Inter-Kingdom Modification of Metabolic Behavior: [GAR+] Prion Induction in Saccharomyces cerevisiae Mediated by Wine Ecosystem Bacteria
- Department of Viticulture and Enology, University of California, Davis, Davis, CA, USA
The yeast Saccharomyces cerevisiae has evolved to dominate grape juice fermentation. A suite of cellular properties, rapid nutrient depletion, production of inhibitory compounds and the metabolic narrowing of the niche, all enable a minor resident of the initial population to dramatically increase its relative biomass in the ecosystem. This dominance of the grape juice environment is fueled by a rapid launch of glycolysis and energy generation mediated by transport of hexoses and an efficient coupling of transport and catabolism. Fermentation occurs in the presence of molecular oxygen as the choice between respiratory or fermentative growth is regulated by the availability of sugar a phenomenon known as glucose or catabolite repression. Induction of the [GAR+] prion alters the expression of the major hexose transporter active under these conditions, Hxt3, reducing glycolytic capacity. Bacteria present in the grape juice ecosystem were able to induce the [GAR+] prion in wine strains of S. cerevisiae. This induction reduced fermentation capacity but did not block it entirely. However, dominance factors such as the rapid depletion of amino acids and other nitrogen sources from the environment were impeded enabling greater access to these substrates for the bacteria. Bacteria associated with arrested commercial wine fermentations were able to induce the prion state, and yeast cells isolated from arrested commercial fermentations were found to be [GAR+] thus confirming the ecological relevance of prion induction. Subsequent analyses demonstrated that the presence of environmental acetic acid could lead to [GAR+] induction in yeast strains under certain conditions. The induction of the prion enabled yeast growth on non-preferred substrates, oxidation and reduction products of glucose and fructose, present as a consequence of bacterial energy production. In native ecosystems prion induction never exceeded roughly 50–60% of the population of yeast cells suggesting that the population retains the capacity for maximal fermentation. Thus, the bacterial induction of the [GAR+] prion represents a novel environmental response: the query of the environment for the presence of competing organisms and the biological decision to temper glucose repression and dominance and enter a metabolic state enabling coexistence.
Introduction
Saccharomyces cerevisiae is a widely studied model research organism and as a consequence much is understood about the biology of this yeast. These investigations have been conducted largely under pure culture laboratory conditions, and analyses in yeast native environments and in the presence of microbial residents of the same ecosystems have been lacking. S. cerevisiae is also an important industrial workhorse responsible for the fermentative stabilization of both foods and beverages across many human cultures. This yeast is considered to be the first domesticated organism as S. cerevisiae is commonly found in man-made environments and mankind has influenced the geographic and biological diversity of this yeast (Legras et al., 2007; Liti et al., 2009; Sicard and Legras, 2011; Bisson, 2012; Almeida et al., 2015; Eberlein et al., 2015).
It is generally accepted that S. cerevisiae evolved to dominate fermentation of sugar-rich environments (Cray et al., 2013; Williams et al., 2015). The inhibition of respiration by sugar substrates in favor of fermentation, particularly of glucose (Crabtree effect), developed early in the evolutionary history of the modern-day yeast species (Pfeiffer et al., 2001; MacLean and Gudelj, 2006; Hagman et al., 2013; Hagman and Piškur, 2015; Williams et al., 2015) and assured fermentative metabolism in sugar-rich environments. This metabolic strategy yields energy faster and enables more rapid growth than respiration (Pfeiffer et al., 2001; MacLean and Gudelj, 2006; Williams et al., 2015).
Several properties of S. cerevisiae enable dominance of grape juice fermentation including rapid depletion of nutrients and molecular oxygen, narrowing of the niche of the juice by changes in the redox status, decrease in pH and production of ethanol, and the production of inhibitory compounds. Nitrogen is the most limiting growth nutrient in grape juice (Ingledew and Kunkee, 1985; Butzke, 1998; Bisson, 1999; Hagen et al., 2008; Bisson and Walker, 2015; Albergaria and Arneborg, 2016) and S. cerevisiae is able to deplete amino acids and ammonium generally within a few to 24 h of being in juice, depending upon the initial cell concentration (Monteiro and Bisson, 1991; Pinu et al., 2014; Gutiérrez et al., 2016). Under these conditions S. cerevisiae uncouples nitrogen uptake from cell growth (Gutiérrez et al., 2016) and this ability to rapidly deplete nitrogen from the environment is thought to be critical to dominance of those environments (Bisson and Walker, 2015; Albergaria and Arneborg, 2016).
The depletion of oxygen rather than the production of ethanol has been shown to be a critical factor in loss of viability of non-Saccharomyces yeasts present on berry surfaces (Holm Hansen et al., 2001). The fermentative environment becomes rapidly chemically reduced making it challenging for microbes that obtain energy mainly from partial oxidation reactions (the acetic acid bacteria) (Matsushita et al., 2005; Deppenmeier and Ehrenreich, 2009) to thrive as well (Bisson and Walker, 2015). S. cerevisiae was found to grow at lower redox potentials than many other yeast species (Visser et al., 1990) and the rapid reduction in redox potential seen in grape juice may reflect the use of metabolism to create a more limiting environment for other organisms by Saccharomyces. S. cerevisiae also produces inhibitory compounds that can be also considered dominance factors: antimicrobial peptides (Comitini et al., 2005; Osbourne and Edwards, 2007; Albergaria et al., 2010; Nehme et al., 2010; Branco et al., 2014), fatty acids (Lafon-Lafourcade et al., 1984), and sulfur dioxide (Hinze and Holzer, 1985; Boulton et al., 1996; Bisson and Walker, 2015). Additional inhibitory compounds have been found to be present although not yet identified chemically (Pérez-Nevado et al., 2006; Wang et al., 2016). Thus, multiple characteristics have evolved in S. cerevisiae to enable domination of the microbiota during the early stages of grape juice fermentation.
