Co-expression of six tightly clustered odorant receptor genes in the antenna of the malaria mosquito Anopheles gambiae
- Institute of Physiology (230), University of Hohenheim, Stuttgart, Germany
The behavior of female malaria mosquitoes, Anopheles gambiae, especially seeking out blood hosts or selecting oviposition sites, highly depends on the detection of relevant odorants by their sense of smell. This is mediated by olfactory sensory neurons (OSNs) which express distinct odorant receptor (OR) types. In the genome of A. gambiae 76 genes have been annotated to encode putative odorant receptors and the majority of these AgOR genes are arranged in clusters. To assess whether clustered AgOR genes are expressed in a characteristic manner we explored the topographic expression pattern of six tightly adjoined AgOR genes in the female antenna. Whole mount fluorescence in situ hybridization experiments were performed to visualize the olfactory neurons which express a distinct AgOR type in order to determine the number and the distribution of the cells. We found that within the 13 antennal segments about 75 cells contain mRNA for the four receptor types AgOR13, AgOR15, AgOR17, and AgOR55. Moreover, about half of these cells also transcribe mRNA for the subtypes AgOR16 and AgOR47. Subsequent RT-PCR experiments with primer pairs spanning the coding regions of adjacent AgOR genes revealed the existence of polycistronic mRNA. This result indicates that individual genes were not transcribed but mRNA was comprised of coding sequence from several genes within the studied cluster. Taken together, the data indicate a unique principle for the expression of odorant receptor genes arranged in a large cluster and suggest that the corresponding olfactory neurons are endowed with a distinct set of odorant receptor types.
Introduction
Blood-sucking insects are dreaded for their capacity to transfer life-threatening diseases. The anthropophilic mosquito, Anopheles gambiae, is the main vector for the perilous malaria parasite, Plasmodium falciparum, affecting millions of people worldwide every year (WHO, 2013). Female malaria mosquitoes find human hosts, oviposition sites and nectar sources mainly by chemical cues recognized by their olfactory system (Takken and Knols, 1999; Zwiebel and Takken, 2004). This is accomplished by olfactory sensory neurons (OSNs) in hair-like sensilla on their antennae, as well as on their maxillary palps and the labellum (McIver, 1982; Kwon et al., 2006; Pitts and Zwiebel, 2006). The pivotal role of the antennae is underpinned by the number of OSNs; female A. gambiae possess about 1500 OSNs per antenna, but only ~200 OSNs per maxillary palp and ~50 OSN per labellar lobe (Kwon et al., 2006; Qiu et al., 2006; Lu et al., 2007).
The responsiveness of OSNs is determined by the odorant receptors (ORs) in their dendritic membrane (Hallem et al., 2004; Jacquin-Joly and Merlin, 2004; Touhara and Vosshall, 2009; Hansson and Stensmyr, 2011; Guidobaldi et al., 2014). In addition, ionotropic receptors (IRs) and gustatory receptor types (GRs) mediate responses of OSNs to odorants and CO2, respectively (Kwon et al., 2007; Benton et al., 2009; Rytz et al., 2013; Tauxe et al., 2013). In the genome of A. gambiae 76 genes have been identified which encode putative odorant receptors (AgORs) (Hill et al., 2002; Pitts et al., 2011), and for most of these genes, expression has been verified in olfactory appendices of adult mosquitoes or larvae (Fox et al., 2002; Iatrou and Biessmann, 2008; Pitts et al., 2011; Rinker et al., 2013). Functional expression of 50 AgOR types in Xenopus oocytes (Wang et al., 2010) or in the Drosophila empty neuron system (Carey et al., 2010) revealed characteristic ligand spectra for about 30 of the AgOR types.
Within the genome of A. gambiae the majority of AgOR genes are arranged in clusters which consist of two to 9 genes (http://metazoa.ensembl.org/Anopheles_gambiae) (Fox et al., 2002; Hill et al., 2002). Such a clustered genomic organization of genes that encode chemosensory receptors appears to be characteristic for insects; for example, it has also been reported for the mosquito Aedes aegypti (Bohbot et al., 2007), the beetle Tribolium castaneum (Engsontia et al., 2008) and the honey bee Apis mellifera (Robertson and Wanner, 2006). Typically such clusters comprise two or three OR genes, but much larger tandem arrays with up to 60 OR genes occur in the honey bee (Robertson and Wanner, 2006). It is largely unknown to what extent OR genes organized in a cluster underlie common control mechanisms which could result in a similar spatial expression pattern or a co-expression in the same cell. A co-expression of clustered OR genes in individual cells of Drosophila melanogaster is demonstrated by the presence of mRNAs for two OR-types within the same cells (Dobritsa et al., 2003; Couto et al., 2005; Ray et al., 2007). For A. gambiae, we have recently found that transcripts for the receptor types AgOR13 and AgOR55 co-exist in the same cells (Schultze et al., 2014). Interestingly, the genes encoding AgOR13 and AgOR55 are immediate neighbors within the largest AgOR gene cluster (hereafter named cluster1) in the genome of A. gambiae; it is comprised of a total of 9 AgOR genes (Figure 1). Within cluster1, the coding regions of OR genes are in the same orientation and are spaced by short stretches of DNA. The first three AgOR genes (AgOR53, AgOR30 and AgOR46) are more separated from the six more downstream located receptor genes (AgOR47, AgOR16, AgOR17, AgOR13, AgOR55, and AgOR15); accordingly, they were designated as cluster1A and cluster1B, respectively (Figure 1A). In transcriptome analysis of female antennae rather low transcript levels were found for the three genes in cluster1A, whereas the expression levels for the six genes in cluster1B were generally quite high (Pitts et al., 2011; Rinker et al., 2013). Based on the similar levels of mRNA for the strikingly tightly clustered OR-genes in the genome, we hypothesized that the genes in cluster1B may underlie a common transcription control. Consequently, combinations of several or even all six AgOR types may be co-expressed in individual cells. Moreover, they may be translated from a large, polycistronic transcript, as reported for a large cluster of genes encoding gustatory receptors in the fruitfly (Slone et al., 2007). Here, we have used whole mount fluorescence in situ hybridization (WM-FISH) to explore the expression patterns of the six genomically adjoined AgOR types in the antenna of female A. gambiae and applied reverse transcription PCR (RT-PCR) to assess the presence of polycistronic mRNA encoding the receptor types.
