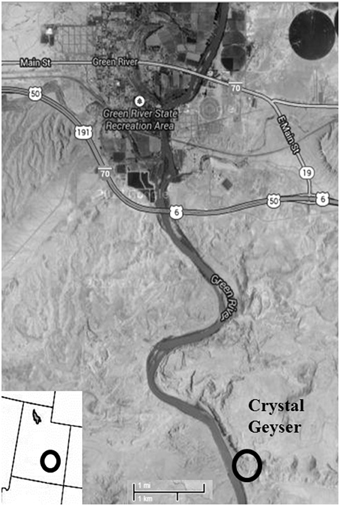
While it is currently difficult to assess the impact of CO2 sequestration on the subsurface biosphere in situ, terrestrial CO2 springs are accessible analogs to determine the long-term impacts of CO2 on subsurface organisms as they can be readily sampled for microbial community composition, biomass, and viability. One such example is Crystal Geyser (CG), Utah (Figure 1), an open well bore that periodically erupts CO2 charged water, likely sourced from carbonate-clay diagenesis at depth (Wilkinson et al., 2009). It originated as an unsuccessful exploratory oil well 800 m deep and spudded in 1935 (Baer and Rigby, 1978; Heath et al., 2009; McPherson and Heath, 2009; Burnside et al., 2013). Previous work by Emerson et al. (2015) have correlated the geochemistry of this field site to both the microbial community and the metagenomics profile from the sampled spring. This study reveals the presence of a microbial community within the geyser and reveals a metagenome with putative metabolisms that are consistent with iron biogeochemistry of the field site.
Using Crystal Geyser as an analog to CO2 sequestration, the goal of this work was to isolate and characterize a capnophilic microorganism to show whether or not subsurface organisms are viable under sequestration conditions. Other field studies have also been performed at CO2 injection sites, all examining microbial community profiles using molecular techniques (Morozova et al., 2011; Mu et al., 2014; Mu and Moreau, 2015) and showing that microbial community changes will occur as a result of CO2perturbation into the subsurface sytem.
We have taken a culture-based approach to studying the microbiology of CO2 sequestration as culture-based assessments can allow for direct observations of microbial growth and activity on laboratory time scales. This in turn, can complement existing genomic and metagenomic studies which can only confirm and infer the presence and activities of microbes based on sequence data. Previous culture-based approaches have also been performed utilizing samples from environmental sites (Mitchell et al., 2008; Kirk et al., 2013; Peet et al., 2015). Our approach was to work with a field site that had been under prolonged CO2 perturbation (Burnside et al., 2013) to find out whether microbial communities under prolonged CO2 exposure would still be viable. We hypothesized that incubation under high CO2 would allow for enrichment and isolation of capnophilic microorganisms from Crystal Geyser.
Materials and Methods
Field Sampling
Sampling of Crystal Geyser was performed in May 2012. Filters, tubing, and containers used for sampling were autoclaved at 122°C and 15 psi for 30 min prior to field work. For field sterilization of large equipment, a 10% sodium hypochlorite solution was applied to surfaces for approximately 30 min followed by treatment with a 10% sodium thiosulfate solution to remove excess chlorine. Materials were then rinsed with approximately 10 liters of discharging spring water prior to any sampling. Filtrate was collected in the field for measurement of pH, temperature, conductivity using an ultrameter (Carlsbad, CA), or for dissolved oxygen, Fe, PO3−4, NO−3, and NH+4 in the field colorimetrically (Chemetrics, Inc., Midland VA). Samples were also collected and characterized for ion chemistry through ion chromatography and inductively coupled plasma mass spectrometry.
Biomass sampling was performed using a peristaltic pump (Geotech, Denver, CO). Approximately 9.7 m of silicone tubing (Masterflex, Vernon Hills, IL) was lowered into the Crystal Geyser outlet and water pumped at a rate of 200 ml/min into a sterile container to allow for excess CO2 to exsolve. We chose this method in comparison to Emerson et al. (2015) in order to reduce the amount of oxidized Fe passing through filters so that clogging would not occur as quickly. Water was then filtered through a 90 mm, 0.1 μm sterile polycarbonate filter (Millipore, Billerica, MA) in a sterile stainless steel filter apparatus. Fifty liters of water was allowed to flow through each filter to collect sufficient biomass. Filtration took approximately 4 h per filter. While the introduction of tubing from the surface may have the potential to add surficial contamination into the subsurface, we believe this contamination was minimal as the water from Crystal Geyser is constantly bubbling upwards, even between eruptions. This constant movement of fluid likely provided little time to allow for contaminants to sink below, though we acknowledge that the fact that the well is open and publicly accessible makes contamination completely difficult to control. In addition, lowering a tube down the well, rather than collection from the well casing allowed us to minimize the entry of surficial contaminants into our filters.
Filters and their collected biomass were designated for phospholipid fatty acid (PLFA) analysis and culturing. Samples for PLFA analysis were preserved at −79°C in a dry dewar (Taylor-Wharton, Theodore, AL) until processing.
Culturing was performed immediately after filtration in order to limit biomass exposure to the atmosphere. Filters used for culturing were placed into serum bottles containing Luria-Bertani broth, covered with sterile aluminum foil (LB, Fisher, Pittsburgh, PA). This media was prepared aerobically and was amended with 15 g/l NaCl to maintain the approximate salinity found at Crystal Geyser (Heath et al., 2009). The inoculated media was immediately placed in a 4 L pressure vessel (Alloy Products Corp, Waukesha, WI) and made anaerobic through the use of a GasPak™ EZ Anaerobe Container System Sachet (BD, Franklin Lakes, NJ). The serum bottles in the vessels were then pressurized to 1.0 MPa in the field with ultrapure CO2. Previous work has shown that CO2 is very effective sterilant (Nakamura et al., 1994; Isenschmid et al., 1995; Ballestra et al., 1996; Shimoda et al., 1998; Hong and Pyun, 1999; Erkmen, 2000; Spilimbergo and Bertucco, 2003; Watanabe et al., 2003; Damar and Balaban, 2006; Oule et al., 2006, 2010; Song et al., 2007). With this in mind, we reasoned that cultures grown at very high CO2 pressures would likely result in a sterilization of most viable microorganisms, especially those accidentally introduced through contamination. Furthermore, we reasoned that the high CO2 environment in our culture vessels would select for the few organisms that have surfaced at depth living in high pressure conditions. To further ensure any growth would come from Crystal Geyser and not from contamination, sterile blank filters were also placed into fresh, uninoculated media.
Culture Experiments
Cultures were allowed to incubate at room temperature under 1.0 MPa of CO2 for approximately 1 month before reculture. Tryptic soy broth (TSB, Fisher, Pittsburgh, PA) with 15 g/L NaCl was used for further enrichments and subsequent culture experiments as cultures appeared to show the best growth in this broth (data not shown). After 3 successive transfers over the course of 3 months, TSB cultures were then diluted to extinction by performing a nine-fold serial dilution to obtain an isolate. Field blanks were also transferred to insure laboratory sterile techniques were adequate. After 1 month of incubation at 1.0 MPa and room temperature, cultures taken from the eight-fold dilution were transferred to fresh TSB broth and recultured. Cultures were then transferred into fresh broth at 0.1 MPa PCO2 periodically to maintain a stock.