Many of these dominance factors are dependent upon ATP availability and require rapid generation of energy from the process of fermentation. Grape juice is a sugar-rich environment and contains ~1.2–1.5 M glucose and an equivalent concentration of fructose (Boulton et al., 1996). Genes not required under these conditions are transcriptionally repressed by glucose and other hexoses and glucose also regulates protein modification, stability and activity post-translationally (reviewed in Carlson, 1987; Gancedo, 1998; Kayikci and Nielsen, 2015). This mobilization of glycolysis in conditions of plentiful sugar substrates allows rapid energy (ATP) generation (Pfeiffer et al., 2001). Uptake of amino acids is energy-requiring (Boulton et al., 1996) and ATP is also needed to generate and sustain a strong proton motive force across the plasma membrane enabling depletion of other nutrients such as oxygen. This ability to rapidly shift to fermentative energy generation is important in the inhibition of competing microorganisms and dominance of the fermentation (Bisson and Walker, 2015).
Prions are self-templating heritable protein conformational states that confer different phenotypes depending upon the specific conformation present within the cell (reviewed in Shorter and Lindquist, 2005; Halfmann and Lindquist, 2010; Halfmann et al., 2010, 2012; Liebman and Chernoff, 2012; Garcia and Jarosz, 2014; Wickner et al., 2015). Prion formation is also dependent upon the presence of protein chaperons or heat shock proteins within the cell. The prion state is curable meaning that cells can spontaneously revert to the prion-negative form. Prion-based phenotypes arise at higher frequencies within the population than those due to genomic mutation and the ability to restore an alternative conformation and lose the prion phenotype serves as a malleable strategy for reversible adaptation to suboptimal environments. Prions show non-Mendelian segregation with all progeny of a prion positive strain crossed against a prion negative strain being prion positive. The establishment of the [GAR+] prion state in yeast cells relaxes glucose repression enabling growth on alternative carbon sources in the presence of high glucose concentrations and reduces the glucose utilization capacity (Brown and Lindquist, 2009; Jarosz et al., 2014a; Walker et al., 2016). Prion-based inheritance appears to be a commonly used strategy in wild yeasts to regulate a variety of reversible phenotypes (Halfmann and Lindquist, 2010; Halfmann et al., 2012; Newby and Lindquist, 2013; Garcia and Jarosz, 2014; Jarosz et al., 2014b).
The [GAR+] prion phenotype is due to the formation of a specific complex between the major plasma membrane proton pump, Pma1, and a transcriptional factor that regulates expression of hexose transporters, Std1 (Brown and Lindquist, 2009). Under normal circumstances Pma1 associates with an alternate regulatory factor, Mth1. When associated with Mth1 glucose repression is fully operational, but when Pma1 associates instead with Std1 this protein is protected against degradation in the presence of glucose and able to interact with a co-repressor protein enabling the complex to decrease expression of hexose transporter Hxt3 (Brown and Lindquist, 2009). The reduction in Hxt3-mediated transport is proposed to reduce the internal glucose signal relieving glucose repression in the presence of glucose and enabling growth on substrates normally blocked during glucose repression. Pma1 is therefore able to exist in two states: a primary state binding to Mth1 or a secondary state binding to Std1. Cellular metabolism responds to the type of complex formed. Thus, the [GAR+] prion represents a switch between these two binding states of Pma1 (Brown and Lindquist, 2009). Pma1 activity is a metabolic pacemaker in S. cerevisiae particularly under fermentative growth at the low pH values seen in grape (pH 3.2–3.9 are typical) playing an essential role in maintenance of cellular proton homeostasis by preventing acidification of the cytoplasm and cell death through the removal of protons (Serrano, 1978; Mason et al., 2014). Protons originate from cellular metabolism of sugars, from symport systems coupled to proton movements, from passive passage of protonated acid species across the cell membrane and from passive proton flux a process that is elevated by the presence of ethanol. The existence of the [GAR+] prion represents one mechanism by which Pma1 activity can not only be coordinated with but control glycolytic flux to prevent saturation of the proton pumping capacity and cytoplasmic acidification.
Induction of the [GAR+] prion appreciably affected regulation of a single gene, the HXT3 hexose transporter. S. cerevisiae contains a multigene family of hexose transporters expressed under differing glucose concentrations and growth stages of the cells (reviewed in Kruckeberg, 1996; Boles and Hollenberg, 1997; Bisson et al., 2016a). Although over 20 genes with glucose transport function have been identified, only a subset (HXT1-HXT7) are required for fermentative growth on glucose and any one of these genes when expressed as the sole transporter enables the cells to ferment and utilize available glucose (Wieczorke et al., 1999). On the surface it would seem that a 30-fold reduction in expression of only one of these genes, HXT3, would have a modest impact on yeast ability to utilize hexose substrates. However, the Hxt3 transporter plays a primary role during grape juice fermentation (Luyten et al., 2002; Perez et al., 2005). Naturally occurring mutations in this gene have been correlated with grape juice fermentative capacity (Zuchowska et al., 2015) and loss of Hxt3, although not impacting growth or substrate utilization, did affect ethanol tolerance (Karpel et al., 2008). The establishment of the [GAR+] prion is therefore proposed to have an impact in the native yeast ecosystem of grape juice fermentation.
