An Increasing Need for Productive and Stress Resilient Festulolium Amphiploids: What Can Be Learnt from the Stable Genomic Composition of Festuca pratensis subsp. apennina (De Not.) Hegi?
- 1Centre of Plant Structural and Functional Genomics, Institute of Experimental Botany, Olomouc-Holice, Czechia
- 2Institute of Biological, Environmental and Rural Sciences, Gogerddan Aberystwyth University, Aberystwyth, Wales, UK
- 3Agroscope INH, Zürich, Switzerland
- 4Department of Earth and Environmental Sciences, University of Pavia, Pavia, Italy
Genome composition of Festuca pratensis subsp. apennina (De Not.) Hegi, a tetraploid fescue species native to the tall forbs communities of south-eastern Europe at altitudes between 1100 and 2200 m a.s.l. has been the subject of some debate by grass taxonomists. Our cytogenetic analyses including fluorescence in situ hybridization with probes for genomic DNA and selected DNA repeats revealed the species to be allotetraploid and derived from interspecific hybridization between F. pratensis Huds., a species confined to grassland at lower altitudes, and a so far unknown Festuca species. Besides tetraploids, triploids, and pentaploids were found growing in Alpine meadows in close association with F. pratensis subsp. apennina. Triploid cytotypes predominated at many sites in Switzerland and Romania, and in some localities, they were the only cytotypes observed. Cytogenetic analyses revealed the triploids to be hybrids between diploid F. pratensis and tetraploid F. pratensis subsp. apennina, while the pentaploid cytotypes originated from hybridization between F. pratensis subsp. apennina and hexaploid F. arundinacea Schreb., a closely-related species growing in a close vicinity to F. pratensis subsp. apennina. Parental genomes of F. pratensis subsp. apennina and of the triploid and pentaploid hybrids showed no evidence of homoeologous chromosome pairing and interspecific recombination, supporting previous observation of a disomic inheritance at meiosis, where chromosome pairing was restricted to bivalent associations. A hypothesis is presented that a chromosome pairing regulator(s), reported previously in other polyploid broad-leaved fescue species of the Festuca subg. Schedonorus, is present and functional in F. pratensis subsp. apennina. It is likely that a common ancestors' genome that carries the chromosome pairing regulator(s) is present in all polyploid broad-leaved fescue species, and its acquisition was a key event that enabled speciation and development of a polyploid series within Festuca. Identification of a functional chromosome pairing regulator capable of stabilizing advantageous genome combinations in hybrids within the Lolium-Festuca complex would greatly assist in development of stable Festulolium cultivars. Its expression within Festulolium amphiploid cultivars would assist strategies aimed at climate-proofing productive European grasslands to combat exposures to stress conditions.
Introduction
Climate-smart strategies are required to ensure sustainability of European-based grassland management systems suitable for livestock agriculture in the face of increasing external environmental pressures. These arrive from increased requirements for improved resilience to extreme weather events and from governments and policy makers seeking some assurance of greater efficiencies and environmental safeguards. Festulolium grasses, defined as hybrids between any ryegrass (Lolium) and fescue (Festuca) species, are considered an effective response by providing combinations of high yields of nutritious fodder from Lolium together with added resilience to abiotic and biotic stress from Festuca (Humphreys et al., 2006, 2014). Whilst Festulolium have hitherto been used quite widely in Central and Eastern Europe, in the case of the UK, the acceptance of the IBERS' bred Festulolium cultivar “AberNiche” onto the UK National Recommended Lists represented a change in mind-set by the commercial sector. There is a growing belief that Festulolium cultivars capable of providing effective and appropriate stress resilience when conditions require, should now become more widely available commercially, to underpin future food security, and grassland agriculture.
Festulolium are marketed either as amphiploids, (where entire Lolium and Festuca genomes are combined), or as introgressive forms [where specific genes from one species (usually from Festuca) are incorporated into the genome of another; Ghesquiere et al., 2010]. Whilst maintaining fertility and genome stability through use of an introgression breeding approach has not thus far presented significant difficulties, the more widely used amphiploid breeding approach which for several quantitative traits may be deemed more appropriate has proved to be more problematic (Ghesquiere et al., 2010). One example where Festulolium amphiploids were required was reported recently where in order to assist efficient ruminant nutrition, the maintenance of complete and balanced Lolium and Festuca genome complements was seen as an essential prerequisite in order to combat plant-mediated proteolysis within ingested fodder (Humphreys et al., 2014). Unfortunately, for such a breeding approach the outcome reported from all cytogenetic studies that involved amphiploid Festulolium varieties was that unbalanced genome complements had arisen over generations of seed multiplication which had resulted in a shift toward a Lolium genome, accompanied by loss of Festuca chromosomes (Canter et al., 1999; Kopecký et al., 2006; Zwierzykowski et al., 2006). This outcome was aided by frequent homoeologous pairing and recombination between Festuca and Lolium chromosomes and possibly by the self-incompatibility of the species involved (Kopecký et al., 2008a, 2010; Harper et al., 2011).
A stable and balanced genome composition could it is thought be achieved by the incorporation of a diploid-like chromosome pairing mechanism in Festulolium hybrids, such as Ph1 found in wheat, capable of encouraging homologous and preventing homoeologous chromosome pairing. However, the agriculturally desirable diploid grass species (such as F. pratensis Huds., Lolium perenne L., and Lolium multiflorum Lam.) widely used in Festulolium amphiploid varieties lack such a functional system (Kopecký et al., 2009). However, an equivalent diploidizing chromosome pairing mechanism to Ph1 has been identified in closely related polyploid broad-leaved fescues of subg. Schedonorus, such as F. arundinacea Schreb. subsp. arundinacea, F. arundinacea var. glaucescens Boiss. (≡F. arundinacea subsp. fenas (Lag.; here-after referred to as F. glaucescens Arcang.), and F. mairei St. Yves (Jauhar, 1975, 1993; Kopecký et al., 2009) and is considered essential in their evolution and speciation. If the chromosome pairing regulator was incorporated successfully into synthetic Festulolium hybrids and its function maintained, it should provide for regular chromosome disjunction, consistent disomic inheritance, and the levels of genome stability over generations required to fully exploit the benefits to be derived from use of Festulolium amphiploids in agriculture, and adherence to the strict requirements for genome uniformity and stability necessary for variety status within the UK.
Unlike the Lolium genus, where species are all diploids, polyploidy is a prominent feature amongst related Festuca species including the broad-leaved fescues of the subg. Schedonorus. The polyploids of this sub-genus, whose genome origins have previously been reported (e.g., Jauhar, 1993), are believed to derive as a consequence of species' hybridization and through assemblies of novel advantageous genome combinations that gave enhanced adaptations absent in their parental species. An excellent example was the result of the hybridization between the North and Central European grass Festuca pratensis Huds. (2n = 2x = 14) and the Mediterranean-based Festuca glaucescens Arcang. (2n = 4x = 28) that gave rise to the now widely dispersed hexaploid tall fescue (Festuca arundinacea Schreb.) (2n = 6x = 42), used widely in grassland agriculture and in particular where abiotic stresses are too severe for less well adapted Lolium varieties (Humphreys et al., 1995).
Key events in the evolution and speciation of these hybrids were their initial chromosome doubling and the subsequent restricted preferential chromosome pairing between homologous partners at meiosis, encouraged by a chromosome pairing regulator (Jauhar, 1975). These events led to hybrid genome stabilization and maintained intact their adaptive advantages, a situation sought but unfortunately not so far replicated to the same extent within current synthetic Festulolium cultivars.
One of the progenitors of F. arundinacea, the tetraploid fescue F. glaucescens is a mid to high altitude species found in the southern part of the Iberian Peninsula and north-western Africa (Devesa et al., 2013). It has been used for the development of three drought-tolerant Festulolium cultivars (L. multiflorum × F. glaucescens), which were released and registered in France (Ghesquiere et al., 2010). The hybrid combination has more recently also been produced independently at IBERS together with an equivalent amphiploid hybrid involving F. glaucescens and L. perenne. Both hybrid combinations have demonstrated excellent potential as fodder and as aides to efficient ruminant nutrition considered likely to reduce greenhouse gas emissions by livestock (Humphreys et al., 2014). Other novel amphiploid Festulolium hybrids with high agronomic potential, large root systems, and significant stress tolerance generated at IBERS include L. multiflorum and L. perenne combinations with tetraploid Atlas fescue (Festuca mairei; Humphreys et al., 2014).
