Genotoxicity of formaldehyde: molecular basis of DNA damage and mutation
- 1Laboratory of Molecular and Cellular Genetics, Department of Biology, Graduate School of Science and Radiation Research Center, Osaka Prefecture University, Sakai, Osaka, Japan
- 2Department of Environmental Quality Management, Research Center for Environmental Quality Management, Kyoto University, Otsu, Japan
- 3Department of Life Science, Dongguk University Seoul, Seoul, Korea
Formaldehyde is commonly used in the chemical industry and is present in the environment, such as vehicle emissions, some building materials, food, and tobacco smoke. It also occurs as a natural product in most organisms, the sources of which include a number of metabolic processes. It causes various acute and chronic adverse effects in humans if they inhale its fumes. Among the chronic effects on human health, we summarize data on genotoxicity and carcinogenicity in this review, and we particularly focus on the molecular mechanisms involved in the formaldehyde mutagenesis. Formaldehyde mainly induces N-hydroxymethyl mono-adducts on guanine, adenine and cytosine, and N-methylene crosslinks between adjacent purines in DNA. These crosslinks are types of DNA damage potentially fatal for cell survival if they are not removed by the nucleotide excision repair pathway. In the previous studies, we showed evidence that formaldehyde causes intra-strand crosslinks between purines in DNA using a unique method (Matsuda et al., 1998). Using shuttle vector plasmids, we also showed that formaldehyde as well as acetaldehyde induces tandem base substitutions, mainly at 5′-GG and 5′-GA sequences, which would arise from the intra-strand crosslinks. These mutation features are different from those of other aldehydes such as crotonaldehyde, acrolein, glyoxal, and methylglyoxal. These findings provide molecular clues to improve our understanding of the genotoxicity and carcinogenicity of formaldehyde.
Introduction
Formaldehyde (Methanal, CAS No.: 50-00-0) is abundantly produced in the chemical industry, and its annual global production is about 20 million tons (IARC, 2012). It is used in industrial and consumer products, and is also generated as a byproduct of biomass and petroleum burning, cigarette smoke and automobile exhaust. Cigarette smoke contains 12–106 μg of formaldehyde per cigarette (IARC, 2012). In medical and biological fields, it has been widely used as a fixative or preservative for pathological specimens. It also occurs as a natural product in most organisms and in the environment. There are several metabolic pathways in humans leading to formaldehyde synthesis, in particular, amino acid and methanol metabolisms, lipid peroxidation, and P450-dependent demethylation (Swenberg et al., 2013).
The major sources of formaldehyde exposure can be roughly classified into three categories: fumes generated from industry, those released in residential housing and those in various occupational indoor settings (Ma and Harris, 1988). Non-occupational exposure in the household is mainly due to the fumes released from formaldehyde-based resin for binding of plywood, particle board, paint, and textile fibers. Formaldehyde fumes have caused health problems across the country. People inhaling formaldehyde fumes indoors often show symptoms such as headache, dizziness, wheezing, runny nose, and nausea. In severe cases, they have sore throat, burning eyes, and skin irritation. These symptoms are called “sick house syndrome” or “sick building syndrome” (Norbäck, 2009). The levels of formaldehyde in the air inside houses are typically 0.02–0.06 mg/m3, but these levels have been declining since the late 1980s as a result of standards for building materials (IARC, 2012). The World Health Organization now recommends a formaldehyde level of 0.1 mg/m3 in indoor air (Kaden et al., 2010).
Formaldehyde also has activities that cause chronic adverse effects, such as genotoxicity, carcinogenicity, and teratogenicity. The health effects have been extensively studied for a long time and reviewed in many articles (Ma and Harris, 1988; Conaway et al., 1996; Swenberg et al., 2013). In this review, we focused on molecular basis of DNA damage and mutations induced by formaldehyde exposure. In this sense, our original data on the mutation spectrum of formaldehyde in human cells, have been clearly showed further details concerning the mechanism of formaldehyde carcinogenesis.
Carcinogenicity
In animal experiments using mice and rats, chronic inhalation of formaldehyde at high concentrations caused cancers, particularly, squamous cell carcinomas of the nasal cavities (IARC, 2006). In other studies, the incidences of lymphoma and leukemia were also increased. In animals administered formaldehyde in drinking water, increased incidences of forestomach papillomas, leukemias, and gastrointestinal tract tumors were reported, but their frequencies varied. These studies have been summarized in IARC Monograph (IARC, 2006).
In the human body, many endogenous and exogenous compounds release formaldehyde through catabolic reactions. The primary metabolic system for formaldehyde involves an initial spontaneous reaction with glutathione to form S-hydroxymethylglutathione, followed by reaction facilitated by alcohol dehydrogenase-3 to convert the intermediate to S-formylglutathione. This intermediate is then further metabolized by S-formylglutathione hydrolase to yield nontoxic formate, which is excreted in the urine (Hedberg et al., 2002). There have been numerous cohort and case-control studies on formaldehyde exposure and an increase in nasopharynx cancer or leukemia in humans (IARC, 2012). The IARC (2006) judged that there was sufficient evidence for the carcinogenicity of formaldehyde, based primarily on its association with nasopharyngeal cancer, and that there was strong but not sufficient evidence for the leukemogenic effects of formaldehyde. Owing to advancements via recent epidemiological studies (Coggon et al., 2003; Hauptmann et al., 2004, 2009; Pinkerton et al., 2004; Marsh and Youk, 2005; Marsh et al., 2007a,b; Beane Freeman et al., 2009) and meta-analyses (Bosetti et al., 2008; Zhang et al., 2009; Bachand et al., 2010), the IARC (2012) concluded that occupational exposure to formaldehyde causes nasopharyngeal cancer and leukemia, and classified formaldehyde as a human carcinogen Group 1.
Genotoxicity
The genotoxicity of formaldehyde has been examined by various methods. In most reports using Salmonella typhimurium strains TA98, TA100, TA1535, TA1537, and TA1538 (Ames test), formaldehyde induced both base change and frame-shift mutations without metabolic activation, but its mutagenicity is not as strong as that of typical environmental mutagens such as 4-nitroquinoline-1-oxide and N-methyl-2-nitro-N'-nitrosoguanidine (Ma and Harris, 1988; Conaway et al., 1996). Formaldehyde also shows mutagenicity in assays using Escherichia coli WP2 (Takahashi et al., 1985; O'Donovan and Mee, 1993).
