Asymmetric Distribution of pl10 and bruno2, New Members of a Conserved Core of Early Germline Determinants in Cephalochordates
- 1Gatty Marine Laboratory, Scottish Oceans Institute, University of St Andrews, St Andrews, UK
- 2Department of Genetics, University of Barcelona, Barcelona, Spain
Molecular fingerprinting of conserved germline and somatic “stemness” markers in different taxa have been key in defining the mechanism of germline specification (“preformation” or “epigenesis”), as well as expression domains of somatic progenitors. The distribution of molecular markers for primordial germ cells (PGCs), including vasa, nanos, and piwil1, as well as Vasa antibody staining, support a determinative mechanism of germline specification in the cephalochordate Branchiostoma lanceolatum, similarly to other amphioxus species. pl10 and bruno2, but not bruno4/6, are also expressed in a pattern consistent with these other germline genes, adding to our repertoire of PGC markers in lancelets. Expression of nanos, vasa, and the remaining markers (musashi, pufA, pufB, pumilio, and piwil2) may define populations of putative somatic progenitors in the tailbud, the amphioxus posterior growth zone, or zones of proliferative activity. Finally, we also identify a novel expression domain for musashi, a classic neural stem cell marker, during notochord development in amphioxus. These results are discussed in the context of germline determination in other taxa, stem cell regulation, and regenerative capacity in adult amphioxus.
Introduction
One of the key innovations coupled to the evolution of multicellularity was the ability to segregate the germline and the soma, with transcriptional repression of a somatic programme being key to maintaining the germ cell fate (Hallmann, 2011). Historically, two main mechanisms of germline specification have been defined in animals: preformation and epigenesis (Extavour and Akam, 2003), or determinative and inductive modes, respectively. In the determinative mode, cytoplasmic determinants associated with the germ plasm in the egg are inherited by a limited number of daughter cells during cleavage, which are thus specified as presumptive germ cells (PGCs), and go on to form the mature adult gametes. In contrast, during induction, inductive cues cause somatic cells to become specified as germline. Studies in mouse, axolotl, and cricket suggest that BMP signaling may be an ancient mechanism for PGC induction from mesoderm in animals (Chatfield et al., 2014; Donoughe et al., 2014). The phylogenetic distribution of these two mechanisms of germ cell specification suggests that the inductive mode may represent the ancestral state, and that germ plasm has evolved independently multiple times (Blackstone and Jasker, 2003; Extavour and Akam, 2003; Johnson et al., 2003a,b; Crother et al., 2007; Extavour, 2007; Ewen-Campen et al., 2010).
Comparative studies in multiple taxa have revealed that the molecular signature of germ cells may often be shared across species that use both determinative and inductive modes of PGC specification, leading to the proposition of a conserved germline multipotency programme (Extavour, 2007; Juliano et al., 2010). Interestingly, some basal metazoans appear to use a combination of mechanisms to specify germ cells, and many of the classic germline markers are in fact also expressed in adult somatic stem cells in these organisms (Alié et al., 2011; Leclère et al., 2012). Germline-associated genes are most often RNA-binding proteins, but there is considerable species-specific variation in the suite employed (see Gazave et al., 2013 for a compilation of much of the recent literature). However, a key core of proteins including Vasa/PL10, Tudor and a PIWI domain containing protein may represent an ancestral “pluripotency module” (Ewen-Campen et al., 2010). Tweaking upstream regulators or downstream targets, combined with the addition of new germ cell genes, such as nanos or bruno, would have generated the diversity in germline specification mechanisms in early metazoans (Ewen-Campen et al., 2010).
Until recently, little was known about germline specification in cephalochordates (lancelets or amphioxus), the sister group to the vertebrates and tunicates, and the best living proxy for the ancestral chordate (Bertrand and Escrivà, 2011). Classic studies suggested that lancelets might employ an inductive mode of PGC specification (reviewed in Extavour and Akam, 2003). Electron microscopy data however showed that in Branchiostoma floridae, the pole plasm localizes to the vegetal cortex soon after fertilization and segregates into a single blastomere during cleavage, putting into question this hypothesis (Holland and Holland, 1992). Although functional data are still lacking, blastomere separation experiments combined with expression data for molecular markers traditionally associated with the germline, including piwi-like1, nanos, vasa, and Vasa protein, strongly support a determinative mode of PGC specification in cephalochordates (Wu et al., 2011; Zhang et al., 2013 and Figure 1). Zygotic expression domains from gastrulation onwards of these genes, as well as piwi-like2 and tudor7, also suggest a function in somatic progenitors/stem cells of the posterior growth zone. Together, these data provide a general framework for understanding how markers for PGCs and posterior progenitors may be expressed in cephalochordates during development (Figure 1).
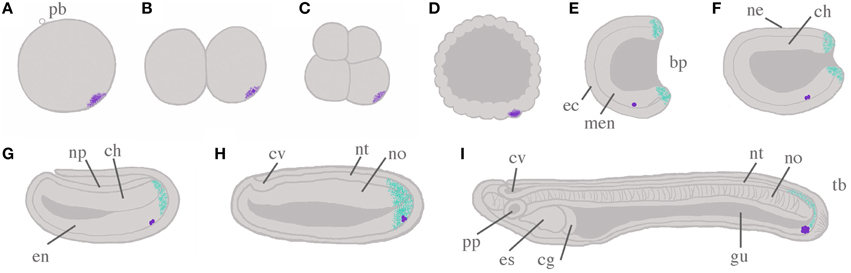
Figure 1. Schema of amphioxus embryogenesis, with hypothesized scenario for germ cell development. The germ plasm and Primitive Germ Cells (PGCs) are indicated by stippled violet and violet dots, respectively. Putative posterior somatic stem cell zones are indicated in cyan. (A) Fertilized egg: the polar body (pb) indicates the animal pole; (B) two cell stage; (C) eight cell stage; (D) blastula; (E) mid-gastrula: ectoderm (ec) and mesendoderm (men) are evident; (F) late gastrula/early neurula: dorsal ectoderm has become neurectoderm (ne), with chordomesoderm (ch) located ventrally in the midline; (G) mid-neurula: ectoderm has grown over the neural plate (np); (H) late neurula: both neural tube (nt) and cerebral vesicle (cv) can be distinguished, as well as notochord (no) and paraxial somites (not shown); (I) pre-mouth stage larva: the notochord has begun to differentiate to form the characteristic “stacked coin” structure. A through-gut (gu) begins to form, and asymmetry becomes apparent with preoral pit (pp) and endostyle (es) on the left side, and club-shaped gland (cg) on the right. With the exception of (A–D), all embryos are illustrated with dorsal upwards, and anterior to the left. All panels show sections through the midline only for simplicity, so somites and axial musculature, located either side of the midline, are by necessity omitted. Additional abbreviations are used to indicate the blastopore (bp), endoderm (en) and the tailbud (tb).
Currently, the most convincing evidence for the existence of somatic stem cells in cephalochordates comes from studies of tail regeneration in the European amphioxus, Branchiostoma lanceolatum, whose adult regenerative ability is comparable to that seen in many ambulacrarians (echinoderms and hemichordates; Somorjai et al., 2012a,b). Unfortunately, no germ cell markers have so far been characterized during development in this species, and few putative somatic stem cell markers exist in cephalochordates. The purpose of this study was therefore threefold: First, to characterize the early expression of candidate amphioxus germline markers nanos, piwil1, vasa, and Vasa protein in B. lanceolatum for comparative purposes with other cephalochordates; second, to determine whether germline markers in other taxa, including pumilio, pufA, pufB, musashi, pl10, bruno2, and bruno4/6, are associated with PGCs in amphioxus; and third, to analyse the late developmental expression of some of these candidates as a prelude to future regeneration studies. Given the considerable conservation in developmental gene expression patterning in cephalochordates demonstrated thus far (e.g., Somorjai et al., 2008; Wu et al., 2011; Zhang et al., 2013), we hypothesize that markers for PGCs and posterior somatic domains will show comparable gene expression profiles in B. lanceolatum to B. belcheri, B. japonicum, and B. floridae.
Here, we present the first analysis of putative germline and somatic stem cell markers in the European amphioxus, B. lanceolatum. We identify a core set of conserved PGC-associated markers in cephalochordates, including piwil1, nanos, and vasa, characterize Vasa protein distribution, and identify two new candidate germ cell markers in cephalochordates, pl10, and bruno2. We also characterize the amphioxus musashi ortholog, whose expression in the notochord represents a novelty in chordates. The highly conserved molecular expression data in the Branchiostomatidae support the view that cephalochordate evolution is strongly constrained, and show that data are broadly transposable across species, even in the context of germline formation. This study also provides the foundation for future studies of regeneration in the amphioxus B. lanceolatum.
