Evolution of Homeobox Gene Clusters in Animals: The Giga-Cluster and Primary vs. Secondary Clustering
- The Scottish Oceans Institute, Gatty Marine Laboratory, University of St Andrews, St Andrews, UK
The Hox gene cluster has been a major focus in evolutionary developmental biology. This is because of its key role in patterning animal development and widespread examples of changes in Hox genes being linked to the evolution of animal body plans and morphologies. Also, the distinctive organization of the Hox genes into genomic clusters in which the order of the genes along the chromosome corresponds to the order of their activity along the embryo, or during a developmental process, has been a further source of great interest. This is known as collinearity, and it provides a clear link between genome organization and the regulation of genes during development, with distinctive changes marking evolutionary transitions. The Hox genes are not alone, however. The homeobox genes are a large super-class, of which the Hox genes are only a small subset, and an ever-increasing number of further gene clusters besides the Hox are being discovered. This is of great interest because of the potential for such gene clusters to help understand major evolutionary transitions, both in terms of changes to development and morphology as well as evolution of genome organization. However, there is uncertainty in our understanding of homeobox gene cluster evolution at present. This relates to our still rudimentary understanding of the dynamics of genome rearrangements and evolution over the evolutionary timescales being considered when we compare lineages from across the animal kingdom. A major goal is to deduce whether particular instances of clustering are primary (conserved from ancient ancestral clusters) or secondary (reassortment of genes into clusters in lineage-specific fashion). The following summary of the various instances of homeobox gene clusters in animals, and the hypotheses about their evolution, provides a framework for the future resolution of this uncertainty.
Introduction
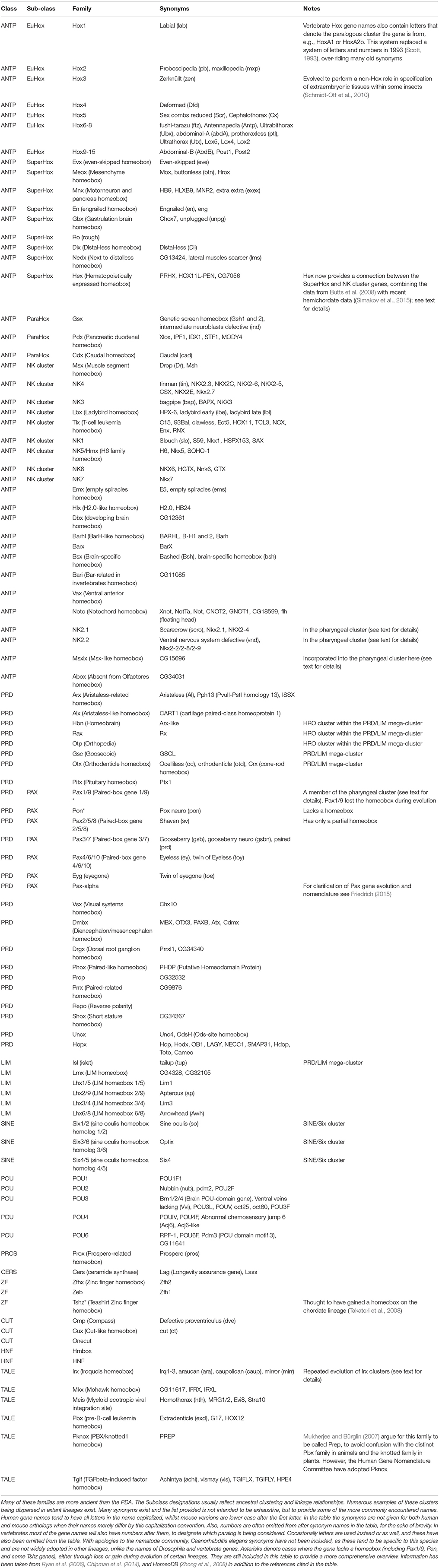
Homeobox genes encode transcription factors that bind DNA in a sequence-specific fashion through the homeodomain motif and control the expression of their target genes in a huge range of developmental processes (Duboule, 1994). It is difficult to find a developmental gene network in animals that does not include a homeobox gene. These genes are taxonomically widespread, being found in animals, plants, fungi, and protists (Derelle et al., 2007; Mukherjee et al., 2009; de Mendoza et al., 2013; Mishra and Saran, 2015) and are thought to have evolved from some sort of Helix-turn-Helix protein similar to those found in prokaryotes (Laughon and Scott, 1984; Kenchappa et al., 2013). Focusing on the homeobox genes of animals, eleven classes of gene families are usually recognized: ANTP, PRD, LIM, POU, HNF, SINE, TALE, CUT, PROS, ZF, and CERS (Holland et al., 2007). Several of these classes are distinct to animals and by implication are likely to be linked to the evolution of aspects of animal-specific biology (see Figure 1; Larroux et al., 2008; Degnan et al., 2009; Suga et al., 2013) [but of course, not all animal-specific biology is entirely attributable to homeobox genes and other animal-specific genes exist (King et al., 2008; Suga et al., 2013)].
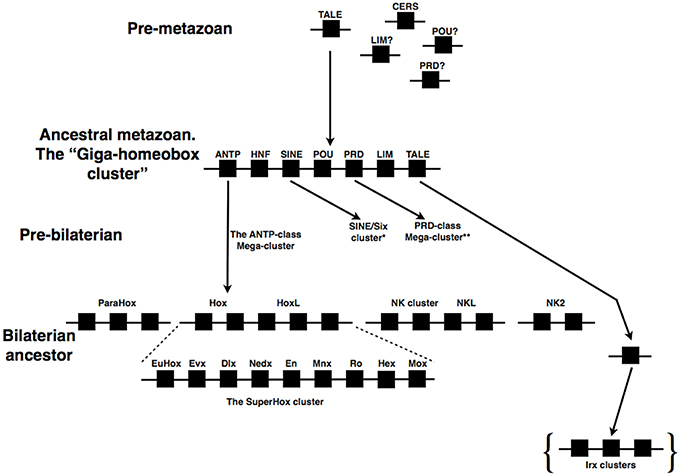
Figure 1. Evolution of the hypothetical metazoan “Giga-homeobox cluster.” Prior to the origin of animals there were only a small number of homeobox genes, including TALE and CERS-class genes along with several further genes of uncertain affinities (Sebé-Pedrós et al., 2011). The genomic arrangement of these genes is also unclear. In an ancestral metazoan a clustered array of homeobox genes likely existed, consisting of the precursors of several classes: the “Giga-homeobox cluster.” The deduction of the composition of this array is described in the text. Further members of this array may be revealed by analyses of additional metazoan genome sequences. Sub-components of this Giga-cluster include the ANTP-class Mega-cluster (see text for details; Pollard and Holland, 2000), which had dispersed into at least four sub-components distributed on distinct chromosomes by the time of the bilaterian (protostome–deuterostome) ancestor (Hui et al., 2012). One of these sub-components was the SuperHox cluster, composed of the true Hox genes (EuHox genes) and several additional ANTP-class genes that were Hox-linked (HoxL). A second sub-component was the “NK cluster” genes with several NK-linked (NKL) genes (see text for details). Further gene clusters deriving from within the Giga-cluster included the SINE/Six cluster and the PRD-class Mega-cluster (see text for details). The Iroquois/Irx cluster is different from the other clusters described here because it expanded to a three-gene cluster independently in several distinct lineages (denoted by the brackets), most likely from a single gene state in the bilaterian ancestor (see text for details). Continuous horizontal lines indicate clustering on the same chromosome. The single asterisk denotes that further details are provided in the text and in Figure 2. The double asterisk denotes that further details are provided in the text and Figure 3.