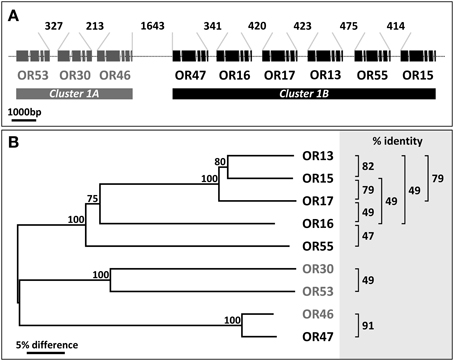
Figure 1. Arrangement of cluster1 genes and relationship between AgORs. (A) Schematic representation of the chromosomal linkage and the intron/exon structure of AgOR genes in cluster1. All genes have the same orientation. The position and relative size of exons and introns were adopted from http://metazoa.ensembl.org/Anopheles_gambiae and drawn to scale. Exons represent protein coding regions only. The length of the intervening DNA strings connecting the exon regions of two adjacent AgORs (in base pairs, bp) is indicated. (B) Neighbor joining tree of cluster1 AgORs. The tree was constructed using MEGA5 based on a Clustal alignment of the AgOR amino acid sequences. Bootstrap support is based on 1000 replicates; support values >75% for branches are indicated. Sequence identities between pairs of AgORs which exceed 45% are given at the right.
Materials and Methods
Animal Rearing
Animals were reared to adults at 28°C with a day-night cycle of 12:12. Eggs and larvae of the Anopheles gambiae (Giles) s.s. strain RSP-H were kindly provided by Bayer CropScience, Monheim, Germany. The laboratory strain was originally derived from the region of Kisumu, Kenya and has been in rearing at Bayer since 2009 (to date: 46th generation). After emergence, animals had access to 10% sucrose ad libitum. For the experiments, one-to-eight-day old animals were used.
Preparation of in situ Hybridization Probes
Digoxigenin (DIG)-labeled and biotin-labeled antisense RNAs for in situ hybridization were transcribed from linearized recombinant plasmids containing AgOR coding regions using the components of the Roche T3/T7/SP6 RNA transcription/labeling system (Roche, Mannheim, Germany) and recommended protocols. To improve tissue penetration, the labeled antisense RNA probes were subsequently fragmented to an average length of about 200 or 800 nucleotides by incubation in carbonate buffer (80 mM NaHCO3, 120 mM Na2CO3, pH 10.2) following the protocol of Angerer and Angerer (1992).
Transcription vectors (pBluescript SK II) carrying the coding sequences for AgOR55 and AgOR68, respectively, were kindly provided by Prof. Kostas Iatrou (NCSR “Demokritos,” Athens, Greece). For AgOR13, AgOR15, AgOR16, AgOR17, and AgOR47, respectively, coding regions were PCR-amplified from cDNA of female A. gambiae heads and cloned into the pGem-T vector (Promega, Puchheim, Germany) using standard protocols. The identities of the AgOR sequences were verified by sequencing.
Head cDNA was prepared by dissecting heads from cold-anesthetized animals and collecting them in a tube cooled on liquid nitrogen. The tissue was crushed in liquid nitrogen using a small mortar and pestle and homogenized in Trizol reagent (Invitrogen). Total RNA was prepared from the Trizol homogenate and poly (A)+ RNA was isolated from total RNA applying oligo (dT)25 magnetic dynabeads (Dynal, Oslo, Norway) according to the suppliers protocols. Poly (A)+ RNA from heads were transcribed into cDNA using the superscript III reverse transcriptase system (Invitrogen) with synthesis at 50°C for 50 min, followed by incubation for 15 min at 70°C.
Whole Mount Fluorescence in situ Hybridization (WM-FISH)
Several receptor proteins encoded by the genes in cluster1 share relatively high sequence identity, higher than 45% (Figure 1B); however, with the other 67 AgOR-types of A. gambiae, sequence identity (10–40%) is rather low (not shown). Within the cluster, the identity between AgOR pairs is generally below 90%, a value that is considered critical with respect to a possible cross hybridization of probes for different AgORs. The exception is the receptor pair AgOR46 and AgOR47, which are 91% identical; however, since the transcript level of AgOR46 in female antennae is very low (Pitts et al., 2011), hybridization signals obtained with the AgOR47 probe most likely represent AgOR47-expressing cells.
