Morphology is dead – long live morphology! Integrating MorphoEvoDevo into molecular EvoDevo and phylogenomics
- Department of Integrative Zoology, University of Vienna, Vienna, Austria
Morphology, the description and analysis of organismal form, is one of the oldest biological disciplines that has significantly contributed to our understanding as to how animals function and how the overwhelming diversity of phenotypes evolved. The early discovery that comparative studies of morphogenesis add to our understanding of the evolutionary history and interrelationships of organisms led to the formulation of highly influential evolutionary principles, including Haeckel's theory of recapitulation or Hatschek's trochozoon-hypothesis, and established the intellectual foundation of a research area today termed EvoDevo. While the rapid integration of molecular techniques into systematics, phylogenetics, and developmental biology from the 1980s onwards made some consider morphology as having little to contribute to evolutionary research, methodological progress together with a revived focus on morphogenesis has resulted in an unexpected renaissance of evolutionary developmental morphology, here termed MorphoEvoDevo. Herein, I briefly summarize some classical landmark contributions and progress achieved by studies using the MorphoEvoDevo approach. I will focus on the role of morphology in modern evolutionary biology, especially with respect to the molecular-driven approaches such as phylogenetics and developmental genetics. I argue that, while MorphoEvoDevo may well survive as an independent field of research, in times of increased competition for funding it will significantly profit from integration of the molecular disciplines into research programs with a strong emphasis on morphology. After all, morphological data are indispensable for reconstruction of phenotypic ground patterns and character evolution, and only a holistic approach incorporating all major subdisciplines of the evolutionary biosciences may ultimately result in a deep understanding, from molecules to ecosystems, of the driving forces that have shaped our organismal world.
A Scientific Discipline of Days Long Gone? The Golden Ages of pre-EvoDevo Morphology
Morphology is one of the classical and oldest disciplines in the biological sciences. From the early naturalists that carried out gross morphological studies on macroscopic taxa, the field has come a long way, thereby generating a number of more or less independent subfields. The invention of light microscopy in the seventeenth century has led scientists the way into comparative analyses of minute specimens including the documentation of embryonic and larval development, thus paving the way into the first “golden age” of developmental morphology.
Groundbreaking and infamous, classical works of this era abound. Every student of biology will be well acquainted with Ernst Haeckel's biogenetic law or theory of recapitulation, which proposes that at least parts of the evolutionary history of a species is conserved in its ontogeny (Haeckel, 1866, 1874a). Although this concept has received some criticism later on (e.g., Gould, 1977), its essence is still the verbalized foundation of the basic principle of modern-day evolutionary developmental biology (EvoDevo), namely, that it is generally possible to reconstruct organismal evolution, ground patterns, and, potentially, phylogenetic relationships, by comparing developmental processes (or characters expressed at any stage of ontogeny) of different organisms to each other. This ontogenetic approach to organismal evolution was also manifested in Charles Darwin's theory of descent with modification, and, accordingly, in The Origin of Species (Darwin, 1859, p. 449), he noted:
“Descent being on my view the hidden bond of connexion which naturalists have been seeking under the term of the natural system. On this view we can understand how it is that, in the eyes of most naturalists, the structure of the embryo is even more important for classification than that of the adult. For the embryo is the animal in its less modified state; and in so far it reveals the structure of its progenitor. In two groups of animal, however much they may at present differ from each other in structure and habits, if they pass through the same or similar embryonic stages, we may feel assured that they have both descended from the same or nearly similar parents, and are therefore in that degree closely related. Thus, community in embryonic structure reveals community of descent. It will reveal this community of descent, however much the structure of the adult may have been modified and obscured; we have seen, for instance, that cirripedes can at once be recognized by their larvae as belonging to the great class of crustaceans.”
From the perspective of a scientist of the twenty-first century, it seems that an important prerequisite for the breakthroughs achieved by these early “pre-EvoDevo” scientists was their unbiased and comparative approach toward organisms unknown—unladen by the burden of concepts such as “model organisms” or “hypothesis-driven” research. Taking Ernst Haeckel's famous gastraea theory (Haeckel, 1874b) as an example, it seems difficult to imagine that he had the calcareous sponge Sycon in mind as an “emerging model organism” for germ layer or blastopore evolution, when he decided to work on the embryology of this animal (Haeckel, 1872). Although we now know that the poriferan outer and inner cell layers are probably not homologous to the eumetazoan ectoderm and endoderm, and neither is the primary opening of the Sycon larva to the bilaterian blastopore (Leys and Eerkes-Medrano, 2005), his far-reaching conclusions of shared germ layers and mouth openings among a great number of animals are now common textbook knowledge. As another showcase of the time, comparative studies on annelids resulted in Berthold Hatschek's trochozoon theory (Hatschek, 1878). Thereby, the finding that a similarly-looking ciliated larva occurs in the lifecycle of various marine invertebrates, irrespective of their highly different adult morphology, led to the conclusion that animals that exhibit such a larva are closely related to each other - a scenario that was confirmed more than a century later by the today globally accepted Lophotrochozoa concept (Halanych et al., 1995; although the dispute remains whether or not all trochozoans or even bilaterians had a trochophore-like larva in their last common ancestor). These examples from the mid- and late nineteenth century illustrate the long history of morphological research with a comparative evolutionary and developmental approach as an important intellectual forerunner of today's EvoDevo.
When electron microscopy entered the stage of the basic organismal sciences in the 1970s, morphological research experienced additional heydays, with an impressive body of submicroscopic data being generated in the following decades on adults and developmental stages of then little-known taxa. This new generation of morphological research culminated in the epic 15-volume landmark treatise Microscopic Anatomy of Invertebrates (1989-1997), edited by Frederick Harrison, which still constitutes an important cross-disciplinary reference work today. The multitude of novel morphological data generated during this time and the emergence of software programs to allow for cladistics analyses using large, coded data matrices, fostered the global breakthrough of phylogenetic systematics, a discipline that had been founded by Willi Hennig a few decades earlier (Hennig, 1950, 1966). Numerous morphology-based phylogenetic trees on all hierarchical levels were produced until the end of the twentieth century.