Another notable feature of animal homeobox genes is that a number of them exist in clusters that are widespread across the animal kingdom. These include clusters of genes from the ANTP-class (e.g., Hox, ParaHox, NK, Mega-homeobox, and SuperHox clusters), the PRD-class (the HRO cluster and its extension), the TALE-class (Irx cluster), and the SINE-class (SIX cluster), as well as an intriguing “pharyngeal”gene cluster composed of different classes of homeobox gene as well as other gene families (Garcia-Fernàndez, 2005; Butts et al., 2008; Mazza et al., 2010; Gómez-Marín et al., 2015; Simakov et al., 2015; and see below). The composition of these clusters and their retention in some animal lineages, but not others, has been the focus of much interest as a possible route to insights into the evolution of animal development as well as genome organization and architecture. Here I provide an overview of animal homeobox gene clusters and the hypotheses linked to their evolution. I focus on gene clusters with deep evolutionary history in the animals that have been conserved across multiple phyla (“primary clustering”), and contrast these with genes being rearranged to form a cluster that was not present ancestrally (“secondary clustering”). I will avoid discussion of lineage-specific instances of gene duplication that have produced, for example, neighboring paralogs of a particular homeobox family (e.g., mammalian examples summarized in Holland, 2013), except for the distinctive case of the Irx gene clusters (see below). Since the evolution of the organization of the Hox cluster has been extensively written about elsewhere (e.g., Monteiro and Ferrier, 2006; Duboule, 2007; Ferrier, 2010, 2012; Ikuta, 2011) and to a lesser extent its evolutionary sister the ParaHox cluster (e.g., Ferrier and Holland, 2001; Ferrier, in press), I will focus on other homeobox clusters here.
The ANTP-class Mega-homeobox Cluster within a Homeobox Superclass Giga-cluster
The Mega-homeobox cluster was first hypothesized by Pollard and Holland (2000) on the basis of an analysis of the then newly available human genome sequence (reviewed in Garcia-Fernàndez, 2005). This hypothesized ancestral cluster of ANTP-class genes includes the well-known Hox genes, as well as the ParaHox genes along with many other ANTP-class families (Pollard and Holland, 2000; Garcia-Fernàndez, 2005). The hypothesis involves the ANTP-class genes evolving via a series of tandem duplications that generated all of the precursors to each of the ANTP-class families, such that there is a clustered array of these family precursor genes together in a Mega-cluster at some point early in animal evolution. Following the origin of this Mega-cluster it is supposed that it started to break apart during evolution, to leave the sub-components now observed in genomes like that of amphioxus (Castro and Holland, 2003), with the Hox cluster and several associated families on one chromosome, the ParaHox cluster on another chromosome and the NK cluster genes on a third chromosome (Pollard and Holland, 2000; Castro and Holland, 2003; Hui et al., 2012).
The Mega-cluster hypothesis was mainly built on the observation that Dlx genes and Msx4 are linked to Hox genes in mammals (Pollard and Holland, 2000). This was thought to be significant because these genes supposedly had greater sequence similarity to the NK cluster genes (see below) than to the Hox genes, which was taken as indicative of an ancestral linkage of all of the Hox and NK cluster genes. The Msx4 data was subsequently excluded when it was found that this gene probably resulted from a retrotransposition event (Castro and Holland, 2003), so that its genomic location in vertebrates cannot necessarily be taken as indicative of the ancestral pre-vertebrate location. This is because such an origin via retrotransposition was distinct from the origins of the other vertebrate Msx paralogs during the two rounds of whole genome duplication events (the so-called 2R events) that occurred at the origin of the vertebrates. Thus, the locations of the other Msx paralogs, rather than Msx4, are more likely to be indicative of an ancestral Msx genomic location. Msx1 and Msx2 (and Msx3 in mouse) are linked to genes of the NK cluster (Pollard and Holland, 2000), which is discussed further below.
The suitability of Dlx as the foundation for the Mega-cluster hypothesis has now also been questioned (Hui et al., 2012). The role of Dlx in the hypothesis hinged on the view that its sequence was closer to those of the NK gene families (placing it within the NK subclass), which led to Dlx sometimes being referred to as an NK-like (NKL) gene (reviewed in Ferrier, 2008). With further taxonomic sampling and a greater diversity of homeobox genes being incorporated into molecular phylogenies and classification analyses, it became clear that the NKL categorization of Dlx was not justified (Ferrier, 2008; Hui et al., 2012). Since the molecular phylogenies of the ANTP-class homeobox genes no longer provided clear support for the Mega-cluster hypothesis, Hui et al. (2012) attempted a different approach, of simply determining the genomic linkage patterns of ANTP-class genes with the aim of determining which are Hox-linked (HoxL) and which are NK-linked (NKL). This change in definition of HoxL and NKL to reflect unambiguous linkage of genes, rather than poorly resolved or unstable phylogenetic relationships of homeobox families, was the precursor to assessing whether distinct animal lineages (such as the deuterostome amphioxus and the protostome Platynereis dumerilii) had distinct remains of the hypothetical Mega-cluster that represented the cluster breaking in different places in independent lineages. If two distinct, but overlapping, patterns of linkage had been found in these two animals then support for the Mega-cluster hypothesis would have been obtained. However, Hui et al. (2012) instead made the surprising discovery that the distribution of the ANTP-class genes across the chromosomes of P. dumerilii is largely identical to the distribution in amphioxus. This may have intriguing implications for potential functional reasons for the retained clustering of some of these homeobox genes across such large evolutionary distances, such as the subsets of NK genes (discussed in Hui et al., 2012). Nevertheless, support for the Mega-cluster hypothesis was not obtained. Instead, it appears that the Mega-cluster had either already broken apart into the distinct linkage groups and patterns that are now present in both P. dumerilii and amphioxus by the time of their last common ancestor (the protostome–deuterostome ancestor), or the Mega-cluster never existed in the first place. Perhaps the various ANTP-class families that are considered within the context of the Mega-cluster hypothesis started to disperse across an ancestral (pre-bilaterian) genome before all of these families had come into existence, such that instead of a single Mega-cluster there were several sub-clusters.
Additional members of the Mega-cluster or “Mega sub-clusters” are now being found as further whole genome sequences become available. These tight linkages and clustering are also now extending beyond the ANTP-class. For example, the sine oculis (So) gene from the SINE-class clusters with the ANTP-class genes Empty spiracles (Ems) and Intermediate neuroblasts defective-b (Ind-b) in the myriapod Strigamia maritima, as well as the Hmbox gene (from the HNF-class) clustering with the ANTP-class genes Exex, Nedx, and Buttonless-a (Btn-a) (Chipman et al., 2014). The first of these two S. maritima examples may in turn relate to the SINE/Six gene clusters (see below), whilst the second example constitutes an extension of a particular sub-component of the Mega-cluster (or one of the “Mega sub-clusters”), the SuperHox cluster (see below). For further discussion of the S. maritima homeobox linkages, see the supplementary text in Chipman et al. (2014).