The [GAR+] prion was identified by selecting for ability of yeast cells to grow on a non-preferred carbon source glycerol in the presence of a non-metabolizable glucose mimetic, glucosamine (Brown and Lindquist, 2009). Glucosamine is able to establish glucose repression when present in the environment but cannot be used as a carbon or energy source by S. cerevisiae (Ball et al., 1976; Kotyk and Knotkova, 1989). Wild type strains are unable to utilize the glycerol and form colonies in the presence of glucosamine. In contrast, cells that have induced the [GAR+] prion are able to grow on this medium (Brown and Lindquist, 2009). In the analysis of the factors impacting [GAR+] prion induction it was discovered that growth on the selective medium could be induced by the presence of bacterial contaminants also growing on the medium. A survey of bacteria revealed that several genera and species could lead to prion induction at higher than normal frequencies in S. cerevisiae (Jarosz et al., 2014a). The inducer was found to be present in the environment and diffusible acting on yeast colonies at a distance from the bacterium (Jarosz et al., 2014a; Walker et al., 2016). The nature of the inducer has not been determined but given the wide range of bacteria capable of inducing this modification of metabolic behavior the inducer would need to be produced by a variety of species. Fermentations of grape juice comparing [GAR+] and [gar−] strains of the same genotype demonstrated that [GAR+] strains fermented more slowly than their non-prion induced counterparts and that bacterial populations differed with evidence of higher populations of the lactic acid bacteria normally dominated by S. cerevisiae in the [GAR+] fermentations (Walker et al., 2016), suggesting that the ability of bacteria to induce the prion in yeast would benefit the bacteria by reducing the ability of the yeast to dominate the environment. To test this hypothesis the goal of this work was to investigate the ability of grape and wine ecosystem bacteria to induce the [GAR+] prion naturally during fermentation and to study the effects of prion induction on yeast strain dominance characteristics during grape juice fermentation. In addition the induction of the [GAR+] prion in populations undergoing commercial grape juice fermentations was assessed to determine if this prion is induced in native ecosystems.
Materials and Methods
Yeast and Bacterial Strains Used
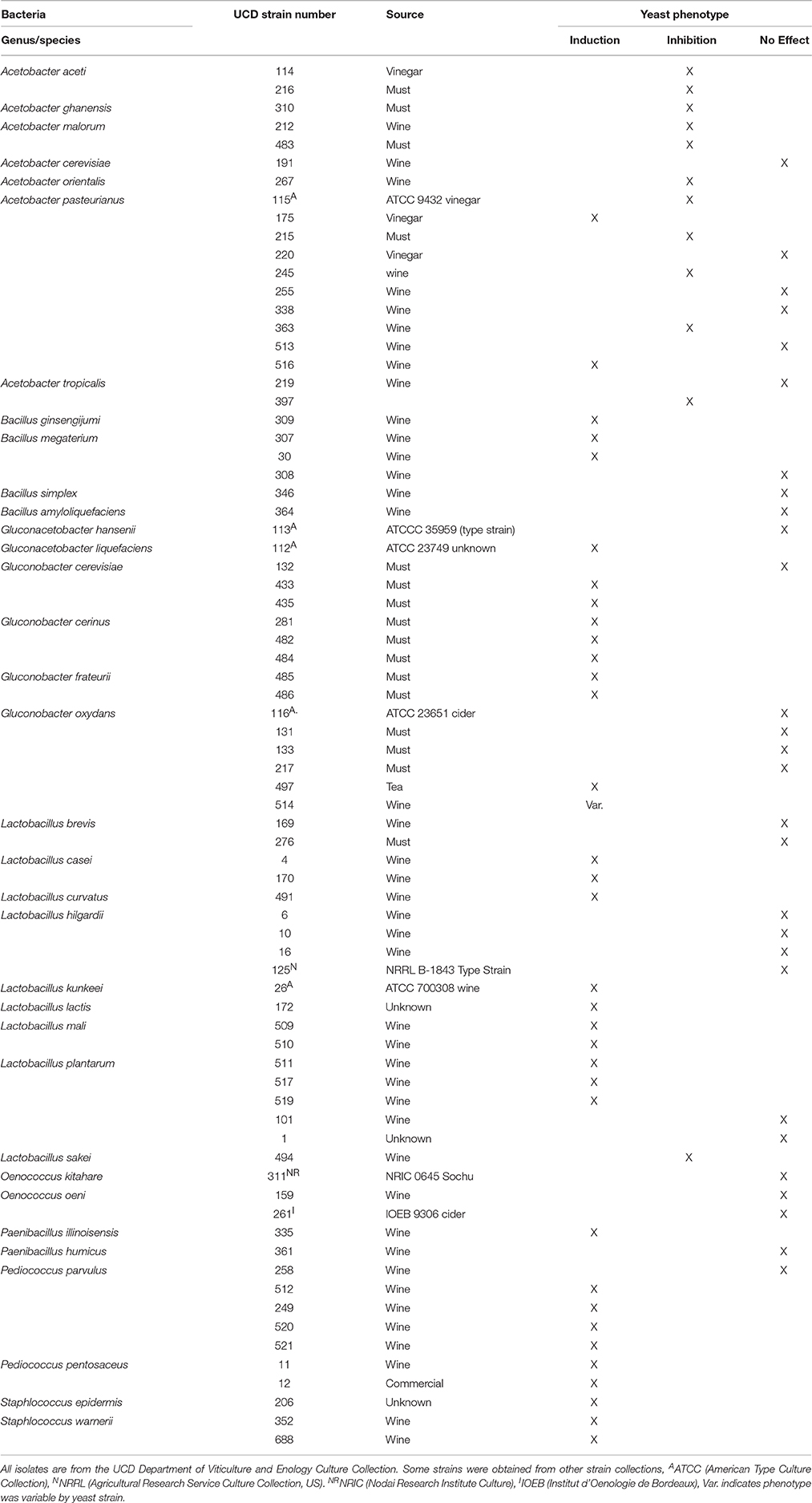
All yeast strains used were obtained from the Department of Viticulture and Enology culture collection. Commercial isolates were purified individual colonies isolated from packets of commercial yeast that were then entered into the collection. Strains used were: UCD932 (Ba2 Lambrusco, Italian vineyard isolate) (Mortimer et al., 1994), UCD777 (isolate of commercial strain EC1118), laboratory strain W303 (MATa/MATα leu2-3,112 trp1-1, can1-100, ade2-1, ura3-1, his3-11,15), laboratory strain Y55 (MATa/MATα), DBVPG1106 (grape isolate, Australia), DBVPG 6040 (fermenting juice isolate, Netherlands). Yeast were maintained as frozen stocks in 50% glycerol solution and propagated on YPD [Yeast extract, peptone, glucose (2%)] medium. The bacterial strains used were obtained from the Department of Viticulture and Enology culture collection and are listed in Table 1. The source of the isolates is also listed. Isolates from natural sources were restreaked to purity and subjected to DNA sequence analysis for unequivocal identification prior to being entered into the collection. In one experiment direct commercial preparations of dried bacteria were used: Viniflora and VP41 as indicted in the text.
