Viral Genome Size Distribution Does not Correlate with the Antiquity of the Host Lineages
- 1Evolutionary Biology, Facultad de Ciencias, Universidad Nacional Autónoma de México, Mexico City, Mexico
- 2Miembro de El Colegio Nacional, Mexico City, Mexico
It has been suggested that RNA viruses and other subcellular entities endowed with RNA genomes are relicts from an ancient RNA/protein World which is believed to have preceded extant DNA/RNA/protein-based cells. According to their proponents, this possibility is supported by the small-genome sizes of RNA viruses and their manifold replication strategies, which have been interpreted as the result of an evolutionary exploration of different alternative genome organizations and replication strategies during early evolutionary stages. At the other extreme are the giant DNA viruses, whose genome sizes can be as large as those of some prokaryotes, and which have been grouped by some authors into a fourth domain of life. As argued here, the comparative analysis of the chemical nature and sizes of the viral genomes reported in GenBank does not reveal any obvious correlation with the phylogenetic history of their hosts. Accordingly, it is somewhat difficult to reconcile the proposal of the putative pre-DNA antiquity of RNA viruses, with their extraordinary diversity in plant hosts and their apparent absence among the Archaea. Other issues related to the genome size of all known viruses and subviral agents and the relationship with their hosts are discussed.
Introduction
Almost as soon as they were discovered and characterized as subcellular entities, viruses were considered by some to be the first forms of life (d'Herelle, 1926). Many were convinced that the small size and apparent simplicity of modern viruses could be interpreted as evidence of their primitiveness, and that they could be considered as operational models of the processes that had led to the emergence of life (Beutner, 1938; Podolsky, 1996; Fisher, 2010).
During the past few years, this virocentric hypothesis has been resurrected based on both the nature and size genome of viruses. Since RNA viruses have small genomes and diverse strategies of replication, it has been suggested they have their roots in the early stages of evolution that preceded the appearance of cellular DNA genomes (Forterre, 2006; Koonin et al., 2006; Agol, 2010; Koonin and Dolja, 2013). On the other extreme are the so-called nucleocytoplasm large DNA viruses (NCLDV) endowed with the largest viral genomes reported so far, which in some cases may be even larger than some small prokaryotic genomes, have been considered by some as a fourth domain of life comparable to the Bacteria, the Archaea and the Eucarya (Raoult et al., 2004; Boyer et al., 2009; Nasir et al., 2012).
There are several studies trying to date the emergence of viruses. In some of them, it has been analyzed the distribution of viral-protein domains at the SCOP database and it has been concluded viruses emerged from primordial segmented RNA proto-virocells and not from modern cellular entities, and also, they suggested that eukaryotic viruses are not descendants from prokaryotic viruses (Nasir et al., 2015). In others, it has been determined the existence of replication-and-structure hallmark genes not found in cell genomes, and therefore it has been proposed an ancient virus world (Koonin et al., 2006). Moreover, in other works, it has been proposed these genes have homologs in cell genomes, and probably they could be horizontally transferred between cells and viruses, and between viruses and other viruses (Caprari et al., 2015). In other studies, it has been suggested the origin of viruses coincides with the appearance of viral capsid. The cell capsid-like genes could be considered an exaptation that emerged from horizontal gene transfer from cells to cellular parasitic templates (Jalasvuori et al., 2015).
However, the ultimate origin of viruses is still unknown and remains an open issue. In the present study, we have studied the correlation of genome size of both RNA- and DNA viruses with the antiquity of the lineages of their prokaryotic and eukaryotic hosts in order to date the possible emergence of viruses after cell origin, and gain some insights on the evolutionary aspects of their phylogenetic relationship. We have used both the genomic information of the reference species of all viral families reported in GenBank as of December 2014, and the molecular, cellular, phylogenetic, and information of their hosts distributed in the three major domains of life.
Materials and Methods
Retrieval of Viral and Host Data
Biological data of RNA- and DNA viruses (species, host, group, family, taxonomic code, genome size, and number of segments) as of December 2014 were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genomes/) on a plain-text file (see Supplementary Material 1) and, in some cases, verified or complemented with the 9th Report of the International Committee on Taxonomy of Viruses (King et al., 2011), the viral web resource ViralZone (http://viralzone.expasy.org/), and from relevant publications. A total of 4182 viral reference strains were obtained from the Genbank. A total of 215 satellite- and 44 viroid reference strains were also retrieved, but treated as additional independent points for this study. Viruses whose host was not identified in the GenBank (n = 31) were excluded.
Classification of Virus Data
All virus reference strains of the database were classified in four categories (see Supplementary Material 1). According to the Baltimore Classification System (Baltimore, 1971), the first category included seven groups: double-stranded DNA (dsDNA, n = 1926), single-stranded DNA (ssDNA, n = 701), double-stranded RNA (dsRNA, n = 205), positive-sense ssRNA [ssRNA(+), n = 966], negative-sense ssRNA [ssRNA(−), n = 253], reverse-transcribing dsDNA (dsDNA-RT, n = 70), and reverse-transcribing ssRNA (ssRNA-RT, n = 62). Depending on the host type, the second category included viruses infecting prokaryotes: Bacteria (n = 1438) and Archaea (n = 69); and viruses infecting eukaryotes: diatoms (n = 5), algae (n = 37), protists (n = 32), plants (n = 1273), fungi (n = 82), plants and invertebrates (n = 58), invertebrates (n = 260), invertebrates and vertebrates (n = 123), vertebrates (n = 1064). According to their level of segmentation, the third category included viruses from 1 to more than 105 segments divided by Baltimore groups and host types. According to the genome type, the four category included 55 families of RNA viruses (n = 1485) and 43 families of DNA viruses (n = 2697).
