Evolutionary reversion of editing sites of ndh genes suggests their origin in the Permian-Triassic, before the increase of atmospheric CO2
- 1Department of Life Science, University of Alcalá, Madrid, Spain
- 2Department of Physical Chemistry, University of Alcalá, Madrid, Spain
The plastid ndh genes have hovered frequently on the edge of dispensability. They are absent in the plastid DNA of many algae and certain higher plants and present editing sites requiring C-to-U corrections of primary transcripts. The evolutionary origin of editing sites and their loss due to C-to-T reversions at the DNA level are unknown and must be related to the dispensability of the ndh genes in specific environments. In order to better understand the evolution of ndh gene editing sites, we have created expandable data banks with the 12 editing sites of the ndhB gene (600 GenBank sequences) and both editing sites of the ndhF gene (1600 GenBank sequences). Since their origin via T-to-C mutations that probably occurred between 300 and 200 Myr BP (Permian-Triassic), ndh editing sites have undergone independent and random C-to-T reversions in the different angiosperm lineages. Some of these reversions appear early in angiosperm diversification. Old C-to-T reversions can be traced back to radiation steps that gave origin to main classes, orders and some families.
Introduction
The plastid ndh genes encode 11 NDH polypeptides of the thylakoid Ndh complex (Maier et al., 1995; Sazanov et al., 1998; Casano et al., 2000; Yukawa et al., 2005) analogous to the mitochondrial complex I (EC 1.6.5.3). The Ndh complex catalyzes the transfer of electrons from NADH to plastoquinone, the first stage of the chlororespiratory reaction chain in which the Mehler reaction, superoxide dismutase and peroxidase activities drain excess electrons to fine-tune the redox level of the cyclic electron transporters (Casano et al., 2000; Rumeau et al., 2007; Martín et al., 2009, 2015). According to this function, the ndh genes are required to optimize photosynthesis rate under fluctuating light and high CO2 concentrations (Martín et al., 2009, 2015).
Among eukaryotic algae, only a few Prasinophyceae and all Charophyceae (the green algae related to higher plants) contain ndh genes (Martín and Sabater, 2010; Fučíková et al., 2014). Most photosynthetic land plants contain the ndh genes that are absent in parasitic non-photosynthetic species of the genera Cuscuta, Epiphagus, Orobanche and the Orchidaceae family (Braukmann et al., 2013; Barrett et al., 2014; Luo et al., 2014), which suggests that the thylakoid Ndh complex encoded by the ndh genes has a role in land plant photosynthesis. However, plastid DNAs of the gymnosperms Pinaceae and Gnetales, as well as a few species of angiosperms in various genera, families and orders (e.g.,: Erodium, Ericaceae, Alismatales, …) lack the ndh genes (Braukmann et al., 2009; Blazier et al., 2011; Braukmann and Stefanovič, 2012; Peredo et al., 2013; Lin et al., 2015; Ruhlman et al., 2015). These particular species may contain ndh gene fragments in either the nucleus or mitochondrion, which suggests that ndh genes could be dispensable in some plants and/or environments.
The 11 ndh genes account for about 50% of all C-to-U editing sites identified in the transcripts of plastid genes of higher plants (Tillich et al., 2005). In contrast, the ndh primary transcripts of lower plants have the appropriate U base and do not require editing. This suggests that the ndh genes accumulated (among other) T-to-C mutations in ancestors (Martín and Sabater, 2010) because these genes were dispensable under the environmental conditions preceding the diversification to seed plants. Later, new environments made the ndh genes useful to improve photosynthesis. Then, the functionality of the ndh genes was recovered by post-transcriptional C-to-U editing or by C-to-T reversion. In other words, the accumulation of editing sites in ndh genes reflects an evolution stage when the ndh genes were dispensable in ancestors of seed plants because they did not significantly improve photosynthetic efficiency.
Transcript editing is carried out by several nuclear encoded proteins (trans-factors) that recognize specific sequences (cis-elements) upstream of the C to be edited (Shikanai, 2006; Tillich et al., 2006; Takenaka et al., 2013). Tillich et al. (2006) proposed that the editing originated in bryophytes as a mechanism to generate variation at the RNA level. In general, RNA editing permits the functional rescue of genes affected by T-to-C mutations (Maier et al., 1996; Martín and Sabater, 2010; Takenaka et al., 2013) and is, therefore, a rapid way to neutralize their effects (Sabater et al., 2002; Tillich et al., 2006; Martín and Sabater, 2010). Over time, random reversions of these mutations restore original T bases and genes recover functionality without the need of editing.
The editing sites in a gene are identified by the comparison of its genomic sequence with that of the DNA complementary (Freyer et al., 1995; Tillich et al., 2005; Chateigner-Boutin and Small, 2007). A number of ndh gene sites are well characterized for undergoing C-to-U editing in at least some seed plants. In species that do not require editing, these sites have a T as opposed to C at the genome level. Therefore, each plant has a distinctive signature of the well-characterized ndh editing sites: a set of sites requires post-transcriptional C-to-U editing whereas the other sites have undergone C-to-T reversion at the genome level. To date, approximately 12 ndhB and two ndhF editing sites have been confirmed in mature transcripts of seed plants (Freyer et al., 1995, 1997; Maier et al., 1995; Tillich et al., 2005; Martín and Sabater, 2010). Although the discovery of additional editing sites cannot be excluded, the high number of genomic sequences of different species provides information regarding the distribution of editable C-to-U and C-to-T reversions of each site among plant lineages. Currently, partial or total genomic sequences of the ndhB (n = 600) and ndhF (n = 1600) genes of seed plants have been deposited in GenBank.
We hypothesize that the status (editable C-to-U or reversed C-to-T) of the editing sites of ndhB and ndhF genes in plants of different lineages must indicate when one site was originated by T-to-C mutation and when it was C-to-T reversed in each phylogenetic branch. Comparison with the environmental changes in past geological eras should explain when the genes became dispensable or beneficial on the basis of their functional role in photosynthesis. To test this hypothesis, we created data bases registering the status of each site in different plants deduced from sequences of ndhB and ndhF genes deposited in GenBank. We describe how the phylogenetic analysis of the data confirms the hypothesis, suggests the origin of massive T-to-C mutations 300 to 200 Myr BP and relates it with the dispensability of ndh genes at very low CO2 concentrations (some 210 ppm). In addition, C-to-T reversions may be traced back to radiation steps that originated main classes, orders and, in some cases, families.
Materials and Methods
Gene Sequences and Editing Sites
The investigation is focused on the 14 editing sites identified in the ndhB and ndhF genes of different seed plants (Freyer et al., 1995, 1997; Maier et al., 1995; Tillich et al., 2005; Martín and Sabater, 2010). All genomic sequences of the plastid ndhB and ndhF genes examined in this study were obtained from GenBank (NCBI) using the BLAST algorithm (optimized for somewhat similar sequences, blastn) in the NCBI website with the corresponding Hordeum vulgare and Arabidopsis thaliana sequences (accession nos. NC_008590.1, AJ002490, and AJ002491). Default parameters of the BLAST algorithm (NCBI) were selected with expected threshold 10 (number of chance matches in a random model). The complete 1533 nucleotides of ndhB (excluding the intron) and the first 414 nucleotides of ndhF were used as subject sequences. Only query sequences from ndhB and ndhF with sequence similarities higher than 85% for gymnosperm and above 97% for angiosperms were selected to display. The printed display of each sequence aligned with that of Hordeum was examined, especially nearby positions (see Table 1) of editing sites, to assure the correct base pairing and to identify the presence of a C or T in the appropriate position in each codon of the potential editing sites. The result was then annotated in an Excel file to construct Supplementary Tables 1, 2 that indicate the status (editable or reversed) of each editing site for each plant tested.