Formaldehyde induced chromosome aberrations (CA) and sister chromatid exchanges (SCE) in cultured Chinese hamster (Natarajan et al., 1983; Basler et al., 1985; Galloway et al., 1985; Merk and Speit, 1998, 1999) and human lymphocyte cells (Kreiger and Garry, 1983). In rats treated with formaldehyde through inhalation (0.5–12 ppm), no increase in the frequencies of CA and SCE was observed in lymphocytes (Kligerman et al., 1984). In rats inhaling 15 ppm formaldehyde, CA was induced in pulmonary lavage cells but not in bone marrow cells (Dallas et al., 1992). In mice treated with formaldehyde by oral administration, no CA or micronucleus (MN) formation was detected in bone marrow and spleen cells (Gocke et al., 1981; Natarajan et al., 1983). In rats administered a single dose of 200 mg/kg formaldehyde by gavage, gastrointestinal organs exhibited local irritation and their cells formed MN (Migliore et al., 1989). In people occupationally exposed to formaldehyde, increases in the frequencies of MN and SCE in the peripheral lymphocytes were reported (Costa et al., 2008), but no statistically significant increases were also reported elsewhere (Ye et al., 2005; Pala et al., 2008).
Formaldehyde induces mutations in human cultured lymphoblastoid TK6 cells and Chinese hamster ovary (CHO) cells, which were detected by resistance to toxic nucleoside analogs, trifluorothymidine (Goldmacher and Thilly, 1983), 8-azaguanine and 6-thioguanine (Liber et al., 1989; Graves et al., 1996; Speit and Merk, 2002).
DNA Damage and Repair
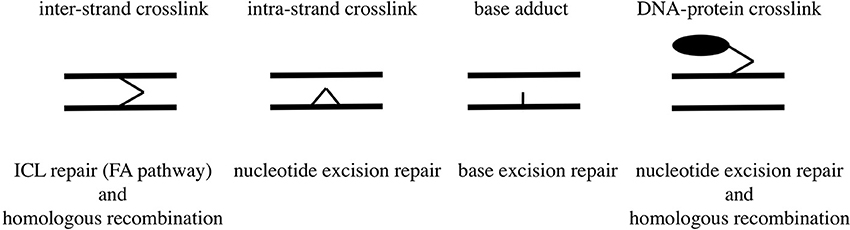
Formaldehyde reacts immediately with primary and secondary amines, thiols, hydroxyls, and amides to form methylol derivatives. It acts as an electrophile and can react with macromolecules such as DNA, RNA, and protein to form adducts and cross-links. Guanine, adenine and cytosine have amino group in their molecules. When deoxyadenosine, deoxyguanosine, deoxy-cytidine, and thymidine were reacted with formaldehyde, the major products determined by high-performance liquid chromatography and electrospray mass spectrometry (LC-MS/MS) were N6-hydroxymethyldeoxyadenosine, N4-hydroxymethyl-deoxycytidine and N2-hydroxymethyldeoxyguanosine (Beland et al., 1984; Cheng et al., 2003). In human and Chinese hamster cells treated with formaldehyde, N6-hydroxymethyldeoxy-adenosine and N2-hydroxymethyldeoxyguanosine were detected in their genome (Beland et al., 1984; Zhong and Que Hee, 2004). N6-hydroxymethyldeoxyadenosine was detected in leukocyte DNA from 29 of 32 heavy smokers (Wang et al., 2009). The reaction of formaldehyde with deoxyadenosine and deoxyguanosine or DNA also produces three major methylene crosslinks: di-(N6-deoxyadenosyl)methane, (N6-deoxyadenosyl-N2-deoxyguanosyl)methane and di-(N2-deoxyguanosyl)methane (Huang et al., 1992; Huang and Hopkins, 1993; Cheng et al., 2003). In a study of the in vitro reactions between formaldehyde and all amino acids, deoxyguanosine crosslinks with cysteine were abundant and stable, while those with lysine were readily formed but rapidly disintegrated (Lu et al., 2010). There are many reports showing that formaldehyde-protein crosslinks were detected in cells treated with formaldehyde (IARC, 2012), but their molecular structures have not been fully clarified. The DNA-protein crosslinks with protein sizes less than 12–14 kDa can be removed by the nucleotide excision repair pathway, whereas oversized DNA-protein crosslinks are processed exclusively by RecBCD-dependent homologous recombination in bacteria (Nakano et al., 2007). In mammalian cells, the upper size limit of crosslinked proteins amendable to the nucleotide excision repair is relatively small (8–10 kDa), therefore, homologous recombination would be the major pathway in tolerance of chromosomal DNA-protein crosslinks (Nakano et al., 2009).
We attempted to determine the likelihood of formaldehyde-induced intra-strand crosslink formation between four kinds of nucleotide in DNA using the unique method developed by Kawanishi (1995) and Matsuda et al. (1998). Single-strand DNA (175 base pairs) and two 20-mer complementary oligonucleotides: one a 5′ [32P]-labeled sequence and the other an unlabeled sequence following the [32P]-labeled sequence, were annealed as shown in Figure 1A. If the oligonucleotides are bound between the 3′-end base of the [32P]-labeled oligonucleotide and the adjacent 5′-end base of the unlabeled oligonucleotide by a formaldehyde crosslink, a 40-mer single-strand oligonucleotide band should appear on the denatured polyacrylamide gel. As shown in Figures 1B,C, formaldehyde formed crosslinks between deoxyguanosines (5′-GG) of the ends of the oligonucleotides. Fewer crosslinks were formed between deoxyguanosine and deoxyadenosine (5′-GA), but no crosslinks were formed between deoxycytidines (5′-CC). Figures 1B,C also shows formaldehyde-produced inter-strand crosslinks. The repair mechanism of the formaldehyde-induced DNA inter-strand crosslinks has not been reported so far, but the inter-strand crosslinks might be removed by the Fanconi anemia (FA) pathway and homologous recombination, which is shown in the studies of mitomycin C-induced inter-strand crosslinks (Kottemann and Smogorzewska, 2013; Unno et al., 2014).