Analysis of Viral Genome Size According to the Baltimore Classification, Host Type, and Their Level of Segmentation
The genome size average of each set of viruses grouped by Baltimore Classification and host type was calculated. All viruses were also classified by genome size, host type, and number of segments. The large ranges of genome sizes were graphed logarithmically with a base-10 log scale in both cases.
Analysis of Viral Genome Size According to the Antiquity of Cell Domains
Data of the 99 families of viruses that have an identified host in the GenBank was compiled according to viral genome nature and host type. The percentage of RNA- and DNA viral families of each host type was estimated. Viral families were double-counted if they infected more than one host type. Viral families were classified according to host types as follows: proteobacteria (n = 8 viral families), other bacteria (n = 7) for the Bacteria domain; Crenarchaeota (n = 9) and Euryarchaeota (n = 4) for the Archaea domain; and protists and algae (n = 7), plants (n = 21), fungi (n = 15), and animals (n = 50) for the Eucarya domain.
The Interactive Tree of Life (IToL) platform was used to generate a phylogenetic tree of cell domains following the online instructions based on Letunic and Bork (2007).
Results
There is a bias in our study. We have determined the number of all viral records according to the Baltimore Classification and host type. Viruses that infect bacteria (34%), plants (31%), and vertebrates (25%) are the most represented viruses in the current database due to medical, agricultural, and historical reasons (see Supplementary Material 1).
Larger and Smaller Viruses Mainly Infect Eukaryotes
DNA viruses exhibit the most diverse-size genomes of all viruses in this study. In our sample, DNA-virus genome sizes vary by approximately four orders of magnitude, with the smallest (0.859 kbp) recorded in Circovirus SFBeef (ssDNA), and the largest one (2473 kbp) in Pandoravirus salinus (dsDNA). RNA viruses have the most-restricted size genomes of all viruses (Figure 1). Interestingly, DNA viruses which infect bacteria have genome sizes slightly larger than those which infect some animals. RNA-virus genome sizes vary by approximately one order of magnitude, from the smallest (1.8 kbp) reported in Saccharomyces cerevisiae killer virus M1 (dsRNA) to the largest one (33.452 kbp) in Ball python nidovirus [ssRNA(+)]. In spite of several major searches, as of June 2015, no RNA viruses infecting Archaea have been yet reported.
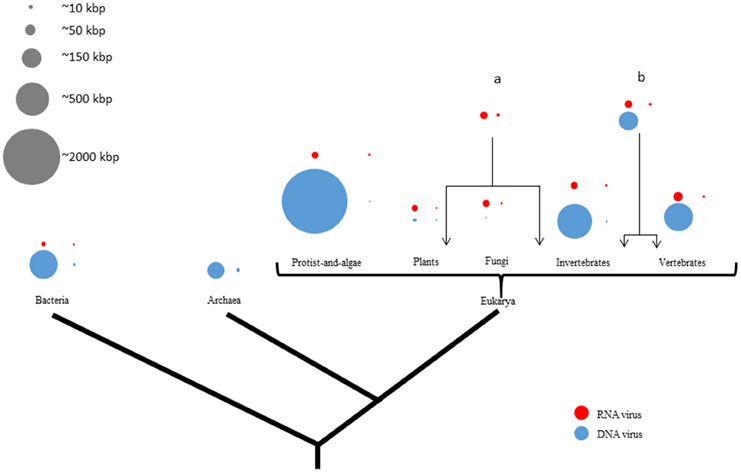
Figure 1. Viral genome sizes are non-randomly distributed in the tree of life representing their hosts. There are four circles by domain or phyla from phylogenetic tree of life which proportionally represent the largest and the smallest genome size from either RNA- (red) or DNA (blue) viruses. Gray circles represent a genome-size scale approximation. Larger and smaller viruses appear to infect mainly eukaryotes. There is more genome-size diversity in DNA viruses than in RNA viruses in all three domains of life. While DNA viral genome exhibits a size range of four orders of magnitude, RNA viral genome sizes are much more restricted. On the contrary, RNA viruses are more widely distributed among eukaryotes than among prokaryotes. Moreover, RNA viruses infect more phylogenetically-distant hosts like both plants and invertebrates (a) or both invertebrates and vertebrates (b).
We have analyzed the genome size distribution of the viruses in our sample following the Baltimore Classification of the seven groups of viruses [dsDNA, ssDNA, dsRNA, ssRNA(+), ssRNA(−), ssRNA-RT, and dsDNA-RT] and host type (Bacteria, Archaea; diatoms, algae, plants, fungi, invertebrates, vertebrates, both plants and invertebrates, and both invertebrates and vertebrates).