Bacterial Species Identification
Bacteria from stuck fermentations were selected by centrifuging 1 mL of wine and resuspending the pellet in 100 μL of wine. This suspension was used to streak for isolation on ½ strength MRS (Difco) (1:1 with water) plates with 25 mg/L of Delvocid (DSM). The bacteria were re-streaked for isolated colonies and separate colonies were selected for identification from each sample. Bacteria were identified either using sequence comparisons of 16s rDNA or via MALDI-TOF using the Bruker Biotyper system according to the manufacturers specifications and matching to the custom database (Sogawa et al., 2011). In cases of unclear identification via MALDI, strains were subjected to 16s rDNA sequencing. DNA from the bacteria was isolated using the Promega Wizard genomic DNA kit (Fischer Scientific). Isolates were identified by PCR of the 16s ribosomal DNA using the 27F and 1492R primers (Frank et al., 2008). PCR was conducted in 50 μL reactions with 1 l of template DNA. Thermocycler conditions were an initial 1 min at 95°C followed by 35 cycles of 45 s at 95°C, 30 s at 56°C, 2 min at 72°C, and a final extension at 72°C for 8 min. PCR products were cleaned with a Promega Wizard or Qiagen PCR clean up kit. Samples were sequenced at the UC Davis DBS sequencing facility and results run on the NIH BLAST database to determine the closest match for taxonomic identification
Assessment of [GAR+] Phenotype
The presence or absence of the [GAR+] phenotype was assessed on GGM medium as described (Brown and Lindquist, 2009). GGM agar contains (per liter): 10 g yeast extract (Becton, Dickinson and Company, United States), 20 g peptone (Becton, Dickinson and Company, United States), 25 mL of sterile 80% glycerol stock solution (Fisher Scientific, United State), 10 mL of filter sterilized 5% glucosamine stock solution (Acros Organics, United State), and 20 g granulated agar (Apex Bioresearch Products, United States). Briefly, yeast were pregrown to an absorbance of 0.4 A580 in YPD and serially diluted five times at 1:5 for a total final dilution of 1:3125 and 3 μL of each dilution plated on GGM agar. Cell concentrations in the final dilution spot were approximately 15–20 cells per 3 μL. Plates were incubated at 30°C and evaluated daily for 7 days. Control platings on fully permissive YPD as well as YP glycerol plates were conducted for a qualitative assessment of growth on permissive and prion inducing media. All growth tests were run in biological triplicates.
Assessment of Bacterial Impact on [GAR+] Prion Formation
Bacterial strains were pre-grown in ½strength MRS broth (Difco) at 30°C to stationary phase. UCD932, UCD777, and W303 yeast strains were pre-grown in YP media (1% Yeast extract and 2% Bacto peptone) with 2% glucose to log phase on a roller drum at room temperature. Yeast cells were harvested at absorbance of 0.4 at A580, washed twice with sterile water and resuspended at approximately 3–4 × 106 cells/mL to be used for dilutions and plating. Bacterial impact on yeast growth on YP media with 2% glycerol and 0.05% glucosamine was measured by plating 3 μL of 1:5 serial dilutions of yeast such that the final dilution contains ~20 cells. Bacteria were grown aerobically in test tubes on a roller drum and diluted to an OD580 of 2.0 and 3 μL plated across all spots at a consistent distance from the diluted yeast cultures. Growth and induction of [GAR+] was assessed after 5 days of incubation at 30°C. All tests were run in triplicate. To assess crossfeeding, control plates of YP no carbon source and YP glycerol, no glucosamine were also run for each sample assessing the level of growth nearest the bacteria as compared to that in the center of the plates. Colonies appearing near the bacteria were isolated, passaged on non-selective media, and then replated on GGM to assess growth phenotype and retention of the ability to grow on GGM in the absence of the bacteria. There were cases where non-wine ecosystem bacteria did lead to either cross-feeding or to apparent degradation of the glucosamine present and “induced” yeast did not retain the prion phenotype following passage on non-selective media, but those organisms were not included in this study as none originated from the wine ecosystem, but served to assure that these controls would identify cases of growth without prion induction.
Spent Medium Prion Induction Analysis
Bacterial strains were grown in YP broth (1% Yeast extract and 2% bactopeptone) with 2% glycerol and 0.05% glucosamine (GGM media) at 30°C. The media with bacteria were clarified by centrifuging and filtered through a 0.22 μM Millex-GV syringe filters (Millipore SLGV033RS) to obtain spent media. UCD932 was pre-grown in YP broth with 2% glucose and cells were harvested at an absorbance of 0.4 at A580 and washed with sterile water and inoculated into the spent media at approximately 6.67 × 105 cells. Samples were taken after 4 h and 1:5 serial dilutions were plated on YP plates with 2% glycerol and 0.05% glucosamine and on YP plates with 2% glucose for dilution controls. Growth and [GAR+] induction were assessed after incubation at 30°C for 5 days. Samples were run in triplicate.
