Insect olfaction and the evolution of receptor tuning
- 1Department of Biology, Lund University, Lund, Sweden
- 2The New Zealand Institute for Plant & Food Research Ltd, Auckland, New Zealand
Insects detect odorants primarily using odorant receptors (OR) housed in the dendritic membrane of olfactory sensory neurons (OSN). Pioneering studies indicated that insects, like mammals, detect odorants in a combinatorial fashion with a specific odor ligand activating several broadly tuned ORs, and each OR being activated by several ligands. Several recent studies, however, challenge this view by providing examples where ecologically relevant odorants are detected by high-specificity ORs activating dedicated neuronal circuits. Here we review these contrasting findings on the ligand selectivity of insect ORs and their neuronal wiring, and outline scenarios describing how adaptive and neutral evolution might shape both narrow and broad receptor tuning. The fact that not all ORs display narrow tuning might partly be due to key ligands having been missed from screens or too high stimuli concentrations being used. However, the birth-and-death model of OR evolution, involving both adaptive and neutral events, could also explain the evolution of broad tuning in certain receptors due to positive selection or relaxed constraint. If the insect olfactory system indeed contains both narrowly and broadly tuned ORs, this suggests that it is a hybrid between dedicated channels and combinatorial coding. The relative extent of the two coding modes is then likely to differ between species, depending on requirements of perceived chemical space and the size of the OR repertoire. We address this by outlining scenarios where certain insect groups may be more likely to have evolved combinatorial coding as their dominant coding strategy. Combinatorial coding may have evolved predominantly in insects that benefit from the ability to discriminate between a larger number of odorants and odor objects, such as polyphagous or social species. Alternatively, combinatorial coding may have evolved simply as a mechanism to increase perceived odor space in species with small OR repertoires.
Introduction
Animals across phyla detect odors from the surrounding environment to find food, hosts, mates and oviposition sites, and to avoid predators and pathogens (Hildebrand and Shepherd, 1997). Thus, the sense of smell is often critically important for survival and arguably linked to fitness, having evolved to fit various ecological lifestyles (Nei et al., 2008; Hansson and Stensmyr, 2011; Cande et al., 2013). Animals detect odorants using several families of receptor proteins, of which the odorant receptors (ORs) are the most thoroughly studied in terms of function. Early studies on mammals suggested that their ORs respond to odorants in a combinatorial fashion, with a specific odor ligand activating multiple ORs, and each OR being activated by multiple ligands (Figure 1A). This in turn results in a combinatorial activation of olfactory sensory neurons (OSN) in the periphery and glomeruli in the primary olfactory center (Figure 1B) (Malnic et al., 1999; but see Wetzel et al., 1999; Keller et al., 2007; Nara et al., 2011; Shirasu et al., 2014). By using promiscuous receptors with overlapping response spectra, an animal is expected to be able to discriminate among more odorants and odor blends through central processing of the signals than predicted by the number of receptors it expresses (e.g., Xu et al., 2000; Bushdid et al., 2014).
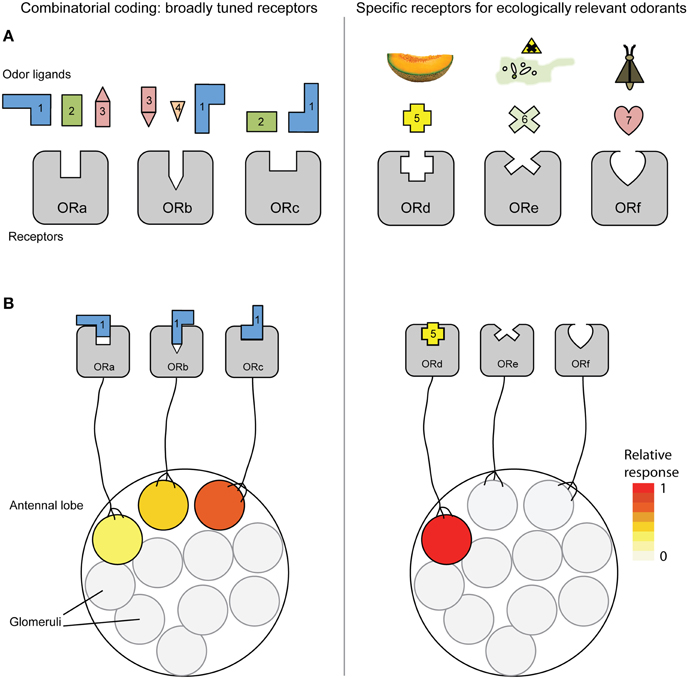
Figure 1. Combinatorial coding including broadly tuned odorant receptors (OR) contrasted with specialized ORs activating dedicated neuronal pathways. (A) With combinatorial coding (left), each odorant binds to several receptors with overlapping ligand affinities, and each OR is activated by multiple ligands. In contrast, specific ORs for ecologically relevant cues are activated by one or a few structurally related odorants, and little cross-activation of ORs occurs (right). (B) With combinatorial coding, a single odorant activates a unique combination of glomeruli in the antennal lobe (left) due to binding to multiple ORs, whereas a single odorant activates a single glomerulus in a system comprised of specialized ORs and dedicated channels (right).
Pioneering functional studies on the OR repertoire of Drosophila melanogaster (Hallem et al., 2004; Kreher et al., 2005; Hallem and Carlson, 2006), and later of mosquitoes (Xia et al., 2008; Carey et al., 2010; Wang et al., 2010), suggested that insect ORs respond to multiple ligands (i.e., are broadly tuned), and that combinatorial coding also likely applies to insect olfaction, with their modest repertoires of receptors and glomeruli. Based on these findings, combinatorial coding has been established as the predominant explanation of how insects “encode” their odor environments (e.g., Touhara and Vosshall, 2009; Wyatt, 2014). However, several recent studies challenge this view by providing examples where certain odors are detected by highly specific receptors (Mathew et al., 2013; Liu et al., 2014) (Figure 1A), activating dedicated neuronal pathways (originally referred to as “labeled-line” channels) (Figure 1B) (Stensmyr et al., 2012; Dweck et al., 2013, 2015; Ronderos et al., 2014). In light of these findings, we review the literature on ligand selectivity of insect ORs, and outline ecological and evolutionary scenarios that, in combination with neurological constraints, we believe are likely to have favored high specificity of the insect ORs and their corresponding sensory neurons. We also discuss how adaptive and neutral evolution might allow some receptors to acquire broad tuning, facilitating combinatorial coding in certain olfactory sub-systems. Finally, we present our views on in which insect groups one would expect combinatorial coding to be more prevalent.
Definition of Combinatorial Coding
Before venturing into the discussion about the different odor coding modes, it is important to clearly state our working definition of “combinatorial coding,” because its meaning is not consistent in the literature. Originally, the definition of combinatorial coding was as described above: each OR is broadly tuned, responding to multiple ligands, and single ligands activate multiple ORs and as a consequence also multiple glomeruli in the primary olfactory center (Malnic et al., 1999). This is the definition we employ here. However, combinatorial coding is also used to describe central processing mechanisms, and can in this case, actually involve receptors specific for single compounds (Wyatt, 2014). For instance, moth pheromone receptors often respond specifically to individual pheromone components, while blend-specific antennal lobe (AL) interneurons and projection neurons might need combinatorial input from several of these OSNs to respond (Christensen et al., 1989; Wu et al., 1996). In general, insects rely on specific combinations and ratios of odorants for behavioral responses, such as host attraction and non-host avoidance (Bruce et al., 2005; Bruce and Pickett, 2011; Binyameen et al., 2014; Riffell et al., 2014; Thoma et al., 2014). Thus, at the level of odor objects, such as host plants releasing a bouquet of odorants, combinatorial coding is operating both at the periphery and in the AL (Semmelhack and Wang, 2009), because the individual constituents of the blend activate different neurons, regardless of whether the ORs they express are specific or promiscuous. This is, however, distinct from combinatorial coding with respect to single odor ligands, which is the focus of the present review.
Experimental Factors Affecting Tuning Widths and Combinatorial Responses
Selection of Test Compounds and Stimuli Concentrations
There are several experimental factors that should be considered when interpreting the tuning width of receptors. For instance, the number and selection of test odorants can influence apparent response specificities, thus also the kurtosis (k) value of ORs, which sometimes is used as a numerical representation of tuning width based on the “peakedness” of the odor response distribution (e.g., Carey et al., 2010). The response profile of a receptor will, in general, appear broader (lower k) if the odor panel contains a larger number of compounds that are chemically related to the primary ligand, as compared to if the test panel contains a smaller number or is devoid of such compounds. Thus, a biased coverage of chemical space in test odor panels will inevitably bias apparent response specificities in a receptor-dependent manner. In our schematic examples of tuning widths (i.e., see Figures 2, 3) we do not consider chemical space coverage when illustrating the difference between narrowly and broadly tuned receptors.
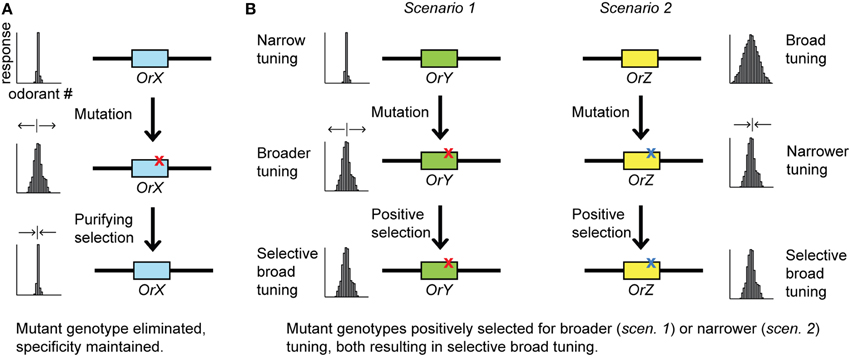
Figure 2. (A) Odorant receptor (Or) genes coding for receptors that detect odorants important for fitness are generally under purifying selection and often narrowly tuned. Mutations to such Ors that change the response specificity will typically be selected against and ultimately eliminated from the population. (B) Selection may favor “selective broad tuning” that allows an OR to detect multiple odorants, such as those emmanating from the same ecological source. Two possible evolutionary scenarios are illustrated. Mutations that widen the response of a narrowly tuned OR (scenario 1), and mutations to a promisquous ORs that narrows the specificity to a certain group of odorants, are positively selected (scenario 2) with the same evolutionary “outcome”. The tuning curves represent crude examples of different tuning widths and, as such, do not imply full coverage of chemical space.
