Mitochondrial isocitrate dehydrogenase is inactivated upon oxidation and reactivated by thioredoxin-dependent reduction in Arabidopsis
- 1Chemical Resources Laboratory, Tokyo Institute of Technology, Midori-ku, Yokohama, Japan
- 2Core Research for Evolutional Science and Technology, Japan Science and Technology Agency, Tokyo, Japan
Regulation of mitochondrial metabolism is essential for ensuring cellular growth and maintenance in plants. Based on redox-proteomics analysis, several proteins involved in diverse mitochondrial reactions have been identified as potential redox-regulated proteins. NAD+-dependent isocitrate dehydrogenase (IDH), a key enzyme in the tricarboxylic acid (TCA) cycle, is one such candidate. In this study, we investigated the redox regulation mechanisms of IDH by biochemical procedures. In contrast to mammalian and yeast counterparts reported to date, recombinant IDH in Arabidopsis mitochondria did not show adenylate-dependent changes in enzymatic activity. Instead, IDH was inactivated by oxidation treatment and partially reactivated by subsequent reduction. Functional IDH forms a heterodimer comprising regulatory (IDH-r) and catalytic (IDH-c) subunits. IDH-r was determined to be the target of oxidative modifications forming an oligomer via intermolecular disulfide bonds. Mass spectrometric analysis combined with tryptic digestion of IDH-r indicated that Cys128 and Cys216 are involved in intermolecular disulfide bond formation. Furthermore, we showed that mitochondria-localized o-type thioredoxin (Trx-o) promotes the reduction of oxidized IDH-r. These results suggest that IDH-r is susceptible to oxidative stress, and Trx-o serves to convert oxidized IDH-r to the reduced form that is necessary for active IDH complex.
Introduction
Mitochondria play a pivotal role in providing ATP required for various cellular events in all eukaryotes. In mitochondrial respiration, the tricarboxylic acid (TCA) cycle generates NADH and FADH2 by metabolizing organic acids. These products are then used to drive electron transport in the respiratory chain and coupled ATP production. In addition to this fundamental energy conversion process, mitochondria host a large number of metabolic pathways. Flexible regulation of these mitochondrial reactions is important for ensuring proper cellular function, particularly in plants, which cannot escape exposure to adverse environmental conditions. Although our knowledge of the regulatory mechanisms of plant mitochondria has advanced, much still remains to be solved, particularly the question of how each mitochondrial enzyme is controlled at the post-translational level (Millar et al., 2011; Tcherkez et al., 2012; Lázaro et al., 2013; Nunes-Nesi et al., 2013).
In the last decade, the progress of redox-proteomics analysis has provided hints that a wide variety of biological processes are governed by the redox state of their responsible enzymes (Hisabori et al., 2007; Montrichard et al., 2009; Lindahl et al., 2011). Thioredoxin (Trx), a small ubiquitous protein, plays a crucial role in redox regulation. Trx has a conserved WCGPC motif at an active site, enabling a dithiol–disulfide exchange reaction with the target enzyme. Based on the subcellular localization and sequence similarity, plant Trxs are classified into seven subtypes (f-, m-, h-, o-, x-, y-, and z-type). Although it has been recognized that the Trx-o resides in plant mitochondria (Laloi et al., 2001), information about the target proteins of Trx-o is limited to date. Using Trx affinity chromatography (Motohashi et al., 2001), we recently performed the systematic screening of Trx-targeted proteins in plant mitochondria and identified a list of target candidates (Yoshida et al., 2013). Redox-proteomics studies by other groups have also suggested that diverse proteins involved in manifold mitochondrial processes are redox-regulated via the interaction with Trx (Balmer et al., 2004; Winger et al., 2007). However, careful biochemical study is needed to determine whether these candidate proteins are actually redox-regulated.