Whole mount fluorescence in situ hybridizations (WM-FISH) with single or combinations of probes were performed as described previously (Schultze et al., 2013, 2014) with a few modifications. The antennae were dissected from the head and transferred directly to fixation solution (4% paraformaldehyde in 0.1 M NaCO3, pH 9.5, 0.03% Triton X-100). After fixation for 20–24 h at 6°C the antennae were washed at room temperature for 1 min in PBS (phosphate-buffered saline = 145 mM NaCl, 1.4 mM KH2PO4, 8 mM Na2HPO4, pH 7.1) containing 0.03% Triton X-100. In a drop of this solution, each antenna was carefully squeezed several times at different positions along the longitudinal axis using fine forceps. This was followed by incubation at room temperature in 0.2 M HCl, 0.03% Triton X-100 for 10 min, two washes for 2 min each in PBS with 0.03% Triton X-100 and a treatment for 10 min with acetylation solution (25% acetic anhydride freshly added to a 0.1 M triethanolamine solution). Subsequently, the antennae were washed three times in PBS with 0.03% Triton X-100 for 3 min and prehybridized in WM-FISH solution (50% formamide, 5xSSC, 1xDenhardt's reagent, 50 μg/ml yeast RNA, 1% Tween 20, 0.1% Chaps, 5 mM EDTA, pH 8.0) at 55°C for 6 h. In WM-FISH experiments with single probes, this step was followed by incubation for at least 48 h at the same temperature in WM-FISH solution containing a DIG-labeled antisense RNA probe. Then the antennae were washed four times for 15 min each in 0.1xSSC, 0.03% Triton X-100 at 60°C. After treatment with 1% blocking reagent (Roche) in TBS (100 mM Tris, 150 mM NaCl, pH 7.5), 0.03% Triton X-100 for 5–6 h at 6°C, DIG-labeled probes were detected by incubation for at least 48 h with an anti-DIG alkaline phosphatase-conjugated antibody (Roche) diluted 1:500 in TBS, 0.03% Triton X-100 with 1% blocking reagent. After five washes for 10 min each in TBS with 0.05% Tween 20 at room temperature, antennae were incubated in the dark for 7–8 h with HNPP (2-hydroxy-3-naphtoic acid-2′-phenylanilide phosphate, Roche) 1:100 in DAP-buffer (100 mM Tris, 100 mM NaCl, 50 mM MgCl2,pH 8.0) at 6°C to visualize hybridization of DIG-labeled probes.
In double WM-FISH experiments, simultaneous hybridization was performed with differentially labeled antisense RNA (DIG- and biotin-labeled, respectively) for two AgOR types. For concurrent detection of DIG- and biotin-labeled probes, the antennae were incubated after the posthybridization washes with the anti-DIG AP-conjugated antibody (Roche, diluted 1:500) and a streptavidin horse radish peroxidase-conjugate (1:100, TSA kit, Perkin Elmer) in TBS, 0.03% Triton X-100, 1% blocking reagent for at least 48 h at 6°C. This was followed by five 10 min washes in TBS, 0.05% Tween 20 at room temperature and incubation with HNPP (1:100 in DAP-buffer) for 7–8 h at 6°C in the dark to visualize the hybridization of DIG-labeled probes. Subsequently, the antennae were washed three times for 5 min in TBS, 0.05% Tween 20, followed by visualization of the biotin-labeled probes using the TSA / FITC development (TSA kit, Perking Elmer) and incubation for 17–18 h at 6°C in the dark. Finally, the antennae were washed three times for 5 min each in TBS with 0.05% Tween 20, briefly rinsed in PBS and mounted in Mowiol solution (10% polyvinylalcohol 4-88, 20% glycerol in PBS).
Analysis of Hybridized Antennae
After the WM-FISH treatment, antennae were analyzed on a Zeiss LSM510 Meta laser scanning microscope (Zeiss, Oberkochen, Germany). Confocal image stacks of the red and green fluorescence channel as well as the transmitted-light channel were taken from single antennal segments. Selected optical planes from image stacks were used to prepare figures, with the fluorescence channels and the transmitted-light channel overlaid or shown separately. Appropriate programs (MS PowerPoint, Adobe Photoshop) were used to arrange the figures and to adjust the brightness or contrast for uniform tone within a single figure.
To examine the distribution and number of AgOR-expressing cells along the 13 antennal segments, the labeled cells in a given antennal segment were counted under fluorescence microscope inspection. Due to breakage of antennae or damage from squeezing, in many cases it was not possible to count the positive cells on all 13 segments of the hybridized antennae. Therefore, the number of different segments (i.e., antennae) that were analyzed to determine the average number of AgOR-expressing cells for distinct segments differ (Table S2).
Reverse Transcription PCR—Detection of Polycistronic mRNA
To investigate whether polycistronic mRNA transcripts encoding several cluster1B AgORs exist, we performed reverse transcription (RT) PCR experiments with cDNA from the head (with appendices) of female A. gambiae and primer pairs spanning the intergenic region of two adjacent AgOR genes. The sense primer was directed against a region located within the second half of the coding sequence of the first AgOR type, whereas the antisense primer matched within the first half of the coding sequence of the AgOR type, which follows downstream in the genome (Figure 4). Primer pairs spanned at least one intron region, thus allowing us to distinguish PCR products obtained from cDNA to that from PCR bands which may have resulted from amplification of genomic DNA. The positions of the primer pairs within the different cluster1B AgORs are indicated in Figure 4; their sequences are listed in Table S1. Standard PCR reactions were performed in a total volume of 50 μl using the High Fidelity PCR Enzyme Mix (Thermo/Fisher Scientific, Schwerte, Germany) and 1 μl of the head cDNA (prepared as described above). The reaction was started at 94°C for 1 min 40 s, followed by 20 cycles with 94°C for 30 s, 55°C for 40 s, and 72°C for 1 min 30 s. In each cycle the annealing temperature was decreased by 0.5°C. Then another 20 cycles at 45°C annealing temperature were conducted, followed by a last elongation step at 72°C for 7 min. PCR products were analyzed on agarose gels and visualized after staining under UV light.
