Reconsidering regeneration in metazoans: an evo-devo approach
- Centre National de la Recherche Scientifique, Sorbonne Universités, UPMC Univ Paris 06, Laboratoire de Biologie du Développement de Villefranche-sur-mer, Observatoire Océanographique, Villefranche-sur-mer, France
Regeneration of body structures is an ability widely but unevenly distributed amongst the animal kingdom. Understanding regenerative biology in metazoans means understanding the multiplicity of the cellular and molecular mechanisms that lead to the differentiation, morphogenesis and ultimately the development of a particular regenerating unit. In this manuscript we critically assess the evolutionary considerations suggesting that regeneration is an ancestral trait rather than a mechanism independently evolved in different taxa. As a general method to test evolutionary hypothesis on regeneration, we propose mechanistically dissecting the regenerative processes according to its conserved chronological steps: wound healing, mobilization of cell precursors and morphogenesis. We then suggest interpreting regenerative biology from an evo-devo perspective, proposing a possible theoretical framework and experimental approaches without necessarily invoking a common origin or only multiple losses of regenerative capabilities.
Introduction
On the 25th November 1740, Abraham Trembley first cut a polyp in two, initiating a modern research program on regeneration, a phenomenon that continues to fascinate and mystify today. Long studied, but poorly understood an enduring question has been the explanation of its phylogenetic distribution, i.e., the uneven distribution among metazoans of the ability to regenerate tissues, organs, parts of or even whole bodies. For a good part of the past century it was thought that regenerative potential was confined to a few invertebrate taxa. Today it is clear that this is not the case, and some sort of regenerative ability is widespread, although unevenly distributed across the animal kingdom (Carlson, 2007).
In the last decades, advances in molecular biology, have renewed interest in the phenomena of regeneration. The literature on this topic has been enriched with new laboratory models thanks to adaptable technical protocols and more affordable access to genomic and transcriptomic approaches (Vickery et al., 2001; Candia Carnevali, 2006; Kürn et al., 2011; Bely, 2014; Jeffery, 2015). Knowledge of more classical models such as planarians, annelids, and urodela has grown, providing important findings on the cellular and molecular dynamics of their regeneration potentials (Alvarado and Tsonis, 2006; Tanaka and Reddien, 2011; Bely, 2014). This renewed interest in regeneration has also included models like Drosophila and C. elegans, where highly developed technical tools present novel channels of enquiry, despite limited regenerative response to injury triggered in their adult form (Ghosh-Roy and Chisholm, 2010; Worley et al., 2012).
Consequently, diverse data on regenerative responses are being collected on a variety of taxa at a great rate (Alvarado and Tsonis, 2006; Brockes and Kumar, 2008; Bely and Nyberg, 2010). Molecular information complements anatomical and morphological descriptions of the regenerative events widespread in the animal kingdom (Vorontsova and Liosner, 1961), and raises the possibility of answering key questions regarding the underlying similarities of these processes. Regeneration is not, however, a universal feature of metazoans. Answering evolutionary questions about regeneration can provide a wider and deeper interpretation of the phenomenon (Brockes et al., 2001), mainly by comparing regenerating versus non-regenerating structures and animals. There is however, a widespread tendency to reduce a complex and highly diverse phenomenon to a unique trait. This trait is then mapped onto animal phylogenies as a single character, and its presence in the last common ancestor of animals appears self-evident, with the variability of “regenerative potential” among the animal kingdom typically explained by invoking adaptive mechanisms of its maintenance and secondary loss (Brockes et al., 2001; Brockes and Kumar, 2008; Bely, 2010; Bely and Nyberg, 2010). Although it is possible, even probable, that the last common ancestor of extant animals possessed some level of broadly defined regenerative ability, and that aspects of the regenerative abilities of contemporary animals are, in some sense, historically continuous with this, the evidence that such a view can be transferred to the level of cellular and molecular mechanisms needs to be critically assessed. If these cellular and molecular mechanisms of regeneration are not conserved, and the parts being regenerated are not conserved, is there anything to be gained by considering the phenomena homologous? The arm of a starfish is not directly comparable to the leg of a salamander. Although it may be true that the phenomenon of regeneration is shared between the two (if cut off, both will grow back), it is unclear what we might mean if we say it is conserved, unless we can identify component mechanisms in common between the two. The problem is loosely analogous to that of deciding whether the eyes of insects and vertebrates are homologous: we can infer from the shared presence of eyes in many animal phyla that their common ancestor had some sort of eye, but we must carefully define and breakdown the developmental and structural components to identify which, if any, are shared by descent from that ancestor. It is precisely this methodology that has allowed the evo-devo field to discuss eye homology in a more nuanced manner than “eyes have evolved x times independently.”
We examine and critically assess the idea that regeneration might be an ancestral trait, as opposed to the possibility that important aspects of regenerative phenomena may have independent origins in different lineages. In particular, we suggest that, as a complex trait, it is unhelpful to consider the distribution of regeneration on phylogenetic trees without first mechanistically dissecting the overall process according to its conserved chronological steps: wound healing, mobilization of cell precursors and morphogenesis. We advocate interpreting regenerative biology from an evo-devo perspective; we propose one such framework and experimental approaches.
Homoplasies and Homologies
The term regeneration is a label attached to many different molecular and cellular phenomena. It covers an extensive and heterogeneous array of processes present in metazoans that have the common result of restoring, partially or totally, a lost body part, at levels covering cells, tissues, or organs, to the extent of the entire body.
We can deconstruct regeneration, comparing it among taxa and following its various evolutionary trajectories, by dividing it into sequential mechanistic events. Our breakdown is biased by events/phases that have been described in the literature as conserved between various taxa. We thus split the regenerative process into wound healing (that is, repair of epithelia), followed by an activation of originators, or precursor, and developing with a series of morphogenetic events that lead to the total or partial reconstitution of the lost portion of the body. This approach is particularly suitable if we consider a level of structural complexity that goes beyond the tissue level, i.e., organ level, body appendages or whole bodies.
Wound Healing
Injuries that lead to the disruption of a certain structure, also involve the disruption of some kind of epithelia. Wound repair mechanisms lead to a prompt re-epithelialization in order to restore a barrier against pathogens, to prevent loss of cells or tissues or, for example in aquatic organisms, to prevent osmotic shocks. The initial phases of wound healing, such as transcription-independent cellular effects like cell shape changes and formation of functional actomyosin structures triggered by the diffusion through the epithelial tissues of Ca2+, hydrogen peroxide (H2O2) and ATP are shared by almost all metazoans (Cordeiro and Jacinto, 2013). Such responses are tightly bound to some sort of “immune response,” i.e., the infiltration in the wound area of immune cells. This varies between organisms, and ranges from the transient intervention of amoeboid cells in cnidarians to the complex adaptive immune response of vertebrates (Kawakami and Nakanishi, 2001; Eming et al., 2009; Nakanishi et al., 2011; Palmer et al., 2011; Gold and Jacobs, 2013; Dubuc et al., 2014; Wenger et al., 2014). The relationship between wound healing, particularly the immune response phase, and the regenerative event is not clear, and is probably not a conserved feature (see next section). Regardless of whether it is followed by full blown regeneration of underlying damaged structures, or simply barrier repair, wound healing is ubiquitous in metazoans, and it is not unreasonable to assume that this is a primitive feature, present in the earliest animals, with aspects conserved in modern animals. Grainyhead transcription factor family members, for instance, are needed for epidermal barrier repair in the fly and in the mouse, controlling expression of proteins involved in cross-linking barrier components. Although this suggests a repair role was present in the bilaterian ancestor, whatever type of epidermis it may have had, the fact that Grainyheads also have a role in epidermal barrier formation during development, means that, on the evidence presented, it is not possible to rule out independent recruitment to repair (Mace et al., 2005; Ting et al., 2005).
Precursors: Origin of Regenerating Elements
The source of the precursors, i.e., the regenerating cells, is one of the clearest processes of regeneration that can be evaluated in an evolutionary framework in order to compare the multiple mechanisms adopted to restore a structure. The source of regenerating cells falls into three main categories: (1) proliferation and differentiation of adult stem cells, either present in the injured tissues or migrating from another region of the body; (2) dedifferentiation or trans-differentiation of somatic cells present in the remaining tissue; or (3) proliferation of cells from the remaining differentiated tissues (Carlson, 2007).
The role of some sort of somatic stem cells, carrying different degrees of potency, has been documented during regenerative events in different taxa across metazoans. Totipotent archeocytes, have been described in demosponges (Funayama, 2013; Alié et al., pers. comm.). These cells express a molecular signature, including DNA-integrity protection systems with homologs of Piwi, Nanos, Vasa, Tudor, which characterize “stemness” in both germline and somatic multi- or pluripotent stem cells across metazoans (Önal et al., 2012; Solana, 2013; Alié, pers. comm.). However there is still a lack of information about the uneven distribution of regenerative capabilities among different species of poriferans and their different, or similar, mechanisms of regeneration (Wulff, 2010, 2013), so the significance of these stem cells for regeneration is still not completely clear.