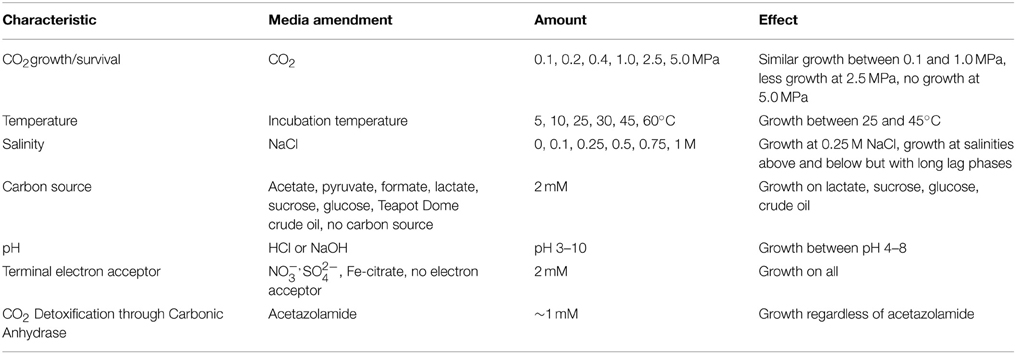
The isolate, designated CG-1, was assessed for growth under various conditions that focused on CO2, temperature, salinity, pH, carbon substrates, electron acceptors, and fermentation capability (Table 1). In all cases, growth was evaluated by taking 200 μl of stock cultures into 10 ml of fresh TSB media and observing growth for 1 week by measuring optical density (OD700) using a LaMotte Smart Spectro spectrophotometer (Chestertown, MD). As the organism appears aerotolerant, growth media was not prepared anaerobically, except when specified. However, cultures were incubated with butyl stoppers to prevent exchange with atmospheric gasses.
CO2
Cultures were grown under CO2 pressures of 0, 0.1, 0.2, 0.4, 1.0, and 2.5 and 5.0 MPa with CO2 as the headspace gas. Cultures grown at 0–0.4 MPa were grown in hungate tubes with butyl stoppers. For cultures grown at 1.0 and 2.5 MPa, samples were incubated in culture tubes inside modified stainless steel Parr reactors (Parr Instruments, Moline, IL). Samples grown at 0.1 MPa and at 1.0 MPa were also harvested for lipid analysis.
Temperature, Salinity, pH, Carbon Source
CG-1 growth at various temperatures was evaluated by incubation at 5, 10, 25, 30, 45, and 60°C containing 0.1 MPa CO2. Cultures tested for salinity were grown in similar conditions but kept at room temperature with media amended with 0, 0.1, 0.25, 0.5, 0.75, and 1 M NaCl. For pH experiments, 1 N HCl or NaOH was used to change media pH to values between 3 and 10. Growth experiments for pH contained no CO2 in the headspace. Carbon sources tested include acetate, pyruvate, formate, lactate, sucrose, glucose, and Teapot Dome crude oil. Cultures were also grown in TSB media without the addition of an extra carbon source beyond the casein and soy present. A set of cultures grown with no CO2 and at 0.1 MPa CO2 were also amended with approximately 1 mM of the carbonic anhydrase inhibitor acetazolamide to determine whether CO2 growth and survival was influenced by the enzyme. These growth experiments are summarized in Table 1.
Electron Acceptor
To determine the isolate's metabolic capabilities, pre-reduced anaerobically sterilized (PRAS) TSB was amended with various electron acceptors including 2 mM NO−3, SO2−4, Fe-citrate, or with no electron acceptors present (Gosden and Ware, 1977). 200 μL of stock culture was then transferred into the different media and grown at 0.4 MPa.
Fermentation
To determine the isolate's fermentative capabilities, cultures were grown in agar slants of triple sugar iron agar (TSI) (e.g., Engel, 2004). Triple sugar iron agar contains 3 sugars, a pH indicator, and sodium thiosulfate and ferrous sulfate to determine fermentative sulfide production. Stab cultures into the agar slants were performed and incubated anaerobically under 0, 0.1, and 1.0 MPa CO2 for 24 h. Samples were left in the tubes for 1 week afterwards to verify organism growth in the slants.
Lipid Analysis
Filters collected from the field were frozen immediately and freeze-dried upon return to the lab. For bacterial cultures grown in the lab, prior to lipid extraction, cultures were centrifuged and then freeze-dried. Lypholized samples were weighed out and then microwave extracted (CEM MARS 100°C, 10 min) using 20 mL of a 9:1 methylene chloride:methanol mixture to provide a total lipid extract. The lipid extract was evaporated under N2 and saponified at 70°C for 1 h using a 0.5 M NaOH solution. The solution was cooled, acidified, and extracted 3 times using methyl tert-butyl ether. Samples were then evaporated and resuspended in methylene chloride for column purification.
Solid phase extraction of fatty acids was performed using an amino-propyl stationary phase (0.5 g of Supelclean-NH−2, Supelco, Bellefonte, PA). Fatty acids were eluted with 50:1 DCM/formic acid. The fraction was then evaporated under N2 and resuspended in hexane.
Fatty acids were converted to fatty acid methyl esters (FAMEs) using previously described methods (Rodriguez-Ruiz et al., 1998). Samples were methylated using a 20:1 methylation mixture of methanol/acetyl chloride. Samples were then heated at 100°C for 10 min and cooled to room temperature. After cooling, distilled water was added to create two phases. The upper hexane phase was extracted, and concentrated prior to analysis.
Samples were characterized and quantitated by gas-chromatography mass–spectrometry. One microliter of the sample in hexane was injected into an Agilent 7890A GC with a split/splitless injector operated in splitless mode at 300°C, a DB-5 column (0.25 mmi.d., 0.25 μm film thickness, 30 m length), 2.0 cm3 min−1 He flow and programmed heating of the oven from 40 to 170°C at 4°C/min then to 240°C at 30°C/min. An Agilent 5973 quadrapole mass spectrometer was used for detection. Compounds were identified by elution time and comparison with published mass- spectra.
Cloning, Sanger Sequencing
Cloning was performed on the CG isolate. DNA from the isolate was extracted using the Mo Bio Power Biofilm DNA extraction kit (Mo Bio Laboratories, Inc., Carlsbad, CA). Extracted DNA from stationary phase cultures were PCR amplified for the 16S rRNA gene using 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGGTTACCTTGTTACGACTT -3′) primers under the following conditions: denaturation at 94°C for 5 min, primer annealing 50°C for 30 s, and chain extension at 72°C for 1.5 min for 45 cycles. Cloning was completed using the TOPO TA Cloning Kit (Life Technologies, Grand Island, NY). PCR product was ligated into pCRII-TOPO vector and heat shocked into chemically competent Escherichia coli. Plasmid DNA from 40 ampicillin-resistant clones was then extracted and sent to the University of Austin ICMB Core Facility for Sanger Sequencing (Austin, TX). Sequences were then subjected to a Basic Local Alignment Search Tool search (BLASTn) search (http://blast.ncbi.nlm.nih.gov/) to determine the organism's taxonomic identity. A phylogenetic tree relating the isolate to related sequences was made using CLUSTALX using the neighbor joining alignment algorithm (Chenna et al., 2003; Larkin et al., 2007).