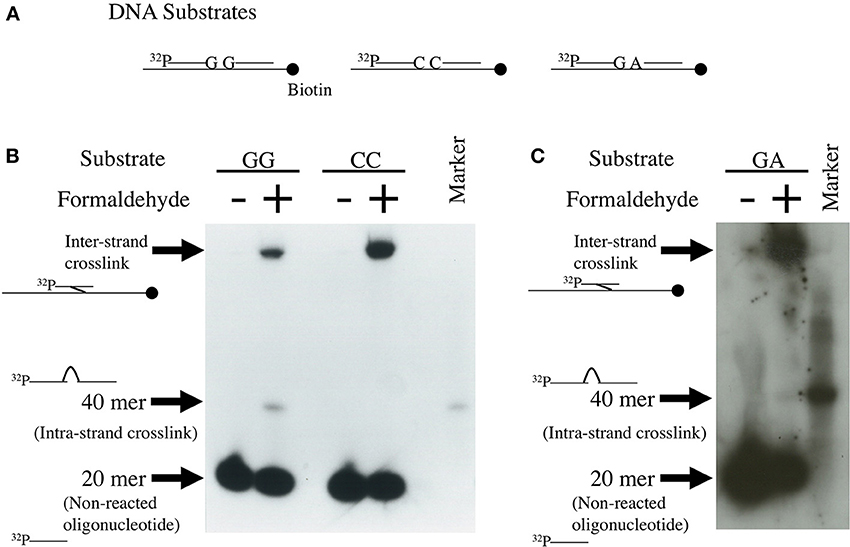
Figure 1. Detection of formaldehyde-induced intra-strand crosslinks. Single-strand 40 mer DNA bands were formed by binding between the 3′-end of 32P-labeled 20-mer DNA and the 5′-end of non-labeled 20-mer DNA by formaldehyde crosslinks. Method: The experimental protocol has been described in Matsuda et al. (1998). Briefly, the 175 base-pair region of pBluescript KS(-) was amplified by polymerase chain reaction (PCR) using 5′-biotinylated and non-biotinylated primers. The biotinylated 175-mer DNA was immobilized to Dynabeads M-280 streptavidin. Four kinds of 20-mer complementary DNA, of which the 5′-ends were labeled with 32P and the 3′-ends had G, A, or C, were prepared. Four kinds of unlabeled 20-mer complementary DNA, the sequence of which followed the 3′-end of the 32P-labeled 20-mer DNA and had G, A, T, or C at the 5′-end, were also prepared. The biotinylated 175-mer DNA, [32P]-labeled complementary 20-mer DNA and the following non-labeled complementary 20-mer DNA were annealed by heating and cooling for detecting GG, CC, and GA-intrastrand crosslinks, as shown in (A). The annealed DNA was treated with 270 mM formaldehyde for 24 h at 4°C. After collecting DNA using a magnet, DNA was purified and subjected to electrophoresis on denatured polyacrylamide gel. Autoradiography images of the gel are shown in (B,C).
Human and Chinese hamster mutant cells deficient in XPF or ERCC1 are hypersensitive to formaldehyde (Kumari et al., 2012), and E. coli uvrA mutant is more sensitive to formaldehyde than wild-type E. coli (Takahashi et al., 1985). These lines of evidence indicate that intra-strand crosslinks constitute a type of damage that could be fatal to cell survival and are removed by the nucleotide excision repair pathway; whereas mono-adducts, hydroxymethyldeoxyadenosines and hydroxymethyldeoxyguanosines, would be removed by the base excision repair pathway. Types of DNA damage induced by formaldehyde and their repair pathways are summarized in Figure 2.
Mutation Spectrum
In E. coli, mutations induced by formaldehyde in the xanthine guanine phosphoribosyl transferase (gpt) gene were characterized (Crosby et al., 1988). Exposure of E. coli to 4 mM formaldehyde induced insertions (41%), deletions (18%), and point mutations (41%). DNA sequencing revealed that most of the point mutations were transversions at the G:C base pair in the gene. In contrast, exposure of E. coli to 40 mM formaldehyde produced point mutations (92%), 62% of which were transitions at the single A:T base-pair in the gene, indicating that there were concentration-dependent variations in mutation spectra. When naked pSV2gpt plasmid was exposed to 3.3 or 10 mM formaldehyde and transfected into E. coli, most mutations were frameshifts, which also suggests the presence of various mechanisms in formaldehyde-induced mutation.
In human lymphoblastoid TK6 cells treated with 150 μM formaldehyde, 30 hprt mutant colonies were characterized by Southern blot analysis (Liber et al., 1989). Fourteen (47%) of these mutants had large deletions, while the other 16 mutants (53%) showed normal restriction patterns. Of these, 12 mutants produced hprt mRNA and 3 mutants produced no detectable mRNA. Sequence analyses of the mRNA showed that 6 mutants had base substitutions occurring at the A:T base pair. Four mutants had A:T to C:G transversions at a specific site that appears to be a mutation hot spot. The remaining 2 mutants lost exon 8 of the hprt gene.
CHO cells treated with 1 mM formaldehyde increased the hprt mutation frequency to 4.7-fold higher than the spontaneous mutation frequency, and mutations of 6 mutant clones were identified. All mutations were single base transversions with 3 A:T to T:A, 2 A:T to C:G, and 1 G:C to T:A (Graves et al., 1996).
Mutations in the p53 gene from rat primary nasal squamous cell carcinomas induced by formaldehyde inhalation (15 ppm up to 2 years) were analyzed (Recio et al., 1992). Five tumors had point mutations, namely, 3 G:C to T:A transversions, 1 G:C to A:T transition, and 1 G:C to C:G transversion.