In cnidarians, regenerative capability has been reported across the whole phylum (Technau and Steele, 2011). However, multipotent somatic cells, called interstitial cells (i-cells), have only been described in hydrozoans (Leclère et al., 2012; Gold and Jacobs, 2013) where they can coexist with other restricted stem cells capable of differentiation into more specialized cell types (Müller et al., 2004; Hobmayer et al., 2012; Gold and Jacobs, 2013). Homologues of i-cells seem to be absent in other classes of cnidarians. Taken together with their phylogenetic position it is likely that i-cells are a derived feature of hydrozoans, rather than ancestral to the cnidarians (Technau and Steele, 2011; Gold and Jacobs, 2013). Instead, other mechanisms seem to provide the regenerative cells in the other classes. Proliferation of cell populations with a restricted fate seems to be the source of the multiple cell types necessary for the regeneration of the lost structure in some species of anthozoans (Passamaneck and Martindale, 2012). Particularly, a population of lineage-restricted multipotent neural precursors, expressing SoxB, have been identified in Nematostella (Richards and Rentzsch, 2014). Regeneration in cnidarians also occurs via cell transdifferentiation as reported for the regeneration of striated muscle in the hydrozoan jellyfish (Galle et al., 2005), and also in Hydra, where this coexists with stem cell based mechanisms (Siebert et al., 2008).
Two of the major clades of regenerating animals are found among flatworms. Within the platyhelminthes, the classic regenerative model Schmidtea mediterranea belongs to Tricladida. Recent phylogenetic work suggests this order is relatively late branching within the phylum as a whole, forming a clade with Bothrioplanida, and Neodermata united by a lack of spiral cleavage and centrosomes (Egger et al., 2015; Laumer et al., 2015), suggesting its derived nature. Neoblasts, however, and regenerative capabilities in general are also present in the more basally branching orders Macrostomorpha and Catenulida (Dirks et al., 2012; Martín-Durán and Egger, 2012). Egger and co-workers note that most flatworm taxa lack the ability to regenerate brain, eyes, pharynx and statocyst (Egger et al., 2007). The other major clade of regenerating flatworms are the acoels, where at least one species shares the regenerative mechanisms of planarians, i.e., the presence of neoblasts, defined by their behavior and by the expression of Piwi, and conserved toolkit of axes formation (De Mulder et al., 2009; Srivastava et al., 2014). The relationship between acoela and platyhelminth regeneration is discussed below.
Among ecdysozoans, cellular and molecular studies on regeneration have been less extensive and have mainly looked at limb regeneration in adult arthropods (Maginnis, 2006), and regeneration of imaginal discs in Drosophila (Worley et al., 2012). A recent study suggests the role of lineage-specific cells for limb regeneration in the crustacean Parhyale hawaiensis. In particular, similarly to vertebrates, a population of Pax3/7-expressing muscle satellite cells seems to be directly involved in muscle regeneration (Konstantinides and Averof, 2014).
In deuterostomes, the origin of regenerative cells appears to be heterogeneous. Colonial ascidians seem to employ totipotent stem cells, expressing Piwi and Vasa markers, during whole body regeneration in botrillidae (Brown et al., 2009a), but in other species cell transdifferentiation is implicated (Kawamura et al., 2013), or even the coexistence of multiple systems, involving the presence and dedifferentiation of multipotent epithelial cells and mobile cells (Kawamura et al., 2008; Tiozzo et al., 2008). In Ciona intestinalis, a solitary species of ascidian, the regeneration of the oral siphon involves the local differentiation of progenitor cells whose identity is uncertain (Auger et al., 2010; Jeffery, 2015). In vertebrates the modus operandi of the regenerative precursor differs. Among teleosts, zebrafish can successfully regenerate heart, retina and fins. Cell dedifferentiation, transdifferentiation and proliferation of cardiomyocytes drives the regeneration of the cardiac muscle (Jopling et al., 2010). Retinal regeneration is tuned to regenerate specific cell types that have been damaged, starting from a subset of glial cells (Müller glia), which dedifferentiate and re-enter mitosis to produce multipotent neural progenitors (Gemberling et al., 2013). Fin regeneration involves the formation of a blastema and while it is not possible to exclude rare cell transdifferentiation events, it seems that the regenerative cells are lineage restricted, i.e., osteoblasts, epidermis, endothelial cells and fibroblasts (Gemberling et al., 2013). Similar lineage restrictions have been observed in mouse digit tip and axolotl limb regeneration (Kragl et al., 2009; Lehoczky et al., 2011).
The cellular nature of the precursors of the different regenerative events seems to fall into three fundamental scenarios, (1) the presence of somatic cells that retain stemness and different degrees of potencies, (2) the ability of differentiated cells to de- or transdifferentiate, or (3) the presence of differentiated cells that replace those missing by proliferation. These different mechanisms are scattered among metazoans, sometimes co-existing in the same species, and their evolutionary history is still far from being resolved.
Morphogenetic Events in Regeneration
The morphogenetic phase of regeneration is the macroscopic reconstruction of the lost part. Although it is not always clearly identifiable, it is convenient to define a boundary between the end of precursor mobilization, and the beginning of the actual phase of morphogenesis. This phase encompasses the processes of cell fate control, fixation/implementation of positional memory, re-patterning and growth control. The final result is the rebuilding of the same, or a similar, functional structure which was present before the injury. Morphogenesis during regeneration often closely resembles the morphogenesis of the same structure during embryonic development and during tissue renewal in normal body homeostasis. For instance retinoic acid (RA), FGFs and Wnts are highly conserved signaling pathways involved in a multitude of cellular processes that range, for example, from regulation of cell proliferation or apoptosis to tuning stem cell behavior (Clevers et al., 2014). It has been shown that Wnts for example, coordinate tissue dedifferentiation and also tune progenitor cells during structure regeneration in cnidarians, planarian, amphibians and teleosts (Guder et al., 2006; Reddien, 2011; Jager et al., 2013; Lin et al., 2013; Wehner et al., 2014). Positional information of blastema cells during limb regeneration in amphibians is affected by retinoids, mainly by influencing the expression of homeobox genes (Gardiner et al., 1995; Mercader et al., 2005). RA also has a role in the transdifferentiation of epithelial cells during gut regeneration, budding, and whole body regeneration in colonial ascidians (Hisata et al., 1998; Kamimura et al., 2000; Rinkevich et al., 2007). By directing intracellular signals, the Hippo pathway intersects directly with several master signaling cascades and acts as an integration point, particularly maintaining the correct organ size during development and homeostasis across many metazoans (Pan, 2007; Mauviel et al., 2011; Varelas and Wrana, 2012). The Hippo pathway, and its effectors YAP and TAZ, act as mediators of other protein signaling pathways that influence cell-fate, proliferation, apoptosis, cell shape and cell movement. However, the same molecules also have a role in regulating stem cell self-renewal and expansion in regenerative processes in different phyla (Zhao et al., 2011; Demircan and Berezikov, 2013; Johnson and Halder, 2014; Lin and Pearson, 2014). Wnt, BMP and Nodal represent the signaling pathways defining the body axes during embryonic development, at least in bilaterians. These pathways were possibly present in the most recent common ancestor of all metazoans (reviewed in Holstein et al., 2011) and their involvement in whole body or body structure re-patterning has been reported in cnidarians, acoel, platyhelminthes, and deuterostomes (Holstein et al., 2003; Yakushiji et al., 2009; Adell et al., 2010; Reddien, 2011; Srivastava et al., 2014; Watanabe et al., 2014).
The use of the canonical developmental signaling pathways during re-patterning and morphogenesis clearly suggests that regeneration is an epiphenomenon of development. It is commonplace to note that repeated use of similar genetic pathways involving pre-existing building blocks is a recurring theme in evolution, and there is abundant evidence that this is also the case in regeneration. As such, the distinctive feature of a regenerative research program is the identification of the molecular mechanisms that activate and regulate such developmental programs in a context other than development. Despite the presence of conserved molecular modules, when looking at regeneration of body and structures in metazoans we come across dramatically different types of morphogenetic phenomena. The cellular and molecular basis of positional memory and their comparative aspects in different taxa are not well understood. In regenerating planarians, for example, expression dynamics of Wnt/β-catenin and BMP signaling pathways, which control anterior-posterior and dorso-ventral axes regeneration, respectively, seem to confirm the establishment of morphogenetic gradients as mechanisms of re-patterning, as suggested originally by Morgan (Morgan, 1901; Adell et al., 2010; Umesono et al., 2013). How neoblasts and cells in the blastema can interpret the gradients and whether these gradients are present in the adult or induced by injury is still uncertain. Similar mechanisms of gradient generation are present in Hydra, during homeostasis, head and foot regeneration (Bode, 2009; Chera et al., 2009; Galliot and Chera, 2010). In a histologically more complex structure, like the axolotl limb, it seems that the proximo-distal positional identity is achieved by the cells present in the blastema, without involving a morphogenetic gradient (Kragl et al., 2009). Very little is known about the morphogenetic mechanisms, i.e., the re-patterning, occurring during structure regeneration in other metazoans, making it essentially impossible to determine whether this key step is likely to be conserved between different phyla, or whether, for instance, different taxa respond to non-homologous positional cues. One possible approach to highlight mechanisms of morphogenesis could be to focus on the degree of restoration of the lost structure by comparing hypomorphic or heteromorphic regeneration in closely related species (Carlson, 2007).