The focus of the research described herein is on another fescue from subg. Schedonorus, the little-studied tetraploid species F. pratensis Huds. subsp. apennina (De Not.) Hegi [also classified as Schedonorus pratensis subsp. apenninus (De Not.) H. Scholtz and Valdès]. The Festuca species henceforth referred to as F. apennina was described by De Notaris (1844) and St. Yves (1913) from the locality of Santo Stefano d'Aveto and is probably the same species as F. pratensis var. megalostachys (Stebler, 1904; Foggi and Müller, 2009). The genomic composition of the species has been a matter of some conjecture. The close relationship between F. apennina and the diploid fescue F. pratensis, a species having wider distribution throughout Eurasia and north-western Africa (Foggi and Müller, 2009), has long been known. Alternative claims have been made that F. apennina is an autotetraploid cytotype of F. pratensis, or now and more widely accepted, an allotetraploid species derived from two ancestral progenitor species (Lewis, 1977; Jauhar, 1993). Borrill et al. (1976) reported a different ecological and altitudinal distribution for the two fescue species with F. apennina absent below 1300 m above sea level (a.s.l.), sometimes accompanied by F. pratensis in regions between 1300 and 1600 m a.s.l., but to its exclusion at higher altitudes. The adaptation of F. apennina to high altitudes derives from its vegetative growth being confined to summer months, followed by a protracted growth quiescence and complete leaf senescence when winter stresses are evident from October to May (Tyler and Chorlton, 1974). Besides diploids and tetraploids, Tyler (1988) discovered the existence of triploid cytotypes and considered them as putative natural hybrids between F. pratensis and F. apennina at the mid-altitude locations where both species coexist.
The current research provides compelling evidence for the genome composition of F. apennina, and for its putative progenitors. Through cytogenetic studies of plant collections abstracted from Alpine meadows, including putative natural hybrids involving F. apennina, a hypothesis is proposed of how the region may have played a pivotal role in events that led to the evolution of polyploidy within the Festuca subg. Schedonorus. Through enhanced understanding of such key events and their genetic controls, it may be possible to replicate in similar fashion in hybrid genomes within Festulolium and thereby achieve desirable and productive grass varieties combined with the essential safeguards required to combat climate change.
Materials and Methods
Plant Materials
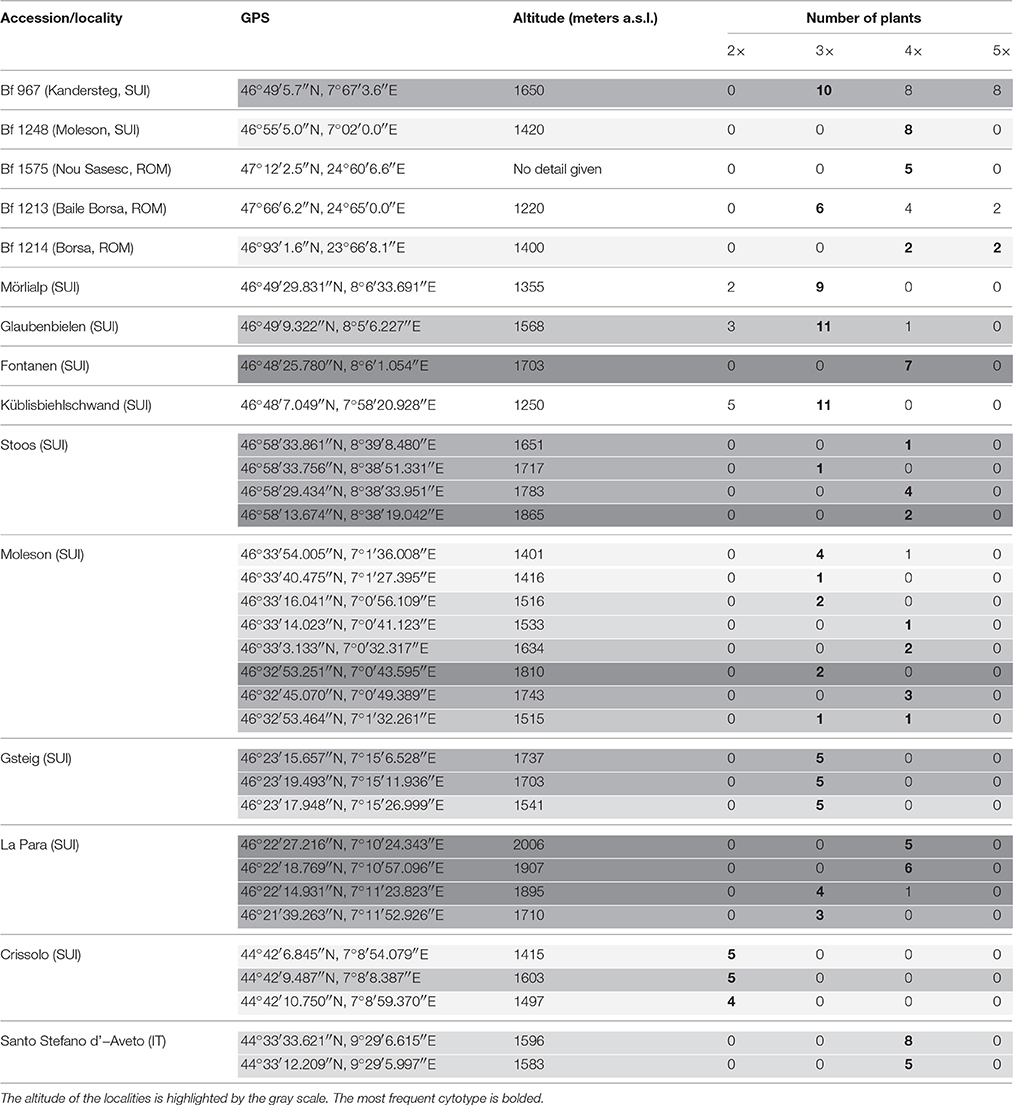
The plant materials derived from two sources: from a genebank maintained at IBERS and from newly sourced plant material. Their origins including GPS coordinates and altitudes from where they were collected are summarized in Table 1 and Figure 1. The initial IBERS F. apennina collection was established from Swiss accessions Bf 967 and Bf 1248 and three accessions from Romania (Bf 1575, Bf 1213, Bf 1214). In addition, an accession of F. pratensis (Bf 970) and of F. arundinacea (Bn 950) were included both collected from locations in close proximity to the sources of Swiss accessions Bf 967 and Bf 1248. Seed storage at IBERS included periods at 2°C with a Relative Humidity of 20% (to maintain viability for an estimated 25 years), but also from unique seed storage facilities where seed was maintained in sealed foil laminate pouches following drying to circa6% moisture content at −20°C (where viability was predicted for circa 100 years).
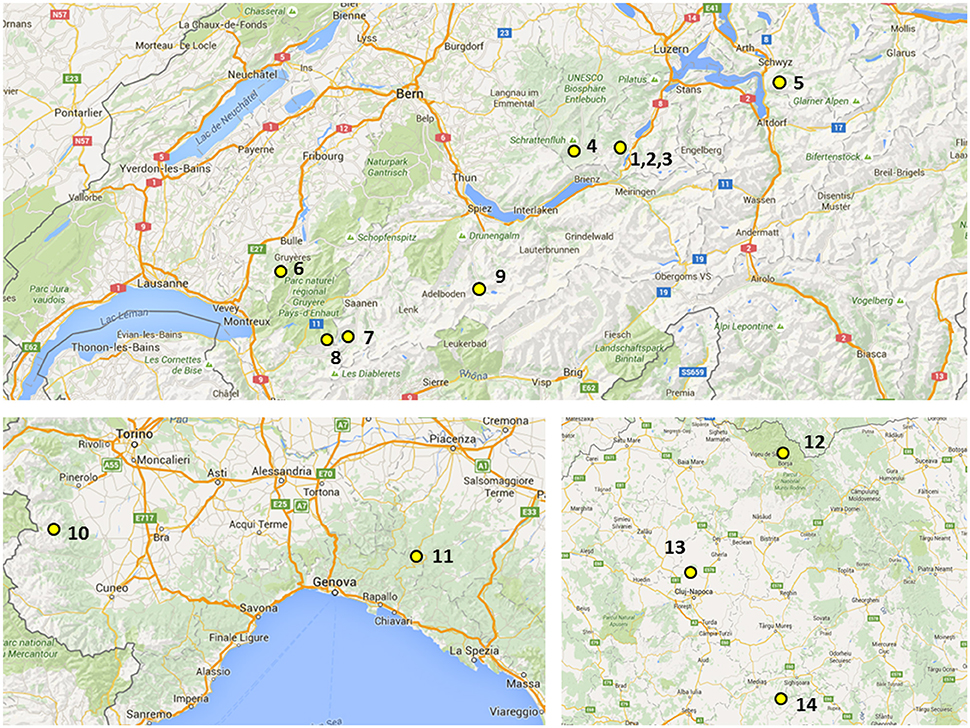
Figure 1. Geographical distribution of localities in Switzerland (up), Italy (bottom, left), and Romania (bottom, right): (1) Mörlialp, (2) Glaubenbielen, (3) Fontanen (4) Küblisbiehlschwand, (5) Stoos, (6) Moleson, (7) Gsteig, (8) La Para, (9) Kandersteg, (10) Crisollo, (11) Santo Stefano d'8FAveto, (12) Baile Borsa, (13) Borsa, and (14) Nou Sasesc.