There are additional examples found in non-bilaterian lineages, such as clustering of a POU-class and ANTP-class gene in a cnidarian (Kamm and Schierwater, 2007). Also, a number of intriguing instances of homeobox clustering involving different gene classes are found in the placozoan Trichoplax adhaerens (Schierwater et al., 2008). These include the PRD-class gene Goosecoid (Gsc) being clustered with ANTP-class genes of NK families, the HNF-class gene (Hnf) being clustered with a PRD-class gene (Prd/Pax-like), there is a cluster of two PRD-class genes (Arx1 and Arx2) with a TALE-class gene (Pknox) and there are two instances of a LIM-class gene being clustered with a TALE-class gene (Lim2/9 with Pbx/PBC, and Lim1/5 with Meis). There is also an instance of a LIM-class cluster that, thus far, seems distinctive for T. adhaerens (Srivastava et al., 2010). These sorts of intriguing single cases of homeobox gene clustering clearly need to be examined more widely, to investigate whether they occur in multiple species. This then will determine how they relate to evolution of primary or secondary clustering, discussed further below. In this vein, there are also a couple of instances of PRD-class gene clustering in T. adhaerens that, in contrast to the LIM cluster, do relate to more taxonomically-widespread clusters (see below).
Since several of these different homeobox gene classes are specific to the animals, it is reasonable to assume that they arose via duplications (probably tandem) from an ancestral metazoan homeobox gene. This likely resulted in an extensive array of different homeobox genes in an early animal ancestor, containing representatives of the precursors for most (perhaps all) of the animal homeobox classes. Some of these genes remained clustered and some of these conserved clusters were retained into modern-day lineages due to functional constraints. These constraints probably included long-range regulatory mechanisms acting across multiple genes, either directly on multiple promoters as occurs in Hox gene regulation (e.g., Tarchini and Duboule, 2006) or indirectly with long-range enhancers spanning bystander genes (Kikuta et al., 2007). Further study of the diversity of homeobox gene clusters across a diversity of animal lineages is thus likely to lead to new insights into the control mechanisms of clustered gene regulation. Furthermore, we can now go beyond the ANTP-class Mega-cluster hypothesis to a homeobox superclass “Giga-cluster” hypothesis (Figure 1).
The SuperHox Cluster
The SuperHox cluster was first described by Butts et al. (2008). This cluster was composed of eight ANTP-class genes that could be deduced as being neighbors of the Hox gene cluster in the bilaterian ancestor, including Mox, Hex, Ro, Mnx, En, Nedx, Dlx, and Evx alongside Hox. The SuperHox cluster was thus seen as a specific sub-component of the hypothetical Mega-cluster and, as with the Mega-cluster, the SuperHox has since been breaking apart during evolution in different places on distinct animal lineages. The 15-gene SuperHox cluster, which contained the eight genes listed above alongside seven true Hox genes (or “EuHox” genes) in the bilaterian ancestor (Balavoine et al., 2002), was deduced from comparisons of the conservatively evolving genomes of amphioxus and the red flour beetle (Tribolium castaneum; Butts et al., 2008). An important assumption underpinned the construction of this cluster from the amphioxus and beetle data; since these genes all belong to the ANTP-class and hence have evolved from each other via duplication, then it is most likely that these duplications were tandem and that the ancestral genes for each family first arose as close genomic neighbors. Thus, ANTP-class genes that are found as close neighbors in extant animals, like amphioxus and the red flour beetle, are more likely to reflect descent from a state in which the genes were neighbors, rather than these genes first evolving as close neighbors, then dispersing around the genome and finally coming back together to be close neighbors secondarily (“close” being taken as <80 kb in the case of the SuperHox deductions; Butts et al., 2008). Whether this assumption is justified will be returned to below, when discussing the NK and pharyngeal clusters.
A further sub-component of the hypothetical Mega-cluster in its initial formulation was the EHGbox cluster, composed of En, HB9, and Gbx (Pollard and Holland, 2000). Given the appealing sounding acronym for this gene cluster it is perhaps unfortunate that the HB9 genes have since been renamed to Motorneuron homeobox (Mnx) (Ferrier et al., 2001). Perhaps in view of this the cluster should also be renamed, to the GEMbox cluster. However, it could also be argued that the idea of an EHGbox/GEMbox cluster can be dispensed with anyway. This is because the molecular distances between the genes are in the order of Megabases in mammals, and hence are much larger than the kilobase distances that constitute the close neighbor relationships since used for deduction of the SuperHox cluster, for instance. Also, the En and Mnx genes of the EHGbox/GEMbox have been subsumed within the SuperHox cluster (Butts et al., 2008).
Further genome sequencing projects have enabled the composition of the SuperHox cluster to be extended slightly. The inclusion of a non-ANTP-class gene, Hmbox (from the HNF-class), has already been mentioned above, in the context of the Mega-cluster and recent data from the myriapod S. maritima (Chipman et al., 2014).
SINE/Six Gene Clusters: CTCF-mediated TADs
If we move out of the ANTP-class we find further examples of homeobox gene clusters. One such cluster is that of the SINE-class genes from the Six1/2, Six4/5, and Six3/6 families. This cluster again is likely to have an ancient ancestry in animal evolution. Six3/6 is clustered with Six1/2 in the non-bilaterian T. adhaerens (which lacks a Six4/5 gene; Schierwater et al., 2008). The full cluster of three genes is found across several bilaterians, including the hemichordates (Simakov et al., 2015), lophotrochozoans (Irimia et al., 2012; Simakov et al., 2013), an echinoderm, and vertebrates (Gómez-Marín et al., 2015), whilst the cluster has dispersed in insects (Figure 2). The situation in vertebrates has been made more complex by the whole genome duplications that occurred at the origin of vertebrates, followed by a further duplication early in teleost evolution. Some gene loss followed each of these whole genome duplications such that in tetrapods there tends to be two SINE clusters, one of Six1, Six6, and Six4 and a second of Six3 and Six2, with a third locus containing only a single SIX gene, Six5 (Figure 2). In a teleost like the zebrafish there are five clusters, two of which contain three genes whilst three clusters possess only two genes (along with a further locus containing one lone SIX gene; Figure 2).
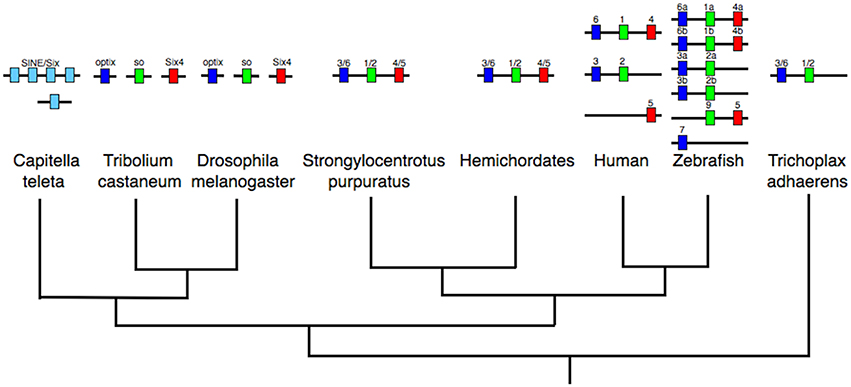
Figure 2. Evolution of the SINE/Six cluster. A cluster of at least two genes existed in non-bilaterians (e.g., T. adhaerens). Gene clusters are widespread across the bilaterians, including deuterostomes (e.g., S. purpuratus, hemichordates, humans, and zebrafish) and at least one lophotrochozoan (the annelid, C. teleta), but not in the insects (e.g., D. melanogaster and T. castaneum) in which the genes are dispersed across separate chromosomes. Closer study is required to resolve the precise orthologs of the C. teleta genes relative to specific gene families (denoted by the pale blue coloration). Dark blue coloration denotes the optix/Six3/6 gene family, green the So/Six1/2 gene family, and red the Six4/5 gene family. All data on gene identification and genomic locations is taken from both Ensembl (http://www.ensembl.org/index.html) and HomeoDB (Zhong et al., 2008; Zhong and Holland, 2011), the Capitella genome portal (http://genome.jgi.doe.gov/Capca1/Capca1.home.html), Gómez-Marín et al. (2015) and Simakov et al. (2015).
