Strategic Design of Synthetic Consortium with Embedded Wastewater Treatment Potential: Deciphering the Competence of Isolates from Diverse Microbiome
- Bioengineering and Environmental Sciences, CSIR-Indian Institute of Chemical Technology, Hyderabad, India
Microorganisms play a vital role in efficient biological treatment. Supplementation of external microorganisms with high degradation rates can enhance the process efficiency significantly. Potential strains were isolated from long term wastewater treating reactors and identified using phylogenetic analysis of 16S rRNA gene fragments with the nearest neighbors extracted during BLAST search. Later the study was designed in two phases which revealed interesting findings. Phase I evaluates the potential of isolated strains viz., Pseudomonas otitidis, Bacillus firmus, Bacillus subtilis, and Bacillus circulans for their individual ability in terms of COD and nutrients removal. B. circulans showed highest carbon (COD) removal (70%; 0.56 kg CODR/m3-day), while maximum nutrients removal (nitrate, 81%; phosphate, 90%) was observed with B. subtilis. B. firmus showed maximum volatile fatty acid (VFA) production. Based on Phase I results, four synthetic consortia were designed in phase II with diverse combination of isolates and evaluated for their remediation efficiencies. Consortium 4 (P. otitidis, B. subtilis, and B. firmus) illustrated higher treatment potential [COD, 86%; SDR (cum): 0.64 kg CODR/m3-day; Nitrate, 87%; Phosphate, 97%]. The exploitation of such consortia can overcome the inefficiencies pre-existing with the biological wastewater treatment plants by acting as prospective candidates for bio-augmenting the native microflora. This communication illustrated development of the efficient consortia using lab isolated strains to improve the performance of wastewater treatment.
Introduction
Biological treatment process assumes importance and relevance due to its sustainable nature which makes this technology as one of the most important biotechnological application. Biological treatment processes have their own design parameters and economic relevance but in these processes there is a common parameter which is its “microbes” or the “microbial consortia.” Microbes as the central drivers have regulatory influence on treatment functioning and efficiency. Wastewater systems may provide diverse conditions for the microbial growth and metabolism. It stimulates development of indigenous microbial populations, toward utilization of the nutrients and specific substrates in the waste as their sole carbon and energy sources thereby degrading the contaminants. There are many strategies applied toward the enhancement of the wastewater treatment efficiency (Venkata Mohan et al., 2001, 2007). Understanding the relationship among microbial communities, individual microbe's role, treatment rate and efficiency is one of the most important criteria in wastewater treatment.
Modern molecular techniques and environmental genomics, explored many microbial key players in wastewater treatment for nutrient removal, sludge bulking and foaming and provided many exhilarating insights into the functions, diversity and niche differentiations of these microorganisms (Daims et al., 2006). Studies have been done to understand the microbial dynamics in wastewater treatment systems (Muszyñski et al., 2005; Shi et al., 2015). Several bacterial species have been identified that have the ability for wastewater degradation. Majorly, Bacillus sps. have been identified as substrate degraders. The genus Bacillus belongs to the phylum Firmicutes and members of this genus are spore-forming aerobic or facultatively anaerobic bacteria (Gander et al., 2000). Bacteria belonging to this genus can easily degrade proteins, starch and lipids. Choi et al. (2002), reported that an advanced wastewater treatment system in Korea operated with Bacillus strains pre-dominating was able to remove nitrogen and phosphorus as well as organic matter efficiently. Iriye and Takatsuka (1999) reported that the wastewater system operated in Japan with Bacillus sps. dominance showed suitable removal rates for waste organic matter combined with high de-nitrification and de-phosphorization activities and less offensive odor. Other than Bacillus sps., Pseudomonas is also considered as a potential species prevailing in wastewater microbiome. Pseudomonas sps. can easily break down carbon and can reduce biological oxygen demand (BOD) levels far more quickly. Since such properties of the bacterial cultures are needed for wastewater treatment, methods have been developed to enhance their growth in the wastewater treatment systems (Iriye and Takatsuka, 1999; Reddy et al., 2012). These microorganisms may be indigenous to contaminated wastewater or they are isolated from various wastewaters and brought to the contaminated area. The success rate of any individual wastewater treatment systems depends on the most abundant microbial culture or the assemblage of the different microbial communities in the system. There are various bacteria which perform treatment efficiently and effectively; but each one is meant for specific function. These competent isolated and studied strains can be added into the treatment system which is known as bioaugmentation. Broadly, it is the general practice of adding specialized microbial strains to a native community for enhancing the ability of the microbial community to degrade certain compounds or to respond to process fluctuations resulting in improved treatment. The characteristics of the microbial community in the wastewater treatment plant can be easily improved through manipulating the already present microbial community by including specific microbes individually or in effective consortia (Raghavulu et al., 2011). Mixed cultures have an apparent advantages as the microbial consortia can communally perform the treatment efficiently which cannot be performed individually. Researchers started developing consortia for increasing the efficiency of wastewater treatment (Venkata Mohan et al., 2009), removal of contaminants like phenol (Azaizeh et al., 2015), nitrogen removal (Chen et al., 2015), to treat tobacco wastewater (Wang et al., 2009) etc. Treatment of different wastewaters differs as it dependent of the different contaminants they contain. Under such stipulation, microbial consortia with different organisms having different specificities can perform the treatment more efficiently which is intricate for individual strains or species. Efficient and effective wastewater treatment can be anticipated when the co-metabolic activities within a microbial consortia complements each other. Hence, the present study was undertaken to explore the wastewater treatment potential of isolated strains and design of synthetic consortia based on carbon and nutrient removal. The study also depicts comprehensive evaluation of both isolates and designed consortia with respect to substrate degradation efficiency, nutrients removal potential, enzymatic analysis, and bioelectrochemical behavior to comprehend the relative suitability of microbiome to the complex wastewater system.
Materials and Methods
Biocatalysts
Facultative aerobic cultures from the long term operated bioreactors used for wastewater treatment were used for isolation of strains by direct plating and enrichment techniques using nutrient broth (pH 7; 24 h under aerobic conditions). One milliliter of the culture suspended in sterile deionized water (9 ml) was serially diluted from 10−1 to 10−8 times with saline water. Diluted sample (0.1 ml) was spread on the nutrient agar plates using spread plating technique and was incubated at 37°C for 24 to 48 h. Pure colonies were obtained after repeated streaking.
DNA Extraction and PCR Amplification
Genomic DNA from the isolated pure colony was extracted and purified using phenol–chloroform method. The concentration of DNA obtained was measured at a wavelength of 260 nm using UV/Vis-spectrophotometer (Thermo Scientific, USA). The variable V3 region of 16S rDNA was amplified by PCR using primers 8F (5′-AGAGTTTGATCCTGGCTCAG- 3′) and 1542R (5′-AAGGAGGTGATCCAGCCGCA- 3′). All PCR amplifications were conducted in 50 μl containing 2 μl of total DNA containing 100 ng/ μl concentrations, 200 μM each of the four deoxynucleotide triphosphates, 15 mM MgCl2, 0.1 μM of individual primers and 1 unit of Taq polymerase. An automated thermal cycler (Eppendorf) was used for PCR amplification with program for an initial denaturation at 96°C for 5 min, 35 cycles of denaturation (40 s at 94°C), annealing (50 s at 52.6°C) and extension (1 min at 72°C) and a final extension at 72°C for 8 min. The PCR amplified product (5 μl) was subjected to agarose gel electrophoresis to confirm product recovery and to estimate product concentration. The amplified PCR product was stored at 4°C until further use.
Sequencing, Characterization, and Phylogenetic Analysis
Amplified PCR product after purification (Fermentas) was sent to MWG Biotech for sequencing. 16S rRNA sequence (1500 bp) was aligned with those of the reference micro organisms existing in the same region of the closest relative strains available in the GenBank database, using the BLASTN facility and was also tested for possible chimera formation with the chimera check program Decipher (online: http://decipher.cee.wisc.edu/FindChimerasOutputs.html). These sequences were further aligned with the closest matches found in the GenBank database with the CLUSTALW function of Molecular Evolutionary Genetics Analysis package (MEGA). Neighbor-joining phylogenetic tree was constructed with the MEGA version. 6.0. A bootstrap analysis with 100 replicates was carried out to check the robustness of the tree. Bootstrap re-sampling analysis for the replicates was performed to estimate the confidence of tree topologies. Strains were named as Bacillus circulans strain SLBEEC1, Bacillus firmus strain IICTSVMHH, Bacillus subtilis strain IICTSVMH13 and Pseudomonas otitidis SVMIICTP2 based on the BLASTN facility that showed 99–100% sequence similarity with the nearest phylogenetic neighbors. The nucleotide sequence identified in this study was deposited in the GenBank database under accession numbers for P. otitidis, B. firmus, B. subtilis, and B. circulans as HE612874, FR849707, FR849706, and KJ501091 respectively.