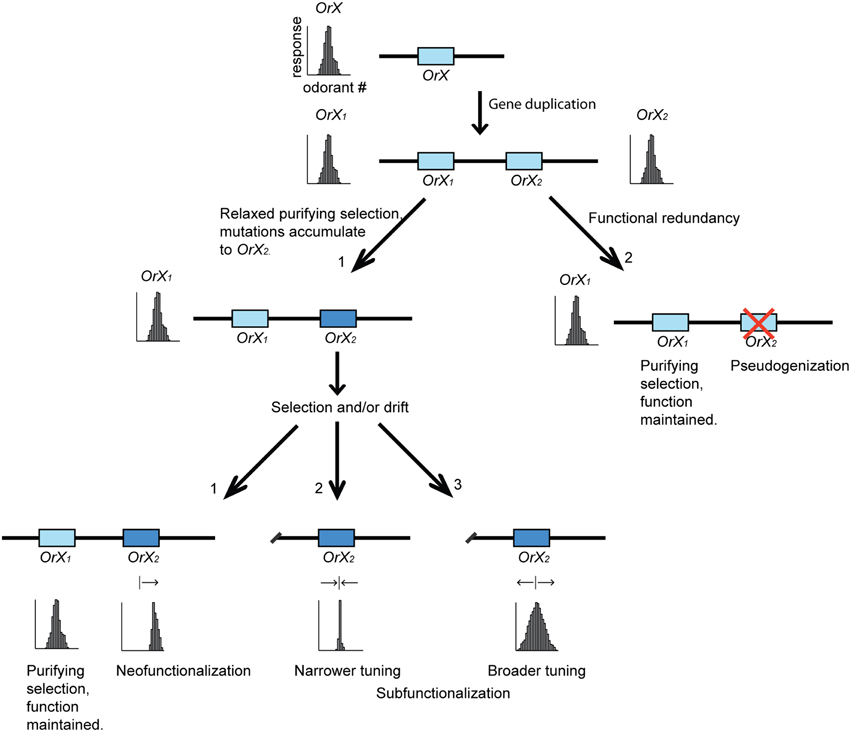
Figure 3. The birth-and-death model of odorant receptor (Or) gene evolution. The hypothetical chain of events, in this example, starts with a gene duplication of OrX giving rise to two paralogous Or genes (OrX1 and OrX2). Frequently, one of the gene copies are pseudogenized due to functional redundancy, but both can also be maintained. In the latter case, mutations may accumulate to one of the genes due to relaxed purifying selection, given that the function of the other receptor remains intact. The mutated OR might acquire a novel function (neofunctionalization), or a similar function as its paralog (subfunctionalization). The tuning width can evolve by natural selection or genetic drift to become narrower or broader as compared to the parental receptor (OrX). The tuning curves represent crude examples of different tuning widths and, as such, do not imply full coverage of chemical space.
The applied stimulus concentration is a second factor that greatly influences response specificities. The first large-scale heterologous screenings of the OR repertoires of D. melanogaster and A. gambiae revealed a wide range of OR specificities, from narrow to broad tuning (Kreher et al., 2005; Hallem and Carlson, 2006; Xia et al., 2008; Carey et al., 2010; Wang et al., 2010). Since most ORs were activated by multiple ligands and most ligands activated multiple ORs, it was concluded that combinatorial coding is the primary coding mode of the insect olfactory system. This conclusion is well supported by the data at that time. For example, of the 24 D. melanogaster ORs studied by Hallem and Carlson (2006), 16 responded strongly (>100 spikes/s) to multiple odorants within a panel of 110 test odorants with variable chemistry. Seven of the other ORs were only weakly (<100 spikes/s) excited, whereas one OR responded strongly only to one odorant. In this study, 67 of the odorants excited more than one OR. However, the stimulus concentrations used (10−4–10−2 dilution) in the early studies on the fly and the mosquito were probably higher than those insects generally would encounter in nature, although the local concentrations close to the sensilla have not been determined. Importantly, it is the natural odor concentrations that exert the selection pressures that shape the OR specificities. Indeed, at lower stimulus loads, the ORs are more narrowly tuned (Hallem and Carlson, 2006; Kreher et al., 2008; Wang et al., 2010), a phenomenon that has also been observed in single-unit electrophysiological recordings from a large variety of insect species (e.g., Hansson et al., 1999; Andersson et al., 2009, 2012a; Yuvaraj et al., 2013). Hence, the broad response of receptors leading to the conclusion of combinatorial activation of insect ORs might in some cases be a pharmacological effect on ORs when exposed to high concentrations of chemicals, thus not always reflecting how the repertoire of receptors is used in nature or how it evolved. This observation has now been recognized by the research group that conducted the pioneering studies on D. melanogaster (Mathew et al., 2013) (see also Section Specific Receptors and Dedicated Neuronal Channels in D. melanogaster).
Measurements of Stimuli Quantities and Purities are Essential
There are additional technical issues that may confound conclusions about receptor tuning. In order to conclude that a receptor or OSN is broadly tuned, it is important to measure the actual amount of stimulus that reaches the sensory cell (or at least the sensory organ), especially if the active compounds are structurally unrelated and have different volatilities (airborne odor delivery systems) or solubilities (liquid-borne systems). These issues are usually not thoroughly considered when analyzing the data from physiological studies. Andersson et al. (2012b) showed that differences between stimuli in their release rates from commonly used Pasteur pipette odor cartridges can significantly confound conclusions about OR specificities, and used the D. melanogaster ab3A neuron, co-expressing Or22a and Or22b (Dobritsa et al., 2003), as an example. The ab3A neuron had previously been shown to respond to several odorants and with similar sensitivity to the three proposed key ligands, methyl hexanoate, ethyl hexanoate, and ethyl butyrate (Stensmyr et al., 2003; Hallem and Carlson, 2006; Pelz et al., 2006). However, these compounds have vastly different release rates from odor cartridges (Andersson et al., 2012b). When correcting for airborne stimuli quantities, the ab3A neuron has the highest sensitivity to ethyl hexanoate, with a response threshold at a dose one order of magnitude lower than for methyl hexanoate and four orders of magnitude lower than for ethyl butyrate, indicating a higher specificity than previously acknowledged (Andersson et al., 2012b). Furthermore, Andersson et al. (2012b) also highlights the importance of the purity of the synthetic test compounds, which is rarely properly analyzed and reported (but see e.g., Andersson et al., 2009). Without careful consideration it is possible to arrive at false conclusions due to the presence of active contaminants. Even compounds of the highest quality are seldom more than 99% pure, and the slightest impurity of an active and highly volatile compound (e.g., 0.001% of ethyl butyrate in the ethyl hexanoate vial) could elicit a significant “false positive” response from a sensitive receptor.
Specific Receptors for Compounds Linked to Fitness
It has long been known that insect OSNs that respond to sex or aggregation pheromone compounds often are highly specific (Mustaparta et al., 1979; Tømmerås, 1985; Hansson et al., 1986; Priesner, 1986; Almaas et al., 1991). More recent screenings of receptors in heterologous systems have confirmed that the specificity generally resides in the pheromone receptors housed in these neurons (Wanner et al., 2007, 2010; Mitchell et al., 2012; Montagné et al., 2012; Zhang and Löfstedt, 2013; Jiang et al., 2014). High specificity of pheromone detection is likely to be maintained due to strong purifying selection on the receptors to keep high fidelity in the mate recognition system (Figure 2A), ensuring reproductive success (e.g., Leary et al., 2012). In other words, mutations in pheromone receptors that widen or alter the ligand specificity would generally be deleterious, potentially leading to reduced mating efficiency or heterospecific mating, thus lower fitness. Moreover, in recent years it has become apparent that OSNs responding to non-pheromonal compounds can also be highly selective, including those responding to ubiquitous plant volatiles (reviewed in e.g., Bruce and Pickett, 2011; Hansson and Stensmyr, 2011). Evidence from a variety of insect species suggests that a high degree of selectivity is common also for receptors that detect non-pheromonal compounds important for survival and reproduction. Discussed below is data indicating a high degree of ligand specificity for over a third of the ORs encoded within the genome of D. melanogaster.
Specific Receptors and Dedicated Neuronal Channels in D. melanogaster
There are several examples of specifically tuned ORs that detect ecologically relevant odorants to activate dedicated neuronal channels in D. melanogaster. Stensmyr et al. (2012) identified a dedicated olfactory circuit in flies for geosmin. This compound signals the presence of toxic molds and bacteria on their breeding substrates, fermenting fruit. By using GC-coupled electrophysiology, the authors tested ca. 3000 compounds that are present in the fly's natural environment, and showed that only the ab4B neuron, which expresses Or56a, is activated exclusively by geosmin. Subsequent recordings from Chinese Hamster Ovary cells showed that OR56a is extremely specific for geosmin. Furthermore, activation of the ab4B neuron by geosmin elicits activity in a single glomerulus (DA2), which is necessary and sufficient for antifeeding activity, inhibition of attraction and oviposition. Geosmin-specific OSNs were identified in seven other fruit-breeding drosophilids, indicating the evolutionary conservation of geosmin detection. It was also shown that the receptor is subjected to purifying selection. However, D. elegans did not respond to geosmin, and the fact that this species breeds on fresh flowers, and therefore is unlikely to encounter the toxic molds, suggests that the link between geosmin and reduced fitness is much weaker in this species (Stensmyr et al., 2012).