The status of the 14 editing sites of each plant was compared with those of species close in reference phylogenetic trees of plants (Givnish et al., 2006; Burleigh et al., 2011; Soltis et al., 2011; http://www2.biologie.fu-berlin.de/sysbot/poster/poster1.pdf). When all species of one branch have corrected one editing site, plausibly, the site reversed C-to-T before diversification of all species of the branch and that is indicated in appropriate nodes of the phylogenetic tree. Alternative explanation (reversions took place independently in all species of the branch) has a very low probability. Recurrent application of this principle allows assigning reverse C-to-T events before diversification of genera, families, orders and classes. Obviously, the validity of the approach depends on the number of species tested in the branch as indicated for specific reversions in Results and in Discussion. Mapping of editing traits in the phylogenetic tree were further confirmed with the Mesquite program (https://mesquiteproject.wikispaces.com/) applied to representative sites and tree branches.
Photosynthetic data were calculated from published results (Martín et al., 2015) of tobacco plants described in detail elsewhere (Martín et al., 2009). For each tobacco plant, photosynthesis rates were determined in leaves fitted on the chamber of the LCpro+ portable photosynthesis system (ADC BioScientific Ltd. Hertfordshire, UK) at different concentrations of CO2 and under abrupt changes of light intensity according to the sequence: 15 min acclimation at 130, 6 min at 870, 6 min at 61, 6 min at 870 and 6 min at 130 μmol m−2 s−1 of photosynthetic active radiation. Data were collected each min and net photosynthesis (in μ mol consumed of CO2 m−2 s−1) was integrated over the last 24 min incubation using the Origin software (Princeton, USA). Photosynthetic efficiency (the fraction of absorbed radiant energy converted to biomass chemical energy) and entropy generated were calculated using Gibbs free energy and entropy values in data banks and conventional thermodynamics formula (Martín et al., 2015).
Results
C-to-T Reversions at the DNA Level of the Editing Sites of ndhB and ndhF Plastid Genes in Seed Plants
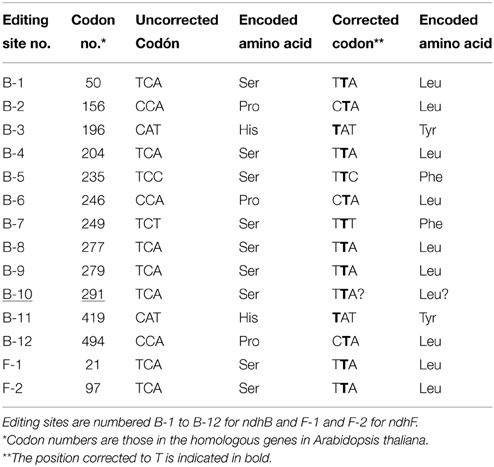
The comparison of genomic and complementary sequences to mRNA in several plants such as Nicotiana tabacum, Arabidopsis thaliana, Zea mays, and Hordeum vulgare, confirmed the existence of 12 and two editing sites in the mature transcripts of the ndhB and ndhF genes, respectively, in angiosperms (Freyer et al., 1995, 1997; Maier et al., 1995; Tillich et al., 2005; Martín and Sabater, 2010). Table 1 lists the codon and encoded amino acid for each site. Most sites are predicted based on a comparison with the sequences of the ndhB and ndhF genes of Marchantia polymorpha as well as from conserved amino acid positions in all plants tested. However, none of the above-mentioned species have all 14 editing sites. Instead, each species has a different set of editing sites due to the fact that certain Cs are substituted by Ts at the DNA level in the other potential sites or because, as occurs with the B-10 site (codon 291: TCA), the C is post-transcriptionally edited to U in Arabidopsis thaliana and other Brassicaceae, but not in other angiospems. This occurs because the cis-elements of the B-10 site in Brassicaceae are similar to those of the second editing site of the matK gene and both are recognized by the same editing trans-factors (Tillich et al., 2005). In Marchantia polymorpha, a species in which no editing has been reported, this site is TCA. B-10 (underlined in Table 1) is edited in some gymnosperms (Serrot et al., 2012) but it is not considered an editing site in angiosperms.
The editing site patterns of angiosperms and gymnosperms are different (Wakasugi et al., 1996; Sabater et al., 2002; Martín and Sabater, 2010). Comparison of complementary and genomic DNA sequences only identified the B-5, B-8, and B-10 editing sites in gymnosperms (Freyer et al., 1997; Chen et al., 2011; Serrot et al., 2012). The codons of the F-1 and F-2 sites are completely different in gymnosperms and there is no evidence for their reversion at the genome level (C-to-T) or of post-transcriptional processing (C-to-U).
In order to establish the occurrence of C-to-T reversions of editing sites at the genome level in seed plants, genomic sequences of all ndhB and ndhF genes deposited to date in GenBank and selected by the BLAST algorithm were examined to identify whether they have a C or T at the appropriate position in each codon of the14 potential editing sites. Data of the genomic correction of the editing sites of the ndhB gene of some 610 different species and of the ndhF gene of some 1650 different species are shown in Supplementary Tables 1, 2. Except for the F-1 site of Lasjia grandis (Proteales), for which the anomalous codon AAC has been reported, all of the approximately 10,000 editing sites identified in GenBank have a C or T in the position of the editable C. This indicates that, although frequently Cs of certain editing sites are only partially corrected to U by the editing machinery (Del Campo et al., 1998; Tillich et al., 2005; Van Den Bekerom et al., 2013), editing sites encode amino acids that are highly conserved in the sequence of NDH-B and NDH-F proteins.
Supplementary Tables 1, 2 reveal that every editing site is C-to-T corrected in a number of species, although the frequency of reversions at each site varies widely among the different plant divisions. Of the total number of angiosperms investigated, the percentages of C-to-T reversions of “true” editing sites range between 2.3% for the B-8 site and 57.2% for the B-5 site. The four editing sites B-2, B-3, B-8, and B-12, that have low percentages of reversion (2–4%), has a mean of 2.9 ± 0.5% reversion, higher than the 0.9% in the “false” B-10 editing site, at which a C-to-T reversion provides no functional advantage (Tillich et al., 2005). Supplementary Figure 1 shows the percentages of C-to-T reversions of the 14 sites in eudicotyledons and monocotyledons. All B-5 and almost all (96.4%) F-1 sites are corrected in eudicotyledons, whereas the percentages drop to 15 and 13%, respectively, in monocotyledons. In contrast, 100 and 74.2% of, respectively, the B-7 and F-2 sites are corrected in monocotyledons and only 6.6 and 24.3% of the B-7 and F-2 sites, respectively, are corrected in eudicotyledons. C-to-T reversions of specific sites are always observed within specific lineages of plants. Hence, it seems obvious that C-to-T reversions of an editing site took place in the ancestor of a lineage when all species of the lineage share the corrected T at the site. This consideration implies that C-to-T reversion events of most editing sites may be traced back both on a plant phylogenetic tree and in geological time. Conversely, it is possible to estimate the evolutionary time when ndh gene editing sites were generated by an accumulation of T-to-C mutations in primitive Marchantia-like ndh genes of the ancestor(s) of seed plants (see Table 1).