As shown in Figures 1, 2, the DNA- and RNA viral entities analyzed in our sample exhibit considerable diversity in their genome sizes but within well-defined limits. Moreover, viral genome sizes are not randomly distributed. Our results show that DNA-virus genomes exhibit a diverse range of sizes, with the dsDNA viruses displaying a more size-flexible genome distribution than ssDNA viruses (Figure 2). The genome sizes of dsDNA viruses vary by approximately two orders of magnitude, and can be divided into three well-defined genome-size groups: (a) those which infect protists (1000 kbp); (b) those which infect algae, invertebrates, and vertebrates (from 160 to 240 kbp); and (c) those which infect prokaryotes and vertebrates (from 40 to 70 kbp). There are no reports of dsDNA viruses infecting plants or fungi. Of the 33 families and 133 unclassified species of dsDNA viruses, the amoeba-infecting P. salinus (unclassified) has the largest genome (2500 kbp), and the vertebrate-infecting Bovine polyomavirus (Polyomaviridae), the smallest one (5 kbp) (Figure 2A and Supplementary Material 2). The majority of the largest-genome sizes of dsDNA, ssDNA, and ssRNA(−) viruses occupies the third quartile (75%) of the data (Figure 2B).
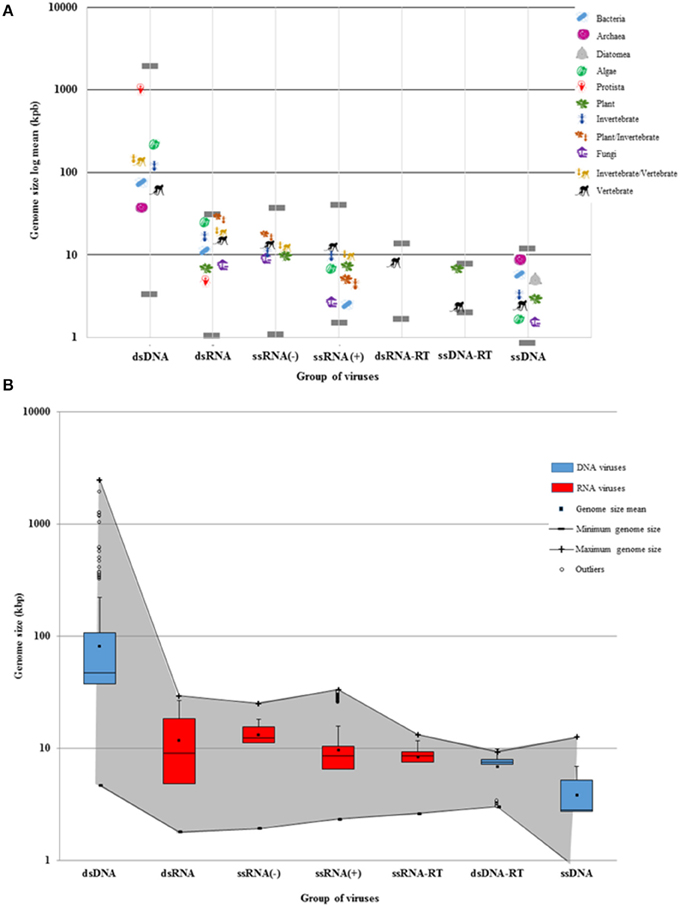
Figure 2. Viral genome sizes and the Baltimore groups. (A) The genome size average of viruses was calculated according to each Baltimore group and host type. Each host is denoted by a representative organism. Gray lines show the largest and the smallest virus of each Baltimore group. DNA viruses are grouped into two size extremes of the log graph (dsDNA and ssDNA). The dsDNA virus genomes show a more extended diversity than ssDNA genomes. Bacteria hosts can also be infected by longer dsDNA viruses, but rarely by RNA viruses. RNA and ssDNA viruses display more restricted genomes. However, RNA virus genomes can be larger than ssDNA genomes. The largest and the smallest RNA viruses are found as parasites infecting eukaryotes. Some families of RNA viruses can infect phylogenetically-distant hosts (plants and invertebrates, or invertebrates and vertebrates). Retroviruses only infect plants and vertebrates. (B) The maximum and minimum genome size of RNA (red) and DNA (blue) viruses are denoted by a gray shadow. While DNA viruses have a more-delineated plasticity of genome size, RNA viruses have more restricted genomes. There are more ds-and-ss DNA viral species that tend to have larger genomes as shown in the third quartile. While ssDNA and ds-RT DNA viruses have as restricted genomes as RNA viruses, dsDNA viruses tend to have larger genomes as shown in the outliers. The mean, the median, and the standard deviation were calculated from the 4182 viral reference strains of GenBank.
Secondly, our results demonstrate that RNA viruses exhibit the most restricted-size genome distribution of all viruses (Figure 2). The dsRNA viral group has, on the average, the largest genomes of all RNA viruses. Of the 12 families and 19 unclassified species of dsRNA viruses, the multihost-infecting Fiji disease virus (Reoviridae) has the largest genome (29 kbp), while the fungi-infecting Saccharomyces cerevisiae killer virus M1 has the smallest one (2 kbp) reported so far (Figure 2B). The majority of dsRNA has genome sizes that range from 4 to 9 kbp (50 and 75%, respectively) (Figure 2B).
