Snowmelt Induced Hydrologic Perturbations Drive Dynamic Microbiological and Geochemical Behaviors across a Shallow Riparian Aquifer
- 1Department of Microbiology, The Ohio State University, Columbus, OH, USA
- 2Hydrology Group, Pacific Northwest National Laboratory, Richland, WA, USA
- 3Lawrence Berkeley National Laboratory, Earth Sciences Division, Berkeley, CA, USA
- 4School of Earth Sciences, The Ohio State University, Columbus, OH, USA
Shallow riparian aquifers represent hotspots of biogeochemical activity in the arid western US. While these environments provide extensive ecosystem services, little is known of how natural environmental perturbations influence subsurface microbial communities and associated biogeochemical processes. Over a 6-month period we tracked the annual snowmelt-driven incursion of groundwater into the vadose zone of an aquifer adjacent to the Colorado River, leading to increased dissolved oxygen (DO) concentrations in the normally suboxic saturated zone. Strong biogeochemical heterogeneity was measured across the site, with abiotic reactions between DO and sulfide minerals driving rapid DO consumption and mobilization of redox active species in reduced aquifer regions. Conversely, extensive DO increases were detected in less reduced sediments. 16S rRNA gene surveys tracked microbial community composition within the aquifer, revealing strong correlations between increases in putative oxygen-utilizing chemolithoautotrophs and heterotrophs and rising DO concentrations. The gradual return to suboxic aquifer conditions favored increasing abundances of 16S rRNA sequences matching members of the Microgenomates (OP11) and Parcubacteria (OD1) that have been strongly implicated in fermentative processes. Microbial community stability measurements indicated that deeper aquifer locations were relatively less affected by geochemical perturbations, while communities in shallower locations exhibited the greatest change. Reactive transport modeling of the geochemical and microbiological results supported field observations, suggesting that a predictive framework can be applied to develop a greater understanding of such environments.
Introduction
Riparian aquifers are key hotspots of biogeochemical activity in regions of the arid inner mountain western US that are predicted to be directly influenced by future climate-change induced shifts in hydrology (Carlyle and Hill, 2001; McClain et al., 2003; Heffernan and Sponseller, 2004). Such aquifers have been implicated in carbon, nitrogen (Jacinthe et al., 2003; Peter et al., 2012), and metal cycling (Williams et al., 2011), and therefore contribute to water quality discharged into stream and river networks. Microbial nitrate reduction has been reported in riparian aquifer materials in laboratory studies (Jacinthe et al., 2003) and in situ (Peter et al., 2012), while microbial attenuation of landfill leachate is an active process in the riparian aquifer at a USGS site near Norman, OK (Scholl et al., 2005). Despite the variety of ecosystem functions contributed by riparian aquifers, the extent to which geochemical and microbiological parameters in these environments fluctuate in response to transient hydrologic perturbations is mostly unknown.
In general, riparian aquifers and river networks are intricately linked through hyporheic flow. In many mountainous regions, seasonal snowmelt controls much of the fluctuation in river discharge, and therefore interactions with groundwater in riparian zones. Future inputs of snowmelt (and other precipitation) in this region are predicted to change across a series of regional climate models, through increases in extreme winter precipitation and mean winter precipitation (Dominguez et al., 2012). In turn, these precipitation shifts will affect hydrologic processes within adjoining aquifers, with implications for ecosystem services provided by these environments (Arora et al., 2015). In a shallow riparian aquifer adjacent to the Colorado River near Rifle, CO, seasonal deviations in water table height have been linked to snowmelt driven changes in river stage. Although little river water intrudes into the aquifer during river stage rise, the hydrologic gradient is lowered across the site, resulting in a rising water table and groundwater incursion into the vadose zone. This incursion drives increased dissolved oxygen (DO) concentrations in groundwater via entrapment of O2-containing gas bubbles within the capillary fringe and vadose zone and likely catalyzes a range of redox sensitive reactions. Currently, this natural hydrologic process represents a predictable, annual perturbation within the normally suboxic aquifer. Under the future climate change scenarios previously described however, this perturbation may occur in a less predictable manner.
While suboxic conditions are frequently found across the majority of the site (Williams et al., 2011), heterogeneously distributed naturally reduced zones (“NRZs”) have been identified where fine grained minerals (e.g., silts, clays) associate with organic carbon deposits, generating highly reducing conditions characterized by an abundance of metal sulfide minerals such as pyrite and mackinawite (Qafoku et al., 2009, 2014; Campbell et al., 2012). Through geophysical measurements, Wainwright et al. (2015) has estimated that ~3% of the aquifer volume is composed of NRZ-type sediments. Coupled to these mineralogical and geochemical observations, community genomic tools have been used to identify key microbial groups likely catalyzing many of the redox reactions that contribute to overall ecosystem functions within the aquifer (Wrighton et al., 2012, 2014; Castelle et al., 2013, 2015; Kantor et al., 2013; Hug et al., 2015). However, most of these studies have occurred over limited temporal scales, and have focused on highly reduced regions of the aquifer. Here, we used a transect of groundwater sampling wells to track the effects of snowmelt driven hydrologic processes at the local floodplain scale on microbial community structure and biogeochemical cycling in riparian subsurface zones over a 6-month period. Our sampling period captured a hydrologic pulse perturbation within the aquifer and enabled an assessment of biogeochemical responses. Such processes are likely to be directly impacted by predicted future climate change scenarios (such as increases in extreme precipitation events and prolonged lowering of groundwater elevations) in the Rocky Mountains.
Materials and Methods
Site Description and Sampling
Groundwater samples were collected from a shallow, alluvial aquifer located near Rifle, CO, USA. An extensive site description can be found in Williams et al. (2011). Materials for geochemical and microbiological analyses were collected across a series of multi-level sampling wells installed on the floodplain. Wells CMT01 (BND location), CMT02 (MID location), and CMT03 (NRZ location) represent a north-northeast to south-southwest transect that follows the seasonally average groundwater flow direction across the site (Figure 1). Each sampling well enabled recovery of groundwater at discrete 13-, 16-, and 19-ft depths below ground surface. Samples were collected weekly over a 6-month period, between 3rd April 2014 and 12th September 2014. One additional set of samples was collected in January 2015. This time period captured the rise and fall of the water table at the site, linked to the changes in river stage in the adjacent Colorado River (Supplementary Figure 1). Prior to sampling, each well was purged of standing water by pumping ~5 liters of groundwater per sampling depth. Samples for anion analysis were filtered through a PTFE 0.2 μm filter (Pall, NY, USA) and stored in refrigerated, no-headspace HDPE vials until analysis. Samples for cation analysis were filtered and preserved with trace metal grade 12 N HNO3. Samples for organic and inorganic carbon analyses were filtered, stored and refrigerated in 40 mL glass vials. Microbial biomass was recovered by passing ~5 liters of groundwater through a Sterivex filter (0.2 μm; Millipore, MA, USA). Filters for microbiological analyses were stored at −80°C until DNA extraction.