Shuttle Vector Mutagenesis
Since no previous reports had described that formaldehyde induces tandem base substitutions that would arise from intra-strand crosslinks, we adopted the shuttle vector plasmid pMY189 (Matsuda et al., 1995; Yagi, 2013) that can be treated with a high concentration of formaldehyde. The plasmid pMY189 can replicate in both human and E. coli cells, and has an ampicillin-resistant marker gene and a mutation marker gene supF that can be selected as colored colonies on bacterial agar plates. The result was published by Kawanishi (1995), and we describe its outline below. The formaldehyde-treated pMY189 propagated in human fibroblast cells caused reduction of the number of ampicillin-resistant bacterial colonies and enhanced the mutation frequency of the supF gene (Figure 3). The mutation frequency of the plasmid was increased 4- to 50-fold following treatment of the plasmid with 70–270 mM formaldehyde. Nucleotide sequences of the supF mutation marker gene of 94 plasmids were determined. Mutated plasmids induced by 200–270 mM formaldehyde were classified and are shown in the third column of Table 1. About half (49%) of the plasmids had tandem base substitutions. Among the mutant plasmids with single base substitutions, 89% were substitutions of G:C base pairs (Table 1). The most common type of single base substitution mutation was G:C to T:A transversions (52%). Types of tandem base substitution are shown in Figure 4A. Thirty-nine percent were 5′-GG to 5′-TT (5′-CC to 5′-AA), and 15% were 5′-GA to 5′-AT (5′-TC to 5′-AT), and 5′-GA to 5′-TT (5′-TC to 5′-TT) substitutions. About half (47%) of them occurred at 5′-GG (5′-CC) sequences and 44% occurred at 5′-GA (5′-TC) sequences. The distribution of the formaldehyde-induced base substitutions in the supF gene is shown in Figure 4B. Underlined letters indicate that these base changes were found in the same mutated plasmid. The “d” or “del” means a deletion. The tandem base substitutions were not randomly distributed in the supF gene, but appeared preferentially at certain sites. About half of the tandem base substitutions occurred at 4 5′-GG (5′-CC) sequences (at base pairs 102–103, 103–104, 109–110, and 159–160) and 2 5′-GA (5′-TC) sequences (107–108 and 171–172).
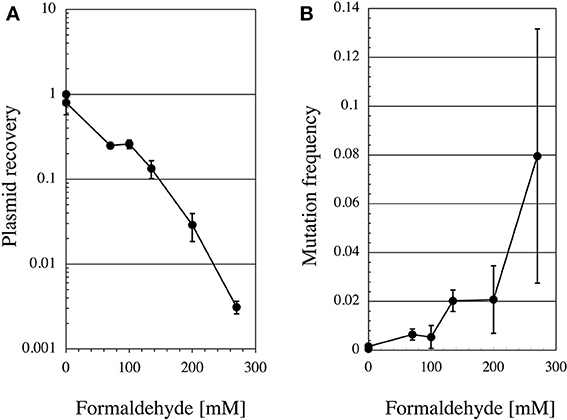
Figure 3. Recovery ratio and mutation frequency of formaldehyde-treated pMY189 plasmid after replication in human cells. Method: Formaldehyde-treated pMY189 in PBS solution (0.2 mL) was introduced into human SV40-transformed fibroblast cells WI38-VA13 by electroporation. The plasmid was also treated with methanol (0.3%) as a control because commercial formaldehyde solution contains methanol. After the cells were incubated for 48 h, the plasmids were extracted from the cells and digested with the restriction endonuclease DpnI to eliminate the non-replicated plasmids that retain the bacterial methylation pattern. The plasmids were introduced into the indicator bacteria KS40/pKY241 (Akasaka et al., 1992) and plated on LB agar containing nalidixic acid, ampicillin and chloramphenicol to select the plasmids containing the mutated supF gene. A portion of the cells were plated on LB agar containing ampicillin and chloramphenicol to measure the total number of transformants. Colonies were counted and plasmid recovery ratio (A) and mutation frequencies (B) were calculated.
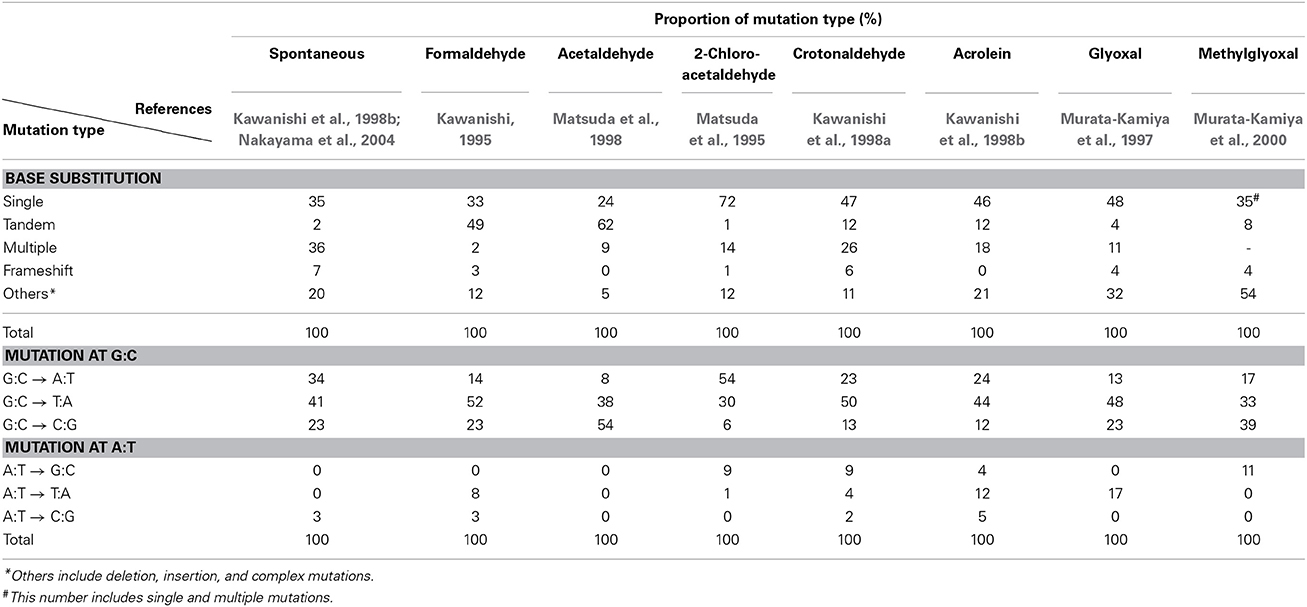
Table 1. Types of mutations induced by various aldehydes in supF shuttle vector plasmids propagated in mammalian cells.