A synthetic triploid hybrid P127/200 (2n = 3x = 21) produced by crossing tetraploid F. apennina and diploid F. pratensis was used to verify the provenance of the unknown triploids collected. Another synthetic hybrid developed from the cross of tetraploid L. multiflorum and F. apennina has been used in this study.
New plant collections were made in May and September 2015 in order to collect new F. apennina genotypes. In May 2015, entire plants were collected from various localities in Switzerland and Italy (Alps and Apennines), including the locus classicus of F. apennina (Santo Stefano d'Aveto; see Ardenghi and Foggi, 2015). In September 2015, two sets of single leaves representing individual plants were collected from three Swiss localities and stored in wet paper towel until the flow cytometry analyses was carried out (10 days after collection). The first set of plants represented a random sample. The second set, were of leaves from selected plants whose morphology (large plants with broad leaves and brown-red bases of the leaves) was considered as consistent with expectations for a tetraploid cytotype. The collection sites in Switzerland and Italy differed; in Switzerland, plants were collected from mountain meadows and grazed-lands near farmsteads; in Italy the collection sites at Santo Stefano d'Aveto were in shaded woodland-cleared and water-logged areas.
Ploidy Level Determination
The ploidy level of each accession from the sampling expedition and plants grown from seed obtained from IBERS gene bank was estimated by flow cytometry (Doležel et al., 2007). Nuclear suspensions were prepared from 50 mg leaf tissues of each grass sample with Pisum sativum cv. Ctirad used as standard having 2C = 9.09 pg (Doležel et al., 1998). The tissues were homogenized with a razor blade in a Petri dish containing 0.5 ml of Otto I solution (0.1M citric acid, 0.5% Tween 20). The nuclear suspension was filtered through 42 μm nylon mesh and stained with 1 ml Otto II solution (0.4M Na2HPO4.12 H2O) containing 2 mg/ml ß-mercaptoethanol, and 2 μg/ml DAPI (4′,6-diamidino-2-phenylindole). Samples were analyzed using a CyFlow Space flow cytometer (Sysmex Partec GmbH., Görlitz, Germany) equipped with a UV led diode array. At least 5000 events were acquired per sample and only measurements with coefficient of variation for G0/G1 peaks <2.5% were accepted. Two representative plants from each tetraploid, triploid, and pentaploid cytotypes were checked by chromosome squash preparations and counting as described in Ahloowalia (1965) to verify the flow cytometric results.
Genomic In situ Hybridization (GISH) and Fluorescence In situ Hybridization (FISH) Analyses
A GISH analysis was undertaken on representative triploid, tetraploid, and pentaploid plants. In total, we analyzed five tetraploid plants: two plants from accession Bf 967, and one each from Bf 1248, Bf 1575, and Bf 1213, three pentaploids: two plants from Bf 967 and one from Bf 1213 and three triploid plants: one each from Bf 967, Bf 1213, and Bf 1214. In addition, the synthetic triploid hybrid F. pratensis × F. apennina P127/200 (2n = 3x = 21) and the synthetic tetraploid hybrid L. multiflorum × F. apennina developed from the cross of autotetraploid L. multiflorum with F. apennina were used for GISH analysis. GISH was done as described in Anamthawat-Jónsson and Reader (1995). In the first experiment, total genomic DNAs (gDNA) of F. pratensis and F. apennina were labeled with tetramethyl-rhodamine-5dUTP and fluorescein-12-dCTP, respectively. Both probes had been preannealed and thereafter hybridized with chromosomes of triploid and tetraploid cytotypes and also the synthetic hybrids squashed on microscopic slides. The same approach was used for GISH with the pentaploid cytotype, except the total gDNAs of F. glaucescens and F. pratensis were used as probes. In the case of the synthetic L. multiflorum × F. apennina hybrid, both probe combinations (F. pratensis–F. apennina and F. pratensis–F. glaucescens) were applied.
In the second experiment, probe made from total gDNA of F. pratensis by Nick-translations with Dig-Nick Translation Mix (Roche) and blocking DNA developed by shearing total gDNA of F. glaucescens were used. Probe hybridization signals were detected by anti-DIG-FITC conjugate.
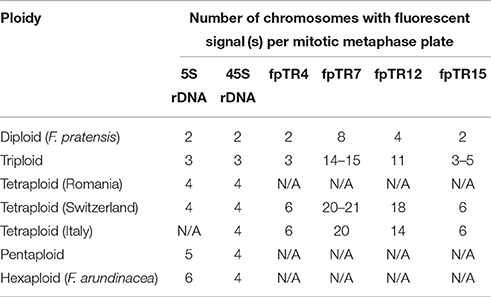
In order to support the GISH results, FISH analyses (as described in Thomas et al., 1996; Harper et al., 2004) were undertaken with probes for 5S and 45S rDNA and tandem repeats (see below). Probes for 5S rDNA and 45S rDNA were prepared as described in Thomas et al. (1997). The rDNA probes were used in an initial FISH experiment with triploid, tetraploid, and pentaploid cytotypes and their possible progenitors represented by F. arundinacea Bn 950 and F. pratensis Bf 970.
Additionally, FISH probes were prepared from four tandem repeats (fpTR4, fpTR7, fpTR12, and fpTR15) identified in our previous work (Kopecký et al., 2013) using PCR labeling with biotin- or digoxigenin-labeled nucleotides (Roche) and pairs of specific primers. In addition, a 45S rDNA probe was used in this subsequent experiment. Probe hybridization signals were detected by anti-DIG-FITC and streptavidin-Cy3 conjugates. In total, 20 plants representing all identified ploidy levels were analyzed, each group comprising five genotypes identified as either having diploid (F. pratensis), triploid, or and tetraploid (F. apennina) genome compositions (localities Fontanen and Küblisbiehlschwand), and an additional five tetraploid F. apennina genotypes from Italy. In all experiments, the chromosomes were counterstained with 1.5 μg/ml DAPI in Vectashield antifade solution (Vector Laboratories). Slides were evaluated with an Olympus AX70 microscope equipped with epi-fluorescence and a SensiCam B/W camera and a Leica DM/RB epifluorescence microscope. ScionImage and Adobe Photoshop software were used for processing of the color pictures.
Results and Discussion
Mitotic chromosome counts in 26 plants randomly selected from Swiss accession Bf 967 indicated large variation in chromosome number. Ten plants had 21 chromosomes (including several plants possessing B chromosomes), eight plants had 35 chromosomes, and only eight plants had the anticipated 28 chromosome constitution. All eight analyzed plants of another accession from Switzerland (Bf 1248) were confirmed as tetraploids. Among 12 plants of Romanian accession Bf 1213, six were triploid, two were pentaploid (2n = 35), whilst the remaining four plants carried the anticipated 28 chromosome complement. In another Romanian Accession (Bf 1214), two plants with 35 chromosomes and another two plants with 28 chromosomes were detected. All five plants of accession Bf 1575 from Romania were tetraploid (Table 1).
Large variation in ploidy level from location to location and at different altitudes has also been found among the plants sampled in Switzerland and Italy. In total, 136 plants were analyzed and 24 diploid (F. pratensis), 64 triploid and 48 tetraploid (F. apennina) genotypes were identified (Figure 2). Surprisingly, unlike the Swiss and Romanian accessions described above, no pentaploid plants were found amongst the newly collected Swiss and Italian ecotypes. The diploids were detected primarily at lower altitudes, and no diploids were detected above 1750 m a.s.l. Plants having triploid chromosome complements were found growing together with diploids in a sympatric manner, but only up to 1900 m a.s.l. Tetraploids were usually detected above 1500 m a.s.l., but one tetraploid plant from the Moleson locality was recovered at 1401 m a.s.l. In most of the Swiss localities, two or three cytotypes were found growing sympatrically. However, in the Italian locality Crissolo, only diploids were detected. In contrast, only tetraploid genotypes were found in the Italian locality Santo Stefano d'Aveto. Triploids predominated in many Swiss localities and in several sites they were the only cytotypes identified (e.g., localities in Gsteig and one La Para locality). Ploidy levels of plants from all sites and localities are given in Table 1.
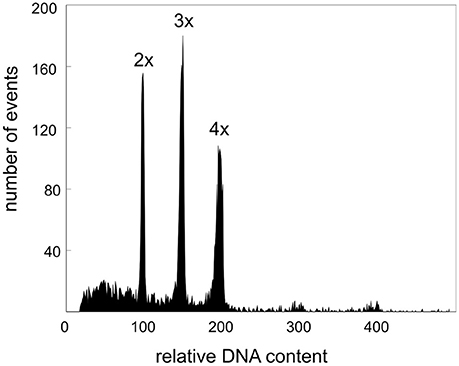
Figure 2. Histogram of relative DNA content (DAPI fluorescence intensity) obtained after simultaneous analysis of nuclei isolated from diploid, triploid, and tetraploid plants collected in Glaubenbielen.