However, with the arrival of molecular techniques in the evolutionary sciences, the ever faster and cheaper generation of gene sequences quickly resulted in an explosion of data useful for phylogenetic and developmental analyses in the past 30 years. As a consequence, morphology received progressively lesser attention, resulting in a noticeable crisis of morphological approaches toward phylogenetic systematics.
Ironically enough, however, it was in the wake of another molecular-driven field, evolutionary developmental biology (EvoDevo), where morphology regained long-sought attention. One reason for that is that it soon became obvious that the growing number of gene expression data is not soundly interpretable without a profound knowledge of the morphology of the very organism under examination (see below). The thus resulting increased interest in morphology, also fuelled by the availability of novel analytical tools for high-throughput analyses including fast, high-resolution microscopy (with confocal microscopy undoubtedly as the major keyplayer) and in silico methods (3D reconstruction software), has led to an astounding renaissance of morphological research, in particular in those approaches dedicated to pattern formation (morphogenesis). Today, this area of research is not only considered a necessary asset of molecular EvoDevo, but is well established as a stand-alone research discipline that produces results valuable in their own right and that I here call evolutionary developmental morphology (MorphoEvoDevo).
In the following, I will highlight some of the significant contributions of morphology to EvoDevo since the turn of the millennium, and I will discuss future perspectives and challenges the discipline may face in the future (for a comprehensive overview of the role of the various other subdisciplines of morphology in modern evolutionary biology, see Richter and Wirkner (2014)). While the impact for MorphoEvoDevo within evolutionary biology is significant enough to justify the discipline as such, I will also point out the potential for synergies that are likely to be generated if morphological research is integrated into a comprehensive intellectual and practical framework that includes the molecular-driven fields of developmental biology and phylogenetics. After all, the expression of morphological phenotypes is the result of underlying molecular and cellular mechanisms. This includes morphodynamic movements and rearrangements of cells during embryogenesis, as well as patterning processes responsible for cell and tissue identity, that generate these morphological characters during ontogeny. The picture becomes even more complicated if the dynamics of gene activities, that are often important to generate entire morphological units, such as the “segmentation clock” in arthropods and vertebrates (e.g., Palmeirim et al., 1997; Sarrazin et al., 2012), or environmentally-driven plasticity of gene expression patterns [e.g., temperature-dependent regulation of segment numbers in arthropods (Vedel et al., 2008) as well as organogenesis and metamorphosis by altered expression levels of heat shock proteins in marine invertebrates (Ueda and Boettcher, 2009; Ueda and Degnan, 2013)], are considered - phenomena that are nowadays often analyzed in a framework termed “EcoEvoDevo” (see Gilbert and Epel, 2015 for a recent, in-depth treatment of the subject).
Given this wide recognition of organismal ontogeny as a highly complex and dynamic, interdependent process on molecular, cellular, tissue, and, hence, morphological level, where given mechanisms often (mutually) regulate each other, it is surprising that the line of demarcation in today's EvoDevo research between those researchers primarily considering molecular data and others largely relying on morphological evidence for reconstructing evolutionary scenarios has not yet been fully overcome. It is thus time to join forces and intellectual approaches as well as methodological expertise if we are to arrive at a holistic view of organismal evolution that has the potential to result in a truly modern evolutionary synthesis (see also Pigliucci and Müller, 2010).
Integrating Morphology, Developmental Biology, Evolutionary Biology, and Phylogenetics
Any organism or taxon, recent or fossil, can be described as a unique assemblage of characters, irrespective of their quality (morphological, physiological, behavioral, molecular, to name but a few) (Figure 1A). In order to delimit taxa from each other on any phyletic level, comparable character sets (or individual variations of given characters, i.e., character states or transformations series) of individuals need to be identified. While this allows for formal descriptions of taxonomic units, these data alone are not sufficient for a sound understanding as to how these organisms may be related to each other (Figure 1B). To this end, phylogenetic analysis is required, whose robustness is commonly assumed to be the higher the more characters are incorporated. Ideally, one (and only one) phylogenetic tree of “maximum likelihood” results from such an analysis, which reveals the assumed interrelationships of the respective taxa (Figure 1C). The such obtained phylogeny may then be used as the starting point, i.e., the framework or backbone, for analyses into the evolution of character (states) and character transformations that occurred along distinct lineages, as well as for the reconstruction of ground patterns at given nodes (Figure 1D). Accordingly, by plotting the characters of interest onto the existing phylogeny, one may speculate as to where a given character evolved for the first time, whether a character is likely to have evolved several times independently, where it was lost along given lineages, or what the potential (minimal) character set of the last common ancestor of a given phylogenetic entity was. Such an analysis may be performed for any character, irrespective of whether or not it was part of the data set used to generate the phylogeny. A widely used principle underlying such analyses is parsimony, which favors a scenario that requires the least number of changes (gains, losses, or transformations of characters) within a lineage in the phylogenetic tree.
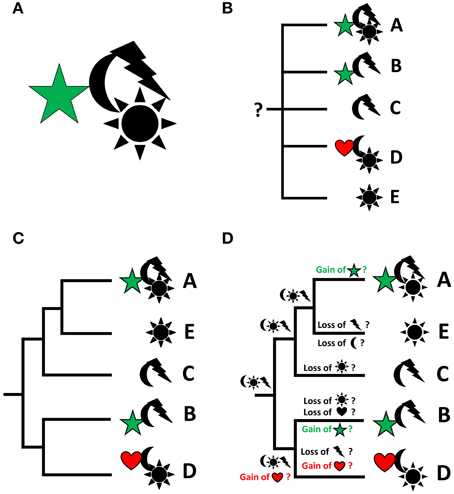
Figure 1. From descriptive science to reconstruction of character evolution by integrating descriptive data and phylogeny. Characters or character states are represented by symbols, taxa by letters. Particular focus is on the evolution of the characters “star” and “heart,” which are therefore depicted in color. (A) Every organism (or taxon) is an assemblage of (known and unknown) characters in specified character states. (B) Describing characters of given taxa is a prerequisite of phylogenetic analyses, but the characters by themselves do not produce a phylogenetic scenario and do thus not allow for ancestral character reconstruction (indicated by the question mark). (C) A phylogenetic analysis results in a hypothesis as to how given (groups of) organisms may be related to each other. (D) The resulting phylogeny allows to project the individual characters of interest onto the phylogenetic tree in order to infer their potential evolutionary emergence, loss, or transformation (usually using the parsimony approach). Thus, potentially, for any given monophyletic unit, a character set of the last common ancestor of the considered taxa can be reconstructed. Thereby, it is irrelevant whether or not the characters that are plotted onto the tree were part of the phylogenetic analysis.