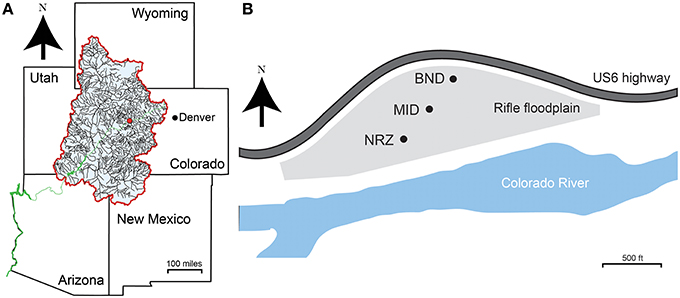
Figure 1. (A) Site location (red dot) in the context of the upper Colorado River basin (blue shaded area). The Colorado River is shown in green. (B) Locations of the three sampling wells across the Rifle aquifer used in this study. CMT01 (BND) and CMT02 (MID) are separated by ~100 m, while CMT02 (MID) and CMT03 (NRZ) are separated by ~70 m.
Geochemical Analyses
Chloride and sulfate were measured using an ion chromatograph (ICS-2100, Dionex, CA, USA) equipped with AS-18 analytical and guard columns. Cation concentrations were determined using inductively coupled plasma mass spectrometry (ICP-MS; Element 2, Thermo Fisher, MA, USA). Water table height was measured using pressure transducers emplaced in all wells. Dissolved oxygen was measured using multi-parameter sondes that were deployed down the wells (YSI, Yellow Springs, OH, USA). The sondes were calibrated at regular intervals throughout the experiment period. Dissolved organic carbon concentrations were measured as non-purgeable organic carbon (NPOC), the difference between total dissolved carbon (TC), and dissolved inorganic carbon (DIC). All measurements were performed on a TOC-LCPN instrument (Shimadzu, MD, USA). Acid volatile- and chromium-reducible sulfide was extracted from sediments collected during well installation using the method of Ulrich et al. (1997) and subsequently quantified using the methylene blue method (Hach Co., Loveland, CO, USA). All geochemical data collected in this study is available in Supplementary Table 1.
16S rRNA Gene Amplicon Sequencing
DNA was extracted from Sterivex filters using the PowerSoil DNA Isolation Kit (MoBio Laboratories, Inc., Carlsbad, CA, USA). The V4 region of 16S rRNA was amplified and sequenced using the bacterial/archaeal primer set 515F/806R at Argonne National Laboratory on the Illumina MiSeq platform. Resulting reads (~9 million) were checked for chimeras (USEARCH 61 algorithm) and remaining reads (~8.5 million) were clustered into OTU classifications at 97% similarities (open-reference picking) using the QIIME pipeline (V1.7.0) and SILVA 16S rRNA database (Caporaso et al., 2010). Sequences classified as unassigned were aligned against a second database, consisting of full-length 16S rRNA sequences from the Candidate Phyla Radiation (CPR). These sequences were reconstructed from Rifle aquifer metagenomic data (Brown et al., 2015; accession number SRP050083). The 16S rRNA gene amplicon library for this study is available through NCBI under SRX1713929.
Data Analyses
Alpha diversity, as defined by multiple diversity and richness indices (Shannon, 1948; Simpson, 1949; Chao, 1984), was determined by utilizing the alpha_diversity.py script within QIIME. Beta diversity was determined using Bray-Curtis dissimilarity (Bray and Curtis, 1957). From these dissimilarities, community differences between well location and depth were examined in R v3.1.2 through NMDS (metaMDS), and compared using global and pairwise ANOSIM (anosim, vegan package v2.2-1; Oksanen et al., 2015). Overall correlative relationships between geochemical parameters (Fe, Mn, U, V, NPOC) were determined in R using a Mantel Test (mantel, vegan package v2.2-1; Oksanen et al., 2015) and subsequently analyzed in more detail using Canonical Correspondence Analysis (CCA) (vegan package v2.2-1; Oksanen et al., 2015). Linear regression correlations were performed using a script written for MATLAB (The MathWorks, Inc., Natick, MA, USA), which allowed for the depth to water table measurement to be compared to the relative abundances of individual family-level groupings of OTUs (>1% relative abundance). The resulting significant correlations were then plotted as a heatmap (gplots package, vegan package, Heatplus package) showing the magnitude of the response rather than that of the correlation. An analysis of similarity percentages (SIMPER) (simper, vegan package v2.2-1; Oksanen et al., 2015) was performed on the microbial community data to determine which members contributed greatly to observed structural differences.
Ecological parameters were measured by calculating the Bray-Curtis dissimilarity between 16S rRNA gene amplicon libraries for each sample, and the Euclidean distance between geochemical measurements (total Fe, Mn, U, V, NPOC) at each time point. In order to test, and then compare stability between well locations and depths, a beta dispersion test for heterogeneity was performed (betadisper, vegan package v2.2-1; Oksanen et al., 2015). Variance among group stability was determined by an ANOVA test while significant differences between groups were determined by pairwise t-tests with a Benjamini-Hochberg correction (vegan package v2.2-1; Oksanen et al., 2015). A rate of change measure was calculated before and after the maximum perturbation by measuring the slope of Bray-Curtis dissimilarities plotted against time (Shade et al., 2012).
Modeling
Modeling results used in this study were taken from a three-dimensional variably saturated flow and reactive transport simulation of the Old Rifle floodplain aquifer using the eSTOMP simulator (Yabusaki et al., 2011). The Richards equation flow modeling is driven by time-dependent water levels measured in the Colorado River and upgradient wells during 2014. The modeled geochemical conditions are based on groundwater chemistry and sediment extractions at monitoring well locations across the floodplain aquifer. A biogeochemical reaction network that includes pH, DO, Fe, sulfate, FeS mineral, organic, and inorganic carbon was employed with functional representation of fermentative bacteria, and sulfate reducing bacteria. Biomass specific to the community metabolic functions was subject to growth and decay similar to those in Yabusaki et al. (2011). Time- and depth-dependent biogeochemical concentrations from model locations corresponding to wells MID and NRZ were used to compare with observed biogeochemistry.
The biogeochemical modeling framework addresses dynamic hydrologic-mineralogic interactions. The biogeochemical response to water table rise and DO incursion reflects the reactivity of the local sediment facies. Two distinctive facies have been identified: (1) a background sandy gravel lithofacies that does not significantly attenuate dissolved oxygen, and (2) highly reactive NRZs associated with elevated organic carbon, Fe2+, and sulfide that likely play a larger role in DO attenuation (Campbell et al., 2012). Probable NRZs have been delineated in the Rifle floodplain aquifer using spectral induced polarization geophysics, groundwater analyses, chemical, and mineralogical characterization of sediment, and biological sampling (Wainwright et al., 2015). The floodplain modeling specification incorporates this three-dimensional distribution of NRZs. In this case, MID is located in the background lithofacies, whereas well NRZ is located in an NRZ (Figure 1).
In the conceptual model, fermentative bacteria are assumed to become active when oxygen concentrations are below 50 μg/L. Where elevated DO is present during the oxygen incursion, fermenter biomass is stable though is subject to decay. Conversely, suboxic conditions, which are more likely at depth, favor growth. Sulfate-reducing bacteria are assumed to tolerate low levels of oxygen allowing them to remain active under most conditions, and grow under suboxic conditions, which are more likely at depth. Sulfate is generally available but carbon can be limiting and biomass is subject to decay. A general biomass turnover rate of 0.015/d is assumed (Yabusaki et al., 2011). Spearman correlations were used to statistically assess similarities and differences between microbiological modeling results and field 16S rRNA relative abundance data (rcorr, HMisc v3.17.1).