NAD+-dependent isocitrate dehydrogenase (IDH) is one of the proteins that were captured by Trx affinity chromatography (Yoshida et al., 2013). IDH catalyzes the oxidative decarboxylation of isocitrate to 2-oxoglutarate coupled to NADH generation, and thus supports the TCA cycle flux. In yeast, it is well established that the minimal functional unit of IDH is a heterodimer comprising regulatory (IDH-r) and catalytic (IDH-c) subunits (Panisko and McAlister-Henn, 2001). Furthermore, detailed biochemical and structural analyses have provided evidence that yeast IDH is allosterically activated by AMP and inactivated by intermolecular disulfide bond formation between IDH-c subunits (Lin and McAlister-Henn, 2003; Taylor et al., 2008; Garcia et al., 2009). In contrast, to our knowledge, there are few reports on the biochemical analysis of plant IDH, although plant IDH has been also suggested to be active in a heterodimeric form based on sequence comparison with its yeast counterpart and the complementation test of yeast IDH mutants with plant IDH genes (Lancien et al., 1998; Lemaitre and Hodges, 2006). The regulatory mechanism of plant mitochondrial IDH thus remains poorly characterized.
In this study, we focused on the molecular basis for controlling IDH activity in Arabidopsis mitochondria with special attention to redox regulation. The results indicate that IDH-r forms intermolecular disulfide bonds upon oxidation, leading to a drastic decrease in IDH activity. We also showed that Trx-o assists in the reduction of oxidized IDH-r. Based on these findings, a novel regulatory mode of plant mitochondrial IDH is discussed.
Materials and Methods
Preparation of Expression Plasmids for IDH-r and IDH-c
Total RNA was isolated from Arabidopsis thaliana as described in Yoshida and Noguchi (2009) and used as a template for RT-PCR. The IDH1 (At4g35260, encoding IDH-r) and IDH5 (At5g03290, encoding IDH-c) gene fragments encoding the mature protein region (Val26-Asp367 and Ile44-Leu374, respectively) were amplified with the following oligonucleotide primer combination; 5′-AACTGCAGCATATGGTGACTTACATGCCCAGACC-3′ (NdeI) and 5′- GCGAATTCAGTCTAGTTTTGCAATGA-3′ (EcoRI) for IDH1, 5′- AACTGCAGCATATGATCACCGCAACTCTCTTCCCT-3′ (NdeI) and 5′-AAGGATCCTCAGAGATGATCACAGATTG-3′ (BamHI) for IDH5. The restriction sites for the enzyme shown in parentheses are underlined. Each of the amplified DNA was ligated into the pET23c expression vector (Novagen). The sequences were confirmed by DNA sequencing (3730xl DNA Analyzer; Applied Biosystems).
Protein Expression and Purification
The IDH1 and IDH5 expression plasmids described above were transformed into E. coli strain BL21 (DE3) to express IDH-r and IDH-c proteins, respectively. The transformed cells were grown at 37°C until A600 = 0.4–0.8. Expression was induced by adding 0.5 mM IPTG, followed by further culture at 21°C overnight. The E. coli cells were disrupted by sonication. After centrifugation (125,000 ×g for 40 min), the resulting supernatant was used to purify the protein of interest. Each protein was purified by a combination of anion exchange chromatography, using a DEAE-Toyopearl 650M column (Tosoh) and Q-Toyopearl 600C column (Tosoh), and hydrophobic interaction chromatography, using a Butyl-Toyopearl 650M column (Tosoh), as described in Yoshida et al. (2013). Purification was performed in a medium containing 25 mM Tris-HCl (pH 7.5–8.1), 1 mM EDTA, and 0.5 mM DTT, but EDTA and DTT were removed by dialysis after purification. All of the procedures during purification were carried out at 4°C. The protein concentration was determined with a BCA protein assay (Pierce).
IDH Activity Measurement
Prior to the assay, 2 μM IDH-r and 2 μM IDH-c were mixed in a medium containing 25 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 20 mM isocitrate. After incubation at 25°C for 30 min, the mixed solution was used for activity measurement. IDH activity was monitored as an increase in absorbance at 340 nm due to NAD+ reduction. The molar extinction coefficient for NADH of 6.22 mM−1 was used for calculation of the amounts of generated NADH. Assays were performed at 25°C in a medium containing 25 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM NAD+, and indicated concentrations of isocitrate. IDH-r and IDH-c were added at 40 nM each.
To test the adenylate effects on IDH activity, each adenylate (AMP, ADP, and ATP) was added at 1 mM to the media described above.