A recurring issue with such analyses, particularly obvious if morphological characters are used, is that the data sets often stem from adult organisms—a problem that may be overcome by incorporating developmental data into the analyses (see Scholtz, 2005, 2010). Thereby, organisms are viewed as lifecycles rather than isolated individual ontogenetic (usually adult) units. Accordingly, the organism constitutes a pool of characters that are expressed at any given time point in its lifecycle. In Figure 2A, for example, two species are depicted, A and C, both exhibiting a distinct set of characters. However, if we look at the entire lifecycle of both species, it becomes obvious that characters appear at different stages in their lifecycles and, what is more important, that some of these characters (“heart” in taxon A and both, “heart” and “star,” in taxon C) do not survive the transition to adulthood – such characters are commonly called “transitory” (Figures 2B,C). For the data matrix, this means that both, adult as well as transitory characters, are part of the body plan of the respective taxa (Figure 2D) and thus need to be incorporated into the phylogenetic analysis. In doing so, parsimony analysis may provide us with a radically different scenario concerning character evolution and ground pattern states (compare Figure 2E and Figure 2F), illustrating the direct relevance of developmental data for the reconstruction of phylogeny-based evolutionary scenarios. Molecular phylogeny partly pays tribute to this phenomenon by using transcriptomic datasets (i.e., RNA sequences of expressed genes) generated from different lifecycle stages to increase the volume of data for phylogenetic analyses.
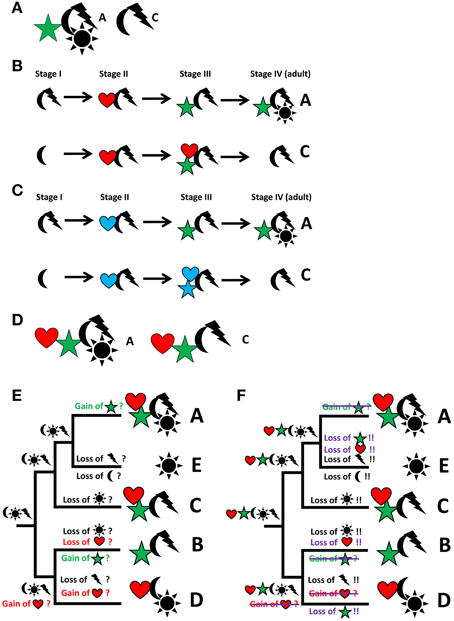
Figure 2. From descriptive science to reconstruction of character evolution by integrating descriptive data, developmental biology, and phylogeny. Particular focus is on the evolution of the characters “star” and “heart,” which are therefore depicted in color. Characters or character states are represented by symbols, taxa by letters. (A) Two individual taxa (A,C) can be discriminated by different sets of characters. (B) Analysis of ontogeny reveals the ontogenetic emergence or loss of characters. (C) Ontogenetic analysis may reveal transitory characters, i.e., characters that appear during ontogeny but are lost prior to adulthood (marked in blue; “heart” in taxon A and both, “heart” and “star,” in taxon C). (D) All characters, including transitory characters, belong to the “body plan” of a taxon. Note that this discussion can be extended into extinct lineages known from the fossil record, thereby increasing taxon sampling and adding the dimension of time to the discussion. (E) Hypothesized scenario of character evolution identical to Figure 1D, i.e., not taking transitory characters into account. (F) Significantly altered hypothetical scenario of the reconstruction of character distribution and gain and loss events at different nodes and along the various lineages due to the incorporation of transitory characters. Events that differ from the scenario in E are either crossed out or added in violet.
Morphology as an Integrated Part of EvoDevo
The technological advances in molecular biology together with dramatically decreasing costs for sequencing have resulted in an explosion of molecular data not only for phylogenetic analyses (thereby sparking the field of phylogenomics) but also on the gene expression level. This has led to the somewhat bizarre situation that RNA (i.e., gene) expression data generated by in situ hybridization have become available for rather cryptic taxa, way beyond the established model systems, for which not even solid morphological data of the respective developmental stages are available. While the expansion of gene expression analyses into such largely unknown taxa is highly welcome (and a prerequisite for any evolutionary interpretations on the level of gene activity and function), the inevitable questions that arise are: How do we interpret these data? What does a positive color reaction somewhere in a 100 μm long embryo or larva tell us, if we don't know its morphology (and, therefore, cannot unambiguously assign its site of expression to any tissue or organ)?
The confrontation with such issues has two major consequences concerning the nature (quality) of RNA expression data and their relationship to organismal morphology: (i) Not only does analysis of gene expression patterns depend on morphological knowledge—gene expression data constitute, by themselves, morphological data. Just like traditional staining methods (e.g., silver impregnation in nineteenth and early twentieth century neurobiology, lead citrate contrasting of ultrathin sections, fluorescence labeling of immunoreactive compounds), in situ hybridization is a means to visualize the location of given structural components (here: RNA) and describes their distribution in a tissue or organism. However, since the location of the labeled RNA is usually within a given superordinate morphological structure (cell, tissue, or organ), interpretation is difficult without further knowledge of the very structure and its position relative to other morphological features in the investigated sample. (ii) As a consequence of this, gene expression analysis needs tissue- and organ system-level morphology for detailed determination of expression sites. In recognition of this, the combination of conventional morphological methods (including histology, immunolabeling, and others) and in situ hybridization studies using novel optical and in silico tools (e.g., reflection confocal laserscanning microscopy, 3D reconstruction; see Jékely and Arendt, 2007; Fritsch et al., in review) have allowed for a much more detailed analysis and reliable interpretation of gene expression studies—a development that will certainly continue in the light of further technological advancements (see below).