Another dedicated olfactory pathway of D. melanogaster is activated by the binding of citrus volatiles (primarily valencene) to the receptor OR19a (Dweck et al., 2013). It was shown that flies prefer citrus fruit as an oviposition substrate, and that the terpenes characteristic of these fruits are detected solely by OSNs expressing Or19a. Activation of these neurons was necessary and sufficient for the oviposition selectivity, but did not induce long-range attraction. Moreover, endoparasitoid wasps that parasitize fly larvae were repelled by citrus odors, and fly larvae had a reduced risk of parasitism in the presence of valencene. Thus, although the OR19a receptor is more broadly tuned than the receptor for geosmin, it still activates a dedicated neuronal circuit because it responds only to terpene compounds associated with the preferred oviposition substrate, and these compounds do not excite other OSN classes. Another recent study showed that DmelOR83c is specific for farnesol, which is released from the peal of citrus fruit (Ronderos et al., 2014). In contrast to the OR19a pathway, activation of farnesol-sensitive OSNs induces attraction. This might suggest two distinct pathways of which one (via OR83c) is essential for citrus attraction and the other (via OR19a) for citrus oviposition preference (Dweck et al., 2013; Ronderos et al., 2014). However, the ecological significance of farnesol and the OR83c pathway was recently questioned by Mansourian and Stensmyr (2015). Farnesol is present in minor quantities only in some citrus varieties, and the fact that the projection neurons from the glomerulus that receives input from Or83c expressing OSNs terminate in the brain area that processes pheromonal information might imply that this receptor simply is an orphan pheromone receptor (as discussed in Mansourian and Stensmyr, 2015).
D. melanogaster also responds to hydroxycinnamic acids (HCAs), potent dietary antioxidants common in fruits, using the detection of ethylphenols (primarily 4-ethylguaiacol and 4-ethylphenol) as a proxy (Dweck et al., 2015). Being able to detect foods containing dietary antioxidants is likely to be beneficial for flies and other generalist insects to counteract acute oxidative stress induced by consumption of entomopathogenic microorganisms (as argued by Dweck et al., 2015). The ethylphenols are produced from HCAs by several yeast species commonly found on fruit, and specifically detected by adult flies solely by the OSN class expressing Or71a. This OSN class was screened with 154 compounds at a high dose (10−2 dilution) to identify its maximum receptive range. In this screen, 4-ethylguaiacol elicited the strongest responses, and only eight other compounds, all structurally similar to the primary ligand, elicited responses at >100 spikes/s. Furthermore, activation of this neuron class was necessary and sufficient for increased feeding, oviposition and attraction. Also, in fly larvae, a similar dedicated neuronal pathway mediates positive chemotaxis to HCA-enriched substrates via proxy detection of the same ethylphenols. Interestingly, this pathway is activated by OSNs expressing the larval receptor OR94b, which has a response spectra very similar to that of OR71a in adults (Dweck et al., 2015).
In addition to the examples above, Mathew et al. (2013) screened the 21 larval-expressed ORs of D. melanogaster with close to 500 odorants at a dilution of 10−4. The test odor panel was comprised of chemically diverse compounds, including aldehydes, ketones, esters, aromatics, alcohols, terpenes, and pyrazines. Many of the compounds are released from natural sources such as yeast, fruits, and fungi. Odor-evoked responses were obtained from 19 of the ORs, and the active compounds elicited little cross-activation of other ORs. It was concluded that naturally occurring concentrations of many of the test odorants are likely to be signaled by single OSN classes, thus not by combinatorial activation of several neurons. Furthermore, the majority of the key ligands for the 19 ORs had effects on larval behavior (various degrees of attraction). However, the ecological relevance of each specific compound was not determined and the high response thresholds for some of the ORs to their key ligands (between 10−2 and 10−3 dilutions) indicate that they are more sensitive to other unidentified ligands (Mathew et al., 2013).
Receptor Specificity in Other Species and Receptor Families
High specificity of ORs for non-pheromonal compounds has also been found in mosquitoes, such as ORs detecting host and oviposition attractants. Lu et al. (2007) screened 97 compounds to identify A. gambiae OR8 as a specific detector for 1-octen-3-ol, a compound characteristic of humans and large mammalian herbivores. In Aedes aegypti, the OR8 ortholog was subsequently shown to primarily respond to the (R)-(–)-enantiomer of the compound (Bohbot and Dickens, 2009). In Culex quinquefasciatus, the oviposition attractants indole (Pelletier et al., 2010) and skatole (Hughes et al., 2010) are detected by specific receptors (OR2 and OR10, respectively), although only 23 compounds were tested in these two studies (see also Bohbot and Dickens, 2012; Bohbot and Dickens, and references therein). In the beet armyworm moth (Spodoptera exigua), SexiOR3 was found to be specific for (E)-(β)-farnesene within a panel of 62 odorants tested (Liu et al., 2014). Only four other compounds with very similar chemical structure elicited minor responses from this receptor. Similarly in the codling moth (Cydia pomonella), the plant compound pear ester ((E,Z)-2,4-decadienoate), which is a powerful pheromone synergist, was detected specifically by CpomOR3 (Bengtsson et al., 2014). However, the test odor panel only included 15 compounds, mostly pheromone compounds and antagonists from related species. Thus, this OR might show a broader response if tested with an expanded and more diverse set of odor stimuli.
In addition to OSNs that harbor ORs, dedicated olfactory pathways, which are fed by input from OSNs expressing ionotropic receptors (IR) or gustatory receptors (GR), have been identified in Drosophila. Examples include avoidance of carbon dioxide via GR21a and GR63a (co-expressed in the same OSN) (Jones et al., 2007; Kwon et al., 2007), aversion toward specific acids via IR64a (Ai et al., 2010), preference for yeast metabolites phenylacetaldehyde and phenylacetic acid via IR84a (Grosjean et al., 2011), and attraction to ammonia and specific amines via IR92a (Min et al., 2013).
Taken together, these studies suggest that odorants linked to important ecological traits of insects are detected by receptors that have evolved a high degree of ligand specificity (Figures 1A, 2A). These include receptors for sex- and host attractants as well as repellents, such as compounds produced by toxic microorganisms. This specificity minimizes cross-activation of receptors for ecologically relevant compounds by other non-relevant ones. Maintaining the high specificity of these receptors makes sense since mutations that reduce the specificity would most likely also reduce the fidelity of corresponding neuronal channels, increasing the risk of, for instance, mating- and oviposition mistakes, or death through inability to specifically detect toxins.
Evolution of Olfactory Receptors and Odor Tuning
Variation in Odorant Receptor Tuning Width
The increasing number of characterized receptors showing narrow tuning raises the possibility that most or even all ORs are specific, with what seems to be broadly tuned receptors only appearing so because key ligands have been missing from test panels or high screening concentrations were used. Despite this, there is still variation in OR tuning widths, even among the most thoroughly characterized ORs, with not all being as specific as the geosmin receptor in Drosophila (Stensmyr et al., 2012). In addition, it is possible that previously identified broadly tuned receptors (Hallem and Carlson, 2006; Carey et al., 2010) will remain broadly tuned even if tested at lower stimulus doses and expanded odor panels. Thus, an alternative scenario is that insect chemosensory systems contain both narrowly and broadly tuned receptors, providing the basis for both specific dedicated channels for certain compounds and combinatorial coding for others. If the insect olfactory system is indeed a hybrid between dedicated channels and combinatorial coding, the question becomes whether dedicated channels have evolved from a combinatorial coding system, or if the opposite is more likely. To address this question we should first discuss how it is thought the receptor families and the receptors themselves evolve at the molecular level.
Birth-and-Death Evolution of Receptor Families and the Ancestral State of Odor Coding
It is currently thought that the rapidly diversifying chemoreceptor gene families evolve according to a birth-and-death evolutionary model, in which gene duplication events represent the births, and pseudogenization and deletion events the deaths (Figure 3) (Nei et al., 2008; Sánchez-Gracia et al., 2009; Ramdya and Benton, 2010; Cande et al., 2013). Within different insect lineages, various groups of receptor genes have expanded, whereas others have contracted or are simply no longer present within a particular insect group. The exception to this pattern is the conserved antennal IRs where orthologs of most genes are found in all insects (Croset et al., 2010), as well as the obligate OR co-receptor, Orco (originally Or83b in D. melanogaster), which is highly conserved among winged insects (Vosshall and Hansson, 2011; Missbach et al., 2014).
Consistent with the birth-and-death model of chemoreceptor evolution, all the receptors within a family will share a common ancestral gene. Thus, under this model, combinatorial coding cannot represent the ancestral state of olfactory coding, unless the appearance of the first receptor was accompanied by one or several duplication events (combinatorial coding requires at least two receptors). Combinatorial coding also requires more sophisticated neuronal wiring and processing, and it therefore seems more parsimonious that the olfactory system started off as simple. But what about the tuning width of the ancestral receptor(s)?
The first receptor with the ability to detect external volatiles had most likely not been pre-selected to bind any particular environmental odor cue. However, once it had gained a role as an odorant detector, its tuning might have evolved toward higher specificity for compounds such as certain mate cues or toxins. Alternatively, selection might have favored a broader response allowing detection of a larger range of general odorants using a single receptor. Can anything be learned from the recent functional studies of OSNs in basal insects? The most ancient family of insect chemoreceptors, the IRs (Croset et al., 2010), are present in primitive insects such as bristletails, silverfish and firebrats, which lack ORs with the exception of Orco in the firebrat (Missbach et al., 2014). Electrophysiological recordings from OSNs expressing IRs in the bristletail Lepismachilis y-signata using 36 stimuli at a high concentration (10−2 dilutions) showed a broader tuning as compared to the response of the IRs in D. melanogaster (Missbach et al., 2014). A similar result was obtained from the firebrat Thermobia domestica. Although these results suggest that primitive insects that lack ORs have olfactory IRs with broader tuning compared with insects that have both IRs and ORs, it is unknown whether this reflects the ancestral state or if the broad responses have evolved from IRs that were more selective hundreds of million years ago. The same reasoning applies to the receptors of the OR family, which are evolutionarily unrelated to the IR family. Missbach et al. (2014) identified 30 Or genes from transcriptomes of antennae and palps, and 23 functional sensillum types in the primitive neopteran leaf insect Phyllium siccifolium. Response profiles ranged from narrow to broad. However, the presence of 30 ORs suggests that the OR repertoire has undergone extensive evolution since the appearance of the first OR in Neoptera. Thus, today's response profiles might not inform us about the ancestral state.
Consistent with the birth-and-death model of OR evolution, all extant insect groups possess expanded OR repertoires. With a repertoire of receptors with different ligand specificities at hand, evolution can narrow down the response profile of a broadly tuned OR, but also broaden the response of a narrowly tuned OR (see also Sections Selection for Broad Receptor Tuning and Relaxed Purifying Selection Might Underlie Broad Receptor Tuning). Our understanding of the exact mutations that may widen or narrow down the ligand selectivity of receptors is still in its infancy. However, a few recent studies have pinpointed certain amino acid sites as being crucial ligand determinants, thus providing molecular insight into the evolution of altered ligand affinity of odorant receptors.