The isolated strains viz., P. otitidis, B. firmus, B. subtilis, and B. circulans were used as parent biocatalyst for the experiments. The stock cultures were maintained on nutrient agar slants overlaid with 20% (v/v) glycerol and kept at −20°C. Prior to inoculation, the organisms were enriched with designed synthetic wastewater (DSW) under aerobic microenvironments (120 rpm; 28°C) to maximize the nutrient removing capacity of the biocatalyst.
Synthetic Consortia
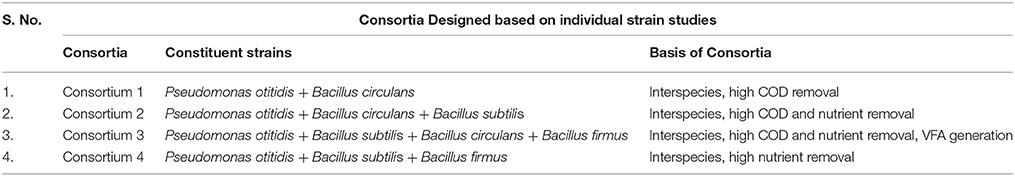
The consortia was developed by aseptically transferring the 12 h grown culture of each individual strains studied in phase I, in different combinations [P. otitidis+ B. circulans (1); P otitidis + B. circulans+ B. subtilis (2); P. otitidis+ B. subtilis + B. circulans + B. firmus (3); P. otitidis+ B. subtilis + B. firmus (4)] in 500 ml Erlenmeyer flasks containing 250 ml of DSW at similar operating conditions for single cultures. Individual strains were inoculated with 2% (v/v) aliquots of 12 hour grown culture, respectively, to maintain the same cell count in the pure culture and in the consortium. All the cultures were added in equal proportion in order to maintain the same initial OD 600.
Experimental Methodology
Prior to inoculation, the biocatalysts were re-suspended in sterilized (15 min; 121°C) designed synthetic wastewater for 12 h at room temperature. Designed synthetic wastewater (DSW; glucose 3 g l−1; NH4Cl 0.5 g l−1, KH2PO4 0.25 g l−1, K2HPO4 0.25 g l−1, MgCl2 0.3 g l−1, CoCl2 l−1, ZnCl2 11.5 mg l−1, CuCl2 10.5 mg l−1, CaCl2 5 mg l−1, MnCl2 15 mg l−1, NiSO4 0.16 g l−1, FeCl3 0.03 g l−1) was used to evaluate the biocatalytic activity. The resulting pellets of P. otitidis, B. subtilis, B. firmus, and B. circulans were inoculated individually into each of the four 500 ml flasks, respectively and were operated for six cycles each with a retention period of 48 h. All the four systems were operated under similar conditions and feed replacement was done with sterilized DSW at an organic load of 3 g COD l−1. Prior to loading and autoclaving, pH of the DSW was adjusted to 7 using 2 N ortho phosphoric acid or 1 M NaOH. After performing the individual strains performance experiment consortia were made using different strains as per discussed in Section 2.2. and studied in the similar condition for individual strain studies for two cycles and average of two was reported in the study, thus sample collection was done more frequently. All the experimental operations were carried out in fed-batch mode. Before every feeding event, the inoculum was allowed to settle down (30 min; settling) and the exhausted feed was decanted (15 min; decanting). The inoculum settled at the bottom was used for subsequent operations. Feeding, decanting and recirculation operations were performed and the operation was properly carried out to ensure that sterile microenvironment is maintained. All the experiments were performed at ambient temperatures (29 ± 2°C).
Analysis
Chemical oxygen demand (COD-closed refluxing titrimetric method), volatile fatty acids (VFA), nitrate, phosphate and pH were estimated by the standard methods (APHA, 1998). Optical density (OD) of the bacterial cultures was measured at 600 nm using spectrophotometer (Genequant 1300). Cyclic voltammetry (CV) was employed to evaluate the electron discharge properties of the biocatalyst, using a potentiostat–galvanostat system (PGSTAT12, Ecochemie). CV was operated and voltammograms were recorded against Ag/AgCl (3.5 M) by applying a potential ramp (+0.5 to −0.5 mV) at a scan rate of 20 mV/s employing platinum wire and carbon rod as counter and working electrodes respectively.
Results and Discussion
Phylogenetic Architecture Analysis of Isolates
The isolated strains based on 16S rDNA method were identified as P. otitidis, B. firmus, B. subtilis, and B. circulans and submitted to the GenBank database under NCBI Accession Numbers HE612874, FR849707, FR849706, and KJ501091, respectively (Table 1). The phylogenetic tree was constructed based on the partial sequences of 16S rDNA gene of four isolates with other related bacterial strains as shown in Figure 1 along with bootstrap values. B. circulans exhibited 92% of bootstrap values with B. circulans strain NBRC 13626 (NR_112632.1). P. otitidis exhibited 82% of bootstrap values with P. otitidis MCC10330 (NR_043289.1). B. subtilis exhibited similarity with B. subtilis subsp. spizizenii TU-B-10 (CP002905.1) while B. firmus exhibited 97% of bootstrap values with B. firmus NBRC 15306T (NR_112635.1). In the phylogenetic tree clear clusters were observed with the bootstrap value of 99–100%. B. firmus, B. circulans, B. subtilis are Gram positive bacteria while P. otitidis is Gram negative bacteria, which can easily be seen in the tree because of their different clusters. Majorly, genus Bacillus is a group of proteolytic, cellulolytic, lipolytic, aerobic or facultative anaerobic bacteria and it is widely used in wastewater treatment (Groudev, 1987). B. circulans is a typical chemo-organo-heterotrophic bacterium, which can use mono-, di- and polysaccharides and polyhydroxylic alcohols as a source of carbon, energy and electrons (Groudev, 1987). B. subtilis possesses multiple enzyme systems producing amylase, lipases, and proteases, which are useful in breakdown of complex molecules and fats in wastewater. B. firmus is capable of producing xylanases (Min-Jen et al., 2002) at alkaline conditions. It widely used of dye removal from textile wastewater (Ogugbue et al., 2012). P. otitidis was studied for its bioplastics production and dye degradation (Wu et al., 2009; Reddy et al., 2012). Pseudomonas sps. is also exploited for pyridine degradation (Venkata Mohan et al., 2003). Hence, these isolated strains mark themselves as the potential candidates for wastewater treatment.
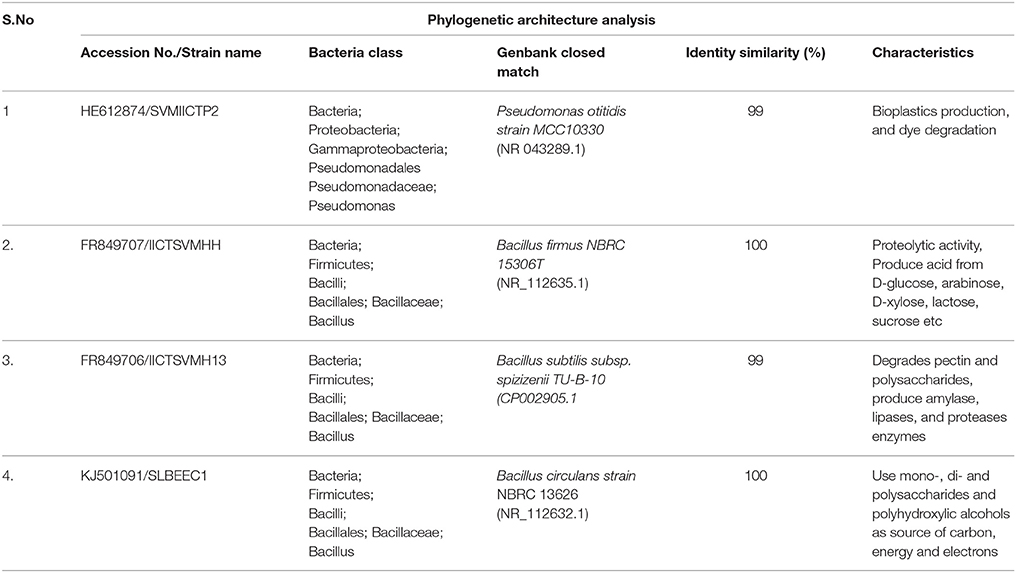
Table 1. List of strains isolated indentified with phylogenetic analysis with accession numbers, classification, GenBank closest match, identity similarities and their properties.