Materials and Methods
Embryos
Ripe adults were collected in Argelès-sur Mer (France) and spawned as previously described (Fuentes et al., 2007). Embryos were fixed at the relevant time points in 4% PFA in MOPS salts (0.1 M MOPS, 2 mM MgSO4, 1 mM EGTA, and 0.5 M NaCL), and stored in 70% ETOH at −20°C. For phalloidin staining, embryos were stored in PBS at 4°C. Embryos were staged according to Hirakow and Kajita (1991, 1994), with modifications as per Zhang et al. (2013).
Phylogenetic Analysis
If not previously published, putative orthologous sequences were identified using a BLASTp search; reciprocal BLAST was used to confirm identity (Camacho et al., 2009). Protein sequences were aligned in Jalview version 2.8.2 (Waterhouse et al., 2009) using MAFFT on default settings, and checked manually. All positions with less than 95% site coverage were eliminated directly in MEGA5 prior to analysis. Evolutionary models considered to best describe the substitution pattern were identified as those with the lowest BIC (Bayesian Information Criterion) scores using MEGA5 (Tamura et al., 2011). Both neighbor joining (NJ) and maximum likelihood (ML) analyses were performed with 500 and 1000 bootstraps, respectively. The Nearest Neighbor Interchange method was used to infer tress in ML. Unless otherwise noted, and since concordant with results from the NJ method, only ML trees are shown. Model details for each analysis are included in the figure legends for ease of reference. All sequences used for phylogenetic analyses, including associated accession numbers, are included in Supplementary File 1.
Cloning and Probe Synthesis
RNA was extracted from embryos and adult tissues using Trizol and phenol chloroform extraction; cDNA was generated using Tetro cDNA Synthesis kit (Bioline). Gene fragments for probe generation were amplified by PCR using gene-specific primers designed against the genome of B. floridae (Supplementary File 2), ligated into PGEMT–Easy (Promega) and transformed into XL10-Gold (Stratagene) or DH5α (Invitrogen) strains of E. coli by heat shock using standard protocols. Selected clones were mini-prepped using peqGOLD or Promega plasmid miniprep kits, and sequence verified. Template was generated by PCR on plasmids using Universal M13F (5′ GTAAACGACGGCCAGT 3′) and M13R (5′AACAGCTATGACCATG 3′) primers. The band was gel-purified using either the QIAquick (Qiagen), GFX (Amersham), or Isolate II (Bioline) gel extraction kits following manufacturers guidelines. DIG-labeled (Roche) antisense probes were in vitro transcribed using T7, T3, and SP6 enzymes as appropriate following standard protocols. Probes were checked by agarose electrophoresis and purified using miniQuick Spin columns (Roche) or via precipitation with sodium acetate (3 M, pH 5.2) and ethanol.
Whole Mount In situ Hybridization (WMISH)
WMISH was performed as previously described (Somorjai et al., 2008). Briefly, fixed embryos were washed in PBT (0.1% Tween), and permeabilized using proteinase K (7.5 mg/ml) for empirically-tested periods based upon embryo stage and enzyme batch. Embryos were postfixed for 40 min in PFA, deacetylated in acetic acid in triethanolamine (0.1 M, pH 8), and pre-hybridized at least 2 h in hybridization solution. Embryos were incubated overnight with shaking at 60–65°C depending on probe. The first post-hybridization washes were performed at the hybridization temperature, with subsequent washes at room temperature in decreasing concentrations of SSC. An RNAse step was included (37°C). Embryos were incubated overnight in primary antibody (anti-DIG AP, Roche), pre-adsorbed at 1:3000, with rocking at 4°C. Copious washing in PBT was performed between each step. For the chromogenic reaction we used either BM Purple (Roche) or NBT/BCIP (Roche); embryos were postfixed in PFA for 20 min when the signal:background was deemed appropriate. At least three WMISH were performed for each gene, on 10–50 embryos per stage in total. Embryos were mounted in 80% glycerol/20% PBS, and photographed under a Leitz DMRB microscope (Leica Microsystems) with Normarski optics. Photographs were taken with the Retiga 2000R camera and the QCapture software suite (QImaging), and processed in Adobe Photoshop CS3.
Immunohistochemistry
Immunohistochemistry and Alexa-fluor 568-labeled phalloidin stainings for F-Actin (Invitrogen, 1:400) were carried out as per Somorjai et al. (2012a). Briefly, after fixation, embryos were washed in PBT (phosphate buffered saline plus 0.1% Tween, pH 7.6), and permeabilized in PBS with 0.2% Triton-X for 40 min. After copious washing in PBT, embryos were incubated overnight at 4°C in primary antibody. Embryos were again washed in PBT and incubated in secondary antibody or phalloidin for 2 h at room temperature, or overnight at 4°C. A specific B. floridae anti-Vasa antibody, generously donated by Dr Jr-Kai Yu, was used at 1:20,000 (Wu et al., 2011). Secondary antibodies were Alexa fluor 488 and 568 diluted at 1:400 (Molecular Probes). Embryos were mounted in Vectashield (VectorLabs) containing Hoescht 33342 dye to stain nuclei (1:2000 of 10 mg/ml). Confocal images were taken on a Lecia TCS SP8 confocal microscope, and processed using NIH ImageJ 1.48 d and Adobe Photoshop CS3.
Results
Identification of Candidate Germline and Somatic Stem Cell Markers in B. lanceolatum
We selected DEAD-box (Vasa, Pl10), Pumilio domain (Pumilio, PufA, PufB), PIWI domain (Piwil1, Piwil2) RRM (Musashi), CELF (Bruno2 and Bruno4/6), and Nanos families as candidate germline and somatic stem cell markers for analysis in B. lanceolatum. When B. floridae orthologs had not been previously characterized in the extensive phylogenetic analyses of Kerner et al. (2011), we identified putative stem cell markers using BLASTp searches against the genomes of B. floridae and B. belcheri, and confirmed the identity of our B. lanceolatum proteins by comparison with published sequences in other cephalochordates (Supplementary File 4), including transcriptomic data from B. lanceolatum in the NCBI TSA database (Oulion et al., 2012). We generated phylogenies that include, where possible, sequences from more than one amphioxus species to support the identity of these proteins (Supplementary File 5; and see below). We then cloned partial sequences of orthologs in B. lanceolatum using primers designed in its sister species B. floridae (Supplementary File 2). Using this approach, we successfully cloned 12 genes (including two piwil1; not shown) with known function in the germline or somatic stem cells (Table 1). While previous phylogenies show that the distinction among Piwi clades is unequivocal (Kerner et al., 2011), the evolutionary history of piwi genes in cephalochordates is more complex. We identified a single piwil2 (piwiA in Kerner et al., 2011) and three piwil1 (piwiB in Kerner et al., 2011) genes in the genomes of B. belcheri and B. floridae. The latter belong to an apparent tandem duplication cluster (not shown and Yue et al., 2015) that appears to be present in all Branchiostoma, as we successfully cloned two of the three paralogs of piwil1 in B. lanceolatum. We also identified an ortholog of piwiX (“piwilike” in Zhang et al., 2013), but have been unable to clone the gene in B. lanceolatum. While EST data collected in NCBI and B. floridae EST databases (Yu et al., 2008) support the expression of piwil1 and piwil2 (Supplementary File 3), we have not identified any expression data for piwiX in any database, including our own tail regenerate transcriptome dataset (Dailey and Somorjai, unpublished).
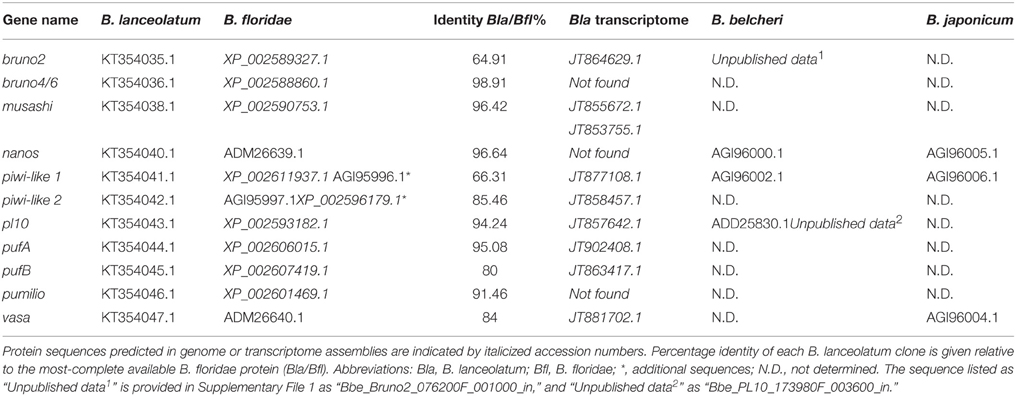
Table 1. Genes cloned in this study and accession numbers for all identified sequences in Branchiostoma species.