Four of the five SINE clusters of zebrafish were recently shown to be subject to long-range regulatory processes that result in Topologically Associated Domains (TADs), the organization of which is similar in both mouse and sea urchin (Gómez-Marín et al., 2015). These TADs are bordered by CCCTC-binding factor (CTCF) sites. This organization, with CTCF-bordered TADs operating over homeobox gene clusters, has also been found for the Hox clusters (Gómez-Díaz and Corces, 2014; Maeda and Karch, 2015; Narendra et al., 2015), and is thus likely to be a rather general mechanism at work in such gene clusters.
The TALE-class Iroquois/Irx Cluster: Independent Cluster Expansions
The TALE-class of homeobox genes is one of the few classes that evolved prior to the origin of the animals (Degnan et al., 2009; Suga et al., 2013; see Figure 1). Within the TALE-class, the Iroquois/Irx genes tend to be clustered in animals. This gene cluster is a little different from the others discussed here. Although, three-gene Irx clusters are widespread across the animal kingdom there appear to be several cases of them having evolved independently, via distinct instances of tandem duplication that in several cases have produced gene clusters of three genes. Thus, although Irx clusters are widespread they are not entirely homologous across all lineages, in the sense that the clusters have been produced from evolutionarily independent gene duplication events. Comparable processes of lineage-specific tandem gene duplication within homeobox gene clusters can be seen in other clusters, such as the Hox (Ferrier, 2012). But the distinctive and intriguing difference about the Irx clusters is that they have repeatedly settled on a three-gene composition. This has happened independently for vertebrates, amphioxus, drosophilids, a myriapod, and an annelid (Irimia et al., 2008; Takatori et al., 2008; Kerner et al., 2009; Maeso et al., 2012; Chipman et al., 2014). Why this might be so still remains a mystery.
PRD-class Clusters: Remains of a PRD-class Mega-cluster?
Mazza et al. (2010) identified the HRO cluster of PRD-class genes in Cnidaria and protostomes, including insects and molluscs. This cluster is composed of the genes Homeobrain (Hbn), Rax/Rx, and Orthopedia (Otp). At least part of the cluster is even more ancient than the cnidarian-bilaterian ancestor as Hbn and Otp are also clustered in the placozoan T. adhaerens (Mazza et al., 2010). Also, elements of the HRO cluster are now known to be more widespread in protostomes than initially described. For example, more recent whole genome sequencing projects like that of the myriapod S. maritima have revealed that this arthropod has also retained the HRO cluster (Chipman et al., 2014).
Intriguingly, this HRO cluster exhibits temporal collinearity in the cnidarian Nematostella vectensis (Mazza et al., 2010). That is, the order of the genes along the chromosome corresponds to the order in which they are activated during development. Temporal collinearity has also been hypothesized to be the main underlying reason for the maintenance of intact, ordered Hox and ParaHox clusters (Ferrier and Holland, 2002; Ferrier and Minguillón, 2003; Monteiro and Ferrier, 2006). Thus, there is the potential that deeper mechanistic understanding of temporal collinearity can be obtained by comparisons across all three homeobox clusters: Hox, ParaHox, and HRO.
Clustering of PRD-class genes is not confined to the HRO cluster. The clustering of Goosecoid (Gsc) and Otx was noted in amphioxus (Putnam et al., 2008; Takatori et al., 2008) and the hemichordate genome sequences analyzed recently, reveal that in one species (Ptychodera flava) Gsc also clusters with Otx, but in another species (Saccoglossus kowalevskii) Gsc instead clusters with Otp, Rx, Hbn, and Islet (Isl) (all of which are PRD-class genes except Isl, which is LIM-class; Simakov et al., 2015). Two things are noteworthy here. First, it will be important to independently check the Saccoglossus gene arrangement, particularly the location of Gsc. Second, the gene nomenclature risks causing confusion and in extended Figure 4 of Simakov et al. (2015), the authors have depicted the cluster containing an Arx gene, when in fact the gene should be named Hbn or Arx-like on the basis of its sequence. Arx is a distinct family from Hbn/Arx-like, as seen in the cnidarian Nematostella vectensis (Ryan et al., 2006; Table 1).
Looking deeper in animal evolution, Schierwater et al. (2008) noted two instances of PRD-class clustering in T. adhaerens: PaxB with Pitx and Ebx/Arx-like with Otp (this second cluster also containing the LIM-class gene Isl). The Ebx/Arx-like gene of Schierwater et al. (2008) is equivalent to the Hbn gene of Mazza et al. (2010). This then, in combination with the new hemichordate data, establishes the clustering of Otp with both Hbn/Arx-like and Isl as an ancient cluster that has been conserved from before the start of the Cambrian, over 541 million years ago. Furthermore, in combination with the data on the HRO PRD-class cluster of cnidarians and selected bilaterians, it is possible to deduce an ancestral extended PRD-LIM class cluster including Hbn, Rx, Otp, Gsc, Otx, and Isl (Figure 3). By comparison to the large ancestral array hypothesized for the ANTP-class (see above), we perhaps should now also view the PRD-class as having evolved via a Mega-cluster array as well (which in turn was also a sub-component of the Giga-cluster outlined above).
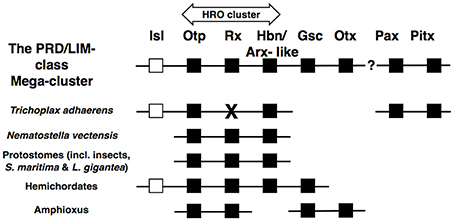
Figure 3. Composition of the PRD/LIM-class Mega-cluster. Specific instances of gene clustering are listed against specific taxa, which when considered together allow the deduction of the PRD/LIM-class Mega-cluster. These animals include non-bilaterians (T. adhaerens and N. vectensis), protostomes, and deuterostomes (hemichordates and amphioxus). Most members of the array are PRD-class genes (black boxes), but there is also a single member of the LIM-class (white box). The Pitx and Pax (PaxB) clustering is found in T. adhaerens, but is not reported for another animal as yet, hence the question mark to denote the ambiguity as to whether these PRD-class genes can be included in the PRD/LIM-class Mega-cluster. The HRO cluster is the PRD-class cluster originally described by Mazza et al. (2010). The figure only shows established instances of clustering arrangements described in the literature (see text for details). Lack of a gene alongside a taxon does not necessarily represent absence of the gene from the genome of that species, except in the case of Rax/Rx for T. adhaerens, which was not found in the placozoan genome by Mazza et al. (2010) (denoted by “X”).
The NK Cluster: An Ancestral Cluster Breaking Apart or Dispersed Genes Coming Together?