In order to achieve a more accurate and representative view of ploidy compositions, as the initial plant numbers sampled were small (up to 15 plants per site), a further collection was taken from the three sites where previously two or three cytotypes had been detected. Random sampling of 85 plants in Mörlialp (1355 m a.s.l.) revealed 11.8% of plants as diploids, 88.2% as triploids, and with no tetraploid genotype detected. Random sampling of one hundred genotypes in Glaubenbielen (1568 m a.s.l.) identified 15% of plants as diploids, 52% as triploids, and 33% as tetraploids. When preferential sampling was undertaken on 20 plants considered from their morphology as likely to be tetraploid cytotypes of F. apennina, three diploids, and 13 triploids were identified, whilst only four plants had an expected tetraploid genome complement. Flow cytometric analysis of 89 randomly sampled plants from Küblisbiehlschwand locality (1250 m a.s.l.) revealed 92.1% diploids, and 7.9% triploids. Attempts at preferential sampling for 20 plants with tetraploid-like growth morphology proved totally inadequate as cytological studies revealed 14 as diploids and 6 as triploids. It is evident that preferential sampling for tetraploid-like plants based only on their morphological characters is not practical as it was impossible to distinguish diploids, triploids and tetraploids at the vegetative stage.
Flow cytometry showed that triploid cytotypes predominated and moreover they were the only cytotype found at some localities. Previous sampling had revealed only diploids and tetraploid cytotypes (Borrill et al., 1976; Tyler et al., 1978), but one report mentioned the recovery of triploid cytotypes (Tyler, 1988). Whilst the fertility of the triploid cytotypes was not assessed, it is expected that they had high sterility relying primarily on vegetative propagation for their widespread establishment. Clarke et al. (1976) developed synthetic triploid hybrids of F. pratensis × F. apennina which they found to be sterile. In this case, fertility was restored by chromosome doubling to encourage preferential homologous chromosome pairing in the resulting hexaploid genotype. The predominance of the triploid cytotype is intriguing, and implies they have an adaptive selection advantage compared to the diploid and tetraploid cytotypes. In the Küblisbiehlschwand and Mörlialp localities, over one hundred individuals were sampled, and no tetraploid detected. A somewhat similar finding was reported by Humphreys and Harper (2008). They reported the presence of triploid hybrids of Festulolium loliaceum (Huds.) P. Fourn., a natural Festulolium hybrid species derived from hybridization between diploid F. pratensis and L. perenne. The triploid cytotypes (which had two alternative genome combinations; either having two genomes of Lolium or two of Festuca) were found growing on heavily waterlogged soils where neither parental species was generally found, suggesting that their enhanced genome dosage and consequent genome interactions had provided them a certain adaptive advantage. However, diploid cytotypes of Festulolium loliaceum are also found growing in waterlogged soils at the same locations as the triploid cytotypes, which might indicate, at least in this instance, that it is more the extent of intergeneric genome activity between the Lolium and Festuca genomes that is responsible for the adaptive advantage found over the parental species. Both diploid and triploid cytotypes of Festulolium loliaceum show high male and female sterility (Humphreys and Harper, 2008).
The triploid cytotypes of F. apennina and of Festulolium loliaceum arose probably from the interspecific hybridization of diploid and tetraploid cytotypes (triploid F. apennina) or as a consequence of the presence of unreduced gametes within their diploid parental (either F. pratensis or L. perenne) species (triploid Festulolium loliaceum). The occurrence of unreduced gametes is believed to be a common feature found in various Festulolium species (Morgan et al., 1988), and is likely within the genus to be an important component part in the events that led to the evolution of a polyploid series and through these, the perpetuation of interspecific hybrids. Through genome duplication in interspecific hybrids preferential intraspecific homologous chromosome pairing becomes possible leading to disomic inheritance, genome stability and to improved fertility. However, on its own, in hybrids between the closely related species of the Lolium-Festuca genome complex, the extent of intraspecific homologous chromosome pairing is insufficient to reliably maintain over generations stable and balanced genome complements. Indeed, if preferential chromosome pairing was sufficient on its own to ensure genome perpetuity, then the consistent difficulties reported by Festulolium breeders in maintaining intact and stable hybrid genomes would not have arisen (Ghesquiere et al., 2010). An additional requirement was necessary in the evolution of Festuca polyploids in order to obtain consistent disomic inheritance in their natural amphiploid hybrid genome complements; the chromosome pairing regulator first identified in F. arundinacea (Jauhar, 1975; Lewis et al., 1980). The event(s) that allowed acquisition of the supplementary diploidising mechanism present in polyploid fescues is likely to have played a pivotal role in the evolution of a polyploid series in Festuca.
It would seem unlikely that such an otherwise unique diploidising mechanism had evolved to function in the same manner through independent events in different polyploid fescue species. The system may have evolved only once and subsequently been distributed by interspecific hybridization events between various fescue species. The mechanism is presumably sited on a common subgenome found in all polyploid fescues. The system in polyploid Festuca species differs from the Ph1 of wheat and the system found in polyploid oats by the haplo-insufficiency or hemizygous-ineffectiveness, which means that the system is inactive in single-dose. From a Festulolium plant breeding perspective strategies need to be developed whereby gene(s) that confer a functional diploid like chromosome -pairing system are present in two copies in order that stable Festulolium genomes are achieved.
Genomic In situ Hybridization (GISH) and Fluorescence In situ Hybridization (FISH) Analysis of Festuca apennina Cytotypes
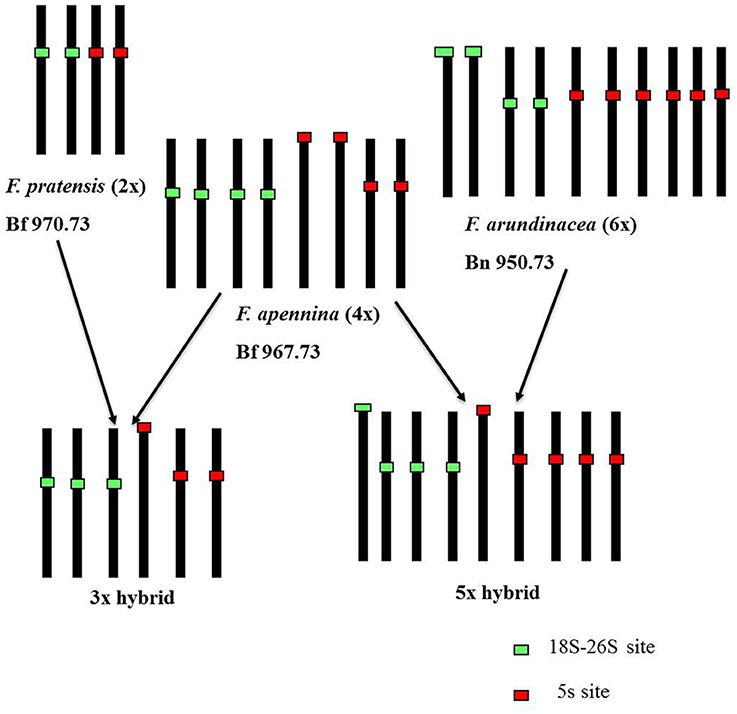
GISH was used to determine the genomic composition of tetraploid F. apennina, and also the triploid and pentaploid cytotypes found growing in close association. Hybridization of F. apennina chromosomes with differentially labeled probes of gDNA of F. pratensis and F. apennina resulted in the painting of 14 chromosomes by gDNA of F. pratensis and of all chromosomes by the gDNA of F. apennina (data not shown). When F. pratensis gDNA probe was used together with blocking DNA from F. glaucescens onto tetraploid F. apennina chromosomes, 14 chromosomes became labeled with 14 chromosomes remaining unlabeled (Figure 3G). From this, it can be concluded that F. apennina is an allotetraploid species having derived from at least two ancestral progenitors, one of which was F. pratensis. The second diploid species progenitor of F. apennina is unknown. This is not surprising as F. pratensis is the only diploid species found in the European clade of Festuca subg. Schedonorus. GISH provided no evidence for any interspecific chromosome recombination between the constituent genomes in F. apennina giving support to previous meiotic studies where strict homologous bivalent chromosome pairing and disomic inheritance of the parental genomes was reported (Lewis, 1977).