MorphoEvoDevo: Evolutionary Developmental Morphology as Stand-Alone Discipline
Although I have argued above that morphology may well be integrated into molecular-driven phylogenetic and developmental approaches, morphogenesis-based research is strong and significant enough to be recognized as an independent scientific discipline. The routine availability of high-resolution and high-throughput methods such as confocal microscopy have opened new horizons for organismal-oriented comparative developmental biologists, allowing for reconstruction of morphogenetic events from early embryonic to juvenile stages. Accordingly, since the late 1980s, the field has seen an unprecedented increase of data on various aspects of invertebrate development, in particular on those organ systems that can be stained by fluorescent dyes such as filamentous actin (musculature), components of the nervous system, or tubulin-containing structures (neurites, ciliary organs, digestive systems) (e.g., Croll and Chiasson, 1989; Dickinson et al., 1999; Wanninger et al., 1999, 2005; Voronezhskaya et al., 2002, 2003; Wanninger and Haszprunar, 2002a,b; Dickinson and Croll, 2003; Croll, 2006; McDougall et al., 2006; Brinkmann and Wanninger, 2008, 2009, 2010a,b; Semmler et al., 2008, 2010; Altenburger and Wanninger, 2009, 2010; Fischer et al., 2010; Semmler and Wanninger, 2010; Fritsch et al., 2013; Hindinger et al., 2013; Jirikowski et al., 2013; Perez et al., 2013; Scherholz et al., 2013; Zieger et al., 2013; Redl et al., 2014; Wurzinger-Mayer et al., 2014; Stegner and Richter, 2015; see also Wanninger, 2007, 2008, 2009, 2015 for reviews). Some of these findings had significant impact on our view of animal development and evolution. For example, the segmented ancestry of two worm-like clades that are unsegmented as adults, the echiurans and sipunculans (Hessling, 2002, 2003; Hessling and Westheide, 2002; Kristof et al., 2008; Wanninger et al., 2009), that had both been suggested to reside within the segmented Annelida by molecular phylogenetic analyses, was confirmed by identification of transitory segmented elements that occur during neurogenesis. Other landmark findings using the MorphoEvoDevo approach include the occurrence of a paired ventral nerve cord in larval phoronids that is absent in adults (Temereva, 2011), or the developmental origin of the simple muscular body plan of a group of aplacophoran (worm-shaped) molluscs from a much more complex larval one as the result of evolutionary simplification (Scherholz et al., 2013). This list could be extended into numerous other taxa and the message these examples convey is obvious: by using developmental approaches to address evolutionary questions, morphological research has expanded its relevance way beyond classical (often functional) morphological research themes. As a consequence, publication of MorphoEvoDevo data have increasingly found their way into scientific journals that are specifically dedicated to EvoDevo, such as Development Genes and Evolution, Evolution and Development, Journal of Experimental Zoology B: Molecular and Developmental Evolution, BMC Developmental Biology, or EvoDevo (for some examples, see the extensive list of papers cited above), thereby considerably broadening the publication spectrum of morphological data. This trend is likely to increase further, as additional tools such as light sheet microscopy, 4D-microscopy, various forms of tomography, or serial transmission electron microscopy are on the rise (e.g., Hejnol et al., 2006; Stach et al., 2008; Laforsch et al., 2012; Randel et al., 2014).
Morphology and Phylogeny in the Age of Phylogenomics
Having addressed the high relevance of morphology for evolutionary, in particular EvoDevo, research in the previous section, I focus here on the role morphology may play in the context of today's molecular phylogenetics. A simple question one may ask is “Will morphology keep contributing significantly to building phylogenetic trees in the era of phylogenomics?” An equally simple answer to that would be “No.” One might further ask whether this is a problem for either morphology or phylogeny. I would, slightly hesitantly, answer “Well, maybe not so much,” at least not with respect to morphology as being an important evolutionary discipline nonetheless, because, as I will outline below, morphology is still of prime importance for other aspects of phylogeny-related research. Concerning phylogenetic tree-building itself, however, one would certainly wish for a morphology-based controlling body and thus an alternative discussion base for the highly diverging phylogenetic scenarios that are often proposed by molecular data (cf. e.g., the varying topologies in recent studies on molluscan intrarelationships by Giribet et al., 2006; Kocot et al., 2011; Smith et al., 2011; Vinther et al., 2012).
As mentioned above, the cheap generation and seemingly endless availability of molecular sequence data has resulted in an inflation of trees produced since molecular cross-animal kingdom phylogenies emerged in the late 1980s (Field et al., 1988). Thereby, the low sequencing costs, together with the decreasing amount of expert manpower that is necessary to produce these raw data – and thus the character set for the phylogenetic analyses – have radically outcompeted morphology-based phylogenetics, and today allow for the generation of large-scale, data-intense, phylogenomic or transcriptomic trees (e.g., Dunn et al., 2008; Hejnol et al., 2009; Misof et al., 2014). In addition, the decrease in taxonomic and morphological expertise for many clades hampers the assembly of large morphological datasets. The sheer amount of molecular phylogenies that still keep emerging has led to increased discussions as to whether or not the sometimes highly unexpected assemblages with little or no morphological support may indeed be the result of real evolutionary events or, rather, the consequence of problems in data acquisition such as sequencing or alignments errors, wrong algorithms used, and the like. As we have seen in the case of EvoDevo, for such in-depth discussions, morphological knowledge is imperative, and the more unexpected phylogenetic scenarios are recovered, the more demand will be for morphologists to make sense of these trees by pointing out the evolution and transformations of key characters, thereby also critically evaluating the results of molecular phylogeny.