Regeneration and Ancestral Animals: Insights from Flatworms
Flatworms have a central role in regeneration studies, and historically have been used as proxies for the ancestral bilaterian. Although once grouped together, modern molecular analysis places acoels (or xenacoelomorphs) and platyhelminthes in quite distinct phylogenetic positions, the xenacoelomorphs being either the sister group of bilaterians (Hejnol et al., 2009) or the deuterostomes (Philippe et al., 2011), whereas the platyhelminthes robustly nest within the spiralia/lophotrochozoa (Dunn et al., 2008). With either position of xenacoelomorphs, if the regenerative abilities they share with Platyhelminthes are homologous, it would suggest that these abilities were present in the last common ancestor of bilaterian animals.
The features of this common ancestor are, however, the subject of much debate. Shared morphological characters and gene expression patterns have led to the view that it may have been a relatively large segmented coelomate, with various other features such as a complex centralized nervous system including a tripartite brain, a regionalized gut (for a brief review see Tweedt and Erwin, 2015) and perhaps an “axochord” (Lauri et al., 2014). As such, the flatworm body type of platyhelminthes and, depending on their phylogenetic position, xenacoelomorphs would be derived. If that is the case, it is not clear that the regenerative capabilities present in extant animals like flatworms would have truly homologous equivalents in the last common bilaterian ancestor, or that this ancestor was adept at regenerating its more complex body structures. This is an area of some uncertainty: recent phylogenetic results have questioned the complexity of the ancestral spiralian, suggesting it may have been a small acoelomate animal like a flatworm (Struck et al., 2014). Depending on the phylogenetic position of the xenacoelomorphs, this does add weight to the argument for a flatworm-like last common ancestor of bilaterians. Regeneration within the major groups of spiralia has been reviewed by Bely (Bely et al., 2014), but the new phylogeny, with paraphyletic platyzoa suggests a focus on gastrotrichs and the gnathiferan groups could shed light on the regenerative properties of the spiralian, and hence the bilaterian common ancestor (Struck et al., 2014).
If, on the other hand, the derived nature of the flatworm body plan were to be confirmed, it would seem more than coincidental that in both cases the simplified morphology comes along with asexual reproduction and exceptional regenerative properties, suggesting that, to make a successful organismal strategy, these traits need to be integrated.
An Evo-Devo Approach to Regeneration
The fact that different taxa adopt different molecular and cellular strategies to rebuild a lost structure does not explain why, even within families or genera, the regenerative response to injury varies so much. We suggest that, in order to understand regeneration in metazoans, either as a general phenomenon or as a mix of phenomena, we compare it to the study of the development of animal forms. The cellular and molecular mechanisms occurring during regeneration can be treated in the same manner as ontogenetic phenomena, and therefore conceptual approaches similar to those employed in comparative and evolutionary developmental biology can be applied. If we accept such parallelism, regeneration is faced with the same set of problems and challenges as evolutionary and developmental biology, for instance the complexities of the genotype-phenotype map, i.e., the degree of phenotypic diversity among metazoans, yet relatively conserved sequence repertoires (Wagner and Zhang, 2011; Bozorgmehr, 2014). Regenerative events do not escape from such developmental and evolutionary dynamics, and concepts like pleiotropy, exaptation, developmental constraints and variational properties can provide a useful framework to better understand the phenomena of regeneration and their evolution. Furthermore, the challenges and the efforts to fit “regeneration” into a phylogenetic tree lead to more general challenges linked to evolutionary biology itself, for instance the definition of homologies, the circumscription of a character, the delineation of units of selection, and the targeting of the level of selection. And ultimately, given the difficulty of pinning down exact origins of regeneration, the comparative study of these events among different species should be outlined in terms of evolutionary trajectories rather than origins (Minelli, 2015), especially if one of the aims directing regenerative biology is to learn something about the limited facility our species has.
Modularity, Canalization and Evolvability of Regenerative Mechanisms
Here we sketch a simplified outline to explain the high variability of regenerative capabilities within relatively phylogenetically close species by invoking theoretical concepts familiar to evo-devo, such as modularity, canalization and evolvability (Bolker, 2000; Hendrikse et al., 2007; Wagner and Zhang, 2011).
We consider a species A (Figure 1) that evolved a robust developmental mechanism (R), e.g., a gene toolkit or a gene network able to produce a cell behavior or a pattern in space. The module R is canalized, i.e., it is resistant to perturbation and it proceeds along certain preferential directions, and therefore it will have a higher chance to be selected and conserved during speciation. Species A also contains a module P (plastic), influencing/interacting with module R. Module P, however, is in a broad sense more plastic and has a higher ability to evolve (Pigliucci, 2008). Let us assume that species A has high ability to regenerate certain structures, competency given by the interaction of R and P. The species B and C, share A as a common ancestor and the same module R, e.g., orthologs of a gene or a gene toolkit. However, selective pressure influenced module P differently in the two species, maintaining the capability to regenerate in B and limiting this capability in C. The species D and E, descending from B and the species F and G, descending from C, all share the ancestral module R but again, different environmental conditions acted on module P modifying the regenerative capabilities, depriving E and conferring G with regenerative ability (Figure 1). Then, observing closely related species D-E we find different regenerative capabilities, which are linked to the evolvability of a particular module (P). The results are both losses and (re)gains of the regenerative phenotype regardless of whether the common ancestor A could or could not regenerate. In the present conceptual framework the modules could assume the most diverse nature, e.g., gene toolkits, signaling pathways or regulatory elements.
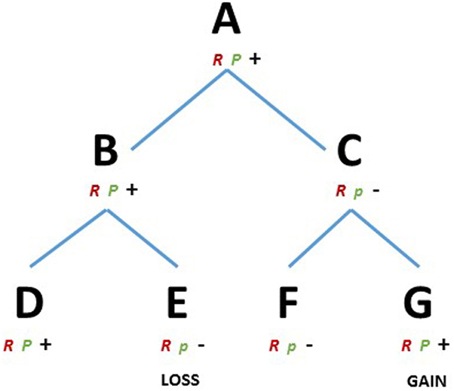
Figure 1. RP theoretical framework. D, E, F, and G represents existing metazoans species and A, B and C their ancestors, respectively. R, robust module; P, plastic module, majuscule, and minuscule represents two variants of the module P. Plus and minus represents the capacity or the lack of regenerative ability, respectively.
An example of this theoretical framework could be the mechanisms suggested to track the evolution of stem cells. Solana (2013), introduced the concept of primordial stem cells (PriSC), an ancestral and evolutionary conserved kind of stem cell that carries germ plasm components from the zygote to the germ cells. The PriSC is characterized by the expression of a set of genes, for instance Vasa, Nanos, Piwi, Tudor, which constitute a conserved germline multipotency program, GMP (Juliano et al., 2010; Solana, 2013; Alié, pers. comm.). However, this program is expressed not only in the germline, but is also essential in organisms that develop asexually and (not coincidentally) possess high regenerative capabilities (Martinez et al., 2005). The GMP is expressed in tissues and cells classically considered to be somatic but that in this case represent the “regenerative cells,” for example in planarians (neoblasts, Guo et al., 2006; Reddien et al., 2005; Rossi et al., 2008), hydrozoan cnidarians (I-cells, Mochizuki et al., 2001; Plickert et al., 2012; Juliano et al., 2014), ctenophores (Alié et al., 2011), polychaetes (in the mesodermal posterior growth zone, Rebscher et al., 2007; Giani et al., 2011) and ascidians circulating putative stem cells, (Brown et al., 2009a,b; Sunanaga et al., 2010). In fact, according to the primordial stem cell hypothesis, PriSCs can exist in a “rudimentary state,” and contribute only to the germline, or in an “unlimited state” and contribute, to differing extents, to somatic tissues (Solana, 2013). The conserved GMP can be considered a robust (R) module that arose in an early multicellular ancestor animal, perhaps in response to selection pressure for preservation of genome integrity (Ghildiyal and Zamore, 2009; Bagijn et al., 2012; Xiol et al., 2014). It is an essential condition for somatic stem cells, but its presence tells us nothing about regenerative capabilities. In organisms where the onset of regeneration is based on some sort of somatic stem cells, the PriSCs change only few characteristics. The mechanisms that determine if a PriSC is “unlimited” or “rudimentary” are not known. Perhaps, the presence of highly evolvable module P, that regulates the activation of the GMP, could provide an evolutionary framework to explain the multiple transitions observed in regeneration. It is worth remarking that preformation (zygotic germ plasm is inherited by primordial germ cells and not somatic cells) and epigenesis (a subpopulation of somatic cells is specified to become primordial germ cells), the two classic models of germ cell specification, have a scattered distribution across metazoans (Extavour and Akam, 2003), reminiscent of regeneration of body structures.
If we hypothesize that, the mechanism that segregates the germ plasm to somatic stem cells rather than exclusively to germline is driven by a highly evolvable module (P), then we can speculate about the existence of a correlation between the scattered distribution of the role of somatic stem cells in regeneration and the modes of germ plasm segregation.