Amino Acid Changes Affecting Receptor Tuning
Alterations of single amino acid residues (non-synonymous nucleotide mutations), or insertions and deletions, can significantly alter the ligand specificity of olfactory receptors (Pellegrino et al., 2011; Leary et al., 2012; Xu and Leal, 2013; Hughes et al., 2014). Mutations can occur within the major ligand binding site (orthosteric site), changing the response to include or preclude structurally similar compounds. Mutations can also occur at potential allosteric binding sites allowing or prohibiting binding of structurally dissimilar compounds (as discussed in Bohbot and Dickens, 2012). For instance, allosteric binding of fruit volatiles to the CO2 receptor complex was implicated as a potential mechanism underlying reduced CO2-mediated avoidance behavior in Drosophila in the presence of these odorants (Turner and Ray, 2009). In addition, mutations outside the ligand-binding site can also change receptor specificity and sensitivity, because they might change the gating equilibrium constant of the receptor (Colquhoun, 1998; Jadey et al., 2011; Hughes et al., 2014), or its structure and stability (Xu and Leal, 2013).
We are only just beginning to understand the positions and types of amino acid substitutions that may change the response of an OR. Mutations to amino acid 148 (located in transmembrane domain 3: TMD3) in OR3 of Ostrinia nubilalis and O. furnacalis have a large effect on the affinity to (E)-11-tetradecenyl acetate (E11), which is a pheromone component in O. nubilalis, but not in O. furnacalis. Wild type OR3 in O. furnacalis responds preferentially to (E)-12- and (Z)-12-tetradecenyl acetate. Changing amino acid residue 148 in O. furnacalis OR3 from threonine (wild type) to alanine (wild type in O. nubilalis) increased the sensitivity to E11 ca. 12-fold, thereby increasing the number of active ligands on this receptor. The converse mutation (A148T) to O. nubilais OR3 reduced the sensitivity to E11 to a similar extent (Leary et al., 2012). Similarly, a single amino acid substitution (V91A) in TMD2 of DmelOR59b, reflecting a natural polymorphism, significantly affected the response to the insect repellent DEET (Pellegrino et al., 2011). Also a few amino acids at the interface between extracellular loop 2 (ECL2) and TMD3 were implicated as determinants of odor sensitivity of DmelOR85b (Nichols and Luetje, 2010). In OR10 of mosquitoes, mutating either of three conserved proline residues in ECL2 to alanine lowered or abolished the response to skatole (Xu and Leal, 2013). The presence of neighboring glycine residues and nearby aromatic residues indicates that ECL2 contains multiple β-turns, and mutations to these residues also abolished the response. The presence of conserved β-turns also in moth ORs responding to structurally different odorants suggests that ECL2 acts as a lid to cover a membrane embedded binding pocket, rather than representing a specificity determinant of the binding pocket (Xu and Leal, 2013). Additionally, Hughes et al. (2014) identified 11 amino acid residues that affected the ligand specificities of A. gambiae OR13 and OR15. Mutations to alanine residue 195 at the interface between ECL2 and TMD4 had the most profound effect on the response to acetophenone, with the sensitivity being correlated to the amino acid side chain length. Together, these findings suggest that odorant receptors contain several amino acid positions located in both ECLs and TMDs in the TMD2-4 region that can be altered for tuning of odor specificity. Based on three-dimensional protein modeling, Hopf et al. (2015) suggest that this region that houses the ligand specificity determinants is likely to be a part of a ligand-binding site, however this needs experimental verification. With a multitude of sites and amino acid types at hand, it appears as if there is substantial opportunity for evolution to shape OR specificity, and in order to evolve a combinatorial odor coding system, broadly tuned receptors are required. We next examine how broad tuning can evolve through positive selection, as well as via neutral evolution under the umbrella of the birth-and-death evolutionary model.
Selection for Broad Receptor Tuning
Natural selection can favor a mutation that results in a more broadly tuned receptor (Figure 2B). It is important to note that this is not the same as selection for a combinatorial coding system per se, which, in addition to broadly tuned receptors, also requires overlap between different receptors' response spectra and potentially subsequent modulation of central processing mechanisms in the brain (Cande et al., 2013). For instance, if several structurally similar compounds emanate from the same or an equally “good” (or bad) source, a broader tuning allowing the OR to detect several of these compounds would be expected to increase the ability to detect the scent of such a source, which might be a host or non-host plant. Evolution of such “selective broad tuning” could also start from the other direction, i.e., mutations to a highly promiscuous receptor that restricts the affinity to include only compounds from the same ecological source might be favored by selection (Figure 2B). Either of these evolutionary pathways is a plausible scenario for how the dedicated neuronal circuit in D. melanogaster for citrus odors came to be activated by a somewhat broadly tuned receptor (Dweck et al., 2013). OR19a detects several terpenoid compounds of citrus fruits, but since these compounds are not abundant in other fruits that flies naturally encounter, they can all be used as a reliable signal from the preferred oviposition substrate. Similarly, the receptor DmelOR22a that responds to several fruit-related compounds (mainly esters) might have a selective broad tuning for the same reason. In addition, this particular OR was used to demonstrate that different ligands may compete for the same binding site (syntopic interactions) in a broadly tuned receptor, providing a means for mixture processing at the periphery (Münch et al., 2013). This may represent an additional advantage for a receptor to be broadly tuned.
Selective broad tuning of receptors for repellents might be favored by selection for the same reason. For instance, attraction to aggregation pheromones by a large number of conifer-feeding bark beetle species is antagonized by green leaf volatiles (GLV) (Zhang and Schlyter, 2004). The antagonizing GLVs are mostly C6 alcohols, including 1-hexanol and monounsaturated analogs such as E2-, E3-, Z2-, and Z3-hexenol. Due to their abundance in green leaves of angiosperm plants and very low levels in conifers, GLVs are supposedly used by coniferous insect species as a trustworthy signal of unsuitable non-host breeding material. Electrophysiological recordings demonstrated that the European spruce bark beetle, Ips typographus, has a single OSN class that is highly sensitive but indiscriminate in its response to three different (1-hexanol, E2-, and Z3-hexenol) behaviorally antagonistic (and behaviorally redundant Unelius et al., 2014) GLV alcohols, and these compounds hardly activate any other OSN class (Andersson et al., 2009). This is in contrast to several species of angiosperm-feeding insects, which have several different OSN classes for GLVs, some specifically tuned to a single compound (Hansson et al., 1999; Larsson et al., 2001; Andersson et al., 2012a). Selective GLV receptors are likely to be important for species that feed on angiosperms, because specific ratios of these ubiquitous compounds can be utilized for host vs. non-host discrimination (Visser and Avé, 1978). However, different GLV compounds would have the same meaning to a conifer specialist, i.e., non-host. Thus, it is likely that broader tuning to detect several GLV compounds has been favored by natural selection in coniferous insect species.
Although most OSNs express only a single Or gene for odorant detection, there are exceptions to this canonical “one-OR-one-OSN rule” (Dobritsa et al., 2003; Hallem et al., 2004; Couto et al., 2005). Thus, another way of broadening the tuning of an OSN would be to co-express several Or genes. This option was recently shown in the European corn borer moth, O. nubilalis, where one OSN was found to co-express up to five pheromone receptor genes and respond broadly to several antagonistic compounds (Koutroumpa et al., 2014). The authors hypothesized that this situation reflects the evolutionary pressure for being able to detect a wide range of heterospecific pheromone compounds to avoid non-specific mating - a mechanism contributing to pre-mating reproductive isolation. Similarly, Karner et al. (2015) found that 75 antennal OSNs in the malaria mosquito, Anopheles gambiae, co-express four Or genes (AgamOr13, 15, 17, and 55) and about half of these OSNs also express AgamOr16 and AgamOr47. Three of these ORs (AgamOR13, 15, and 16) have partly overlapping response spectra, including volatiles released by humans (Carey et al., 2010; Wang et al., 2010). It was therefore suggested that their co-expression mediates broad OSN tuning to improve the detection of complex host odor blends (Karner et al., 2015). These two recent examples suggest that co-expression of Or genes in the same neurons might be more common among insects than previously thought.
Relaxed Purifying Selection Might Underlie Broad Receptor Tuning
When a gene duplication event gives birth to a new Or gene, purifying selection is expected to be relaxed for one of the gene copies, given that the function of the other paralog is maintained (Ramdya and Benton, 2010). Frequently one of the copies will become pseudogenized due to functional redundancy (Figure 3). However, in some instances the second copy will be maintained due to this gene acquiring a new role (neofunctionalization), or sharing some of the role of the ancestral gene (subfunctionalization) or by gene conversion. These processes are thought to be important for the rapid diversification of chemoreceptor gene families (Nei et al., 2008).
Broad tuning of an olfactory receptor could be one of the possible outcomes of the relaxation of purifying selection (Figure 3). An example is found in the pheromone receptor OR7 from the leafroller moth, Ctenopseustis herana. This receptor is likely not required in sex pheromone communication in this species, but is capable of detecting a greater range of related pheromone components compared to its ortholog in C. obliquana that is specifically tuned to (Z)-8-tetradecenyl acetate and is required in pheromone communication (Steinwender et al., 2014). Broad tuning in this example is likely to be the derived state. Thus, broad tuning can in this case not be thought to be adaptive but rather a consequence of relaxed constraint. However, if the new detection capability of a broadly tuned receptor is beneficial, it might be maintained and positively selected, permitting the evolution of new signaling systems and ecological adaptations (Ramdya and Benton, 2010; Wyatt, 2014). In the case of pheromone receptors in moths, a number of species have been reported where at least one of the receptors displays broad tuning, detecting several of the sex pheromone components alongside receptors more selectively tuned to individual components (Miura et al., 2010; Zhang and Löfstedt, 2013). It would be interesting to investigate whether these broadly tuned pheromone receptors represent the ancestral or the derived state. In the turnip moth, Agrotis segetum, analysis of the selection pressures (i.e., the ratio of non-synonymous to synonymous mutations, dN/dS) acting on the pheromone receptors indicated that two of its broadly tuned receptors, AsegOR1 and AsegOR7, have evolved under relaxed constraint (dN/dS = 1.02, indicative of neutral evolution) (Zhang and Löfstedt, 2013). However, since the OR (AsegOR10) that roots the clade containing the two broadly tuned receptors was unresponsive to the seven test compounds, it is not possible to infer whether or not broad tuning is the derived state in this example (Zhang and Löfstedt, 2013).