Accordingly, at present, I see the prime contribution of morphology to phylogeny in making sense of trees rather than building them, although this, of course, does not question the principal suitability of morphological datasets to produce valuable phylogenetic trees (see Jenner, 2004). While it would certainly be refreshing and inspiring to see a revival of morphology-based phylogeny (because, as Scholtz, 2010 puts it, “if morphological and molecular results clash, there is no logical necessity to dismiss morphological data”), current developments in funding policy, scientific expertise, and efficiency in data generation and analysis are, unfortunately, most likely to prevent such efforts for a while. Recent initiatives in making morphological characters accessible to a wider audience by online databases such as MorphDBase (www.morphdbase.de) or MorphoBank (www.morphobank.org) are promising first steps into the right direction, but even if the proposed acceleration in generating and coding of morphological characters and resolving problems related to homology assessments were implemented (see Scotland et al., 2003; Giribet, 2010, in press), I have little hope that the situation will change anytime soon. At present, it seems as if incorporation of selected morphological data into matrices dominated by molecular characters (total evidence approach) are the main contribution of morphology to phylogenetic tree-building today, and even these efforts remain the exception rather than the rule, at least on higher taxonomic levels (e.g., Zrzavy et al., 1998; Peterson and Eernisse, 2001; Glenner et al., 2004; Sørensen and Giribet, 2006; Eklöf et al., 2007; see also Giribet, in press).
Is it all in vain, then, to engage oneself as a morphologist in phylogenetic issues these days? Certainly not. Rather, it is a question of re-adjusting the focus of morphology with respect to phylogeny. Using given phylogenetic scenarios as a backbone to reconstruct major evolutionary events and ground patterns of given clades is a key ability only morphology can provide. Whether or not we will arrive at broadly accepted phylogenetic trees in the future may ultimately depend on an agreement of the in silico tools (software, algorithms) used, rather than on the organisms or the sequences that provide the raw data for these analyses. While this may mean that phylogenetic analysis will move even further away from the biosciences, the fact that only organismal knowledge can fill these trees with life and make evolutionary sense of them may offer at least some relief to organismal biologists. Or, in other words, once all organisms have been sequenced and once we have agreed on the “true” phylogenetic tree, we will still need morphology to understand how phenotypic diversity evolved.
The Future of MorphoEvoDevo
Morphology is alive and thriving. Descriptive and experimental functional morphology has found its way into the applied (physical) sciences (biomaterials engineering, bionics, nanoscience), where basic morphological research is expected to receive even further attention in the future. The future of comparative and, especially, developmental morphology looks equally promising. MorphoEvoDevo is strong and generates results significant enough to sustain itself as a stand-alone discipline in evolutionary biology. However, unlike our scientific ancestors, we live in a largely hypothesis-driven scientific environment where competition for funding is high and cross-disciplinary approaches often outcompete proposals with too specific research foci. Accordingly, morphologists should further and proactively embrace the evolutionary disciplines that are currently dominated by molecular approaches, especially phylogenetics and EvoDevo, and integrate these into their own research programs, in order to avoid becoming a mere add-on to these and degenerate to a “shrinking or even vanishing field” (Scholtz, 2010). The foreseeable further increase of gene expression data for morphologically poorly known taxa, together with novel molecular technologies such as genome editing using the CRISPR/Cas9 system, which are rapidly evolving and are on the verge of being established beyond the classical non-model systems (e.g., Perry and Henry, 2015), will provide a multitude of possibilities for synergies between morphology and molecular biology.
The continued increase of molecular EvoDevo data from across the animal kingdom will call for well-trained morphologists to contribute to data generation and interpretation of gene functions during development and evolution. Novel high-resolution micro-morphological techniques keep developing, including serial transmission electron microscopy, focused ion beam scanning electron microscopy (FIB-SEM), stimulated emission depletion microscopy (STED), micro-computed tomography (Micro-CT), synchrotron X-ray tomography, magnetic resonance tomography (MRT), and light sheet microscopy, which will open new avenues for modern morphological research, especially if combined with powerful 3D reconstruction software. The possibilities offered by these techniques and the impressive and detailed visual data produced should also aid in developing better marketing strategies for morphological research, such that this landmark biological research area, that may still be perceived as old-fashioned by some, will continue to be viewed as an indispensable discipline within the twenty-first century biosciences.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Many of the thoughts presented here were developed when preparing a talk for the 3rd International Congress on Invertebrate Morphology, held from August 3rd–7th, 2014 in Berlin. I thank the congress hosts and, in particular, Jo Wolfe (New York), who organized and kindly invited me to present my views in the symposium “The role of ontogeny in phylogenetic reconstruction.” Current research in my lab is funded by the Austrian Science Foundation (FWF, grant number P 24276). I thank Alessandro Minelli (Padova) for his kind invitation to submit this review to the Specialty Section Evolutionary Developmental Biology of Frontiers in Ecology and Evolution.