Results
Expression of Cluster1B AgORs in the Antenna of Female A. gambiae
We analyzed the expression of six AgORs genes that belong to the largest AgOR cluster in the genome of A. gambiae. Using labeled riboprobes and the WM-FISH method, we visualized the cells that express the AgOR-types and determined their topographic distribution pattern throughout the female antenna (Figure 2). AgOR16 and AgOR47 are expressed in about the same number of cells within each of the studied antennal segments (Figure 2A). Moreover, for both receptor types, the distribution of the labeled cells along the 13 antennal segments was similar with the highest number of cells on antennal segments 4 and 5 and only in rare cases did labeled cells appear in segment 1 (Figure 2B). Evaluating the distribution of cells positive for the receptor types AgOR13, AgOR15, AgOR17, and AgOR55 revealed a pattern that was similar to that obtained for cells expressing AgOR16 and AgOR47 (Figure 2B). However, the number of labeled cells on each of the 13 antennal segments was twice as high as the number of cells for AgOR16 and AgOR47 (Figures 2A,B, Table S2). This 2:1 ratio was clearly reflected in the total number of labeled cells per antenna. A quantification of all labeled cells on the 13 antennal segments (Table S2) resulted in similar numbers across various receptor types; in detail we revealed 78 AgOR13-, 77 AgOR15-, 70 AgOR17-, and 76 AgOR55-cells per antenna as well as 37 AgOR16- and 36 AgOR47-cells per antenna.
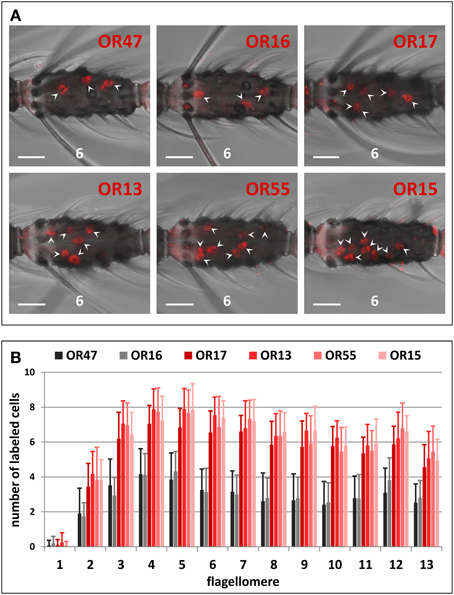
Figure 2. Topography of cluster1B AgOR gene expression in the female antenna. (A) WM-FISH using DIG-labeled antisense RNA probes for the six cluster1B AgORs. Cells bearing AgOR transcripts have been visualized by a detection system leading to red fluorescence. The same (6th) flagellomere of the antenna from different animals is shown. Labeled cells are marked by arrowheads. Pictures have been arranged according to the order of the six AgORs in the genome (Figure 1). Probes for AgOR16 and AgOR47 label about half the number of cells compared to probes for AgOR13, AgOR15, AgOR16, and AgOR55, respectively. Scale bars: 20 μm. (B) Number and distribution of cells expressing cluster1B AgORs on the 13 flagellomeres of the female antenna. Segments are numbered 1–13 from proximal (head) to distal (tip of antenna). The average number of cells (± S.D.) shown for the different antennal segments has been determined by counting the cells from up to 28 antenna; details are given in Table S2.
Co-Expression Pattern of Clustered AgORs
The similarities in the number and distribution of antennal cells which express the AgOR-subtypes encoded by genes arranged in cluster1B led us to suggest that these receptor types may be co-expressed by the same cells, or alternatively, may be expressed in adjacent cells of the same sensillum. In two-color WM-FISH experiments with combinations of receptor-specific probes (Figure 3) the two probes for the AgOR16/AgOR47 labeled the same cells (Figure 3A). This result indicates that AgOR16 and AgOR47 are indeed co-expressed in the same cells and not in adjacent cells within a sensillum. Similar results were obtained in WM-FISH experiments with various combinations of probes for AgOR13, AgOR15, AgOR17, and AgOR55. Results are exemplarily shown for the combination AgOR13/AgOR17 (Figure 3G) and AgOR15/AgOR17 (Figure 3H). Each pair of the four AgOR probes revealed a 100% match of the red- and green-labeled cells (Table 1). These results led us to conclude that all four AgOR types are co-expressed in the same sensory neurons of the female antenna.