Acetic Acid Prion Induction Analysis
For the liquid induction assays, a culture of wild type [gar−] strain of UCD932 cells were grown to an OD580 of 0.6–0.8 in YPD, washed, and diluted to an OD580 of 0.4 in GGM with acetic acid at the concentrations indicated and incubated at room temperature on roller drum for 4 h. OD580 was measured again after incubation and cultures were diluted back to OD580 0.4 to make five-fold serial dilution series. Serial dilutions were then plated on GGM agar as described above. Samples were run in triplicate.
End Product Induction Spot Assays
Yeast cultures in log phase (about 104 cells/mL) were spread plated on GGM media. Assay discs were saturated with 10 μl of different concentrations of acetic acid (0, 0.5, 1, 1.5, 2, 3, 4%) or lactic acid (0, 1, 2, 3%) or pyruvic acid (0, 0.5, 1, 2%) which corresponds to 0, 50, 100, 150, 200, 300, or 400 μg of acetic acid, 0, 100, 200, or 300 μg of lactic acid, and 0, 50, 100, or 200 μg of pyruvic acid respectively per spot. These were placed on each quadrant of the petri dish. Plates were incubated at 30°C and growth pattern was monitored for 2–5 days. Samples were run a minimum of four times.
Analysis of Bacterial End Products
UCD 26 (Lactobacillus kunkeii), UCD 175 (Acetobacter pasturianus), UCD215 (Acetobacter pasteurianus) were grown in ½ strength MRS media, 0.45 μM filtered Chardonnay juice, YP (1% Yeast extract and 2% bactopeptone) media with different carbon sources including 2% glucose, 2% fructose, 1% each of glucose and fructose and 2% glycerol, to stationary phase, then centrifuged and filtered through a 0.22 μm Millex-GV syringe filters (Millipore SLGV033RS). Acetic acid and lactic acid levels were measured by Anion analysis kit using Beckman Coulter ProteomeLab PA800 capillary electrophoresis system. Biological replicates were run for each strain per condition and technical replicates were done for each analysis.
Amino Acid Depletion Analysis
Pre-inocula of UCD932 without and with the [GAR+] prion were grown to stationary phase (48 h) in 10 mL sterile filtered Chardonnay juice on a roller drum at room temperature. Fermentations were carried out in triplicates in filtered Chardonnay juice with starting inoculum of approximately 3.3 × 105 cells. Flasks were incubated with agitation at 120 rpm at 28°C. Fermentation progress was monitored by measuring weight loss due to carbon dioxide release. Samples were taken routinely through the fermentation for 10 days, clarified by centrifugation and filtered through 0.22 μM Millex-GV syringe filters (Millipore SLGV033RS) prior to storing at 4°C for amino acid analysis. Amino acid analysis was performed by the UC Davis Proteomics Core (University of California, Davis).
Screening for the [GAR+] Phenotype in Arrested Commercial Wines
Samples of arrested wine fermentations were directly plated onto WLN medium (Difco) and S. cerevisiae colonies identified by colony morphology (Pallmann et al., 2001) and confirmed by cell morphology. Up to 8 isolated colonies were selected per wine sample and each colony inoculated into one well of a 96 well plate containing 100 μL of YPD broth and grown for 24 h with shaking at room temperature. A multipronged replica plater was used to transfer liquid cultures from the 96 well plates to GGM and YPD media plates and incubated at 30°C. Growth of the spots was assessed at 1 day for YPD and after 5 days for GGM. Growth on the GGM plates was compared to [GAR+] and [gar−] control strains: UCD932 [GAR+], UCD932 [gar−], UCD777 [GAR+], and UCD777 [gar−]. Isolates showing similar growth to the [GAR+] stains were selected as presumptive [GAR+] isolates.
Curing of the [GAR+] Prion by Desiccation
Curing of the [GAR+] prion was adapted from Tapia and Koshland (2014). Isolated colonies were selected from the YPD plate of the screening process and inoculated into 200 μL of YPD broth using one isolate per well of a 96 well plate. Cells were grown for 24 h with shaking at room temperature. The absorbance of the wells were taken using a plate reader at A580. Cell suspensions were washed by spinning the plate in a centrifuge and extracting excess water without disturbing cell pellet. Two hundred microliters of water was added, the cells resuspended and recentrifuged in the plate twice. Cell pellets were then resuspended in 100 μL of dilute phosphate buffered saline (1.25 mM , 17.125 mM NaCl, and 0.3375 mM KCl). Ten microliters of each isolate were added to 100 μL of sterile Milli Q water to be tip plated for viability count on both YPD and GGM. After day 1 for YPD and day 5 for GGM the number of isolated colonies bigger than 0.3 mM diameter were counted to obtain initial total viability and population penetrance of the [GAR+] prion levels. The remainder of the cells in the 96 well plate were placed in an area with approximately 60% relative humidity at 23°C for 1 month with the top cover slightly open by holding up the corners of the plate with stiff paper. After 1 month cells in the wells were rehydrated in 100 μL of dilute phosphate buffer saline and tip plated for viability after desiccation on both YPD and GGM.
Analysis of Growth on Different Substrates
Single colonies of UCD932 [gar−] and [GAR+] were picked from YPD plates, and inoculated into 5 mL of 2% YPD liquid and left to grown overnight on a rotary drum at room temperature (~25°C). Overnight cultures were inoculated into fresh 5 mL 2% YPD liquid cultures and allowed to reach mid-late exponential phase (~0.8 A580). Cells were washed in sterile DI H2O and concentrated to 1.2 A580. A 96 well plate was filled with 250 μL of YP liquid base containing the corresponding percentage of added carbon stock solution. Washed cells were inoculated into wells at 0.05 A580 using a Rannin LTS multichannel pipet (Mettler Toledo, Oakland, CA). A Breath Easy® sealing membrane was placed on the 96 well plate (Sigma-Aldrich, St. Louis, MO). The plate was then placed in a Synergy 2 BioTek plate reader, incubating at 30°C sampling every 30 min preceded by 40 s of variable shaking (BioTek, Winooski, VT). Samples were run in 8 biological replicates. Data represent the average of 8 independent wells.