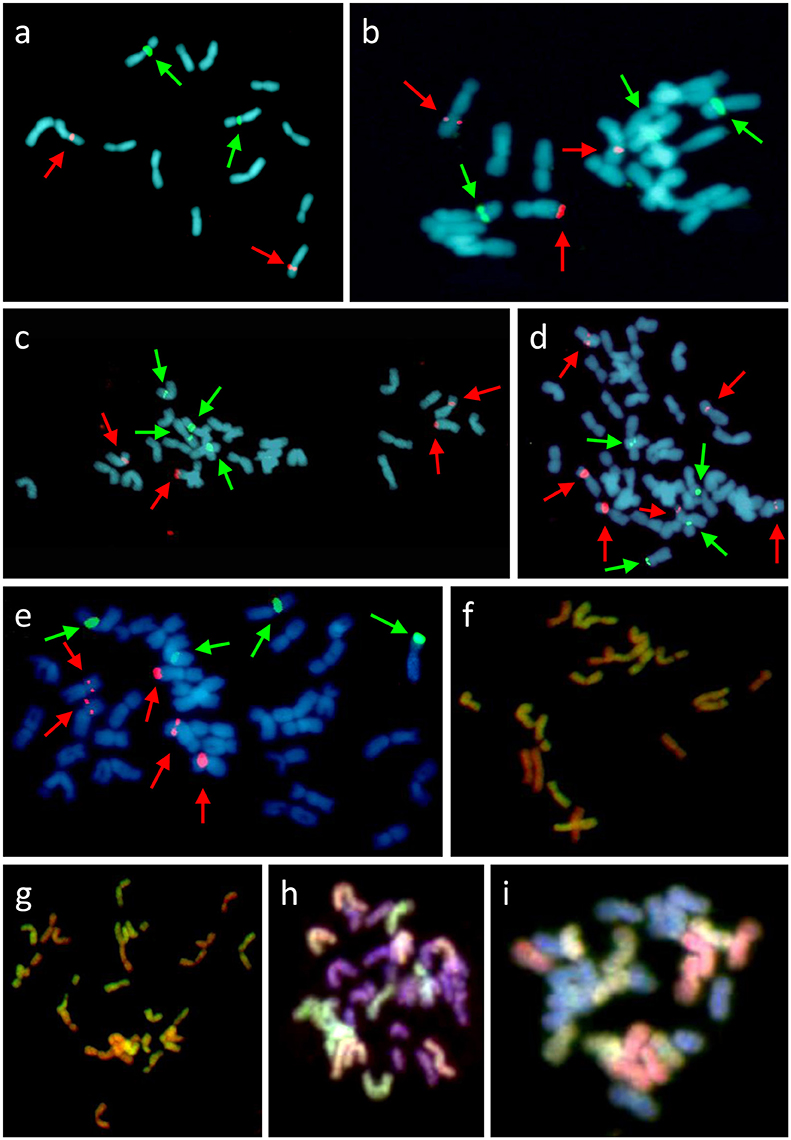
Figure 3. Cytogenetic analysis of F. pratensis/F. apennina/F. arundinacea complex (A–G) and L. multiflorum × F. apennina hybrid (H,I). FISH with probes for 5S rDNA (red color, highlighted with red arrows) and 45S rDNA (green color, highlighted with green arrows) on metaphase spreads of diploid F. pratensis (A), triploid hybrid (B), tetraploid F. apennina (C), pentaploid hybrid (D), and hexaploid F. arundinacea (E). GISH with probe made from labeled gDNA of F. pratensis and blocking DNA from sheared gDNA of F. glaucescens in triploid (F) and tetraploid (G) revealed hybrid origin of both cytotypes. Additional GISH analysis of tetraploid L. multiflorum × F. apennina hybrid with two probe combinations: (H) gDNA of F. pratensis (red color) and gDNA of F. apennina (green color) and (I) gDNA of F. pratensis (green color) and F. glaucescens (red color). Chromosomes were counterstained with DAPI (blue color).
Conclusions drawn from the GISH results over the genomic constitution of F. apennina gained further support from the FISH study where rDNA and tandem repeats were used as probes. In F. pratensis, two 45S rDNA loci were detected each at homologous sites on the short arm of chromosome 3. Two 5S rDNA loci were located at a pericentromeric region on the short arm of chromosome 2 (Figure 3A). These findings are in agreement with previous results of Thomas et al. (1997) and Kopecký et al. (2008a). In F. apennina, four 45S rDNA loci were detected on two pairs of chromosomes. In addition, a pair of chromosomes carried 5S rDNA loci at proximal regions, whilst another pair of chromosomes carried 5S rDNA loci in telomeric or subtelomeric regions (Figure 3C). This supports expectations that F. pratensis was one of the progenitor species found in F. apennina. However, FISH studies where four tandem repeats were used as probes revealed that in the years subsequent to speciation, the subgenome of F. pratensis present in F. apennina had undergone substantial chromosome reconstruction (Figure 4).
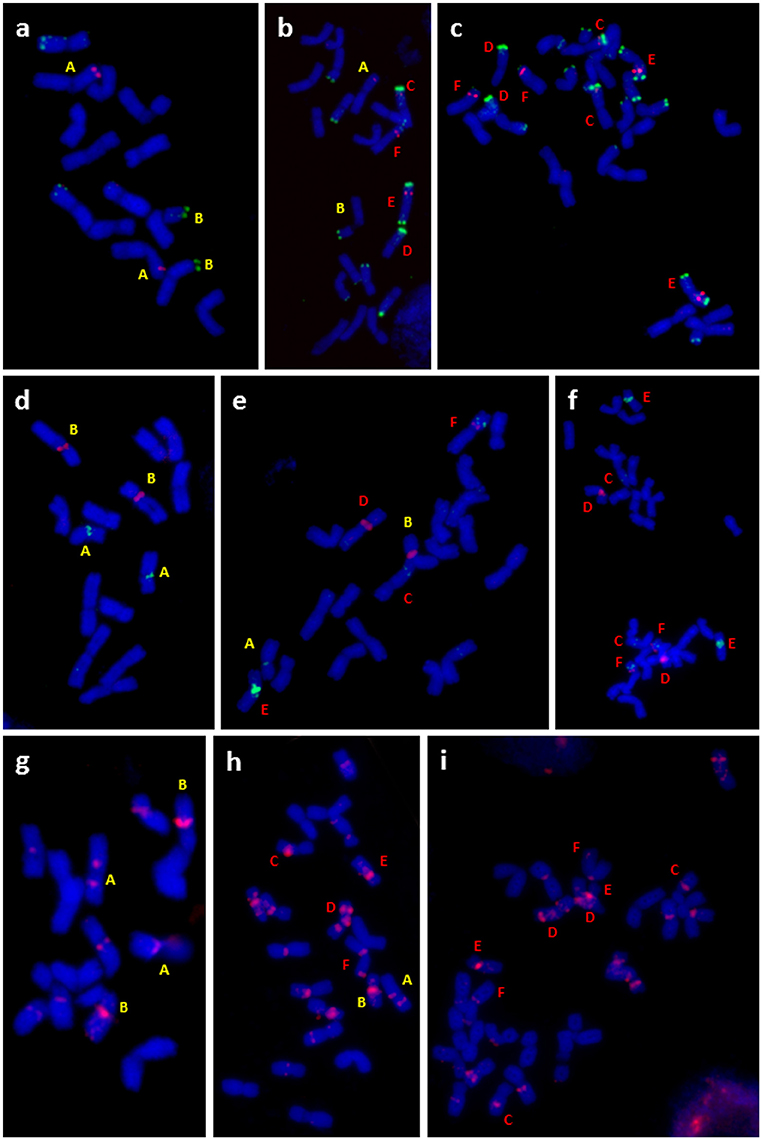
Figure 4. Cytogenetic analysis of diploid F. pratensis (A,D,G), triploid hybrid (B,E,H), and tetraploid F. apennina (C,F,I). FISH was done on metaphase spreads with probes for: (A,B,C) tandem repeats fpTR12 (green color) and fpTR4 (red color), (D,E,F) fpTR15 (green color) and 45S rDNA (red color) and (G,H,I) fpTR7 (red color). Chromosomes with unique fluorescent signals present in diploid F. pratensis, tetraploid F. apennina and their triploid hybrid are marked by letters. Chromosomes were counterstained with DAPI (blue color).
A similar approach was applied to the triploid and pentaploid cytotypes. In triploid plants (2n = 3x = 21), 14 chromosomes were found to be labeled by a gDNA probe of F. pratensis and the other seven chromosomes by a gDNA probe of F. apennina. When F. pratensis gDNA probe was applied together with blocking DNA from F. glaucescens again 14 chromosomes were labeled (Figure 3F). This was evidence that the triploid cytotype was a consequence of the hybridization of tetraploid F. apennina and diploid F. pratensis. The numbers and position of rDNA loci (Figure 3B) and tandem repeats (Figure 4) detected were consistent with expectations based on their number and positions in F. apennina and F. pratensis and that these were the parental species supporting the GISH results. Given the same GISH and FISH treatments, the synthetic triploid hybrid P217/200 (F. apennina × F. pratensis) produced an identical result to those obtained in the natural triploid hybrids. No evidence for homoeologous recombination events was detected, which implies either that the triploids were sterile F1 hybrids, or that they were de novo and had not yet passed through a flowering stage.