References
Altenburger, A., and Wanninger, A. (2009). Comparative larval myogenesis and adult myoanatomy in the rhynchonelliform (articulate) brachiopods Argyrotheca cordata, A. cistellula, and Terebratalia transversa. Front. Zool. 6:3. doi: 10.1186/1742-9994-6-3
Altenburger, A., and Wanninger, A. (2010). Neuromuscular development in Novocrania anomala: evidence for the presence of serotonin and a spiralian-like apical organ in lecithotrophic brachiopod larvae. Evol. Dev. 12, 16–24. doi: 10.1111/j.1525-142X.2009.00387.x
Brinkmann, N., and Wanninger, A. (2008). Larval neurogenesis in Sabellaria alveolata reveals plasticity in polychaete neural patterning. Evol. Dev. 10, 606–618. doi: 10.1111/j.1525-142X.2008.00275.x
Brinkmann, N., and Wanninger, A. (2009). Neurogenesis suggests independent evolution of opercula in serpulid polychaetes. BMC Evol. Biol. 9:270. doi: 10.1186/1471-2148-9-270
Brinkmann, N., and Wanninger, A. (2010a). Integrative analysis of polychaete ontogeny: cell proliferation patterns and myogenesis in trochophore larvae of Sabellaria alveolata. Evol. Dev. 12, 5–15. doi: 10.1111/j.1525-142X.2009.00386.x
Brinkmann, N., and Wanninger, A. (2010b). Capitellid connections: contributions from neuromuscular development of the maldanid polychaete Axiothella rubrocincta (Annelida). BMC Evol. Biol. 10:168. doi: 10.1186/1471-2148-10-168
Croll, R. P. (2006). Development of embryonic and larval cells containing serotonin, catecholamines, and FMRFamide-related peptides in the gastropod mollusc Phestilla sibogae. Biol. Bull. 211, 232–247. doi: 10.2307/4134546
Croll, R. P., and Chiasson, B. J. (1989). Postembryonic development of serotoninlike immunoreactivity in the central nervous system of the snail, Lymnaea stagnalis. J. Comp. Neurol. 280, 122–142. doi: 10.1002/cne.902800109
Dickinson, A. J. G., and Croll, R. P. (2003). Development of the larval nervous system of the gastropod Ilyanassa obsoleta. J. Comp. Neurol. 466, 197–218. doi: 10.1002/cne.10863
Dickinson, A. J. G., Nason, J., and Croll, R. P. (1999). Histochemical localization of FMRFamide, serotonin and catecholamines in embryonic Crepidula fornicata (Gastropoda, Prosobranchia). Zoomorphology 119, 49–62. doi: 10.1007/s004350050080
Dunn, C. W., Hejnol, A., Matus, D. Q., Pang, K., Browne, W. E., Smith, S. A., et al. (2008). Broad phylogenomic sampling improves resolution of the Animal Tree of Life. Nature 452, 745–749. doi: 10.1038/nature06614
Eklöf, J., Pleijel, F., and Sundberg, P. (2007). Phylogeny of benthic Phyllodocidae (Polychaeta) based on morphological and molecular data. Mol. Phylogenet. Evol. 45, 261–271. doi: 10.1016/j.ympev.2007.04.015
Field, K. G., Olsen, G. J., Lane, D. J., Giovannoni, S. J., Ghiselin, M. T., Raff, E. C., et al. (1988). Molecular phylogeny of the animal kingdom. Science 239, 748–753. doi: 10.1126/science.3277277
Fischer, A. H. L., Henrich, T., and Arendt, D. (2010). The normal development of Platynereis dumerilii (Nereididae, Annelida). Front. Zool. 7:31. doi: 10.1186/1742-9994-7-31
Fritsch, M., Kaji, T., Olesen, J., and Richter, S. (2013). The development of the nervous system in Laevicaudata (Crustacea, Branchiopoda): insights into the evolution and homologies of branchiopod limbs and ‘frontal organs’. Zoomorphology 132, 163–181. doi: 10.1007/s00435-012-0173-0
Gilbert, S. F., and Epel, D. (2015). Ecological Developmental Biology: The Environmental Regulation of Development, Health, and Evolution. Sunderland: Sinauer.
Giribet, G. (2010). A new dimension in combining data? The use of morphology and phylogenomic data in metazoan systematics. Acta Zool. 91, 11–19. doi: 10.1111/j.1463-6395.2009.00420.x
Giribet, G. (in press). Morphology should not be forgotten in the era of genomics—a phylogenetic perspective. Zool. Anz. doi: 10.1016/j.jcz.2015.01.003
Giribet, G., Okusu, A., Lindgren, A. R., Huff, S. W., Schrödl, M., and Nishiguchi, M. K. (2006). Evidence for a clade composed of molluscs with serially repeated structures: monoplacophorans are related to chitons. Proc. Natl. Acad. Sci. U.S.A. 103, 7723–7728. doi: 10.1073/pnas.0602578103
Glenner, H., Hansen, A. J., Sørensen, M. V., Ronquist, F., Huelsenbeck, J. P., and Willerslev, E. (2004). Bayesian inference of the metazoan phylogeny: a combined molecular and morphological approach. Curr. Biol. 14, 1644–1649. doi: 10.1016/j.cub.2004.09.027
Haeckel, E. (1866). Generelle Morphologie der Organismen. Berlin: Georg Reimer. doi: 10.1515/9783110848281
Haeckel, E. (1874b). Die Gastraea-Theorie, die phylogenetische Classification des Thierreiches und die Homologie der Keimblätter. Jenaische Z. f. Naturwiss. 8, 1–55.
Halanych, K. M., Bacheller, J. D., Aguinaldo, A. M., Liva, S. M., Hillis, D. M., and Lake, J. A. (1995). Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science 267, 1641–1643. doi: 10.1126/science.7886451
Hatschek, B. (1878). Studien über die Entwicklungsgeschichte der Anneliden. Ein Beitrag zur Morphologie der Bilateralien. Arb. Zool. Inst. Univ. Wien, Zool. Stat. Triest. 1, 57–129.
Hejnol, A., Obst, M., Stamatakis, A., Ott, M., Rouse, G. W., Edgecombe, G. D., et al. (2009). Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. Lond. B 276, 4261–4270. doi: 10.1098/rspb.2009.0896
Hejnol, A., Schnabel, R., and Scholtz, G. (2006). A 4D-microscopic analysis of the germ band in Porcellio scaber (Malacostraca, Peracarida) – developmental and phylogenetic implications. Dev. Genes Evol. 216, 755–767. doi: 10.1007/s00427-006-0105-4
Hennig, W. (1950). Grundzüge einer Theorie der phylogenetischen Systematik. Berlin: Deutscher Zentralverlag.