Results
Induction of the [GAR+] Prion Occurs across Wine Strains
Extensive analysis of one yeast strain vineyard isolate, UCD932, demonstrated that this yeast can induce the [GAR+] phenotype which displays all of the hallmark traits of a prion (Walker et al., 2016). Therefore, the ability of a broader array of wine strains to induce the prion was assessed. To evaluate prion induciblity of wine strains, cells were plated on glycerol glucosamine medium and on control media lacking glucosamine. All strains evaluated grew well on the basal medium containing glucose (YPD) as sole carbon and energy source which also served to quantify the number of viable cells in the original culture based upon number of colonies formed at the two greatest dilutions (Figure 1A). The second control medium was YP glycerol to assess ability to grow on glycerol as a carbon and energy source, Glycerol is in general a poorer energy source than glucose at 2% (both substrates were present at this concentration). One of the strains tested, S288C, was not able to grow well on the medium with glycerol as sole carbon and energy source without addition of glucosamine (YPG, Figure 1B), but all other strains tested were able to grow on this medium. Strains induce the [GAR+] prion at different frequencies on glycerol glucosamine medium (GGM) as seen in Figure 1C. This medium contains the same level of glycerol as Figure 1B but in addition contains the glucose mimetic glucosamine a non-metabolizable analog of glucose that induces glucose repression. Juice isolate strain DBVPG6040 showed the best induction of the prion with growth to the terminal dilution plated. Two other wine strains, UCD932 (grape isolate) and EC1118 (UCD777, commercial isolate) also showed growth at the terminal dilution on GGM plates (inoculated with [gar−] starting cultures) in previously published work (Walker et al., 2016). The two laboratory strains, Y55 and W303 displayed moderate induction on the GGM medium. Strain DBVPG1106, also a vineyard isolate, showed weak induction of the [GAR+] phenotype.
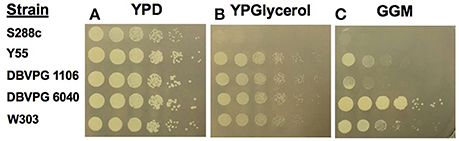
Figure 1. [GAR+] prion induction by wine strains. Growth of the evaluated yeast strains on the control media, YPD (YP media with 2% glucose) (A). Growth of yeast strains on basal YP media with 2% glycerol (B). Growth on prion selective GGM medium (C).
A hallmark of prion induction is the heritability of the associated phenotype. Colonies of UCD932 and EC1118 obtained from the terminal dilutions on GGM were grown on YPD for several generations and then replated onto GGM and compared to the parental strain that had not previously been plated on GGM (Figure 2), That the [GAR+] derivatives show full growth upon replating as compared to [gar−] parental strains after passage on non-selective media indicates that the ability to grow on alternative carbon sources in the presence of inducers of glucose repression represents a heritable change within the population.
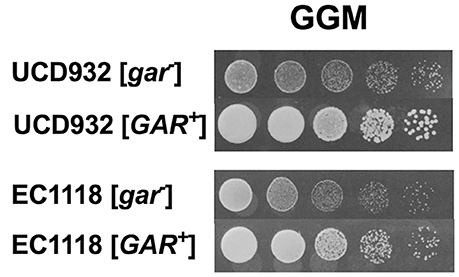
Figure 2. The [GAR+] phenotype displays heritability. Growth of [GAR+] yeast isolates from the selective (GGM) on GGM medium following passage on permissive YPD medium as compared to the respective parent [gar−] strain previously maintained only on YPD. Spot assays represent five-fold serial dilutions.
Induction of the [GAR+] prion has been associated with a decrease in expression of the major hexose transporter, Hxt3, present during the active phase of wine fermentations (Brown and Lindquist, 2009). This modification of glucose utilization slows fermentation progression in juices such as Chardonnay consistent with a decrease in sugar uptake capacity (Walker et al., 2016). This observation was repeatable using Chardonnay from a different vintage (Supplemental Figure 1). Under these pure culture conditions the [GAR+] yeast are fully capable of completing the fermentation, catabolizing all available sugar, but the onset of fermentation is delayed and the fermentation curve differs in shape from that of the wildtype strain consistent with the observed impact of the prion on induction of the Hxt3 transporter.
Saccharomyces Ecosystem Bacteria Induce the [GAR+] Prion
Previous work demonstrated that bacteria in co-cultivation with S. cerevisiae on GGM medium could accelerate the induction of the [GAR+] prion (Jarosz et al., 2014a). A wide array of bacteria were able to induce on this medium (Jarosz et al., 2014a). However, many of these bacteria are not found naturally in the same ecosystems as S. cerevisiae. We therefore screened our collection of bacterial isolates from grapes, musts and wines to determine if native ecosystem organisms could also induce the formation of the prion. For this analysis strains were categorized as having one of three effects: (1) induction as evidenced by enhanced growth on GGM plates in the yeast columns nearest to the bacteria, (2) inhibition as evidenced by the absence of growth of yeast columns nearest to the bacteria, and (3) no effect, characterized by yeast growth near the bacteria matching that of columns in the middle of the plate (Figure 3). Inhibition can be observed for the acetic acid bacterium Acetobacter malorum (Figure 3A) and induction for the acetic acid bacterium Gluconobacter cerinus (Figure 3A) and for two commercial lactic acid bacterium preparations of Oenoccus oeni (Figure 3B). The commercial preparations were pregrown directly from the packets and purity of the commercial preparations was not assessed. Both of the commercial preparations showed induction (Figure 3B) in contrast to pure cultures of O. oeni (Table 1).