The RP conceptual framework could also be used to approach the last step of the regenerative process: morphogenesis. Once again, planarians are important models to understand variability of regenerative potential. It has been shown that the limited regenerative capabilities of the species Procotyla fluviatilis can be “reversed” by knocking down components of the Wnt/β-catenin pathway (Sikes and Newmark, 2013). Blocking the same pathway in the non-regenerating planaria Dendrocoelum lacteum and Phagocata kawakatsui triggers their head regeneration where naturally it does not occur (Liu et al., 2013; Umesono et al., 2013). Therefore, interfering with one single signaling pathway, particularly with its gradient of expression, can reverse evolutionary loss of regeneration. Although the ultimate causes leading to gain or loss of regeneration capabilities in planarians are at the moment purely speculative, these studies in different regenerating and non-regenerating species represent a milestone for the understanding of molecular mechanism.
This proposed dual-layer of different evolvability between two modules can be compared with the kernel theory of gene regulatory networks (GRN, Davidson and Erwin, 2015), which propose that the different components of the GRN evolve at different rates and in response to different selective pressures. The kernel theory proposes that the establishment of the basic phyla bauplan is partially due to the evolution of robust “kernels,” i.e., nodes of the network, which represent the robust layers and the diversity of body forms within phyla and these are driven by the plasticity of other layers of the network. A similar approach, also at the level of transcriptional regulation might explain the scatter of regenerative capability within phyla (Garfield et al., 2013).
Goss and Morgan before him, and most of the common literature, suggest, or imply, a monophyletic origin of regenerative abilities, followed by multiple losses (Morgan, 1901; Goss, 1992; Brockes and Kumar, 2008; Bely and Nyberg, 2010; Bely, 2010), each pleiotropically linked to adaptations more beneficial than regeneration (Goss, 1992). According to the proposed theoretical framework, the scattered capacity of regeneration among metazoans could be seen as losses, but also as gains, particularly when viewed at the intra-phylum level and considered alongside a complex ancestor to the bilateria (Figure 1). Such a view is made more likely if we consider regeneration as recruitment of pre-existing modules that have been conserved for other reasons during the course of evolution.
Constraints and Exaptations in Regeneration
Clearly, the heuristic and over-simplistic nature of the presented framework is far from explaining the distribution of regeneration, particularly among different phyla. Whereas it is true that there are only few comparative studies focused on loss, and gain, of regeneration capabilities (Bely and Nyberg, 2010), besides the wnt/β-catenin in planarians (Liu et al., 2013; Sikes and Newmark, 2013; Umesono et al., 2013), there is no evidence of the existence of master regulators of regeneration, i.e., genes or gene pathways that simply switch on or off the ability of a structure to regenerate. Regeneration is clearly a complex multistate character. Once again we can borrow some concepts from evo-devo and instrumentally use them to try to approach regeneration and its distribution. Together with the idea of modularity, comes the concept of developmental constraint, i.e., restraints on phenotype production due to limited interactions among existing modules (Gilbert, 2000; Richardson and Chipman, 2003; Gilbert and Epel, 2009). In this context we can consider the evolution of different developmental trajectories as potential constraints to regeneration. This may seem a circular argument but, as the characteristics of a particular developmental system impact the type of phenotypic space the descendants of that species can occupy, the same effects can apply to regeneration. For instance, increasing body complexity can generate physical and morphogenetic constraints to regenerative mechanisms. The presence of an exoskeleton, as in many ecdysozoan phyla could constrain the regeneration of a body structure just as, for example, it constitutes a limit for the increase of body size in insects (Nijhout and Emlen, 1998). Soft bodied animals, where the histological and cytological structure is more homogeneous could facilitate or restrain the propagation of morphogenetic gradients and bioelectrical signals through voltage gradients across the plasma membrane of cells (Levin, 2012; Levin and Stevenson, 2012; Beane et al., 2013). In fact, among metazoans, the evolution of particular body structures seems to interact at different levels during the regenerative processes. For example, in many regenerative contexts, both in vertebrates and invertebrates, the transient presence of regenerating nerves or axons is required for the correct behavior of progenitor cells and their niches (Kumar and Brockes, 2012). Recently, Witchley and collaborators showed that the set of genes that control positional information during regeneration in planarians, are expressed not in neoblasts, but in muscle cells, showing the role of musculature during re-patterning of the planarian body (Witchley et al., 2013). Vascular networks, and in particular endothelial cells, are emerging as important signaling centers, essential to coordinate whole body regeneration in ascidians (Brown et al., 2009a; Rinkevich et al., 2010) and in vertebrate organs (reviewed in Ramasamy et al., 2015). Evolution of different components of the extracellular matrix can also directly foster or inhibit regenerative processes. For instance, the metalloproteinases, a family of proteinases conserved across metazoans, seem to have been co-opted to different roles in regeneration, e.g., maintaining cell identity during Hydra foot regeneration (Leontovich et al., 2000), sea cucumber intestine regeneration (Quiñones et al., 2002) and planarian body regeneration (Isolani et al., 2013). In vertebrates, Calve and collaborators suggested hyaluronic acid, tenascin-C and fibronectin as part of a regeneration-specific transitional extracellular matrix during muscle regeneration in newt (Calve et al., 2010).
One innovation of vertebrates that has been related to their limited ability to regenerate complex structures is the evolution of the adaptive immune system. For example, appendage regeneration in anuran amphibians is inversely correlated with development, but is maintained in the adult life of urodeles. Tadpoles of the anuran Xenopus levis, during the so called “refractory period,” transiently lose their ability to regenerate the whole tail (Beck et al., 2003). Fukazawa and colleagues showed a transitory expression of immune-related genes in this period, the suppression of these genes or the depletion of immune cells restored regenerative abilities (Fukazawa et al., 2009). In anurans, the development of a complex adult adaptive immunity starts with metamorphosis and it is associated with a progressive loss of patterned regeneration. In urodeles, metamorphosis is linked to relatively minor changes in adaptive immunity (Godwin and Rosenthal, 2014). The adaptive immune response is an intricate process itself, and how it affects regeneration is largely uninvestigated. There is evidence for a correlation between the loss of regenerative capabilities and the immune response (Martin and Leibovich, 2005; Eming et al., 2009). The innate immune system, just like the evolution of any developmental feature that defines a particular body plan, can interfere with potential regenerative responses. In the process of tracking and comparing regenerative events among different taxa, it is difficult to avoid considering the probable independent evolution of these other constraints.
Adaptive Significance of Variations in Patterns of Regeneration
Treating regeneration(s) as a developmental process does not mean that we have to embrace the rigid view of this processes as a strictly pleiotropic epiphenomenon associated with development (Goss, 1992). Even if regenerative events often co-opt mechanisms and molecular tools from development and homeostasis, it does not mean that such events proceed irrespectively of their adaptive value (Morgan, 1901; Goss, 1992).
Studying the adaptability of regeneration is certainly not a simple task, however, in organisms where regeneration is tightly linked to asexual propagation, coloniality and their life history strategies (Berrill, 1951; Bely and Wray, 2001; Kawamura et al., 2008; Tiozzo et al., 2008; Burton and Finnerty, 2009; Brown and Swalla, 2012). It is likely that regeneration traits are subject to selection, mainly because they may be directly linked to reproductive success. For example, among ascidians the phylogenetic distribution of solitary species, which reproduce only sexually, and colonial species that can propagate both sexually and asexually, is scattered among different clades (Zeng and Swalla, 2005). Kott suggested that evolutionary transitions to asexual propagation in many ascidian lineages represent a derived condition in response to selective pressures for coloniality rather than an ancestral condition (Kott, 1981). If instead of focusing on its possible common origin, we consider the scattered distribution of regenerative traits, it is clear that, at least within particular clades like ascidians, we have to look at its adaptive role. Moreover, if we consider that regeneration capabilities can not only be lost, but also gained multiple times, for example by mechanisms like those proposed here, then direct selection can possibly act not only on its maintenance (Bely, 2010; Bely and Nyberg, 2010) but also on its evolutionary trajectories.
Another possible approach is to look at the trade-offs between regeneration of particular structures versus living without. For instance, in populations among many taxa, there is a well-documented range of structures that can be lost relatively frequently (reviewed by Bely and Nyberg, 2010). While the advantages of regenerating damaged portions of the body may seem clear, the costs of re-developing it are not. In order to attempt explaining the actual variations we have to take into account the “cost and benefits” of structure loss and regeneration. For example, the comparison of the costs of appendage regeneration and autotomy shown among a range of amphibians, reptiles, teleosts and arthropods suggests that the distribution of resources to restore lost appendages negatively affects both somatic growth and reproduction (Maginnis, 2006). It is likely that multiple factors influence the trade-offs, and hence the selective pressures associated with appendage regeneration. In order to understand the evolution of regenerative tendencies in term of adaptations it is essential to consider the ecological context and perhaps to focus on closely related species. For example, among species of Demospongiae chosen to represent four different orders, the susceptibility to damage by predators or by physical disturbance was inversely related to regeneration rate, suggesting trade-offs between sets of traits that promote regeneration and others that dispense with the need for regeneration (Hoppe, 1988; Wulff, 2010). Comparing similar wounds among different species showed that the variation in regeneration was linked to morphological characters and to the vulnerability to the damaging agents, but not to the phylogenetic relationships among the species studied (Wulff, 2010). There are only a few studies focusing on the possible adaptive role of regeneration, trying to identify the effective environmental agents of selection for a given taxon. Besides comparing closely related species, it is probably necessary to focus also on microevolutionary events, such as comparing individuals experiencing autotomy with or without regeneration.