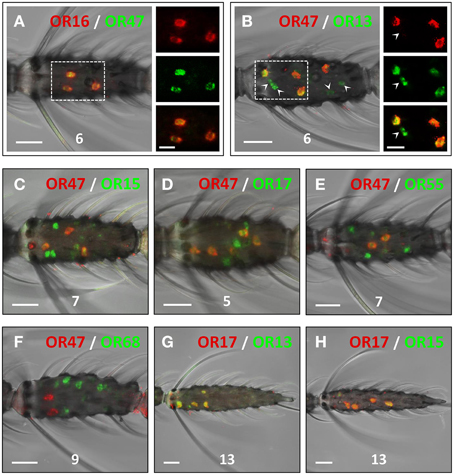
Figure 3. Co-expression of AgORs in antennal cells. Double WM-FISH with female antennae using combinations of differentially labeled AgOR probes and visualization of cells bearing distinct AgOR transcripts by green (biotin) and red (DIG) fluorescence, respectively. Co-labeling of cells by both AgOR probes appears as yellow/orange color in the overlay of the red and green fluorescence channels. Numbers indicate the antennal segment shown. Scale bars: 20 μm. (A,B) WM-FISH using the combinations AgOR47/AgOR16 and AgOR47/AgOR13. The boxed areas are shown at higher magnification (right; scale bars: 10 μm), with the fluorescence channels displayed separately or superimposed. The AgOR47 and AgOR16 probes label the same cells (A). AgOR47 labeled cells are also positive for the receptor types AgOR13 (B), AgOR15 (C), AgOR17 (D) and AgOR55 (E), which in each case have transcripts in additional cells (green-labeled cells in B–E; arrowheads in B). (F) The probe combination AgOR47/AgOR68 (control) clearly labels different populations of cells. (G,H) The probe combinations AgOR17/AgOR13 and AgOR17/AgOR15 label the same cells.
We next examined the relative spatial segregation of cells that co-express AgOR16 and AgOR47 vs. the cells that co-express the other four AgOR genes in cluster1B. In WM-FISH experiments with the AgOR47 probe in pairwise combination with probes for AgOR13, AgOR15, AgOR17, and AgOR55, for each pair a partial overlap of the labeled cells was found (Figures 3B–E, Table 1). This is shown in detail for the combination AgOR47/AgOR13 (Figure 3B); all AgOR47-positive cells (red) were also labeled by a probe for AgOR13 (green). In addition, several green-labeled cells were found (Figure 3B, arrowheads, Figures 3C–E). In control experiments, application of a AgOR47 probe in combination with a biotin-labeled probe for the non-cluster1 receptor AgOR68 resulted in only clearly separated red- or green-labeled cells (Figure 3F), demonstrating that the two AgORs are expressed in different cell populations. Together, the results of the WM-FISH experiments indicate that a rather large number of sensory neurons (75 of about 1500 = 5%) in the female antenna co-transcribe the four cluster1B genes AgOR13, AgOR15, AgOR17, and AgOR55. Moreover, about half of these cells also comprise mRNA of AgOR16 and AgOR47 which is indicative for an expression of all six cluster1B AgORs in the cells.
Evidence for Polycistronic mRNA Encoding Cluster1B AgORs
The identification of transcripts for several AgOR genes of cluster1B in the same cells suggests that their transcription may be controlled by common regulatory mechanisms. A thorough assessment of the complete cluster1B sequence using the Eukaryotic Promotor Databases (EPD and EPDnew) (Dreos et al., 2013) did not lead to an identification of promotor motifs or common sequences in the 5′upstream regions of the six AgOR genes. In addition, the intervening sequences of two adjacent AgOR genes in cluster1B are very short (Figure 1). As a consequence of these observations, we hypothesized that the group of genes may be transcribed as polycistronic mRNA. To scrutinize this idea we performed RT-PCR experiments using five primer pairs each matching the coding regions of two adjacent AgOR genes and spanning the intervening regions OR47-16, OR16-17, OR17-13, OR13-55, and OR55-15 (Figure 4). In each case, the size of the PCR band corresponded well to the size expected if the exon sequences (but no introns) of the adjacent AgOR genes as well as the intervening regions are transcribed. Sequencing of the amplicons obtained with the primers for OR13-55 as well as for OR55-15 verified in both cases the exon sequences of the adjacent AgOR genes with a correct intron splicing. Moreover, the intervening sequences of the adjacent AgOR genes in the genome were found. From this region a stretch of 55 bp (of 475 bp) was missing for OR13-55 and 87 bp (of 414 bp) for the OR55-15 PCR products which most likely indicates a splicing of the precursor mRNA. Together the results demonstrate that individual genes in cluster1B are not transcribed but rather that polycistronic mRNA is generated encoding multiple AgORs.
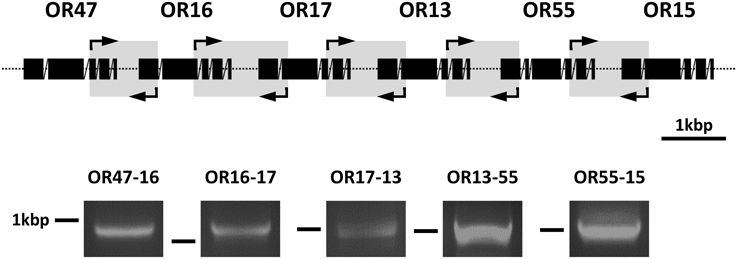
Figure 4. Detection of polycistronic transcripts for cluster1B AgORs. Poly (A)+ RNA prepared from the heads of female A. gambiae were reverse transcribed into cDNA and used in PCR reactions with primer pairs spanning parts of the coding regions of two adjacent AgORs as well as at least one intron and the region connecting the two AgOR genes being investigated. The position of the five sense and antisense primer pairs used are indicated by arrows framing gray boxes in the graphic representation of the exon-intron structure of the AgOR genes in cluster1B. Reverse transcription PCR products were analyzed on agarose gels. The obtained bands are shown below the region of the corresponding primer pair. The position of a 1 kbp marker band is indicated. Based on the primer design and assuming that the DNAs intervening the coding regions are transcribed, the predicted band sizes of the appropriately spliced cDNA products were: 938 bp for OR47-16, 1188 bp for OR16-17, 1030 bp for OR17-13, 992 bp for OR13-55, and 1027 bp for OR55-15.