Peptide Mapping Analysis
After separation by non-reducing SDS-PAGE, stained protein bands of interest were excised from the gel and fully destained with 50 mM NH4HCO3 and 50% (v/v) acetonitrile. The gel slice was then incubated in 55 mM iodoacetamide and 100 mM NH4HCO3 for protecting free Cys residues. The gel slice was dried completely and then incubated with 50 mM NH4HCO3 containing 20 ng μl−1 trypsin at 37°C overnight. Tryptic peptides were extracted from the gel with 0.1% (v/v) trifluoroacetic acid with 50 and 75% (v/v) acetonitrile, continuously. Whole extracts were concentrated using a centrifugal concentrator and desalted using Solid Phase Extraction C-TIP (Nikkyo Technos). The peptide sample was spotted onto the matrix (α-cyano-4-hydroxycinnamic acid) and air-dried on a MALDI plate (MTP 384 target plate ground steel BC, Bruker Daltonics). MALDI mass spectra were obtained using an UltrafleXtreme-TK2 spectrometer (Bruker Daltonics). Results were queried with the Mascot search engine (http://www.matrixscience.com/) to identify matched peptides.
Trx-Dependent Reduction of Oxidized IDH-r
For oxidation treatment, IDH-r was incubated in 50 μM CuCl2 for 15 min at 25°C. After dialysis for the removal of CuCl2, the oxidized IDH-r was incubated in a medium containing 25 mM Tris-HCl (pH 7.5), indicated concentrations of DTT, and indicated concentrations of Trx-o1 for 30 min at 25°C. The IDH-r redox state was assayed by non-reducing SDS-PAGE. IDH-r equivalent to 1 μg was loaded into each lane. The Arabidopsis recombinant Trx-o1 protein was prepared and confirmed to be efficient in dithiol–disulfide exchange reaction in our previous study (Yoshida et al., 2013).
Results
Arabidopsis IDH is Insensitive to Adenylates
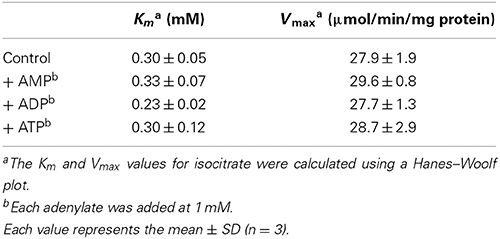
In Arabidopsis, there are five genes encoding mitochondrial IDH subunits. Based on the similarity with yeast IDH, three genes (IDH1, IDH2, and IDH3) are regarded as the genes encoding the IDH-r subunit, whereas two genes (IDH5 and IDH6) encode the IDH-c subunit (Supplementary Fig. S1). Primary amino acid sequences of these gene products show high identity among each isoform (IDH1-3; >84%, IDH5-6; 90%, mature protein region except for targeting peptide). Given that IDH1 and IDH5 gene products appear to be more abundantly expressed in Arabidopsis (Yoshida et al., 2013), we prepared recombinant proteins from these genes as representative IDH-r and IDH-c, respectively (Figure 1A). NAD+-reducing activity was observed in the presence of both subunits, whereas neither subunit alone showed any catalytic activity (Figure 1B). This finding clearly shows that both IDH-r and IDH-c are essential for functional IDH in Arabidopsis.
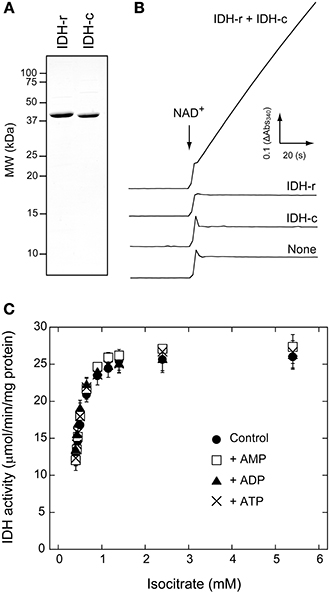
Figure 1. Activity measurements of Arabidopsis IDH recombinant protein. (A) SDS-PAGE profiles of purified recombinant IDH-r and IDH-c. (B) Monitoring of NAD+-reducing activity of IDH. Time course of absorbance change at 340 nm (Abs340) is shown. IDH-r and/or IDH-c (40 nM each) were added to a reaction medium. Isocitrate concentration was 5.4 mM. The reaction was initiated by the addition of NAD+ (2 mM) at the time point indicated by arrow. See Materials and Methods for details. (C) The effect of adenylate on IDH activity under several concentrations of isocitrate. Each adenylate (AMP, ADP, ATP) was added at 1 mM. Each value represents the mean ± SD (n = 3).