More generally speaking, any mutation that changes the specificity of a pre-existing or newly born (and expressed) receptor will escape selection if the fitness of the individuals carrying the mutation is unaffected. For instance, if a mutation widens the response of a narrowly tuned receptor, fitness will not be affected if the insect in its natural environment never encounters the additional compounds that the receptor can pick up. In this scenario, the frequency of the mutated receptor allele in the population can be influenced randomly by genetic drift. The population might remain polymorphic at the mutated receptor locus, which is seen among ORs in natural populations of Drosophila (Rollmann et al., 2010; Pellegrino et al., 2011), but drift can also ultimately lead to fixation (or elimination) of the mutant receptor. If such a receptor is heterologously screened for responses to a large panel of ecologically relevant and irrelevant compounds, it might show a broad tuning, whereas in the natural environment it still acts as a specialist detector of high fidelity. Large-scale systematic screenings of ORs including both naturally occurring odorants and odorants that the insect is expected to never encounter are needed to test if this scenario is common in nature.
Having outlined evolutionary scenarios that may explain how some receptors can become broadly tuned, the question remains whether natural selection also could have acted to favor combinatorial coding in insects due to the associated benefits of this system?
Evolution of Combinatorial Coding in Insects
Advantages with Combinatorial Coding
While an olfactory system comprised solely of receptors for single compounds and dedicated neuronal pathways would provide high fidelity detection, it would also restrict the number of odorants that can be detected. In comparison, a lower level of fidelity is intuitively expected with a combinatorial coding system due to the overlapping responses of the receptors. However, there are also advantages associated with this system. Firstly, it renders the olfactory system more robust in response to disturbances. For instance, if the expression of a certain receptor gene fails or a sensillum is mechanically damaged, there would still be other receptors that respond to the compound, thus potentially rescuing the behavioral response (as implicated in Fishilevich et al., 2005; Keller and Vosshall, 2007). Secondly, broad receptor tuning increases the number of odorants and odor blends that can be detected by a receptor repertoire of a given size (Malnic et al., 1999; Bushdid et al., 2014), thus increasing perceived odor space and possibly allowing a higher degree of flexibility and more olfactory behaviors to come about. Thus, the different advantages associated with the two contrasting modes of odor coding suggest that natural selection could have favored both, with either one of them dominating in certain species or olfactory sub-systems. We return to this in Section In which Insects can We Expect Combinatorial Coding to be the Dominant Coding 582 Strategy?, after addressing the neurodevelopmental constraints that possibly restrict the evolution of combinatorial coding in insects.
Neurological Constraints Acting Against the Evolution of Combinatorial Coding in Insects?
Though there might be advantages with a combinatorial system, it seems unlikely that natural selection originally acted in favor of combinatorial coding of single odorants in insects. This is because it would require “synchronized” mutations to multiple receptors in the periphery, alterations in regulatory genes controlling receptor gene expression (Ray et al., 2008), as well as potential modulation of neuronal circuits, structures, and processing mechanisms in the brain (Couto et al., 2005; Ramdya and Benton, 2010; Silbering et al., 2011; Cande et al., 2013). Rather, and as outlined above (Sections Selection for Broad Receptor Tuning and Relaxed Purifying Selection Might Underlie Broad Receptor Tuning), we believe it is more likely that the individual receptor genes respond to natural selection or relaxation of purifying selection, although an altered response profile of one receptor might change the selection pressures acting on another. Once receptors have acquired broad tuning, natural selection may secondarily start to mold a combinatorial coding system due to its associated benefits or, alternatively, to allow the animal to decipher the complex non-adaptive input from promiscuous receptors with overlapping ligand affinities (i.e., evolution builds on the system at hand).
However, it has been argued that constraints in developmental circuit patterning programs in insects might restrict their response to selection (Cande et al., 2013). For instance, glomerular development is fundamentally different between insects and mammals, and it is possible that neurological constraints in insects impede the evolution of combinatorial coding in this taxon compared to mammals. For instance, in mammals the ORs themselves guide axons to their glomeruli, which in simplified terms means that if a new OR is “born” a new glomerulus forms, and if an OR “dies” its glomerulus disappears (Mombaerts, 2006; Zou et al., 2009). In contrast, hard-wired genetic programs produce the insect AL, and the ORs play no role in glomerular formation (Imai et al., 2010; Ramdya and Benton, 2010). Likewise, the sensilla also develop under precise genetic control ensuring a stereotypical pattern of functional OSN classes across the sensory organs (Imai et al., 2010; Ramdya and Benton, 2010). Thus, insects might also require alterations in genetic control mechanisms to express new ORs (Wyatt, 2014). It is not known whether these constraints act against the evolution of combinatorial coding in insects, but intuitively it appears as if they are at least not facilitated. While the neurological constraints may be stronger in insects compared to mammals, nematodes are even more constrained with an extremely hard-wired nervous system. The nematode worm, Caenorhabditis elegans, has about 500 chemoreceptor genes, but only three pairs of neurons that detect volatile chemicals (Krieger and Breer, 1999; Troemel, 1999). Two of the neuron pairs induce attraction in response to activation of their many co-expressed receptors, and the other pair induces avoidance behavior (Troemel, 1999). This hard-wiring principally restricts the evolution of the olfactory sense to the chemoreceptors, leaving little room for combinatorial coding to evolve. Neurological constraints aside, broad receptor tuning provides the fundamental material for natural selection to start molding a combinatorial system. Due to the context-dependent advantages of this system, one might expect combinatorial coding to be more prevalent in certain insect groups than others.
In Which Insects can We Expect Combinatorial Coding to be the Dominant Coding Strategy?
One obvious advantage of broadly tuned receptors and a combinatorial coding system is the increase in the number of odorants that can be detected, thereby increasing perceived odor space. In insects, one could envision that being able to perceive a large odor space would be especially important for polyphagous species, those that have highly evolved semiochemical communication systems (e.g., social hymenopterans and termites), and for good learners that benefit from a greater flexibility in olfactory guided behaviors (e.g., honeybees and ants). For instance, honeybees have a remarkable ability to discriminate and learn hundreds of complex odor mixtures (Laska et al., 1999), and they also communicate using a large variety of pheromonal compounds. Early calcium imaging studies of honeybee AL activity suggested a combinatorial activation of glomeruli in response to high compound doses (undiluted compounds and 10−2 dilutions) (Joerges et al., 1997; Sachse et al., 1999). Similarly, recent recordings from honeybee projection neurons showed that all the 27 pheromonal compounds that were tested (including queen-, brood-, alarm-, and aggregation compounds) elicit a combinatorial activation pattern (Carcaud et al., 2015). In this study, compounds were tested either undiluted or at 50 μg/μl dilutions in isopropanol (5 μl loads in both cases). However, whether the observed combinatorial activation pattern is reflected in the response profiles of the ORs at lower odor concentrations is unknown because functional studies on honeybee ORs are largely lacking. In addition, little insight can be gained from single sensillum recordings from bees because their sensilla contain too many OSNs to be able to discern them based on spike amplitudes (Getz and Akers, 1994). Only a few hymenopteran ORs have been functionally described in heterologous systems. In the honeybee, the queen substance 9-oxo-2-decenoic acid is detected by a highly specific OR, AmelOR11 (Wanner et al., 2007). Moreover, AmelOR151 responds primarily to linalool, and AmelOR152 to a small set of other floral compounds (Claudianos et al., 2014). However, since the test odor panel in the latter study was modest (14 compounds), tuning widths are difficult to conclude. Functional studies of ORs in ants are also too scarce to conclude if ants mostly employ combinatorial coding or dedicated channels. To our knowledge only two ant ORs have been characterized. Camponotus floridanus OR263 responds specifically to 2,4,5-trimethylthiazole and Harpegnathos saltator OR55 to 4-methoxyphenylacetone, both naturally occurring odorants (Zhou et al., 2012).
In addition, hymenopteran genomes contain larger numbers of Or genes than genomes from other insect orders, with for example honeybees having about 170 Or genes and some ant species over 400 (Robertson and Wanner, 2006; Robertson et al., 2010; Zhou et al., 2012). It is possible that this could facilitate the evolution of a combinatorial coding system, because a larger number of ORs increases the probability that the responses of their many ORs overlap. Alternatively, expressing many ORs might allow for a larger number of dedicated olfactory pathways without too much compromise of perceived odor space. In line with the latter reasoning are the results from Behrens et al.'s (2014) study on the response profiles of avian and amphibian bitter taste receptors (Tas2Rs) across a wide range of receptor repertoire sizes (2-ca. 50). The Tas2R receptors were always broadly tuned in species with two receptors, whereas individual receptors generally were more ligand-specific in species with a larger number of receptors.
So although the recordings from the honeybee brain suggest that combinatorial coding is operating in this species at high stimulus doses, there remains too little evidence to conclude whether or not this coding strategy would be more prevalent in polyphagous species, social insects and good learners, or species with large OR repertoires. This is, however, mainly due to lack of functional characterization of the ORs of such species. Studies should include a large proportion of the ORs encoded by the genome as well as a large set of ecologically relevant stimuli at natural concentrations in combination with measurements of glomerular activation in the AL.
In stark contrast to the hymenopterans, psyllid (Sternorrhyncha: Psyllidae) species are equipped with a truly minimalistic olfactory system. In three species studied using single sensillum electrophysiology, as few as 12 OSNs, grouped into four sensilla, appear to be devoted to plant odor detection (Kristoffersen et al., 2008b; Yuvaraj et al., 2013; Coutinho-Abreu et al., 2014), and these OSNs project to atypical, aglomerular, ALs (Kristoffersen et al., 2008a). Is there evidence of combinatorial coding in these species or do they mostly use narrowly tuned receptors? In fact, it appears as if the OSNs of psyllids are either very narrowly tuned or quite broadly tuned. Thus, the tuning ranges show a bimodal distribution and not a continuum as seen in most other insect species, at least at high stimulus concentrations (10−2 dilutions) (Coutinho-Abreu et al., 2014). This might indicate that the olfactory system of psyllids is hybrid between dedicated channels and combinatorial coding, possibly as a means of increasing odor space while retaining specificity for certain important odor cues. It remains to be seen whether or not this holds true also when challenged with larger odor panels and lower stimulus doses.