The available data show that ssRNA(−) viruses have a limited range of genome sizes (from 10 to 15 kbp) independently of their eukaryotic hosts. Of the nine families and eight unclassified species of ssRNA(−) viruses, the plant-and-invertebrate-infecting Rice grassy stunt virus (unclassified) has the largest genome (25 kbp), while the plant-infecting Blueberry mosaic associated virus (Ophioviridae) has the smallest one (2 kbp) (Figure 2A). The majority of ssRNA(−) has genome sizes that range from 1 to 3 kbp (50 and 75%, respectively) (Figure 2B). On the other hand, dsRNA viruses and some ssRNA(−) that infect either plants or vertebrates (some of them via a vector) have rather large genomes (Figure 2 and Supplementary Material 2).
On the average, the smallest RNA viral genomes are found in the ssRNA(+) viruses. They can be divided in two genome-size groups: (a) those which infect bacteria and fungi (4 kbp) and (b) those which infect algae, plants, invertebrates, and vertebrates (from 6 to 12 kbp) (Figure 2). Of the 33 families and 61 unclassified species of ssRNA(+) studied here, the vertebrate-infecting Ball python nidovirus (Nidoviridae) has the largest genome (33 kbp), and the fungi-infecting Ophiostoma mitovirus 6 (Narnaviridae), the smallest one (2 kbp). The majority of ssRNA(+) has genome sizes that range from 1 to 2 kbp (75 and 50%, respectively) (Figure 2B).
Thirdly, the retro-transcribing viruses (ssRNA-RT and dsDNA-RT) have the most limited habitats of all viruses. The ssRNA-RT viruses only infect vertebrates and have genome sizes of 2–13 kbp. Similarly, the dsDNA-RT viruses described so far only infect plants or vertebrates and have a genome of ~9 kbp (Figure 2). While the majority of ssRNA-RT has genome sizes that range from 1 to 0.8 kbp (75 and 50%, respectively) (Figure 2B), most of dsDNA-RT have genome sizes that range from 0.3 to 7 kbp (75 and 50%, respectively) (Figure 2B).
Quite surprisingly, the smallest viral genomes are found in the ssDNA viruses (Circoviridae, ~1 to 2 kbp). The genomes of ssDNA viruses are clearly more size-restricted than those of dsDNA viruses, and in our sample the upper-size limit of ssDNA genome sizes is 10 kbp on average (Figure 2A). The available data show that ssDNA viruses are the only ones that infect diatomea, and the smallest known genomes of ssDNA viruses (and, indeed, of all viruses) are found in algae on average. There is only one ssDNA viral species with a genome size of 2 kbp that infects fungi. In the sample reported here, of all nine families of ssDNA viruses and the 54 unclassified species of ssDNA viruses, the invertebrate-infecting Bombyx mori bidensovirus (Bidnaviridae) has the largest genome (12 kbp), while the invertebrate-infecting Circoviridae SFBeef (Circoviridae) has the smallest one (< 1 kbp). The majority of ssDNA viruses has genome sizes that range from 0.8 to 2 kbp (50 and 75%, respectively) (Figure 2B).
Quite remarkably, the analysis of the distribution of viral sample studied here indicates that viruses endowed with the largest and the smallest genomes only infect eukaryotes (see Supplemented Material 2).
RNA Viruses Mostly Infect Eukaryotes
The sample reported here has 44% of DNA viruses (44 families) and 56% of RNA viruses (55 families) (Supplementary Material 3). There are 18 DNA viral families that infect prokaryotes, and 26 DNA viral families that infect eukaryotes. On the other side, there are only two RNA viral families that infect prokaryotes, and 53 RNA viral families that infect eukaryotes. Hence, RNA viral families are found mostly infecting the eukaryotic domain (Figure 3).
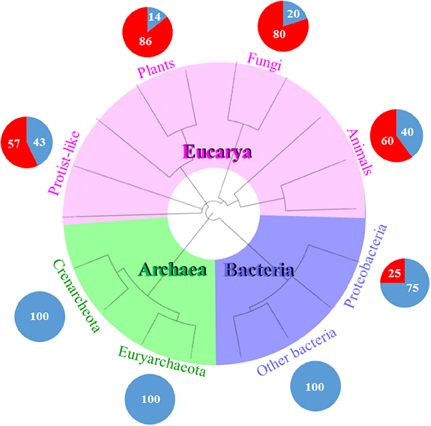
Figure 3. RNA viruses mostly infect eukaryotes. This schematic tree represents the three domains of life (Bacteria in purple, Archaea in green, and Eucarya in pink). Each pie chart represents the percentage of DNA or RNA viral families infecting each domain. For example, there are only two RNA and six DNA viral families representing the 25 and 75%, respectively, which infect Proteobacteria and no other bacteria. Proteobacteria is a phyla which is phylogenetically and physiologically related to eukaryotes. It seems that the evolutionary history of RNA viruses is linked to the phylogeny of the recent eukaryotes. The phylogenetic tree was generated and visualized using iTOL (2). The circular tree was constructed following the online instructions based on Letunic and Bork (2007).
Ten DNA- and RNA viral families have been described that infect Bacteria. There are eight DNA- and only two RNA families of bacterial viruses. Seven of the eight DNA viral families infect the Actinobacteria, Deinococcus-Thermus, Firmicutes, and Tenericutes clades. Of all the eight viral families that infect Proteobacteria, only two of them are RNA viruses.
Of the 12 DNA viral families that are known to infect Archaea, four of them infect Euryarchaeota, and nine infect Crenarchaeota, while the Fuselloviridae infect both archeal clades. The Myoviridae and the Siphoviridae have viral species that cross-infect bacteria and archaea. As of today, there is not a single record of an RNA virus infecting an archaea (Figure 3).