It has been well documented that ADP and AMP act as allosteric activators of IDH in mammal and yeast (Nunes-Nesi et al., 2013). For example, the Km value for isocitrate in yeast IDH is drastically lowered in the presence of AMP (Lin and McAlister-Henn, 2003). However, it remains unclear whether this regulatory mechanism is common for plant IDH. We accordingly investigated the effects of adenylates on IDH activity. No adenylates (AMP, ADP, and ATP) affected the saturation velocity curve (Figure 1C) or correspondingly the Km and Vmax values (Table 1) for isocitrate. These results imply that changes in the mitochondrial adenylate energy charge have no direct impact on IDH activity in Arabidopsis.
IDH-r Forms Intermolecular Disulfides, Leading to a Loss of IDH Catalytic Activity
We next investigated whether IDH activity is controlled by the redox state of the enzyme molecule itself. Prior to the assay, IDH-r or IDH-c was incubated in the presence of 50 μM CuCl2 to oxidize the possible thiols on the protein molecule. While the oxidation treatment of IDH-c exerted no significant effect, the IDH-r oxidation nearly completely suppressed enzymatic activity when the oxidized IDH-r was mixed with untreated IDH-c (Figure 2A). As revealed by non-reducing SDS-PAGE, IDH-r was shifted to dimeric, trimeric, and higher-order oligomeric forms mediated by intermolecular disulfide bonds under oxidative conditions (Figure 2B). The reduction of oxidized IDH-r restored IDH activity to approximately half of the control level (Figure 2A), accompanied by the cleavage of the intermolecular disulfide bonds (Figure 2C). It thus appeared that Arabidopsis IDH is reversibly inactivated in response to oxidative stress via oligomer formation of IDH-r.
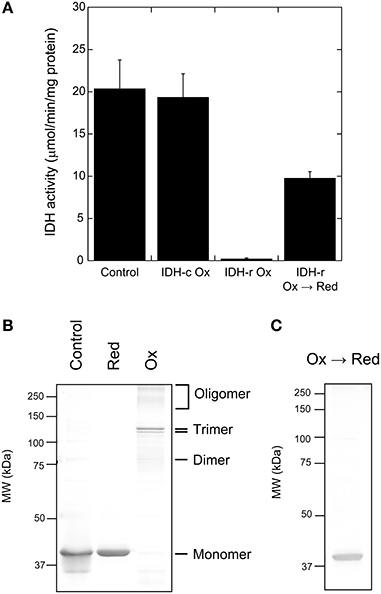
Figure 2. Redox regulation of Arabidopsis IDH recombinant protein. (A) Redox-dependent change in IDH activity. For oxidation (Ox) treatment, IDH-r or IDH-c was incubated in 50 μM CuCl2 for 15 min. For re-reduction (Ox → Red) treatment, IDH-r after Ox treatment was incubated in 50 mM DTT for 30 min. Each value represents the mean ± SD (n = 4). (B) Non-reducing SDS-PAGE profiles of IDH-r after reduction (Red) treatment (5 mM DTT for 15 min) or Ox treatment. (C) Non-reducing SDS-PAGE profiles of IDH-r after Ox → Red treatment.
Cys128 and Cys216 Conserved in IDH-r Play a Critical Role in the Redox Regulation of IDH
In plant IDH-r, six Cys residues are commonly conserved (Supplementary Fig. S1). We attempted to identify the Cys residues involved in intermolecular disulfide bond formation of IDH-r. For this purpose, proteins in the reduced monomeric and the oxidized trimeric forms (the prevalent form of IDH-r oligomer) were in-gel digested using trypsin after non-reducing SDS-PAGE (Figure 2B). Mass spectra of the resulting peptides were then acquired (Figure 3). Three major peptides (m/z: 1280.6, 2276.2, and 2404.3) were specifically detected in the reduced monomeric form but not in the oxidized trimeric form. By searching for matching peptides using Mascot, the peptides were determined to correspond to Leu207-Arg217, Glu118-Arg137, and Lys117-Arg137 (calculated masses: 1279.6, 2275.2, and 2403.3, respectively) in IDH-r, respectively. The peptide Leu207-Arg217 contained Cys216, whereas Glu118-Arg137 and Lys117-Arg137 contained Cys128. These Cys residues are thus likely to be primarily involved in redox changes in the Arabidopsis IDH-r molecule.