Hessling, R. (2002). Metameric organization of the nervous system in developmental stages of Urechis caupo (Echiura) and its phylogenetic implications. Zoomorphology 121, 221–234. doi: 10.1007/s00435-002-0059-7
Hessling, R. (2003). Novel aspects of the nervous system of Bonellia viridis (Echiura) revealed by the combination of immunohistochemistry, confocal laser-scanning microscopy and three-dimensional reconstruction. Hydrobiologia 496, 225–239. doi: 10.1023/A:1026153016913
Hessling, R., and Westheide, W. (2002). Are Echiura derived from a segmented ancestor? Immunohistochemical analysis of the nervous system in developmental stages of Bonellia viridis. J. Morphol. 252, 100–113. doi: 10.1002/jmor.1093
Hindinger, S., Schwaha, T., and Wanninger, A. (2013). Immunocytochemical studies reveal novel neural structures in nemertean pilidium larvae and provide evidence for incorporation of larval components into the juvenile nervous system. Front. Zool. 10:31. doi: 10.1186/1742-9994-10-31
Jékely, G., and Arendt, D. (2007). Cellular resolution expression profiling using confocal detection of NBT/BCIP precipitate by reflection microscopy. Biotechniques 42, 751–755. doi: 10.2144/000112462
Jenner, R. A. (2004). Accepting partnership by submission? Morphological phylogenetics in a molecular millennium. Syst. Biol. 53, 333–342. doi: 10.1080/10635150490423962
Jirikowski, G., Richter, S., and Wolff, C. (2013). Myogenesis of Malacostraca – the “egg-nauplius” concept revisited. Front. Zool. 10:76. doi: 10.1186/1742-9994-10-76
Kocot, K. M., Cannon, J. T., Todt, C., Citarella, M. R., Kohn, A. B., Meyer, A., et al. (2011). Phylogenomics reveals deep molluscan relationships. Nature 447, 452–456. doi: 10.1038/nature10382
Kristof, A., Wollesen, T., and Wanninger, A. (2008). Segmental mode of neural patterning in Sipuncula. Curr. Biol. 18, 1129–1132. doi: 10.1016/j.cub.2008.06.066
Laforsch, C., Imhof, H. K., Sigl, R. M., Settles, M., He,ß, M., and Wanninger, A. (2012). “Applications of computational 3D-modeling in organismal biology,” in Modeling and Simulation in Engineering, ed A. Catalin (Rijeka: InTech open access publisher), 117–142.
Leys, S. P., and Eerkes-Medrano, D. (2005). Gastrulation in calcareous sponges: in search of Haeckel's gastraea. Integr. Comp. Biol. 45, 342–351. doi: 10.1093/icb/45.2.342
McDougall, C., Chen, W. C., Shimeld, S., and Ferrier, D. E. K. (2006). The development of the larval nervous system, musculature and ciliary bands of Pomatoceros lamarckii (Annelida): heterochrony in polychaetes. Front. Zool. 3:16. doi: 10.1186/1742-9994-3-16
Misof, B., Liu, S., Meusemann, K., Peters, R. S., Donath, A., Mayer, C., et al. (2014). Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. doi: 10.1126/science.1257570
Palmeirim, I., Henrique, D., Ish-Horowicz, D., and Pourquié, O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648. doi: 10.1016/S0092-8674(00)80451-1
Perez, Y., Rieger, V., Martin, E., Müller, C., and Harzsch, S. (2013). Neurogenesis in an early protostome relative: progenitor cells in the ventral nerve centre of chaetognath hatchlings are arranged in a highly organized geometrical pattern. J. Exp. Zool. 320, 179–193. doi: 10.1002/jez.b.22493
Perry, K. J., and Henry, J. Q. (2015). CRISPR/Cas9-mediated genome modification in the mollusc, Crepidula fornicata. Genesis 53, 237–244. doi: 10.1002/dvg.22843
Peterson, K. J., and Eernisse, D. J. (2001). Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences. Evol. Dev. 3, 170–205. doi: 10.1046/j.1525-142x.2001.003003170.x
Pigliucci, M., and Müller, G. B. (eds.) (2010). Evolution, The Extended Synthesis. Cambridge: MIT Press. doi: 10.7551/mitpress/9780262513678.001.0001
Randel, N., Asadulina, A., Bezares-Calderón, L. A., Verasztó, C., Williams, E. A., Conzelmann, M., et al. (2014). Neuronal connectome of a sensory-motor circuit for visual navigation. eLife 3:e02730. doi: 10.7554/eLife.02730
Redl, E., Scherholz, M., Todt, C., Wollesen, T., and Wanninger, A. (2014). Neurogenesis in Solenogastres (Mollusca) reveals putative ancestral lophotrochozoan features. EvoDevo 5:48. doi: 10.1186/2041-9139-5-48
Richter, S., and Wirkner, C. S. (2014). A research program for evolutionary morphology. J. Zoolog. Syst. Evol. Res. 52, 338–350. doi: 10.1111/jzs.12061
Sarrazin, A. F., Peel, A. D., and Averof, M. (2012). A segmentation clock with two-segment periodicity in insects. Science 336, 338–341. doi: 10.1126/science.1218256
Scherholz, M., Redl, E., Wollesen, T., Todt, C., and Wanninger, A. (2013). Aplacophoran molluscs evolved from ancestors with polyplacophoran-like features. Curr. Biol. 23, 2130–2134. doi: 10.1016/j.cub.2013.08.056
Scholtz, G. (2005). Homology and ontogeny: pattern and process in comparative developmental biology. Theory Biosci. 124, 121–143. doi: 10.1007/BF02814480
Scholtz, G. (2010). Deconstructing morphology. Acta Zool. 91, 44–63. doi: 10.1111/j.1463-6395.2009.00424.x
Scotland, R. W., Olmstead, R. G., and Bennett, J. R. (2003). Phylogeny reconstruction: the role of morphology. Syst. Biol. 52, 539–548. doi: 10.1080/10635150309309
Semmler, H., Bailly, X., and Wanninger, A. (2008). Myogenesis in the basal bilaterian Symsagittifera roscoffensis (Acoela). Front. Zool. 5:14. doi: 10.1186/1742-9994-5-14
Semmler, H., Chiodin, M., Bailly, X., Martinez, P., and Wanninger, A. (2010). Steps towards a centralized nervous system in basal bilaterians: insights from neurogenesis of the acoel Symsagittifera roscoffensis. Dev. Growth Diff. 52, 701–713. doi: 10.1111/j.1440-169X.2010.01207.x
Semmler, H., and Wanninger, A. (2010). Myogenesis in two indirect developing platyhelminths, Pseudoceros canadensis and Stylostomum sanjuania. Evol. Dev. 12, 208–219. doi: 10.1111/j.1525-142X.2010.00405.x