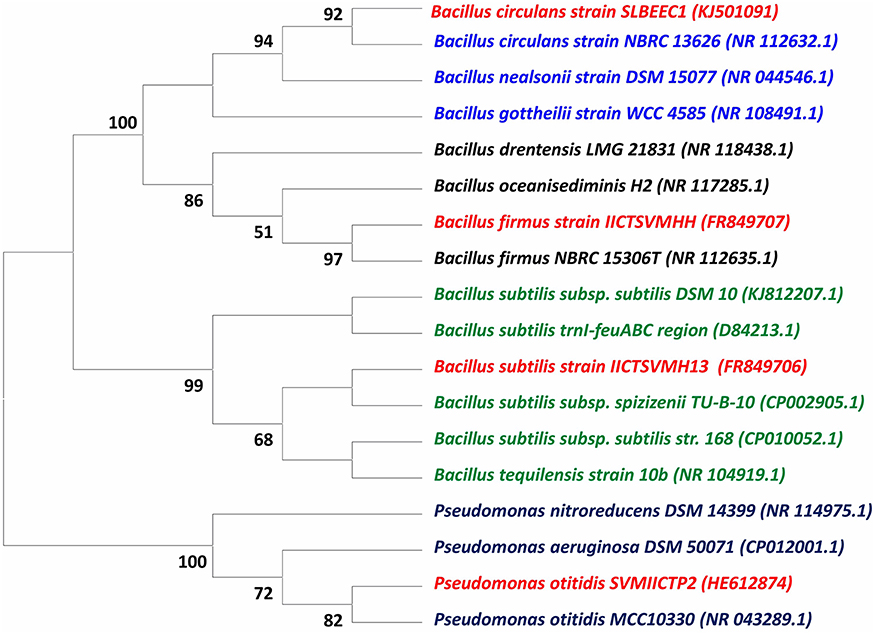
Figure 1. Evolutionary relationship analysis of 16S rDNA sequence of wastewater treating Strains (Isolated strains are marked in red color).
Evaluation of Isolates for Treatment Efficiency
Biomass Growth
The pattern of growth of the cultures was observed in the form of growth curves (OD 600; Figure 2A). For all isolates, cells were growing well for the wastewater composition. B. subtilis showed maximum growth of about 0.4 OD at the end of exponential phase followed by B. circulans (0.37 OD) and P. otitidis of around 0.35 OD in the third cycle. B. firmus showed a long lag phase compared to all other strains. Interestingly, floc formation in B. firmus operated system was observed after 12 h, which may be attributed to production of polysaccharides by this culture. Formation of polysaccharides by B. firmus was already being reported (Salehizadeh and Shojaosadati, 2002). Microbial activities in the system allow the mineralization of pollutants into carbon dioxide and water, and microbial transformation is measured as an important route for complete degradation of pollutants in the wastewater. Hence, efficiency of a treatment system is dependent on the growth of the culture. If a viable culture is inoculated into a solution which contains a suitable carbon nitrogen source, other micro and macro-nutrients necessary for growth, and physiological temperature and pH are maintained, it will grow as seen in B. subtilis. However, if the conditions are adverse the growth can be withheld, while some cultures can withstand the adverse conditions. Similarly in the wastewater systems, if the conditions are not suitable then growth of bacteria cannot be observed. If the subjected inoculum can endure the adverse conditions prevailing in the system and grows well, the treatment efficiency will automatically increase. The rate of biomass synthesis is proportional to biomass present in the system.
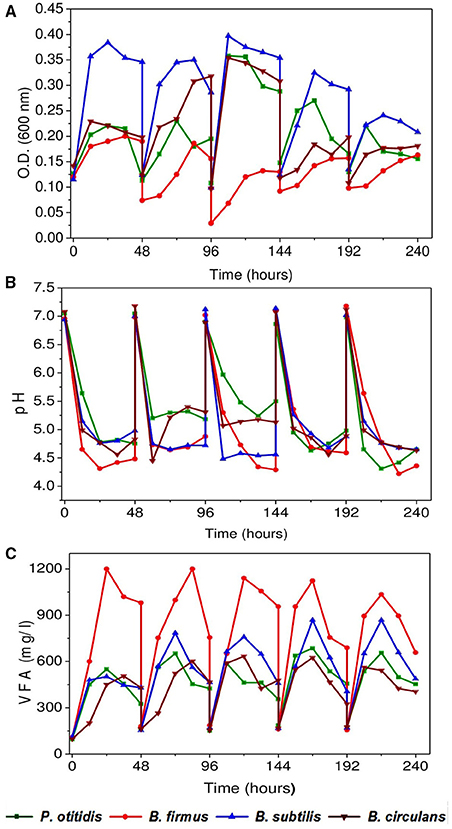
Figure 2. (A) Bacterial growth of isolated strains in synthetic wastewater (B) pH variation with time (C) Variation in Volatile fatty acids (VFA) with time.
Wastewater Treatment
Carbon load removal
COD removal is considered as a critical parameter to assess the wastewater treatment efficiency. A gradual depletion in substrate (COD concentration) was observed with operation time among all strains. COD removal efficiency (Figures 3A,B), cumulative (Figure 3C) and non-cumulative substrate degradation rate (SDR) (Figure 4) were calculated to track the decrement in the carbon load. Observation revealed that COD (%) and SDR (%) varied depending on the inherent capabilities of strains. Among the isolates, higher average COD removal efficiency (CODR) was observed for B. circulans (CODR: 70%) followed by B. subtilis (CODR: 60%) and least with B. firmus (CODR: 44%). B. circulans showed highest SDR (cumulative) with 0.56 kg CODR/m3-day followed by B. subtilis (0.48 kg CODR/m3-day). and B. firmus (0.32 kg CODR/m3-day). The SDR (non-cumulative; SDRT) of all the strains increased till the 24th hour and later decreased with time. Growth of all isolates directly supported the SDRT while, in stationary phase there was some decrement in SDRT (Figure 5). Lowest SDRT at 48th hour was observed in B. firmus and highest rates in B. circulans. B. circulans can produce metabolites which have specific functions such as acidolysis, alkalysis and complexolysis (Dragutinovi et al., 2012). Tannery wastewater treatment was reported with B. circulans (Renitha et al., 2014) where good COD removal was observed. B. subtilis was also exploited for COD removal from tannery wastewater (Yusuf et al., 2013), where COD removal efficiency of 87.6% was observed compared to P. fragii (85.2%).
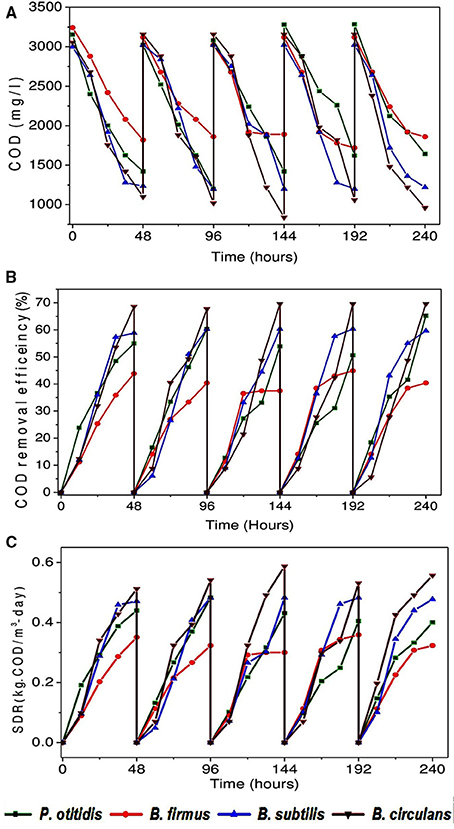
Figure 3. (A) Variation in COD concentration removal (mg/l) with isolated strains (B) Variation in COD removal efficiency (%) with isolated strains (C) Variation in substrate degradation rate (cumulative) (kg· CODR/m3-day) with isolated strains.
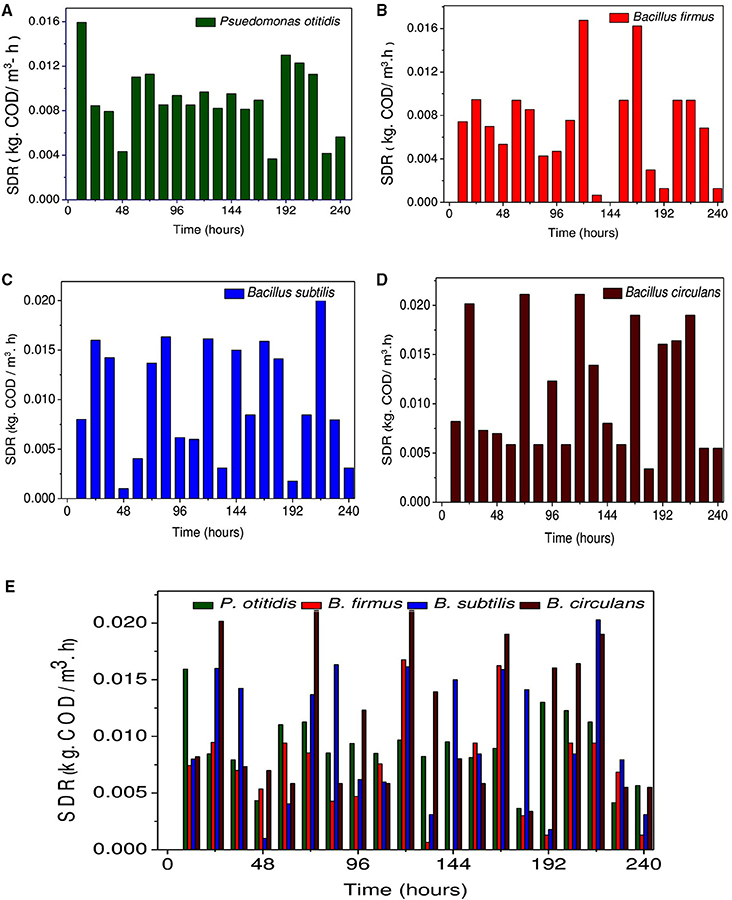
Figure 4. Variation in substrate degradation rate (non-cumulative) (kg· CODT/m3-h) for (A) Pseudomonas otitidis, (B) Bacillus firmus, (C) Bacillus subtilis, (D) Bacillus circulans, and (E) Comparative evaluation of each isolated strains.