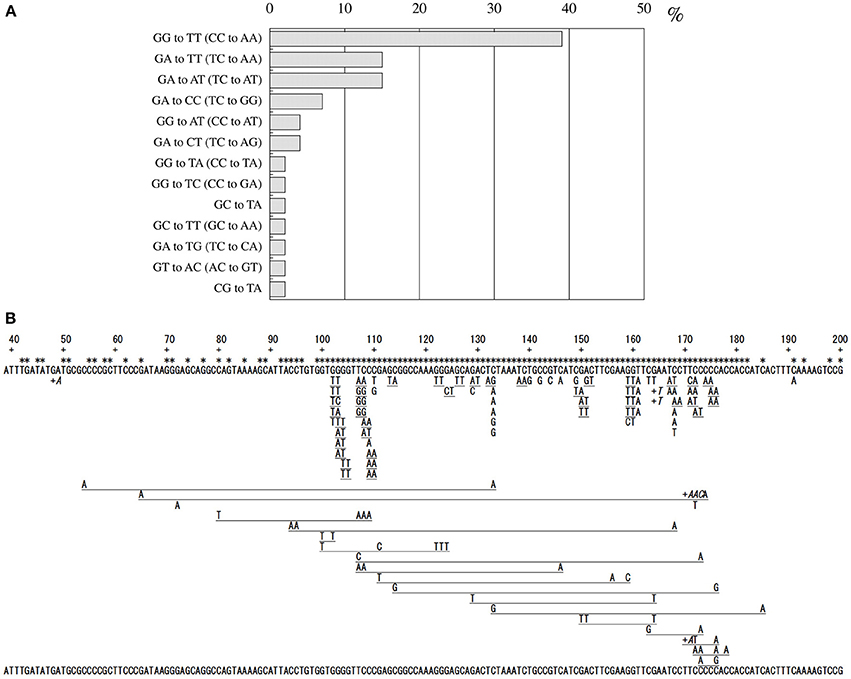
Figure 4. Classification of formaldehyde-induced tandem base substitution mutations (A), and Location of formaldehyde-induced base substitution and frameshift mutations in supF gene (B). Method: Mutated plasmids were extracted and purified, and the size of each plasmid was checked by agarose gel electrophoresis. The nucleotide sequences of the supF gene of the plasmids of a normal size were determined with the ABI DNA sequencer. Each letter under the sequence of the supF gene represents a base substitution. Underlined adjacent letters mean tandem base substitutions found in a plasmid. The “+” and adjacent letters (in italics) mean base insertion at the position just to the right of “+.” Multiple letters with long underline show multiple base substitutions found in a plasmid. Star symbols mean positions of the targets of mutations that can inactivate the suppressor function.
Comparison of Mutation Spectra with Other Aldehydes
Mutagenesis of various aldehydes such as acetaldehyde, 2-chloroacetaldehyde, crotonaldehyde, acrolein, glyoxal, and methylglyoxal has been examined using supF shuttle vector plasmids (Matsuda et al., 1995, 1998; Murata-Kamiya et al., 1997, 2000; Kawanishi et al., 1998a,b). These aldehydes react with DNA and form exocyclic guanines, ethenoguanine, and propanoguanine, as major adducts (Minko et al., 2009; Voulgaridou et al., 2011). These DNA lesions generate mutations mainly at G:C base pairs, but the spectra of base substitutions differ among aldehydes with different chemical structures. Table 1 summarizes mutation spectra of various aldehydes, which are induced in the shuttle vector plasmids propagated in mammalian cells. All aldehydes induced base substitutions manly at G:C base pairs. It should be noted that acetaldehyde induced tandem base substitutions at 5′-GG sequences like formaldehyde, suggesting that acetaldehyde forms intra-strand crosslinks between adjacent guanines, probably through propanoguanine formation (Matsuda et al., 1998). Glyoxal is an endogenous mutagen produced abundantly in the process of DNA oxidation by reactive oxygen free radicals that have been thought to contribute to the pathogenesis of many age-related human diseases including cancer. Features of glyoxal-induced mutations are similar to those of spontaneous mutations (Table 1), which may suggest that glyoxal is a cause of spontaneous mutations.
Conclusion
Formaldehyde is one of the most abundantly exposed environmental mutagens for humans, and shown to be carcinogenic in experimental animals. Formaldehyde causes MN, SCE, and CA in mammalian cells and induces gene mutations in mammalian and bacterial cells. Formaldehyde acts as an electrophile to react mostly with guanine and adenine in DNA, and forms various kinds of DNA lesions. The DNA lesions are classified into DNA adducts, DNA intra-strand crosslinks, DNA inter-strand crosslinks and DNA-protein crosslinks. Organisms have mechanisms to remove these DNA lesions, which are specific to each type of DNA lesions. If these DNA lesions are not repaired, mutations are induced during DNA replication. Formaldehyde-induced mutations in human cells were analyzed using shuttle vector plasmids (Kawanishi, 1995). Interestingly, about half of the formaldehyde-induced mutations were tandem base substitutions, and most of them occurred at 5′-GG (or 5′-CC) and 5′-GA (or 5′-TC) sequences. These mutation features agree with the predominance of formaldehyde-induced 5′-GG and 5′-GA crosslinks, indicating that the intra-strand crosslinks lead to tandem base substitutions. The prevalence of N2-hydroxymethyl adduct to guanine would lead to the G to T transversion that is the major single base substitution mutation. In early studies using bacteria, cultured cells and animals, exposure concentrations of formaldehyde would not have been sufficiently high to manifest the features of the mutation spectrum because formaldehyde mutagenicity is relatively weak and its acute cytotoxic effect is strong compared with those of other environmental mutagens. Our findings as well as the accumulation of molecular data provide clues to promote our understanding of the mechanisms behind the genotoxicity and carcinogenicity of formaldehyde.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported in part by Grants-in-Aid for Scientific Research (B) (KAKENHI Grants 24310047) from the Japan Society for the Promotion of Science (JSPS).