Smith, S., Wilson, N. G., Goetz, F., Feehery, C., Andrade, S. C. S., Rouse, G. W., et al. (2011). Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480, 364–367. doi: 10.1038/nature10526
Sørensen, M. V., and Giribet, G. (2006). A modern approach to rotiferan phylogeny: combining morphological and molecular data. Mol. Phylogenet. Evol. 40, 585–608. doi: 10.1016/j.ympev.2006.04.001
Stach, T., Winter, J., Bouquet, J.-M., Chourrout, D., and Schnabel, R. (2008). Embryology of a planktonic tunicate reveals traces of sessility. Proc. Natl. Acad. Sci. U.S.A. 105, 7229–7234. doi: 10.1073/pnas.0710196105
Stegner, M. E. J., and Richter, S. (2015). Development of the nervous system in Cephalocarida (Crustacea): early neuronal differentiation and successive patterning. Zoomorphology 134, 183–209. doi: 10.1007/s00435-014-0248-1
Temereva, E. N. (2011). Ventral nerve cord in Phoronopsis harmeri larvae. J. Exp. Zool. B Mol. Dev. Evol. 318, 26–34. doi: 10.1002/jez.b.21437
Ueda, N., and Boettcher, A. (2009). Differences in heat shock protein 70 expression during larval and early spat development in the Eastern oyster, Crassostrea virginica (Gmelin, 1791). Cell Stress Chaperones 14, 439–443. doi: 10.1007/s12192-008-0096-3
Ueda, N., and Degnan, S. M. (2013). Nitric Oxide acts as a positive regulator to induce metamorphosis of the ascidian Herdmania momus. PLoS ONE 8:e72797. doi: 10.1371/journal.pone.0072797
Vedel, V., Chipman, A. D., Akam, M., and Arthur, W. (2008). Temperature-dependent plasticity of segment number in an arthropod species: the centipede Strigamia maritima. Evol. Dev. 10, 487–492. doi: 10.1111/j.1525-142X.2008.00259.x
Vinther, J., Sperling, E. A., Briggs, D. E. G., and Peterson, K. J. (2012). A molecular palaeobiological hypothesis for the origin of aplacophoran molluscs and their derivation from chiton-like ancestors. Proc. R. Soc. B 279, 1259–1268. doi: 10.1098/rspb.2011.1773
Voronezhskaya, E. E., Tyurin, S. A., and Nezlin, L. P. (2002). Neuronal development in larval chiton Ischnochiton hakodadensis (Mollusca: Polyplacophora). J. Comp. Neurol. 444, 25–38. doi: 10.1002/cne.10130
Voronezhskaya, E. E., Tsitrin, E. B., and Nezlin, L. P. (2003). Neuronal development in larval polychaete Phyllodoce maculata (Phyllodocidae). J. Comp. Neurol. 455, 299–309. doi: 10.1002/cne.10488
Wanninger, A. (2007). “The application of confocal microscopy and 3D imaging software in functional, evolutionary, and developmental zoology: Reconstructing myo- and neurogenesis in space and time,” in Modern Research and Educational Topics in Microscopy, 3rd edition, Vol.1: Applications in Biology and Medicine, eds A. Méndez-Vilas and J. Diaz J (Bardajoz: Formatex), 353–361.
Wanninger, A. (2008). Comparative lophotrochozoan neurogenesis and larval neuroanatomy: recent advances from previously neglected taxa. Acta Biol. Hung. 59(Suppl.), 127–136. doi: 10.1556/ABiol.59.2008.Suppl.21
Wanninger, A. (2009). Shaping the things to come: ontogeny of lophotrochozoan neuromuscular systems and the Tetraneuralia concept. Biol. Bull. 216, 293–306.
Wanninger, A. (ed). (2015). Evolutionary Developmental Biology of Invertebrates, Vols. I-VI. Wien: Springer (in press).
Wanninger, A., and Haszprunar, G. (2002a). Chiton myogenesis: perspectives for the development and evolution of larval and adult muscle systems in molluscs. J. Morphol. 251, 103–113. doi: 10.1002/jmor.1077
Wanninger, A., and Haszprunar, G. (2002b). Muscle development in Antalis entalis (Mollusca, Scaphopoda) and its significance for scaphopod relationships. J. Morphol. 254, 53–64. doi: 10.1002/jmor.10004
Wanninger, A., Koop, D., Bromham, L., Noonan, E., and Degnan, B. M. (2005). Nervous and muscle system development in Phascolion strombus (Sipuncula). Dev. Genes Evol. 215, 509–518. doi: 10.1007/s00427-005-0012-0
Wanninger, A., Kristof, A., and Brinkmann, N. (2009). Sipunculans and segmentation. Commun. Integr. Biol. 2, 56–59. doi: 10.4161/cib.2.1.7505
Wanninger, A., Ruthensteiner, B., Lobenwein, S., Salvenmoser, W., Dictus, W. J. A. G., and Haszprunar, G. (1999). Development of the musculature in the limpet Patella (Mollusca, Patellogastropoda). Dev. Genes Evol. 209, 226–238. doi: 10.1007/s004270050247
Wurzinger-Mayer, A., Shipway, R., Kristof, A., Schwaha, T., Cragg, M. S., and Wanninger, A. (2014). Developmental dynamics of myogenesis in the shipworm Lyrodus pedicellatus (Mollusca: Bivalvia). Front. Zool. 11:90. doi: 10.1186/s12983-014-0090-9
Zieger, E., Bräunig, P., and Harzsch, S. (2013). A developmental study of serotonin-immunoreactive neurons in the embryonic brain of the Marbled Crayfish and the Migratory Locust: evidence for a homologous protocerebral group of neurons. Arthropod Struct. Dev. 42, 507–520. doi: 10.1016/j.asd.2013.08.004
Keywords: developmental biology, evolution, evolutionary synthesis, zoology, Haeckel, Hatschek, phylogeny, bioscience
Citation: Wanninger A (2015) Morphology is dead – long live morphology! Integrating MorphoEvoDevo into molecular EvoDevo and phylogenomics. Front. Ecol. Evol. 3:54. doi: 10.3389/fevo.2015.00054
Received: 28 February 2015; Accepted: 06 May 2015;
Published: 27 May 2015.
Edited by:
Angelika Stollewerk, Queen Mary University of London, UKReviewed by:
Cedric Boeckx, Universitat de Barcelona, SpainStuart A. Newman, New York Medical College, USA
Copyright © 2015 Wanninger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Wanninger, Department of Integrative Zoology, University of Vienna, Althanstrasse 14, 1090 Vienna, Austria, andreas.wanninger@univie.ac.at
 Andreas Wanninger
Andreas Wanninger