Conclusions and Future Directions
The established theory of combinatorial odor coding in insects has become challenged by several studies showing that ecologically relevant compounds are detected by highly specific receptors activating dedicated olfactory circuits. This has now been demonstrated for more than a third of the ORs encoded by the D. melanogaster genome, and for some ORs from other insect species and other classes of insect chemosensory receptors. The conclusion of combinatorial activation of broadly tuned receptors that was arrived at from earlier studies might be partly explained by experimental issues such as the use of high stimuli concentrations, the lack of ecologically relevant ligands in the screen, or the lack of quantification of airborne stimuli amounts.
We suggest that insect chemosensory systems are hybrids between combinatorial coding and dedicated circuits, with combinatorial coding likely evolving from dedicated circuits. The birth-and-death model of chemoreceptor evolution, including both adaptive and neutral changes, could allow some receptors and olfactory sub-systems to become broadly tuned, resulting in hybrid insect olfactory systems. In some cases, natural selection might have acted to favor broader tuning perhaps to increase the perceivable odor space, while in other instances, relaxed constraint might have allowed mutations to accumulate in receptors. Irrespective of the underlying evolutionary scenarios, once a subset of broadly tuned receptors is present, neurological or developmental mechanisms to process the more complex odor input might evolve in response to natural selection due to the associated benefits of combinatorial coding.
In different insects, one or the other system (dedicated vs. combinatorial) may come to dominate based on ecological context. Further studies are required to test this hypothesis and look at the nature of coding systems in non-drosophilid insects. Such studies should also reveal whether there is any correlation between OR repertoire size and the proportion of broadly tuned receptors, and therefore the extent of combinatorial coding in different insect groups. Future studies should also reveal whether more of the currently described broadly tuned receptors are actually dedicated detectors for yet to be discovered specific ligands. Finally, it will be interesting to learn to what extent the mechanisms that provide olfactory plasticity, such as regulation via differential Or gene expression (Fox et al., 2001; Reinhard and Claudianos, 2012; Claudianos et al., 2014) and neuromodulatory hormones (Flecke and Stengl, 2009; Ignell et al., 2009; Root et al., 2011), are operating in systems dominated by dedicated circuits or combinatorial systems.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the two reviewers for useful critique of the manuscript. We are also grateful to the Swedish Foundation for International Cooperation in Research and Higher Education (STINT; grants IB2013-5256 and IG2013-5483) for the funds supporting the collaboration between the Swedish and New Zealand partners. Also MNA acknowledges funding from the Swedish Research Council FORMAS (grant 217-2014-689), and CL funding from the Swedish Research Council VR (grant VR-621-2013-4355).
References
Ai, M., Min, S., Grosjean, Y., Leblanc, C., Bell, R., Benton, R., et al. (2010). Acid sensing by the Drosophila olfactory system. Nature 468, 691–695. doi: 10.1038/nature09537
Almaas, T. J., Christensen, T. A., and Mustaparta, H. (1991). Chemical communication in heliothine moths. J. Comp. Physiol. A 169, 249–258. doi: 10.1007/BF00206989
Andersson, M. N., Larsson, M. C., and Schlyter, F. (2009). Specificity and redundancy in the olfactory system of the bark beetle Ips typographus: single-cell responses to ecologically relevant odors. J. Insect Physiol. 55, 556–567. doi: 10.1016/j.jinsphys.2009.01.018
Andersson, M. N., Larsson, M. C., Svensson, G. P., Birgersson, G., Rundlöf, M., Lundin, O., et al. (2012a). Characterization of olfactory sensory neurons in the white clover seed weevil, Apion fulvipes (Coleoptera: Apionidae). J. Insect Physiol. 58, 1325–1333. doi: 10.1016/j.jinsphys.2012.07.006
Andersson, M. N., Schlyter, F., Hill, S. R., and Dekker, T. (2012b). What reaches the antenna? How to calibrate odor flux and ligand–receptor affinities. Chem. Senses 37, 403–420. doi: 10.1093/chemse/bjs009
Behrens, M., Korsching, S., and Meyerhof, W. (2014). Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol. Biol. Evol. 31, 3216–3227. doi: 10.1093/molbev/msu254
Bengtsson, J. M., Gonzalez, F., Cattaneo, A. M., Montagné, N., Walker, W. B., Bengtsson, M., et al. (2014). A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2:33. doi: 10.3389/fevo.2014.00033
Binyameen, M., Jankuvova, J., Blaženec, M., Jakuš, R., Song, L., Schlyter, F., et al. (2014). Co-localization of insect olfactory sensory cells improves the discrimination of closely separated odour sources. Funct. Ecol. 28, 1216–1223. doi: 10.1111/1365-2435.12252
Bohbot, J. D., and Dickens, J. C. (2009). Characterization of an enantioselective odorant receptor in the yellow fever mosquito Aedes aegypti. PLoS ONE 4:e7032. doi: 10.1371/journal.pone.0007032
Bohbot, J. D., and Dickens, J. C. (2012). Selectivity of odorant receptors in insects. Front. Cell. Neurosci. 6:29. doi: 10.3389/fncel.2012.00029
Bruce, T. J. A., Wadhams, L. J., and Woodcock, C. M. (2005). Insect host location: a volatile situation. Trends Plant Sci. 10, 269–274. doi: 10.1016/j.tplants.2005.04.003
Bruce, T. J., and Pickett, J. A. (2011). Perception of plant volatile blends by herbivorous insects – finding the right mix. Phytochemistry 72, 1605–1611. doi: 10.1016/j.phytochem.2011.04.011
Bushdid, C., Magnasco, M. O., Vosshall, L. B., and Keller, A. (2014). Humans can discriminate more than 1 trillion olfactory stimuli. Science 343, 1370–1372. doi: 10.1126/science.1249168
Cande, J., Prud'homme, B., and Gompel, N. (2013). Smells like evolution: the role of chemoreceptor evolution in behavioral change. Curr. Opin. Neurobiol. 23, 152–158. doi: 10.1016/j.conb.2012.07.008
Carcaud, J., Giurfa, M., and Sandoz, J.-C. (2015). Differential combinatorial coding of pheromones in two olfactory subsystems of the honey bee brain. J. Neurosci. 35, 4157–4167. doi: 10.1523/JNEUROSCI.0734-14.2015
Carey, A. F., Wang, G., Su, C.-Y., Zwiebel, L. J., and Carlson, J. R. (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71. doi: 10.1038/nature08834
Christensen, T. A., Mustaparta, H., and Hilderbrand, J. G. (1989). Discrimination of sex pheromone blends in the olfactory system of the moth. Chem. Senses 14, 463–477. doi: 10.1093/chemse/14.3.463
Claudianos, C., Lim, T., Young, M., Yan, S., Cristino, A. S., Newcomb, R. D., et al. (2014). Odor memories regulate olfactory receptor expression in the periphery. Eur. J. Neurosci. 39, 1642–1654. doi: 10.1111/ejn.12539
Colquhoun, D. (1998). Binding, gating, affinity and efficacy: the interpretation of structure−activity relationships for agonists and of the effects of mutating receptors. Br. J. Pharmacol. 125, 923–947. doi: 10.1038/sj.bjp.0702164
Coutinho-Abreu, I. V., Mcinally, S., Forster, L., Luck, R., and Ray, A. (2014). Odor coding in a disease-transmitting herbivorous insect, the Asian Citrus psyllid. Chem. Senses 39, 539–549. doi: 10.1093/chemse/bju023
Couto, A., Alenius, M., and Dickson, B. J. (2005). Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15, 1535–1547. doi: 10.1016/j.cub.2005.07.034
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6:e1001064. doi: 10.1371/journal.pgen.1001064
Dobritsa, A. A., Van Der Goes Van Naters, W., Warr, C. G., Steinbrecht, R. A., and Carlson, J. R. (2003). Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. doi: 10.1016/S0896-6273(03)00094-1
Dweck, H. K. M., Ebrahim, S. A. M., Farhan, A., Hansson, B. S., and Stensmyr, M. C. (2015). Olfactory proxy detection of dietary antioxidants in Drosophila. Curr. Biol. 25, 455–466. doi: 10.1016/j.cub.2014.11.062
Dweck, H. K. M., Ebrahim, S. A. M., Kromann, S., Bown, D., Hillbur, Y., Sachse, S., et al. (2013). Olfactory preference for egg laying on Citrus substrates in Drosophila. Curr. Biol. 23, 2472–2480. doi: 10.1016/j.cub.2013.10.047
Fishilevich, E., Domingos, A. I., Asahina, K., Naef, F., Vosshall, L. B., and Louis, M. (2005). Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr. Biol. 15, 2086–2096. doi: 10.1016/j.cub.2005.11.016
Flecke, C., and Stengl, M. (2009). Octopamine and tyramine modulate pheromone-sensitive olfactory sensilla of the hawkmoth Manduca sexta in a time-dependent manner. J. Comp. Physiol. A 195, 529–545. doi: 10.1007/s00359-009-0429-4
Fox, A. N., Pitts, R. J., Robertson, H. M., Carlson, J. R., and Zwiebel, L. J. (2001). Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc. Natl. Acad. Sci. U.S.A. 98, 14693–14697. doi: 10.1073/pnas.261432998
Getz, W. M., and Akers, R. P. (1994). Honeybee olfactory sensilla behave as integrated processing units. Behav. Neural Biol. 61, 191–195. doi: 10.1016/S0163-1047(05)80075-5
Grosjean, Y., Rytz, R., Farine, J.-P., Abuin, L., Cortot, J., Jefferis, G. S., et al. (2011). An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature 478, 236–240. doi: 10.1038/nature10428
Hallem, E. A., and Carlson, J. R. (2006). Coding of odors by a receptor repertoire. Cell 125, 143–160. doi: 10.1016/j.cell.2006.01.050