We also cloned partial pl10, vasa, nanos, bruno2 (brunoB or CELF2 in Kerner et al., 2011), bruno4/6 (brunoA or CELF4/5/6 in Kerner et al., 2011), pufA, pufB, and pumilio sequences. The phylogenetic analyses broadly confirm previous studies (Kerner et al., 2011), though we could only confirm the existence of single A-type and B-type Bruno sequences. In most cases we could identify B. belcheri orthologs for the B. floridae proteins, in addition to several B. lanceolatum sequences (Supplementary Files 4, 5). EST data in B. floridae also supported the expression of these putative germline and somatic stem cell markers (Supplementary File 3).
As we were interested specifically in stem cell-related Musashi, and relationships among Musashi-related protein families are complex (Gasparini et al., 2011) we generated phylogenies utilizing the available full length B. floridae and B. belcheri sequences, and included putative Saccoglossus kowalevski orthologs. We clearly identified sequences belonging to the TARDBP43 and hnrpA3/hnrpD clades (Figure 2). The close relationship between Musashi-like and DAZAP proteins is also strongly supported by this analysis, although the branching order is unclear particularly within DAZAP sequences and in basal metazoans. Notably, we were unable to find an amphioxus sequence with convincing affinity to DAZAP/hnrp27 genes in either species. We did however identify a Musashi-like sequence in both B. belcheri and B. floridae (Figure 2, Supplementary File 4). In spite of the relatively low support for the Musashi clade, most likely due to the inclusion of non-bilaterian metazoan sequences and the divergent insect “Musashi” proteins, the amphioxus sequence groups with vertebrate and hemichordate sequences with strong support (85), in addition to the recently identified “real” Drosophila Musashi-related protein Rbp6 (Siddall et al., 2012). Insect “musashi” and Rbp6 may therefore represent clade-specific duplications in this group from a musashi-like ancestor. We therefore propose that the Rbp6/Msi sequences be referred to as Musashi-like (blue boxed region in Figure 2), and all others outside the clade as DAZAP. Based on this nomenclature and the firm position of amphioxus musashi among deuterostome sequences, we are therefore confident that we identified a musashi gene orthologous to vertebrate musashi1 and musashi2.
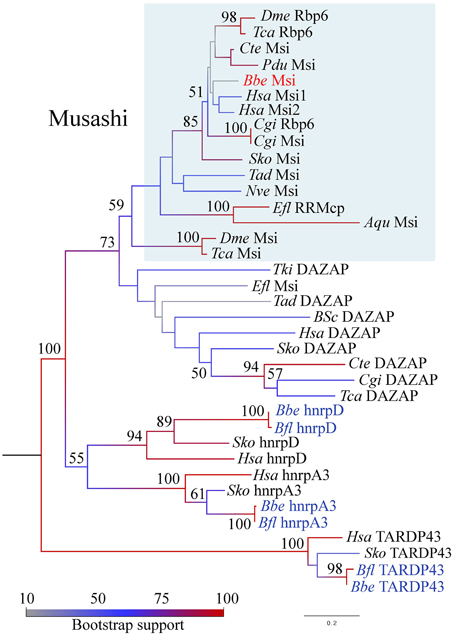
Figure 2. Phylogenetic analysis of the RRM domain containing protein family in animals, including Musashi-like, DAZAP, hnrpD, hnrpA, and TARDP43 clades. Maximum likelihood analysis was performed in MEGA5 with 1000 bootstrap replicates, indicated as a percentage at each node. The model used was rtREV + G with five rate categories on 173 sites. Branches are colored according to the level of node support; amphioxus species names are highlighted in blue and red, with the clade representing musashi genes boxed in blue. Protein names are taken directly from the literature where available. See text for details. Species name abbreviations are as follows: Aqu, Amphimedon queenslandica; Bfl, Branchiostoma floridae; Bbe, Branchiostoma belcheri; Bsc, Botryllus schlosseri; Cgi, Crassostreas gigas; Cte, Capitella teleta; Dme, Drosophila melanogaster; Efl, Ephydatia fluviatilis; Hsa, Homo sapiens; Nve, Nematostella vectensis; Pdu, Platynereis dumerilii; Sko, Saccoglossus kowalevski; Tad, Trichoplax adherens; Tca, Tribolium castaneum; Tki, Thelohanellus kitauei; Xtr, Xenopus tropicalis.
Candidate Marker Expression in Putative PGCs
Recently, expression patterns for putative germline markers have been described in three other species of amphioxus: B. floridae, B. belcheri, and B. japonicum (Wu et al., 2011; Zhang et al., 2013). We therefore performed WMISH for piwil1, piwil2, vasa, and nanos orthologs in early developmental stages of B. lanceolatum. Figures 3A–D show the characteristic expression in single “points” from the two cell stage to the gastrula stage in all four genes with the exception of piwil2. In some cases, the morulae or gastrulae contained up to three points (not shown). By the early neurula stages, the punctate distribution may be masked by the zygotic tailbud expression (discussed below).
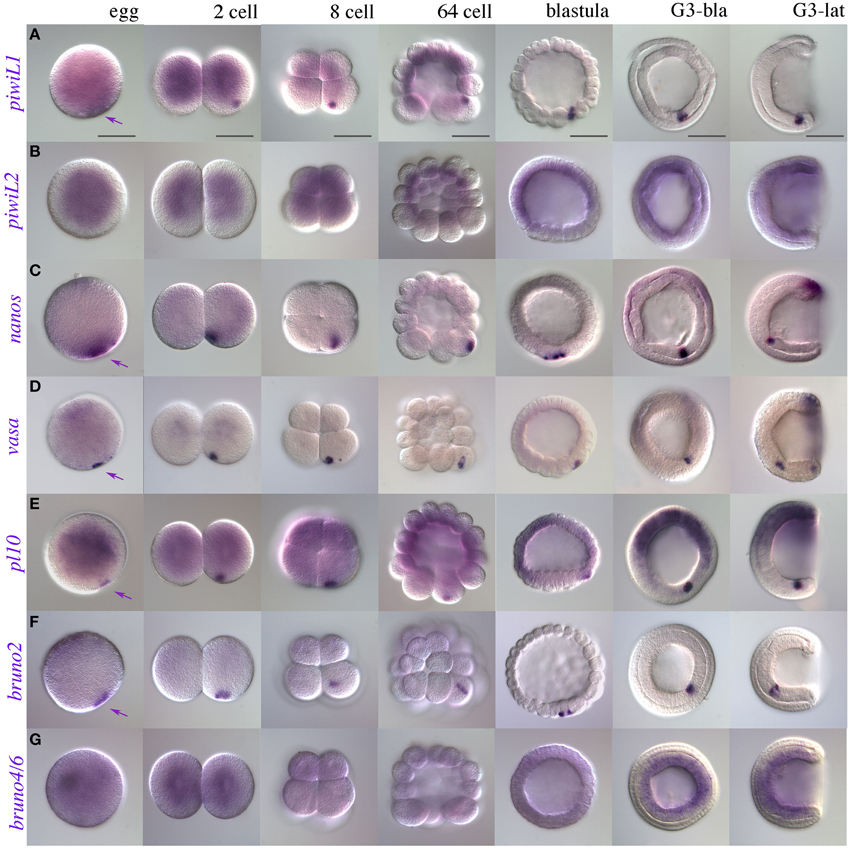
Figure 3. Early expression of putative somatic stem cell and germline markers in B. lanceolatum. (A) piwi-like1; (B) piwi-like2; (C) nanos; (D) vasa; (E) pl10; (F) bruno2; (G) bruno4/6. Purple arrowheads indicate putative PGC-associated expression, as early as the fertilized egg, of piwi-like1, nanos, vasa, pl10, and bruno2. Stages are indicated along the top of the figure in panel (A), from egg to mid-gastrula stages (G3). In morula (ca 64-cell) and blastula stages, macromeres are oriented downwards. G3-bla, blastopore view; G3-lat, lateral view. Scale bars = 50 microns.