Discussion
A close chromosomal linkage of odorant receptor genes is a common phenomenon in various insect species (Robertson and Wanner, 2006; Bohbot et al., 2007; Engsontia et al., 2008). However, very little is known about how the closely linked OR genes are transcribed and to what extent they may be co-expressed in the OSNs. The result of the present study indicates that in the malaria mosquito, A. gambiae, the transcription of six tightly clustered AgOR genes leads to polycistronic mRNA and to co-expression of several receptor types in OSNs in the female antenna.
This mode of gene expression is remarkable, since in eukaryotes the transcription of multiple genes as polycistronic mRNA is supposed to be quite rare, except for the nematode Caenorhapditis elegans, where around 15% of the genes are co-transcribed as operons (Blumenthal, 2004; Pi et al., 2009). For insect species, the first indication resulted from genome analysis of Drosophila species where, among the total of about 17,000 genes, around 100 dicistronic genes were predicted (Lin et al., 2007). In addition, polycistronic polypeptide coding RNAs (ppcRNAs) were identified in Drosophila and other insect species (Galindo et al., 2007). With respect to genes encoding chemosensory receptors of D. melanogaster, three cases of two genomically linked OR genes that are transcribed as dicistronic mRNA and co-expressed in OSNs have been reported (Ray et al., 2007). Surprisingly, a dicistronic transcript has been identified that underlies the co-expression of an odorant receptor type, DmOR10a, and a gustatory receptor type, DmGR10a, in the same cells (Fishilevich and Vosshall, 2005; Ray et al., 2007).
Interestingly, a case with similar features to the cluster1B genes of A. gambiae analyzed in this study was reported for six gustatory receptor genes of the fruitfly (Dahanukar et al., 2007; Slone et al., 2007). Similar to the six cluster1B odorant receptor genes, the genes for the sugar receptors DmGR64a – GR64f are tightly arranged and co-expressed in sensory neurons. Moreover, RT-PCR experiments indicated that the coding sequences of adjacent GR genes are on the same mRNA and that all six DmGR genes may be transcribed as a single polycistronic mRNA (Slone et al., 2007). Together, the actual data indicate co-expression of clustered chemosensory receptor genes in sensory neurons based on polycistronic mRNA. These recent findings suggest that co-expression of multiple receptor types from the same mRNAs transcript may be more widespread among insect species than previously thought.
For the co-transcription of the genes in cluster1B, the results of this study suggest two principles; it was found that one population of antennal OSNs contains transcripts for all six AgOR genes and another population has transcripts for four AgOR genes. This observation indicates that in the two cell populations different polycistronic mRNAs are transcribed from cluster1B. Based on the arrangement of genes in cluster1B, it is conceivable that one population of OSNs generates a polycistronic mRNA encoding AgOR17, AgOR13, AgOR55 and AgOR15, while the second population would generate a longer transcript, which also includes AgOR47 and AgOR16. Bioinformatic analyses of the sequences upstream and downstream of each of the cluster1B AgOR genes for promotor motifs and polyadenylation signals (AATAAA, ATTAAA) did not reveal any clue for explaining the transcription of mRNA comprising either four or six coding regions for AgORs, respectively. Thus, further investigation is needed to understand how differential transcription of the receptor genes in cluster1B in the two sensory neuron populations is controlled.
Our results show that a relatively high number of OSNs on a female antenna (about 5%) have transcripts from at least four cluster1B genes and half of these cells have transcripts from six genes. If all transcripts are in fact translated, the two populations of sensory neurons would be endowed with multiple receptor types. The high number of receptor types may render these cells responsive to a broad spectrum of odorants. With respect to the ligand reactivity of these cells, functional analyses of some of the AgOR types encoded by genes in cluster1B in Xenopus oocytes and in the Drosophila empty neuron system have shown that the receptors AgOR13, AgOR15, and AgOR16 respond to distinct but partly overlapping spectra of ligands, which include volatiles found in human emanations (Carey et al., 2010; Wang et al., 2010).