Transmission Electron Microscopy
Samples were prepared for transmission electron microscopy (TEM) following methods outlined by Romanovicz, personal communication. Pelleted samples were first fixed overnight in a 2.5% glutaraldehyde solution. After 2 washes and centrifugations with a 0.1 N Na-cacodylic buffer, cultures were stained using a 1 mL mixture of 1% OsO4solution and 2% K-ferrocyanide for 1 h. Stained pellets were washed with cacodylic buffer and suspended in thin (approximately 0.5 cm) 3% agarose strings. Agarose-suspended samples were incubated in 50% ethanol for 15 min and transferred to a solution containing 2% uranyl acetate in 70% ethanol for 2 h. After incubation in uranyl acetate, samples were sequentially dehydrated in ethanol solutions starting with 90–100% for 15 min each. Samples were twice rinsed in LR White resin and embedded in the same resin.
Embedded samples were sliced into 70 nm thin sections with a diamond knife (DiATOME, Hatfield, PA) using a Leica Ultracut UTC Ultramicrotome at the University of Texas ICMB Core Facility (Austin, TX). Sections were viewed using a FEI Tecnai Transmission Electron Microscope at the same facility using an accelerating voltage of 80 kV on bright-field.
Results
Crystal Geyser Composition
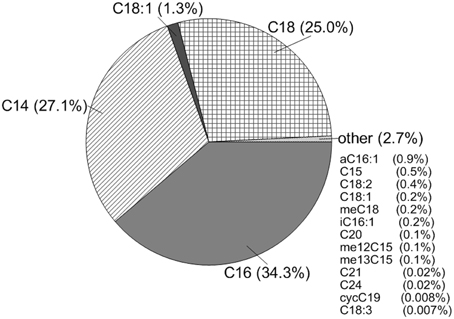
The total amount of PLFA extracted at CG was 1.0 ng/ml. Using a conversion factor of 2×104 cells/ng of PLFA, CG had approximately 2×104 cells/ml (Stroes-Gascoyne et al., 2007). The dominant fatty acids present at this site were even-numbered, saturated fatty acids, specifically C12 (9.8%), C14 (27.1%), C16 (34.3%), and C18 (25.0%) with some unsaturated C18 present (1.3%). This overwhelming contribution of saturated fatty acids is consistent with observations of microorganisms that have decreased in cell sizes due to stress (e.g., Veerkamp, 1971; Brown et al., 1997; Chang and Cronan, 1999). Other fatty acids were present, but only made a contribution of 2.7% of the total fatty acid content, including the cyclopropyl C19 lipid characteristic of Lactobacillus which made less than 0.1% of the fatty acid content (Figure 2).
Isolate Identity and Morphology
Sanger sequence data for CG-1 indicates that of the 40 clones selected, 39 clones were 97–99% identical to Lactobacillus casei with one clone being only 95% similar likely due to problems with sequence quality. There was also an errant sequence due to an incomplete transformation into E. coli (Table S1). Figure 3 shows the inferred phylogeny of the 16S rRNA gene of CG-1 in relation to other Lactobacillus species. The isolate is approximately 99% identical to Lactobacillus casei based on the phylogenetic tree, suggesting we have isolated a new strain of Lactobacillus casei. Furthermore, negative controls showed no cloudiness, suggesting the organism isolated likely did not originate from contamination.
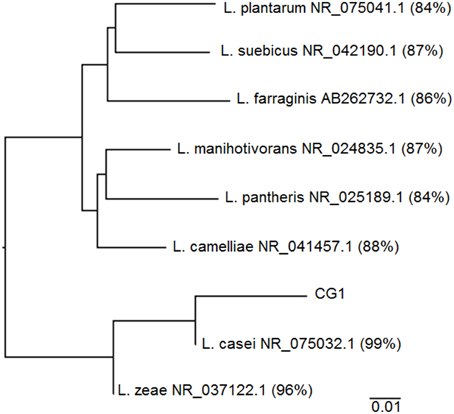
Figure 3. Phylogenetic tree showing CG-1 in relation to other organisms belonging to the genus Lactobacillus. Lactobacillus species include Genbank accession numbers as well as percent alignment to CG-1 using the neighbor joining method on ClustalX.
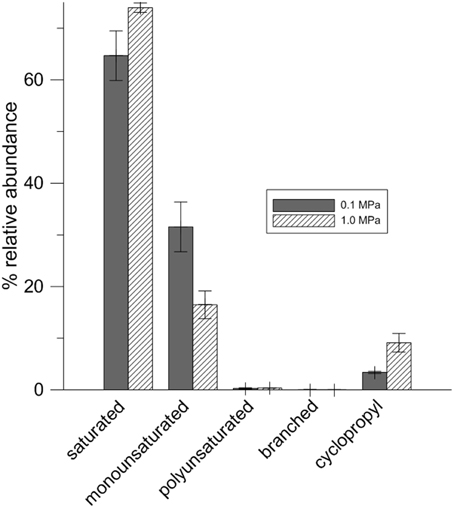
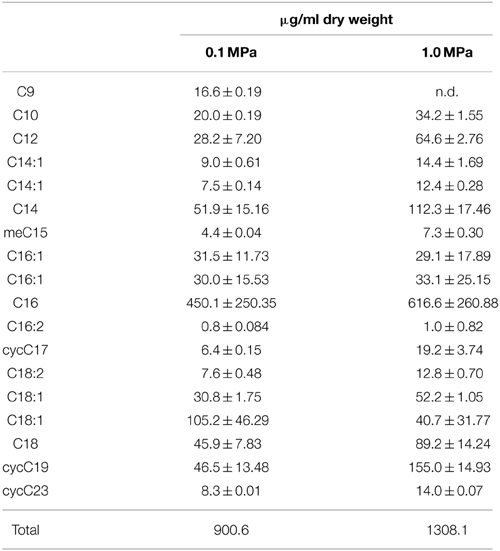
Table 1 lists fatty acids expressed at 0.1 MPa and 1.0 MPa PCO2. While there are more cells per ml of solution present at low PCO2, the amount of fatty acids per mg of biomass is lower than at high PCO2 (Table 1). At low PCO2, cells express a higher relative abundance of monounsaturated fatty acids (MUFA). At high PCO2, this relative abundance decreases and an increase in cyclopropyl (CFA) and saturated fatty acids (SFA) is seen (Figure 4) with cyclopropyl C19 (cycC19) fatty acids showing the largest increase in abundance.