Tracing Back C-to-T Reversions of ndh Gene Editing Sites on a Phylogenetic Tree
Assuming that all species in one branch of the phylogenetic tree share a C-to-T corrected site, the reversion must have occurred in the common ancestor of that branch. Figures 1, 2 show the distribution of major reversion events (the editing site affected in red within a box) along the phylogeny of eudicotyledons and monocotyledons, respectively. The phylogenetic trees used were those of references (Givnish et al., 2006; Burleigh et al., 2011; Soltis et al., 2011) and the angiosperm Phylogeny page available at http://www2.biologie.fu-berlin.de/sysbot/poster/poster1.pdf with slight modifications to accommodate data of C-to-T reversions of editing sites. Hence, the correction of the B-5 site found in all eudicotyledons (Figure 1) must have occurred before the radiation of eudicotyledons. Similarly, the correction of the F-1 site probably occurred before the splitting (Figure 1) of the Trochodendrales, Buxales, Gunnerales and Core eudicot lineages. On the other hand, the correction of the B-7 site must have occurred before the early radiation leading to the different monocotyledonean lineages (Figure 2). The B-5, B-7, and F-1 sites were previously TCC, TCT, and TCA, respectively, a result of primary mutations of the original TTC, TTT, and TTA codons. To further support the results, phylogenetic trees of eudicotyledons and monocotyledons were constructed with the Mesquite program (https://mesquiteproject.wikispaces.com/), the 12 B editing sites as characters (editable or corrected) and all respective plants. Supplementary Figure 2 (eudicotyledons) and Supplementary Figure 3 (monocotyledons) show the color traces in the trees indicating corrections of, respectively, B-5 and B-7 editing sites. It must be noted that a few plants whose editing site sequence is unknown appear non-colored. Except for these, in the two figures, all plants are colored and rooted in one common ancestor, indicating that corrections of B-5 and B-7 sites occurred early in the origin of, respectively, eudicotyledons, and monocotyledons. In contrast, for the other 11 editing sites (see as supplementary.nex archives eudicotyledons and monocotyledons matrices) tree branches included many color discontinuities indicating unrelated, independent corrections events of the editing site.
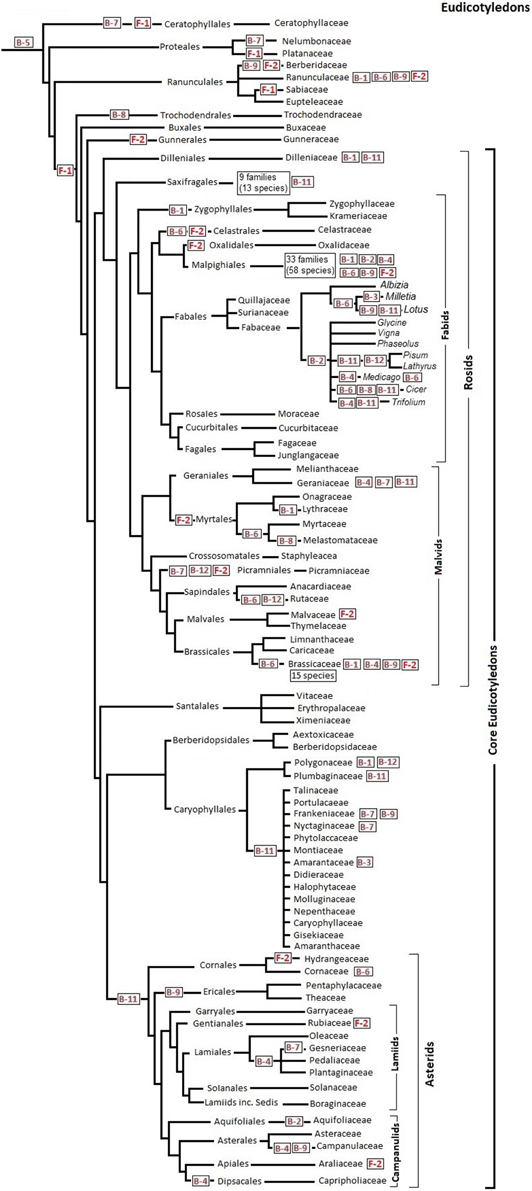
Figure 1. C-to-T reversion of editing sites of ndhB and ndhF genes during eudicotyledon phylogeny. Reference phylogenetic trees are slightly modified for insertion of the main editing site correction stages. Specific editing sites (in red squared) are C-to-T corrected in all species tested that belong to the order, family or genus that follow it at right in the phylogenetic tree. Editing sites placed at far right indicate that some but not all species of the family or the genus affected have C-to-T back mutations at that site.
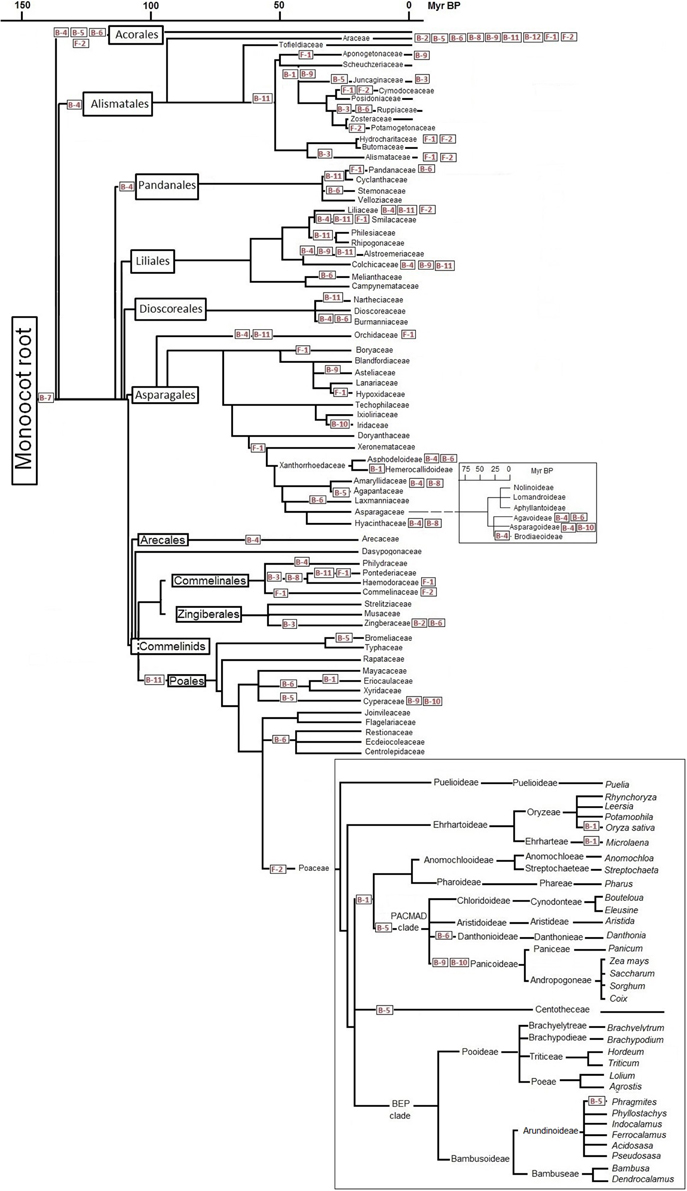
Figure 2. C-to-T reversion of editing sites of ndhB and ndhF genes during monocotyledon phylogeny. Reference phylogenetic trees are slightly modified for insertion of the main editing site correction stages. Some approximate time scales are included on the basis of references (Bremer, 2000, 2002). Specific editing sites (in red squares) are C-to-T corrected in all species tested that belong to the order, family or genus that follow it at right in the phylogenetic tree. Editing sites placed at far right indicate that some but not all species of the family or genus affected have a C-to-T back mutation at that site.