Emanations from humans or oviposition sites consist of complex blends of various chemical compounds (Dormont et al., 2013; Himeidan et al., 2013). Thus, the first step to finding targets relevant to A. gambiae is to register a blend of volatile compounds with no need to discriminate distinct odorous compounds. Therefore, broadly tuned sensory neurons are particularly suitable for sensing a complex blend of components emitted from a relevant odor source. This scenario would be reminiscent of the bitter responsive cells in the taste buds of mice, which express many of the bitter receptor T2R types (Adler et al., 2000; Chandrashekar et al., 2000) making taste cells responsive to a large spectrum of potentially harmful compounds without discriminating among distinct molecules. Therefore, it is possible that antennal sensory neurons with multiple receptors may serve as “non-specific” sensors for odor blends and thus elicit the attention of female A. gambiae toward a relevant odor source and initiate directed behaviors, like host seeking or searching for oviposition sites. In this regard, targeting large gene clusters such as cluster1B of A. gambiae may be an interesting option for novel strategies toward a control of blood sucking mosquitoes and thereby reduce the danger of a transmission of life-threatening diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Günther Nentwig, Jörg Egger, Melanie Nolden and Dr. Maike Hink (Bayer CropScience, Monheim, Germany) for support and providing Anopheles gambiae. Prof. Kostas Iatrou (NCSR ”Demokritos,” Athens, Greece) is acknowledged for providing plasmids containing AgOR cDNAs.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fevo.2015.00026/abstract
References
Adler, E., Hoon, M. A., Mueller, K. L., Chandrashekar, J., Ryba, N. J., and Zuker, C. S. (2000). A novel family of mammalian taste receptors. Cell 100, 693–702. doi: 10.1016/S0092-8674(00)80705-9
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Angerer, L. M., and Angerer, R. C. (1992). “In situ hybridization to cellular RNA with radiolabelled RNA probes,” in In Situ Hybridization, ed D. G. Wilkinson (Oxford: IRL Press), 15–32.
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Blumenthal, T. (2004). Operons in eukaryotes. Brief. Funct. Genomic. Proteomic. 3, 199–211. doi: 10.1093/bfgp/3.3.199
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bohbot, J., Pitts, R. J., Kwon, H. W., Rutzler, M., Robertson, H. M., and Zwiebel, L. J. (2007). Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect. Mol. Biol. 16, 525–537. doi: 10.1111/j.1365-2583.2007.00748.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Carey, A. F., Wang, G., Su, C. Y., Zwiebel, L. J., and Carlson, J. R. (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71. doi: 10.1038/nature08834
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chandrashekar, J., Mueller, K. L., Hoon, M. A., Adler, E., Feng, L., Guo, W., et al. (2000). T2Rs function as bitter taste receptors. Cell 100, 703–711. doi: 10.1016/S0092-8674(00)80706-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Couto, A., Alenius, M., and Dickson, B. J. (2005). Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15, 1535–1547. doi: 10.1016/j.cub.2005.07.034
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dahanukar, A., Lei, Y. T., Kwon, J. Y., and Carlson, J. R. (2007). Two Gr genes underlie sugar reception in Drosophila. Neuron 56, 503–516. doi: 10.1016/j.neuron.2007.10.024
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dobritsa, A. A., van der Goes van Naters, W., Warr, C. G., Steinbrecht, R. A., and Carlson, J. R. (2003). Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. doi: 10.1016/S0896-6273(03)00094-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dormont, L., Bessiere, J. M., and Cohuet, A. (2013). Human skin volatiles: a review. J. Chem. Ecol. 39, 569–578. doi: 10.1007/s10886-013-0286-z
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dreos, R., Ambrosini, G., Cavin, P. R., and Bucher, P. (2013). EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 41, D157–D164. doi: 10.1093/nar/gks1233
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Engsontia, P., Sanderson, A. P., Cobb, M., Walden, K. K., Robertson, H. M., and Brown, S. (2008). The red flour beetle's large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 387–397. doi: 10.1016/j.ibmb.2007.10.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fishilevich, E., and Vosshall, L. B. (2005). Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol. 15, 1548–1553. doi: 10.1016/j.cub.2005.07.066
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fox, A. N., Pitts, R. J., and Zwiebel, L. J. (2002). A cluster of candidate odorant receptors from the malaria vector mosquito, Anopheles gambiae. Chem. Senses 27, 453–459. doi: 10.1093/chemse/27.5.453
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Galindo, M. I., Pueyo, J. I., Fouix, S., Bishop, S. A., and Couso, J. P. (2007). Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5:e106. doi: 10.1371/journal.pbio.0050106
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Guidobaldi, F., May-Concha, I. J., and Guerenstein, P. G. (2014). Morphology and physiology of the olfactory system of blood-feeding insects. J. Physiol. Paris 108, 96–111. doi: 10.1016/j.jphysparis.2014.04.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. doi: 10.1016/j.cell.2004.05.012
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hansson, B. S., and Stensmyr, M. C. (2011). Evolution of insect olfaction. Neuron 72, 698–711. doi: 10.1016/j.neuron.2011.11.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hill, C. A., Fox, A. N., Pitts, R. J., Kent, L. B., Tan, P. L., Chrystal, M. A., et al. (2002). G protein-coupled receptors in Anopheles gambiae. Science 298, 176–178. doi: 10.1126/science.1076196
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Himeidan, Y. E., Temu, E. A., El rayah, E. A., Munga, S., and Kweka, E. J. (2013). Chemical cues for malaria vectors oviposition site selection: challenges and opportunities. J. Insects 2013:685182. doi: 10.1155/2013/685182
Iatrou, K., and Biessmann, H. (2008). Sex-biased expression of odorant receptors in antennae and palps of the African malaria vector Anopheles gambiae. Insect Biochem. Mol. Biol. 38, 268–274. doi: 10.1016/j.ibmb.2007.11.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jacquin-Joly, E., and Merlin, C. (2004). Insect olfactory receptors: contributions of molecular biology to chemical ecology. J. Chem. Ecol. 30, 2359–2397. doi: 10.1007/s10886-004-7941-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kwon, H. W., Lu, T., Rutzler, M., and Zwiebel, L. J. (2006). Olfactory responses in a gustatory organ of the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 103, 13526–13531. doi: 10.1073/pnas.0601107103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kwon, J. Y., Dahanukar, A., Weiss, L. A., and Carlson, J. R. (2007). The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 104, 3574–3578. doi: 10.1073/pnas.0700079104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lin, M. F., Carlson, J. W., Crosby, M. A., Matthews, B. B., Yu, C., Park, S., et al. (2007). Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 17, 1823–1836. doi: 10.1101/gr.6679507
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lu, T., Qiu, Y. T., Wang, G., Kwon, J. Y., Rutzler, M., Kwon, H. W., et al. (2007). Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 17, 1533–1544. doi: 10.1016/j.cub.2007.07.062
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pi, H., Lee, L. W., and Lo, S. J. (2009). New insights into polycistronic transcripts in eukaryotes. Chang Gung. Med. J. 32, 494–498.