If we now return to the ANTP-class, a cluster of NK homeobox genes has been known in insects like Drosophila melanogaster for a number of years, with a prominent role in patterning mesoderm development (Jagla et al., 2001). The composition of the ancestral insect NK cluster has been deduced by consideration of a range of species, such that the “NK cluster” genes can be considered to be a selection from Msx/Drop, tin/NK4, bap/NK3, Lbx, Tlx/C15, slou/NK1, and Hmx/NK5, with subsets of this group forming clusters in particular extant species (Luke et al., 2003; Wotton et al., 2009). Combining this insect data with chordate information has led to the hypothesis that the NK cluster in the bilaterian ancestor included all of the insect “NK cluster” genes as well as NK6 and NK7 (Wotton et al., 2009; Holland, 2013). An NK cluster has also been described for the sponge Amphimedon queenslandica (Larroux et al., 2007). More recently an NK cluster has been identified in hemichordate deuterostomes, with the composition of Hmx/Nkx5-Msx-Nkx3.2-Nkx4-Lbx-Hex when both Saccoglossus kowalevskii and Ptychodera flava are considered together (see Supplementary Extended Figure 4 in Simakov et al., 2015). This is the most extensive deuterostome NK cluster known, and it intriguingly includes the Hex gene. This gene is also a member of the SuperHox cluster as well as the Mega- and Giga-clusters (see above), thus possibly helping to tie all of these clusters together.
In many other species, sub-components of the NK cluster are found as “fragments” of the canonical cluster defined from the insect–chordate comparisons. The assumption is that an ancestral animal had an intact NK cluster and this cluster largely remained intact on the lineage leading to insects, but on the lophotrochozoan and deuterostome lineages the cluster started to break apart. Intriguingly, these breaks are often in similar places, such that the same sub-groups of “NK cluster” genes are found as close genomic neighbors across phylogenetically disparate species (Luke et al., 2003; Wotton et al., 2009; Hui et al., 2012). A likely explanation for the retention of certain sub-components of the NK cluster is that multigenic or shared regulatory elements existed in the ancestral cluster which have been retained into extant lineages. This then restricts the locations within the cluster at which viable breaks can be made. Evidence for ordered enhancers and insulator elements across a subset of NK cluster genes in insects (Cande et al., 2009) lends support to this hypothesis.
Gene nomenclature is complicated and often confusing for the NK genes. This hinders comparisons across species (but see Table 1 for an overview of many of the commonly used names and synonyms for the NK genes). A further problem is that some genes are not easily identified as belonging to a particular gene family due to low node support values in the phylogenetic trees used to classify the genes. This has been particularly troublesome for the NK subclass of genes. One relevant example in the current context is the difficulty with which the sponge NK cluster genes are identified as particular homologs of bilaterian counterparts (Larroux et al., 2007). The A. queenslandica NK cluster is without doubt an NK cluster, but the precise composition of this sponge cluster relative to the bilaterian NK clusters is still open to some debate due to the lack of robust, clearly resolved molecular phylogenies (Larroux et al., 2007; Fortunato et al., 2014). Thus, it is difficult to determine the precise composition of the NK cluster in the earliest stages of animal evolution, before the origin of the bilaterians.
The NK cluster also presents one of the clearest examples yet of the uncertainty that we have about the dynamics and polarity of evolutionary change in homeobox gene clusters: ancient clusters breaking apart vs. dispersed genes coming together (perhaps multiple times independently such that clusters might not be homologous). A recent analysis of NK gene locations across the densely sampled drosophilids revealed that these genes can come together secondarily by multiple intrachromosomal rearrangements over relatively short evolutionary periods, i.e., within genera rather than across phyla, at least for genes that are already linked on the same chromosome (Chan et al., 2015). In contrast, the presence of NK clusters in sponges, insects and now hemichordates pushes us to assume that there was an ancestral NK cluster formed via the types of tandem duplications and cluster retention invoked in hypotheses of the evolution of other homeobox clusters, and then this ancestral cluster simply disperses (at least to a certain degree) in distinct lineages. How then can the two opposing scenarios be reconciled? There is insufficient data and too poor an understanding of genome evolutionary dynamics to provide a definitive answer. However, one relevant fact is clear: not all animal genomes are equal in their evolutionary behavior, with some genomes evolving and rearranging at much higher rates than others (Irimia et al., 2012). This is most clearly exemplified by comparisons of synteny across animals, which reveal that some species exhibit high (statistically significant) levels of conserved synteny across large evolutionary timescales [e.g., between cnidarians, chordates (Putnam et al., 2007, 2008), some arthropods (Chipman et al., 2014), and lophotrochozoans (Simakov et al., 2013)] whilst other lineages show high rates of rearrangements such that little, if any, conserved synteny can be seen even between members of the same phylum [e.g., tunicates (Denoeud et al., 2010) or some insects (Zdobnov and Bork, 2007)]. Consequently, it is clear that this evolutionary diversity must be taken into account and more homeobox linkage data is required from a taxonomically widespread selection of species in order to distinguish generalities from lineage-specific oddities.
Two further NK genes are not commonly considered as part of the NK cluster, namely Nkx2.1 and Nkx2.2 (for synonyms see Table 1). Furthermore, these NK genes tend not to be linked on the same chromosome as the NK cluster genes (Hui et al., 2012), which is taken as a further ancient interchromosomal split of the ancestral Mega-cluster (if this ancestral cluster did actually exist; see above). These genes have now been found to be components of a “pharyngeal” gene cluster in some deuterostomes, which has important implications for our understanding of the evolution of gene clusters more generally.
The Pharyngeal Gene Cluster
The pharyngeal gene cluster was first identified in vertebrates, but has recently been described in other deuterostomes, including hemichordates and an echinoderm (Simakov et al., 2015). This gene cluster gains its name from several of the genes being expressed in the pharyngeal regions of several species in which the cluster is found. It consists of six genes; Nkx2.1, Nkx2.2, Pax1/9, FoxA, mipol1, and slc25A21 (Simakov et al., 2015). Four of the genes are transcription factor-encoding genes, two of which contain homeoboxes (Nkx2.1 and Nkx2.2) and one of which is derived from an ancestral homeobox-containing gene [Pax1/9, which lacks a homeobox whilst other Pax genes have retained some or all of their homeoboxes (Takatori et al., 2008)]. FoxA is the fourth transcription factor-encoding gene, but is a forkhead domain-encoding gene rather than being from the homeobox superclass. The clustering of these genes seems to be due, at least in part, to the location of regulatory elements of some of the transcription factor-encoding genes (Pax1/9 and FoxA) within the introns of the two non-transcription factor genes (mipol1 and slc25A21) (Simakov et al., 2015).
One of the distinctive features of this cluster, relative to the clusters discussed above, is that it is not composed of genes that are all related to each other by gene duplication. Also, Simakov et al. (2015) report that although the cluster can be found in several different deuterostomes, it has not yet been found in any non-deuterostome and thus is likely to have evolved specifically in the deuterostome lineage. It will be important to continue investigating whether the pharyngeal cluster is indeed deuterostome-specific, as further genome sequences become available, as discussed further below.
Since orthologs of these pharyngeal cluster genes do exist in non-deuterostome animals then it seems this gene cluster constitutes an example of a cluster being assembled secondarily during evolution. How this then impacts on our understanding of the homeobox gene clusters described above remains to be seen. Much of the thinking on homeobox clusters has included assumptions that tight physical linkage reflects an ancestral genomic juxtaposition, as described for several of the clusters mentioned above. This has always seemed reasonable due to the genes being in the same class or superclass and hence being related via gene duplication. Since the most common form of gene duplication is tandem duplication (Mendivil Ramos and Ferrier, 2012) then it seems reasonable to suppose that closely neighboring homeobox genes first arose as gene neighbors that have stayed as neighbors in some lineages. This is in contrast to the less parsimonious alternative that such genes first arose as tandem duplicate neighbors, were then dispersed around the genome during evolution, but secondarily came back together again to be close neighbors only in some lineages.
However, perhaps we need to revise our assumptions about such evolution of genome architecture. The assembly of a functional gene cluster such as the pharyngeal cluster by “pulling genes together” during evolution, rather than tandemly duplicating genes and then co-regulating them, provides an important contrast to the homeobox gene clusters.