References
Akasaka, S., Takimoto, K., and Yamamoto, K. (1992). G:C→T:A and G:C→C:G transversions are the predominant spontaneous mutations in the Escherichia coli supF gene: an improved lacZ(am) E. coli host designed for assaying pZ189 supF mutational specificity. Mol. Gen. Genet. 235, 173–178. doi: 10.1007/BF00279358
Bachand, A. M., Mundt, K. A., Mundt, D. J., and Montgomery, R. R. (2010). Epidemiological studies of formaldehyde exposure and risk of leukemia and nasopharyngeal cancer: a meta-analysis. Crit. Rev. Toxicol. 40, 85–100. doi: 10.3109/10408440903341696
Basler, A., van der Hude, W., and Scheutwinkel-Reich, M. (1985). Formaldehyde-induced sister chromatid exchanges in vitro and the influence of the exogenous metabolizing systems S9 mix and primary hepatocytes. Arch. Toxicol. 58, 10–13. doi: 10.1007/BF00292609
Beane Freeman, L. E., Blair, A., Lubin, J. H., Stewart, P. A., Hayes, R. B., Hoover, R. N., et al. (2009). Mortality from lymphohematopoietic malignancies among workers in formaldehyde industries: the National Cancer Institute Cohort. J. Natl. Cancer Inst. 101, 751–761. doi: 10.1093/jnci/djp096
Beland, F. A., Fullerton, N. F., and Heflich, R. H. (1984). Rapid isolation, hydrolysis and chromatography of formaldehyde-modified DNA. J. Chromatogr. 308, 121–131. doi: 10.1016/0378-4347(84)80202-9
Bosetti, C., McLaughlin, J. K., Tarone, R. E., Pira, E., and La Vecchia, C. (2008). Formaldehyde and cancer risk: a quantitative review of cohort studies through 2006. Ann. Oncol. 19, 29–43. doi: 10.1093/annonc/mdm202
Cheng, G., Shi, Y., Sturla, S. J., Jalas, J. R., McIntee, E. J., Villalta, P. W., et al. (2003). Reactions of formaldehyde plus acetaldehyde with deoxyguanosine and DNA: formation of cyclic deoxyguanosine adducts and formaldehyde cross-links. Chem. Res. Toxicol. 16, 145–152. doi: 10.1021/tx025614r
Coggon, D., Harris, E. C., Poole, J., and Palmer, K. T. (2003). Extended follow-up of a cohort of British chemical workers exposed to formaldehyde. J. Natl. Cancer Inst. 95, 1608–1615. doi: 10.1093/jnci/djg046
Conaway, C. C., Whysner, J., Verna, L. K., and Williams, G. M. (1996). Formaldehyde mechanistic data and risk assessment: endogenous protection from DNA adduct formation. Pharmacol. Ther. 71, 29–55. doi: 10.1016/0163-7258(96)00061-7
Costa, S., Coelho, P., Costa, C., Silva, S., Mayan, O., Santos, L. S., et al. (2008). Genotoxic damage in pathology anatomy laboratory workers exposed to formaldehyde. Toxicology 252, 40–48. doi: 10.1016/j.tox.2008.07.056
Crosby, R. M., Richardson, K. K., Craft, T. R., Benforado, K. B., Liber, H. L., and Skopek, T. R. (1988). Molecular analysis of formaldehyde-induced mutations in human lymphoblasts and E. coli. Environ. Mol. Mutag. 12, 155–166. doi: 10.1002/em.2860120202
Dallas, C. E., Scott, M. J., Ward, J. B. Jr., and Theiss, J. C. (1992). Cytogenetic analysis of pulmonary lavage and bone marrow cells of rats after repeated formaldehyde inhalation. J. Appl. Toxicol. 12, 199–203. doi: 10.1002/jat.2550120309
Galloway, S. M., Bloom, A. S., Resnick, M., Margolin, B. H., Nakamura, F., Archer, P., et al. (1985). Development of a standard protocol for in vitro cytogenetic testing with Chinese hamster ovary cells: comparison of results for 22 compounds in two laboratories. Environ. Mutagen. 7, 1–51. doi: 10.1002/em.2860070102
Gocke, E., King, M.-T., Eckhardt, K., and Wild, D. (1981). Mutagenicity of cosmetic ingredients licensed by the European communities. Mutat. Res. 90, 91–109. doi: 10.1016/0165-1218(81)90072-0
Goldmacher, V. S., and Thilly, W. G. (1983). Formaldehyde is mutagenic for cultured human cells. Mutat. Res. 116, 417–422. doi: 10.1016/0165-1218(83)90080-0
Graves, R. J., Trueman, P., Jones, S., and Green, T. (1996). DNA sequence analysis of methylene chloride-induced HPRT mutations in Chinese hamster ovary cells: comparison with the mutation spectrum obtained for 1,2-dibromoethane and formaldehyde. Mutagenesis 11, 229–233. doi: 10.1093/mutage/11.3.229
Hauptmann, M., Lubin, J. H., Stewart, P. A., Hayes, R. B., and Blair, A. (2004). Mortality from solid cancers among workers in formaldehyde industries. Am. J. Epidemiol. 159, 1117–1130. doi: 10.1093/aje/kwh174
Hauptmann, M., Stewart, P. A., Lubin, J. H., Beane Freeman, L. E., Hornung, R. W., Herrick, R. F., et al. (2009). Mortality from lymphohematopoietic malignancies and brain cancer among embalmers exposed to formaldehyde. J. Natl. Cancer Inst. 101, 1696–1708. doi: 10.1093/jnci/djp416
Hedberg, J. J., Hoog, J. O., and Grafstrom, R. C. (2002). “Assessment of formaldehyde metabolizing enzymes in human oral mucosa and cultured oral keratinocytes indicate high capacity for detoxification of formaldehyde,” in Crucial Issues in Inhalation Research—Mechanistic, Clinical and Epidemiologic (INIS Monographs), eds U. Heinrich and U. Mohr (Stuttgart: Fraunhofer IRB Verlag), 103–115.
Huang, H., and Hopkins, P. B. (1993). DNA interstrand cross-linking by formaldehyde: nucleotide sequence preference and covalent structure of the predominant cross-link formed in synthetic oligonucleotides. J. Am. Chem. Soc. 115, 9402–9408. doi: 10.1021/ja00074a005
Huang, H., Solomon, M. S., and Hopkins, P. B. (1992). Formaldehyde preferentially interstrand cross-links duplex DNA through deoxyguanosine residues at the sequence 5′-d(AT). J. Am. Chem. Soc. 114, 9240–9241. doi: 10.1021/ja00049a097
IARC. (2006). “Formaldehyde, 2-butoxyethanol and 1-tertbutoxypropan-2-ol,” in International Agency for Research on Cancer Monographs on the Evaluation of Carcinogenic Risks to Human (Lyon: International Agency for Research on Cancer, WHO), 1–478. Available online at: http://monographs.iarc.fr/ENG/Monographs/vol88/index.php
IARC. (2012). “A review of human carcinogens: chemical agents and related occupations,” in International Agency for Research on Cancer Monographs on the Evaluation of Carcinogenic Risks to Human (Lyon: International Agency for Research on Cancer, WHO), 401–435. Available online at: http://monographs.iarc.fr/ENG/Monographs/vol100F/index.php
Kaden, D. A., Mandin, C., Nielsen, G. D., and Wolkoff, P. (2010). “Formaldehyde” in WHO Guidelines for Indoor Air Quality: Selected Pollutants (Copenhagen: World Health Organization Regional Office for Europe), 103–156.