At 0.1 MPa CO2, cells are rod-shaped and elongated, averaging 2 μm long and 400 nm wide with cytoplasm evenly distributed throughout the cells (Figure 5). The cell wall structure suggests that they are Gram-positive, which was verified with a Gram stain. In contrast, TEM micrographs of cultures at grown at 1.0 MPa reveal amorphous cell structures that are rod shaped but trending toward cocci. These cells appear surrounded by extensive capsular material. Additionally, cells contain visible invaginations, suggesting growth and division occurs even under the high PCO2 conditions (Figure 5).
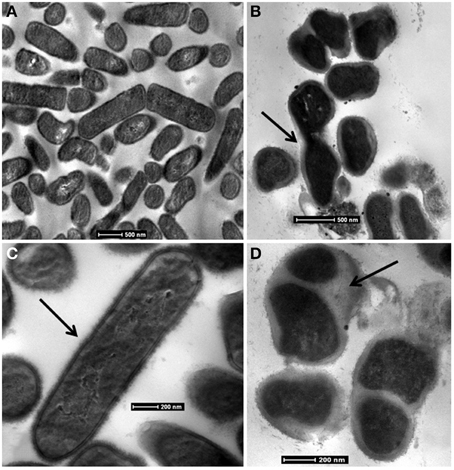
Figure 5. Bright-field TEM images of CG-1 at 0.1 MPa CO2 (A,C) and 1.0 MPa CO2 (B,D). Arrows in: (B) show invaginations in CO2 exposed cells that may suggest cell division; (C) show the intact cell wall for organisms at low CO2exposure; (D) show the capsular material present for CO2 exposed cells.
Isolate Growth and Metabolism
Table 1 also shows a brief summary of the effects the different media amendments had on growth. Growth was similar between atmospheric to 0.4 MPA CO2 pressure Increased CO2 pressures resulted in decreased growth. Cultures grown at 0.1 MPa and amended with the carbonic anhydrase inhibitor acetazolamide appeared unchanged in comparison to cultures grown without inhibitor.
During survival tests, most stationary phase cultures incubated at pressures from 0.2 to 5.0 MPa survived beyond the 24 h limit tested. Cultures at 1.0 MPa have since been recultured and remain viable after 3 weeks of incubation. We also observed cultures exposed to 2.5 MPa to survive at least 5 days of exposure. At 5.0 MPa, only one of the two cultures tested showed viability after 24 h of exposure.
Cultures grew only at temperatures between 25 and 45°C with fastest growth and highest cell density occurring at 25°C (Figure 6B). While there was variability in stationary phase cell counts, similar growth curves were observed between pH 4 and 8 at atmospheric CO2with fastest growth between pH 5 and 8 (Figure 6F). CG-1 also grew under a wide range of salinities. Lag phase increased, and growth decreased at salinities above and below 0.25 M NaCl (Figure 6C). Growth was variable for different carbon sources. However, the most growth was seen with sugars and no growth was seen with organic acids. No measurable growth was found when no carbon source was supplied (Figure 6E). Growth curves look similar for all the electron acceptors tested, though there was an approximate 17%- increase in growth with Fe-citrate (Figure 6D). Color change occurred in TSI slants, indicating pH in the slant cultures decreased and that cultures were producing acid and fermenting the glucose, lactose, and sucrose present in the agar. Furthermore, there was no black precipitate present, indicating no sulfide was produced.
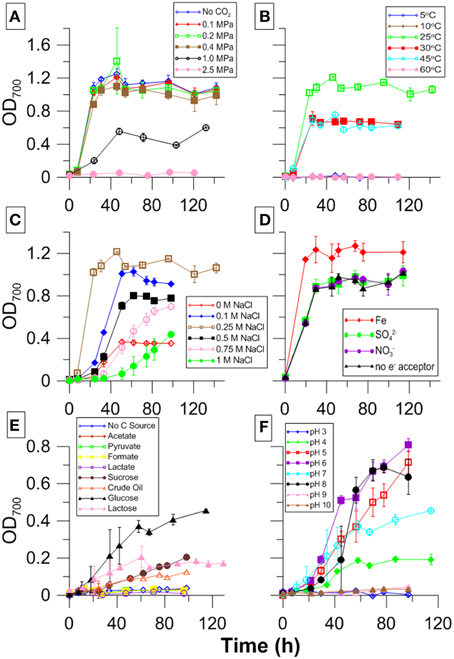
Figure 6. CG-1 physiological data showing growth with varying (A) PCO2, (B) temperature, (C) salinity, (D) electron acceptor, (E) carbon source, and (F) pH.
Discussion
Table S2 lists the composition of CG as well as a calculation of the PCO2 when the water was collected from the surface. Based on calculations, the microbial community sampled at CG was exposed to approximately 0.583 MPa of CO2 though we have been able to culture an organism at 20 times that pressure. The characteristic cycC19 lipid produced by Lactobacilli was detected at CG, though this amount constituted less than 0.1% of the total lipids present. While selection of this organism in the lab indicates a culture bias, enrichment of CG-1 at high CO2 pressures, elevated salinity, high Fe content (through Fe-citrate experiments), and increased temperature suggests this organism can survive in the environmental conditions present within the well. Because CG-1 constitutes only a minor component of the CG microbial community, it is difficult to determine its contribution to the overall geochemistry of CG. However, the microorganisms examined within CG are composed of organisms outwashing from the geyser and do not include organisms found as biofilms along the well casing or attached to pore spaces in the subsurface. As such, it is possible that CG-1 constitutes a greater proportion of the subsurface microbial community than is represented by the sample taken from the well. The source of water coming from Crystal Geyser is a combination of deeply sourced saline water as well as recharge from the nearby San Rafael swell (Baer and Rigby, 1978; Heath et al., 2009; McPherson and Heath, 2009; Burnside et al., 2013). Because recharge to Crystal Geyser may be seasonal, the microbial community of the site is likely dynamic and this could explain why CG-1 was not detected by Emerson et al. (2015). Nevertheless, the detection and cultivation of CG-1 validates the findings of Morozova et al. (2011), Mu et al. (2014), Emerson et al. (2015), Mu and Moreau (2015) showing that organisms present in CO2 environments are viable and can potentially impact geochemistry.
A comparison of CG-1 to its closest cultured relative shows it is 99% identical to Lactobacillus casei strain ATCC 334, indicating this capnophilic isolate represents a new strain of Lactobacillus casei (Figure 3). CG-1′s relation to Lactobacillus casei makes sense in that this organism has demonstrated remarkable adaptability to diverse habitats including raw and fermented dairy products, plant products, intestinal tracts, and reproductive systems (Cai et al., 2007). To this date, CG-1 is the first reported Lactobacillus strain coming from an underground, CO2-rich environment largely because there has been no previous drive to identify and isolate subsurface strains of this organism (Cai et al., 2007). Its minor presence within the greater CG community may also mean other Lactobacilli are present, but in small amounts and may be overlooked. While it is impossible to completely rule out that we have selected for a contaminant due to the unpredictability of fieldwork, we believe our isolate comes from CG itself as negative controls showed no turbidity throughout the course of the experiment, even in the lab. The isolate is also closely related to Lactobacillus zeae, and most distantly related to Lactobacillus plantarum and Lactobacillus suebicus, among the Lactobacillus species.