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. doi: 10.1016/j.cell.2004.05.012
Hansson, B. S., and Stensmyr, M. C. (2011). Evolution of insect olfaction. Neuron 72, 698–711. doi: 10.1016/j.neuron.2011.11.003
Hansson, B. S., Larsson, M. C., and Leal, W. S. (1999). Green leaf volatile-detecting olfactory receptor neurones display very high sensitivity and specificity in a scarab beetle. Physiol. Entomol. 24, 121–126. doi: 10.1046/j.1365-3032.1999.00121.x
Hansson, B. S., Löfstedt, C., Löfqvist, J., and Hallberg, E. (1986). Spatial arrangement of different types of pheromone-sensitive sensilla in a male moth. Naturwissenschaften 73, 269–270. doi: 10.1007/BF00367782
Hildebrand, J. G., and Shepherd, G. M. (1997). Mechanisms of olfactory discrimination: Converging evidence for common principles across phyla. Annu. Rev. Neurosci. 20, 595–631. doi: 10.1146/annurev.neuro.20.1.595
Hopf, T. A., Morinaga, S., Ihara, S., Touhara, K., Marks, D. S., and Benton, R. (2015). Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat. Commun. 6:6077. doi: 10.1038/ncomms7077
Hughes, D. T., Pelletier, J., Luetje, C. W., and Leal, W. S. (2010). Odorant receptor from the southern house mosquito narrowly tuned to the oviposition attractant skatole. J. Chem. Ecol. 36, 797–800. doi: 10.1007/s10886-010-9828-9
Hughes, D. T., Wang, G., Zwiebel, L. J., and Luetje, C. W. (2014). A determinant of odorant specificity is located at the extracellular loop 2-transmembrane domain 4 interface of an Anopheles gambiae odorant receptor subunit. Chem. Senses 39, 761–769. doi: 10.1093/chemse/bju048
Ignell, R., Root, C. M., Birse, R. T., Wang, J. W., Nässel, D. R., and Winther, A. M. E. (2009). Presynaptic peptidergic modulation of olfactory receptor neurons in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 106, 13070–13075. doi: 10.1073/pnas.0813004106
Imai, T., Sakano, H., and Vosshall, L. B. (2010). Topographic mapping—the olfactory system. Cold Spring Harb. Perspect. Biol. 2:a001776. doi: 10.1101/cshperspect.a001776
Jadey, S. V., Purohit, P., Bruhova, I., Gregg, T. M., and Auerbach, A. (2011). Design and control of acetylcholine receptor conformational change. Proc. Natl. Acad. Sci. U.S.A. 108, 4328–4333. doi: 10.1073/pnas.1016617108
Jiang, X.-J., Guo, H., Di, C., Yu, S., Zhu, L., Huang, L.-Q., et al. (2014). Sequence similarity and functional comparisons of pheromone receptor orthologs in two closely related Helicoverpa species. Insect Biochem. Mol. Biol. 48, 63–74. doi: 10.1016/j.ibmb.2014.02.010
Joerges, J., Küttner, A., Galizia, C. G., and Menzel, R. (1997). Representations of odours and odour mixtures visualized in the honeybee brain. Nature 387, 285–288. doi: 10.1038/387285a0
Jones, W. D., Cayirlioglu, P., Kadow, I. G., and Vosshall, L. B. (2007). Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90. doi: 10.1038/nature05466
Karner, T., Schneider, I., Schultze, A., Breer, H., and Krieger, J. (2015). Co-expression of six tightly clustered odorant receptor genes in the antenna of the malaria mosquito Anopheles gambiae. Front. Ecol. Evol. 3:26. doi: 10.3389/fevo.2015.00026
Keller, A., and Vosshall, L. B. (2007). Influence of odorant receptor repertoire on odor perception in humans and fruit flies. Proc. Natl. Acad. Sci. U.S.A. 104, 5614–5619. doi: 10.1073/pnas.0605321104
Keller, A., Zhuang, H., Chi, Q., Vosshall, L. B., and Matsunami, H. (2007). Genetic variation in a human odorant receptor alters odour perception. Nature 449, 468–472. doi: 10.1038/nature06162
Koutroumpa, F. A., Kárpáti, Z., Monsempes, C., Hill, S. R., Hansson, B. S., Jacquin-Joly, E., et al. (2014). Shifts in sensory neuron identity parallel differences in pheromone preference in the European corn borer. Front. Ecol. Evol. 2:65. doi: 10.3389/fevo.2014.00065
Kreher, S. A., Kwon, J. Y., and Carlson, J. R. (2005). The molecular basis of odor coding in the Drosophila larva. Neuron 46, 445–456. doi: 10.1016/j.neuron.2005.04.007
Kreher, S. A., Mathew, D., Kim, J., and Carlson, J. R. (2008). Translation of sensory input into behavioral output via an olfactory system. Neuron 59, 110–124. doi: 10.1016/j.neuron.2008.06.010
Krieger, J., and Breer, H. (1999). Olfactory reception in invertebrates. Science 286, 720–723. doi: 10.1126/science.286.5440.720
Kristoffersen, L., Hansson, B. S., Anderbrant, O., and Larsson, M. C. (2008a). Aglomerular hemipteran antennal lobes—basic neuroanatomy of a small nose. Chem. Senses 33, 771–778. doi: 10.1093/chemse/bjn044
Kristoffersen, L., Larsson, M. C., and Anderbrant, O. (2008b). Functional characteristics of a tiny but specialized olfactory system: olfactory receptor neurons of carrot psyllids (Homoptera: Triozidae). Chem. Senses 33, 759–769. doi: 10.1093/chemse/bjn034
Kwon, J. Y., Dahanukar, A., Weiss, L. A., and Carlson, J. R. (2007). The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 104, 3574–3578. doi: 10.1073/pnas.0700079104
Larsson, M. C., Leal, W. S., and Hansson, B. S. (2001). Olfactory receptor neurons detecting plant odours and male volatiles in Anomala cuprea beetles (Coleoptera: Scarabaeidae). J. Insect Physiol. 47, 1065–1076. doi: 10.1016/S0022-1910(01)00087-7
Laska, M., Galizia, C. G., Giurfa, M., and Menzel, R. (1999). Olfactory discrimination ability and odor structure–activity relationships in honeybees. Chem. Senses 24, 429–438. doi: 10.1093/chemse/24.4.429
Leary, G. P., Allen, J. E., Bunger, P. L., Luginbill, J. B., Linn, C. E., Macallister, I. E., et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A. 109, 14081–14086. doi: 10.1073/pnas.1204661109
Liu, C., Liu, Y., Guo, M., Cao, D., Dong, S., and Wang, G. (2014). Narrow tuning of an odorant receptor to plant volatiles in Spodoptera exigua (Hübner). Insect Mol. Biol. 23, 487–496. doi: 10.1111/imb.12096
Lu, T., Qiu, Y. T., Wang, G., Kwon, J. Y., Rutzler, M., Kwon, H. W., et al. (2007). Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 17, 1533–1544. doi: 10.1016/j.cub.2007.07.062
Malnic, B., Hirono, J., Sato, T., and Buck, L. B. (1999). Combinatorial receptor codes for odors. Cell 96, 713–723. doi: 10.1016/S0092-8674(00)80581-4
Mansourian, S., and Stensmyr, M. C. (2015). The chemical ecology of the fly. Curr. Opin. Neurobiol. 34, 95–102. doi: 10.1016/j.conb.2015.02.006
Mathew, D., Martelli, C., Kelley-Swift, E., Brusalis, C., Gershow, M., Samuel, A. D. T., et al. (2013). Functional diversity among sensory receptors in a Drosophila olfactory circuit. Proc. Natl. Acad. Sci. U.S.A. 110, E2134–E2143. doi: 10.1073/pnas.1306976110
Min, S., Ai, M., Shin, S. A., and Suh, G. S. B. (2013). Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 110, E1321–E1329. doi: 10.1073/pnas.1215680110
Missbach, C., Dweck, H. K. M., Vogel, H., Vilcinskas, A., Stensmyr, M. C., Hansson, B. S., et al. (2014). Evolution of insect olfactory receptors. eLife 3:e02115. doi: 10.7554/eLife.02115
Mitchell, R. F., Hughes, D. T., Luetje, C. W., Millar, J. G., Soriano-Agatón, F., Hanks, L. M., et al. (2012). Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae. Insect Biochem. Mol. Biol. 42, 499–505. doi: 10.1016/j.ibmb.2012.03.007
Miura, N., Nakagawa, T., Touhara, K., and Ishikawa, Y. (2010). Broadly and narrowly tuned odorant receptors are involved in female sex pheromone reception in Ostrinia moths. Insect Biochem. Mol. Biol. 40, 64–73. doi: 10.1016/j.ibmb.2009.12.011
Mombaerts, P. (2006). Axonal wiring in the mouse olfactory system. Annu. Rev. Cell Dev. Biol. 22, 713–737. doi: 10.1146/annurev.cellbio.21.012804.093915
Montagné, N., Chertemps, T., Brigaud, I., François, A., François, M. C., De Fouchier, A., et al. (2012). Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci. 36, 2588–2596. doi: 10.1111/j.1460-9568.2012.08183.x
Münch, D., Schmeichel, B., Silbering, A. F., and Galizia, C. G. (2013). Weaker ligands can dominate an odor blend due to syntopic interactions. Chem. Senses 38, 293–304. doi: 10.1093/chemse/bjs138
Mustaparta, H., Angst, M. E., and Lanier, G. N. (1979). Specialization of olfactory cells to insect and host produced volatiles in the bark beetle Ips pini (Say). J. Chem. Ecol. 5, 109–123. doi: 10.1007/BF00987692
Nara, K., Saraiva, L. R., Ye, X., and Buck, L. B. (2011). A large-scale analysis of odor coding in the olfactory epithelium. J. Neurosci. 31, 9179–9191. doi: 10.1523/JNEUROSCI.1282-11.2011
Nei, M., Niimura, Y., and Nozawa, M. (2008). The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9, 951–963. doi: 10.1038/nrg2480
Nichols, A. S., and Luetje, C. W. (2010). Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interactions. J. Biol. Chem. 285, 11854–11862. doi: 10.1074/jbc.M109.058321
Pellegrino, M., Steinbach, N., Stensmyr, M. C., Hansson, B. S., and Vosshall, L. B. (2011). A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature 478, 511–514. doi: 10.1038/nature10438
Pelletier, J., Hughes, D. T., Luetje, C. W., and Leal, W. S. (2010). An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS ONE 5:e10090. doi: 10.1371/journal.pone.0010090
Pelz, D., Roeske, T., Syed, Z., De Bruyne, M., and Galizia, C. G. (2006). The molecular receptive range of an olfactory receptor in vivo (Drosophila melanogaster Or22a). J. Neurobiol. 66, 1544–1563. doi: 10.1002/neu.20333
Priesner, E. (1986). “Correlating sensory and behavioural responses in multichemical pheromone systems of Lepidoptera,” in Mechanisms in Insect Olfaction, eds T. L. Payne, M. C. Birch and C. E. J. Kennedy (Oxford: Clarendon press), 225–241.