Results and Discussion
Fluctuating River Stage Drives Biogeochemical Shifts in the Aquifer
Through spatial and temporal groundwater sampling within the aquifer, we were able to track biogeochemical responses to water table fluctuation. Over the first half of the monitoring period, increasing river stage lowered the hydraulic gradient across the Rifle aquifer, resulting in increases in water table height (~4-ft. increase) across all three sampling locations (BND, MID, NRZ) at the site (Figure 2). As the water table moved through the capillary fringe into the vadose zone, entrapment of air bubbles within the newly saturated pores led to oxygenation of previously suboxic groundwater. This resulted in a series of geochemical responses that were dependent on local geochemical conditions, and contributed to each well (all depths) having a distinct geochemical signature (ANOSIM = 0.5952, p = 0.001).
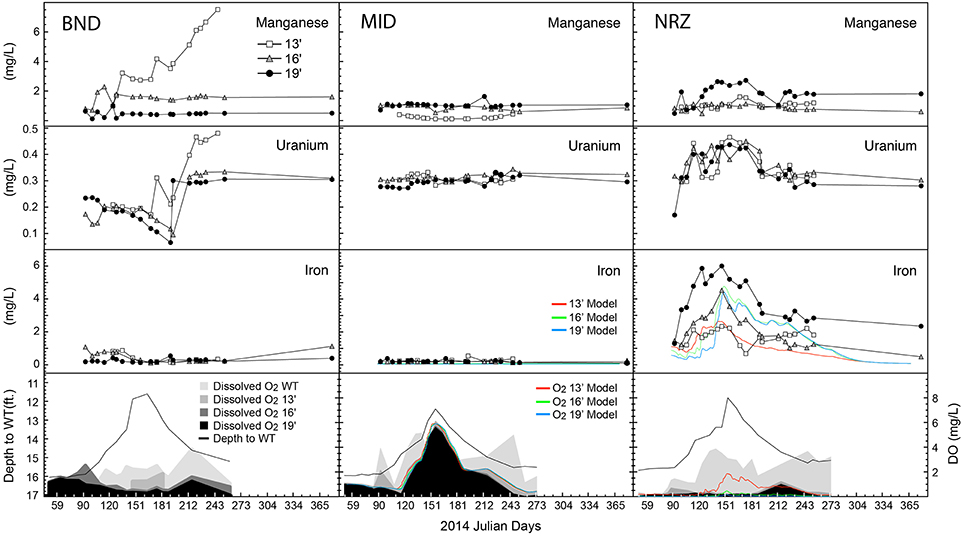
Figure 2. Geochemical parameters and model output plotted over time for each of the well-depth combinations. Depth to water table is plotted alongside dissolved oxygen (DO) content. WT, water table. All metal measurements are provided as total dissolved species.
The NRZ well was the most reduced, located in a region of the aquifer characterized by elevated abundances of iron sulfide minerals at 16- and 19-ft. depths (Supplementary Figure 2; Campbell et al., 2012). Any DO within this location was rapidly consumed at these depths, likely through abiotic oxidation reactions with solid phase iron sulfides that liberated Fe2+(aq) into groundwater. Laboratory experiments using native NRZ materials have revealed their ability to rapidly consume added DO (Supplementary Figure 3). Further supporting the role of iron sulfide minerals as a major sink for DO in the aquifer, no consistent increase in Fe2+(aq) was observed where fewer reactive sulfide minerals were present (13-ft., NRZ location, all MID depths; Campbell et al., 2012; Figure 2). Due to these largely abiotic reactions, increasing DO concentrations in the NRZ location were only observed at the top of the water table and not deeper in the aquifer.
With less-reduced sediments surrounding wells MID and BND, increasing DO concentrations coupled to water table rise were observed at greater depths in these areas, with an especially pronounced DO signal across the whole vertical profile in the least-reduced MID location (Figure 2). In the absence of abundant iron sulfide mineral phases that might liberate ferrous iron via reactions with DO, Fe2+(aq) concentrations remained relatively low over the duration of the sampling period in BND and MID groundwater samples, although some Fe2+(aq) oxidation may have occurred in the BND location where Fe2+(aq) concentrations decreased from ~1 to 0.2 mg L−1 during water table rise (Figure 2). Such reactions represent a smaller pool of redox buffering capacity in this portion of the aquifer as compared with solid phase sulfides in the NRZ location. During water table rise, the elevated DO in the largely non-reactive background aquifer sediments (MID location) was used as a model input to estimate the DO provided to the entire floodplain aquifer. The model accurately predicted little Fe2+(aq) mobilization in the MID location along with elevated and vertically heterogeneous Fe2+(aq) concentrations in the NRZ location (Figure 2). In the NRZ location, the model assumed DO consumption via relatively fast abiotic iron sulfide mineral oxidation (leading to Fe2+(aq) mobilization), and slower background biotic oxidation via chemolithoautotrophy.
In the shallowest location in well BND, we observed concurrent increases in both U6+(aq) and Mn2+(aq) during water table fall, which may be indicative of abiotic manganese-dependent uranium oxidation (Plathe et al., 2013). Via this reaction, solid phase reduced uranium (as U4+) is oxidized by manganese oxides. Aqueous uranium increases were also detected across the remaining depths at location BND, and all depths in the NRZ well (Figure 2), although the exact mechanisms for mobilization remain unclear. Two possible mechanisms include (1) DO-dependent re-oxidation of reduced uranium phases in the saturated portion of the aquifer (Bargar et al., 2013; Alessi et al., 2014), and (2) the mobilization of U6+(aq) located in the vadose zone during water table rise. In the NRZ location, U6+(aq) concentrations increased rapidly during water table rise, while in the BND location increasing U6+(aq) concentrations occurred during water table fall. These variable responses suggest that different mechanisms may be responsible for uranium mobilization between these two locations.
Despite these specific examples of abiotic responses to water table rise and DO intrusion, many geochemical parameters between depths at a specific location were remarkably similar, indicative of (1) greater horizontal rather than vertical heterogeneity across the saturated subsurface (Figure 2), and (2) some level of geochemical stability within a particular well. Despite being separated by only 150 ft., samples from the MID wells exhibited greater geochemical stability than samples from the BND location (Figure 3). Conversely, samples from the 13-ft. BND location were statistically less geochemically stable than samples from all depths in the MID well. The relatively oxidized nature of the MID location (relative to NRZ and BND) may explain the geochemical stability of this portion of the aquifer in response to O2 intrusion, with few reduced species that could react with increasing DO. In more reducing subsurface regions (e.g., NRZ), reactions between DO and abundant reduced species (e.g., FeS) generated significant geochemical deviations from baseline conditions. Such deviations contributed to less geochemically stable environments over the period of water table rise and fall (Figure 3).