There are 79 DNA- and RNA viral families that infect Eucarya. Six of them infect protist-and-algae hosts, and 74 families infect fungi, plants, and animals. The Reoviridae is the only family that infects both protist-and-algae and multicellular eukaryotes. Of the 55 RNA viral families, the 96% of them belong to eukaryotic viruses, i.e., most RNA viral families are found in eukaryotes and not in prokaryotes (Figure 3).
The dsRNA Viruses with Larger and Segmented Genomes Infect a Wide Range of Phylogenetically-Distant Eukaryotes
In our sample, 89% of all known viruses are non-segmented. Of all the DNA- and RNA viruses described so far, 9% of them have two or three segments, and only 2% of them have more than four segments. There are twice as much segmented RNA viruses than segmented DNA viruses. There is a clear bias in their distribution. Of all segmented DNA- and RNA viruses, 60% infect plants, 21% infect several phylogenetically-distant eukaryotes (either plants and invertebrates, or invertebrates and vertebrates), 18% infect vertebrates, and only 1% infects bacteria.
The Polydnaviridae is the only dsDNA viral group with segmented genomes, and it exhibits the largest genome size of all known invertebrate viruses. Their genomes range from 15 to 105 segments (Figure 4). The second most-segmented genomes of all known viruses are found in the plant-infecting Nanoviridae (ssDNA), and range from 6 to 14 segments. The plant-infecting Geminiviridae (two segments), and the vertebrate-infecting Birdnaviridae (two segments) are the only other ssDNA viral families endowed with segmented genomes.
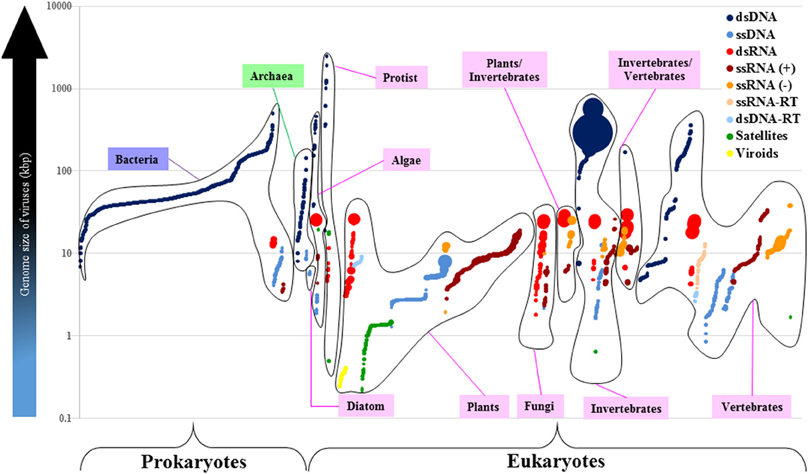
Figure 4. The dsRNA viruses with larger-and-segmented genomes infect different groups of eukaryotes. Each viral group based on the Baltimore Classification is identified by a different color. Each closed-line group refers to either a prokaryote or a eukaryote host. Each circle represents one of all 4182 viral reference strains distributed in the x axis according to the host they infect (Bacteria in purple, Archaea in green, and Eucarya in pink). The volume of circles is directly proportional to the number of genome segments of each virus. The log scale shows the genome size of viruses in kilobase-pair units. There are no segmented-dsDNA genomes in viruses (only one exception: the Polydnaviridae that exceeds the 100 segments; big dark-blue circles). Genome size of RNA viruses cannot surpass the 40-kbp boundary. Most of the segmented viruses infect plants and animals (vertebrates and invertebrates).
On the other hand, there are 20 RNA viral families with segmented genomes. The third most-segmented genomes (2–12 segments) of all known viruses are found in the Reoviridae (dsRNA) which also exhibit, on the average, the largest genomes of all RNA viruses, but do not seem to surpass the 40-kbp size limit. The Reoviridae is the only viral family that infects several eukaryotic hosts including algae, plants, fungi, invertebrates, and vertebrates (Figure 4 and Supplementary Material 3). There are seven dsRNA-viral families whose genomes range from 2 to 12 segments, and have sizes that range from 3 to 29 kbp. There are five ssRNA(−) viral families whose genomes range from 1 to 8 segments, and vary from 1 to 38 kbp (the Filoviridae have the largest genome of all known RNA viruses, with two segments that add up to 38 kbp). There are eight ssRNA(+) viral families whose genomes range from 1 to 4 segments, and whose sizes vary from 3 to 19 kbp.
Therefore, viruses endowed with the smallest genomes, like ssDNA and RNA viruses, appear to have the most segmented genomes (with an extreme exception in dsDNA viruses from the Polydnaviridae). Moreover, the largest RNA genomes are segmented and are found in dsRNA viruses that infect a wide range of phylogenetically-distant eukaryotes.
Discussion
In this report we have investigated the relationship between viral genome sizes and the antiquity of their host lineages, and have shown that the available data demonstrate that genome sizes do not exhibit a random distribution. In our sample, genome sizes of dsDNA viruses have the highest diversity and also surpass by far the more restricted genome sizes of RNA and ssDNA viruses. On the contrary, RNA viruses exhibit a wider range of eukaryotic hosts, but infect relatively few bacterial lineages compared to DNA viruses.