With the exception of Nympheales, an order in which there are extensive C-to-T reversions of sites B-3, B-7, B-11, F-1, and F-2 in poorly diversified angiosperms that may be considered similar to the oldest ones most of the 14 sites have a C in the place of the corrected T (Supplementary Tables 1, 2). In fact, in the three Austrobaileyales, four of the six Chloranthales and three of the four Laurales reported, none of the 12 ndhB sites have undergone C-to T reversions. Similarly, in the five Chloranthales, 73 Magnoliales and 47 of the 48 Laurales reported, neither the F-1 nor the F-2 sites are corrected, they are both TCA. Supplementary Table 3 summarizes the frequency of ndhB and ndhF site corrections in ANITA GRADE and Magnoliid species. Therefore, excluding the “false” B-10 site, the 13 editing sites investigated were probably originated by T-to-C mutations far earlier than angiosperm radiation, although later than the separation of the angiosperm ancestor from a hypothetical common root with some gymnosperms. The alternative possibility, which is that T-to-C mutations created the same 13 editing sites in the different angiosperm branches, is obviously highly improbable.
A first glimpse of Figures 1, 2 suggests that C-to-T reversions occurred randomly in the different editing sites during the evolution of angiosperms, with different frequencies in the different orders and families. At least in the case of C-to-T reversions that occurred around 100 Myr BP and affected all of the large number of species of the group analyzed (B-5 of eudicotyledons, F-1 of Core eudicotyledons and B-7 of monocotyledons), there was no event of a further mutation of the T to a C (reversion), A or G. Therefore, genomic C-to-T back mutations of editing sites of the ndh genes must confer an evolutionary advantage to plants. The ndhB and ndhF genes are very unevenly represented in GenBank: in the case of ndhB, there are many orders and families with a low number of species and in the case of ndhF many genera and species of a low number of orders and families. Therefore, the information in Figures 1, 2 is based primarily on the ndhB data. At present, several orders and genera are represented in GenBank by a low number of sequences. In these cases, the proposal that certain C-to-T reversions affect an entire order or genus must be confirmed with further data entries.
The sequences of all 37 Myrtales species reported have a corrected F-2 site as indicated in Figure 1 and the sequences of all 40 asterids species reported have a corrected B-11 site (Figure 1). Significantly, the B-11 site is also corrected in a high number of species (32 of a total of 41) distributed among different families of Caryophyllales, an order closely related to the asterids (Figure 1). Within monocotyledons, the 33 sequences of Alismatales reported to date have a corrected B-4 site and the 74 sequences of Poales reported to date have a corrected B-11 site. Therefore, the B-4 and B-11 sites were probably corrected in the ancestor of, respectively, Alismatales and Poales as shown in Figure 2. In Poales, the ndhF gene has only been extensively sequenced in the Poaceae family in which all sequences of the 517 species reported have a corrected F-2 site and only 15 have a C-to-T reversion of the F-1 site (Supplementary Table 2). Hence, the F-2 site was corrected in an ancestor of Poaceae as shown in Figure 2. The amount of data available at present is insufficient to establish the reversion of the F-2 site in an ancestor of all Poales, but, surely, the correction event occurred specifically in the Poales (before or after its families branched) because none of the total of 51 Arecales and Zingiberales species reported and only three of 19 Commelinales species reported (all within the Commelinids clade) have a corrected F-2 site (Supplementary Table 2).
The different number of editing sites in the ndhB and ndhF genes and the very different distribution within the plant kingdom of the deposited sequences of the two genes make the tentative identification of early C-to-T reversions of ndhB sites feasible in the case of events foregoing the radiation of division, order and even some families within an order. On the other hand, the sequences available at present are useful in identifying reversions of ndhF sites foregoing the radiation of several families, genera and, less frequently, orders. Further sequence data should improve the assignation of sites at which C-to-T reversions occurred in both phylogenetic trees and on a time scale and, eventually, the tracing of editing sites could be a helpful tool in solving certain phylogenetic enigma. At present, the numbers of species whose sequence data are available permit us to postulate the timing of early reversion events that affected a division, several orders, and a few families as in the example of the Poaceae F-2 site. Within a few genera, the deposited ndhF gene sequences of a high number of species permit the tracing of recent correction events affecting several species of a genus.
The ndhB editing sites of 54 angiosperm genera are represented in Supplementary Table 1 by at least 2 species each with a maximum of 10 species in the case of Gossypium, which total 152 species. We found only four intra-genus differences, consisting of one C-to-T reversion in one but not in other species of the same genus: Piper bettle but not P. houttuynia has a corrected B-9 site, Allium cepa and A. fistulosum but not A. textile have a corrected B-8 site, Medicago truncatula but not M. sativa has a corrected B-6 site, and Erodium carvifolium, but not the other four deposited Erodium sequences, has a corrected B-4 site. Three species of this genus (E. gruinum, E. guicciardii and E. chrysanthum) have strongly modified ndhB genes that are in fact pseudogenes but conserve the uncorrected TCA at the B-4 site. The fifth specie, E. texanum, has a true ndhB gene but in contrast to E. carvifolium, conserves the uncorrected TCA at the B-4 site. Significantly, extensive differences were reported in the chloroplast DNA organization among Erodium species (Weng et al., 2014).
The ndhF editing sites of 222 angiosperm genera are represented in Supplementary Table 2 by at least 2 species each, with a maximum of 43 species of Magnolia, totaling 922 species. Only five intra-genus differences indicating one C-to-T reversion in one but not in the other species of the same genus were found: Trithuria filamentosa and T. inconspicua but not T. submerse have a corrected F-2 site, Hydrocleys martii but not H. sp. Givnish s.n has a corrected F-1 site, Gymnopogon foliosus but not G. brevifolius has a corrected F-1 site, Urochloa foliosa, but none of the other 28 Urochloa species reported, has a corrected F-1 site, and Psychotria kirkii but not P. poeppigiana has a corrected F-2 site.