Figure 3. Mass spectrometry of IDH-r tryptic peptides. The reduced monomeric form (Upper panel) and the oxidized trimeric form (Lower panel) were analyzed. The specific signals for monomeric form are indicated by arrows and the corresponding peptides are shown. Experiments were repeated five times and representative results are shown.
Trx-o Promotes the Reduction of Oxidized IDH-r
Finally, we addressed the involvement of Trx-o in redox regulation of IDH-r (Figure 4). The reduction patterns of oxidized IDH-r under several concentrations of DTT (0–500 μM) were compared in the presence and absence of 5 μM Trx-o1 (an isoform of Trx-o). When Trx-o1 was added to a reaction medium, IDH-r was reduced back to monomer even at lower concentrations of DTT (Figure 4A). We further analyzed the reduction of oxidized IDH-r with varying Trx-o1 concentration (0–5 μM) under low concentration of DTT (50 μM). The efficiency of IDH-r reduction was highly dependent on Trx-o1 concentration (Figure 4B). These findings suggest that Trx-o1 can efficiently reduce oxidized IDH-r.
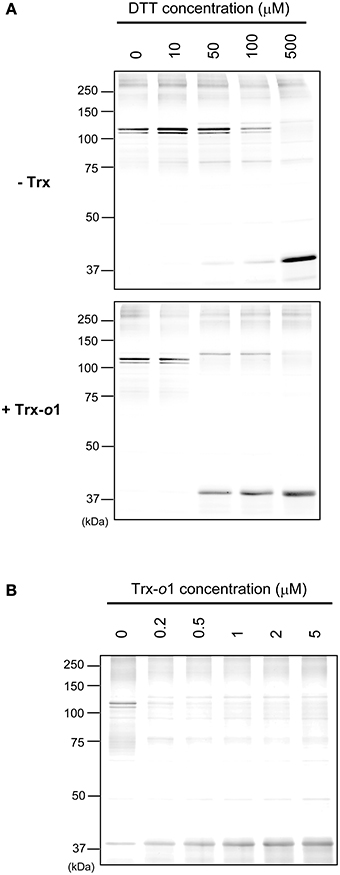
Figure 4. Trx-dependent reduction of oxidized IDH-r. (A) Oxidized IDH-r was incubated in several concentrations of DTT in the absence (upper panel) or presence (lower panel) of 5 μM Trx-o1 for 30 min. (B) Oxidized IDH-r was incubated in several concentrations of Trx-o1 in the presence of 50 μM DTT for 30 min. The redox state of IDH-r was assayed by non-reducing SDS-PAGE.
Discussion
Mitochondrial respiration is controlled at multiple levels from transcriptional to post-translational to enzyme function levels (Millar et al., 2011). It has been reported that several TCA cycle enzymes are regulated by mitochondrial NAD(P)H/NAD(P)+ ratio, adenylate energy charge, and TCA cycle intermediates (Noctor et al., 2007; Nunes-Nesi et al., 2013). Using partially purified IDH from pea leaves, Igamberdiev and Gardeström (2003) demonstrated that IDH activity is negatively regulated by NAD(P)H. However, further biochemical studies of plant IDH have not been performed to date, and accordingly the regulatory mechanisms of this enzyme at the molecular level remain to be fully characterized. Previous studies using Trx affinity chromatography have raised the possibility that IDH activity is redox-regulated via the Trx system in plant mitochondria (Balmer et al., 2004; Yoshida et al., 2013). Based on this research background, we addressed in this study the biochemical characteristics of Arabidopsis IDH, focusing particularly on redox regulation.