Conclusion
The evidence against a universal and conserved program behind regeneration is compelling. Understanding regenerative biology in metazoans clearly means understanding the multiplicity of cellular and molecular mechanisms that lead to the activation of progenitor cells, morphogenesis and ultimately the development of a particular regenerating unit.
We suggest that in a modern context, Morgan's classical regeneration concepts of epimorphosis and morphallaxis may not be useful if we are trying to find evolutionary relationships among regenerative processes. Freshwater Planarians (Gurley et al., 2010) and even in Hydra (Galliot and Chera, 2010), two classical textbook example of pure morphallaxis, show how both processes can coexist. Not only can morphallaxis and epimorphosis occur in the same species, but different species belonging to the same phylum may adopt very different ways of regeneration, as is the case with cnidarians (Chera et al., 2009; Passamaneck and Martindale, 2012) or different species of polychaetes (Hill, 1970; Bely, 2014). Also, transient structures like the blastema are not always useful for comparative studies across distant taxa. Where blastemas can be characterized by a mass of cells clustering in the area of the wound after closure, leading to growth and regeneration into new organs or new body parts, it has been shown that the histological nature of this mass of cells is different in different regenerative structures (Shibata et al., 1999; Cebrià et al., 2002; Bosch et al., 2008; Kragl et al., 2009; Wenemoser and Reddien, 2010; Aboobaker, 2011; Reddien, 2013; Bely et al., 2014). Additionally in some epimorphic regenerative events, i.e., where local stimulation of cell proliferation precedes the development of the new part, there is a lack of structures that resemble a blastema, such as during the Ciona intestinalis siphon regeneration (Auger et al., 2010) or the whole body regeneration in colonial ascidians (Brown et al., 2009b).
To track evolutionary trajectories of regenerative events it may be more convenient to dissect the conserved chronological event that occurs during the restoration of a particular structure, i.e., wound healing, nature and mobilization of the precursors, and morphogenesis, and compare them in clades that at least come from the same phylum, or that belong to an even more restrained taxonomical clade, e.g., class, order or family. From this starting point regeneration is still a convenient term that covers a class of diverse molecular and cellular events, and it is useful to consider it recapitulating normal development, but it is the modifications and adaptations operating on top of this that are of central interest to regenerative biologists. Evolutionary novelties often arise through combinatorial processes (Sanetra et al., 2005). Therefore, the co-option of conserved genes and signaling pathways, together with the trans-acting regulatory factors and their epigenetic regulation (Yakushiji et al., 2007, 2009), could explain why different species have a regenerative capability without necessarily invoking a common origin or only multiple losses.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Alexandre Alié, Lucas Leclère, Chiara Serra, and Patrick Chang for their suggestions and the constructive discussions. Financial support was provided by AFMTelethon grant (#16611), IRG Marie Curie grant (#276974), ANR (ANR-14-CE02-0019-01) and IDEX Super (INDIBIO) to ST.
References
Aboobaker, A. A. (2011). Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol. 21, 304–311. doi: 10.1016/j.tcb.2011.01.005
Adell, T., Cebrià, F., and Saló, E. (2010). Gradients in planarian regeneration and homeostasis. Cold Spring Harb. Perspect. Biol. 2:a000505. doi: 10.1101/cshperspect.a000505
Alié, A., Leclère, L., Jager, M., Dayraud, C., Chang, P., Le Guyader, H., et al. (2011). Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of “germline genes” with stemness. Dev. Biol. 350, 183–197. doi: 10.1016/j.ydbio.2010.10.019
Alvarado, A. S., and Tsonis, P. A. (2006). Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet. 7, 873–884. doi: 10.1038/nrg1923
Auger, H., Sasakura, Y., Joly, J.-S., and Jeffery, W. R. (2010). Regeneration of oral siphon pigment organs in the ascidian Ciona intestinalis. Dev. Biol. 339, 374–389. doi: 10.1016/j.ydbio.2009.12.040
Bagijn, M. P., Goldstein, L. D., Sapetschnig, A., Weick, E.-M., Bouasker, S., Lehrbach, N. J., et al. (2012). Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578. doi: 10.1126/science.1220952
Beane, W. S., Morokuma, J., Lemire, J. M., and Levin, M. (2013). Bioelectric signaling regulates head and organ size during planarian regeneration. Development 140, 313–322. doi: 10.1242/dev.086900
Beck, C. W., Christen, B., and Slack, J. M. W. (2003). Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev. Cell 5, 429–39. doi: 10.1016/S1534-5807(03)00233-8
Bely, A. E., and Wray, G. A. (2001). Evolution of regeneration and fission in annelids: insights from engrailed- and orthodenticle-class gene expression. Development 128, 2781–2791.
Bely, A. E. (2014). Early events in annelid regeneration: a cellular perspective. Integr. Comp. Biol. 54, 688–699. doi: 10.1093/icb/icu109
Bely, A. E. (2010). Evolutionary loss of animal regeneration: pattern and process. Integr. Comp. Biol. 50, 515–527. doi: 10.1093/icb/icq118
Bely, A. E., and Nyberg, K. G. (2010). Evolution of animal regeneration: re-emergence of a field. Trends Ecol. Evol. 25, 161–170. doi: 10.1016/j.tree.2009.08.005
Bely, A. E., Zattara, E. E., and Sikes, J. M. (2014). Regeneratino in spiralians: evolutionary patterns and developmental processes. Int. J. Dev. Biol. 10, 187. doi: 10.1007/s12519-014-0487-8
Berrill, N. J. (1951). Regeneration and budding in tunicates. Biol. Rev. 26, 456–475. doi: 10.1111/j.1469-185X.1951.tb01207.x
Bode, H. R. (2009). Axial patterning in hydra. Cold Spring Harb. Perspect. Biol. 1, 1:a000463. doi: 10.1101/cshperspect.a000463
Bolker, J. A. (2000). Modularity in development and why it matters to evo-devo. Integr. Comp. Biol. 40, 770–776. doi: 10.1093/icb/40.5.770
Bosch, M., Baguñà, J., and Serras, F. (2008). Origin and proliferation of blastema cells during regeneration of Drosophila wing imaginal discs. Int. J. Dev. Biol. 52, 1043–1050. doi: 10.1387/ijdb.082608mb
Bozorgmehr, J. E. H. (2014). The role of self-organization in developmental evolution. Theory Biosci. 133, 145–163. doi: 10.1007/s12064-014-0200-4
Brockes, J. P., Kumar, A., and Velloso, C. P. (2001). Regeneration as an evolutionary variable. J. Anat. 199, 3–11. doi: 10.1046/j.1469-7580.2001.19910003.x
Brockes, J. P., and Kumar, A. (2008). Comparative aspects of animal regeneration. Annu. Rev. Cell Dev. Biol. 24, 525–549. doi: 10.1146/annurev.cellbio.24.110707.175336
Brown, F. D., Keeling, E. L., Le, A. D., and Swalla, B. J. (2009a). Whole body regeneration in a colonial ascidian, Botrylloides violaceus. J. Exp. Zool. B Mol. Dev. Evol. 312, 885–900. doi: 10.1002/jez.b.21303
Brown, F. D., and Swalla, B. J. (2012). Evolution and development of budding by stem cells: ascidian coloniality as a case study. Dev. Biol. 369, 151–162. doi: 10.1016/j.ydbio.2012.05.038
Brown, F. D., Tiozzo, S., Roux, M. M., Ishizuka, K., Swalla, B. J., and De Tomaso, A. W. (2009b). Early lineage specification of long-lived germline precursors in the colonial ascidian Botryllus schlosseri. Development 136, 3485–3494. doi: 10.1242/dev.037754
Burton, P. M., and Finnerty, J. R. (2009). Conserved and novel gene expression between regeneration and asexual fission in Nematostella vectensis. Dev. Genes Evol. 219, 79–87. doi: 10.1007/s00427-009-0271-2
Calve, S., Odelberg, S. J., and Simon, H.-G. (2010). A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev. Biol. 344, 259–271. doi: 10.1016/j.ydbio.2010.05.007
Candia Carnevali, M. D. (2006). Regeneration in Echinoderms: repair, regrowth, cloning. Rev. Lit. Arts Am. 3, 64–76.