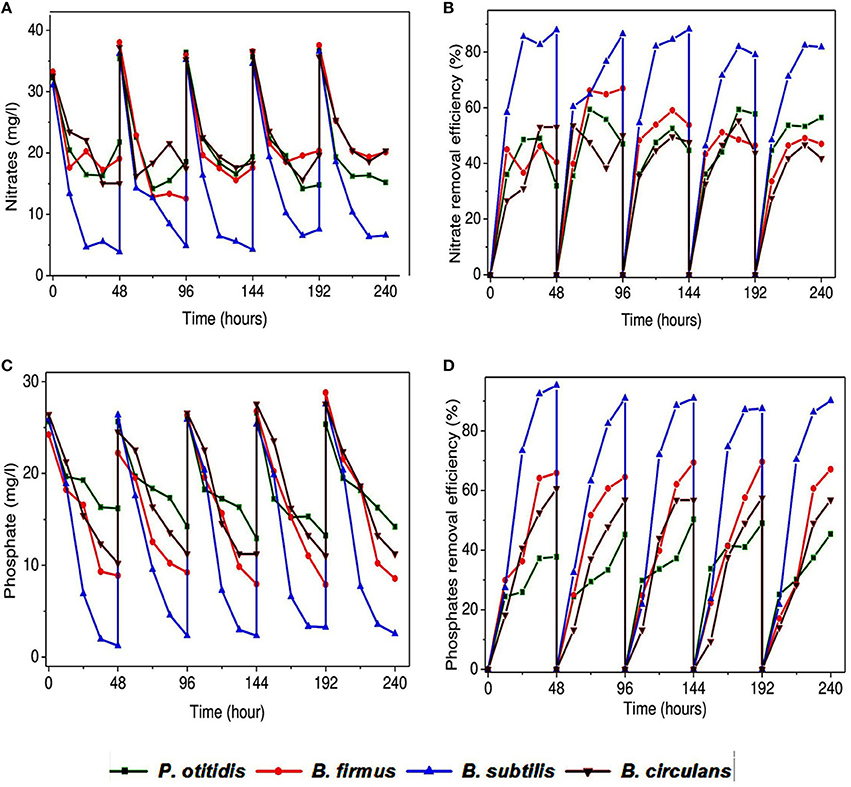
Figure 5. Variation in (A) nitrate removal (mg/l), (B) nitrate removal efficiency (%), (C) phosphate removal (mg/l), and (D) phosphate removal efficiency (%) with time in presence of the different isolated strains.
Nitrate removal
Nitrogen-containing compounds when present in environment can cause serious problems, hence its removal becomes necessary from wastewater. Therefore in this study nitrate removal was considered as an important parameter. All isolates documented good nitrate removal but was found to depend on the individual strain (Figures 5A,B). Operation with B. subtilis showed highest removal efficiency of nitrate accounting for nearly 80%. Subsequently in the system operated with P. otitidis showed higher nitrate removal efficiency (56%) than B. firmus (46%). Least nitrate removal was found in the system operated with B. circulans (41%). Biological treatment includes both anaerobic and aerobic treatment operations where nitrate is metabolized by microorganisms in three different ways, i.e., assimilation to ammonia under aerobic conditions, dissimilation to nitrogen or to ammonia under micro aerobic or anaerobic conditions. Thus, aerobic nitrate-respiring bacteria may be particularly important in environments with fluctuating or minimal oxygen availability as mostly studies were done with anaerobic nitrate reduction. These bacteria may be obligate aerobe, anaerobe or facultative aerobes or anaerobes. In wastewater, microbes with the complete denitrification pathway can catalyze the dissimilatory reduction of nitrate to nitrogen. The aerobic metabolism of Bacillus and Pseudomonas sps. have revealed many fascinating features of assimilatory nitrate reduction to ammonia via nitrite. Few microbes, can support anaerobic growth or dissipate excess reducing power using dissimilatory reduction of nitrate to ammonia via nitrite like B. subtilis viz. a facultative aerobe can utilize nitrate or nitrite as sole nitrogen source, and nitrate and nitrite are also utilized as electron acceptors for anaerobic respiration under anaerobic conditions. Karunai Selvi and Ravindran (2012) observed that B. subtilis can effectively utilize nitrate as nitrogen source under aerobic conditions. In a nitrogen cycle, nitrate are converted to nitrite by assimilatory or dissimilatory nitrate reductases. Nitrate reduction takes place by the action of a membrane-bound nitrate reductase, which has an active site in the cytoplasm and is prevalent amongst nitrate-respiring bacteria, including nitrate-denitrifying and ammonifying species (Carter et al., 1995). Nitrate reductases are also reported in periplasmic compartment in various organisms like T. pantotropha, P. stutzeri and many more (Carter et al., 1995). Reduction of nitrate in the periplasm is insensitive to the oxygen inhibition of nitrate transport across the cytoplasmic membrane that stops the reduction by the membrane-bound enzyme (Carter et al., 1995). B. subtilis has two reductases the nitrate and nitrite reductases and both with two different physiological functions. Nitrogen limitation subjects these enzymes to catalyze the reduction of nitrate to ammonia via nitrite for the anabolic integration of nitrogen into biomolecules and under anaerobic respiration they use nitrate and nitrite as terminal electron acceptors (Nakano et al., 1998). These two distinct nitrate reductases, encoded by nasBC and narGHI, functions in catabolic and anabolic nitrogen metabolism, respectively. B. subtilis can exploit nitrate or nitrite as sole nitrogen source, and nitrate and nitrite are also utilized as electron acceptors for anaerobic respiration. Nakano and co-workers described the aerobic assimilatory function by NADH-dependent reductase (NasDE) in B. subtilis (Nakano et al., 1998). Kim and co-workers used dominant isolates viz. B.cereus, B. subtilis and B. licheniform in flask experiment for nitrogen removal under aerobic growth conditions. They supported that the Bacillus strains were able to perform simultaneously aerobic nitrification/denitrification process (Kim et al., 2005). Pseudomonas also showed relatively higher nitrate removal efficiency. For studying nitrogen removal Ayyasamy et al., attained assimilatory denitrification process using Pseudomonas sp. (RS7) in the ground water system under aerobic conditions. Pseudomonas species possess periplasmic nitrate reductase providing ability to respire nitrate in the presence of oxygen (Ayyasamy et al., 2007). Studies were done with Pseudomonas, Aeromonas, and Moraxella, which have expressed a nitrate reductase with an active site located in the periplasmic compartment (Carter et al., 1995). These interesting facts can support the nitrate removal in aerobic conditions.
Phosphate removal
Phosphate is one of the major nutrients in wastewater. In this study, phosphate removal was observed and found to be varied with the type of strain used (Figures 5C,D). Observation showed that the maximum phosphate removal till the end of the cycle was observed in the system operated with B. subtilis (91%) followed by B. firmus (58%), and B. circulans (54%). P. otitidis showed least phosphate removal (47%). Phosphate removal here becomes an important factor as phosphorus is one of the most essential nutrients and is an important component of different cellular structures, like cell membranes and nucleic acids. However, when present in excess it can cause eutrophication. Phosphate from wastewater bodies are biologically removed by absorption of dissolved orthophosphate, organic phosphate and polyphosphate by microorganisms. The phosphorus removal biological techniques are based by co-metabolism wherein the microorganisms involved could be able to remove solubilized phosphate in the wastewater by accumulating them intra-cellularly in the form of polyphosphates or solubilizing the phosphate. Phosphate removal varies with different conditions. Along with the carbon source, pH also plays a crucial role in phosphate removal as acidic pH favors acid phosphatase which helps in the removal of phosphate. Increase in biomass concentrations of the B. subtilis showed a greater phosphate uptake capacity. Torriani-Gorini (1987) observed that the genes and proteins of microbial cells involved in the hydrolysis of organic phosphate with oxygen as the terminal electron acceptor. Karunai Selvi and Ravindran (2012) studied B. subtilis for its phosphate solubilisation activity in different carbon and nitrogen sources. They observed B. subtilis showed maximum phosphate solubilisation when glucose and ammonium sulfate were used as carbon and nitrogen source, respectively. The bacterial species like Pseudomonas are well studied for polyphosphate accumulations. It uses the stored carbon reserves to produce energy for growth and to replenish their stores polyphosphate resulting the net removal of phosphate (Krishnaswamy et al., 2009). The phosphate is taken up by cells for growth and to reform polyphosphate under aerobic condition. Studies also revealed that presence of both carbon and phosphate under aerobic conditions at same times can have negative effects on phosphorus uptake. When carbon sources are present it will be utilized for PHA formation, which is property of P. otitidis and only when carbon sources gets exhausted the phosphorus uptakes occurs (Mino et al., 1998). This may be the reason why less phosphate removal was observed with P. otitidis.