The biochemical properties of IDH, including action mechanisms and regulatory factors, have been best characterized in yeast (Panisko and McAlister-Henn, 2001; Lin and McAlister-Henn, 2003; Taylor et al., 2008; Garcia et al., 2009). The minimal functional unit of yeast IDH is reported to be a heterodimer comprising IDH-r and IDH-c. This form is considered to be common in plant IDH, but there has been only indirect supporting evidence to date in the form of growth restoration of yeast IDH mutants by complementation with plant IDH genes (Lancien et al., 1998; Lemaitre and Hodges, 2006). By using the recombinant subunits, we clearly showed that both IDH-r and IDH-c are essential for ensuring the catalytic activity of Arabidopsis IDH (Figure 1B). An intriguing finding is that, in contrast to the animal and yeast counterparts, Arabidopsis IDH activity is unaffected by any adenylates (Figure 1C, Table 1). In yeast IDH, critical residues for allosteric activation by AMP have been determined by a survey using site-directed mutagenesis (Lin and McAlister-Henn, 2003). Alignment between plant and yeast IDH indicated that plant IDH does not conserve some of these residues (Supplementary Fig. S1), and the lack of these critical residues may account for the insensitivity of Arabidopsis IDH to adenylates.
Redox-proteomics studies using Trx affinity chromatography have provided a comprehensive list of potentially redox-regulated proteins in plant mitochondria (Balmer et al., 2004; Yoshida et al., 2013). It has been shown that, among these candidate proteins, two stress-related proteins, namely alternative oxidase and peroxiredoxin IIF, are reduced and activated in a Trx-o-dependent manner (Barranco-Medina et al., 2008; Martí et al., 2009; Yoshida et al., 2013). In addition, the TCA cycle enzyme citrate synthase has recently been reported to be redox-regulated by Trx-o (Schmidtmann et al., 2014). However, most plant mitochondrial proteins suggested as Trx targets remain to be further analyzed by detailed biochemical assays. In the present study, IDH was newly confirmed to be a Trx-o-targeted redox-regulated protein in Arabidopsis mitochondria. Upon oxidation of IDH-r, IDH activity was largely diminished via intermolecular disulfide-mediated oligomer formation of IDH-r (Figures 2A,B). Trx-o was effective in the reduction of oxidized IDH-r, likely leading to a recovery of IDH activity (Figures 2, 4). It should be noted that IDH activity was restored to only half of the control level, even after the reduction and monomerization of oxidized IDH-r (Figure 2). This result implies that IDH-r also undergoes irreversible oxidative modifications, which could not be revealed in the present study. Further investigation is required for concluding this possibility.
Yeast IDH activity is also controlled in a redox-dependent manner (Garcia et al., 2009). However, the molecular basis of IDH redox regulation is different between Arabidopsis and yeast. In the case of yeast IDH, IDH-c forms intermolecular disulfide bonds under oxidative conditions, leading to the inactivation of IDH (Garcia et al., 2009). The critical Cys residue for the intermolecular disulfide bond formation of yeast IDH-c is not conserved in plant IDH-c (Supplementary Fig. S1). In contrast, we identified Cys128 and Cys216 as the Cys residues responsible for oxidative modification of Arabidopsis IDH-r (Figure 3). These Cys residues are highly conserved in the plant kingdom but not in yeast (Supplementary Fig. S1). This fact implies that the mode of IDH redox regulation disclosed in this study is unique to plants.
As summarized in Figure 5, our biochemical analysis suggested that IDH-r is a redox-sensitive protein and that the redox change affects IDH activity. This finding implies the significance of the mitochondrial Trx system for the regulation of TCA cycle performance. However, for several redox-regulated protein candidates identified by redox-proteomics studies, it remains to be determined whether they are “pseudo” or “true” redox-regulated proteins. In order to understand the mitochondrial redox network in more detail, biochemical studies of the individual enzymes should be performed one by one. Another key step in dissecting the mitochondrial redox regulation system is the elucidation of the working dynamics or biological significance of the mitochondrial Trx system in living plants. Future studies addressing the visualization of in vivo redox states of redox-regulated proteins, as shown in our recent study (Yoshida et al., 2014), and effects of Trx-o deficiency on mitochondrial performance will provide physiological insights into the mitochondrial redox regulation system in plants.