Cebrià, F., Nakazawa, M., Mineta, K., Ikeo, K., Gojobori, T., and Agata, K. (2002). Dissecting planarian central nervous system regeneration by the expression of neural-specific genes. Dev. Growth Differ. 44, 135–146. doi: 10.1046/j.1440-169x.2002.00629.x
Chera, S., Ghila, L., Dobretz, K., Wenger, Y., Bauer, C., Buzgariu, W., et al. (2009). Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev. Cell 17, 279–289. doi: 10.1016/j.devcel.2009.07.014
Clevers, H., Loh, K. M., and Nusse, R. (2014). An integral program for tissue renewal and regeneration: wnt signaling and stem cell control. Science 346, 1248012–1248012. doi: 10.1126/science.1248012
Cordeiro, J. V., and Jacinto, A. (2013). The role of transcription-independent damage signals in the initiation of epithelial wound healing. Nat. Rev. Mol. Cell Biol. 14, 249–262. doi: 10.1038/nrm3541
Davidson, E. H., and Erwin, D. H. (2015). Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800. doi: 10.1126/science.1113832
Demircan, T., and Berezikov, E. (2013). The Hippo pathway regulates stem cells during homeostasis and regeneration of the flatworm Macrostomum lignano. Stem Cells Dev. 22, 2174–2185. doi: 10.1089/scd.2013.0006
Dirks, U., Gruber-Vodicka, H. R., Egger, B., and Ott, J. A. (2012). Proliferation pattern during rostrum regeneration of the symbiotic flatworm Paracatenula galateia: a pulse-chase-pulse analysis. Cell Tissue Res. 349, 517–525. doi: 10.1007/s00441-012-1426-4
Dubuc, T. Q., Traylor-knowles, N., and Martindale, M. Q. (2014). Initiating a regenerative response; cellular and molecular features of wound healing in the cnidarian Nematostella vectensis. BMC Biol. 12:24. doi: 10.1186/1741-7007-12-24
Dunn, C. W., Hejnol, A., Matus, D. Q., Pang, K., Browne, W. E., Smith, S. A., et al. (2008). Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749. doi: 10.1038/nature06614
Egger, B., Gschwentner, R., and Rieger, R. (2007). Free-living flatworms under the knife: past and present. Dev. Genes Evol. 217, 89–104. doi: 10.1007/s00427-006-0120-5
Egger, B., Lapraz, F., Tomiczek, B., Müller, S., Dessimoz, C., Girstmair, J., et al. (2015). A transcriptomic-phylogenomic analysis of the evolutionary relationships of flatworms. Curr. Biol. 25, 1347–1353. doi: 10.1016/j.cub.2015.03.034
Eming, S. A., Hammerschmidt, M., Krieg, T., and Roers, A. (2009). Interrelation of immunity and tissue repair or regeneration. Semin. Cell Dev. Biol. 20, 517–527. doi: 10.1016/j.semcdb.2009.04.009
Extavour, C. G., and Akam, M. (2003). Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869–5884. doi: 10.1242/dev.00804
Fukazawa, T., Naora, Y., Kunieda, T., and Kubo, T. (2009). Suppression of the immune response potentiates tadpole tail regeneration during the refractory period. Development 2327, 2323–2327. doi: 10.1242/dev.033985
Funayama, N. (2013). The stem cell system in demosponges: suggested involvement of two types of cells: archeocytes (active stem cells) and choanocytes (food-entrapping flagellated cells). Dev. Genes Evol. 223, 23–38. doi: 10.1007/s00427-012-0417-5
Galle, S., Yanze, N., and Seipel, K. (2005). The homeobox gene Msx in development and transdifferentiation of jellyfish striated muscle. Int. J. Dev. Biol. 49, 961–967. doi: 10.1387/ijdb.052009sg
Galliot, B., and Chera, S. (2010). The Hydra model: disclosing an apoptosis-driven generator of Wnt-based regeneration. Trends Cell Biol. 20, 514–523. doi: 10.1016/j.tcb.2010.05.006
Gardiner, D. M., Blumberg, B., Komine, Y., and Bryant, S. V. (1995). Regulation of HoxA expression in developing and regenerating axolotl limbs. Development 121, 1731–1741.
Garfield, D. A., Runcie, D. E., Babbitt, C. C., Haygood, R., Nielsen, W. J., and Wray, G. A. (2013). The impact of gene expression variation on the robustness and evolvability of a developmental gene regulatory network. PLoS Biol. 11:e1001696. doi: 10.1371/journal.pbio.1001696
Gemberling, M., Bailey, T. J., Hyde, D. R., and Poss, K. D. (2013). The zebrafish as a model for complex tissue regeneration. Trends Genet. 29, 611–620. doi: 10.1016/j.tig.2013.07.003
Ghildiyal, M., and Zamore, P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108. doi: 10.1038/nrg2504
Ghosh-Roy, A., and Chisholm, A. D. (2010). Caenorhabditis elegans: a new model organism for studies of axon regeneration. Dev. Dyn. 239, 1460–1464. doi: 10.1002/dvdy.22253
Giani, V. C., Yamaguchi, E., Boyle, M. J., and Seaver, E. C. (2011). Somatic and germline expression of piwi during development and regeneration in the marine polychaete annelid Capitella teleta. Evodevo 2:10. doi: 10.1186/2041-9139-2-10
Gilbert, S. F. (2000). Developmental Biology. Available online at: http://www.ncbi.nlm.nih.gov/books/NBK9983 (Accessed March 23, 2015).
Gilbert, S. F., and Epel, D. (2009). Ecological Developmental Biology: Integrating Epigenetics, Medicine and Evolution. Sunderland: Sinauer As.
Godwin, J. W., and Rosenthal, N. (2014). Scar-free wound healing and regeneration in amphibians: immunological in fl uences on regenerative success. Differentiation 87, 66–75. doi: 10.1016/j.diff.2014.02.002
Gold, D. A., and Jacobs, D. K. (2013). Stem cell dynamics in Cnidaria: are there unifying principles? 223, 53–66. doi: 10.1007/s00427-012-0429-1
Goss, R. J. (1992). The evolution of regeneration: adaptive or inherent? J. Theor. Biol. 159, 241–260. doi: 10.1016/S0022-5193(05)80704-0
Guder, C., Philipp, I., Lengfeld, T., Watanabe, H., Hobmayer, B., and Holstein, T. W. (2006). The Wnt code: cnidarians signal the way. Oncogene 25, 7450–7460. doi: 10.1038/sj.onc.1210052
Guo, T., Peters, A. H., and Newmark, P. A. (2006). A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159–169. doi: 10.1016/j.devcel.2006.06.004
Gurley, K. A., Elliott, S. A., Simakov, O., Schmidt, H. A., Holstein, T. W., and Sánchez Alvarado, A. (2010). Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev. Biol. 347, 24–39. doi: 10.1016/j.ydbio.2010.08.007
Hejnol, A., Obst, M., Stamatakis, A., Ott, M., Rouse, G. W., Edgecombe, G. D., et al. (2009). Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. Biol. Sci. 276, 4261–4270. doi: 10.1098/rspb.2009.0896
Hendrikse, J. L., Parsons, T. E., and Hallgrímsson, B. (2007). Evolvability as the proper focus of evolutionary developmental biology. Evol. Dev. 9, 393–401. doi: 10.1111/j.1525-142X.2007.00176.x
Hill, S. (1970). Origin of regeneration blastema in polychaete annelids. Am. Zool. 10, 101–112. doi: 10.1093/icb/10.2.101
Hisata, K., Fujiwara, S., Tsuchida, Y., Ohashi, M., and Kawamura, K. (1998). Expression and function of a retinoic acid receptor in budding ascidians. Dev. Genes Evol. 208, 537–46. doi: 10.1007/s004270050213
Hobmayer, B., Jenewein, M., Eder, D., Eder, M., Glasauer, S., Gufler, S., et al. (2012). Stemness in Hydra - a current perspective. 517, 509–517. doi: 10.1387/ijdb.113426bh
Holstein, T. W., Hobmayer, E., and Technau, U. (2003). Cnidarians: an evolutionarily conserved model system for regeneration? Dev. Dyn. 226, 257–267. doi: 10.1002/dvdy.10227
Holstein, T. W., Watanabe, H., and Suat, Ö. (2011). Signaling Pathways and Axis Formation in the Lower Metazoa, 1st Edn. Elsevier Inc. doi: 10.1016/B978-0-12-385975-4.00012-7
Hoppe, W. (1988). Growth, regeneration and predation in three species of large coral reef sponges. Mar. Ecol. Prog. Ser. 50, 117–125. doi: 10.3354/meps050117
Isolani, M. E., Abril, J. F., Saló, E., Deri, P., Bianucci, A. M., and Batistoni, R. (2013). Planarians as a model to assess in vivo the role of matrix metalloproteinase genes during homeostasis and regeneration. PLoS ONE 8:e55649. doi: 10.1371/journal.pone.0055649
Jager, M., Dayraud, C., Mialot, A., Quéinnec, E., Le Guyader, H., and Manuel, M. (2013). Evidence for involvement of Wnt signalling in body polarities, cell proliferation, and the neuro-sensory system in an adult ctenophore. PLoS ONE 8:e84363. doi: 10.1371/journal.pone.0084363
Jeffery, W. R. (2015). Closing the wounds: one hundred and twenty five years of regenerative biology in the ascidian Ciona intestinalis. Genesis 53, 48–65. doi: 10.1002/dvg.22799
Johnson, R., and Halder, G. (2014). The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 13, 63–79. doi: 10.1038/nrd4161
Jopling, C., Sleep, E., Raya, M., Martí, M., Raya, A., and Belmonte, J. C. I. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609. doi: 10.1038/nature08899
Juliano, C. E., Reich, A., Liu, N., Götzfried, J., Zhong, M., Uman, S., et al. (2014). PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, 337–342. doi: 10.1073/pnas.1320965111
Juliano, C. E., Swartz, S. Z., and Wessel, G. M. (2010). A conserved germline multipotency program. Development 137, 4113–4126. doi: 10.1242/dev.047969
Kamimura, M., Fujiwara, S., Kawamura, K., and Yubisui, T. (2000). Functional retinoid receptors in budding ascidians. Dev. Growth Differ. 42, 1–8. doi: 10.1046/j.1440-169x.2000.00478.x
Kawakami, M., and Nakanishi, N. (2001). The role of an endogenous PKA inhibitor, PKIalpha, in organizing left-right axis formation. Development 128, 2509–2515
Kawamura, K., Shiohara, M., Kanda, M., and Fujiwara, S. (2013). Retinoid X receptor-mediated transdifferentiation cascade in budding tunicates. Dev. Biol. 384, 343–355. doi: 10.1016/j.ydbio.2013.10.004
Kawamura, K., Sugino, Y., Sunanaga, T., and Fujiwara, S. (2008). Multipotent epithelial cells in the process of regeneration and asexual reproduction in colonial tunicates. Dev. Growth Differ. 50, 1–11. doi: 10.1111/j.1440-169X.2007.00972.x
Konstantinides, N., and Averof, M. (2014). A common cellular basis for muscle regeneration in arthropods and vertebrates. Science 343, 788–791. doi: 10.1126/science.1243529
Kott, P. (1981). “Replication in the ascidiacea: an adaptive strategy in the coral reef environment,” in Proceedings of the Fourth International Coral Reef Symposium (Manila), 725–733.