Another dead-box containing gene, pl10, has been implicated in germ cell specification, and in some cases regeneration, from sponges to annelids (Alié et al., 2011; Rebscher et al., 2012; Leininger et al., 2014; Kozin and Kostyuchenko, 2015). PL10 is closely related phylogenetically to the Vasa protein (Kerner et al., 2011), but expression of pl10 has so far not been described in any cephalochordate. We therefore cloned a clear pl10 ortholog in B. lanceolatum (Supplementary Files 4, 5) and determined its expression using WMISH. Like vasa, pl10 is expressed in a punctate pattern from fertilization until gastrula stages, consistent with a role in PGC specification or maintenance (Figure 3E).
We also determined early expression of members of three other classes of RNA-binding proteins that we might expect to have a stem cell association based on reports in other species: the Pumilio domain containing genes pumilio, pufA, and pufB; the CELF/Bruno genes bruno2 and bruno4/6, and musashi (Gazave et al., 2013 and references therein). Up to gastrulation, pumilio, pufA, and pufB show no clear localization in the presumptive germline (Supplementary Files 6A–C). Interestingly, pufA ESTs are found in blastula-stage embryos, and we observe several independent but convincing instances in which pufA appeared to be expressed in a punctate distribution reminiscent of our other PGC-associated patterns in some cleavage stage embryos (Supplementary File 6A). Similarly to PUM domain containing genes, bruno4/6 was absent in B. floridae EST databases, and showed no convincing expression until gastrulation (Figure 3G). In contrast, bruno2 showed clear and strong localization to nuage or PGCs (Figure 3F). No other marker analyzed had specific expression in the presumptive PGCs (Supplementary File 6), including musashi, which had diffuse ubiquitous expression at early stages (Supplementary File 6D; see Supplementary File 7 for sense control).
Vasa Protein Distribution is Consistent with PGCs and Somatic Progenitor Cell Domains in B. lanceolatum
Along with transcript expression, localization of Vasa is a hallmark of primordial germ cells (PGCs) in multiple species. In order to confirm the identity of PGCs in B. lanceolatum, we took advantage of the recent generation of an antibody against B. floridae Vasa (Wu et al., 2011) to perform immunohistochemistry. Given its clear cross-reaction in several amphioxus species (Zhang et al., 2013), we reasoned that α-BfVasa should also label PGCs in the European amphioxus, confirming our expression data. The protein distribution resembles that of vasa transcripts (Figure 3D), with a pattern reminiscent of germplasm in fertilized eggs and cleavage stages (Figures 4A–D). In the late gastrula/early neurula, the protein is perinuclear in small clusters of cells within the ventral endoderm (Figures 4E,F); although variable in number (or at least detection), we could clearly identify as many as eight cells by the careful analysis of series of confocal image z-sections (Figure 4F). Such clusters could be identified even in some mid-neurula stage embryos, either on one side in the ventral mesoderm (Figure 4G and inset), or in most cases posteriorly congruent with the zygotic tailbud domain (Figure 4H and inset). Vasa expression was however most conspicuous in the posterior neural tube throughout neurulation (Figures 4I,J). Only in premouth stage and later larvae was it possible to again more easily identify posterior clusters of Vasa-expressing cells as distinct from posterior neural and tailbud expression (Figures 4K,L). Vasa also appeared to demarcate the posteriormost somites (not shown), similarly to vasa transcripts (Figure 5C, see below).
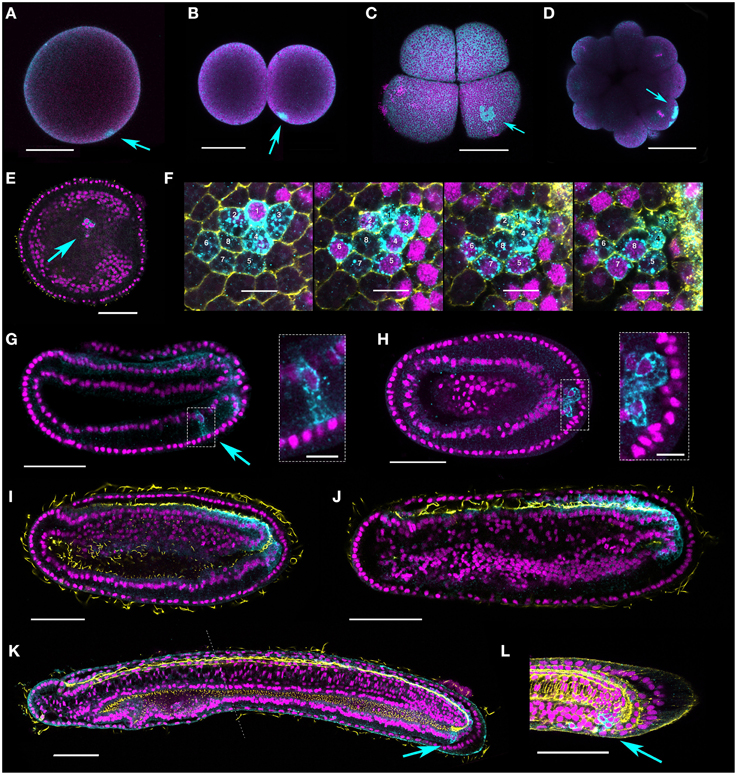
Figure 4. Vasa protein expression during B. lanceolatum embryogenesis. Vasa is expressed in a single point in fertilized eggs (A), two cell (B), eight-cell (C), and morula (D) stages (arrows). (E,F) Perinuclear punctae are evident in late gastrula/early neurula (N0–N1) stages in up to eight cells (two different embryos shown, E and F). (E) shows a cluster of cells of which two clearly express Vasa (arrow). In (F), a series of z-stacks shows a cluster of eight cells expressing Vasa. Mid-neurula (N2) stage embryos (G,H) are variable, with some individuals showing a cluster of Vasa-positive cells in the ventral endoderm on one side (G, arrow and boxed inset); in others the putative germ cells are just ventral to and contiguous with the posterior Vasa expression domain (H, arrow and boxed inset). (I,J) germline expression is difficult to distinguish in late neurula (N3) stage embryos; zygotic expression is most apparent in the posterior neural tube and posteriormost ventral endoderm. (K,L) Clusters of cells are again evident in some pre-mouth stage and late larvae (L1, L2 arrows), and neural expression is much reduced. The dotted line in (K) indicates where two confocal images have been combined. Nuclei stained with Hoescht are false-colored magenta, and Vasa protein is show in cyan. Acetylated tubulin marks axons and cilia in yellow in (E), (I–K). Phalloidin stains F-actin yellow in the membranes in (F) and (L). With the exception of scale bars in (F) and in boxed insets (G,H), which are 10 microns, all scale bars are 50 microns. Orientations are as follows: (E–L) anterior, left; (E,F,H), ventral views; (G,I–L), lateral, dorsal is to the top.
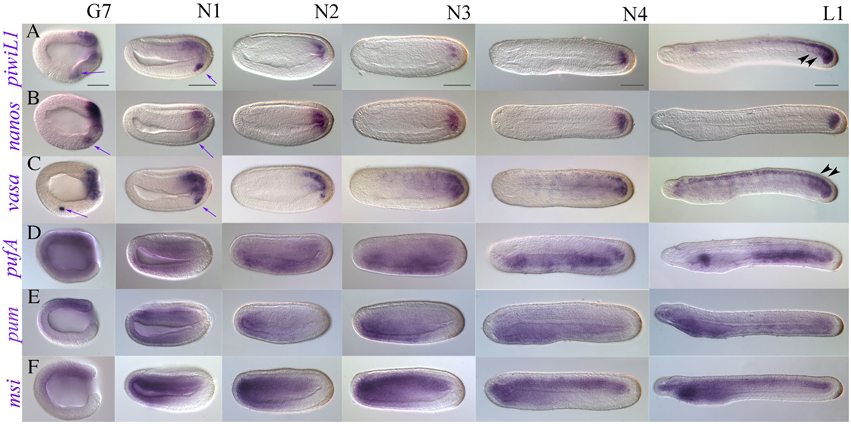
Figure 5. Late expression of a representative subset of putative somatic stem cell and germline makers in B. lanceolatum (lateral views). (A) piwi-like1; (B) nanos; (C) vasa; (D) pufA; (E) pumilio; (F) musashi. Embryonic stages are indicated along the top of panel (A), from late gastrula/early neurula G7 to the premouth larval stage L1. Purple arrows indicate PGC-like expression that continues to be detectable for piwi-like1, nanos, and vasa into the early neurula stage N1. Black arrowheads in (A) and (C) indicate expression demarcating somite boundaries. All views are lateral, and all panels are oriented with anterior to the left and dorsal up. Scale bars = 50 microns.