Figure 3. Effect of Saccharomyces ecosystem bacteria on growth and induction of the [GAR+] prion. Growth of Gluconobacter cerinus (top) and Acetobacter malorum (bottom) adjacent to [gar −] Saccharomyces UCD932 strain on GGM media induces and inhibits growth respectively (A). Both strains of Oenococcus oeni adjacent to [gar−] Saccharomyces UCD932 strain induce growth of yeast on GGM media (B). Spot assays represent five-fold serial dilutions.
All three classes of impacts on yeast growth were observed among the bacteria evaluated with, in some cases, all members of a species showing the identical effect and in others the effect varying by strain within a species (Table 1). This analysis was conducted using yeast strain UCD932. Two bacterial classes, the acetic acid bacteria and the lactic acid bacteria, are commonly found on the surfaces of grapes and may persist in the wine. Many of these organisms are associated with arrested or slow fermentations. In general the majority of the species assessed in the genus Acetobacter were inhibitory on GGM media. Of the 19 strains tested in this genus, 11 were inhibitory (58%), 6 showed no effect and 2 (10%) were inducing. The two strains showing induction were isolates of A. pasteurianus one of which was isolated from wine. Strains of multiple species of Gluconobacter, also classified as an acetic acid bacterium, were evaluated. Of the 14 strains from 4 species, 8 (57%) were clearly inducing and 5 demonstrated no effect. One strain showed a variable response meaning that some yeast strains were induced and others showed no effect (strain 514). Two species showed strain variability: two G. cerevisiae isolates were inducing and the third showed no effect. Similarly for G. oxydans 4 of the 6 strains evaluated were neutral and two were inducing with one isolate showing a strain effect in induction (514). All isolates from the other two species, G. cerinus and G. frateurii were inducing. None of the Gluconobacter strains isolated from wines was inhibitory of yeast growth on the GGM plates. Gluconacetobacter is rarely isolated from grapes or wine but the ability of type strains to induce the prion was evaluated (Table 1). One species, Gluconacetobacter liquefaciens induced and the other, Gluconacetobacter hansenii showed no effect.
Three genera of lactic acid bacteria, Oenococcus, Lactobacillus, and Pediococcus, are commonly found on grapes and in wines and can be associated with arrest of fermentation. All but one isolate of Pediococcus, P. parvulus 258, were capable of induction of the [GAR+] prion. The two Oenococcus species evaluated also were unable to induce the prion in contrast to the two commercial preparations of Oenococcus used in Figure 3. Nine species of Lactobacillus were analyzed: L. brevis, L. casei, L. curvatus, L. hilgardii, L. kunkeei, L. lactis, L. mali, L. plantarum, and L. sakei, all of which have been isolated from grapes and/or wine. The L. sakei strain, which came from an arrested wine fermentation, was inhibitory similar to the acetic acid bacteria. None of the isolates of L. brevis and L. hilgardii showed an effect. All isolates tested of L. casei, L. curvatus, L kunkeei, L. lactis, and L mali were able to induce the prion state. Three of the five isolates of L. plantarum showed induction of the prion and the other two isolates showed no effect. Interestingly the 3 strains showing an effect were all isolated from arrested wine fermentations. Thus, although the ability to induce prion formation is widespread it is not universal among isolates of the same species.
Other genera of bacteria can be isolated from wines although much less frequently. Isolates of Bacillus megaterium and B. gingsengijumi obtained from wines were both able to induce the prion as were isolates of Staphylococcus. One isolate of Paenibacillus from wine was also able to induce the prion. Thus, the ability to induce the prion spans the spectrum of bacterial isolates obtained from wines.
The spot plate assay and diffusible nature of the inducer leaves open the possibility that what is being observed is not prion induction but cross-feeding of substrate-limited colonies of yeast. Inducing bacteria make end products such as pyruvate, lactate, and acetic acid that could serve as growth substrates. To differentiate between cross-feeding and prion induction two types of experiments were conducted. Given the heritable nature of prion induction, simple restreaking of the growth adjacent to the bacterial isolates on GGM agar would lead either to growth in the absence of the bacteria if induction had occurred or to a lack of growth in the absence of the bacteria if cross-feeding were responsible for the observations of growth on the initial plate. In all cases growth on GGM occurred in the absence of bacteria. A colony of UCD932 induced by Acetobacter pastuerianus (UCD 175) was obtained from a GGM plate adjacent to spots of the inducing bacterium A. pasteurianus. The colony was restreaked onto the non-selective medium, YPD, and retested on GGM. The retested strain grew well at all dilutions on the GGM medium as compared to the original [gar−] strain (Figure 4).
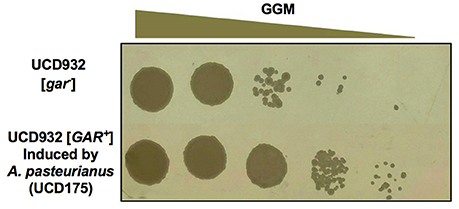
Figure 4. Induction of [GAR+] prion by bacteria is heritable. Growth of retested Saccharomyces UCD932 strain from a colony adjacent to Acetobacter pasteurianus UCD175 strain on GGM plate after passage through nonselective YPD.
The second type of experiment performed was to conduct the induction during liquid growth in spent medium. A. pasteurianus UCD175 was grown in liquid GGM media to an absorbance of 2 at 580 nm, removed by filtration and the yeast incubated in the spent medium for a period of 4 h. Following incubation yeast cells were collected and washed twice then serial dilutions plated onto GGM. Incubation of UCD932 in the spent medium (top panel, Figure 5) resulted in growth on GGM to the terminal dilution. Incubation in liquid GGM alone or in synthetic must Triple M (Spiropoulos et al., 2000) did not result in better growth on GGM plates (Figure 5). Washing of the cells did not impact ability to grow on GGM indicating that a heritable change had been induced in the yeast during the incubation period in the spent medium. Thus, Saccharomyces ecosystem bacteria induce the prion state in this yeast via diffusible factors.