Kawanishi, M. (1995). Molecular Analysis of Mutations Induced by Formaldehyde and Acetaldehyde in Human Cells using Shuttle Vectors. Master's thesis No. GE-M-1995-42. Graduate School of Engineering, Kyoto University.
Kawanishi, M., Matsuda, T., Sasaki, G., Yagi, T., Matsui, S., and Takebe, H. (1998a). A spectrum of mutations induced by crotonaldehyde in shuttle vector plasmids propagated in human cells. Carcinogenesis 19, 69–72. doi: 10.1093/carcin/19.1.690
Kawanishi, M., Matsuda, T., Nakayama, A., Takebe, H., Matsui, S., and Yagi, T. (1998b). Molecular analysis of mutations induced by acrolein in human fibroblast cells using supF shuttle vector plasmids. Mutat. Res. 417, 65–73. doi: 10.1016/S1383-5718(98)00093-X
Kligerman, A. D., Phelps, M. C., and Erexson, G. L. (1984). Cytogenetic analysis of lymphocytes from rats following formaldehyde inhalation. Toxicol. Lett. 21, 241–246. doi: 10.1016/0378-4274(84)90079-1
Kottemann, M. C., and Smogorzewska, A. (2013). Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493, 356–363. doi: 10.1038/nature11863
Kreiger, R. A., and Garry, V. F. (1983). Formaldehyde-induced cytotoxicity and sister-chromatid exchanges in human lymphocyte cultures. Mutat. Res. 120, 51–55. doi: 10.1016/0165-7992(83)90073-8
Kumari, A., Lim, Y. X., Newell, A. H., Olson, S. B., and McCullough, A. K. (2012). Formaldehyde-induced genome instability is suppressed by an XPF-dependent pathway. DNA Repair 11, 236–246. doi: 10.1016/j.dnarep.2011.11.001
Liber, H. L., Benforado, K., Crosby, R. M., Simpson, D., and Skopek, T. R. (1989). Formaldehyde-induced and spontaneous alterations in human hprt DNA sequence and mRNA expression. Mutat. Res. 226, 31–37. doi: 10.1016/0165-7992(89)90089-4
Lu, K., Ye, W., Zhou, L., Collins, L. B., Chen, X., Gold, A., et al. (2010). Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 132, 3388–99. doi: 10.1021/ja908282f
Ma, T. H., and Harris, M. M. (1988). Review of the genotoxicity of formaldehyde. Mutat. Res. 196, 37–59. doi: 10.1016/0165-1110(88)90027-9
Marsh, G. M., and Youk, A. O. (2005). Reevaluation of mortality risks from nasopharyngeal cancer in the formaldehyde cohort study of the National Cancer Institute. Regul. Toxicol. Pharmacol. 42, 275–283. doi: 10.1016/j.yrtph.2005.05.003
Marsh, G. M., Youk, A. O., Buchanich, J. M., Erdal, S., and Esmen, N. A. (2007a). Work in the metal industry and nasopharyngeal cancer mortality among formaldehyde-exposed workers. Regul. Toxicol. Pharmacol. 48, 308–319. doi: 10.1016/j.yrtph.2007.04.006
Marsh, G. M., Youk, A. O., and Morfeld, P. (2007b). Mis-specified and non-robust mortality risk models for nasopharyngeal cancer in the National Cancer Institute formaldehyde worker cohort study. Regul. Toxicol. Pharmacol. 47, 59–67. doi: 10.1016/j.yrtph.2006.07.007
Matsuda, T., Kawanishi, M., Yagi, T., Matsui, S., and Takebe, H. (1998). Specific tandem GG to TT base substitutions induced by acetaldehyde are due to intra-strand crosslinks between adjacent guanine bases. Nucleic Acids Res. 26, 1769–1774. doi: 10.1093/nar/26.7.1769
Matsuda, T., Yagi, T., Kawanishi, M., Matsui, S., and Takebe, H. (1995). Molecular analysis of mutations induced by 2-chloroacetaldehyde, the ultimate carcinogenic form of vinyl chloride, in human cells using shuttle vectors. Carcinogenesis 16, 2389–2394. doi: 10.1093/carcin/16.10.2389
Merk, O., and Speit, G. (1998). Significance of formaldehyde-induced DNA–protein crosslinks for mutagenesis. Environ. Mol. Mutag. 32, 260–268. doi: 10.1002/(SICI)1098-2280(1998)32:3<260::AID-EM9>3.0.CO;2-M
Merk, O., and Speit, G. (1999). Detection of crosslinks with the comet assay in relationship to genotoxicity and cytotoxicity. Environ. Mol. Mutag. 33, 167–172. doi: 10.1002/(SICI)1098-2280(1999)33:2<167::AID-EM9>3.0.CO;2-D
Migliore, L., Ventura, L., Barale, R., Loprieno, N., Castellino, S., and Pulci, R. (1989). Micronuclei and nuclear anomalies induced in the gastro-intestinal epithelium of rats treated with formaldehyde. Mutagenesis 4, 327–334. doi: 10.1093/mutage/4.5.327
Minko, I., G., Kozekov, I. D., Harris, T. M., Rizzo, C. J., Lloyd, R. S., and Stone, M. P. (2009). Chemistry and biology of DNA containing 1,N(2)-deoxyguanosine adducts of the alpha,beta-unsaturated aldehydes, acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol. 22, 759–778. doi: 10.1021/tx9000489
Murata-Kamiya, N., Kamiya, H., Kaji, H., and Kasai, H. (1997). Glyoxal, a major product of DNA oxidation, induces mutations at G:C sites on a shuttle vector plasmid replicated in mammalian cells. Nucleic Acids Res. 25, 1897–1902. doi: 10.1093/nar/25.10.1897
Murata-Kamiya, N., Kamiya, H., Kaji, H., and Kasai, H. (2000). Methyl glyoxal induces G:C to C:G and G:C to T:A transversions in the supF gene on a shuttle vector plasmid replicated in mammalian cells. Mutat. Res. 468, 173–182. doi: 10.1016/S1383-5718(00)00044-9
Nakano, T., Katafuchi, A., Matsubara, M., Terato, H., Tsuboi, T., Masuda, T., et al. (2009). Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J. Biol. Chem. 284, 27065–27076. doi: 10.1074/jbc.M109.019174
Nakano, T., Morishita, S., Katafuchi, A., Matsubara, M., Horikawa, Y., Terato, H., et al. (2007). Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol. Cell 28, 147–158. doi: 10.1016/j.molcel.2007.07.029
Nakayama, A., Noguchi, Y., Mori, T., Morisawa, S., and Yagi, T. (2004). Comparison of mutagenic potentials and mutation spectra of benzene metabolites using supF shuttle vectors in human cells. Mutagenesis 19, 91–97. doi: 10.1093/mutage/geh009
Natarajan, A. T., Darroudi, F., Bussman, C. J. M., and van Kesteren-van Leeuwen, A. C. (1983). Evaluation of the mutagenicity of formaldehyde in mammalian cytogenetic assays in vivo and in vitro. Mutat. Res. 122, 355–360. doi: 10.1016/0165-7992(83)90019-2
Norbäck, D. (2009). An update on sick building syndrome. Curr. Opin. Allergy Clin. Immunol. 9, 55–59. doi: 10.1097/ACI.0b013e32831f8f08
O'Donovan, M. R., and Mee, C. D. (1993). Formaldehyde is a bacterial mutagen in a range of Salmonella and Escherichia indicator strains. Mutagenesis 8, 577–581.