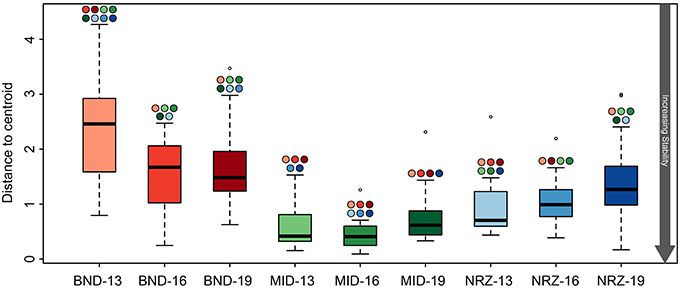
Figure 3. Boxplots illustrating the test for homogeneity of geochemical parameters at the nine well-depth combinations. The lower the average “distance to centroid” the more stable the system is whereas a higher number denotes greater variation from the average, and therefore a less geochemically stable environment. Significant differences between locations are indicated by the colored circles located above the whiskers, with colors corresponding to other boxes. For example, BND-13 is significantly less stable than BND-16.
Microbial Community Compositional Shifts Are Linked to Water Table Rise
Coupled to these abiotic reactions, we detected and modeled clear microbial community responses to this hydrologic perturbation and associated geochemical deviations. Using 16S rRNA gene amplicon sequencing, microbial communities defined by well location, and depth were examined via non-metric multidimensional scaling (NMDS) for emergent properties, and CCA to determine potential geochemical drivers of dissimilarity. Differences identified via NMDS were compared using ANOSIM (Figure 4). Similar to the previously described geochemical and mineralogical heterogeneity, there also existed significant and pronounced differences in microbial community structure across all locations and depths (ANOSIM: 0.8812, p-value: 0.001). Generally, the largest community composition differences existed between the deepest NRZ depth and any other well-depth combinations (Supplementary Table 2). Conversely, the most similar community structure existed between NRZ-13 and BND-13 samples (ANOSIM: 0.507, p-value: 0.001), potentially reflecting somewhat similar geochemical conditions (i.e., more reduced relative to MID) in these locations. Of the measured geochemical parameters, dynamic Fe2+(aq) and U6+(aq) behaviors in the NRZ location covaried strongly with differences between microbial communities across the depth profile. Similarly, fluctuating V(aq) concentrations were inferred to play a role in community structures separation within the MID location (Figure 4B).
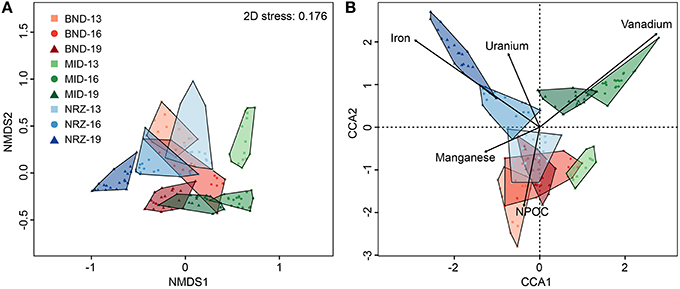
Figure 4. (A) A non-metric multidimensional scaling (NMDS) graph of the microbial community data. Distances between samples indicated in this graph are explained by their Bray-Curtis dissimilarity. (B) A canonical correspondence analysis (CCA) of the microbial community data and key geochemical parameters. Distances in this graph are explained by variation in the provided geochemical parameters. Proximity to the parameter lines indicates co-linearity, while line length represents the magnitude of the effect. For example, NRZ-19 linearizes very closely to iron, due to the oxidation of iron sulfides by dissolved oxygen, and subsequent release of Fe2+(aq) in this location.
To determine temporal responses to the hydrologic perturbation in the microbial communities, we split the 16S rRNA gene datasets into two groups, comprising those samples collected during water table rise, and those collected during water table fall across the site. OTU abundances within these groups were correlated against water table rise and fall, enabling the assessment of OTUs that responded either positively or negatively to geochemical conditions during these time periods (Figure 5).
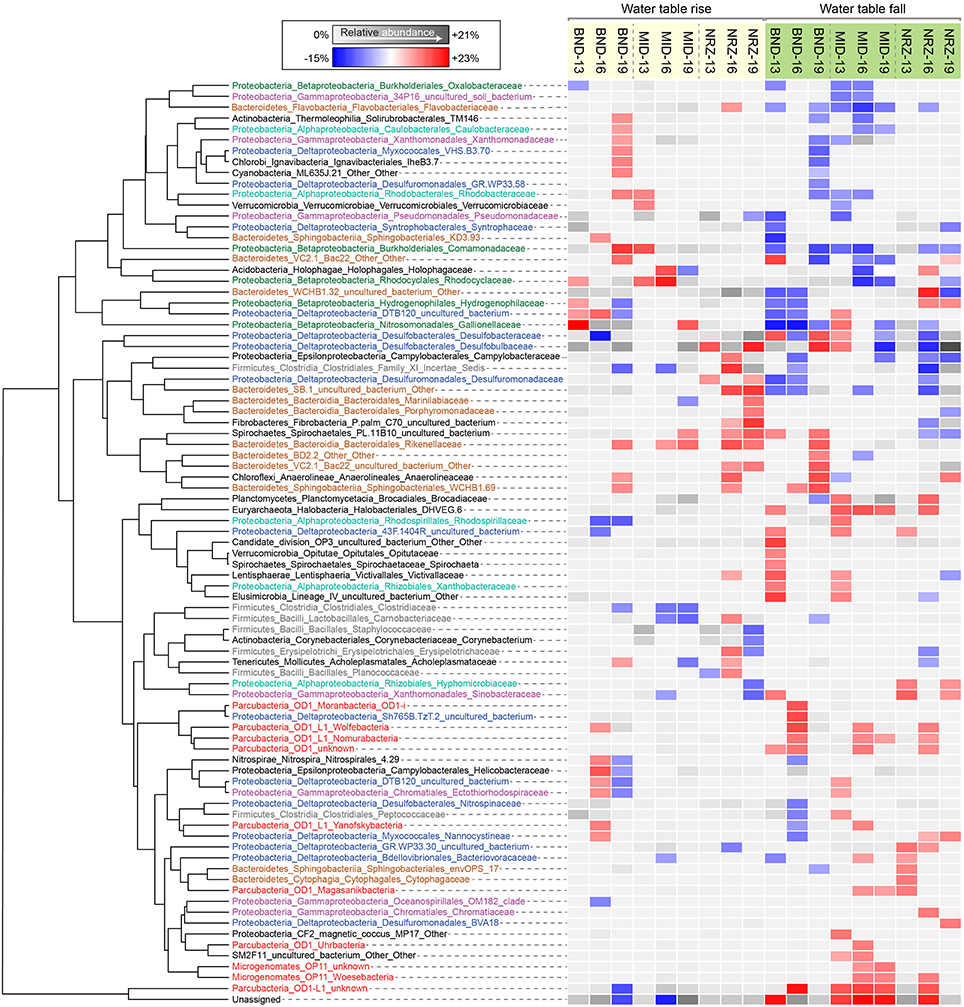
Figure 5. Heatmap illustrating family-level relative abundance responses to fluctuations in water table height, clustered using Euclidean distances. Each of the families displayed had a significant level of correlation (p < 0.05) to water table rise or fall in at least one sampling location. Either a relative abundance increase or a decrease associated with the significant correlation is indicated by the red-blue color scheme. Not every family responded the same at each location, thus the average relative abundance is indicated by the gray scale when correlations were not significant.