The identification of a pentaploid cytotype involving F. apennina was a new discovery. It may have arisen from an unreduced gamete of the triploid cytotype that hybridized with a normally reduced gamete of tetraploid F. apennina. Alternatively, the pentaploid may have arisen following interspecific hybridization of tetraploid F. apennina and a closely related hexaploid fescue, probably F. arundinacea subsp. arundinacea. This species was found growing nearby and could have inter-pollinated with tetraploid F. apennina. The latter scenario was tested. F. arundinacea subsp. arundinacea Shreb. is an allohexaploid species developed from hybridization of F. pratensis and tetraploid F. glaucescens (Humphreys et al., 1995). Consequently, labeled gDNAs of F. glaucescens and F. pratensis were selected as probes for GISH and applied to denatured chromosomes of the pentaploid plants. The results supported expectation that the pentaploid genotypes had arisen following the hybridization of F. apennina by F. arundinacea subsp. arundinacea. The F. pratensis probe labeled 14 chromosomes expected to have derived from one genome of F. pratensis chromosome complement from F. arundinacea and another from F. apennina. The F. glaucescens gDNA probe hybridized to 14 chromosomes, expected to have derived from F. arundinacea subsp. arundinacea. The remaining seven chromosomes in the pentaploid hybrid were weakly hybridized to the F. glaucescens gDNA probe (Figure 4). Through further detailed GISH analysis of the pentaploid hybrid its origin was confirmed (Figure 5). In this case two probe combinations were used: (1) gDNA of F. pratensis in combination with 45S rDNA with sheared DNA from F. glaucescens used as blocking gDNA; (2) gDNA of F. glaucescens with sheared F. pratensis used as blocking DNA. Chromosomes were counterstained with DAPI. The F. pratensis gDNA probe hybridized onto 14 chromosomes (representing one F. pratensis genome from its putative F. apennina parent and one from F. arundinacea). The remaining 21 chromosomes were unlabeled. The gDNA probe of F. glaucescens hybridized onto 21 chromosomes. These represented the two ancestral genomes of F. glaucescens found in F. arundinacea together with the single F. glaucescens genome in F. apennina. The signal intensity of the F. glaucescens probe over the pentaploid hybrid chromosomes varied with 14 chromosomes having a stronger signal compared to the other seven chromosomes. It is proposed that the stronger hybridization was onto the two chromosome sets derived from F. arundinacea whilst the inferior signal derived from the chromosomes of the so far unknown subgenome of F. apennina. It is likely that this subgenome has a common origin to that of one of the subgenomes of F. glaucescens. As current F. glaucescens shares greater genome homology to the two F. glaucescens subgenomes of F. arundinacea than to the F. glaucescens genome present in F. apennina it can be speculated that the speciation events that gave rise to the allotetraploid F. apennina may have occurred at an earlier date than those that led to the hexaploid F. arundinacea. In that event, it can be further speculated, the speciation of F. apennina may represent a key intervening stage during the evolution of F. arundinacea.
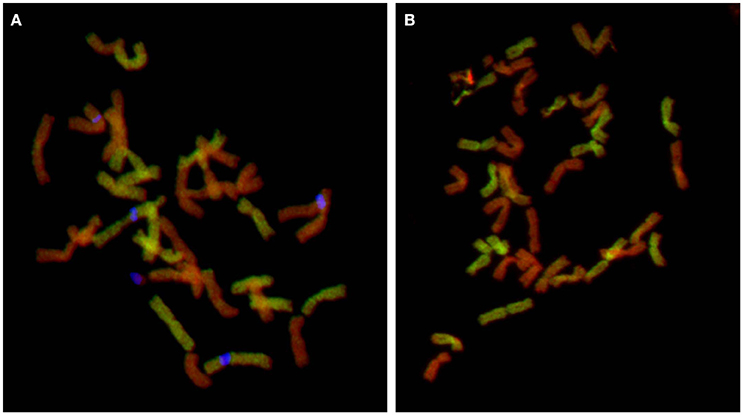
Figure 5. Cytogenetic analysis of pentaploid hybrid. GISH with two probe combinations: (A) gDNA of F. pratensis (green color) and 45S rDNA (blue pseudocolor) and blocking DNA from sheared gDNA of F. glaucescens and (B) gDNA of F. glaucescens (green color) and blocking DNA from sheared gDNA of F. pratensis. Chromosomes were counterstained with DAPI (red pseudocolor). In the first combination, F. pratensis probe produced signal over 14 chromosomes (representing two sets of F. pratensis chromosomes: one from F. apennina and other from F. arundinacea) and other 21 chromosomes remained unlabeled (one set from the second subgenome of F. apennina and two sets representing subgenomes of F. glaucescens present in F. arundinacea). The second probe combination supports the previous findings: 14 chromosomes were unlabeled and represent two sets of F. pratensis chromosomes (one coming from F. arundinacea and the second from F. apennina) and 21 chromosomes were labeled with F. glaucescens probe. However, there was a variation in the signal intensity—signal over 14 chromosomes was much stronger (representing two chromosome sets of F. glaucescens coming from F. arundinacea) than other seven poorly labeled chromosomes, which are probably from the so far unknown subgenome of F. apennina. This subgenome is probably related to F. glaucescens, but has diversified during its evolution.
The 5S rDNA loci found in F. arundinacea subsp. arundinacea accession Bf 950 (Figure 3E) differed in their position as compared to those reported previously in another genotype of F. arundinacea subsp. arundinacea (Thomas et al., 1997). In accession Bf 950, all three pairs of 5S rDNA loci were located proximally. This contrasted with the previous report where one pair of 5S rDNA loci was located in telomeric region of long arm of one chromosome pair. The number and distribution of 45S rDNA loci found in accession Bf 950 agreed with previous reports (Thomas et al., 1997). As in the case of the triploids, the distribution and number of rDNA sites in the pentaploid plants was completely consistent with expectations based on their presumed parentage (F. apennina and F. arundinacea subsp. arundinacea) (Figure 3D) drawn from the GISH analyses. All FISH results are summarized in Table 2 and in Figure 6.
GISH analysis of a synthetic tetraploid hybrid L. multiflorum × F. apennina (2n = 4x = 28) supported the conclusions drawn from cytogenetic studies involving the natural F. apennina cytotypes. GISH analysis using gDNA probes of F. pratensis and of F. apennina identified seven chromosomes when labeled by the F. pratensis probe (one of the two genomes of F. apennina) and 14 chromosomes when labeled by the F. apennina probe (both genomes of F. apennina). Fourteen chromosomes remained unlabeled and were the chromosome complement derived from L. multiflorum (Figure 3H). Similarly when gDNA probes of F. pratensis and of F. glaucescens were applied in conjunction to chromosomes of the synthetic L. multiflorum × F. apennina tetraploid hybrid, the F. pratensis probe labeled seven chromosomes, and the F. glaucescens gDNA probe labeled further seven chromosomes (the two species' genomes in combination believed to represent the complete F. apennina haplotype). Fourteen chromosomes representing L. multiflorum genome remained unlabeled (Figure 3I). The GISH study of the synthetic Festulolium hybrid revealed the close relationship between a subgenome of tetraploid F. glaucescens and of F. apennina and it is hypothesized here that they share a common progenitor.
The development of the cytogenetic techniques GISH and FISH for applications with the grasses of the Lolium-Festuca complex has enabled the effective discrimination between genomes of several species (Thomas et al., 1994; reviewed in Kopecký et al., 2008b) and allowed determination of the progenitors of certain polyploid Festuca species. A good example was the determination of the ancestry of F. arundinacea Schreb. (Humphreys et al., 1995). Using FISH with rDNA probes, Ezquerro-Lopez et al. (personal communication) identified F. mairei and F. glaucescens to be potential progenitors of octoploid F. arundinacea subsp. atlantigena (St.-Yves) Auquier and decaploid F. arundinacea var. letourneuxiana (St.-Yves) Torrec. & Catalán. In a similar approach, the combined GISH and FISH analysis reported here reveals for the first time the genomic constitution of F. apennina.
In addition to the hybridization that has occurred naturally between species within Festuca subg. Schedonorus, there are equivalent occurrences between these Festuca species and representatives of genus Lolium. An example is Festulolium holmbergii (Dőrfl.) P. Fourn., a tetraploid grass hybrid formed between Lolium perenne and F. arundinacea subsp. arundinacea. Similarly, Festulolium braunii (K.Richt.) A.Camus (L. multiflorum × F. pratensis) and the previously mentioned Festulolium loliaceum (Huds.) P.Fourn. (L. perenne × F. pratensis). All these Festulolium hybrids are well known and as synthetic combinations have been bred for agricultural production with the aim to combine the attributes of their parent species (Ghesquiere et al., 2010).
How Can Plant Breeders Benefit from Improved Understanding of the Genomic Constitution of F. apennina and Its Hybrids?