Perhaps the pharyngeal cluster can be viewed as an extreme version of the co-regulated gene “clusters” such as muscle or house-keeping genes loosely co-localizing in some animal genomes (Hurst et al., 2004), or groups of genes regulated by the same transcription factors or localizing in the same nuclear domains of transcriptional activity then coming to lie in the same regions of genomes following rearrangements during evolution (Janga et al., 2008; Zhang et al., 2012; Farré et al., 2015; Vieux-Rochas et al., 2015). An extension of this evolutionary process might then have involved the pharyngeal cluster being “driven” toward the more extreme, tighter clustering by further consolidation under overlapping or pan-cluster regulatory mechanisms. Consolidation under long-range, multigenic regulatory mechanisms has been hypothesized for the evolution of vertebrate Hox gene clusters (Duboule, 2007). Also, the evolutionary stabilization of genome neighbors can be linked to long-range regulatory elements acting on developmental control genes across genomic distances that also happen to harbor neighboring bystander genes, as also seems to be happening for the pharyngeal cluster (Simakov et al., 2015). However, how “difficult” or “easy” it is for such arrangements to evolve, and tight clusters of functionally related genes be assembled secondarily, still needs to be examined more widely across the animals. Also, if such a “secondary” evolutionary process is to be invoked for homeobox clusters such as the Hox, ParaHox, NK, and so on, then it will be necessary to establish the additional likelihood of tandemly duplicated genes dispersing prior to then coming together again secondarily in a process comparable to the assembly of the pharyngeal cluster.
There is an additional gene that should perhaps also be considered in the context of the pharyngeal cluster: Msxlx. Although Simakov et al. (2015) do not formally include this homeobox gene in the pharyngeal cluster, they do show that it is present in the clusters of hemichordates and the echinoderm Acanthaster planci. Msxlx is also clustered with Nkx2.2 in the protostome Lottia gigantea (Simakov et al., 2015). This is intriguing, and indicates that it is definitely necessary to look more closely across a wider range of species before we conclude that the pharyngeal cluster definitely does represent a deuterostome-specific entity (rather than simply a cluster that has dispersed in the limited range of non-deuterostomes examined to date). Examination of the expression of Msxlx in a range of species is also required. The expression has been studied in the invertebrate chordate amphioxus (Branchiostoma floridae; Butts et al., 2010). Butts et al. (2010) focused on Msxlx because it is one of a small handful of homeobox genes that have been lost during the evolution of the Olfactores (i.e., the urochordates plus vertebrates). This accounts for why it is not found in the pharyngeal clusters of vertebrates, but, more importantly, the expression in amphioxus exhibits an intriguing association with the pharyngeal region (as do the other “lost” homeobox genes investigated by Butts et al., 2010). Amphioxus Msxlx is expressed in the region of the anterior endoderm that constitutes Hatschek's left diverticulum, and develops into the pre-oral pit by fusing with the ectoderm. This is thought to be homologous to the vertebrate adenohypophysis. The genes of the pharyngeal cluster, including Msxlx, are thus an interesting group of genes to investigate further for two main reasons. Firstly, the evolution of their genomic organization is intriguing, for the potential for improving our understanding of gene cluster evolution. Secondly, the evolution of their expression is interesting in the context of understanding the evolution of the pharyngeal region.
Conclusion
The instances of homeobox gene clustering discussed above are focused on those that are already described in, or can be gleaned from, the literature. There are likely to be additional instances of homeobox clustering to be found in the ever-increasing number of whole genome sequences that are becoming available, which will enable further refinement of the clusters described here as well as possibly providing new examples of clusters that had ancient origins but have thus far been overlooked. It is valuable to continue to search for such clusters as they provide important insights into evolutionary transitions, both in terms of animal development as well as genome organization. Such links between genome organization, as represented by cluster organization, and the evolution of animal development have been the focus of much attention for the renowned Hox genes, almost ever since their discovery in the 1980s. The further homeobox clusters discussed here provide a whole new suite of opportunities to expand the study systems available to us for such evolutionary developmental genomics research. Such research is also vital if we are to understand the evolutionary dynamics of animal genomes and distinguish primary from secondary clustering.
Author Contributions
DF conceived and wrote the manuscript.
Funding
Work in the author's lab is funded by BBSRC DTP studentships and the School of Biology, University of St. Andrews.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to thank past and present members of the lab for discussions as well as colleagues in the community. The referees also provided a number of helpful comments that improved the manuscript.
References
Balavoine, G., de Rosa, R., and Adoutte, A. (2002). Hox clusters and bilaterian phylogeny. Mol. Phyl. Evol. 24, 366–373. doi: 10.1016/S1055-7903(02)00237-3
Butts, T., Holland, P. W. H., and Ferrier, D. E. K. (2008). The urbilaterian SuperHox cluster. Trends Genet. 24, 259–262. doi: 10.1016/j.tig.2007.09.006
Butts, T., Holland, P. W. H., and Ferrier, D. E. K. (2010). Ancient homeobox gene loss and the evolution of chordate brain and pharynx development: deductions from amphioxus gene expression. Proc. Biol. Sci. 277, 3381–3389. doi: 10.1098/rspb.2010.0647
Cande, J. D., Chopra, V. S., and Levine, M. (2009). Evolving enhancer-promoter interactions within the tinman complex of the flour beetle, Tribolium castaneum. Development 136, 3153–3160. doi: 10.1242/dev.038034
Castro, F. L., and Holland, P. W. H. (2003). Chromosomal mapping of ANTP class homeobox genes in amphioxus: piecing together ancestral genomes. Evol. Dev. 5, 459–465. doi: 10.1046/j.1525-142X.2003.03052.x
Chan, C., Jayasekera, S., Kao, B., Pàramo, M., von Grotthuss, M., and Ranz, J. M. (2015). Remodelling of a homeobox gene cluster by multiple independent gene reunions in Drosophila. Nat. Commun. 6:6509. doi: 10.1038/ncomms7509
Chipman, A. D., Ferrier, D. E. K., Brena, C., Qu, J., Hughes, D. S. T., Schröder, R., et al. (2014). The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12:e1002005. doi: 10.1371/journal.pbio.1002005
Degnan, B. M., Vervoort, M., Larroux, C., and Richards, G. S. (2009). Early evolution of metazoan transcription factors. Curr. Opin. Genet. Dev. 19, 591–599. doi: 10.1016/j.gde.2009.09.008
de Mendoza, A., Sebé-Pedrósa, A., Sestak, M. S., Matejcic, M., Torruella, G., Domazet-Loso, T., et al. (2013). Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc. Natl. Acad. Sci. U.S.A. 110, E4858–E4866. doi: 10.1073/pnas.1311818110
Denoeud, F., Henriet, S., Mungpakdee, S., Aury, J-M., Da Silva, C., Brinkmann, H., et al. (2010). Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330, 1381–1385. doi: 10.1126/science.1194167
Derelle, R., Lopez, P., Le Guyader, H., and Manuel, M. (2007). Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evol. Dev. 9, 212–219. doi: 10.1111/j.1525-142X.2007.00153.x
Duboule, D. (2007). The rise and fall of Hox gene clusters. Development 134, 2549–2560. doi: 10.1242/dev.001065
Farré, M., Robinson, T. J., and Ruiz-Herrera, A. (2015). An Integrative Breakage Model of genome architecture, reshuffling and evolution: the Integrative Breakage Model of genome evolution, a novel multidisciplinary hypothesis for the study of genome plasticity. BioEssay 37, 479–488. doi: 10.1002/bies.201400174
Ferrier, D. E. K. (2008). “When is a Hox gene not a Hox gene? The importance of gene nomenclature,” in Evolving Pathways: Key Themes in Evolutionary Developmental Biology, eds A. Minelli and G. Fusco (Cambridge: Cambridge University Press), 175–193.