Our results show that dsDNA viral genomes display a much more diverse size range than RNA- and ssDNA viruses. This can be explained as a result of the enhanced chemical stability of the Watson-Crick helices. The available data indicate that dsDNA viral genome sizes can be divided in three major groups. The largest dsDNA genomes are those of the so-called nucleo-cytoplasmic large DNA viruses (NCLDV) that infect amoeba, such as Pandoravirus and Pithovirus (Raoult et al., 2004; Pennisi, 2013; Philippe et al., 2013; Legendre et al., 2014), whose genomes appear to have major contributions from other viruses as well as from prokaryotic and eukaryotic microbes, due to accretion processes in which horizontal gene transfer may have played a significant role (Filée et al., 2007; Colson and Raoult, 2010; Filée, 2013).
The second group is that of dsDNA viruses that infect algae, invertebrates and vertebrates, and that have genome sizes that range from 150 to 240 kbp. This second genome-size group includes giant viruses like the Phycodnaviridae, the Iridoviridae, and the Asfarviridae. It has been suggested that these eukaryotic viral families share a common ancestor with the largest-genome giant viruses, supporting the idea of an additional branch of life (Iyer et al., 2006; Boyer et al., 2009; Yutin and Koonin, 2012; Nasir et al., 2015). However, it has been argued that the giant viruses, like the Marseilleviridae, have in fact increased their genomes with eukaryotic sequences through horizontal gene transfer (Moreira and Brochier-Armanet, 2008; Boyer et al., 2009; Filée, 2014) and do not constitute a fourth domain of life (Yutin et al., 2014).
Finally, the third and also the smallest genome-size group includes dsDNA viruses that infect prokaryotes and vertebrates. It is possible that the small size of prokaryotic viruses is constrained by their small-size capsids (Krupovic et al., 2011). However, the Myoviridae and the Siphoviridae, the only two families that cross-infect both Bacteria and Archaea, have genomes that range from 10 to 500 kbp. Together with the Podoviridae, these two-tailed viral families of bacteriophages appear to be an ancient and genetically connected viral group (Hendrix, 2002). There is no evidence of cross-infection between prokaryotes and eukaryotes, which may suggest a domain-specific origin of viruses. The smallest genomes of dsDNA viruses (5–7 kbp) are those of the Polyomaviridae and the Papillomaviridae, which infect mammals and birds (de Villiers et al., 2004; Crandall et al., 2006). Therefore, the largest genomes of dsDNA viruses are found in those which infect eukaryotes.
It is somewhat surprising that the smallest genomes are found not only in RNA viruses but also in ssDNA viruses. Although both viral types exhibit a difference of one magnitude in their genome sizes, the smallest viral genomes are found in ssDNA viruses. It thus appears that the genomes of ssDNA viruses are subjected to the same restrictions that hinder the size increase in RNA viral genomes, most likely due to the lack of repair mechanisms (Reanney, 1982). Both viral types exhibit comparable behavior, including high mutation rate, large population sizes, small levels of horizontal gene transfer, little gene duplication, overlapping reading frames and, often, little recombination (Duffy and Holmes, 2008; Holmes, 2009).
Unlike DNA viruses, RNA viral families infect a wide range of phylogenetically diverse eukaryotic hosts, an evolutionary dispersal that may explain why some of them have coevolved with their invertebrate vectors (Gray and Banerjee, 1999; Lobo et al., 2009; Obbard and Dudas, 2014). One of the viral families that infect multiple hosts is the Reoviridae, which also exhibit multiple segmentation and large genomes (see Figure 3). It has been suggested that segments of dsRNA genome of Reoviridae probably recombine through complementation when two or more viruses co-infect a single cell (Reanney, 1982; Froissart et al., 2004; Holmes, 2009). It has been argued that the Reoviridae cannot undergo major increases in the genome size, since this would require a complex molecular machinery including unwinding proteins, DNA-dependent ATPases, and nucleases which are not encoded by RNA viruses (Reanney, 1982).
It is somewhat surprising that with the exception of only two known examples, all RNA viral families appear to be restricted to eukaryotic hosts. The only two families that infect bacteria (Proteobacteria) are the Leviviridae [ssRNA(+)] and the Cystoviridae (dsRNA). It has been speculated that the latter could be derived from eukaryotic viruses (Holmes, 2009).
The wide range of RNA viral parasites infecting nucleated cells very likely explains the eukaryotic defense mechanisms that include degradation of viral RNA, presence of microRNAs, and RNAi mechanisms against viruses (Berkhout and Haasnoot, 2006; Lodish et al., 2008; Obbard et al., 2009; Parameswaran et al., 2010; Obbard and Dudas, 2014). RNA-mediated silencing is a highly conserved mechanism that was probably present in the last common ancestor of eukaryotes (Cerutti and Casas-Mollano, 2006), which may indicate an ancient evolutionary relationship between nucleated cells and RNA viruses, whose origin could thus be placed some time near the actual emergence of eukaryotic microbes.