Candidate Stem Cell Marker Expression in Developing Somatic Tissues
We performed WMISH for selected genes from gastrulation onwards, reasoning that they should show expression patterns with possible roles in late developmental processes (Figure 5 and Supplementary File 8). We thus identified two classes: “tailbud-enriched” and, broadly speaking, “anterior endoderm-associated.” We found that piwil1, nanos, and vasa have strong tailbud expression throughout development (Figures 5A–C). piwil1 and nanos also show clear posterior neural tube expression in N4 neurulae and L1 stage premouth larvae, as well as expression outlining the posterior somites (black arrowheads, Figure 5A; Supplementary File 8A). Though weaker, piwil2 and pl10 both show tailbud expression at later stages, and pl10 is clearly expressed in the neural tube (Supplementary File 9).
In contrast, Pumilio domain containing genes appear enriched in anterior endoderm (Figures 5D,E and Supplementary Files 8D,E). During gastrulation, pumilio shows weak expression around the blastopore. In early and mid-neurula stages, stronger expression is evident in the neural plate and anterior ventral endoderm, as well as anterior mesoderm. As neurulation proceeds, pumilio appears mostly restricted to the anterior endoderm, with expression much weaker in the last third of the embryo (Figure 5E). Expression continues to be strongest in the future pharyngeal domain until the pre-mouth larval stage. Weaker expression is evident in the rest of the endoderm, with some conspicuous staining in the mesoderm and endoderm of the tailbud region. Expression of pufA is broadly mesendodermal until N3 neurula stages, when it becomes stronger in an anterior domain that resolves into the club shaped gland in premouth L1 larval stages, as well as in most of the posterior endoderm (Figure 5D). pufB expression was very difficult to evaluate as long staining exposures were required for stages post-gastrulation, but quite closely matched that of pufA (not shown). In addition to its posterior expression, pl10 shows diffuse but clear staining in anterior endoderm in N4 and L1 stages (Supplementary Files 9G–I), and clearly resolves to a domain encompassing the presumptive first gill slit in 2–3 day-old larvae (not shown).
We cloned the amphioxus musashi ortholog with the expectation that it would have neural expression. During early stages of development, musashi is ubiquitously expressed (Supplementary File 6D), paralleling B. floridae EST data (Supplementary File 3). However, in the gastrula stage, musashi resolves to a chordomesodermal domain of expression (Supplementary Files 6D, 7), which broadens in the early neurula N1 (Figure 5F). musashi is strongly expressed in the anteriormost endoderm and mesoderm from mid-neurula onwards, with weak expression in the neural floorplate and strong expression throughout the chordal plate. By the late neurula stage (30 h, N4) patches of expression can be seen in the neural tube as well as weakly in the cerebral vesicle. Expression is high and stable throughout the forming notochord as well as in the anterior endoderm. Strong notochordal and weak neural expression domains persist in the premouth L1 larva, with strongest expression in the anterior and posteriormost domains of the notochord. The presumptive pharynx also expresses musashi.
Discussion
Germline-Associated Gene Expression Conservation in Cephalochordates
Recent work in B. floridae, B. japonicum, and B. belcheri has suggested that germline specification occurs by the asymmetric segregation of cytoplasmic determinants during cleavage, with expression of key conserved germline markers such as vasa and nanos, as well as piwil1 and tudor-related7, in the germ plasm and PGCs (Wu et al., 2011; Zhang et al., 2013). We set out here to characterize the expression of germline-associated markers in the European amphioxus, B. lanceolatum, for which there were until now no data. Similarly to other species, our results also argue against an inductive mechanism for PGC specification: we demonstrate here that B. lanceolatum expresses nanos, piwil1, and vasa in the putative PGCs, as well as Vasa protein, suggesting the presence of a conserved core of germline-associated transcripts in cephalochordates. Stasis in developmental gene expression over millions of years of evolution is considered typical of Branchiostoma (Somorjai et al., 2008), paralleling the genus' relative genomic and morphological conservativeness. The apparent conservation in germline-associated gene expression in amphioxus species is in stark contrast to hypotheses derived in vertebrates that suggest that the evolution of germ plasm is coupled to increased speciation in this lineage (Johnson et al., 2011; Evans et al., 2014). Data in Asymmetron, the earliest diverging and most slowly evolving of the three extant amphioxus lineages (Kon et al., 2007; Yue et al., 2014), will be invaluable in evaluating the degree of conservation of germline specification mechanisms in cephalochordates.
Our research also identifies pl10, a DEAD-box gene related to vasa, and bruno2 as putative PGC markers in amphioxus. Accumulating evidence suggests that pl10 often plays a role in the germline in metazoans. In addition to Drosophila, pl10 orthologs are expressed in the germinal cells or their derivatives in the annelid Platynereis dumerilii (Rebscher et al., 2007; Gazave et al., 2013), the platyhelminth Dugesia japonicum (Shibata et al., 1999; reported as vasa-related genes) several hydrozoan cnidarian species (Leclère et al., 2012; Siebert et al., 2015), the ctenophore Mnemopsis leydii (Alié et al., 2011), the sponge Sycon ciliatum (Leininger et al., 2014), and in the colonial urochordate B. schlosseri (Rosner et al., 2009). vasa is coexpressed with pl10 in the latter, similarly to our results in B. lanceolatum. In contrast, data are sparse for the second gene identified, bruno2. Homologs of bruno are expressed in PGCs and/or germline derivatives in ctenophores (Alié et al., 2011), but not in Platynereis (Gazave et al., 2013). Interestingly, using RNAi, the Bruno-like gene bruli was shown to be required for maintenance of a subset of neoblasts in the asexual planarian Schmidtea mediterranea (Guo et al., 2006), but this gene is not homologous to canonical bruno genes. Confirmation of expression of pl10 and bruno2 in other species, and Tudor related tdrd7 in B. lanceolatum, will further expand this repertoire.
We also identified several markers with weak ubiquitous expression during early development. For instance, musashi, piwil2, and genes of the Pumilio domain family do not appear to be associated specifically with PGCs in B. lanceolatum or B. floridae (this study; Yue et al., 2015). A possible exception is pufA, which we found to be concentrated in a PGC-like domain in some cleavage stage embryos (two-cell to morula) in several independent experiments. Given the variability of the expression observed, we hesitate to classify this as bone fide expression in PGCs. However, pufA is expressed in germ cells in other species, including zebrafish (Kuo et al., 2009) and P. dumerilii (Gazave et al., 2013). Interestingly, a global search of germline and reproduction-associated genes using the transcriptome and genome of Asymmetron lucayanum and B. floridae, respectively, identified a pumilio/puf gene with expression in oocytes (Yue et al., 2015). Similar studies in maturing gonads in B. lanceolatum may also reveal functions for some of our candidates during germline maturation.
Evolution of Musashi Related RRM-Containing Proteins and Novel Expression of Amphioxus musashi
The Musashi related proteins belong to a larger superfamily of RRM containing proteins, including Musashi, DAZAP, hnrp, and TARDBP clades. Although the evolutionary history of RRM domain containing proteins is complex, orthologs of musashi-related genes have been identified from sponge to human (Gasparini et al., 2011; Okamoto et al., 2012), including lancelets (Gasparini et al., 2011, this study). One of the principal findings of this study is that cephalochordates appear to have lost the ortholog of DAZAP, as we were unable to identify the gene in either the genomes of B. floridae or B. belcheri. Considerable confusion exists in the nomenclature in the literature due to the difficulty in distinguishing between musashi and DAZAP related genes. This is particularly evident in basal metazoans, where phylogenetic signal is weak (Okamoto et al., 2012, this study). Gasparini et al. (2011) first suggested that previously identified musashi-like genes in Halocynthia roretzi and Ciona intestinalis (Kawashima et al., 2000) are in fact DAZAP. This gene is expressed in the brain and nerve cord, as might be expected from musashi-like genes (Kawashima et al., 2000). However, the bona fide DAZAP1 in Botryllus schlosseri is expressed both during asexual (blastogenesis) and sexual (embryonic) development in many proliferating cell types, including the new growing vessels of the colonial circulatory system and the embryonic nerve cord, and is not restricted to neural stem cells as in other systems. Likewise, in the planarian D. japonicum DAZAP/musashi-like gene Djdmlg is expressed in differentiated tissues as well as X-ray sensitive neoblasts (Higuchi et al., 2008). In this context, it would be particularly interesting to determine whether the cephalochordate musashi is taking on any of the DAZAP functions, or whether a different functional homolog might be involved.