A series of studies at Rifle have previously revealed high abundances of microorganisms from the candidate phyla radiation (CPR) present in sediments and groundwater (Wrighton et al., 2012; Brown et al., 2015). Using phylogenetic classifications developed by Brown et al. (2015) for OTU assignments, we identified numerous groups within the Microgenomates (OP11) and Parcubacteria (OD1) over the course of this experiment. As trends in Figure 5 illustrate, members representing these groups (putative members of OP11 and OD1) responded positively to decreasing water table height and disappearance of DO across all three well locations. These temporal shifts in relative abundance are supported by extensive genomic inferences from ~800 genomes that indicate obligate anaerobic fermentative lifestyles for members of the CPR (Wrighton et al., 2012; Castelle et al., 2013; Kantor et al., 2013; Rinke et al., 2013; Brown et al., 2015). Some of the largest relative abundance increases were associated with OTUs affiliated with the Woesebacteria within the Microgenomates (OP11) and the Magasanikbacteria, Moranbacteria, Wolfebacteria, and Nomurabacteria within the Parcubacteria (OD1; Figure 5). Modeled shifts in fermentative microorganism abundances supported these experimental data, with biomass predicted to increase significantly during water table fall in both MID and NRZ locations (Figure 6). Strongest correlations between experimental 16S rRNA gene relative abundances for Parcubacteria and model output (r > 0.52, p < 0.025) were observed at both MID (16- and 19-ft.), and NRZ (16-ft.) locations. However, decreased relative abundances of putative fermenters were initially observed in all MID depths and the 13-ft. NRZ depth, where relatively few abiotic DO-consuming reactions were predicted to occur. Similar enrichments during the disappearance of DO were also observed for the remaining unassigned reads, suggesting that many of these sequences may be from more poorly characterized families within the CPR that cannot currently be assigned to taxonomic groups (Figure 5). Via analysis of similarity percentages (SIMPER), unassigned sequences contributed ~7% to distinguishing MID location samples from other wells, suggesting that more oxidized regions of the aquifer may harbor more phylogenetic novelty relative to other portions of the aquifer. Further community genomic sampling at the site will better resolve the placement of these OTUs.
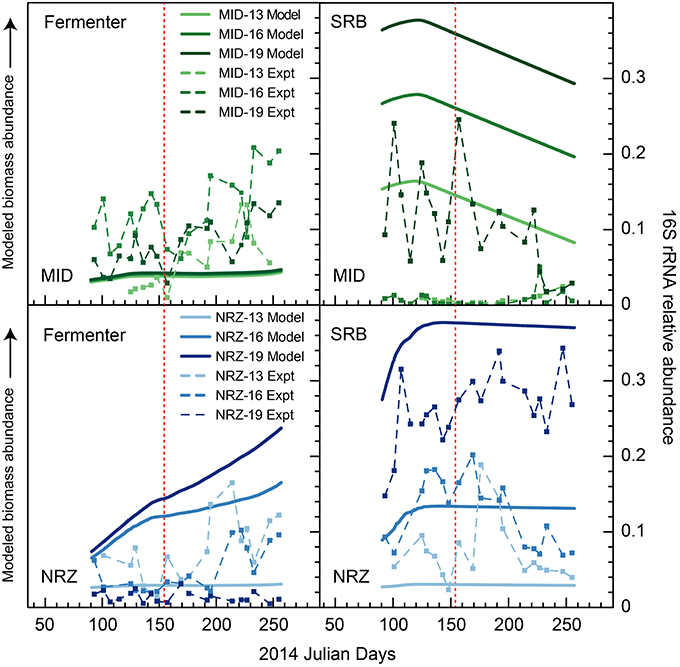
Figure 6. Modeled biomass and 16S rRNA gene relative abundance (“experimental”) during the monitoring period in wells MID and NRZ for two key functional groups—inferred fermenters, and SRB. For temporal shifts in 16S rRNA gene relative abundances, fermenters are represented by members of candidate phyla OD1 (Parcubacteria), while SRB are represented by members of the Desulfobulbaceae and Desulfobacteraceae. Modeled biomass is presented to illustrate relative differences between depths within a sample, and changes over time. Scales for each modeled dataset are different and so should not be directly compared between functional groups or wells. The red line marks the date of peak water table height in the aquifer.
While OTUs belonging to microbial groups that are generally characterized as anaerobes (e.g., members of the Clostridaceae, Rhodospirillaceae) decreased in relative abundance during water table rise and DO intrusion into the aquifer, other taxa were able to respond positively over these same geochemical conditions, with OTUs assigned to putative chemolithoautotrophs (e.g., members of the Gallionellaceae, Hydrogenophilaceae) increasing in relative abundance over the first half of the experiment. The strongest signal was assigned to OTUs matching members of the Gallionellaceae in the shallowest location at well BND (13-ft.), where increasing DO could potentially be used as a terminal electron acceptor during enzymatic iron-oxidation. Such a model would support recent metatranscriptomic analysis at Rifle (Jewell et al., 2016) and previous functional characterization of the family Gallionellaceae (Hallbeck and Pedersen, 2014). SIMPER analyses indicated that these Gallionellaceae OTUs contributed ~5–6% of the differences between shallower and deeper depths at this location. In addition, these analyses indicated that OTUs from this group were the largest consistent contributor distinguishing well BND from other locations over the course of this experiment (~4% contribution to differences). Relative abundance increases were also observed in OTUs matching members of the Rhodocyclaceae and Comamonadaceae (e.g., metabolically versatile groups such as Dechloromonas and Albidiferax) as DO concentrations increased in the BND and MID locations. Supporting the inference that geochemical parameters (e.g., O2) exert a strong control on microbial community dynamics, OTUs from the Gallionellaceae, Rhodocyclaceae, and Comamonadaceae nearly all decreased in relative abundance during the subsequent water table fall in the latter samples (Figure 5).
Groups representing known sulfate reducing bacteria (SRB) previously identified as active at the site (Miletto et al., 2011; Handley et al., 2013)—including members of the Desulfobacteraceae and Desulfobulbaceae—were present in all samples collected during this study, and exhibited dynamic relative abundance fluctuations in many locations. In many instances, relative abundance changes varied over orders of magnitude (Figure 6) despite relatively constant groundwater sulfate concentrations (typically between 8 and 10 mM with slightly elevated concentrations in BND between 8 and 15 mM). For example, OTUs affiliated with members of the Desulfobulbaceae increased in relative abundance during water table fall in BND (19-ft.) and MID (13-ft.), and yet decreased in abundance over the same period in wells MID (19-ft.) and NRZ (16-ft.; Figure 5). Model output was unable to capture this dynamic behavior, likely due to unknown driving factors that were not implemented in the model framework. One such factor might be the generation of other oxidized sulfur species (e.g., thiosulfate) from rapid sulfur cycling that can support the activity of SRB (Zopfi et al., 2004). However, the model was able to resolve the vertical differences in SRB relative abundance in both MID and NRZ locations, where deeper depths are characterized by greater abundances of putative SRB (as inferred from OTUs; Figure 6). The key role played by SRB in generating reducing conditions in NRZs was supported by SIMPER analyses that identified OTUs matching members of the Desulfobulbaceae (~6% contribution to differences) and Desulfobacteraceae (~3–4% contribution to differences) as the groups distinguishing microbial communities in the NRZ location from other regions of the aquifer. Members of these groups have been previously detected at Rifle under a range of geochemical conditions, including during biostimulation experiments in some regions of the aquifer (Anderson et al., 2003; Vrionis et al., 2005; Wrighton et al., 2014). The persistence of these microorganisms under the fluctuating geochemical conditions reported here suggests some tolerance to oxygen stress (Kjeldsen et al., 2004), and additionally reflects the high concentrations (~8–10 mM) of sulfate present in groundwater at the site (Williams et al., 2011). The oxidation of iron sulfide minerals in more reduced portions of the aquifer during DO intrusion generates more oxidized sulfur species that may be available for enzymatic reduction, while the recent identification of cryptic sulfur cycling in freshwater sediments (Hansel et al., 2015) represents a further mechanism that would support persistent sulfate reduction.