Redox Condition
Volatile fatty acids (VFA) and pH are the active intrinsic expressers of acid and base reactions of the system. Inlet pH of all the reactors were adjusted to 7 and a consistent drop in pH was observed in all the systems throughout the operation (Figure 2B). Maximum pH drop was observed with B. firmus corresponding to maximum VFA generation followed by B. subtilis and P. otitidis (Figure 2C). The reduction in pH may be due to the production of various organic acids in the culture medium which was observed in B. firmus. Higher VFA generation was observed with in B. firmus (1200 mg/l) followed by B. subtilis (800 mg/l) and P. otitidis (500 mg/l). Least VFA production was observed with B. circulans (400 mg/l). Phosphate utilizing microorganisms produced various organic acids and consequently a fall in pH of the medium (Kundu and Gaur, 1984; Satar and Gaur, 1984).
Bioelectrochemical Analysis
For understanding the electrochemical activity of individual bacterial strain during the process, CV analysis was carried out (Figure 6). CV helps to elucidate the appropriate information of the nature and the rate of metabolic activities by evaluating bioelectrochemical activity of bacteria in whole cell form. It gives detailed electrochemical reaction occurring on the electrode surface and measures redox activities of the components which are involved in solution as well as components bound to the bacteria (Naresh Kumar et al., 2014). In this study, CV revealed observations related to the electron discharge (ED) pattern as a function of bacterial strain used. In biological metabolism protons and electrons are generated and utilized continuously in the bacterial cells. Voltammograms visualized significant variation in the bioelectrochemical behavior with respect to particular organism. Voltammetric signatures of individual culture illustrated marked variation with the function of microbial cycle indicating the variation in electron transfer mechanism. In case of B. firmus reduction currents (RC) were significantly higher than the oxidation currents (OC) which were more or less similar (RC: −7.0 μA and OC: 1.7 μA) during all the time intervals. The RC distinctively increased during 24th hour (RC: −7.0 μA) from 0th hour and slightly decreased at 48th hour. The RC currents were found to be almost equal at both 0th and 48th hour but OC increased with time and was found to be marginaly higher at 48th hour than the 0th hour depicting decline phase with slight activity of the bacterial activity. Like B. firmus, in B. subtilis also higher RC was observed than the OC, which were more or less similar during all the time intervals. The RC increased with time (0th hour −3.8 μA; 24th hour, −6.5 μA) and marginally increased till 48th hour (RC: −6.7 μA). The OC currents also increased from 0th hour and were found to be almost equal at both 24th and 48th hour which shows that B. subtilis got well adapted to the system which was supported by the highest growth rate. For B. circulans, RC decreased and OC increased from 0th hour to 24th hour, but the RC and OC were distinctively equal (RC: −3.0 μA and OC: 3.0 μA) depicting its electrogenic activity that aids in effective discharge and transfer of the redox equivalents as well as their reduction at a higher rate. For comparative evaluation, voltammograms were also compared for each strain at 24th hour (active cultures; Figure 6E). Maximum RC was observed for B. firmus (RC −9.0 μA) followed by P. otitidis (RC: -7.0 μA). B. circulans showed almost equal OC and RC (OC/ RC: 3/-3 μA). The increment in both the oxidation and reduction catalytic currents at 24th hour indicates the biochemical reactions of substrate utilization where the role of oxygen is negligible as electron acceptor. Highest COD removal by B. circulans can be supported by cyclic voltametry where equal oxidation reduction currents were observed.
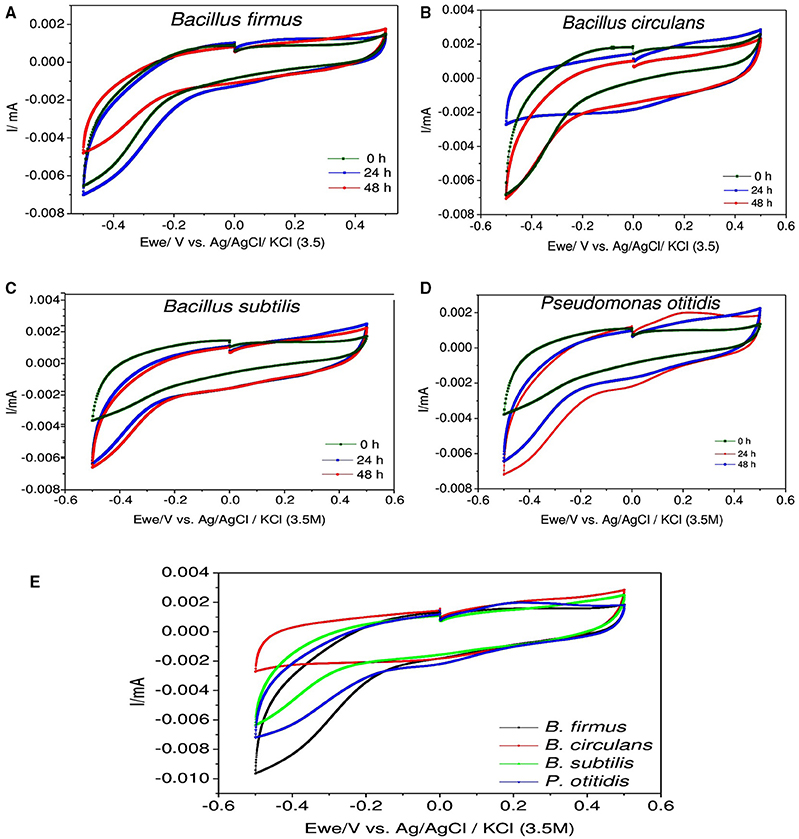
Figure 6. Cyclic voltammograms depicting the redox catalytic currents in (A) Bacillus firmus, (B) Bacillus circulans, (C) Bacillus subtilis, (D) Pseudomonas otitidis, and (E) All isolated strains at specific 24 h recorded at a scan rate of 20 mV s−1.
Performance of Designed Synthetic Consortia
Individual strains were studied and based on their performance four different consortia were developed and studied for their effective wastewater treatment potential as depicted in Table 2.
Biomass Growth
All the consortia showed good biomass growth and depicted that the cultures got well adapted to the conditions implemented and have some positive association. Highest biomass at OD 600 was observed in consortium 4 constituted with P. otitidis, B. subtilis, and B. firmus (OD 600/1.2) followed by consortium 2 constituted with P. otitidis + B. circulans+ B. subtilis (OD 600/1). Least biomass was observed in consortium 1 constituted with P. otitidis + B. circulans. Highest growth in consortium 4 may attribute to high COD and nutrient removal. Interestingly, consortium 4 showed maximum stationary phase till 36 h while others showed decline phase after 24th hour itself (Figure 7). Growth of the cultures in any environment is the indication of favorable condition for the cultures to nurture themselves. Growth of the individual microorganism is relatively easy when compared to growth of consortium having different organisms. The bacteria can form various complex associations with other organisms present in the system. These interactions may be positive, negative or may be neutral. In neutral associations, two different bacterial species occupy the same environment without affecting each other. Such association might be transitory; as conditions change in the environment or might be the change in relationship. Positive associations help the species to grow with mutual benefits which might have happened in consortium system 4. Negative association inhibits or adversely effects the growth of other organism. This might have happened in the consortium 3 where although four strains were present but the growth was less when compared to consortium system 2 and 4. Mutualism helps bacteria to form close spatial associations with the others that are essential for their survival. Hence, growth is also an important parameter in consortia study. Along with growth, pH drop in consortia system was observed (Figure 7B) but the VFA production was negligible (data not shown) which may be attributed to the utilization of electrons and protons in the co-metabolism of the consortium (Annie Modestra and Venkata Mohan, 2014).
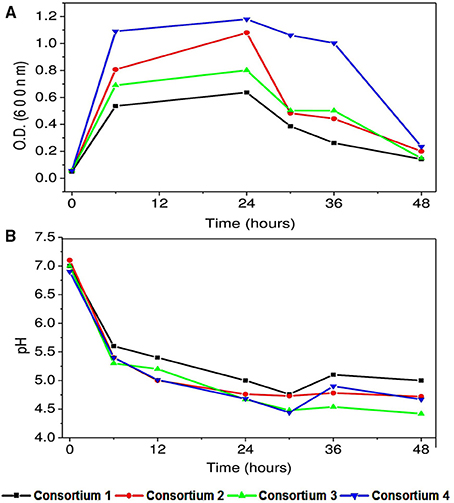
Figure 7. (A) Bacterial growth of synthetic consortia in synthetic wastewater (B) pH variation with time with synthetic consortia.