Our observation that neural cells within the developing CNS of amphioxus express musashi is broadly consistent with data in bilaterians. For instance, in the flatworm Dugesia japonica, three musashi-like genes have been identified with expression in the brain primordia (Higuchi et al., 2008). Similarly, in zebrafish, musashi1 is expressed in neural tissues during early development, and knockdown by morpholino results in aberrant CNS formation (Shibata et al., 2012). Surprisingly however, amphioxus does not express musashi in a pattern consistent with a role in PGC specification or maintenance, in contrast to many other taxa. In Drosophila, musashi is required to maintain stem cell identity in GSCs (Siddall et al., 2006), and Rbp6, which is more closely related to vertebrate musashi1/2 (Siddall et al., 2012; this study), also may play some function in the germline (Siddall et al., 2012). In mice, the msi1 and msi2 orthologs appear to have sub-functionalized such that msi1 is required to maintain stem cell identity during early spermatogenesis, whereas msi2 plays a role in differentiation (Siddall et al., 2006). The generation of specific antibodies will be critical to gaining an understanding of the distribution of Musashi protein during amphioxus development and stem cell regulation.
Given its known neural and germline functions, the finding that musashi is predominantly expressed in the developing notochord in amphioxus was unexpected. We are not aware of any data demonstrating a specific function for musashi in the notochord in any chordate. However, the ancestral function of these RRM containing proteins may simply be in the switch between undifferentiated/stem cell and differentiated cell types and in the regulation of proliferation (Potten et al., 2003; MacNicol et al., 2011; Hochgreb-Hägele et al., 2014). Supporting this, the anterior endoderm encompassing the zones that will form the mouth and gill slits in amphioxus larvae, which has conspicuous musashi expression, is a zone of extensive proliferation and remodeling (Holland and Holland, 2006). The expression in developing notochord described here, which is unique to amphioxus, might also reflect a role in differentiation of this structure. Functional studies will help elucidate the role of Musashi in this and other structures.
Posterior Stem Cells and Implications for Amphioxus Regeneration
The zygotic expression of several markers in the tailbud, including nanos, vasa, piwil1, and piwil2 among others, combined with circumstantial evidence that PGCs may migrate at the neurula stage toward the posterior (this study; Wu et al., 2011), suggest that the tailbud may be a source of progenitors or stem cells in larval amphioxus. Posterior elongation in amphioxus involves budding of somites directly from the tailbud, a source of Wnt ligand (Holland et al., 2000; Schubert et al., 2000, 2001; Somorjai et al., 2008). Although architecturally different, the tailbuds of vertebrates like mouse and chick are also sources of multipotent stem cells for embryonic elongation whose fate is Wnt signaling-dependent (Wilson et al., 2009; Garriock et al., 2015). The posterior growth zone may also act as a niche for progenitor cells even into adulthood, particularly in animals that add segments throughout their lives, such as many arthropods and most annelids (Bely and Wray, 2001; de Rosa et al., 2005; Seaver et al., 2005). The observation that the Vasa-positive PGCs lie within a stem cell marker-expressing posterior growth zone in amphioxus larvae (Wu et al., 2011; this study), representing a “mosaic” of PGCs and somatic stem cells, is however not unique to amphioxus. Gazave et al. (2013) have proposed the existence of an RNA binding protein signature for a new type of animal stem cell, termed “posterior stem cells,” in P. dumerilii. Lineage analysis and EdU labeling have also revealed that the 4 presumptive PGCs, which appear during gastrulation, are derived from a mesoderm posterior growth zone (MPGZ; Rebscher et al., 2007, 2012). While the mechanisms employed by these annelids and cephalochordates to specify the germline are somewhat different, the use of such techniques in amphioxus will be instrumental in elucidating the origin and fate of different cell types during posterior elongation.
The existence of a posterior stem cell in the tailbud, or any other resident stem cell population that could be activated following tail amputation, has clear implications for regeneration in amphioxus. Although it has recently been demonstrated that the European amphioxus has considerable regenerative ability, most notably of the tail (Somorjai et al., 2012a,b), we still know next to nothing about the molecular signature or function of the somatic stem cells/progenitor pools involved in the process. This study represents the first step toward identifying a putative posterior stem cell pool in B. lanceolatum. Our prediction is that somatic stem cell markers that are normally expressed during tailbud development, such as vasa, nanos, piwil1, or piwil2, will also be expressed during the adult tail regeneration process. We are currently analysing blastema transcriptomes and proteomes to test this hypothesis (Dailey and Somorjai, unpublished). We might also expect to find genes traditionally associated with the germline to be expressed during tail regeneration, if common expression of “stemness” markers in PGCs and somatic stem cells reflect broader roles in developmental regulation, as has recently been demonstrated for Vasa in the sea urchin (Yajima and Wessel, 2015). Although functional experiments are lacking, comparative expression data in annelids are beginning to provide compelling evidence for this. In P. dumerilii, a number of RNA binding protein genes are expressed in PGCs as well as in putative posterior mesodermal and ectodernal stem cells during caudal regeneration, including vasa, pl10, piwi, pufA, pufB, nanos, and several tudor related genes (Rebscher et al., 2007; Gazave et al., 2013). Of these, several markers are also differentially expressed both in the germline and terminal growth zone during normal development and regeneration in the polychaetes Alitta virens and Capitella sp I (Dill and Seaver, 2008; Giani et al., 2011; Kozin and Kostyuchenko, 2015). However, the most striking example of a germline-independent redeployment of classic PGC markers in somatic tissues has been shown in the freshwater annelid Pristina leidyi, which reproduces exclusively asexually in the laboratory via paratomic fission. As might be expected, nanos, piwi1, and vasa are expressed in the posterior growth zone and developing (but unused) gonads. Notably, transcripts are also detected following amputation in the anterior blastema as well as the fission zone (Bely and Sikes, 2010; Özpolat and Bely, 2015), highlighting a more general role in tissues undergoing proliferation and remodeling. This phenomenon is not restricted to invertebrates or basal metazoans, as piwil1 and piwil2 are expressed in a complex spatiotemporal sequence during axolotl limb regeneration, with knockdown of either gene resulting in retardation of the regenerate outgrowth (Zhu et al., 2012). Future work in amphioxus will assess the tissue-specific expression pattern of some of the candidates identified here during adult tail regeneration. Development of knockdown tools and lineage analysis will be indispensable to elucidate their functional role during the regeneration process. Moreover, these methodologies will permit the comparative analyses of cellular and molecular processes necessary to understand the evolution of regeneration mechanisms in deuterostomes. More broadly, these types of studies should add to the growing body of literature aimed at understanding the link between soma and germline evolution.
Author Contributions
SD, RF, and AR performed experiments. JG discussed experiments and contributed reagents. IS conceived the study, performed experiments, contributed reagents, analyzed the data and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was carried out sporadically over the course of several years in various countries. We would like to thank Jr-Kai Sky Yu (Academia Sinica, Taiwan) for contributing the Vasa antibody, and Irene Garcia for help in the laboratory (Barcelona). SD is funded through a MASTS PhD studentship (St Andrews, UK). IS gratefully acknowledges previous funding from Marie Curie IEF postdoctoral fellowship, FP7 People Programme (Barcelona, Spain); as well as MASTS (Marine Alliance for Science and Technology Scotland) laboratory start-up funds (St Andrews, UK). Embryo collection was made possible in part through the ASSEMBLE access programme (grant agreement no. 227799). We thank the Laboratoire Océanologique de Banyuls-sur-Mer, and most especially Dr. Hector Escrivà and Dr. Stéphanie Bertrand for hosting us. The University of St Andrews Library fund for open access supported the article publishing fee.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fevo.2015.00156
References
Alié, A., Leclère, L., Jager, M., Dayraud, C., Chang, P., Le Guyader, H., et al. (2011). Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of “germline genes” with stemness. Dev. Biol. 350, 183–197. doi: 10.1016/j.ydbio.2010.10.019
Bely, A. E., and Sikes, J. M. (2010). Latent regeneration abilities persist following recent evolutionary loss in asexual annelids. Proc. Natl. Acad. Sci. U.S.A. 107, 1464–1469. doi: 10.1073/pnas.0907931107
Bely, A. E., and Wray, G. A. (2001). Evolution of regeneration and fission in annelids: insights from engrailed- and orthodenticle-class gene expression. Development 128, 2781–2791.