Monitoring of vadose zone geochemistry at this site has indicated sharp gradients in nitrate concentrations that develop between the base of the vadose zone and the transition to permanently saturated conditions (Jewell et al., 2016). While nitrate mobilization and transport to deeper portions of the aquifer during groundwater fluctuations is possible, we were unable to measure any consistent nitrate increases in geochemical samples collected at 13-ft. depths across all the wells, with concentration fluctuating between 0 and 100 μM over the course of the experiment. This may indicate that any nitrate inputs into the system during water table rise were rapidly reduced prior to sampling. 16S rRNA gene amplicon data may support this hypothesis, with nitrate reducing metabolisms represented within microbial groups that responded positively in certain locations to water table rise (e.g., members of the Rhodocyclaceae). Further evidence of nitrogen cycling within the aquifer is evidenced by detection of OTUs matching members of the Brocadiaceae, which are implicated in anaerobic ammonium oxidation (ANAMMOX), and are mostly enriched as DO concentrations decrease during water table fall (Figure 5).
Measuring Stability and Rate of Change in the Microbial Community
Microbial community structure stability was interrogated through the use of a test for homogeneity, and revealed variable linkages with geochemical trends (Anderson et al., 2006). Similar to the trends in geochemical stability described earlier, microbial community metrics (Bray-Curtis dissimilarity distances) indicated that the microbial population at the shallowest BND location (13-ft.) was statistically less stable than all MID locations, and NRZ locations at 16- and 19-ft. depths (Figure 7). Via these measurements, we infer that interdependent shifts in geochemistry and microbiology were strongest in the shallowest location at the northern site boundary (BND 13-ft.). A similar effect was also observed for the microbial community at the shallowest depth in the NRZ location (NRZ 13-ft.), although with fewer statistically significant comparisons (Figure 7). Such stability interdependencies were less evident at the MID location, where despite stable geochemical profiles (with the exception of DO; Figure 2), microbial community structure was not statistically more stable than many of the other locations. Contrasting with the other locations, DO incursion across the vertical profile in the MID well was likely an important parameter driving shifts in microbial community composition.
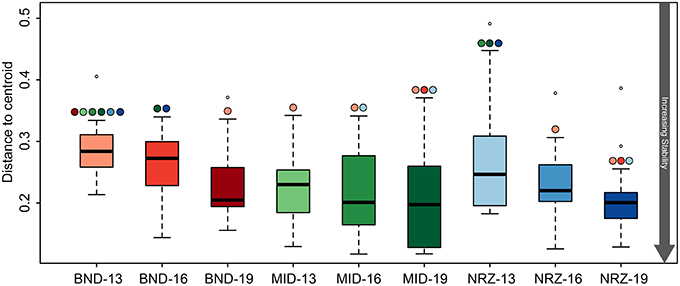
Figure 7. Boxplots illustrating the test for homogeneity for the 16S rRNA gene microbial community data. Like the geochemical test for homogeneity (Figure 3), a lower “distance to centroid” value indicates a more stable system while a larger value is less stable. The colored circles above the whiskers indicate where significant differences exist between the average values. For example BND-13 is significantly less than stable than MID-13.
Deeper subsurface locations are traditionally thought of as being more stable than shallower near-surface ecosystems (Anderson et al., 2013). While both the species richness (chao1) and diversity (Shannon's) within the microbial community generally decreased with depth at the Rifle site, stability measurements increased in BND and NRZ locations (Table 1), suggesting that the geochemical perturbations driven by the rising water table in these regions are less strongly felt by the deeper (i.e., 19-ft.) microbial communities. Contrastingly, the MID location was exposed to the greatest DO perturbation across the vertical profile (Figure 2) and is characterized by the highest diversity and richness measures relative to the other two well locations, potentially reflecting the range of microbial metabolisms that can be supported under such fluctuating redox conditions.
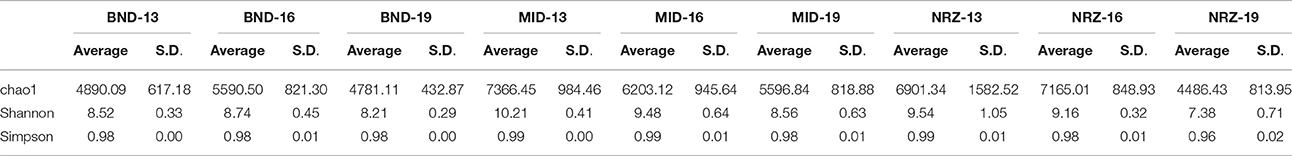
Table 1. Diversity measurements for the nine subsurface locations sampled over the course of water table rise and fall.
Community fluctuation rates were also examined in order to conceptualize variations in overall stability. Calculations indicated that the microbial community at the BND 13-ft. location was the most rapidly changing during water table rise, in addition to being the least stable (Supplementary Figure 4). Such a result implies that the microorganisms present were able to rapidly respond to DO increases and any subsequent geochemical shifts. Overall, rates of change across all depths and locations were slower during water table fall, suggesting that longer periods of time were required for microbial communities to revert to “baseline” conditions.
Conclusions
The results presented here illustrate the clear impact of geochemical heterogeneity across this riparian aquifer on responses to environmental perturbations. Summarizing the results, the abundance of reduced phases within a given region of the aquifer controls the geochemical response to water table rise and oxygen entrapment within the saturated zone. The predominance of reduced species and compounds in NRZ locations (and to a lesser extent the BND location) results in a dynamic geochemical response to DO intrusion, with extensive re-oxidation of iron sulfide minerals and reduced uranium phases. The relative lack of reduced mineral phases in the MID location limits the extent of DO consumption via abiotic reactions, with the result that DO penetrates across the whole vertical profile in this region. Despite these geochemical differences across the site, there is always a biotic response to water table rise, linked to either direct (increased DO) or indirect (e.g., increased Fe2+(aq) concentrations) perturbations associated with DO incursion.
The potential for inhibition of some anaerobic metabolisms during water table rise and DO intrusion has implications for the functioning of the microbial communities within the riparian aquifer. As described by Wrighton et al. (2014) the conceptual model for microbial community interdependencies at this site involves microbial members within the CPR and more well characterized fermentative microorganisms (e.g., members of the Clostridiaceae) degrading relatively recalcitrant carbon substrates, generating labile compounds as fermentation by-products for respiratory processes such as enzymatic sulfate reduction. While we were unable to accurately identify the key controls on fluctuating abundances of SRB (Figure 6), these data revealed the importance of this metabolic group in deeper aquifer locations in maintaining abundances of reduced species that then act as reductant during DO incursion. Broader trends were clearer for putative fermentative groups (e.g., members of the Parcubacteria), which became more enriched during water table fall and disappearance of DO. However, more subtle abundance increases during the intrusion of DO into the aquifer (e.g., in MID location—see Figure 6) suggest that at least some of these groups can tolerate the presence of DO. Recently published work identifying members of the Parcubacteria in the aerobic Hanford formation in eastern Washington State further supports this inference (Nelson and Stegen, 2015). This same analysis suggested a symbiotic lifestyle for members of the Parcubacteria. Further analyses are clearly required to better define ecological and metabolic roles for members of the CPR in shallow subsurface environments.