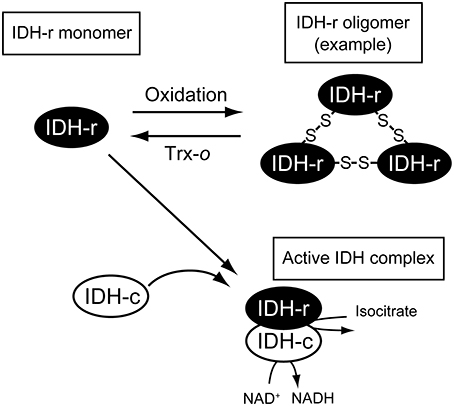
Figure 5. Possible scheme of IDH redox regulation. IDH-r forms oligomer mediated by intermolecular disulfide bonds upon oxidation. Oxidized IDH-r can be reduced back to the monomer by Trx-o, which is essential for forming functional IDH complex (see the main text for details).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by the Core Research of Evolutional Science and Technology program (CREST) from the Japan Science and Technology Agency (JST) and a Grant-in-Aid for Scientific Research (grant number 24870010 to Keisuke Yoshida) from the Japan Society for the Promotion of Science.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fenvs.2014.00038/abstract
References
Balmer, Y., Vensel, W. H., Tanaka, C. K., Hurkman, W. J., Gelhaye, E., Rouhier, N., et al. (2004). Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 101, 2642–2647. doi: 10.1073/pnas.0308583101
Barranco-Medina, S., Krell, T., Bernier-Villamor, L., Sevilla, F., Lázaro, J.J., and Dietz, K.J. (2008). Hexameric oligomerization of mitochondrial peroxiredoxin PrxIIF and formation of an ultrahigh affinity complex with its electron donor thioredoxin Trx-o. J. Exp. Bot. 59, 3259–3269. doi: 10.1093/jxb/ern177
Garcia, J. A., Minard, K. I., Lin, A. P., and McAlister-Henn, L. (2009). Disulfide bond formation in yeast NAD+-specific isocitrate dehydrogenase. Biochemistry 48, 8869–8878. doi: 10.1021/bi900968a
Hisabori, T., Motohashi, K., Hosoya-Matsuda, N., Ueoka-Nakanishi, H., and Romano, P.G. (2007). Towards a functional dissection of thioredoxin networks in plant cells. Photochem. Photobiol. 83, 145–151. doi: 10.1562/2006-02-27-IR-816
Igamberdiev, A.U., and Gardeström, P. (2003). Regulation of NAD- and NADP-dependent isocitrate dehydrogenases by reduction levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim. Biophys. Acta 1606, 117–125. doi: 10.1016/S0005-2728(03)00106-3
Laloi, C., Rayapuram, N., Chartier, Y., Grienenberger, J.M., Bonnard, G., and Meyer, Y. (2001). Identification and characterization of a mitochondrial thioredoxin system in plants. Proc. Natl. Acad. Sci. U.S.A. 98, 14144–14149. doi: 10.1073/pnas.241340898
Lancien, M., Gadal, P., and Hodges, M. (1998). Molecular characterization of higher plant NAD-dependent isocitrate dehydrogenase: evidence for a heteromeric structure by the complementation of yeast mutants. Plant J. 16, 325–333. doi: 10.1046/j.1365-313x.1998.00305.x
Lázaro, J.J., Jiménez, A., Camejo, D., Iglesias-Baena, I., Martí, M.C., Lázaro-Payo, A., et al. (2013). Dissecting the integrative antioxidant and redox systems in plant mitochondria. Effect of stress and S-nitrosylation. Front. Plant Sci. 4:460. doi: 10.3389/fpls.2013.00460
Lemaitre, T., and Hodges, M. (2006). Expression analysis of Arabidopsis thaliana NAD-dependent isocitrate dehydrogenase genes shows the presence of a functional subunit that is mainly expressed in the pollen and absent from vegetative organs. Plant Cell Physiol. 47, 634–643. doi: 10.1093/pcp/pcj030
Lin, A.P., and McAlister-Henn, L. (2003). Homologous binding sites in yeast isocitrate dehydrogenase for cofactor (NAD+) and allosteric activator (AMP). J. Biol. Chem. 278, 12864–12872. doi: 10.1074/jbc.M300154200
Lindahl, M., Mata-Cabana, A., and Kieselbach, T. (2011). The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid. Redox Signal. 14, 2581–2642. doi: 10.1089/ars.2010.3551