Wastewater Treatment
Carbon load removal
COD removal for the consortia systems interestingly was higher when compared to the individual strains (Figures 8A,B). Highest COD removal was observed in consortium 4 with CODR efficiency of 86%. This may be attributed to the availability of carbon source to the culture and the co-metabolic activity of the culture is favorable for wastewater treatment. Lowest COD removal in consortium 1 (CODR: 50%) may be attributed to the combination of the isolates with which it is constituted. This combination may not be favorable for substrate degradation, which can be supported with cyclic voltammogram of consortium 1. Cumulative SDR of 0.64 kg CODR/m3-day was found to be maximum with consortium 4 (Figures 8C,D). Interestingly, when SDR per hour (non-cumulative) (SDRT) with consortia was calculated it showed variation form the strains. Sudden increase in SDRT was observed in all systems and thereafter a drop in SDRT at 24th hour was observed. Consortium 1 constituted with P. otitidis and B. circulans showed minimum SDRT at individual hours. At 36th hour and 48th hour SDRT for consortium 1 was 0.0025 kg CODT/m3·h and 0.001 kg CODT/m3·h, respectively which was very less when compared to other consortia. Sudden increment at 6th hour is attributed to growth of the strains in the culture. Later decrement at 24th hour may be attributed to the competition among the strains in the consortia to uptake the substrate or transformation of the substrate towards metabolite production. At 30th hour again increase in SDR% was observed which may be because of the survival of the fittest. At 36th hour slight decrement was observed for all consortia except for consortium 2. The reason for this may be either the surviving strains were trying to uptake the residual COD, or may be the strains were adapting to the depleted carbon and nutrients condition in the wastewater. The consortia study gave different perspective when compared to the studies with individual strains. For better understanding, it was found that the consortia where P. otitidis and B. subtilis were present it showed enhanced treatment efficiency. Various studies also reported higher COD removal using B. subtilis in different consortium using different wastewaters (Sharma et al., 2010).
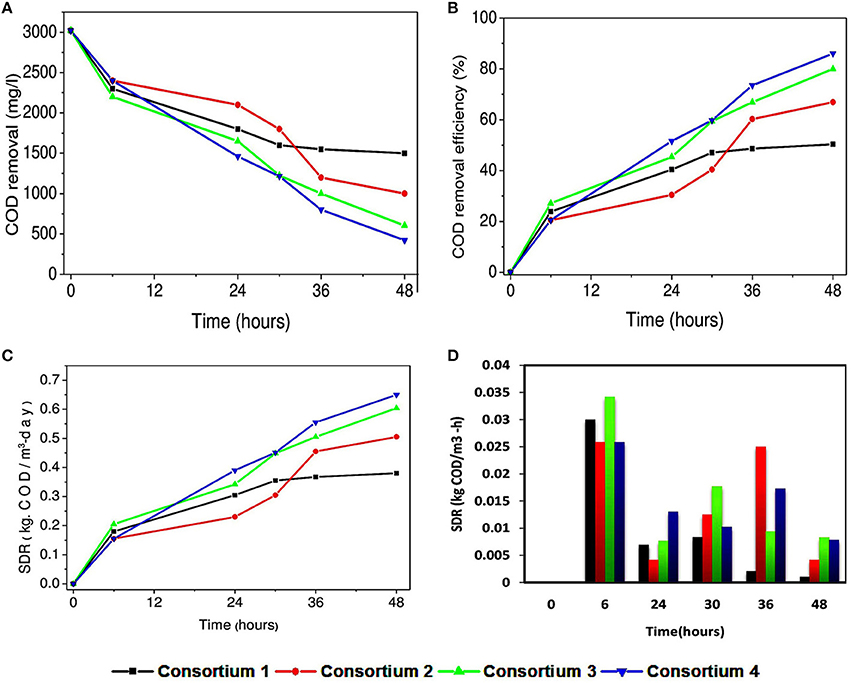
Figure 8. (A) Variation in COD concentration removal (mg/l), (B) COD removal efficiency (%) with synthetic consortia. (C) Variation in substrate degradation rate (cumulative) (kg· CODR/m3-day) and (D) Substrate degradation rate (non-cumulative) (kg· CODT/m3-h) with synthetic consortia.
Nitrate removal
In the consortia operated systems, nitrate removal was also observed to be higher compared to the individual strains (Figures 9A,B). Highest nitrate removal was observed in system 4 (88%). It is interesting to find the consortia having P. otitidis and B. subtilis (consortia 2, 3, and 4) showed higher nutrient removal (75–86%). This can be supported with the previous study where Pseudomonas sps and B. subtilis consortia reported higher nitrate removal (Rajakumar et al., 2008). Literature reveals that B. subtilis is considered as the one of the best candidate for nitrate removal with Pseudomonas sps. Nitrogen removal where microbial community cycled through a series of aerobic, anoxic, and anaerobic stages enables complete removal of nitrogen (N) and phosphorus (P) from wastewater (Rittmann, 2006). This removal involves aerobic chemolithoautotrophic ammonia-oxidizing bacteria (AOB), nitrite-oxidizing bacteria (NOB), chemolithoautotrophic bacteria, polyphosphate-accumulating organisms (PAO), and the glycogen-accumulating organisms (GAO) (Daims et al., 2006). Out of these communities very few microorganism are efficient enough to carry out mechanism, among which B. subtilis is one of the major organism. Many of these organisms were isolated and studied for its individual efficiencies for wastewater treatment (Hamza et al., 2009; Sharma et al., 2010).
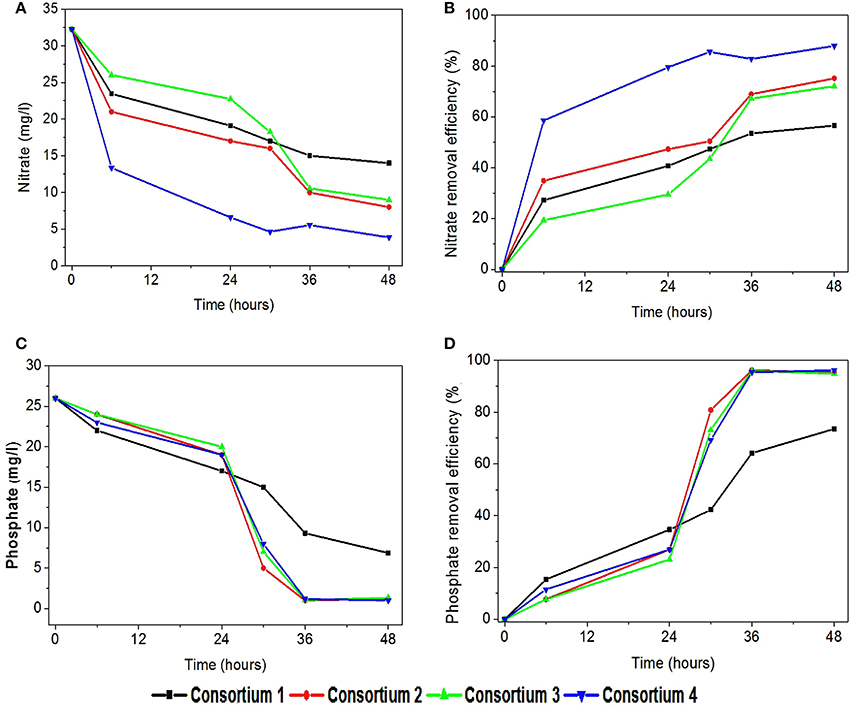
Figure 9. Variation in (A) nitrate removal (mg/l), (B) nitrate removal efficiency (%), (C) phosphate removal (mg/l), and (D) phosphate removal efficiency (%) with synthetic consortia.
Phosphate removal
In consortia operated systems phosphate removal was also observed higher when compared to the individual strains operated systems (Figures 9C,D). Apart from consortia 1 (73%) other consortia showed high phosphate removal (95–97%) supporting the fact that the presence of B. subtilis and P. otitidis helps in higher phosphate solubilisation. Mino et al. (1998) reported that the enhanced biological phosphorus removal capacity also relies on the ability of some organisms that can convert volatile fatty acids to polyhydroxyalkanoate (PHA) under aerobic conditions. Presence of P. otitidis might have lead to lower volatile fatty acids concentrations in all the systems, as its ability for PHA synthesis is well exploited through augmentation (Reddy et al., 2012). The presence of organic acids releases protons which involves in biological ammonium assimilation that enhances the utilization of phosphate.
Bioelectrochemical Analysis
For better understanding CV analysis for the consortia was carried out regularly at different time intervals (Figure 10). Analysis revealed that comparatively redox currents were less compared to individual strains but interestingly RC were high in all the systems. In case of consortium 1, RC was significantly higher than the OC which were more or less similar during all the time intervals. The RC was found to increase with time till 36 h (−2.6 μA) but sudden drop was observed at 48 h (−1.6 μA) which was almost similar to 0th hour depicting a decline in the activity of the culture. In consortium 2, RC was found to have increased till 24 h (RC: −1.8 μA) then dropped from 30 h till 48 h. Interestingly, at 6th hour high RC (−1.2 μA) and OC (0.6 mA) were observed. In consortium 3, interestingly redox currents were found to be relatively similar with RC (−1.6 μA) except OC at 6th hour (0.6 μA) as it was higher compared to others. In consortium 4, the RC increased till 36th hour (RC: −1.7 μA) and later decrement was observed at 48th hour. Interestingly, consortium containing B. subtilis showed higher OC specifically at 6th hour than the consortium lacking it. In this study, the increment in both the oxidation and reduction catalytic currents in aerobic systems indicated the initial biochemical reactions of substrate utilization, where the oxygen is playing negligible role as electron acceptor. Broadly, drop in redox current were observed in later hours which indicates that the metabolically released protons and electrons were consumed toward substrate degradation instead of discharging toward counter and working electrodes.