Kragl, M., Knapp, D., Nacu, E., Khattak, S., Maden, M., Epperlein, H. H., et al. (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65. doi: 10.1038/nature08152
Kumar, A., and Brockes, J. P. (2012). Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci. 35, 691–699. doi: 10.1016/j.tins.2012.08.003
Kürn, U., Rendulic, S., Tiozzo, S., and Lauzon, R. J. (2011). Asexual propagation and regeneration in colonial ascidians. Biol. Bull. 221, 43–61.
Laumer, C. E., Hejnol, A., and Giribet, G. (2015). Nuclear genomic signals of the “microturbellarian” roots of platyhelminth evolutionary innovation. Elife 4:e05503. doi: 10.7554/eLife.05503
Lauri, A., Brunet, T., Handberg-Thorsager, M., Fischer, A. H. L., Simakov, O., Steinmetz, P. R. H., et al. (2014). Development of the annelid axochord: insights into notochord evolution. Science 345, 1365–1368. doi: 10.1126/science.1253396
Leclère, L., Jager, M., Barreau, C., Chang, P., Le Guyader, H., Manuel, M., et al. (2012). Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev. Biol. 364, 236–248. doi: 10.1016/j.ydbio.2012.01.018
Lehoczky, J. A., Robert, B., and Tabin, C. J. (2011). Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 108, 20609–20614. doi: 10.1073/pnas.1118017108
Leontovich, A. A., Zhang, J., Shimokawa, K., Nagase, H., and Sarras, M. P. (2000). A novel hydra matrix metalloproteinase (HMMP) functions in extracellular matrix degradation, morphogenesis and the maintenance of differentiated cells in the foot process. Development 127, 907–920.
Levin, M. (2012). Molecular bioelectricity in developmental biology: new tools and recent discoveries: control of cell behavior and pattern formation by transmembrane potential gradients. Bioessays 34, 205–217. doi: 10.1002/bies.201100136
Levin, M., and Stevenson, C. G. (2012). Regulation of cell behavior and tissue patterning by bioelectrical signals: challenges and opportunities for biomedical engineering. Annu. Rev. Biomed. Eng. 14, 295–323. doi: 10.1146/annurev-bioeng-071811-150114
Lin, A. Y. T., and Pearson, B. J. (2014). Planarian yorkie/YAP functions to integrate adult stem cell proliferation, organ homeostasis and maintenance of axial patterning. Development 141, 1197–1208. doi: 10.1242/dev.101915
Lin, G., Chen, Y., and Slack, J. M. W. (2013). Imparting regenerative capacity to limbs by progenitor cell transplantation. Dev. Cell 24, 41–51. doi: 10.1016/j.devcel.2012.11.017
Liu, S.-Y., Selck, C., Friedrich, B., Lutz, R., Vila-Farré, M., Dahl, A., et al. (2013). Reactivating head regrowth in a regeneration-deficient planarian species. Nature 500, 81–84. doi: 10.1038/nature12414
Mace, K. A., Pearson, J. C., and McGinnis, W. (2005). An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 308, 381–385. doi: 10.1126/science.1107573
Maginnis, T. L. (2006). The costs of autotomy and regeneration in animals: a review and framework for future research. Behav. Ecol. 17, 857–872. doi: 10.1093/beheco/arl010
Martin, P., and Leibovich, S. J. (2005). Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol. 15, 599–607. doi: 10.1016/j.tcb.2005.09.002
Martín-Durán, J. M., and Egger, B. (2012). Developmental diversity in free-living flatworms. Evodevo 3:7. doi: 10.1186/2041-9139-3-7
Martinez, V. G., Menger, G. J., and Zoran, M. J. (2005). Regeneration and asexual reproduction share common molecular changes: upregulation of a neural glycoepitope during morphallaxis in Lumbriculus. Mech. Dev. 122, 721–732. doi: 10.1016/j.mod.2004.12.003
Mauviel, A., Nallet-Staub, F., and Varelas, X. (2011). Integrating developmental signals: a Hippo in the (path)way. Oncogene 31, 1743–1756. doi: 10.1038/onc.2011.363
Mercader, N., Tanaka, E. M., and Torres, M. (2005). Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development 132, 4131–4142. doi: 10.1242/dev.01976
Minelli, A. (2015). Grand challenges in evolutionary developmental biology. Front. Ecol. Evol. 2:85. doi: 10.3389/fevo.2014.00085
Mochizuki, K., Nishimiya-Fujisawa, C., and Fujisawa, T. (2001). Universal occurrence of the vasa -related genes among metazoans and their germline expression in Hydra. Dev. Genes Evol. 211, 299–308. doi: 10.1007/s004270100156
Morgan, T. H. (1901). Regeneration. Macmillan. Available online at: http://books.google.com/books?id=QAYFAAAAYAAJ&pgis=1 (Accessed February 24, 2015).
De Mulder, K., Pfister, D., Kuales, G., Egger, B., Salvenmoser, W., Willems, M., et al. (2009). Stem cells are differentially regulated during development, regeneration and homeostasis in flatworms. Dev. Biol. 334, 198–212. doi: 10.1016/j.ydbio.2009.07.019
Müller, W. A., Teo, R., and Frank, U. (2004). Totipotent migratory stem cells in a hydroid. Dev. Biol. 275, 215–224. doi: 10.1016/j.ydbio.2004.08.006
Nakanishi, Y., Nagaosa, K., and Shiratsuchi, A. (2011). Phagocytic removal of cells that have become unwanted: implications for animal development and tissue homeostasis. Dev. Growth Differ. 53, 149–160. doi: 10.1111/j.1440-169X.2010.01224.x
Nijhout, H. F., and Emlen, D. J. (1998). Competition among body parts in the development and evolution of insect morphology. Proc. Natl. Acad. Sci. U.S.A. 95, 3685–3689. doi: 10.1073/pnas.95.7.3685
Önal, P., Grün, D., Adamidi, C., Rybak, A., Solana, J., Mastrobuoni, G., et al. (2012). Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J. 31, 2755–2769. doi: 10.1038/emboj.2012.110
Palmer, C. V., Traylor-knowles, N. G., Willis, B. L., and Bythell, J. C. (2011). Corals use similar immune cells and wound-healing processes as those of higher organisms. PLoS ONE 6:e23992. doi: 10.1371/journal.pone.0023992
Pan, D. (2007). Hippo signaling in organ size control. Genes Dev. 21, 886–897. doi: 10.1101/gad.1536007
Passamaneck, Y. J., and Martindale, M. Q. (2012). Cell proliferation is necessary for the regeneration of oral structures in the anthozoan cnidarian Nematostella vectensis. BMC Dev. Biol. 12:34. doi: 10.1186/1471-213X-12-34
Philippe, H., Brinkmann, H., Copley, R. R., Moroz, L. L., Nakano, H., Poustka, A. J., et al. (2011). Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature 470, 255–258. doi: 10.1038/nature09676
Plickert, G., Frank, U., and Müller, W. A. (2012). Hydractinia, a pioneering model for stem cell biology and reprogramming somatic cells to pluripotency. Int. J. Dev. Biol. 56, 519–534. doi: 10.1387/ijdb.123502gp
Quiñones, J. L., Rosa, R., Ruiz, D. L., and García-Arrarás, J. E. (2002). Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima. Dev. Biol. 250, 181–197. doi: 10.1006/dbio.2002.0778
Ramasamy, S. K., Kusumbe, A. P., and Adams, R. H. (2015). Regulation of tissue morphogenesis by endothelial cell-derived signals. Trends Cell Biol. 25, 148–157. doi: 10.1016/j.tcb.2014.11.007
Rebscher, N., Zelada-González, F., Banisch, T. U., Raible, F., and Arendt, D. (2007). Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev. Biol. 306, 599–611. doi: 10.1016/j.ydbio.2007.03.521
Reddien, P. W. (2011). Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 27, 277–285. doi: 10.1016/j.tig.2011.04.004
Reddien, P. W. (2013). Specialized progenitors and regeneration. Development 140, 951–957. doi: 10.1242/dev.080499
Reddien, P. W., Oviedo, N. J., Jennings, J. R., Jenkin, J. C., and Sánchez Alvarado, A. (2005). SMEDWI-2 Is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330. doi: 10.1126/science.1116110
Richards, G. S., and Rentzsch, F. (2014). Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis. Development 141, 4681–4689. doi: 10.1242/dev.112029
Richardson, M. K., and Chipman, A. D. (2003). Developmental constraints in a comparative framework: a test case using variations in phalanx number during amniote evolution. J. Exp. Zool. B Mol. Dev. Evol. 296, 8–22. doi: 10.1002/jez.b.13