Discussion
Estimating Reversion Time Frames and the Origin of the Editing Sites of the ndh Genes
Despite considerable sample differences of the available sequences of the ndhB and ndhF genes, there are similarities in the patterns in plant phylogeny of C-to-T reversions that could be characterized for the ndh genes. As pointed out above, the low number of intra-genus differences in C to T substitutions suggests that the frequency of reversions could have declined relatively recently, say between 2 and 10 Myr BP. Certainly, the time span for the recent diversification of species must vary enormously between the different genera and the range of the time scale proposed, 2–12 Myr BP, although supported by some estimations (Jakob and Blattner, 2006), could in fact be longer. The low number of intra-genus differences regarding C-to-T reversions in the nine editing sites found in 1074 species sequenced could also be biased because only two species have been analyzed in many genera. However, the scant number of C-to-T reversions at the genus level strongly contrasts with the high frequency of corrections differentially affecting the comparatively low number of divisions, orders and families, even taking into account the longer time span, which would be around 200 Myr for the last.
Data pertaining to editing sites in gymnosperms and the fact that the 13 editing sites remain uncorrected in most species of primitive angiosperm orders such as Austrobaileyales, Chloranthales and Laurales (Supplementary Tables 1–3) suggest that sites B-5 and B-8 were created by T-to-C mutations in the hypothetical common ancestor of angiosperms and most, if not all, extant gymnosperms before the splitting of the branch leading to angiosperms, approximately 300 Myr BP (Savart et al., 1994; Herron et al., 2009). This common ancestor also has the “false” B-10 editing site with a C in the place of a T. Therefore, the other nine editing sites of the ndhB gene of angiosperms were originated by T-to-C mutations after the split which led to angiosperms but before the radiation leading to extant orders of angiosperms, probably around 170 Myr BP (Moore et al., 2007) as shown in Figure 3. Later, the correction of the B-7 site in monocotyledons preceded the early radiation of monocotyledon orders (Figures 2, 3) by some 130 Myr BP (Moore et al., 2007). The C-to-T reversion of editing sites, starting with B-5, B-7, and F-1 in the ancestors of extant species, should have begun at an uncertain time between 200 and 140 Myr BP, after which the recovery of some of the editing sites by T-to-C back mutations was unlikely.
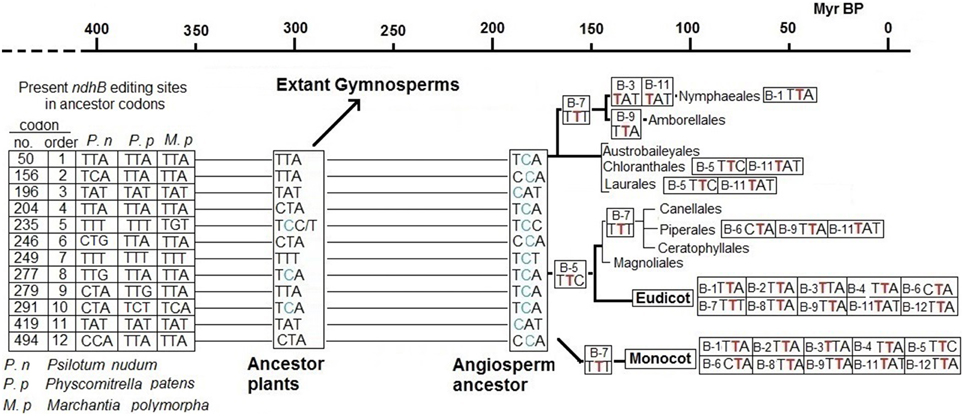
Figure 3. Timing for the creation and correction of editing sites of ndh genes. Specific Cs of codons B-5, B-8, and B-10 of non-flowering plants were T-to-C mutated (new C marked in blue) in a common ancestor of gymnosperms and angiosperms around 300 Myr BP. Additional Ts of the other nine editing sites were mutated to C (blue) between 300 and 170 Myr ago in the ancestor of extant angiosperms completing the 12 editing sites of the ndhB gene. Probably also between 300 and 170 Myr BP, similar T-to-C mutations created the ndhF sites F-1 and F-2 (not represented). After 170 Myr BP, C-to-T reversions successively corrected editing sites in specific plant lines. The number of each corrected editing site and codon involved (with the new T marked in red) is indicated at left when it affects all species comprised at right. The number of each corrected editing site and the codon involved (with the new T marked in red) is indicated at right when it affects some but not all species belonging to the indicated order or family.
Significantly, it is estimated that the gymnosperms Pinaceae and Gnetales lost the ndh genes more than 150 Myr ago (Braukmann et al., 2009). The ndh genes probably began to accumulate mutations more than 200 Myr ago and the genes were pseudogenized or completely lost in some gymnosperms in the common ancestor of Pinaceae (a family of Conifers) as well as the Gnetales, Gnetaceae, and Welwitschiaceae, whose branches lay close in the gymnosperm phylogenetic tree (Chaw et al., 2000), in the Permian-Triassic epochs. Most of the T-to-C mutations that generated the editing sites of ndh genes occurred between 330 and 200 Myr BP, from the end of the Carboniferous to the end of the Triassic. Therefore, it seems plausible that the functional role of the ndh genes was dispensable under the environmental conditions on Earth between 330 and 200 Myr BP. Pinaceae, a family of Conifers that were the predominant seed plants early in this period, were progressively displaced by Cycads and, later, by angiosperms (both of which have ndh genes) in the subsequent Jurassic and Cretaceous epochs (Figure 4).
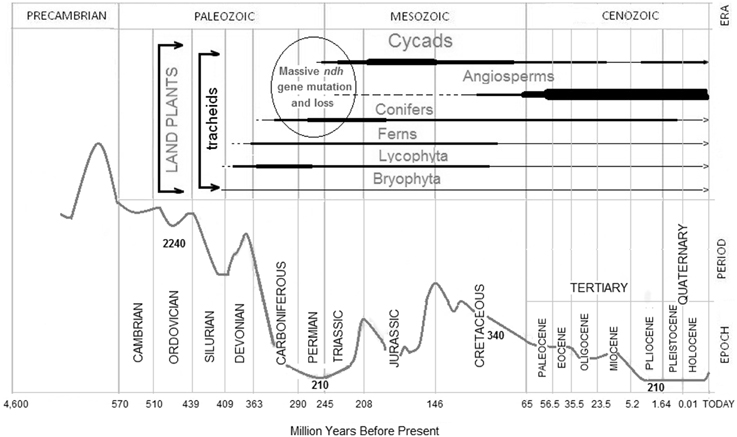
Figure 4. Expansion of selected plant groups and changes in the atmospheric CO2 concentration. Atmospheric CO2 (gray curve) and geological eras, periods, and epochs were redrawn from Nasif Nahle, Biology Cabinet (2009, http://www.biocab.org/carbon_dioxide_geological_timescale.html). The abundance of different plant groups along geological eras is represented by the relative thickness of the corresponding horizontal bars (in black below each plant group) as deduced from classical and recent references (Lowry et al., 1980; Taylor et al., 2009; Nagalingum et al., 2011). Representative CO2 concentrations are indicated (in ppm) below the gray curve. The ellipsoid grouping Cycads, angiosperms, and Conifers in the Permian and Triassic indicates the period during which the proposed main massive ndh mutations occurred on the basis of data of plants lacking ndh genes or requiring ndh transcript editing. At present, there are limited data on the ndh genes of Ferns, Lycophyta, and Bryophyta, which were probably also affected by massive mutations.