The Rifle aquifer site is the previous location of a uranium and vanadium milling operation. Although significant efforts have been made to remediate the site through removal of surface contaminated soils and sediments, persistent low-level uranium concentrations have been detected in the subsurface. Given that previous studies have demonstrated that NRZs are “hot spots” of immobilized uranium within the aquifer (Campbell et al., 2012), the annual intrusion of DO and oxidation of reduced species such as monomeric U4+ in these regions may at least partly account for the observed persistence of uranium in groundwater (Bargar et al., 2013; Alessi et al., 2014). Indeed, our geochemical measurements showed a sharp spike in U6+ concentrations during DO intrusion into the NRZ location sampled here (Figure 2). As many former milling sites are located across the US west (e.g., Shiprock, NM; Moab, UT) in hydrologically similar environments, the presence of NRZs and the effects of annual hydrologic perturbations may need to be considered when developing long-term models for natural flushing and realistic assessments of uranium attenuation.
Overall, these data revealed dynamic heterogeneous biogeochemistry in response to seasonal hydrologic perturbations in a riparian aquifer. The ability to model such responses suggests that a predictive framework could be applied to understand subsurface responses to future climate change scenarios in the intermountain west, where increasing extreme precipitation events and potentially increasing mean winter precipitation may increase the intensity of hydrologic perturbations (Dominguez et al., 2012). The implications for regional biogeochemical cycles remain to be determined.
Author Contributions
RD, KW, CH, and MW performed sample collection. KW and CH performed geochemical analyses. MW and RD performed microbiological analyses. SY and YF performed modeling studies using these data. All authors were involved in drafting the manuscript.
Funding
This work was supported as part of the Genomes to Watershed Scientific Focus Area at Lawrence Berkeley National Laboratory, which is funded by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DEAC02-05CH11231.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
A portion of the work described here was performed at Pacific Northwest National Laboratory, which is operated by Battelle for the United States Department of Energy under Contract DE-AC05-76RL01830. Massively parallel processing simulations were performed on the Cascade supercomputer at the Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/feart.2016.00057
Supplementary Table S1. Geochemical parameters measured over the course of this study.
References
Alessi, D. S., Lezama-Pacheco, J. S., Janot, N., Suvorova, E. I., Cerrato, J. M., Giammar, D. E., et al. (2014). Speciation and reactivity of uranium products formed during in Situ bioremediation in a shallow alluvial aquifer. Environ. Sci. Technol. 48, 12842–12850. doi: 10.1021/es502701u
Anderson, M. J., Ellingsen, K. E., and McArdle, B. H. (2006). Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 9, 683–693. doi: 10.1111/j.1461-0248.2006.00926.x
Anderson, R. E., Brazelton, W. J., and Baross, J. A. (2013). The deep viriosphere: assessing the viral impact on microbial community dynamics in the deep subsurface. Rev. Miner. Geochem. 75, 649–675. doi: 10.2138/rmg.2013.75.20
Anderson, R. T., Vrionis, H. A., Ortiz-Bernad, I., Resch, C. T., Long, P. E., Dayvault, R., et al. (2003). Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69, 5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003
Arora, B., Spycher, N. F., Steefel, C. I., Molins, S., Bill, M., Conrad, M. E., et al. (2015). Influence of hydrological, biogeochemical and temperature transients on subsurface carbon fluxes in a flood plain environment. Biogeochemistry. 127, 367–396. doi: 10.1007/s10533-016-0186-8
Bargar, J. R., Williams, K. H., Campbell, K. M., Long, P. E., Stubbs, J. E., Suvorova, E. I., et al. (2013). Uranium redox transition pathways in acetate-amended sediments. Proc. Natl. Acad. Sci. U.S.A. 110, 4506–4511. doi: 10.1073/pnas.1219198110
Bray, J. R., and Curtis, J. T. (1957). An ordination of upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Brown, C. T., Hug, L. A., Thomas, B. C., Sharon, I., Castelle, C. J., Singh, A., et al. (2015). Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211. doi: 10.1038/nature14486
Campbell, K. M., Kukkadapu, R. K., Qafoku, N. P., Peacock, A. D., Lesher, E., Williams, K. H., et al. (2012). Geochemical, mineralogical and microbiological characteristics of sediment from a naturally reduced zone in a uranium-contaminated aquifer. Appl. Geochem. 27. 1499–1511. doi: 10.1016/j.apgeochem.2012.04.013
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carlyle, G. C., and Hill, A. R. (2001). Groundwater phosphate dynamics in a river riparian zone: effects of hydrologic flowpaths, lithology and redox chemistry. J. Hydrol. 247, 151–168. doi: 10.1016/S0022-1694(01)00375-4
Castelle, C. J., Hug, L. A., Wrighton, K. C., Thomas, B. C., Williams, K. H., Wu, D., et al. (2013). Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat. Commun. 4:2120. doi: 10.1038/ncomms3120
Castelle, C. J., Wrighton, K. C., Thomas, B. C., Hug, L. A., Brown, C. T., Wilkins, M. J., et al. (2015). Genomic expansion of domain Archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr. Biol. 25, 690–701. doi: 10.1016/j.cub.2015.01.014
Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270.
Dominguez, F., Rivera, E., Lettenmaier, D. P., and Castro, C. L. (2012). Changes in winter precipitation extremes for the western United States under a warmer climate as simulated by regional climate models: precipitation extremes Western US. Geophys. Res. Lett. 39. doi: 10.1029/2011gl050762
Hallbeck, L., and Pedersen, K. (2014). “The family gallionellaceae,” in The Prokaryotes, eds E. Rosenberg, E. DeLong, S. Lory, E. Stackebrandt, F. Thompson (Berlin; Heidelberg: Springer), 853–858.
Handley, K. M., VerBerkmoes, N. C., Steefel, C. I., Williams, K. H., Sharon, I., Miller, C. S., et al. (2013). Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J. 7, 800–816. doi: 10.1038/ismej.2012.148
Hansel, C. M., Lentini, C. J., Tang, Y., Johnston, D. T., Wankel, S. D., and Jardine, P. M. (2015). Dominance of sulfur-fueled iron oxide reduction in low-sulfate freshwater sediments. ISME J. 9, 2400–2412. doi: 10.1038/ismej.2015.50
Heffernan, J. B., and Sponseller, R. A. (2004). Nutrient mobilization and processing in Sonoran desert riparian soils following artificial re-wetting. Biogeochemistry 70, 117–134. doi: 10.1023/B:BIOG.0000049339.74391.14
Hug, L. A., Thomas, B. C., Brown, C. T., Frischkorn, K. R., Williams, K. H., Tringe, S. G., et al. (2015). Aquifer environment selects for microbial species cohorts in sediment and groundwater. ISME J. 9, 1846–1856. doi: 10.1038/ismej.2015.2
Jacinthe, P.-A., Groffman, P. M., and Gold, A. J. (2003). Dissolved organic carbon dynamics in a riparian aquifer. J. Environ. Qual. 32, 1365. doi: 10.2134/jeq2003.1365
Jewell, T. N. M., Karaoz, U., Brodie, E. L., Williams, K. H., and Beller, H. R. (2016). Metatranscriptomic evidence of pervasive and diverse chemolithoautotrophy relevant to C, S, N and Fe cycling in a shallow alluvial aquifer. ISME J. doi: 10.1038/ismej.2016.25. [Epub ahead of print].