Many Lactobacilli are defined as capnophilic and fermentative, growing syntrophically with CO2 and acid-producing organisms (Bringel et al., 2008). CO2 enriched environments for cultured Lactobacilli are between 4 and 25% CO2(0.004–0.025 MPa)(Song et al., 2007; Bringel et al., 2008; Arioli et al., 2009). In contrast, CG-1 can grow at 1.0 MPa, or 40 times the CO2 utilized by L. plantarum. Stationary phase cultures can also survive at pressures 200 times that of known capnophilic culture conditions (Figure 6A).
Our isolate grows equally well under CO2 pressures ranging from 0 to 0.4 MPa, shows approximately half that growth at 1.0 MPa, and demonstrates at least one doubling at 2.5 MPa. Its ability to grow non-selectively between 0 and 0.4 MPa suggests the organism may be capnotolerant rather than capnophilic. However, when 3 week-old cultures, which are at long term stationary phase, are recultured, there is a much longer lag phase for samples grown with no CO2 present suggesting CO2 may have some importance in its metabolism. Its versatility in handling multiple CO2 pressures or no CO2 suggests the organism is more capnotolerant than capnophilic.
The Lactobacilli appear to be fairly capnotolerant bacteria and have been rigorously tested as model organisms in the food and biomedical industries in order to determine effective ways to sterilize products. However, to this point, experiments have focused on the CO2 pressures required to kill the organisms using the model organism Lactobacillus plantarum in pressures between 0.04 and 2.5 MPa and beyond. Our work is the first work to examine long-term growth and survival under CO2 stress in high aqueous CO2 concentrations rather than just those of gaseous and supercritical CO2 pressures (e.g., Hong and Pyun, 1999; Guerzoni et al., 2001; Spilimbergo and Bertucco, 2003; Damar and Balaban, 2006; De Vries et al., 2006).
The organism demonstrates a remarkable resilience to CO2 pressures. While growth and cell division appears to slow at 1.0 MPa, the organism can still survive at 2.5 and 5.0 MPa for some time, showing CG-1 and other similar organisms could potentially reside deep in Crystal Geyser. Its survival at these high pressures also shows that under sequestration conditions, many microorganisms can survive The pressures examined in these work only constitute conditions with high aqueous CO2 concentrations and do not consider organisms that are living within the supercritical CO2. It should also be noted that the CO2 used in these experiments was pure CO2. During actual sequestration, the CO2 introduced into the subsurface will contain many other impurities, such as H2S and O2 (Daraboina et al., 2013)which will also have very important consequences to the geochemistry of the subsurface. One possibility for the organism's ability to survive CO2 is through the use of the enzyme carbonic anhydrase, which catalyzes the hydration of CO2 to H2CO3 and then eventually to HCO−3 (Maren, 1967). The enzyme is widespread in nature, found in all domains of life, and is used for multiple reasons. Cyanobacteria utilize it for carbon fixation while organisms like Neisseria and E. coli use it as the first step in amino acid synthesis (Smith and Ferry, 2000). Carbonic anhydrases are also inhibited by sulfonamides such as acetazolamide (Smith and Ferry, 2000). However, when grown under high PCO2 and supplemented with 1 mM of acetazolamide, CG-1 showed no change in growth, suggesting either this organism's form of carbonic anhydrase is resistant to acetazolamide or that this organism contains another method for CO2 utilization not yet previously described. Some other possible enzymes that may aid in CO2 survival are phosphoenolpyruvate carboxylase and carbamoyl-phosphate synthetase, both enzymes previously described for metabolite synthesis in bacteria (Arioli et al., 2009; Banks et al., 2010).
Based on culture experiments, it appears acidity caused by CO2 dissolution may be one component that can inhibit growth. The pH of growth media used to culture CG-1 in equilibrium with 2.5 MPa CO2 can reach pH 4.3 as inferred by geochemical modeling. CG-1 showed optimal growth between pH values of 5 and 8, though growth is still observed at pH values of 4 (Figure 6F). At CO2 pressures between 0.1 and 1.0 MPa, pH ranges between 5.6 and 4.7. At greater than 2.5 MPa, no growth was seen possibly as a combination of CO2 and acid stress, although the pH differences between 1.0 and 2.5 MPa do not vary substantially, and stay between pH values of 4 and 5, as shown by geochemical modeling (Bethke, 1996). Pressure may also have an influence in cell survival, though previous work has shown organisms to be piezotolerant under extremely high pressures (Sharma et al., 2002; Santillan et al., 2013).
Many Lactobacilli are fermentative, and CG-1 shows evidence of this. We had initially thought that CG-1 had a remarkable versatility in electron acceptor utilization as it grew in PRAS media containing Fe3+, SO2−4, and NO−3. However, the organism did not reduce any of these TEAs and also grew in media without TEAs showing its growth in anaerobic conditions is fermentative. Its inability to grow in media without a carbon source indicates it is not performing peptone catabolism. Evidence from TSI slants corroborate its fermentative capabilities through its ability to ferment glucose, lactose, and sucrose. Furthermore, the color change from red to yellow throughout the slants suggests it produces acid during fermentation, showing it is likely performing lactic acid fermentation. In addition, CG-1 is unable to grow in many of the byproducts resultant from fermentation including actetate, formate, and lactate (Figure 6E).
Growth was best on glucose, sucrose, and Teapot Dome crude oil, all long-chained carbon molecules (Figure 6E). Its ability to grow on complex carbon sources is consistent with other Lactobacilli, which ferment molecules like sugars and starches (De Vries et al., 2006; Cai et al., 2007). Its ability to grow well with crude oil also further indicates its environmental relevance to CG as many oil seeps are present in the surrounding environment that may provide a carbon source to the subsurface microbial populations. Many fermentative organisms have been found and isolated from oil fields degrading hydrocarbons and contributing to the indigenous microbial communities they are part of Onstott et al. (1998), Winderl et al. (2008), Duncan et al. (2009).
Unstressed CG-1 cells have an appearance similar to other Lactobacilli. CG-1 is rod-shaped and contains a distinct cell wall (Figure 5). It is Gram-positive and produces no endospores based on TEM observations. Cells grown at 0.1 MPa PCO2 appear elongated and turgid. At 1.0 MPa PCO2, they are slightly amorphous in appearance. The images at 1.0 MPa capture cells in multiple life stages, with some cells appearing like they are degrading while many other cells are attached together and contain invaginations. These invaginations have previously been observed as stress responses, and many factors such as cytoplasmic CO2 disruption, media acidification, or hydrostatic pressure, may contribute to this appearance (e.g., Zobell and Oppenheimer, 1950). Cells grown at 1.0 MPa are surrounded by a capsule material, likely composed of extracellular polymeric substances (EPS). This could have multiple purposes. Many cells responding to environmental stresses, such as acidity, produce EPS to create a physical barrier to slow the diffusion of the toxic substance into the cell, which in this case would be CO2 (Cotter and Hill, 2003; Kidd, 2011). The presence of EPS could also be a means for cells to maintain cell integrity and structure in the presence of a toxic substance (Flemming and Wingender, 2010).