As reviewed above, it has been argued that viruses were the first living entities and RNA viruses or viroids may be direct descendants of the RNA World. It has also been suggested that retroviral-like elements are relics of the early evolutionary transition from an RNA/protein world into the extant DNA/RNA/protein cells, and that the ancestor of dsDNA giant viruses was an ancient cell (Podolsky, 1996; Daròs et al., 2006; Koonin et al., 2006; Flores et al., 2014). Our results suggest that these schemes may be mistaken. This alternative possibility is supported by phylogenetic analyses that indicate that all known viral monomeric RNA polymerases are derived from cellular DNA polymerases A and B (Jácome et al., 2015). Although the results presented here may be severely affected by methodological issues that include biased representations of viral diversity, our data show that in terms of their genome size and organization RNA viruses are not endowed with the simpler and smallest genomes of all known viruses as is generally believed, and in fact that they may be more closely related to the evolutionary history of their eukaryotic hosts. Our results also suggest that since retroviruses appear to be restricted to plants and vertebrates, they could not have played a role in the evolutionary transition from primitive cellular RNA genomes to the extant DNA-based genetic systems of extant cells, nor the viral reverse transcriptase can be considered an evolutionary vestige of the polymerase that played a role in this transition. The results presented here demonstrate that viral genome sizes are not randomly distributed, but do not appear to be correlated with the antiquity of their hosts. Therefore, viruses may be ancient, but not primitive.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are indebted to Dr. León Patricio Martínez Castilla for several useful discussions. JC is supported by the Consejo Nacional de Ciencia y Tecnología (CONACyT), scholarship number 165264. The support of the Posgrado en Ciencias Biológicas, UNAM, to JC is gratefully acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fevo.2015.00143
References
Agol, V. I. (2010). Which came first, the virus or the cell? Paleontol. J. 44, 728–736. doi: 10.1134/S0031030110070038
Berkhout, B., and Haasnoot, J. (2006). The interplay between virus infection and the cellular RNA interference machinery. FEBS Lett. 580, 2896–2902. doi: 10.1016/j.febslet.2006.02.070
Boyer, M., Yutin, N., Pagnier, I., Barrassi, L., Fournous, G., Espinosa, L., et al. (2009). Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. U.S.A. 106, 21848–21853. doi: 10.1073/pnas.0911354106
Caprari, S., Metzler, S., Lengauer, T., and Kalinina, O. V. (2015). Sequence and structure analysis of distantly-related viruses reveals extensive gene transfer between viruses and hosts and among viruses. Viruses 10, 5388–5409. doi: 10.3390/v7102882
Cerutti, H., and Casas-Mollano, J. A. (2006). On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 50, 81–99. doi: 10.1007/s00294-006-0078-x
Colson, P., and Raoult, D. (2010). Gene repertoire of amoeba-associated giant viruses. Intervirology 53, 330–343. doi: 10.1159/000312918
Crandall, K. A., Pérez-Losada, M., Christensen, R. G., McClellan, D. A., and Viscidi, R. P. (2006). Phylogenomics and molecular evolution of polyomaviruses. Adv. Exp. Med. Bio. 577, 46–59. doi: 10.1007/0-387-32957-9_3
Daros, J.-A., Elena, S. F., and Flores, R. (2006). Viroids: an Ariadne's thread into the RNA labyrinth. EMBO Rep. 7, 593–598. doi: 10.1038/sj.embor.7400706
de Villiers, E. M., Fauquet, C., Broker, T. R., Bernard, H. U., and Zur Hausen, H. (2004). Classification of papillomaviruses. Virology 324, 17–27. doi: 10.1016/j.virol.2004.03.033
d'Herelle, F. (1926). The Bacteriophage and its Behavior. Baltimore, MD: The Williams & Wilkins Company.
Duffy, S., and Holmes, E. C. (2008). Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus tomato yellow leaf curl virus. J. Virol. 82, 957–965. doi: 10.1128/JVI.01929-07
Filee, J. (2013). Route of NCLDV evolution: the genomic accordion. Curr. Opin. Virol. 3, 595–599. doi: 10.1016/j.coviro.2013.07.003
Filee, J. (2014). Multiple occurrences of giant virus core genes acquired by eukaryotic genomes: the visible part of the iceberg? Virology 466, 53–59. doi: 10.1016/j.virol.2014.06.004
Filée, J., Siguier, P., and Chandler, M. (2007). I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet. 23, 10–15. doi: 10.1016/j.tig.2006.11.002
Fisher, S. (2010). Are RNA viruses vestiges of an RNA world? J. Gen. Philos. Sci. 41, 121–141. doi: 10.1007/s10838-010-9119-8
Flores, R., Gago-Zachert, S., Serra, P., Sanjuán, R., and Elena, S. F. (2014). Viroids: Survivors from the RNA World? Annu. Rev. Microbiol. 68, 395–414. doi: 10.1146/annurev-micro-091313-103416
Forterre, P. (2006). The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 117, 5–16. doi: 10.1016/j.virusres.2006.01.010
Froissart, R., Wilke, C. O., Montville, R., Remold, S. K., Chao, L., and Turner, P. E. (2004). Co-infection weakens selection against epistatic mutations in RNA viruses. Genetics 168, 9–19. doi: 10.1534/genetics.104.030205
Gray, S. M., and Banerjee, N. (1999). Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63, 128–148.
Hendrix, R. W. (2002). Bacteriophages: evolution of the majority. Theor. Popul. Biol. 61, 471–480. doi: 10.1006/tpbi.2002.1590
Holmes, E. (2009). The Evolution and Emergence of RNA Viruses. New York, NY: Oxford University Press Inc.