Pitts, R. J., Rinker, D. C., Jones, P. L., Rokas, A., and Zwiebel, L. J. (2011). Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics 12:271. doi: 10.1186/1471-2164-12-271
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pitts, R. J., and Zwiebel, L. J. (2006). Antennal sensilla of two female anopheline sibling species with differing host ranges. Malaria J. 5:26. doi: 10.1186/1475-2875-5-26
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Qiu, Y. T., van Loon, J. J., Takken, W., Meijerink, J., and Smid, H. M. (2006). Olfactory coding in antennal neurons of the malaria mosquito, Anopheles gambiae. Chem. Senses 31, 845–863. doi: 10.1093/chemse/bjl027
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ray, A., van Naters, W. G., Shiraiwa, T., and Carlson, J. R. (2007). Mechanisms of odor receptor gene choice in Drosophila. Neuron 53, 353–369. doi: 10.1016/j.neuron.2006.12.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rinker, D. C., Pitts, R. J., Zhou, X., Suh, E., Rokas, A., and Zwiebel, L. J. (2013). Blood meal-induced changes to antennal transcriptome profiles reveal shifts in odor sensitivities in Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 110, 8260–8265. doi: 10.1073/pnas.1302562110
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Robertson, H. M., and Wanner, K. W. (2006). The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16, 1395–1403. doi: 10.1101/gr.5057506
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rytz, R., Croset, V., and Benton, R. (2013). Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 43, 888–897. doi: 10.1016/j.ibmb.2013.02.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schultze, A., Breer, H., and Krieger, J. (2014). The blunt trichoid sensillum of female mosquitoes, Anopheles gambiae: odorant binding protein and receptor types. Int. J. Biol. Sci. 10, 426–437. doi: 10.7150/ijbs.8754
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schultze, A., Pregitzer, P., Walter, M. F., Woods, D. F., Marinotti, O., Breer, H., et al. (2013). The co-expression pattern of odorant binding proteins and olfactory receptors identify distinct trichoid sensilla on the antenna of the malaria mosquito Anopheles gambiae. PLoS ONE 8:e69412. doi: 10.1371/journal.pone.0069412
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Slone, J., Daniels, J., and Amrein, H. (2007). Sugar receptors in Drosophila. Curr. Biol. 17, 1809–1816. doi: 10.1016/j.cub.2007.09.027
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Takken, W., and Knols, B. G. (1999). Odor-mediated behavior of Afrotropical malaria mosquitoes. Annu. Rev. Entomol. 44, 131–157. doi: 10.1146/annurev.ento.44.1.131
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tauxe, G. M., MacWilliam, D., Boyle, S. M., Guda, T., and Ray, A. (2013). Targeting a dual detector of skin and CO2 to modify mosquito host seeking. Cell 155, 1365–1379. doi: 10.1016/j.cell.2013.11.013
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Touhara, K., and Vosshall, L. B. (2009). Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 71, 307–332. doi: 10.1146/annurev.physiol.010908.163209
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, G., Carey, A. F., Carlson, J. R., and Zwiebel, L. J. (2010). Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 107, 4418–4423. doi: 10.1073/pnas.0913392107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zwiebel, L. J., and Takken, W. (2004). Olfactory regulation of mosquito-host interactions. Insect Biochem. Mol. Biol. 34, 645–652. doi: 10.1016/j.ibmb.2004.03.017
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: insect olfaction, antenna, odorant receptor, expression, polycistronic mRNA
Citation: Karner T, Kellner I, Schultze A, Breer H and Krieger J (2015) Co-expression of six tightly clustered odorant receptor genes in the antenna of the malaria mosquito Anopheles gambiae. Front. Ecol. Evol. 3:26. doi: 10.3389/fevo.2015.00026
Received: 19 January 2015; Accepted: 27 February 2015;
Published: 17 March 2015.
Edited by:
William Benjamin Walker, Swedish University of Agricultural Sciences, SwedenReviewed by:
Maya Louise Evenden, University of Alberta, CanadaNicolas Montagné, Université Pierre et Marie Curie, France
Copyright © 2015 Karner, Kellner, Schultze, Breer and Krieger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jürgen Krieger, Institute of Physiology (230), University of Hohenheim, Garbenstrasse 30, 70599 Stuttgart, Germany juergen.krieger@uni-hohenheim.de
†Present Address: Jürgen Krieger, Department of Animal Physiology, Institute of Biology/Zoology, Martin-Luther-University Halle-Wittenberg, Halle (Saale), Germany
 Tim Karner
Tim Karner  Heinz Breer
Heinz Breer Jürgen Krieger
Jürgen Krieger