Ramdya, P., and Benton, R. (2010). Evolving olfactory systems on the fly. Trends Genet. 26, 307–316. doi: 10.1016/j.tig.2010.04.004
Ray, A., Van Naters, W. V. D. G., and Carlson, J. R. (2008). A regulatory code for neuron-specific odor receptor expression. PLoS Biol. 6:e125. doi: 10.1371/journal.pbio.0060125
Reinhard, J., and Claudianos, C. (2012). “Molecular insights into honey bee brain plasticity,” in Honeybee Neurobiology and Behavior, eds C. G. Galizia, D. Eisenhardt and M. Giurfa. (New York, NY: Springer), 359–372.
Riffell, J. A., Shlizerman, E., Sanders, E., Abrell, L., Medina, B., Hinterwirth, A. J., et al. (2014). Flower discrimination by pollinators in a dynamic chemical environment. Science 344, 1515–1518. doi: 10.1126/science.1251041
Robertson, H. M., and Wanner, K. W. (2006). The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16, 1395–1403. doi: 10.1101/gr.5057506
Robertson, H. M., Gadau, J., and Wanner, K. W. (2010). The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol. Biol. 19, 121–136. doi: 10.1111/j.1365-2583.2009.00979.x
Rollmann, S. M., Wang, P., Date, P., West, S. A., Mackay, T. F. C., and Anholt, R. R. H. (2010). Odorant receptor polymorphisms and natural variation in olfactory behavior in Drosophila melanogaster. Genetics 186, 687–697. doi: 10.1534/genetics.110.119446
Ronderos, D. S., Lin, C.-C., Potter, C. J., and Smith, D. P. (2014). Farnesol-detecting olfactory neurons in Drosophila. J. Neurosci. 34, 3959–3968. doi: 10.1523/JNEUROSCI.4582-13.2014
Root, C. M., Ko, K. I., Jafari, A., and Wang, J. W. (2011). Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 145, 133–144. doi: 10.1016/j.cell.2011.02.008
Sachse, S., Rappert, A., and Galizia, C. G. (1999). The spatial representation of chemical structures in the antennal lobe of honeybees: steps towards the olfactory code. Eur. J. Neurosci. 11, 3970–3982. doi: 10.1046/j.1460-9568.1999.00826.x
Sánchez-Gracia, A., Vieira, F. G., and Rozas, J. (2009). Molecular evolution of the major chemosensory gene families in insects. Heredity 103, 208–216. doi: 10.1038/hdy.2009.55
Semmelhack, J. L., and Wang, J. W. (2009). Select Drosophila glomeruli mediate innate olfactory attraction and aversion. Nature 459, 218–223. doi: 10.1038/nature07983
Shirasu, M., Yoshikawa, K., Takai, Y., Nakashima, A., Takeuchi, H., Sakano, H., et al. (2014). Olfactory receptor and neural pathway responsible for highly selective sensing of musk odors. Neuron 81, 165–178. doi: 10.1016/j.neuron.2013.10.021
Silbering, A. F., Rytz, R., Grosjean, Y., Abuin, L., Ramdya, P., Jefferis, G. S. X. E., et al. (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31, 13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011
Steinwender, B., Thrimawithana, A. H., Crowhurst, R. N., and Newcomb, R. D. (2014). Pheromone receptor evolution in the cryptic leafroller species, Ctenopseustis obliquana and C. herana. J. Mol. Evol. 80, 42–56. doi: 10.1007/s00239-014-9650-z
Stensmyr, M. C., Dweck, H. K. M., Farhan, A., Ibba, I., Strutz, A., Mukunda, L., et al. (2012). A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357. doi: 10.1016/j.cell.2012.09.046
Stensmyr, M. C., Giordano, E., Balloi, A., Angioy, A.-M., and Hansson, B. S. (2003). Novel natural ligands for Drosophila olfactory receptor neurones. J. Exp. Biol. 206, 715–724. doi: 10.1242/jeb.00143
Thoma, M., Hansson, B. S., and Knaden, M. (2014). Compound valence is conserved in binary odor mixtures in Drosophila melanogaster. J. Exp. Biol. 217, 3645–3655. doi: 10.1242/jeb.106591
Tømmerås, B.A. (1985). Specialization of the olfactory receptor cells in the bark beetle Ips typographus and its predator Thanasimus formicarius to bark beetle pheromones and host tree volatiles. J. Comp. Physiol. A 157, 335–342. doi: 10.1007/BF00618123
Touhara, K., and Vosshall, L. (2009). Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 71, 307–332. doi: 10.1146/annurev.physiol.010908.163209
Turner, S. L., and Ray, A. (2009). Modification of CO2 avoidance behaviour in Drosophila by inhibitory odorants. Nature 461, 277–281. doi: 10.1038/nature08295
Unelius, R. C., Schiebe, C., Bohman, B., Andersson, M. N., and Schlyter, F. (2014). Non-host volatile blend optimization for forest protection against the european spruce bark beetle, Ips typographus. PLoS ONE 9:e85381. doi: 10.1371/journal.pone.0085381
Visser, J. H., and Avé, D. A. (1978). General green leaf volatiles in the olfactory orientation of the Colorado beetle, Leptinotarsa decemlineata. Entomol. Exp. Appl. 24, 538–549. doi: 10.1111/j.1570-7458.1978.tb02838.x
Vosshall, L. B., and Hansson, B. S. (2011). A unified nomenclature system for the insect olfactory coreceptor. Chem. Senses 36, 497–498. doi: 10.1093/chemse/bjr022
Wang, G., Carey, A. F., Carlson, J. R., and Zwiebel, L. J. (2010). Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 107, 4418–4423. doi: 10.1073/pnas.0913392107
Wanner, K. W., Nichols, A. S., Allen, J. E., Bunger, P. L., Garczynski, S. F., Linn, C. E. Jr., et al. (2010). Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS ONE 5:e8685. doi: 10.1371/journal.pone.0008685
Wanner, K. W., Nichols, A. S., Walden, K. K. O., Brockmann, A., Luetje, C. W., and Robertson, H. M. (2007). A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc. Natl. Acad. Sci. U.S.A. 104, 14383–14388. doi: 10.1073/pnas.0705459104
Wetzel, C. H., Oles, M., Wellerdieck, C., Kuczkowiak, M., Gisselmann, G., and Hatt, H. (1999). Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus laevis Oocytes. J. Neurosci. 19, 7426–7433.
Wu, W., Anton, S., Löfstedt, C., and Hansson, B. S. (1996). Discrimination among pheromone component blends by interneurons in male antennal lobes of two populations of the turnip moth, Agrotis segetum. Proc. Natl. Acad. Sci. U.S.A. 93, 8022–8027. doi: 10.1073/pnas.93.15.8022
Wyatt, T. D. (2014). Pheromones and Animal Behavior: Chemical Signals and Signatures. New York, NY: Cambridge University Press.
Xia, Y., Wang, G., Buscariollo, D., Pitts, R. J., Wenger, H., and Zwiebel, L. J. (2008). The molecular and cellular basis of olfactory-driven behavior in Anopheles gambiae larvae. Proc. Natl. Acad. Sci. U.S.A. 105, 6433–6438. doi: 10.1073/pnas.0801007105
Xu, F., Greer, C. A., and Shepherd, G. M. (2000). Odor maps in the olfactory bulb. J. Comp. Neurol. 422, 489–495. doi: 10.1002/1096-9861(20000710)422:4<489::AID-CNE1>3.0.CO;2-#
Xu, P., and Leal, W. S. (2013). Probing insect odorant receptors with their cognate ligands: insights into structural features. Biochem. Biophys. Res. Commun. 435, 477–482. doi: 10.1016/j.bbrc.2013.05.015
Yuvaraj, J. K., Andersson, M. N., Steinbauer, M., Farnier, K., and Anderbrant, O. (2013). Specificity and sensitivity of plant odor-detecting olfactory sensory neurons in Ctenarytaina eucalypti (Sternorrhyncha: Psyllidae). J. Insect Physiol. 59, 542–551. doi: 10.1016/j.jinsphys.2013.03.004
Zhang, D.-D., and Löfstedt, C. (2013). Functional evolution of a multigene family: orthologous and paralogous pheromone receptor genes in the turnip moth, Agrotis segetum. PLoS ONE 8:e77345. doi: 10.1371/journal.pone.0077345
Zhang, Q.-H., and Schlyter, F. (2004). Olfactory recognition and behavioural avoidance of angiosperm nonhost volatiles by conifer-inhabiting bark beetles. Agric. For. Entomol. 6, 1–19. doi: 10.1111/j.1461-9555.2004.00202.x
Zhou, X., Slone, J. D., Rokas, A., Berger, S. L., Liebig, J., Ray, A., et al. (2012). Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet. 8:e1002930. doi: 10.1371/journal.pgen.1002930
Keywords: combinatorial coding, evolution, olfaction, odorant receptor, selectivity, specificity
Citation: Andersson MN, Löfstedt C and Newcomb RD (2015) Insect olfaction and the evolution of receptor tuning. Front. Ecol. Evol. 3:53. doi: 10.3389/fevo.2015.00053
Received: 23 March 2015; Accepted: 05 May 2015;
Published: 20 May 2015.
Edited by:
Sharon Rose Hill, Swedish University of Agricultural Sciences, SwedenReviewed by:
Michel Renou, Institut National de la Recherche Agronomique, FranceNicolas Montagné, Université Pierre et Marie Curie, France
Copyright © 2015 Andersson, Löfstedt and Newcomb. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin N. Andersson, Department of Biology, Lund University, Sölvegatan 37, SE-223 62 Lund, Sweden, martin_n.andersson@biol.lu.se
 Martin N. Andersson
Martin N. Andersson Christer Löfstedt
Christer Löfstedt Richard D. Newcomb
Richard D. Newcomb