Rinkevich, Y., Paz, G., Rinkevich, B., and Reshef, R. (2007). Systemic bud induction and retinoic acid signaling underlie whole body regeneration in the urochordate Botrylloides leachi. PLoS Biol. 5:e71. doi: 10.1371/journal.pbio.0050071
Rinkevich, Y., Rosner, A., Rabinowitz, C., Lapidot, Z., Moiseeva, E., and Rinkevich, B. (2010). Piwi positive cells that line the vasculature epithelium, underlie whole body regeneration in a basal chordate. Dev. Biol. 345, 94–104. doi: 10.1016/j.ydbio.2010.05.500
Rossi, L., Salvetti, A., Batistoni, R., Deri, P., and Gremigni, V. (2008). Planarians, a tale of stem cells. Cell. Mol. Life Sci. 65, 16–23. doi: 10.1007/s00018-007-7426-y
Sanetra, M., Begemann, G., Becker, M.-B., and Meyer, A. (2005). Conservation and co-option in developmental programmes: the importance of homology relationships. Front. Zool. 2:15. doi: 10.1186/1742-9994-2-15
Shibata, N., Umesono, Y., Orii, H., Sakurai, T., Watanabe, K., and Agata, K. (1999). Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev. Biol. 206, 73–87. doi: 10.1006/dbio.1998.9130
Siebert, S., Anton-erxleben, F., and Bosch, T. C. G. (2008). Cell type complexity in the basal metazoan Hydra is maintained by both stem cell based mechanisms and transdifferentiation. Dev. Biol. 313, 13–24. doi: 10.1016/j.ydbio.2007.09.007
Sikes, J. M., and Newmark, P. A. (2013). Restoration of anterior regeneration in a planarian with limited regenerative ability. Nature 500, 77–80. doi: 10.1038/nature12403
Solana, J. (2013). Closing the circle of germline and stem cells: the primordial stem cell hypothesis. Evodevo 4:2. doi: 10.1186/2041-9139-4-2
Srivastava, M., Mazza-Curll, K. L., Van Wolfswinkel, J. C., and Reddien, P. W. (2014). Whole-body acoel regeneration is controlled by Wnt and Bmp-Admp signaling. Curr. Biol. 24, 1107–1113. doi: 10.1016/j.cub.2014.03.042
Struck, T. H., Wey-Fabrizius, A. R., Golombek, A., Hering, L., Weigert, A., Bleidorn, C., et al. (2014). Platyzoan paraphyly based on phylogenomic data supports a noncoelomate ancestry of spiralia. Mol. Biol. Evol. 31, 1833–1849. doi: 10.1093/molbev/msu143
Sunanaga, T., Inubushi, H., and Kawamura, K. (2010). Piwi-expressing hemoblasts serve as germline stem cells during postembryonic germ cell specification in colonial ascidian, Botryllus primigenus. Dev. Growth Differ. 52, 603–614. doi: 10.1111/j.1440-169X.2010.01196.x
Tanaka, E. M., and Reddien, P. W. (2011). The cellular basis for animal regeneration. Dev. Cell 21, 172–185. doi: 10.1016/j.devcel.2011.06.016
Technau, U., and Steele, R. E. (2011). Evolutionary crossroads in developmental biology: cnidaria. 138, 1447–1458. doi: 10.1242/dev.090472
Ting, S. B., Caddy, J., Hislop, N., Wilanowski, T., Auden, A., Zhao, L.-L., et al. (2005). A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 308, 411–413. doi: 10.1126/science.1107511
Tiozzo, S., Brown, F. D., and De Tomaso, A. W. (2008). “Regeneration and stem cells in ascidians,” in Stem Cells: From Hydra to Man, ed T. C. G. Bosh (London: Springer), 95–112. doi: 10.1007/978-1-4020-8274-0-6
Tweedt, S. M., and Erwin, D. H. (2015). “Origin of metazoan developmental toolkits and their expression in the fossil record,” in Evolutionary Transitions to Multicellular Life Advances in Marine Genomics, eds I. Ruiz-Trillo and A.M. Nedelcu (Dordrecht: Springer), 47–77.
Umesono, Y., Tasaki, J., Nishimura, Y., Hrouda, M., Kawaguchi, E., Yazawa, S., et al. (2013). The molecular logic for planarian regeneration along the anterior-posterior axis. Nature 500, 73–76. doi: 10.1038/nature12359
Varelas, X., and Wrana, J. L. (2012). Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 22, 88–96. doi: 10.1016/j.tcb.2011.10.002
Vickery, M. C., Vickery, M. S., McClintock, J. B., and Amsler, C. D. (2001). Utilization of a novel deuterostome model for the study of regeneration genetics: molecular cloning of genes that are differentially expressed during early stages of larval sea star regeneration. Gene 262, 73–80. doi: 10.1016/S0378-1119(00)00554-0
Vorontsova, M. A., and Liosner, L. D. (1961). Asexual Propagation and Regeneration New York, NY: Pergamon Press.
Wagner, G. P., and Zhang, J. (2011). The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 12, 204–213. doi: 10.1038/nrg2949
Watanabe, H., Schmidt, H. A., Kuhn, A., Höger, S. K., Kocagöz, Y., Laumann-Lipp, N., et al. (2014). Nodal signalling determines biradial asymmetry in Hydra. Nature 515, 112–115. doi: 10.1038/nature13666
Wehner, D., Cizelsky, W., Vasudevaro, M. D., Ozhan, G., Haase, C., Kagermeier-Schenk, B., et al. (2014). Wnt/β-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep. 6, 467–481. doi: 10.1016/j.celrep.2013.12.036
Wenemoser, D., and Reddien, P. W. (2010). Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344, 979–991. doi: 10.1016/j.ydbio.2010.06.017
Wenger, Y., Buzgariu, W., Reiter, S., and Galliot, B. (2014). Seminars in immunology injury-induced immune responses in Hydra. Semin. Immunol. 26, 277–294. doi: 10.1016/j.smim.2014.06.004
Witchley, J. N., Mayer, M., Wagner, D. E., Owen, J. H., and Reddien, P. W. (2013). Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633–641. doi: 10.1016/j.celrep.2013.07.022
Worley, M. I., Setiawan, L., and Hariharan, I. K. (2012). Regeneration and transdetermination in Drosophila imaginal discs. Annu. Rev. Genet. 46, 289–310. doi: 10.1146/annurev-genet-110711-155637
Wulff, J. (2010). Regeneration of sponges in ecological context: is regeneration an integral part of life history and morphological strategies? Integr. Comp. Biol. 50, 494–505. doi: 10.1093/icb/icq100
Wulff, J. (2013). Integrative and comparative biology recovery of sponges after extreme mortality events: morphological and taxonomic patterns in regeneration versus recruitment. Integr. Comp. Biol. 53, 512–523. doi: 10.1093/icb/ict059
Xiol, J., Spinelli, P., Laussmann, M. A., Homolka, D., Yang, Z., Cora, E., et al. (2014). RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157, 1698–1711. doi: 10.1016/j.cell.2014.05.018
Yakushiji, N., Suzuki, M., Satoh, A., Sagai, T., Shiroishi, T., Kobayashi, H., et al. (2007). Correlation between Shh expression and DNA methylation status of the limb-specific Shh enhancer region during limb regeneration in amphibians. Dev. Biol. 312, 171–182. doi: 10.1016/j.ydbio.2007.09.022
Yakushiji, N., Yokoyama, H., and Tamura, K. (2009). Repatterning in amphibian limb regeneration: a model for study of genetic and epigenetic control of organ regeneration. Semin. Cell Dev. Biol. 20, 565–574. doi: 10.1016/j.semcdb.2008.12.007
Zeng, L., and Swalla, B. J. (2005). Molecular phylogeny of the protochordates: chordate evolution. Can. J. Zool. 83, 24–33. doi: 10.1139/z05-010
Keywords: regeneration, development, phylogenesis, evo-devo, developmental modularity, robustness, evolvability, developmental constraints
Citation: Tiozzo S and Copley RR (2015) Reconsidering regeneration in metazoans: an evo-devo approach. Front. Ecol. Evol. 3:67. doi: 10.3389/fevo.2015.00067
Received: 07 May 2015; Accepted: 11 June 2015;
Published: 23 June 2015.
Edited by:
Giuseppe Fusco, University of Padova, ItalyReviewed by:
Francesc Cebrià, University of Barcelona, SpainUri Frank, National University of Ireland, Galway, Ireland
Copyright © 2015 Tiozzo and Copley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefano Tiozzo, Observatoire Océanologique de Villefranche-sur-Mer Quai de la Darse, 06234 Villefranche-sur-Mer Cedex, France, tiozzo@obs-vlfr.fr
 Stefano Tiozzo
Stefano Tiozzo Richard R. Copley
Richard R. Copley