Ferrier, D. E. K. (2010). “Evolution of Hox complexes,” in Hox Genes: Studies from the 20th to the 21st Century, ed J. S. Deutsch (Austin, TX; New York, NY: Landes Bioscience and Springer Science and Business Media), 91–100.
Ferrier, D. E. K. (2012). Evolution of the Hox Gene Cluster. Chichester: eLS. John Wiley & Sons, Ltd.
Ferrier, D. E. K. (in press). The origin of the Hox/ParaHox genes, the Ghost Locus hypothesis the complexity of the first animal. Brief. Funct. Genomics. doi: 10.1093/bfgp/elv056.
Ferrier, D. E. K., Brooke, N. M., Panopoulou, G., and Holland, P. W. H. (2001). The Mnx homeobox gene class defined by HB9, MNR2 and amphioxus AmphiMnx. Dev. Genes Evol. 211, 103–107. doi: 10.1007/s004270000124
Ferrier, D. E. K., and Holland, P. W. H. (2001). Ancient origin of the Hox gene cluster. Nat. Rev. Genetics 2, 33–38. doi: 10.1038/35047605
Ferrier, D. E. K., and Holland, P. W. H. (2002). Ciona intestinalis ParaHox genes: evolution of Hox/ParaHox cluster integrity, developmental mode and temporal colinearity. Mol. Phylogenet. Evol. 24, 412–417. doi: 10.1016/S1055-7903(02)00204-X
Ferrier, D. E. K., and Minguillón, C. (2003). Evolution of the Hox/ParaHox gene clusters. Int. J. Dev. Biol. 47, 605–611.
Fortunato, S. A., Adamski, M., Mendivil Ramos, O., Leininger, S., Liu, J., Ferrier, D. E. K., et al. (2014). Calcisponges have a ParaHox gene and dynamic expression of dispersed NK homeobox genes. Nature 514, 620–623. doi: 10.1038/nature13881
Friedrich, M. (2015). Evo-devo gene toolkit update: at least seven Pax transcription factor subfamilies in the last common ancestor of bilaterian animals. Evol. Dev. 17, 255–257. doi: 10.1111/ede.12137
Garcia-Fernàndez, J. (2005). The genesis and evolution of homeobox gene clusters. Nat. Rev. Genetics 6, 881–892. doi: 10.1038/nrg1723
Gómez-Díaz, E., and Corces, V. G. (2014). Architectural proteins: regulators of 3D genome organization in cell fate. Trends Cell Biol. 24, 703–711. doi: 10.1016/j.tcb.2014.08.003
Gómez-Marín, C., Tena, J. J., Acemel, R. D., López-Mayorga, M., Naranjo, S., de la Calle-Mustienes, E., et al. (2015). Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc. Natl. Acad. Sci. U.S.A. 112, 7542–7547. doi: 10.1073/pnas.1505463112
Holland, P. W. H. (2013). Evolution of homeobox genes. WIREs Dev. Biol. 2, 31–45. doi: 10.1002/wdev.78
Holland, P. W. H., Booth, H. A. F., and Bruford, E. A. (2007). Classification and nomenclature of all human homeobox genes. BMC Biol. 5:47. doi: 10.1186/1741-7007-5-47
Hui, J. H. L., McDougall, C., Monteiro, A. S., Holland, P. W. H., Arendt, D., Balavoine, G., et al. (2012). Extensive chordate and annelid macrosynteny reveals ancestral homeobox gene organization. Mol. Biol. Evol. 29, 157–165. doi: 10.1093/molbev/msr175
Hurst, L. D., Pál, C., and Lercher, M. J. (2004). The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genetics 5, 299–310. doi: 10.1038/nrg1319
Ikuta, T. (2011). Evolution of invertebrate deuterostomes and Hox/ParaHox genes. Genomics Proteomics Bioinformatics 9, 77–96. doi: 10.1016/S1672-0229(11)60011-9
Irimia, M., Maeso, I., and Garcia-Fernàndez, J. (2008). Convergent evolution of clustering of Iroquois homeobox genes across metazoans. Mol. Biol. Evol. 25, 1521–1525. doi: 10.1093/molbev/msn109
Irimia, M., Tena, J. J., Alexis, M. S., Fernandez-Miñan, A., Maeso, I., Bogdanovic, O., et al. (2012). Extensive conservation of ancient microsynteny across metazoans due to cis-regulatory constraints. Genome Res. 22, 2356–2367. doi: 10.1101/gr.139725.112
Jagla, K., Bellard, M., and Frasch, M. (2001). A cluster of Drosophila homeobox genes involved in mesoderm differentiation programs. BioEssays 23, 125–133. doi: 10.1002/1521-1878(200102)23:2<125::AID-BIES1019>3.0.CO;2-C
Janga, S. C., Collado-Vides, J., and Babu, M. M. (2008). Transcriptional regulation constrains the organization of genes on eukaryotic chromosomes. Proc. Natl. Acad. Sci. U.S.A. 105, 15761–15766. doi: 10.1073/pnas.0806317105
Kamm, K., and Schierwater, B. (2007). Ancient linkage of a POU class 6 and an anterior Hox-like gene in Cnidaria: implications for the evolution of homeobox genes. J. Exp. Zool. B Mol. Dev. Evol. 308, 777–784. doi: 10.1002/jez.b.21196
Kenchappa, C. S., Heidarsson, P. O., Kragelund, B. B., Garrett, R. A., and Poulsen, F. M. (2013). Solution properties of the archaeal CRISPR DNA repeat-binding homeodomain protein Cbp2. Nucl. Acid Res. 41, 3424–3435. doi: 10.1093/nar/gks1465
Kerner, P., Ikmi, A., Coen, D., and Vervoort, M. (2009). Evolutionary history of the Iroquois/Irx genes in metazoans. BMC Evol. Biol. 9:74. doi: 10.1186/1471-2148-9-74
Kikuta, H., Laplante, M., Navratilova, P., Komisarczuk, A. Z., Engström, P. G., Fredman, D., et al. (2007). Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 17, 545–555. doi: 10.1101/gr.6086307
King, N., Westbrook, M. J., Young, S. L., Kuo, A., Abedin, M., Chapman, J., et al. (2008). The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788. doi: 10.1038/nature06617
Larroux, C., Fahey, B., Degnan, S. M., Adamski, M., Rokhsar, D. S., and Degnan, B. M. (2007). The NK homeobox gene cluster predates the origin of hox genes. Curr. Biol. 17, 706–710. doi: 10.1016/j.cub.2007.03.008
Larroux, C., Luke, G. N., Koopman, P., Rokhsar, D. S., Shimeld, S. M., and Degnan, B. M. (2008). Genesis and expansion of metazoan transcription factor gene classes. Mol. Biol. Evol. 25, 980–996. doi: 10.1093/molbev/msn047
Laughon, A., and Scott, M. P. (1984). Sequence of a Drosophila segmentation gene–protein-structure homology with DNA binding proteins. Nature 310, 25–31. doi: 10.1038/310025a0
Luke, G. N., Castro, L. F., McLay, K., Bird, C., Coulson, A., and Holland, P. W. H. (2003). Dispersal of NK homeobox gene clusters in amphioxus and humans. Proc. Natl. Acad. Sci. U.S.A. 100, 5292–5295. doi: 10.1073/pnas.0836141100
Maeda, R. K., and Karch, F. (2015). The open for business model of the bithorax complex in Drosophila. Chromosoma 124, 293–307. doi: 10.1007/s00412-015-0522-0