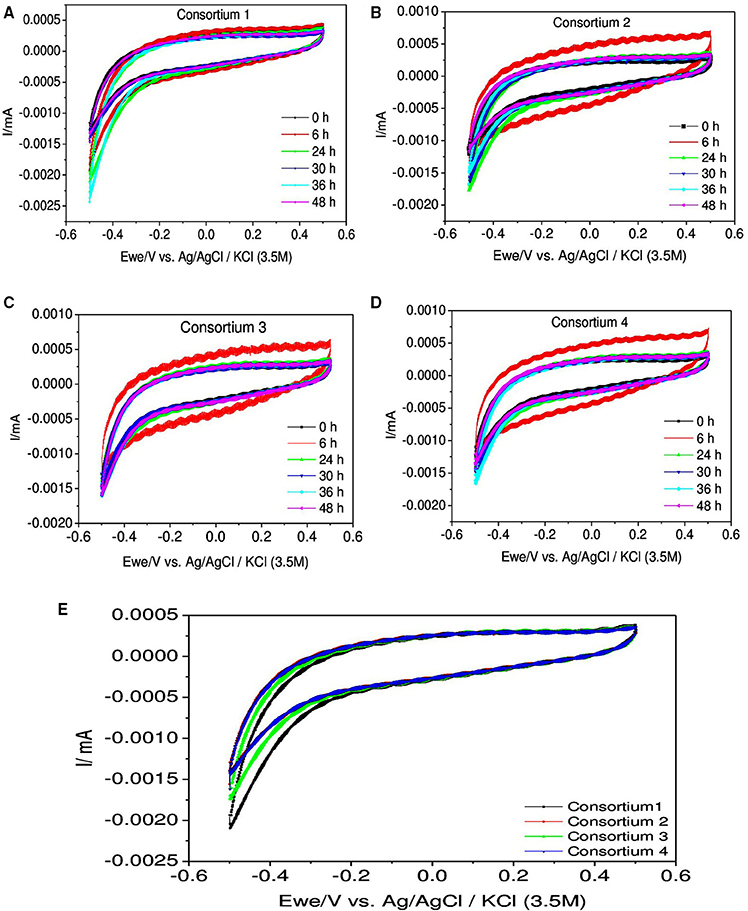
Figure 10. Cyclic voltammograms depicting the redox catalytic currents in (A) Consortium 1, (B) Consortium 2, (C) Consortium 3, (D) Consortium 4, and (E) All Consortia at 24 h recorded at a scan rate of 20 mV s−1.
The comparative evaluation of both the strains and consortia studies revealed that the consortia were effective than the individual strains, suggesting that the potential strains can be used for synthesizing a defined consortium for the effective wastewater treatment (Table 3). When consortia were designed with multiple strains, the performance varied from the individual strains (either positively or negatively) due to the competition/synergy among the strains within the microenvironment. When B. circulans and B. subtilis along with P. otitidis were present in the same microenvironment, the treatment efficiency was low but supplementation of B. firmus has lead to the increment in the wastewater treatment efficiency. Interestingly, B. firmus and B. subtilis along with P. otitidis showed higher wastewater treatment.
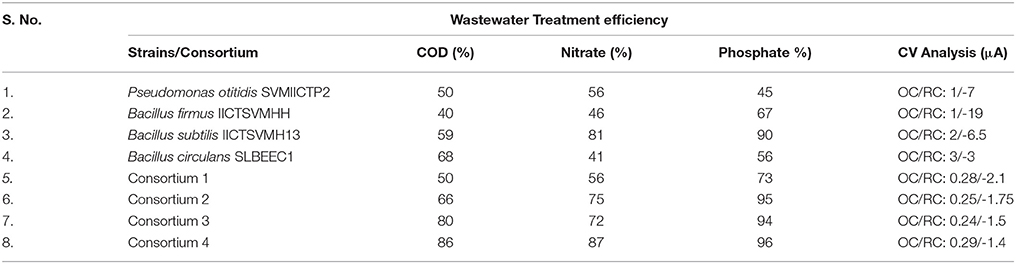
Table 3. Comparative evaluation of synthetic consortia with the single strains for their wastewater treatment potential.
Conclusions
In this study four different strains viz., P. otitidis, B. firmus, B. subtilis, and B. circulans were isolated, identified and further selected for the wastewater treatment. Highest efficiency of carbon removal was observed with B. circulans while nutrients removal was observed for B. subtilis. Four consortia with these isolated and studied strains were designed using different combinations. Synthetic consortium constituted with P. otitidis, B. circulans and B. firmus showed highest treatment efficiency viz. is 86%, 87%, and 96% in terms of carbon, nitrate and phosphate removal, respectively. Bio-electrochemical analysis helped in understanding the release and utilization of electrons and protons toward substrate degradation. Functioning of nitrate reductase in Bacillus and Pseudomonas might have lead to nitrate reduction under aerobic conditions. Lower catalytic currents in consortia studies than the individual strains depicted utilization of the electrons and protons toward enhancing the wastewater treatment. Presence of interspecies bacterial strains and their synergetic relationship enhanced the wastewater treatment efficiency.
Author Contributions
Both the authors have Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; Drafting the work or revising it critically for important intellectual content; final approval of the version to be published; and Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to the Director, CSIR-IICT for his kind support in carrying out this work. This research was funded by CSIR in the form of XII five year plan project on “Sustainable Waste Management Technologies for Chemical and Allied Industries” (SETCA; CSC 0113).
References
Annie Modestra, J., and Venkata Mohan, S. (2014). Bio-electrocatalyzed electron efflux in Gram positive and Gram negative bacteria: an insight into disparity in electron transfer kinetics. RSC Adv. 4, 34045–34055. doi: 10.1039/C4RA03489A
APHA. (1998). Standard Methods for the Examination of Water and Wastewater, 20th Edn. Washington, DC: American Public Health Association/American Water Works Association/Water Environment Federation.
Ayyasamy, P. M., Shanthi, K., Lakshmanaperumalsamy, P., Lee, S. J., Choi, N. C., and Kim, D. J. (2007). Two-stage removal of nitrate from groundwater using biological and chemical treatments. J. Biosci. Bioeng. 104, 129–134. doi: 10.1263/jbb.104.129
Azaizeh, H., Kurzbaum, E., Said, O., Jaradat, H., and Menashe, O. (2015). The potential of autochthonous microbial culture encapsulation in a confined environment for phenol biodegradation. Environ. Sci. Pollut. R. 22, 15179–15187. doi: 10.1007/s11356-015-4981-x
Carter, J. P., Hsaio, Y. H., Spiro, S., and Richardson, D. J. (1995). Soil and sediment bacteria capable of aerobic nitrate respiration. Appl. Environ. Microbiol. 61, 2852–2858
Chen, Q., Ni, J., Ma, T., Liu, T., and Zheng, M. (2015). Bioaugmentation treatment of municipal wastewater with heterotrophic-aerobic nitrogen removal bacteria in a pilot-scale SBR. Bioresource Technol. 183, 25–32. doi: 10.1016/j.biortech.2015.02.022
Choi, Y. S., Hong, S. W., Kim, S. J., and Chung, I. H. (2002). Development of a biological process for livestock wastewater treatment using a technique for predominant outgrowth of Bacillus species. Water Sci. Technol. 45, 71–78. Available online at: http://wst.iwaponline.com/content/45/12/71
Daims, H., Taylor, M. W., and Wagner, M. (2006). Wastewater treatment: a model system for microbial ecology. Trends in Biotechnol. 24, 483–489. doi: 10.1016/j.tibtech.2006.09.002
Dragutinovi, V., Vrvi, M. M., Swiecicka, I., Cvetkovi, O., Beri, T., and Stankovi, S. (2012). Characterisation of new bacillus circulans strain isolated from oil shale. Food Technol. Biotechnol. 50, 123–127. Available online at: http://www.ftb.com.hr/index.php/archives/77-volume-50-issue-no-1/412-characterisation-of-new-bacillus-circulans-strain-isolated-from-oil-shale
Gander, M., Jefferson, B., and Judd, S. (2000). Aerobic MBRs for domestic wastewater treatment: a review with cost considerations. Sep. Purif. Technol. 18, 119–130. doi: 10.1016/S1383-5866(99)00056-8
Groudev, S. N. (1987). Use of heterotrophic microorganisms in mineral biotechnology. Acta Biotechnol. 7, 299–306. doi: 10.1002/abio.370070404
Hamza, D., Mohammed, A., and Ibrahim, S. (2009). Kinetics of biological reduction of chemical oxygen demand from petroleum refinery wastewater. J. Res. 1, 17–23. doi: 10.7537/marsrsj010209.04
Iriye, R., and Takatsuka, H. (1999). “Studies on Sewage Treatment Improvement by Increase/Domination of Genus Bacillus Bacteria”, Bokin Bobai. J. Antibact. Antifung. Agents. 27, 431–440.