Possible Relationships Among the Inactivation and Restoration of the ndh Genes, Past Atmospheric Concentrations of CO2 and the Functional Role of the Thylakoid Ndh Complex
The concentration of atmospheric CO2 drastically decreased during the Carboniferous (http://www.biocab.org/carbon_dioxide_geological_timescale.html) to extraordinarily low levels of approximately 210 ppm during the Permian and Triassic (Figure 4). By the middle Triassic (some 220 Myr BP), the concentration of CO2 initiated an increase that, with oscillations, remained high until the middle Pliocene (some 4 Myr BP), when it again decreased to about 210 ppm until the recent agricultural and industrial revolutions.
The combination of sequence data with geological and paleontological records suggests that ancestor(s) of extant Pinaceae, Gnetaceae and Welwitschiaceae lost their ndh genes at the same time that ancestor(s) of other extant plants accumulated mutations, among other, T-to-C mutations, in the ndh genes during the Permian-Triassic period of low CO2 levels. As shown in Figure 4, the atmospheric CO2 concentrations decreased significantly (in the range of 200–250 ppm) twice: in the Permian-Triassic (200–300 Myr BP) and after the Pliocene (5 Myr BP) and it is also estimated that ndh genes were again lost in a few Erodium clades diverging 5 Myr BP (Fiz et al., 2008; Blazier et al., 2011). Therefore, it seems that the functional role of the ndh genes became dispensable under conditions of low CO2 concentrations and the genes were definitively lost in Pinnaceae, Gnetaceae and Welwitschiaceae.
In agreement with this hypothesis, we recently found that, under fluctuating light mimicking environmental conditions, photosynthetic performance is impaired in tobacco transgenics defective in ndh genes at high but not at low concentrations of CO2. Table 2, representing the aggregate measurements of more extensive experiments (Martín et al., 2015), shows that when the concentration of CO2 increases from 360 to 500 ppm, the efficiency of the photosynthetic conversion of light energy to biomass chemical energy increases in a range of 31–38% in tobacco plants with functional ndh genes (wt, ndhF FC and T181D) and only 8 to 11% in ndh-deficient tobacco (T181A and ΔndhF). Accordingly, the entropy production associated to the photosynthetic process (per unit of chemical energy stored as biomass) decreases 9–10% in ndh-deficient and 25–29% in tobacco plants with functional ndh genes when the concentration of CO2 increases from 360 to 500 ppm. These results strongly suggest that the presence of functional ndh genes contributes positively to plant fitness (Sabater, 2006) by improving photosynthetic productivity at high concentrations of CO2 and that ndh genes could become dispensable at low concentrations of CO2.
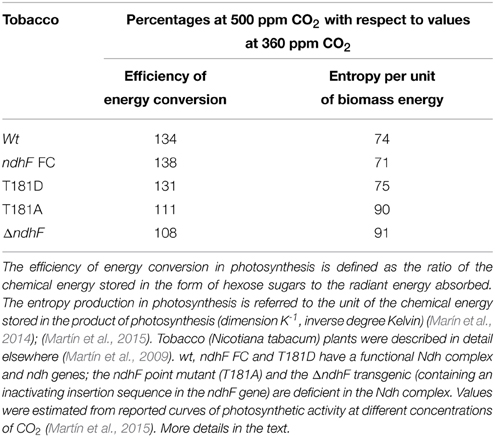
Table 2. Effect of ndh gene deficiency on the photosynthetic efficiency and the entropy production at high CO2 concentrations.
The role of the ndh genes improving photosynthetic efficiency at high CO2 concentrations and fluctuating light intensities explains the accumulation of nearly 50% of all chloroplast gene editing sites in their transcripts and their absence in some photosynthetic land plants. When the atmospheric CO2 concentration was below 300 ppm, the ndh genes provided no obvious advantages and became dispensable, accumulated mutations that rendered them non-functional and, over time, were deleted. However, at CO2 concentrations significantly higher than actual (Berner, 1997; Pearson and Palmer, 2000), the ndh genes contributed to vigorous photosynthesis with high photosynthetic efficiencies and low rates of entropy production (Martín et al., 2015). The loss of ndh genes in some species of aquatic Alismatales (Iles et al., 2013; Peredo et al., 2013) could be related to their lifestyle in submersed environments with no exposure to the frequent and broad range of light intensity fluctuations. Dependence on efficient photosynthetic activity is moderate in epiphytic plants, which could permit the loss of ndh genes. Hence, some Orchidaceae (such as those of the genus Dendrobium) have pseudo-ndh genes, but others (such as those of the genus Cypripedium) retain complete plastid ndh genes (Luo et al., 2014). However, ndh deletion is not always correlated with epiphytic habit and all orchids analyzed contain ndh gene fragments in the mitochondrial genomes (Lin et al., 2015). Probably, the Ndh complex is also dispensable in mild environments (Martín and Sabater, 2010; Ruhlman et al., 2015), where CO2 concentrations could be high, and further genomic and functional investigations in Orchidaceae must provide key information.
Accordingly, the absence of ndh genes was probably a factor contributing to the relative decline of Conifers during the Jurassic (Figure 4) when the concentration of CO2 increased. Gnetales, supposedly abundant in the Permian, are now represented by only a few species (Chaw et al., 2000). Conifers, the predominant vegetation during the Permian-Triassic, were progressively displaced by Cycads and, later, by angiosperms (both of which have ndh genes) in the subsequent Jurassic and Cretaceous epochs when, with oscillations, the concentrations of CO2 increased (Figure 4) (Nagalingum et al., 2011). Common ancestors of angiosperms and many gymnosperms have been traced back to 280 Myr BP (Savart et al., 1994) to the transition from the Carboniferous to the Permian when the concentration of CO2 could have decreased and the accumulation of mutations in ndh genes did not impair the photosynthetic efficiency. Conceivably, sites B-5, B-8, and B-10, common to both gymnosperms and angiosperms, were generated at this time by T-to-C mutations. Shortly after, in the middle Permian-Triassic when the low concentration of CO2 persisted, massive mutations (including T-to-C) accumulated in the ndh genes of the specific angiosperm line (Martín and Sabater, 2010). Later, when the concentration of CO2 increased again, ndh genes only affected by T-to-C mutations recuperated their functionality by C-to-U transcript editing. The functional rescue by C to T reversions would be slow in the highly polyploidal background of chloroplasts (Sabater et al., 2002) whereas editing provided the remedy for deleterious point mutations in transcripts of all plastid DNAs of a plant cell. Subsequent slower occurring C to T reversions progressively dispenses the requirement of editing of specific sites.
Concluding Remarks and Further Prospects
Most, if not all, editing sites of the plastid ndh genes were originated by T-to-C mutations between 300 and 200 Myr BP (when certain gymnosperms completely lost the ndh genes) and are being corrected by C-to-T reversions since about 200 Myr BP, coinciding with the main radiations of angiosperms. C-to-T back mutations are essentially irreversible and occur randomly at the different editing sites and among the different angiosperm branches. Hence, when one site is C-to-T corrected in all species of, say, an order, the reversion can reasonably be traced back to the common ancestor of all species of the order. The wide range of frequencies of C-to-T reversions in angiosperms (from 57.2% for the B-5 site to 2.3% for the B-8 site) reflects (from the high to the low percentage) the antiquity of the correction event and the extent and diversification of descendants. From this perspective, the identification of the corrected and uncorrected editing sites allows dating of past reversion events and could provide a complementary tool for determining phylogenetic relationships. With this objective, we have created data bases of the editing sites of the ndhB and ndhF genes, the actual states of which are provided in the Supplementary Tables 1, 2 which we intend to update, include other ndh genes and make accessible over the coming years.