CG-1 exhibited variable lipid compositions at 0.1 and 1.0 MPa of CO2, demonstrating a physiological response to CO2 pressures (Figure 4, Table 2). Interestingly, while cell density at 1.0 MPa was lower than at 0.1 MPa CO2, there was a higher lipid to biomass ratio for cells at 1.0 MPa (Table 1). This ratio could either mean cells are creating thicker cell membranes or that cell sizes are decreasing. Observations by TEM would be more consistent with a cell size decrease. Bacteria demonstrate cell size decrease during starvation (Chien et al., 2012) and smaller cell sizes for CO2 stressed CG-1 may indicate a state similar to starvation. While the same nutrients at 1.0 MPa are present at 0.1 MPa, increased CO2 toxicity may be inhibiting nutrient uptake, resulting in smaller cells.
Cells grown at 0.1 and 1.0 MPa shared similar lipid compositions, with the highest variability coming from C14, C16, and C18 fatty acids in both treatments, possibly owing to the variability of fatty acid compositions present for cells in stationary phase. However, the relative abundance of different fatty acids change with increased PCO2. At low PCO2, lipid composition favors MUFA. At high PCO2, this relative abundance decreases and an increase in SFA and CFA occurs. The difference in lipid composition at different CO2 pressures may reflect structural changes cells must make to maintain cell shape or fluidity under stress. SFAs, for example, pack together much more tightly than MUFAs, which would be consistent with a decrease in cell size under high PCO2. Such responses have been documented for temperature, pH, hydrostatic pressure, and osmotic stresses but not for CO2 (To and Etzel, 1997; De Vries et al., 2006; Bringel et al., 2008).
Cyclopropyl C19 fatty acids, or lactobacillic acid, is characteristic of Lactobacillus species (Veerkamp, 1971). Previous work with different Lactobacilli has shown that under stress, Lactobacillus converts unsaturated fatty acids into CFA (Guerzoni et al., 2001). With CG-1 at 1.0 MPa, there is a decrease in the relative abundance of unsaturated fatty acids along with an increase in the relative abundance of CFA. CFAs are energetically costly for cells to produce and are often found during the transition from growth to stationary phase, as a response to starvation, and as a response to acidic conditions (Brown et al., 1997; Chang and Cronan, 1999). Here, we document another stress response that generates an increase in CFA.
CFAs increase rigidity in cell membranes (Veerkamp, 1971; Brown et al., 1997; Chang and Cronan, 1999; Guerzoni et al., 2001). TEM micrographs show that cultures at high PCO2 appear smaller, more amorphous, and less turgid than cultures exposed to lower PCO2. The presence of molecules such as CFA in the cell membrane may be a way to maintain some form of rigidity for cell shape. In high PCO2 cultures, this relative increase suggests cells are actively responding to CO2 despite the energy costs possibly by increasing the efficiency of maintaining cell structure at the expense of other energetically costly activities such as cell division (Santillan et al., 2013).
Although CG-1 makes up a minor component of the overall Crystal Geyser microbial community, its isolation and cultivation demonstrates that capnotolerant organisms can exist in the subsurface environment. Our work demonstrates that microorganisms can survive in CO2 rich environments. Using an alternate approach to other studies, we are can confirm that that organisms that have been detected through sequencing and molecular methods are also viable and can potentially impact the geochemistry of CO2 seqeustration.
Organism Description
Lactobacillus casei strain CG-1 (Genbank accession number KJ493942) is a Gram-positive, non-sporulating, facultative anaerobic bacterium isolated from Crystal Geyser, Utah. The organism is fermentative, putatively converting glucose, sucrose, or long-chained hydrocarbons into lactate and producing no CO2, acid, or sulfide, during this process. The bacterium grows optimally at 25°C in 100% CO2 atmospheres, and pressures ranging from 0.1 to 0.4 MPa CO2, with less growth at 1.0 MPa, and no growth but continued survival at 2.5 MPa. Stock cultures recultured after long-term stationary phase exhibit shorter lag phase when supplied with CO2 indicating a metabolic need for CO2. The organism is pH sensitive, growing between pH values of 6 and 8, and exhibits a longer lag phase under acidic and basic conditions. Its optimal salinity is 0.25 M NaCl, though it shows versatility in that it can grow between 0 and 1 M NaCl. It expresses a suite of fatty acids, but most notably cycC19, especially under extreme CO2 conditions. The organism is rod-shaped at 0.1 MPa PCO2 but begins to surround itself with a thick capsule structure and exhibits a somewhat amorphous morphology when cultured under 1.0 MPa CO2 pressures. The organism is currently stored at the University of Texas at Austin, Jackson School of Geosciences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This material is based upon work supported as part of the Centers for Frontiers in Subsurface Energy Security, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC0001114. We would also like to thank Kim Gilbert, Bayani Cardenas, Susan Altman, and Matt Kirk for their consultation on high pressure geochemistry; Will Wolfe and Tiffany Kocis for their invaluable contribution to the field work at Crystal Geyser; Veronica Anderson and Vera Stoynova for their assistance in lipid extraction and mass spectrometry; as well as the members of UT's Microbial Geochemistry group for their support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/feart.2015.00041
Table S1. Sequences and BLAST percent identities of CG-1 clones.
Table S2. Crystal Geyser fluid composition. All values are in mmol/L unless stated otherwise.
References
Arioli, S., Roncada, P., Salzano, A. M., Deriu, F., Corona, S., Guglielmetti, S., et al. (2009). The relevance of carbon dioxide metabolism in Streptococcus thermophilus. Microbiology 155, 1953–1965. doi: 10.1099/mic.0.024737-0
Baer, J. L., and Rigby, J. K. (1978). Geology of the Crystal Geyser and environmental implications of its effluent, Grand County, Utah. Utah Geol. 5, 125–130.