Maeso, I., Irimia, M., Tena, J. J., González-Pérez, E., Tran, D., Ravis, V., et al. (2012). An ancient genomic regulatory block conserved across bilaterians and its dismantling in tetrapods by retrogene replacement. Genome Res. 22, 642–655. doi: 10.1101/gr.132233.111
Mazza, M. E., Pang, K., Reitzel, A. M., Martindale, M. Q., and Finnerty, J. R. (2010). A conserved cluster of three PRD-class homeobox genes (homeobrain, rx and orthopedia) in the Cnidaria and Protostomia. EvoDevo 1:3. doi: 10.1186/2041-9139-1-3
Mendivil Ramos, O., and Ferrier, D. E. K. (2012). Mechanisms of gene duplication and translocation and progress towards understanding their relative contributions to animal genome evolution. Int. J. Evol. Biol. 2102:846421. doi: 10.1155/2012/846421
Mishra, H., and Saran, S. (2015). Classification and expression analyses of homeobox genes from Dictyostelium discoideum. J. Biosci. 40, 241–255. doi: 10.1007/s12038-015-9519-3
Monteiro, A. S., and Ferrier, D. E. K. (2006). Hox genes are not always collinear. Int. J. Biol. Sci. 2, 95–103. doi: 10.7150/ijbs.2.95
Mukherjee, K., Brocchieri, L., and Bürglin, T. R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26, 2775–2794. doi: 10.1093/molbev/msp201
Mukherjee, K., and Bürglin, T. R. (2007). Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J. Mol. Evol. 65, 137–153. doi: 10.1007/s00239-006-0023-0
Narendra, V., Rocha, P. P., An, D., Raviram, R., Skok, J. A., Mazzoni, E. O., et al. (2015). CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021. doi: 10.1126/science.1262088
Pollard, S. L., and Holland, P. W. H. (2000). Evidence for 14 homeobox gene clusters in human genome ancestry. Curr. Biol. 10, 1059–1062 doi: 10.1016/S0960-9822(00)00676-X
Putnam, N. H., Butts, T., Ferrier, D. E. K., Furlong, R. F., Hellsten, U., Kawashima, T., et al. (2008). The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071. doi: 10.1038/nature06967
Putnam, N. H., Srivastava, M., Hellsten, U., Dirks, B., Chapman, J., Salamov, A., et al. (2007). Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94. doi: 10.1126/science.1139158
Ryan, J. F., Burton, P. M., Mazza, M. E., Kwong, G. K., Mullikin, J. C., and Finnerty, J. R. (2006). The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the startlet sea anemone, Nematostella vectensis. Genome Biol. 7:R64. doi: 10.1186/gb-2006-7-7-r64
Schierwater, B., Kamm, K., Srivastava, M., Rokhsar, D., Rosengarten, R. D., and Dellaporta, S. L. (2008). The early ANTP gene repertoire: insights from the placozoan genome. PLoS ONE 3:e2457. doi: 10.1371/journal.pone.0002457
Schmidt-Ott, U., Rafiqi, A. M., and Lemke, S. (2010). “Hox3/zen and the evolution of extraembryonic epithelia in insects,” in Hox Genes: Studies from the 20th to the 21st Century, ed J. S. Deutsch (Austin, TX; New York, NY: Landes Bioscience and Springer Science+Business Media).
Scott, M. P. (1993). A rational nomenclature for vertebrate homeobox (HOX) genes. Nucl. Acids Res. 21, 1687–1688. doi: 10.1093/nar/21.8.1687
Sebé-Pedrós, A., de Mendoza, A., Lang, B. F., Degnan, B. M., and Ruiz-Trillo, I. (2011). Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol. Biol. Evol. 28, 1241–1254. doi: 10.1093/molbev/msq309
Simakov, O., Kawashima, T., Marlétaz, F., Jenkins, J., Koyanagi, R., Mitros, T., et al. (2015). Hemichordate genomes and deuterostome origins. Nature 527, 459–465. doi: 10.1038/nature16150
Simakov, O., Marlétaz, F., Cho, S. J., Edsinger-Gonzales, E., Havlak, P., Hellsten, U., et al. (2013). Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526–531. doi: 10.1038/nature11696
Srivastava, M., Larroux, C., Lu, D. R., Mohanty, K., Chapman, J., Degnan, B. M., et al. (2010). Early evolution of the LIM homeobox gene family. BMC Biol. 8:4. doi: 10.1186/1741-7007-8-4
Suga, H., Chen, Z., de Mendoza, A., Sebé-Pedrós, A., Brown, M. W., Kramer, E., et al. (2013). The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 4, 2325. doi: 10.1038/ncomms3325
Takatori, N., Butts, T., Candiani, S., Pestarino, M., Ferrier, D. E. K., Saiga, H., et al. (2008). Comprehensive survey and classification of homeobox genes in the genome of amphioxus, Branchiostoma floridae. Dev. Genes Evol. 218, 579–590. doi: 10.1007/s00427-008-0245-9
Tarchini, B., and Duboule, D. (2006). Control of Hoxd genes' collinearity during early limb development. Dev. Cell 10, 93–103. doi: 10.1016/j.devcel.2005.11.014
Vieux-Rochas, M., Fabre, P. J., Leleu, M., Dubole, D., and Noordermeer, D. (2015). Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc. Natl. Acad. Sci. U.S.A. 112, 4672–4677. doi: 10.1073/pnas.1504783112
Wotton, K. R., Weierud, F. K., Juárez-Morales, J. L., Alvares, L. E., Dietrich, D., and Lewis, K. E. (2009). Conservation of gene linkage in dispersed vertebrate NK homeobox clusters. Dev. Genes Evol. 219, 481–496. doi: 10.1007/s00427-009-0311-y
Zdobnov, E. M., and Bork, P. (2007). Quantification of insect genome divergence. Trends Genet. 23, 16–20. doi: 10.1016/j.tig.2006.10.004
Zhang, Y., McCord, R. P., Ho, Y-J., Lajoie, B. R., Hildebrand, D. G., Simon, A. C., et al. (2012). Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148, 908–921. doi: 10.1016/j.cell.2012.02.002
Zhong, Y-F., Butts, T., and Holland, P. W. H. (2008). HomeoDB: a database of homeobox gene diversity. Evol. Dev. 10, 516–518. doi: 10.1111/j.1525-142X.2008.00266.x
Keywords: SuperHox, collinearity, SIX genes, Pax genes, Nkx genes, pharyngeal gene cluster, genome evolution
Citation: Ferrier DEK (2016) Evolution of Homeobox Gene Clusters in Animals: The Giga-Cluster and Primary vs. Secondary Clustering. Front. Ecol. Evol. 4:36. doi: 10.3389/fevo.2016.00036
Received: 22 December 2015; Accepted: 27 March 2016;
Published: 14 April 2016.
Edited by:
Alistair Peter McGregor, Oxford Brookes University, UKReviewed by:
Ralf Janssen, Uppsala University, SwedenNico Posnien, University of Göttingen, Germany
Ignacio Maeso, Centro Andaluz de Biología del Desarrollo, Spain
Copyright © 2016 Ferrier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David E. K. Ferrier, dekf@st-andrews.ac.uk
 David E. K. Ferrier
David E. K. Ferrier