Martí, M.C., Olmos, E., Calvete, J.J., Díaz, I., Barranco-Medina, S., Whelan, J., et al. (2009). Mitochondrial and nuclear localization of a novel pea thioredoxin: identification of its mitochondrial target proteins. Plant Physiol. 150, 646–657. doi: 10.1104/pp.109.138073
Millar, A. H., Whelan, J., Soole, K. L., and Day, D. A. (2011). Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62, 79–104. doi: 10.1146/annurev-arplant-042110-103857
Montrichard, F., Alkhalfioui, F., Yano, H., Vensel, W. H., Hurkman, W. J., and Buchanan, B.B. (2009). Thioredoxin targets in plants: the first 30 years. J. Proteomics 72, 452–474. doi: 10.1016/j.jprot.2008.12.002
Motohashi, K., Kondoh, A., Stumpp, M.T., and Hisabori, T. (2001). Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 11224–11229. doi: 10.1073/pnas.191282098
Noctor, G., De Paepe, R., and Foyer, C.H. (2007). Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 12, 125–134. doi: 10.1016/j.tplants.2007.01.005
Nunes-Nesi, A., Araújo, W.L., Obata, T., and Fernie, A.R. (2013). Regulation of the mitochondrial tricarboxylic acid cycle. Curr. Opin. Plant Biol. 16, 335–343. doi: 10.1016/j.pbi.2013.01.004
Panisko, E.A., and McAlister-Henn, L. (2001). Subunit interactions of yeast NAD+-specific isocitrate dehydrogenase. J. Biol. Chem. 276, 1204–1210. doi: 10.1074/jbc.M005056200
Schmidtmann, E., König, A.C., Orwat, A., Leister, D., Hartl, M., and Finkemeier, I. (2014). Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol. Plant 7, 156–169. doi: 10.1093/mp/sst144
Taylor, A. B., Hu, G., Hart, P. J., and McAlister-Henn, L. (2008). Allosteric motions in structures of yeast NAD+-specific isocitrate dehydrogenase. J. Biol. Chem. 283, 10872–10880. doi: 10.1074/jbc.M708719200
Tcherkez, G., Boex-Fontvieille, E., Mahé, A., and Hodges, M. (2012). Respiratory carbon fluxes in leaves. Curr. Opin. Plant Biol. 15, 308–314. doi: 10.1016/j.pbi.2011.12.003
Winger, A. M., Taylor, N. L., Heazlewood, J. L., Day, D. A., and Millar, A. H. (2007). Identification of intra- and intermolecular disulphide bonding in the plant mitochondrial proteome by diagonal gel electrophoresis. Proteomics 7, 4158–4170. doi: 10.1002/pmic.200700209
Yoshida, K., Matsuoka, Y., Hara, S., Konno, H., and Hisabori, T. (2014). Distinct redox behaviors of chloroplast thiol enzymes and their relationships with photosynthetic electron transport in Arabidopsis thaliana. Plant Cell Physiol. 55, 1415–1425. doi: 10.1093/pcp/pcu066
Yoshida, K., and Noguchi, K. (2009). Differential gene expression profiles of the mitochondrial respiratory components in illuminated Arabidopsis leaves. Plant Cell Physiol. 50, 1449–1462. doi: 10.1093/pcp/pcp090
Keywords: Arabidopsis, isocitrate dehydrogenase, mitochondria, redox regulation, thioredoxin
Citation: Yoshida K and Hisabori T (2014) Mitochondrial isocitrate dehydrogenase is inactivated upon oxidation and reactivated by thioredoxin-dependent reduction in Arabidopsis. Front. Environ. Sci. 2:38. doi: 10.3389/fenvs.2014.00038
Received: 30 July 2014; Accepted: 08 September 2014;
Published online: 23 September 2014.
Edited by:
Adriano Sofo, Università degli Studi della Basilicata, ItalyReviewed by:
Shailendra Pratap Singh, Michigan State University, USAAran Incharoensakdi, Chulalongkorn University, Thailand
Copyright © 2014 Yoshida and Hisabori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toru Hisabori, Chemical Resources Laboratory, Tokyo Institute of Technology, Nagatsuta 4259-R1-8, Midori-ku, Yokohama 226-8503, Japan e-mail: thisabor@res.titech.ac.jp
 Keisuke Yoshida
Keisuke Yoshida Toru Hisabori
Toru Hisabori