Ballestra, P., Dasilva, A. A., and Cuq, J. L. (1996). Inactivation of Escherichia coli by carbon dioxide under pressure. J. Food Sci. 61, 829–836. doi: 10.1111/j.1365-2621.1996.tb12212.x
Banks, E. D., Taylor, N. M., Gulley, J., Lubbers, B. R., Giarrizo, J. G., Bullen, H. A., et al. (2010). Bacterial calcium carbonate precipitation in cave environments: a function of calcium homeostasis. Geomicrobiol. J. 27, 444–454. doi: 10.1080/01490450903485136
Bringel, F., Hammann, P., Kugler, V., and Arsène-Ploetze, F. (2008). Lactobacillus plantarum response to inorganic carbon concentrations: PyrR2-dependent and -independent transcription regulation of genes involved in arginine and nucleotide metabolism. Microbiology 154, 2629–2640. doi: 10.1099/mic.0.2008/018184-0
Brown, J. L., Ross, T., McMeekin, T. A., and Nichols, P. D. (1997). Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance. Int. J. Food Microbiol. 37, 163–173. doi: 10.1016/S0168-1605(97)00068-8
Burnside, N. M., Shipton, Z. K., Dockrill, B., and Ellam, R. M. (2013). Man-made versus natural CO2 leakage: a 400 k.y. history of an analogue for engineered geological storage of CO2. Geology 41, 471–474. doi: 10.1130/g33738.1
Cai, H., Rodriguez, B. T., Zhang, W., Broadbent, J. R., and Steele, J. L. (2007). Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology 153, 2655–2665. doi: 10.1099/mic.0.2007/006452-0
Chang, Y. Y., and Cronan, J. E. Jr. (1999). Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 33, 249–259. doi: 10.1046/j.1365-2958.1999.01456.x
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G., et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. doi: 10.1093/nar/gkg500
Chien, A.-C., Hill, N. S., and Levin, P. A. (2012). Cell size control in bacteria. Curr. Biol. 22, R340–R349. doi: 10.1016/j.cub.2012.02.032
Cotter, P. D., and Hill, C. (2003). Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67, 429–453. doi: 10.1128/MMBR.67.3.429-453.2003
Damar, S., and Balaban, M. O. (2006). Review of dense phase CO2 technology: microbial and enzyme inactivation, and effects on food quality. J. Food Sci. 71, R1–R11. doi: 10.1111/j.1365-2621.2006.tb12397.x
Daraboina, N., Ripmeester, J., and Englezos, P. (2013). The impact of SO2 on post combustion carbon dioxide capture in bed of silica sand through hydrate formation. Int. J. Greenhouse Gas Control 15, 97–103. doi: 10.1016/j.ijggc.2013.02.008
De Vries, M. C., Vaughan, E. E., Kleerebezem, M., and De Vos, W. M. (2006). Lactobacillus plantarum—survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 16, 1018–1028. doi: 10.1016/j.idairyj.2005.09.003
Duncan, K. E., Gieg, L. M., Parisi, V. A., Tanner, R. S., Tringe, S. G., Bristow, J., et al. (2009). Biocorrosive thermophilic microbial communities in Alaskan Worth Slope Oil Facilities. Environ. Sci. Technol. 43, 7977–7984. doi: 10.1021/es9013932
Emerson, J. B., Thomas, B. C., Alvarez, W., and Banfield, J. F. (2015). Metagenomic analysis of a high carbon dioxide subsurface microbial community populated by chemolithoautotrophs and bacteria and archaea from candidate phyla. Environ. Microbiol. doi: 10.1111/1462-2920.12817. [Epub ahead of print].
Engel, A. S. (2004). Geomicrobiology of Sulfuric Acid Speleogenesis: Microbial Diversity, Nutrient Cycling, and Controls on Cave Formation. Ph.D. The University of Texas at Austin.
Erkmen, O. (2000). Effect of carbon dioxide pressure on Listeria monocytogenes in physiological saline and foods. Food Microbiol. 17, 589–596. doi: 10.1006/fmic.2000.0361
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Gosden, P. E., and Ware, G. C. (1977). Method for production of pre-reduced anaerobically sterilized culture media. J. Appl. Bacteriol. 42, 77–79. doi: 10.1111/j.1365-2672.1977.tb00671.x
Guerzoni, M. E., Lanciotti, R., and Cocconcelli, P. S. (2001). Alteration in cellular fatty acid composition as a response to salt, acid, oxidative and thermal stresses in Lactobacillus helveticus. Microbiology 147, 2255–2264. doi: 10.1099/00221287-147-8-2255
Heath, J. E., Lachmar, T. E., Evans, J. P., Kolesar, P. T., and Williams, A. P. (2009). Hydrogeochemical characterization of leaking, carbon dioxide-charged fault zones in east-central Utah, with implications for geological carbon storage. Geophys. Monogr. 183, 147–158. doi: 10.1029/2006GM000407
Hertzberger, R. Y., Pridmore, R. D., Gysler, C., Kleerebezem, M., and De Mattos, M. J. T. (2013). Oxygen relieves the CO2 and acetate dependency of Lactobacillus johnsonii NCC 533. PLoS ONE 8:e57235. doi: 10.1371/journal.pone.0057235
Hong, S. I., and Pyun, Y. R. (1999). Inactivation kinetics of Lactobacillus plantarum by high pressure carbon dioxide. J. Food Sci. 64, 728–733. doi: 10.1111/j.1365-2621.1999.tb15120.x
Isenschmid, A., Marison, I. W., and Von Stockar, U. (1995). The influence of pressure and temperature of compressed CO2 on the survival of yeast cells. J. Biotechnol. 39, 229–237. doi: 10.1016/0168-1656(95)00018-L
Kaszuba, P. J., and Janecky, R. D. (2009). Geochemical Impacts of Sequestering Carbon Dioxide in Brine Formations. Washington, DC, ETATS-UNIS: American Geophysical Union.
Kharaka, Y. K., Cole, D. R., Hovorka, S. D., Gunter, W. D., Knauss, K. G., and Freifeld, B. M. (2006). Gas-water-rock interactions in Frio Formation following CO2 injection: implications for the storage of greenhouse gases in sedimentary basins. Geology 34, 577–580. doi: 10.1130/G22357.1
Kirk, M. F., Santillan, E. F. U., Sanford, R. A., and Altman, S. J. (2013). CO2-induced shift in microbial activity affects carbon trapping and water quality in anoxic bioreactors. Geochim. Cosmochim. Acta 122, 198–208. doi: 10.1016/j.gca.2013.08.018
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Little, M. G., and Jackson, R. B. (2010). Potential impacts of leakage from deep CO2 geosequestration on overlying freshwater aquifers. Environ. Sci. Technol. 44, 9225–9232. doi: 10.1021/es102235w
Maren, T. H. (1967). Carbonic anhydrase - chemistry, physiology, and inhibition. Physiol. Rev. 47, 595–781.
McPherson, B. J., and Heath, J. (2009). Self-sealing of faults by CO2-rich fluids. Geochim. Cosmochim. Acta 73, A861–A861.