It has long been an aim for grass breeders to combine the complementary characters found in Lolium (high growth rates and forage quality) with those found in Festuca (stress tolerance, deeper rooting) species (Ghesquiere et al., 2010) to ensure that grass varieties are both productive and resilient to climate-change. The additional novel ecosystem service properties found recently in certain Festulolium hybrids, such as for flood mitigation (Macleod et al., 2013), or for improved ruminant nutrition with potential for lowering greenhouse gas emissions by livestock (Humphreys et al., 2014) only serve to reinforce the view. Genetically stable, productive, and resilient high quality Festulolium varieties are necessary to ensure we have in place future safeguards in grassland agriculture sufficient to withstand extreme weather events and to achieve consistent and sustainable fodder for use by livestock to help feed an increasing global population. Introgressive forms of Festulolium, where limited numbers of targeted genes were transferred from a wild-relative into the genomes of existing high quality varieties have been successful. They have proven to be genetically stable and have led to advances in the breeding of drought tolerant or disease resistant cultivars (Humphreys et al., 2006).
However, evolution and speciation within the broad-leaved fescues of the Festuca subgenus Schedonorus has demonstrated consistently that hybridization followed by the retention of intact species' genomes in combination is an essential prerequisite for ensuring consistent adaptations to stresses encountered regularly by grasses that grow in extreme climates. It is clearly of advantage that once a hybrid form and desirable genome combination has been achieved, that it is stabilized and perpetuated into subsequent generations. Incidents of heterosis abound within the species of the Lolium-Festuca complex capable of providing adaptive traits that otherwise are unavailable to their parental genotypes. The hexaploid species F. arundinacea is a good example (Humphreys et al., 1995). The hexaploid fescue, as shown here, shares two common genomes with tetraploid F. apeninna. Their progenitors are the northern European and cold-tolerant species F. pratensis and an ancestral southern European or Mediterranean based drought and heat-tolerant species also found in F. glaucescens. These species' genomes of F. apennina in combination provide the appropriate stress-tolerance adaptations necessary for grasses to grow and perpetuate at high altitude in Alpine meadows. The species F. glaucescens in Mediterranean regions avoids extreme temperatures and desiccation by undergoing complete growth quiescence (Humphreys et al., 1997) during the summer months. F. apennina undergoes an equivalent growth cessation and induced senescence stage that offers it protection against freezing temperatures and desiccation throughout the winter months (Tyler and Chorlton, 1974). From a plant breeding perspective, any prolonged cessation in foliar growth is far from ideal from the perspective of satisfying requirements for achieving sustainable agricultural production and sufficient fodder provision for livestock. However, the incorporation of genes for drought and heat tolerance from F. glaucescens into Lolium has been achieved without recourse to summer growth cessation (Humphreys et al., 2005) showing that drought avoidance alone is not the only strategy available to F. glaucescens to combat drought stresses. As it shares common ancestral genomes, the findings by Humphreys et al. (2005) provide hope that equivalent physiological mechanisms for stress-tolerance may be available to F. apennina that may provide benefits to Lolium spp. in certain synthetic hybrid combinations.
The evolution of the polyploid series of Festuca species within the subgenus Schedonorus provides us with an insight as to how optimal Lolium and Festuca species' genome combinations may be retained over generations. An essential prerequisite appears to be the incorporation of an intact and functional diploidising mechanism present in polyploid Festuca spp. within the genomes of a synthetic amphiploid Festulolium hybrid. As far as is known diploid species within the Lolium/Festuca complex lack such a functional system (Kopecký et al., 2009). As F. pratensis provides a subgenome for F. apennina, its chromosome regulator must reside on the other species' subgenome, one that may well be common in all polyploid species within the subgenus Schedonorus. From a practical Festulolium plant breeding perspective, this provides F. apennina with an advantage over all other Festuca polyploids. The location of the chromosome pairing regulator in these other polyploid fescues is unknown as it may reside anywhere in two or more subgenomes making its selection in breeding programmes in a functional homozygous form very problematic. The substitution of the F. pratensis genome of F. apennina by one from a Lolium spp. whilst also maintaining intact the integrity of the second F. apennina subgenome should provide opportunities to select genotypes that contain a functional chromosome pairing regulator to thereby restrict chromosome pairing to intraspecific homologous associations. Using conventional plant breeding technologies and interpollination of Lolium spp. × F. apennina amphiploid (2n = 4x = 28) hybrids and incorporating selections over generations for disomic inheritance, aided hopefully by future inclusion of relevant gene marker technologies, the attributes of both Lolium and Festuca may be combined in a stable and consistent form with the full potential for assemblies of complementary Lolium and Festuca traits within a single Festulolium genome be achieved.
Future genomic investigations may reveal candidate gene(s) for the chromosome pairing regulator known to occur in F. apennina and other related polyploid fescues. Its successful transfer and operation in a homozygous form in Lolium × Festuca hybrids would mark a significant progress into achieving climate-smart grasses and for procedures to safeguard future grassland agriculture by the creation of Festulolium cultivars with fescue-derived adaptations to abiotic stresses.
Author Contributions
DK, JH, JD, and MH designed the project and experiments; JH, DG, and MH established the initial seed collection; DK, BB, and NA realized sampling expeditions; NA checked taxonomic and nomenclatural aspects; JB, JV, and DŠ made cytometric analyses; JH performed chromosome counting; JH and DG made GISH experiments and FISH with rDNAs; DK, EH, and DŠ performed FISH with tandem repeats. DK, JH, and MH drafted the manuscript. All authors revised the text and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer DP and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
DK, JB, JV, EH, DŠ, and JD has been partially supported by the grant LO1204 from the National Program of Sustainability I. MH, JH, and DG are funded by the Biotechnology and Biological Sciences Research Council. MH is also part funded by the BBSRC-LINK Programme SUREROOT which in addition to BBSRC is supported by partners from the UK's grass seed, grassland, and livestock industries. MH is part of the Climate-Smart Grass Consortium. Funding to support this work was provided by the Welsh Government and HEFCW through the Sêr Cymru National Research Network for Low Carbon, Energy, and Environment.
References
Ahloowalia, B. S. (1965). A root tip squash technique for screening chromosome number in Lolium. Euphytica 14, 170–172. doi: 10.1007/BF00038983
Anamthawat-Jónsson, K., and Reader, S. M. (1995). Pre-annealing of total genomic DNA probes for simultaneous genomic in situ hybridization. Genome 38, 814–816. doi: 10.1139/g95-104
Ardenghi, N. M. G., and Foggi, B. (2015). Lectotypification and combination of Festuca apennina (Poaceae). Taxon 64, 1038–1041. doi: 10.12705/645.14
Borrill, M., Tyler, B. F., and Morgan, W. G. (1976). Studies in Festuca 7. Chromosome atlas (Part 2), an appraisal of chromosome race distribution and ecology, including F. pratensis var. apennina (De Not.) Hack– tetraploid. Cytologia 41, 219–236. doi: 10.1508/cytologia.41.219
Canter, P. H., Pašakinskiene, I., Jones, R. N., and Humphreys, M. W. (1999). Chromosome substitution and recombination in the amphiploid Lolium perenne x Festuca pratensis cv. Prior (2n = 4x = 28). Theor. Appl. Genet. 98, 809–814. doi: 10.1007/s001220050087
Clarke, J., Chandrasekharan, P., and Thomas, H. (1976). Studies in Festuca 9. Cytological studies of Festuca pratensis var. apennina (De Not) Hack. (2n = 28). Z. Pflanzenzüchtg. 77, 205–214.
Devesa, J. A., Catalán, P., Müller, J., Cebolla, C., and Ortúñez, E. (2013). Checklist of Festuca L. (Poaceae) in the Iberian Peninsula. Lagascalia 33, 183–274.
Doležel, J., Greilhuber, J., Lucretti, S., Meister, A., Lysák, M., Nardi, L., et al. (1998). Plant genome size estimation by flow cytometry: interlaboratory comparison. Ann. Bot. 82, 17–26. doi: 10.1006/anbo.1998.0730
Doležel, J., Greilhuber, J., and Suda, J. (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protocols 2, 2233–2244. doi: 10.1038/nprot.2007.310
Foggi, B., and Müller, J. (2009). (last updated Apr 2015). “Schedonorus,” in Poaceae. Euro+Med Plantbase - The Information Resource for Euro-Mediterranean Plant Diversity, eds B. Valdés and H. Scholz, Available online at: http://ww2.bgbm.org/EuroPlusMed/ (Accessed on May 28, 2016).
Ghesquiere, M., Humphreys, M. W., and Zwierzykowski, Z. (2010). “Festulolium,” in Fodder Crops and Amenity Grasses, Book Series: Handbook of Plant Breeding, Vol. 5, eds B. Boller, U. K. Posselt, and F. Veronesi (New York, NY; Dordrecht; Heidelberg; London: Springer Science+Business Media), 293–316.