Iyer, L. A., Balaji, S., Koonin, E. V., and Aravind, L. (2006). Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117, 156–184. doi: 10.1016/j.virusres.2006.01.009
Jácome, R., Becerra, A., Ponce De León, S., and Lazcano, A. (2015). Structural analysis of monomeric RNA-dependent polymerases: evolutionary and therapeutic implications. PLoS ONE 10:e0139001. doi: 10.1371/journal.pone.0139001
Jalasvuori, M., Mattila, S., and Hoikkala, V. (2015). Chasing the origin of viruses: capsid-forming genes as a life-saving preadaptation within a community of early replicators. PLoS ONE 10:e0126094. doi: 10.1371/journal.pone.0126094
King, A. M. Q., Lefkowitz, E., Adams, M. J., and Carstens, E. B. (2011). Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic Press.
Koonin, E. V., and Dolja, V. V. (2013). A virocentric perspective on the evolution of life. Curr. Opin. Virol. 3, 546–557. doi: 10.1016/j.coviro.2013.06.008
Koonin, E. V., Senkevich, T. G., and Dolja, V. V. (2006). The ancient virus world and evolution of cells. Biol. Direct 1, 1–27. doi: 10.1186/1745-6150-1-1
Krupovic, M., Prangishvili, D., Hendrix, R. W., and Bamford, D. H. (2011). Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol. Mol. Biol. Rev. 75, 610–635. doi: 10.1128/MMBR.00011-11
Legendre, M., Bartoli, J., Shmakova, L., Jeudy, S., Labadie, K., Adrait, A., et al. (2014). Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. U.S.A. 111, 4274–4279. doi: 10.1073/pnas.1320670111
Letunic, I., and Bork, P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Lobo, F. P., Mota, B. E. F., Pena, S. D. J., Azevedo, V., Macedo, A. M., Tauch, A., et al. (2009). Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PLoS ONE 4:e6282. doi: 10.1371/journal.pone.0006282
Lodish, H. F., Zhou, B., Liu, G., and Chen, C. Z. (2008). Micromanagement of the immune system by microRNAs. Nat. Rev. Immunol. 8, 120–130. doi: 10.1038/nri2252
Moreira, D., and Brochier-Armanet, C. (2008). Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol. Biol. 8:12. doi: 10.1186/1471-2148-8-12
Nasir, A., Kim, K. M., and Caetano-Anolles, G. (2012). Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya. BMC Evol. Biol. 12:156. doi: 10.1186/1471-2148-12-156
Nasir, A., Sun, F.-J., Kim, K. M., and Caetano-Anollés, G. (2015). Untangling the origin of viruses and their impact on cellular evolution. Ann. N.Y. Acad. Sci. 1341, 61–74. doi: 10.1111/nyas.12735
Obbard, D. J., and Dudas, G. (2014). The genetics of host-virus coevolution in invertebrates. Curr. Opin. Virol. 8, 73–78. doi: 10.1016/j.coviro.2014.07.002
Obbard, D. J., Gordon, K. H. J., Buck, A. H., and Jiggins, F. M. (2009). The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. B Biol. Sci. 364, 99–115. doi: 10.1098/rstb.2008.0168
Parameswaran, P., Sklan, E., Wilkins, C., Burgon, T., Samuel, M. A., Lu, R., et al. (2010). Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6:e1000764. doi: 10.1371/journal.ppat.1000764
Pennisi, E. (2013). Ever-bigger viruses shake tree of life. Science 341, 226–227. doi: 10.1126/science.341.6143.226
Philippe, N., Legendre, M., Doutre, G., Couté, Y., Poirot, O., Lescot, M., et al. (2013). Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341, 281–286. doi: 10.1126/science.1239181
Raoult, D., Audic, S., Robert, C., Abergel, C., Renesto, P., Ogata, H., et al. (2004). The 1.2-megabase genome sequence of mimivirus. Science 306, 1344–1350. doi: 10.1126/science.1101485
Reanney, D. C. (1982). The evolution of RNA viruses. Annu. Rev. Microbiol. 36, 47–73. doi: 10.1146/annurev.mi.36.100182.000403
Yutin, N., and Koonin, E. V. (2012). Hidden evolutionary complexity of nucleocytoplasmic large DNA viruses of eukaryotes. Virol. J. 9:161. doi: 10.1186/1743-422X-9-161
Keywords: viral genome sizes, origin of viruses, viruses and the RNA-protein world
Citation: Campillo-Balderas JA, Lazcano A and Becerra A (2015) Viral Genome Size Distribution Does not Correlate with the Antiquity of the Host Lineages. Front. Ecol. Evol. 3:143. doi: 10.3389/fevo.2015.00143
Received: 04 July 2015; Accepted: 07 December 2015;
Published: 23 December 2015.
Edited by:
Johann Peter Gogarten, University of Connecticut, USAReviewed by:
Youn-Sig Kwak, Gyeongsang National University, South KoreaSoo Rin Kim, Kyungpook National University, South Korea
Copyright © 2015 Campillo-Balderas, Lazcano and Becerra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arturo Becerra, abb@ciencias.unam.mx
 José A. Campillo-Balderas
José A. Campillo-Balderas Antonio Lazcano
Antonio Lazcano Arturo Becerra
Arturo Becerra