Mitchell, A. C., Phillips, A. J., Hamilton, M. A., Gerlach, R., Hollis, W. K., Kaszuba, J. P., et al. (2008). Resilience of planktonic and biofilm cultures to supercritical CO2. J. Supercritical Fluids 47, 318–325. doi: 10.1016/j.supflu.2008.07.005
Morozova, D., Zettlitzer, M., Let, D., and Würdemann, H. (2011). Monitoring of the microbial community composition in deep subsurface saline aquifers during CO2 storage in Ketzin, Germany. Energy Procedia 4, 4362–4370. doi: 10.1016/j.egypro.2011.02.388
Mu, A., Boreham, C., Leong, H. X., Haese, R. R., and Moreau, J. W. (2014). Changes in the deep subsurface microbial biosphere resulting from a field-scale CO2 geosequestration experiment. Front. Microbiol. 5:209. doi: 10.3389/fmicb.2014.00209
Mu, A., and Moreau, J. W. (2015). The Geomicrobiology of CO2 geosequestration: a focused review on prokaryotic community responses to field-scale CO2 injection. Front. Microbiol. 6:263. doi: 10.3389/fmicb.2015.00263
Nakamura, K., Enomoto, A., Fukushima, H., Nagai, K., and Hakoda, M. (1994). Disruption of microbial-cells by the flash discharge of high-pressure carbon dioxide. Biosci. Biotechnol. Biochem. 58, 1297–1301. doi: 10.1271/bbb.58.1297
Onstott, T. C., Phelps, T. J., Colwell, F. S., Ringelberg, D., White, D. C., Boone, D. R., et al. (1998). Observations pertaining to the origin and ecology of microorganisms recovered from the deep subsurface of Taylorsville Basin, Virginia. Geomicrobiol. J. 15, 353–385. doi: 10.1080/01490459809378088
Oule, M., Dickman, M., and Arul, J. (2010). Microbicidal effect of pressurized CO2 and the influence of sensitizing additives. Eur. J. Sci. Res. 41, 569–581.
Oule, M. K., Tano, K., Bernier, A. M., and Arul, J. (2006). Escherichia coli inactivation mechanism by pressurized CO2. Can. J. Microbiol. 52, 1208–1217. doi: 10.1139/w06-078
Peet, K. C., Freedman, A. J. E., Hernandez, H. H., Britto, V., Boreham, C., Ajo-Franklin, J. B., et al. (2015). Microbial growth under supercritical CO2. Appl. Environ. Microbiol. 81, 2881–2892. doi: 10.1128/AEM.03162-14
Rodriguez-Ruiz, J., Belarbi, E. H., Sanchez, J. L. G., and Alonso, D. L. (1998). Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol. Tech. 12, 689–691. doi: 10.1023/A:1008812904017
Santillan, E. U., Kirk, M. F., Altman, S. J., and Bennett, P. C. (2013). Mineral influence on microbial survival during carbon sequestration. Geomicrobiol. J. 30, 578–592. doi: 10.1080/01490451.2013.767396
Sharma, A., Scott, J. H., Cody, G. D., Fogel, M. L., Hazen, R. M., Hemley, R. J., et al. (2002). Microbial activity at gigapascal pressures. Science 295, 1514–1516. doi: 10.1126/science.1068018
Shimoda, M., Yamamoto, Y., Cocunubo-Castellanos, J., Tonoike, H., Kawano, T., Ishikawa, H., et al. (1998). Antimicrobial effects of pressured carbon dioxide in a continuous flow system. J. Food Sci. 63, 709–712. doi: 10.1111/j.1365-2621.1998.tb15819.x
Smith, K. S., and Ferry, J. G. (2000). Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24, 335–366. doi: 10.1111/j.1574-6976.2000.tb00546.x
Song, H., Lee, J. W., Choi, S., You, J. K., Hong, W. H., and Lee, S. Y. (2007). Effects of dissolved CO2 levels on the growth of Mannheimia succiniciproducens and succinic acid production. Biotechnol. Bioeng. 98, 1296–1304. doi: 10.1002/bit.21530
Spilimbergo, S., and Bertucco, A. (2003). Non-thermal bacteria inactivation with dense CO2. Biotechnol. Bioeng. 84, 627–638. doi: 10.1002/bit.10783
Stroes-Gascoyne, S., Schippers, A., Schwyn, B., Poulain, S., Sergeant, C., Simonoff, M., et al. (2007). Microbial community analysis of Opalinus Clay drill core samples from the Mont Terri Underground Research Laboratory, Switzerland. Geomicrobiol. J. 24, 1–17. doi: 10.1080/01490450601134275
To, B. C. S., and Etzel, M. R. (1997). Spray drying, freeze drying, or freezing of three different lactic acid bacteria species. J. Food Sci. 62, 576–585. doi: 10.1111/j.1365-2621.1997.tb04434.x
Ueda, K., Tagami, Y., Kamihara, Y., Shiratori, H., Takano, H., and Beppu, T. (2008). Isolation of bacteria whose growth is dependent on high levels of CO2 and implications of their potential diversity. Appl. Environ. Microbiol. 74, 4535–4538. doi: 10.1128/AEM.00491-08
Veerkamp, J. H. (1971). Fatty acid composition of Bifidobacterium and Lactobacillus strains. J. Bacteriol. 108, 861–867.
Watanabe, T., Furukawa, S., Hirata, J., Koyama, T., Ogihara, H., and Yamasaki, M. (2003). Inactivation of Geobacillus stearothermophilus spores by high-pressure carbon dioxide treatment. Appl. Environ. Microbiol. 69, 7124–7129. doi: 10.1128/AEM.69.12.7124-7129.2003
Wilkinson, M., Gilfillan, S. V. M., Haszeldine, R. S., and Ballentine, C. J. (2009). “Plumbing the depths: testing natural tracers of subsurface CO2 origin and migration, Utah,” in Carbon Dioxide Sequestration in Geological Media-State of the Science, Volume AAPG Studies in Geology, eds M. Grobe, J. C. Pashin and R. L. Dodge (Tulsa, OK: American Association of Petroleum Geologists), 619–634.
Winderl, C., Anneser, B., Griebler, C., Meckenstock, R. U., and Lueders, T. (2008). Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar oil contaminant plume. Appl. Environ. Microbiol. 74, 792–801. doi: 10.1128/AEM.01951-07
Wynne, E. S., and Foster, J. W. (1948). Physiological studies on spore germination, with special reference to Clostridium botulinum: 3. Carbon dioxide and germination, with a note on carbon dioxide and aerobic spores. J. Bacteriol. 55, 331–339.
Keywords: CO2 sequestration, Lactobacillus, capnophile, CO2, Crystal Geyser, fermenter
Citation: Santillan E-FU, Shanahan TM, Omelon CR, Major JR and Bennett PC (2015) Isolation and characterization of a CO2-tolerant Lactobacillus strain from Crystal Geyser, Utah, U.S.A. Front. Earth Sci. 3:41. doi: 10.3389/feart.2015.00041
Received: 06 March 2015; Accepted: 10 July 2015;
Published: 23 July 2015.
Edited by:
Jennifer Glass, Georgia Institute of Technology, USAReviewed by:
John W. Moreau, University of Melbourne, AustraliaJoanne B. Emerson, University of California, Berkeley, USA
Copyright © 2015 Santillan, Shanahan, Omelon, Major and Bennett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eugenio-Felipe U. Santillan, Smithsonian Environmental Research Center, 647 Contees Wharf Rd., Edgewater, MD 21037, USA, efu.santillan@gmail.com
 Eugenio-Felipe U. Santillan
Eugenio-Felipe U. Santillan Timothy M. Shanahan
Timothy M. Shanahan Christopher R. Omelon
Christopher R. Omelon Jonathan R. Major
Jonathan R. Major Philip C. Bennett1
Philip C. Bennett1