Kantor, R. S., Wrighton, K. C., Handley, K. M., Sharon, I., Hug, L. A., Castelle, C. J., et al. (2013). Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. mBio 4. doi: 10.1128/mBio.00708-13
Kjeldsen, K. U., Joulian, C., and Ingvorsen, K. (2004). Oxygen tolerance of sulfate-reducing bacteria in activated sludge. Environ. Sci. Technol. 38, 2038–2043. doi: 10.1021/es034777e
McClain, M. E., Boyer, E. W., Dent, C. L., Gergel, S. E., Grimm, N. B., Groffman, P. M., et al. (2003). Biogeochemical hot spots and hot moments at the interface of terrestrial and aquatic ecosystems. Ecosystems 6, 301–312. doi: 10.1007/s10021-003-0161-9
Miletto, M., Williams, K. H., N'Guessan, A. L., and Lovley, D. R. (2011). Molecular analysis of the metabolic rates of discrete subsurface populations of sulfate reducers. Appl. Environ. Microbiol. 77, 6502–6509. doi: 10.1128/AEM.00576-11
Nelson, W. C., and Stegen, J. C. (2015). The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 6:713. doi: 10.3389/fmicb.2015.00713
Oksanen, J., Guillaume Blanchet, F., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., et al. (2015). Vegan: Community Ecology Package. R. Package Version 2, 2–1.
Peter, S., Rechsteiner, R., Lehmann, M. F., Brankatschk, R., Vogt, T., Diem, S., et al. (2012). Nitrate removal in a restored riparian groundwater system: functioning and importance of individual riparian zones. Biogeosciences 9, 4295–4307. doi: 10.5194/bg-9-4295-2012
Plathe, K. L., Lee, S.-W., Tebo, B. M., Bargar, J. R., and Bernier-Latmani, R. (2013). Impact of microbial Mn oxidation on the remobilization of bioreduced U(IV). Environ. Sci. Technol. 47, 3606–3613. doi: 10.1021/es3036835
Qafoku, N. P., Gartman, B. N., Kukkadapu, R. K., Arey, B. W., Williams, K. H., Mouser, P. J., et al. (2014). Solid phase characterization of uranium in a multi-element contaminated, subsurface naturally reduced zone. Appl. Geochem. 42, 77–85. doi: 10.1016/j.apgeochem.2013.12.001
Qafoku, N. P., Kukkadapu, R. K., McKinley, J. P., Arey, B. W., Kelly, S. D., Wang, C., et al. (2009). Uranium in framboidal pyrite from a naturally bioreduced alluvial sediment. Environ. Sci. Technol. 43, 8528–8534. doi: 10.1021/es9017333
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J.-F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Scholl, M., Christenson, S., Cozzarelli, I., Ferree, D., and Jaeschke, J. (2005). Recharge Processes in an Alluvial Aquifer Riparian Zone, Norman Landfill, Oklahoma, 1998-2000. Oklahoma City: USGS.
Shade, A., Read, J. S., Youngblut, N. D., Fierer, N., Knight, R., Kratz, T. K., et al. (2012). Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J. 6, 2153–2167. doi: 10.1038/ismej.2012.56
Shannon, C. (1948). A mathematical theory of communication. Bell. Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Ulrich, G. A., Krumholz, L. R., and Suflita, J. M. (1997). A rapid and simple method for estimating sulfate reduction activity and quantifying inorganic sulfides. Appl. Environ. Microbiol. 63, 1627–1630.
Vrionis, H. A., Anderson, R. T., Ortiz-Bernad, I., O'Neill, K. R., Resch, C. T., Peacock, A. D., et al. (2005). Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl. Environ. Microbiol. 71, 6308–6318. doi: 10.1128/AEM.71.10.6308-6318.2005
Wainwright, H. M., Flores-Orozco, A., Bücker, M., Dafflon, B., Chen, J., Hubbard, S. S., et al. (2015). Hierarchical Bayesian method for mapping biogeochemical hot spots using induced polarization imaging. Water Resour. Res. 52, 533–551. doi: 10.1002/2015WR017763
Williams, K. H., Long, P. E., Davis, J. A., Wilkins, M. J., N'Guessan, A. L., Steefel, C. I., et al. (2011). Acetate availability and its influence on sustainable bioremediation of uranium-contaminated groundwater. Geomicrobiol. J. 28, 519–539. doi: 10.1080/01490451.2010.520074
Wrighton, K. C., Castelle, C. J., Wilkins, M. J., Hug, L. A., Sharon, I., Thomas, B. C., et al. (2014). Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J. 8, 1452–1463. doi: 10.1038/ismej.2013.249
Wrighton, K. C., Thomas, B. C., Sharon, I., Miller, C. S., Castelle, C. J., VerBerkmoes, N. C., et al. (2012). Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated Bacterial phyla. Science 337, 1661–1665. doi: 10.1126/science.1224041
Yabusaki, S. B., Fang, Y., Williams, K. H., Murray, C. J., Ward, A. L., Dayvault, R. D., et al. (2011). Variably saturated flow and multicomponent biogeochemical reactive transport modeling of a uranium bioremediation field experiment. J. Contam. Hydrol. 126, 271–290. doi: 10.1016/j.jconhyd.2011.09.002
Zopfi, J., Ferdelman, T. G., and Fossing, H. (2004). “Distribution and fate of sulfur intermediates—sulfi te, tetrathionate, thiosulfate, and elemental sulfur—in marine sediments,” in Sulfur Biogeochemistry—Past and Present, eds J. P. Amend, K. J. Edwards, and T. W. Lyons (Boulder, CO: Geological Society of America), 97–116.
Keywords: aquifer, reactive transport modeling, fermentation, candidate phyla, Rifle
Citation: Danczak RE, Yabusaki SB, Williams KH, Fang Y, Hobson C and Wilkins MJ (2016) Snowmelt Induced Hydrologic Perturbations Drive Dynamic Microbiological and Geochemical Behaviors across a Shallow Riparian Aquifer. Front. Earth Sci. 4:57. doi: 10.3389/feart.2016.00057
Received: 24 March 2016; Accepted: 26 April 2016;
Published: 11 May 2016.
Edited by:
David Emerson, Bigelow Laboratory for Ocean Sciences, USAReviewed by:
Jeffrey M. Dick, Wattanothaipayap School, ThailandAnirban Chakraborty, University of Calgary, Canada
Copyright © 2016 Danczak, Yabusaki, Williams, Fang, Hobson and Wilkins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael J. Wilkins, wilkins.231@osu.edu
 Robert E. Danczak1
Robert E. Danczak1  Michael J. Wilkins
Michael J. Wilkins