Karunai Selvi, B., and Ravindran, A. D. (2012). Influence of different carbon and nitrogen sources on insoluble inorganic phosphate solubilization by bacillus subtilis. IJABR 2, 441–445. Available online at: http://scienceandnature.org/IJABR_Vol2(3)2012.php
Kim, J. K., Park, K. J., Cho, K. S., Nam, S. W., Park, T. J., and Bajpai, R. (2005). Aerobic nitrification-denitrification by heterotrophic Bacillus strains. Bioresour Technol. 96, 1897–1906. doi: 10.1016/j.biortech.2005.01.040
Krishnaswamy, U., Muthusamy, M., and Perumalsamy, L. (2009). Studies on the efficiency of the removal of phosphate using bacterial consortium for the biotreatment of phosphate wastewater. European J. Appl. Sci. 1, 06–15. doi: 10.1128/genomeA.00251-12
Kundu, B. S., and Gaur, A. C. (1984). Rice response to inoculation with N-fixing and p-solubilizing microorganism. Plant Soil 79, 227–234. doi: 10.1007/BF02182344
Min-Jen, T., Mee-Nagan, Y., Ratanakhanokchai, K., Kyu, K. L., and Chen, S.-T. (2002). Purification and characterization of two cellulase free xylanases from an alkaliphilic Bacillus firmus. Enz. Microb. Technol. 30, 590–595. doi: 10.1016/S0141-0229(02)00018-2
Mino, T., Van Loosdrecht, M. C. M., and Heijnen, J. J. (1998). Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32, 3193–3207. doi: 10.1016/S0043-1354(98)00129-8
Muszyñski, A., and Tabernacka, A., Miłobêdzka. (2005). Long-term dynamics of the microbial community in a full-scale wastewater treatment plant. Int. Biodeter. Biodegr. 100, 44–51. doi: 10.1016/j.ibiod.2015.02.008
Nakano, M. M., Hoffmann, T., Zhu, Y., and Jahn, D. (1998). Nitrogen and oxygen regulation of bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J. Bacteriol. 180, 5344–5350.
Naresh Kumar, A., Nagendranatha Reddy, C., Hari Prasad, R., and Venkata Mohan, S. (2014). Azo dye load-shock on relative behaviour of biofilm and suspended growth configured periodic discontinuous batch mode operations: critical evaluation with enzymatic and bio-electrocatalytic analysis. Water Res. 60, 182–196. doi: 10.1016/j.watres.2014.04.027
Ogugbue, C. J., Morad, N., Sawidis, T., and Oranusi, N. A. (2012). Decolorization and partial mineralization of a polyazo dye by Bacillus firmus immobilized within tubular polymeric gel. Biotech. 2, 67–78. doi: 10.1007/s13205-011-0035-3
Raghavulu, S. V., Veer., Sarma, P. N., and Venkata Mohan, S. (2011). Comparative bioelectrochemical analysis of Pseudomonas aeruginosa and Escherichia coli with anaerobic consortia as anodic biocatalyst for biofuel cell application. J. Appl. Microbiol. 110, 666–674. doi: 10.1111/j.1365-2672.2010.04916.x
Rajakumar, S., Ayyasamy, P. M., Shanthi, K., Thavamani, P., Velmurugan, P., Song, Y. C., et al. (2008). Nitrate removal efficiency of bacterial consortium (Pseudomonas sp. KW1 and Bacillus sp. YW4) in synthetic nitrate-rich water. J. Hazard. Mater. 15, 553–563. doi: 10.1016/j.jhazmat.2008.01.020
Reddy, M. V., Nikhil, G. N., Venkata Mohan, S., Swamy, Y. V., and Sarma, P. N. (2012). Pseudomonas otitidis as a potential biocatalyst for polyhydroxyalkanoates (PHA) synthesis using synthetic wastewater and acidogenic effluents. Bioresour. Technol. 123, 471–479. doi: 10.1016/j.biortech.2012.07.077
Renitha, T. S., Ramesh, B., Kayalvizhi, M., Sundarapandiyan, S., and Saravanan, P. (2014). Aerobic digestion of tannery soaking wastewater and reuse of recovered salt. J. Environ. App. Biores. 2, 36–40. Available online at: http://jeab.scienceresearchlibrary.com/view_issue.php?id=Mjg=
Rittmann, B. E. (2006). Microbial ecology to manage processes in environmental biotechnology. Trends in Biotechnol. 24, 261–266. doi: 10.1016/j.tibtech.2006.04.003
Salehizadeh, H., and Shojaosadati, S. A. (2002). Isolation and characterisation of a bioflocculant produced by Bacillus firmus. Biotechnol Lett. 24, 35–40. doi: 10.1023/A:1013853115624
Satar, M. A., and Gaur, A. C. (1984). “Solubilization of insoluble and fixed phosphates in crop rhizobium by phosphomicroorganisms,” in Proc. Int. Symposium on Soil Test Crop Responses Correlation Studies. International Congress Meeting of Soil Science (Dhaka), 237–251.
Sharma, P., Singh, L., and Mehta, J. (2010). COD reduction and colour removal of simulated textile mill wastewater by mixed bacterial consortium. Rasayan J. Chem. 3, 731–735. Available online at: http://rasayanjournal.co.in/vol-3/issue-4/21.pdf
Shi, S., Qu, Y., Ma, Q., Zhang, X., Zhou, J., and Ma, F. (2015). Performance and microbial community dynamics in bioaugmented aerated filter reactor treating with coking wastewater. Bioresour Technol. 190, 159–166. doi: 10.1016/j.biortech.2015.04.075
Torriani-Gorini, A. (1987). “The birth and growth of the Pho regulon,” in Phosphate Metabolism and Cellular Regulation in Microorganisms, eds A. Torriani-Gorini, F. G. Rothman, S. Silver, A. Wright and E. Yagil (Washington, DC: American Society for Microbiology), 3–11.
Venkata Mohan, S., Babu, V. L., Bhaskar, Y. V., and Sarma, P. N. (2007). Influence of recirculation on the performance of anaerobic sequencing batch biofilm reactor (AnSBBR) treating hypersaline composite chemical wastewater. Bioresour. Technol. 98, 1373–1379. doi: 10.1016/j.biortech.2006.05.036
Venkata Mohan, S., Falkentoft, C., Nancharaiah, Y. V., Sturm, B. M., Wattiau, P., Wilderer, P. A., et al. (2009). Bioaugmentation of microbial communities in laboratory and pilot scale sequencing batch biofilm reactors using the TOL plasmid. Bioresour. Technol. 100, 1746–1753. doi: 10.1016/j.biortech.2008.09.048
Venkata Mohan, S., Prakasham, R. S., Satyavathi, B., Annapurna, J., and Ramakrishna, S. V. (2001). Biotreatability studies of pharmaceutical wastewater using an anaerobic suspended film contact reactor. Water Sci. Technol. 43, 271–276. Available online at: http://wst.iwaponline.com/content/43/2/271
Venkata Mohan, S., Sistla, S., Kumar Guru, R., Krishna Prasad, K., Suresh Kumar, C., Ramakrishna, S. V., et al. (2003). Microbial degradation of pyridine using Pseudomonas sp. and isolation of plasmid responsible for degradation. Waste Manage. 23, 167–171. doi: 10.1016/S0956-053X(02)00150-2
Wang, M., Yang, G., Min, H., Lv, Z., and Jia, X. (2009). Bioaugmentation with the nicotine-degrading bacterium Pseudomonas sp. HF-1 in a sequencing batch reactor treating tobacco wastewater: degradation study and analysis of its mechanisms. Water Res. 43, 4187–4196. doi: 10.1016/j.watres.2009.07.012
Wu, J., Jung, B. G., Kim, K. S., Lee, Y. C., and Sung, N. C. (2009). Isolation and characterization of Pseudomonas otitidis WL-13 and its capacity to decolorize triphenylmethane dyes. J. Environ. Sci. 21, 960–964. doi: 10.1016/S1001-0742(08)62368-2
Keywords: phylogenetic analysis, carbon load, nutrients, bacterial interactions, bioelectrochemical analysis
Citation: Dahiya S and Venkata Mohan S (2016) Strategic Design of Synthetic Consortium with Embedded Wastewater Treatment Potential: Deciphering the Competence of Isolates from Diverse Microbiome. Front. Environ. Sci. 4:30. doi: 10.3389/fenvs.2016.00030
Received: 09 February 2016; Accepted: 01 April 2016;
Published: 04 May 2016.
Edited by:
Mohiuddin Md. Taimur Khan, Washington State University, USAReviewed by:
Srikanth Sandipam, Indian Oil Corporation Limited, IndiaDipti Prakash Mohapatra, National Research Council, Canada
Copyright © 2016 Dahiya and Venkata Mohan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: S. Venkata Mohan, vmohan_s@yahoo.com
 Shikha Dahiya
Shikha Dahiya S. Venkata Mohan
S. Venkata Mohan