Evidences for the functional role of the ndh genes improving photosynthesis under rapidly fluctuating light intensities, at high but not at low CO2 concentrations, and the analysis of the editing sites of ndh genes allow us to propose that the concentration of CO2 was the key environmental factor which selected or dispensed ndh genes during gymnosperm and angiosperm evolution. Probably, when enough sequences are available, a similar approach with non-vascular and first vascular plants could help in understanding the phylogeny and the functional adaptation of plants to land.
Author Contributions
BS and MM conceived and designed research. BS, MM and PS conducted data screening and analyzed results. DM revised data and performed thermodynamics comparison. BS wrote the manuscript. PS English edited the manuscript. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Grant BFU2010-15916 of the Spanish Dirección General de Investigación (Ministerio de Economía y Competitividad).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fevo.2015.00081
Abbreviations
BP, before present; Myr, million years.
References
Barrett, C. F., Freudenstein, J. V., Li, J., Mayfield-Jones, D. R., Perez, L., and Santos, C. (2014). Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 31, 3095–3112. doi: 10.1093/molbev/msu252
Berner, R. A. (1997). The rise of plants and their effect on weathering and atmospheric CO2. Science 276, 544–546.
Blazier, J. C., Guisinger, M. M., and Jansen, R. K. (2011). Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol. Biol. 76, 263–272. doi: 10.1007/s11103-011-9753-5
Braukmann, T., Kuzmina, M., and Stefanovič, S. (2013). Plastid genome evolution across the genus Cuscuta (Convolvulaceae): two clades within subgenus Grammica exhibit extensive gene loss. J. Exp. Bot. 64, 977–989. doi: 10.1093/jxb/ers391
Braukmann, T., and Stefanovič, S. (2012). Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol. Biol. 79, 5–20. doi: 10.1007/s11103-012-9884-3
Braukmann, T. W. A., Kuzmina, M., and Stefanovíc, S. (2009). Loss of all ndh genes in Gnetales and conifers: extent and evolutionary significance for seed plant phylogeny. Curr. Genet. 55, 323–337. doi: 10.1007/s00294-009-0249-7
Bremer, K. (2000). Early Cretaceus lineages of monocot flowering plants. Proc. Natl. Acad. Sci. U.S.A. 97, 4707–4711. doi: 10.1073/pnas.080421597
Bremer, K. (2002). Gondwanan evolution of the grass alliance of families (Poales). Evolution 56, 1374–1387. doi: 10.1111/j.0014-3820.2002.tb01451.x
Burleigh, J. G., Bansal, M. S., Eulenstein, O., Hartmann, S., Wehe, A., and Vision, T. J. (2011). Genome-scale phylogenetics: inferring the plant tree of life from 18,896 Gene Trees. Syst. Biol. 60, 117–125. doi: 10.1093/sysbio/syq072
Casano, L. M., Zapata, J. M., Martín, M., and Sabater, B. (2000). Chlororespiration and poising of cyclic electron transport: plastoquinone as electron transporter between thylakoid NADH dehydrogenase and peroxidase. J. Biol. Chem. 275, 942–948. doi: 10.1074/jbc.275.2.942
Chateigner-Boutin, A. L., and Small, I. (2007). A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 35, e114. doi: 10.1093/nar/gkm640
Chaw, S. M., Parkinson, C. L, Cheng, Y., Vincent, T. M., and Palmer, J. D. (2000). Seed plant phylogeny inferred from all three plant genomes: monophyly of extant gymnosperms and origin of Gnetales from conifers. Proc. Natl. Acad. Sci. U.S.A. 97, 4086–4091. doi: 10.1073/pnas.97.8.4086
Chen, H., Deng, L., Jiang, Y., Lu, P., and Yu, J. (2011). RNA editing sites exist in protein-coding genes in the chloroplast genome of Cycas taitungensis. J. Integr. Plant Biol. 53, 961–970. doi: 10.1111/j.1744-7909.2011.01082.x
Del Campo, E. M., Albertazzi, F., Freyer, R., Maier, R. M., Sabater, B., and Martin, M. (1998). Sequence and transcript editing of ndhB gene of Arabidopsis thaliana L. Plastid (Accession Nos. AJ002490 & AJ002491). Plant Physiol. (PGR98-093) 117, 718.
Fiz, O., Vargas, P., Alarcón, M., Aedo, C., García, J. L., and Juan José Aldasoro, J. J. (2008). Phylogeny and historical biogeography of Geraniaceae in relation to climate changes and pollination ecology. Sys. Bot. 33, 326–342. doi: 10.1600/036364408784571482
Freyer, R., Kiefer-Meyer, M. C., and Kossel, H. (1997). Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl. Acad. Sci. U.S.A. 94, 6285–6290.
Freyer, R., Lopez, C., Maier, R. M., Martin, M., Sabater, B., and Kossel, H. (1995). Editing of the chloroplast ndhB encoded transcript shows divergence between closely related members of the grass family (Poaceae). Plant Mol. Biol. 29, 679–684.
Fučíková, K., Leliaert, F., Cooper, E. D., Škaloud, P., D'Hondt, S., De Clerck, O., et al. (2014). New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front. Ecol. Evol. 2: 63. doi: 10.3389/fevo.2014.00063
Givnish, T. J., Pires, J. C. H., Graham, S. W., McPeherson, M. A., Prince, L. M., Patterson, T. B, et al. (2006). Phylogenetic relationships of monocots based on the highly informative plastid gene ndhF: evidence for widespread concerted convergence. Aliso 22, 28–51.
Herron, M. D., Hackett, J. D., Aylward, F. O., and Michod, R. E. (2009). Triassic origin and early radiation of multicellular volvocine algae. Proc. Natl. Acad. Sci. U.S.A. 106, 3254–3258. doi: 10.1073/pnas.0811205106
Iles, W. J. D., Smith, S. Y., and Graham, S. W. (2013). “A well-upported phylogenetic fremwork for the monocot order Alismatales reveals multiple losses of the plastid NADH dehydrogenase complex and strong long-branch effect,” in Early Events in Monocot Evolution, eds P. Wilkin and S. J. Mayo (Cambridge, UK: Cambridge University Press), 1–28.
Jakob, S. S., and Blattner, F. R. (2006). A Chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol. Biol. Evol. 23, 1602–1612. doi: 10.1093/molbev/msl018
Lin, C. S., Chen, J. J. W., Huang, Y. T., Chan, M. T., Daniell, H., Chang, W. J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Reports 5:9040. doi: 10.1038/srep09040
Lowry, B., Lee, D., and Hébant, C. (1980). The origin of land plants: a new look at an old problem. Taxon 29, 183–197.
Luo, J., Hou, B.-W., Niu, Z.-T, Liu, W., Xue, Q.-Y., and Ding, X.-Y. (2014). Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS ONE 9:e99016. doi: 10.1371/journal.pone.0099016
Maier, R. M., Neckermann, K., Igloi, G. L., and Kössel, H. (1995). Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 251, 614–628.