Harper, J. A., Thomas, A., Armstead, I. P., James, C., Gasior, D., Bisaga, M., et al. (2011). Alien introgression in the grasses Lolium perenne (perennial ryegrass) and Festuca pratensis (meadow fescue) - the development of seven monosomic substitution lines and their molecular and cytological characterisation. Ann. Bot. 107, 1313–1321. doi: 10.1093/aob/mcr083
Harper, J. A., Thomas, I. D., Lovatt, J. A., and Thomas, H. M. (2004). Physical mapping of rDNA sites in possible diploid progenitors of polyploid Festuca species. Plant Syst. Evol. 245, 163–168. doi: 10.1007/s00606-003-0110-2
Humphreys, J., Harper, J. A., Armstead, I. P., and Humphreys, M. W. (2005). Introgression-mapping of genes for drought resistance transferred from Festuca arundinacea var. glaucescens into Lolium multiflorum. Theor. Appl. Genet. 110, 579–587. doi: 10.1007/s00122-004-1879-2
Humphreys, M. W., and Harper, J. A. (2008). Festulolium loliaceum, an understudied natural UK grass hybrid species that may provide benefits to UK grasslands withstanding the onsets of climate change. Crop Wild Rel. 6, 7–9.
Humphreys, M. W., O'Donovan, S. A., Farrell, M. S., Gay, A. P., and Kingston-Smith, A. H. (2014). The potential of novel Festulolium (2n = 4x = 28) hybrids as productive, nutrient-use-efficient fodder for ruminants. Food Ener. Security 3, 98–110. doi: 10.1002/fes3.50
Humphreys, M., Thomas, H. M., Harper, J., Morgan, G., James, A., Zare, A. G., et al. (1997). Dissecting drought- and cold-tolerance traits in the Lolium-Festuca complex by introgression mapping. New Phytol. 137, 55–60. doi: 10.1046/j.1469-8137.1997.00832.x
Humphreys, M. W., Thomas, H. M., Morgan, W. G., Meredith, M. R., Harper, J. A., Thomas, H., et al. (1995). Discriminating the ancestral progenitors of hexaploid Festuca arundinacea using genomic in situ hybridization. Heredity 75, 171–174. doi: 10.1038/hdy.1995.120
Humphreys, M. W., Yadav, R. S., Cairns, A. J., Turner, L. B., Humphreys, J., and Skøt, L. (2006). A changing climate for grassland research. New Phytol. 169, 9–26. doi: 10.1111/j.1469-8137.2005.01549.x
Jauhar, P. P. (1975). Genetic control of diploid-like meiosis in hexaploid tall fescue. Nature 254, 595–597. doi: 10.1038/254595a0
Jauhar, P. P. (1993). Cytogenetics of the Festuca-Lolium Complex. Relevance to Breeding. Monographs on Theoretical and Applied Genetics No. 18. Berlin: Springer-Verlag.
Kopecký, D., Bartoš, J., Zwierzykowski, Z., and Doležel, J. (2009). Chromosome pairing of individual genomes in tall fescue (Festuca arundinacea Schreb.), its progenitors, and hybrids with Italian ryegrass (Lolium multiflorum Lam.). Cytogenet. Genome Res. 124, 170–178. doi: 10.1159/000207525
Kopecký, D., Havránková, M., Loureiro, J., Castro, S., Lukaszewski, A. J., Bartoš, J., et al. (2010). Physical distribution of homoeologous recombination in individual chromosomes of Festuca pratensis in Lolium multiflorum. Cytogenet. Genome Res. 129, 162–172. doi: 10.1159/000313379
Kopecký, D., Loureiro, J., Zwierzykowski, Z., Ghesquière, M., and Doležel, J. (2006). Genome constitution and evolution in Lolium x Festuca hybrid cultivars (Festulolium). Theor. Appl. Genet. 113, 731–742. doi: 10.1007/s00122-006-0341-z
Kopecký, D., Lukaszewski, A. J., and Doležel, J. (2008a). Meiotic behavior of individual chromosomes of Festuca pratensis in tetraploid Lolium multiflorum. Chromosome Res. 16, 987–998. doi: 10.1007/s10577-008-1256-0
Kopecký, D., Lukaszewski, A. J., and Doležel, J. (2008b). Cytogenetics of Festulolium (Festuca × Lolium hybrids). Cytogenet. Genome Res. 120, 370–383. doi: 10.1159/000121086
Kopecký, D., Martis, M., Cíhalíková, J., Hřibová, E., Vrána, J., Bartoš, J., et al. (2013). Flow sorting and sequencing meadow fescue chromosome 4F. Plant Physiol. 163, 1323–1337. doi: 10.1104/pp.113.224105
Lewis, E. J. (1977). Studies in Festuca, I. V. A phyletic study of Festuca pratensis var. apennina (De Not.) Hack., hybridization with synthetic tetraploid F. pratensis Huds. Genetica 47, 59–64. doi: 10.1007/BF00122440
Lewis, E. J., Humphreys, M. W., and Caton, M. P. (1980). Disomic inheritance in Festuca arundinacea Schreb. Z. Pflanzenzüchtg. 84, 335–341.
Macleod, C. J. A., Humphreys, M. W., Whalley, W. R., Turner, L., Binley, A., Watts, C. W., et al. (2013). A novel grass hybrid to reduce flood generation in temperate regions. Sci. Rep. 3:1683. doi: 10.1038/srep01683
Morgan, W. G., Thomas, H., and Lewis, E. J. (1988). Cytogenetic studies of hybrids between Festuca gigantea Vill. and Lolium multiflorum Lam. Plant Breed. 101, 335–343. doi: 10.1111/j.1439-0523.1988.tb00306.x
Stebler, F. G. (1904). Jahresbericht der Schweizerischen Samenuntersuchungs- und Kontrollstation Zürich. Schweiz. Landw. Jahrbuch 18, 43.
Thomas, H. M., Harper, J. A., Meredith, M. R., Morgan, W. G., and King, I. P. (1997). Physical mapping of ribosomal DNA sites in Festuca arundinacea and related species by in situ hybridization. Genome 40, 406–410. doi: 10.1139/g97-054
Thomas, H. M., Harper, J. A., Meredith, M. R., Morgan, W. G., Thomas, I. D., Timms, E., et al. (1996). Comparison of ribosomal DNA sites in Lolium species by fluorescence in situ hybridization. Chromosome Res. 4, 486–490. doi: 10.1007/BF02261775
Thomas, H. M., Morgan, W. G., Meredith, M. R., Humphreys, M. W., and Leggett, J. M. (1994). Identification of parental and recombined chromosomes in hybrid derivatives of Lolium multiflorum x Festuca pratensis by genomic in situ hybridization. Theor. Appl. Genet. 88, 909–913. doi: 10.1007/BF00220795
Tyler, B. F. (1988). “Description and distribution of natural variation in forage grasses,” in Proceedings of the Eucarpia Fodder Crops Section Meeting (Lusignan), 13–22.
Tyler, B. F., Borrill, H., and Chorlton, K. H. (1978). Studies in Festuca pratensis and tetraploid F. pratensis var. apennina in relation to their altitudinal distribution. J. Appl. Ecol. 15, 219–226. doi: 10.2307/2402932
Tyler, B. F., and Chorlton, K. H. (1974). Growth rhythm studies on Festuca collected in Western Europe in 1971. Rep. Welsh Pl. Breed. 1973, 21–22.
St. Yves, A. (1913). Les Festuca de la section Eu-Festuca et leurs variations dans les Alpes Maritimes. Annuaire Conservatoire Jardin Botaniques Geneve 42, 1–218.
Keywords: amphiploidy, Festuca pratensis subsp. apennina (De Not.) Hegi, chromosome pairing, diploidization, Festulolium breeding
Citation: Kopecký D, Harper J, Bartoš J, Gasior D, Vrána J, Hřibová E, Boller B, Ardenghi NMG, Šimoníková D, Doležel J and Humphreys MW (2016) An Increasing Need for Productive and Stress Resilient Festulolium Amphiploids: What Can Be Learnt from the Stable Genomic Composition of Festuca pratensis subsp. apennina (De Not.) Hegi? Front. Environ. Sci. 4:66. doi: 10.3389/fenvs.2016.00066
Received: 01 June 2016; Accepted: 26 September 2016;
Published: 14 October 2016.
Edited by:
Mauro Centritto, National Research Council, ItalyReviewed by:
Luigi Frusciante, University of Naples Federico II, ItalyDomenico Pignone, National Research Council, Italy
Copyright © 2016 Kopecký, Harper, Bartoš, Gasior, Vrána, Hřibová, Boller, Ardenghi, Šimoníková, Doležel and Humphreys. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mike W. Humphreys, mkh@aber.ac.uk
 David Kopecký
David Kopecký John Harper
John Harper Jan Bartoš1
Jan Bartoš1  Beat Boller
Beat Boller Jaroslav Doležel
Jaroslav Doležel Mike W. Humphreys
Mike W. Humphreys