Bertrand, S., and Escrivà, H. (2011). Evolutionary crossroads in developmental biology: amphioxus. Development 138, 4819–4830. doi: 10.1242/dev.066720
Blackstone, N. W., and Jasker, B. D. (2003). Phylogenetic considerations of clonality, coloniality, and mode of germline development in animals. J. Exp. Zool. B Mol. Dev. Evol. 297, 35–47. doi: 10.1002/jez.b.16
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chatfield, J., O'Reilly, M. A., Bachvarova, R. F., Ferjentsik, Z., Redwood, C., Walmsley, M., et al. (2014). Stochastic specification of primordial germ cells from mesoderm precursors in axolotl embryos. Development 141, 2429–2440. doi: 10.1242/dev.105346
Crother, B. I., White, M. E., and Johnson, A. D. (2007). Inferring developmental constraint and constraint release: primordial germ cell determination mechanisms as examples. J. Theor. Biol. 248, 322–330. doi: 10.1016/j.jtbi.2007.05.035
de Rosa, R., Prud'homme, B., and Balavoine, G. (2005). Caudal and even-skipped in the annelid Platynereis dumerilii and the ancestry of posterior growth. Evol. Dev. 7, 574–587. doi: 10.1111/j.1525-142X.2005.05061.x
Dill, K. K., and Seaver, E. C. (2008). Vasa and nanos are coexpressed in somatic and germ line tissue from early embryonic cleavage stages through adulthood in the polychaete Capitella sp. I. Dev. Genes Evol. 218, 453–463. doi: 10.1007/s00427-008-0236-x
Donoughe, S., Nakamura, T., Ewen-Campen, B., Green, D. A. I. I., Henderson, L., and Extavour, C. G. (2014). BMP signaling is required for the generation of primordial germ cells in an insect. Proc. Natl. Acad. Sci. U.S.A. 111, 4133–4138. doi: 10.1073/pnas.1400525111
Evans, T., Wade, C. M., Chapman, F. A., Johnson, A. D., and Loose, M. (2014). Acquisition of germ plasm accelerates vertebrate evolution. Science 344, 200–203. doi: 10.1126/science.1249325
Ewen-Campen, B., Schwager, E. E., and Extavour, C. G. (2010). The molecular machinery of germ line specification. Mol. Reprod. Dev. 77, 3–18. doi: 10.1002/mrd.21091
Extavour, C. G. (2007). Evolution of the bilaterian germ line: lineage origin and modulation of specification mechanisms. Integr. Comp. Biol. 47, 770–785. doi: 10.1093/icb/icm027
Extavour, C. G., and Akam, M. (2003). Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869–5884. doi: 10.1242/dev.00804
Fuentes, M., Benito, E., Bertrand, S., Paris, M., Mignardot, A., Godoy, L., et al. (2007). Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum). J. Exp. Zool. B Mol. Dev. Evol. 308, 484–493. doi: 10.1002/jez.b.21179
Garriock, R. J., Chalamalasetty, R. B., Kennedy, M. W., Canizales, L. C., Lewandoski, M., and Yamaguchi, T. P. (2015). Lineage tracing of neuromesodermal progenitors reveals novel Wnt-dependent roles in trunk progenitor cell maintenance and differentiation. Development 142, 1628–1638. doi: 10.1242/dev.111922
Gasparini, F., Shimeld, S. M., Ruffoni, E., Burighel, P., and Manni, L. (2011). Expression of a Musashi-like gene in sexual and asexual development of the colonial chordate Botryllus schlosseri and phylogenetic analysis of the protein group. J. Exp. Zool. B Mol. Dev. Evol. 316, 562–573. doi: 10.1002/jez.b.21431
Gazave, E., Béhague, J., Laplane, L., Guillou, A., Préau, L., Demilly, A., et al. (2013). Posterior elongation in the annelid Platynereis dumerilii involves stem cells molecularly related to primordial germ cells. Dev. Biol. 382, 246–267. doi: 10.1016/j.ydbio.2013.07.013
Giani, V. C. Jr., Yamaguchi, E., Boyle, M. J., and Seaver, E. C. (2011). Somatic and germline expression of piwi during development and regeneration in the marine polychaete annelid Capitella teleta. Evodevo 2:10. doi: 10.1186/2041-9139-2-10
Guo, T., Peters, A. H., and Newmark, P. A. (2006). A bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159–169. doi: 10.1016/j.devcel.2006.06.004
Hallmann, A. (2011). Evolution of reproductive development in the volvocine algae. Sex. Plant Reprod. 24, 97–112. doi: 10.1007/s00497-010-0158-4
Higuchi, S., Hayashi, T., Tarui, H., Nishimura, O., Nishimura, K., Shibata, et al. (2008). Expression and functional analysis of musashi-like genes in planarian CNS regeneration. Mech. Dev. 125, 631–645. doi: 10.1016/j.mod.2008.03.002
Hirakow, R., and Kajita, N. (1991). Electron microscopic study of the development of amphioxus, Branchiostoma belcheri tsingtauense: the gastrula. J. Morphol. 207, 37–52. doi: 10.1002/jmor.1052070106
Hirakow, R., and Kajita, N. (1994). Electron microscopic study of the development of amphioxus, Branchiostoma belcheri tsingtauense: the neurula and larva. Acta Anat. Nippon 69, 1–13.
Hochgreb-Hägele, T., Koo, D. E., Das, N. M., and Bronner, M. E. (2014). Zebrafish stem/progenitor factor msi2b exhibits two phases of activity mediated by different splice variants. Stem Cells 32, 558–571. doi: 10.1002/stem.1583
Holland, L. Z., and Holland, N. N. (1992). Early development in the lancelet (= Amphioxus) Branchiostoma floridae from sperm entry through pronuclear fusion: presence of vegetal pole plasm and lack of conspicuous ooplasmic segregation. Biol. Bull. 182, 77–96. doi: 10.2307/1542182
Holland, L. Z., Holland, N. N., and Schubert, M. (2000). Developmental expression of AmphiWnt1, an amphioxus gene in the Wnt1/wingless subfamily. Dev. Genes Evol. 210, 522–524. doi: 10.1007/s004270000089
Holland, N. D., and Holland, L. Z. (2006). Stage- and tissue-specific patterns of cell division in embryonic and larval tissues of amphioxus during normal development. Evol. Dev. 8, 142–149. doi: 10.1111/j.1525-142X.2006.00085.x
Johnson, A. D., Crother, B. I., White, M. E., Patient, R., Bachvarova, R. F., Drum, M., et al. (2003b). Regulative germ cell specification in axolotl embryos: a primitive trait conserved in the mammalian lineage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 1371–1379. doi: 10.1098/rstb.2003.1331
Johnson, A. D., Drum, M., Bachvarova, R. F., Masi, T., White, M. E., and Crother, B. I. (2003a). Evolution of predetermined germ cells in vertebrate embryos: implications for macroevolution. Evol. Dev. 5, 414–431. doi: 10.1046/j.1525-142X.2003.03048.x
Johnson, A. D., Richardson, E., Bachvarova, R. F., and Crother, B. I. (2011). Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction 141, 291–300. doi: 10.1530/REP-10-0474
Juliano, C. E., Swartz, S. Z., and Wessel, G. M. (2010). A conserved germline multipotency program. Development 137, 4113–4126. doi: 10.1242/dev.047969
Kawashima, T., Murakami, A. R., Ogasawara, M., Tanaka, K., Isoda, R., Sasakura, Y., et al. (2000). Expression patterns of musashi homologs of the ascidians, Halocynthia roretzi and Ciona intestinalis. Dev. Genes Evol. 210, 162–165. doi: 10.1007/s004270050024
Kerner, P., Degnan, S. M., Marchand, L., Degnan, B. M., and Vervoort, M. (2011). Evolution of RNA-binding proteins in animals: insights from genome-wide analysis in the sponge Amphimedon queenslandica. Mol. Biol. Evol. 28, 2289–2303. doi: 10.1093/molbev/msr046
Kon, T., Nohara, M., Yamanoue, Y., Fujiwara, Y., Nishida, M., and Nishikawa, T. (2007). Phylogenetic position of a whale-fall lancelet (Cephalochordata) inferred from whole mitochondrial genome sequences. BMC Evol. Biol. 7:127. doi: 10.1186/1471-2148-7-127
Kozin, V. V., and Kostyuchenko, R. P. (2015). Vasa, PL10, and Piwi gene expression during caudal regeneration of the polychaete annelid Alitta virens. Dev. Genes Evol. 225, 129–138. doi: 10.1007/s00427-015-0496-1
Kuo, M. W., Wang, S. H., Chang, J. C., Chang, C. H., Huang, L. J., Lin, H. H., et al. (2009). A novel puf-A gene predicted from evolutionary analysis is involved in the development of eyes and primordial germ-cells. PLoS ONE 4:e4980. doi: 10.1371/journal.pone.0004980