Maier, R. M., Zeltz, P., Kossel, H., Bonnard, G., Gualberto, J. M., and Grienenberger, J. M. (1996). RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 32, 343–365.
Marín, D., Martín, M., Serrot, P., and Sabater, B. (2014). Thermodynamic balance of photosynthesis and transpiration at increasing CO2 concentrations and rapid light fluctuations. BioSystems 116, 21–26. doi: 10.1016/j.biosystems.2013.12.003
Martín, M., Funk, H. T., Serrot, P. H., Poltnigg, P., and Sabater, B. (2009). Functional characterization of the thylakoid Ndh complex phosphorylation by site-directed mutations in the ndhF gene. Biochim. Biophys. Acta. 1787, 920–928. doi: 10.1016/j.bbabio.2009.03.001
Martín, M., Marín, D., Serrot, P. H., and Sabater, B. (2015). The rise of the photosynthetic rate when light intensity increases is delayed in ndh gene-defective tobacco at high but not at low CO2 concentrations. Front Plant Sci. 6:34. doi: 10.3389/fpls.2015.00034
Martín, M., and Sabater, B. (2010). Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 48, 636–645. doi: 10.1016/j.plaphy.2010.04.009
Moore, M. J., Bell, C. D., Soltis, P. S., and Soltis, D. E. (2007). Using plastid genome-scala data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. U.S.A. 104, 19363–19368. doi: 10.1073/pnas.0708072104
Nagalingum, N. S., Marshall, C. R., Quental, T. B., Rai, H. S., Little, D. P., and Mathews, S. (2011). recent synchronous radiation of a living fossil. Science 334, 796–799. doi: 10.1126/science.1209926
Pearson, P. N., and Palmer, M. R. (2000). Atmospheric carbon dioxide concentrations over the past 60 million years. Nature 406, 695–700. doi: 10.1038/35021000
Peredo, E. L., King, U. M., and Les, D. H. (2013). The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH Complex in an Aquatic Angiosperm. PLoS ONE 8:e68591. doi: 10.1371/journal.pone.0068591
Ruhlman, T. A., Chang, W. J., Chen, J. J. W., Huang, Y. T., Chan, M. T., Zhang, J., et al. (2015). NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 15:100. doi: 10.1186/s12870-015-0484-7
Rumeau, D., Peltier, G., and Courmac, L. (2007). Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 30, 1041–1051. doi: 10.1111/j.1365-3040.2007.01675.x
Sabater, B. (2006). Are organisms committed to lower their rates of entropy production? Possible Relevance to evolution of the Prigogine theorem and the ergodic hypothesis. BioSystems 83, 10–17. doi: 10.1016/j.biosystems.2005.06.012
Sabater, B., Martín, M., Schmitz-Linneweber, C., and Maier, R. M. (2002). Is clustering of plastid10. RNA editing sites a consequence of transitory loss of gene function? Implications for past environmental and evolutionary events in plants. Persp. Plant Ecol. Evol. Syst. 5, 81–90. doi: 10.1078/1433-8319-00024
Savart, L., Li, P., Strauss, S. H., Chase, M. W., Michaud, M., and Bousquet, J. (1994). Chloroplast and nuclear gene sequences indicate late Pennsylvanian time for the last common ancestor of extant seed plants. Proc. Natl. Acad. Sci. U.S.A. 91, 5163–5167.
Sazanov, L. A., Burrows, P. A., and Nixon, P. J. (1998). The plastid ndh genes code for an NADH-specific dehydrogenase: purification and characterization of a mitochondrial-like complex I from pea thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 95, 1319–1324.
Serrot, P. H, Sabater, B., and Martín, M. (2012). Activity, polypeptide and gene identification of thylakoid Ndh complex in trees: potential physiological relevance of fluorescence assays. Physiol. Plant. 146, 110–120. doi: 10.1111/j.1399-3054.2012.01598.x
Shikanai, T. (2006). RNA editing in plant organelles: machinery, physiological function and evolution. Cell. Mol. Life Sci. 63, 698–708. doi: 10.1007/s00018005-5449-9
Soltis, D. E., Smith, S. A., Cellinese, N., Wurdack, K. J., Tank, D. C., Brockington, S. F., et al. (2011). Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 98, 704–730. doi: 10.3732/ajb.1000404
Takenaka, M., Zehrmann, A., Verbitskiy, D., Härtel, B., and Brennicke, A. (2013). RNA editing in plants and its evolution. Ann Rev Genet. 47, 335–352. doi: 10.1146/annurev-genet-111212-133519
Taylor, T. N., Taylor, E. L., and Krings, M. (2009). Paleobotany: the Biology and Evolution of Fossil Plants. Amsterdam: Burlington Academic Press.
Tillich, M., Funk, H. T, Schmitz-Linneweber, C., Poltnigg, P., Sabater, B., Martin, M., et al. (2005). Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 43, 708–715. doi: 10.1111/j.1365-313X.2005.02484.x
Tillich, M., Lehwark, P., Morton, B. R., and Maier, U. G. (2006). The evolution of chloroplast RNA editing. Mol. Biol. Evol. 23, 1912–1921. doi: 10.1093/molbev/msl054
Van Den Bekerom, R. J. M., Dix, P. J., Diekmann, K., and Barth, S. (2013). Variations in efficiency of plastidial RNA editing within ndh transcripts of perennial ryegrass (Lolium perenne) are not linked to differences in drought tolerance. AoB Plants 5:plt035. doi: 10.1093/aobpla/plt035
Wakasugi, T., Hirose, T., Horihata, M., Tsudzuki, T., Kossel, H., and Sugiura, M. (1996). Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc. Natl. Acad. Sci. U.S.A. 93, 8766–8770.
Weng, M. L., Blazier, J. C., Govindu, M., and Jansen, R. K. (2014). Reconstruction of the ancestral plastid genome in geraniaceae reveals a correlation between genome rearrangements, repeats and nucleotide substitution rates. Mol. Biol. Evol. 31, 645–659. doi: 10.1093/molbev/mst257
Keywords: angiosperm radiation, atmospheric CO2, chloroplast DNA, editing phylogeny, ndhB gene, ndhF gene, Permian-Triasic, transcript editing
Citation: Martín M, Marín D, Serrot PH and Sabater B (2015) Evolutionary reversion of editing sites of ndh genes suggests their origin in the Permian-Triassic, before the increase of atmospheric CO2. Front. Ecol. Evol. 3:81. doi: 10.3389/fevo.2015.00081
Received: 04 February 2015; Accepted: 07 July 2015;
Published: 22 July 2015.
Edited by:
Laura M. Boykin, The University of Western Australia, AustraliaReviewed by:
Rodney L. Honeycutt, Pepperdine University, USARobert K. Jansen, The University of Texas at Austin, USA
Copyright © 2015 Martín, Marín, Serrot and Sabater. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bartolomé Sabater, Department of Life Science, University of Alcalá, Alcalá de Henares, 28805 Madrid, Spain, bartolome.sabater@uah.es
 Mercedes Martín
Mercedes Martín Dolores Marín
Dolores Marín Patricia H. Serrot
Patricia H. Serrot Bartolomé Sabater
Bartolomé Sabater