Pala, M., Ugolini, D., Ceppi, M., Rizzo, F., Maiorana, L., Bolognesi, C., et al. (2008). Occupational exposure to formaldehyde and biological monitoring of research Institute workers. Cancer Detect. Prev. 32, 121–126. doi: 10.1016/j.cdp.2008.05.003
Pinkerton, L. E., Hein, M. J., and Stayner, L. T. (2004). Mortality among a cohort of garment workers exposed to formaldehyde: an update. Occup. Environ. Med. 61, 193–200. doi: 10.1136/oem.2003.007476
Recio, L., Sisk, S., Pluta, L., Bermudez, E., Gross, E. A., Chen, Z., et al. (1992). p53 Mutations in formaldehyde-induced nasal squamous cell carcinomas in rats. Cancer Res. 52, 6113–6116.
Speit, G., and Merk, O. (2002). Evaluation of mutagenic effects of formaldehyde in vitro: detection of crosslinks and mutations in mouse lymphoma cells. Mutagenesis 17, 183–187. doi: 10.1093/mutage/17.3.183
Swenberg, J. A., Moeller, B. C., Lu, K., Rager, J. E., Fry, R. C., and Starr, T. B. (2013). Formaldehyde carcinogenicity research: 30 years and counting for mode of action, epidemiology, and cancer risk assessment. Toxicol. Pathol. 41, 181–189. doi: 10.1177/0192623312466459
Takahashi, K., Morita, T., and Kawazoe, Y. (1985). Mutagenic characteristics of formaldehyde on bacterial systems. Mutat. Res. 156, 153–161. doi: 10.1016/0165-1218(85)90058-8
Unno, J., Itaya, A., Taoka, M., Sato, K., Tomida, J., Sakai, W., et al. (2014). FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 7, 1039–1047. doi: 10.1016/j.celrep.2014.04.005
Voulgaridou, G. P., Anestopoulos, I., Franco, R., Panayiotidis, M. I., and Pappa, A. (2011). DNA damage induced by endogenous aldehydes: current state of knowledge. Mutat. Res. 711, 13–27. doi: 10.1016/j.mrfmmm.2011.03.006
Wang, M., Cheng, G., Balbo, S., Carmella, S. G., Villalta, P. W., and Hecht, S. S. (2009). Clear differences in levels of a formaldehyde-DNA adduct in leukocytes of smokers and nonsmokers. Cancer Res. 69, 7170–7174. doi: 10.1158/0008-5472.CAN-09-1571
Yagi, T. (2013). The achievement of shuttle vector techniques in mammalian cell mutation research. Genes Environ. 35, 93–98. doi: 10.3123/jemsge.2013.001
Ye, X., Yan, W., Xie, H., Zhao, M., and Ying, C. (2005). Cytogenetic analysis of nasal mucosa cells and lymphocytes from high level long-term formaldehyde exposed workers and low-level short-term exposed waiters. Mutat. Res. 588, 22–27. doi: 10.1016/j.mrgentox.2005.08.005
Zhang, L., Steinmaus, C., Eastmond, D. A., Xin, X. K., and Smith, M. T. (2009). Formaldehyde exposure and leukemia: a new meta-analysis and potential mechanisms. Mutat. Res. 681, 150–168. doi: 10.1016/j.mrrev.2008.07.002
Keywords: genotoxicity, mutation spectrum, supF, DNA intra-strand crosslink, DNA damage
Citation: Kawanishi M, Matsuda T and Yagi T (2014) Genotoxicity of formaldehyde: molecular basis of DNA damage and mutation. Front. Environ. Sci. 2:36. doi: 10.3389/fenvs.2014.00036
Received: 09 June 2014; Accepted: 23 August 2014;
Published online: 16 September 2014.
Edited by:
Eduardo Alves Almeida, São Paulo State University, BrazilReviewed by:
Jamel Jebali, Higher Institute of Biotechnology of Monastir, TunisiaCamilo Dias Seabra Pereira, Federal University of São Paulo, Brazil
Copyright © 2014 Kawanishi, Matsuda and Yagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Yagi, Laboratory of Molecular and Cellular Genetics, Department of Biology, Graduate School of Science and Radiation Research Center, Osaka Prefecture University, 1-2, Gakuen-cho, Naka-ku, Sakai, Osaka 599-8570, Japan e-mail: yagi-t@riast.osakafu-u.ac.jp
 Masanobu Kawanishi
Masanobu Kawanishi Tomonari Matsuda2
Tomonari Matsuda2  Takashi Yagi
Takashi Yagi