Leclère, L., Jager, M., Barreau, C., Chang, P., Le Guyader, H., Manuel, M., et al. (2012). Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev. Biol. 364, 236–248. doi: 10.1016/j.ydbio.2012.01.018
Leininger, S., Adamski, M., Bergum, B., Guder, C., Liu, J., Laplante, M., et al. (2014). Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nat. Commun. 5:3905. doi: 10.1038/ncomms4905
MacNicol, M. C., Cragle, C. E., and MacNicol, A. M. (2011). Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle 10, 39–44. doi: 10.4161/cc.10.1.14388
Okamoto, K., Nakatsukasa, M., Alié, A., Masuda, Y., Agata, K., and Funayama, N. (2012). The active stem cell specific expression of sponge Musashi homolog EflMsiA suggests its involvement in maintaining the stem cell state. Mech. Dev. 129, 24–37. doi: 10.1016/j.mod.2012.03.001
Oulion, S., Bertrand, S., Belgacem, M. R., Le Petillon, Y., and Escrivà, H. (2012). Sequencing and analysis of the Mediterranean amphioxus (Branchiostoma lanceolatum) transcriptome. PLoS ONE 7:e36554. doi: 10.1371/journal.pone.0036554
Özpolat, B. D., and Bely, A. E. (2015). Gonad establishment during asexual reproduction in the annelid Pristina leidyi. Dev. Biol. 405, 123–136. doi: 10.1016/j.ydbio.2015.06.001
Potten, C. S., Booth, C., Tudor, G. L., Booth, D., Brady, G., Hurley, P., et al. (2003). Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation 71, 28–41. doi: 10.1046/j.1432-0436.2003.700603.x
Rebscher, N., Lidke, A. K., and Ackermann, C. F. (2012). Hidden in the crowd: primordial germ cells and somatic stem cells in the mesodermal posterior growth zone of the polychaete Platynereis dumerillii are two distinct cell populations. Evodevo 3:9. doi: 10.1186/2041-9139-3-9
Rebscher, N., Zelada-González, F., Banisch, T. U., Raible, F., and Arendt, D. (2007). Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev. Biol. 306, 599–611. doi: 10.1016/j.ydbio.2007.03.521
Rosner, A., Moiseeva, E., Rinkevich, Y., Lapidot, Z., and Rinkevich, B. (2009). Vasa and the germ line lineage in a colonial urochordate. Dev. Biol. 331, 113–128. doi: 10.1016/j.ydbio.2009.04.025
Schubert, M., Holland, L. Z., and Holland, N. D. (2000). Characterization of an amphioxus wnt gene, AmphiWnt11, with possible roles in myogenesis and tail outgrowth. Genesis 27, 1–5. doi: 10.1002/1526-968X(200005)27:13.3.CO;2-V
Schubert, M., Holland, L. Z., Stokes, M. D., and Holland, N. D. (2001). Three amphioxus Wnt genes (AmphiWnt3, AmphiWnt5, and AmphiWnt6) associated with the tail bud: the evolution of somitogenesis in chordates. Dev. Biol. 240, 262–273. doi: 10.1006/dbio.2001.0460
Seaver, E. C., Thamm, K., and Hill, S. D. (2005). Growth patterns during segmentation in the two polychaete annelids, Capitella sp. I and Hydroides elegans: comparisons at distinct life history stages. Evol Dev. 7, 312–326. doi: 10.1111/j.1525-142X.2005.05037.x
Shibata, N., Umesono, Y., Orii, H., Sakurai, T., Watanabe, K., and Agata, K. (1999). Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev. Biol. 206, 73–87. doi: 10.1006/dbio.1998.9130
Shibata, S., Umei, M., Kawahara, H., Yano, M., Makino, S., and Okano, H. (2012). Characterization of the RNA-binding protein Musashi1 in zebrafish. Brain Res. 1462, 162–173. doi: 10.1016/j.brainres.2012.01.068
Siddall, N. A., Kalcina, M., Johanson, T. M., Monk, A. C., Casagranda, F., Been, R. P., et al. (2012). Drosophila Rbp6 is an orthologue of vertebrate Msi-1 and Msi-2, but does not function redundantly with dMsi to regulate germline stem cell behaviour. PLoS ONE 7:e49810. doi: 10.1371/journal.pone.0049810
Siddall, N. A., McLaughlin, E. A., Marriner, N. L., and Hime, G. R. (2006). The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc. Natl. Acad. Sci. U.S.A. 103, 8402–8407. doi: 10.1073/pnas.0600906103
Siebert, S., Goetz, F. E., Church, S. H., Bhattacharyya, P., Zapata, F., Haddock, S. H. D., et al. (2015). Stem cells in Nanomia bijuga (Siphonophora), a colonial animal with localized growth zones. EvoDevo 6, 1. doi: 10.1186/s13227-015-0018-2
Somorjai, I., Bertrand, S., Camasses, A., Haguenauer, A., and Escrivà, H. (2008). Evidence for stasis and not genetic piracy in developmental expression patterns of Branchiostoma lanceolatum and Branchiostoma floridae, two amphioxus species that have evolved independently over the course of 200 Myr. Dev. Genes Evol. 218, 703–713. doi: 10.1007/s00427-008-0256-6
Somorjai, I. M., Escrivà, H., and Garcia-Fernàndez, J. (2012b). Amphioxus makes the cut-Again. Commun. Integr. Biol. 5, 499–502. doi: 10.4161/cib.21075
Somorjai, I. M., Somorjai, R. L., Garcia-Fernàndez, J., and Escrivà, H. (2012a). Vertebrate-like regeneration in the invertebrate chordate amphioxus. Proc. Natl. Acad. Sci. U.S.A. 109, 517–522. doi: 10.1073/pnas.1100045109
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wilson, V., Olivera-Martinez, I., and Storey, K. G. (2009). Stem cells, signals and vertebrate body axis extension. Development 136, 1591–1604. doi: 10.1242/dev.021246
Wu, H. R., Chen, Y. T., Su, Y. H., Luo, Y. J., Holland, L. Z., and Yu, J. K. (2011). Asymmetric localization of germline markers Vasa and Nanos during early development in the amphioxus Branchiostoma floridae. Dev. Biol. 353, 147–159. doi: 10.1016/j.ydbio.2011.02.014
Yajima, M., and Wessel, G. M. (2015). Essential elements for translation: the germline factor Vasa functions broadly in somatic cells. Development 142, 1960–1970. doi: 10.1242/dev.118448
Yu, J. K., Wang, M. C., Shin, I. T., Kohara, Y., Holland, L. Z., Satoh, N., et al. (2008). A cDNA resource for the cephalochordate amphioxus Branchiostoma floridae. Dev Genes Evol. 218, 723–727. doi: 10.1007/s00427-008-0228-x
Yue, J. X., Li, K. L., and Yu, J. K. (2015). Discovery of germline-related genes in Cephalochordate amphioxus: a genome wide survey using genome annotation and transcriptome data. Mar. Genomics 24, 147–157. doi: 10.1016/j.margen.2015.03.010
Yue, J. X., Yu, J. K., Putnam, N. H., and Holland, L. Z. (2014). The transcriptome of an amphioxus, Asymmetron lucayanum, from the Bahamas: a window into chordate evolution. Genome Biol. Evol. 6, 2681–2696. doi: 10.1093/gbe/evu212
Zhang, Q. J., Luo, Y. J., Wu, H. R., Chen, Y. T., and Yu, J. K. (2013). Expression of germline markers in three species of amphioxus supports a preformation mechanism of germ cell development in cephalochordates. Evodevo 4:17. doi: 10.1186/2041-9139-4-17
Keywords: amphioxus, posterior stem cells, evolution, regeneration, PGCs, preformation
Citation: Dailey SC, Febrero Planas R, Rossell Espier A, Garcia-Fernàndez J and Somorjai IML (2016) Asymmetric Distribution of pl10 and bruno2, New Members of a Conserved Core of Early Germline Determinants in Cephalochordates. Front. Ecol. Evol. 3:156. doi: 10.3389/fevo.2015.00156
Received: 01 September 2015; Accepted: 28 December 2015;
Published: 22 January 2016.
Edited by:
Sylvain Marcellini, University of Concepcion, ChileReviewed by:
Brian I. Crother, Southeastern Louisiana University, USAEduardo E. Zattara, Indiana University, USA
Copyright © 2016 Dailey, Febrero Planas, Rossell Espier, Garcia-Fernàndez and Somorjai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ildikó M. L. Somorjai, imls@st-andrews.ac.uk
†These authors have contributed equally to this work.
 Simon C. Dailey
Simon C. Dailey Roser Febrero Planas
Roser Febrero Planas Ariadna Rossell Espier2
Ariadna Rossell Espier2  Jordi Garcia-Fernàndez
Jordi Garcia-Fernàndez Ildikó M. L. Somorjai
Ildikó M. L. Somorjai