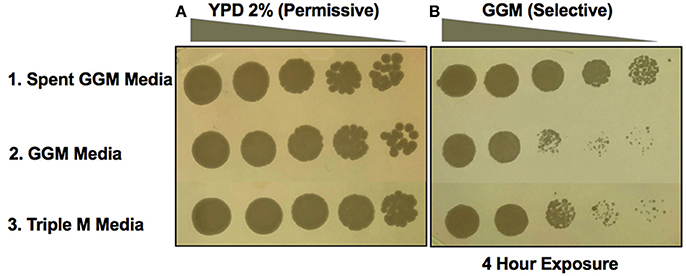
Figure 5. Bacterial spent media induction of [GAR+] in Saccharomyces. Growth of UCD932 in different conditions on the control media, YP with 2% glucose (A). Growth following exposure to Acetobacter pasteurianus (UCD175) spent GGM media for 4 h compared to growth in control media or synthetic grape juice (Triple M) (B). Spent GGM media was filter sterilized before exposure to UCD932 strains. Spot assays represent five-fold serial dilutions.
Prion Induction Is Observed during Commercial Wine Fermentation
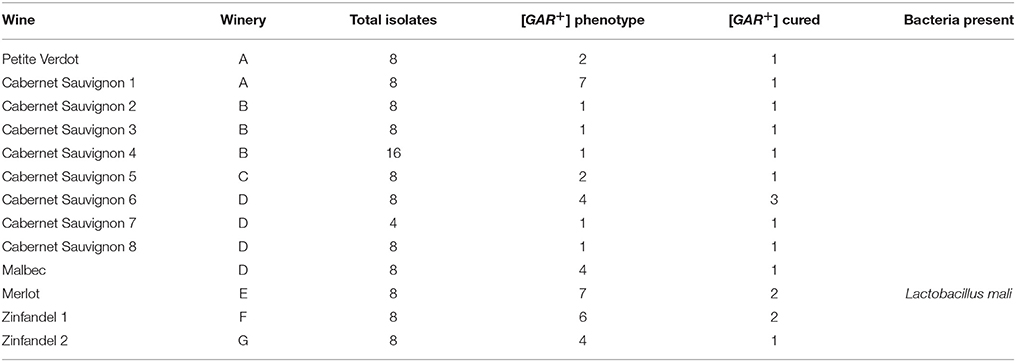
The bacteria listed in Table 1 as isolated from wine were isolated from arrested or protracted wine fermentations and several (20) were found to be capable of inducing the [GAR+] prion as were bacteria isolated from grape must (7 isolates). However, in order to understand the ecological impact of the phenomenon of bacterial induction it is important to determine if it can occur during typical batch fermentation of grape juice. Arrested wine samples were sought for the 2014 vintage and used to isolate both bacteria and yeast. These samples were solicited as simply being arrested fermentations and bacterial inhibition represents only one class of arrest (Bisson, 1999). Our goal was to assess these samples for the presence of inhibitory or inducing bacteria and to isolate viable yeast if possible to assess the presence of the [GAR+] phenotype in these yeasts. Samples of arrested fermentations were received from 40 wineries from which viable yeast were isolated. Not all of the wines had viable yeast and some had been reinoculated with commercial strains which could potentially interfere in the analysis for the prion but all were included in the survey for prion induction. Of the 127 wines received 59 yielded isolatable viable yeast cells on WLN medium and 1–8 colonies were selected from each plate for a total of 400 yeast isolates analyzed. Seventy-one of the 400 isolates from 28 wines showed viable yeast capable of growth on GGM media matching that of known [GAR+] controls (Figure 6). Thus, of the original 127 wines 22% contained yeast populations with evidence of induction of the [GAR+] prion phenotype. Only one of the commercial wine samples containing [GAR+] yeast also contained a viable inducing bacterial species, L. mali suggesting that if induction were solely mediated by bacteria in the natural environment those bacteria may become non-viable or non-culturable later in the fermentation.
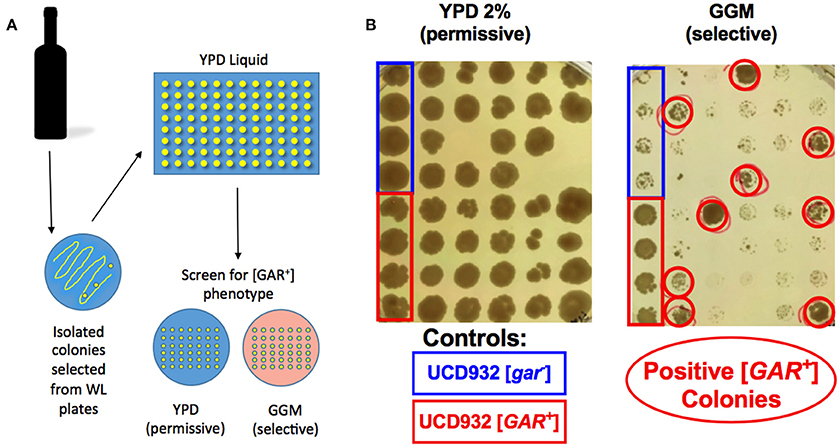
Figure 6. Screen for [GAR+] yeast from stuck fermentations. Isolates of Saccharomyces cerevisiae from stuck fermentations were arrayed in 96 well plates such that four replicates of [gar−] and [GAR+] controls and 40 isolates would be included in each of the two petri plates created from the 96 well plate (A). Replicate plating of each half of the 96 well plate was done onto non-selective or permissive medium (YPD) and selective medium (GGM) to screen for possible [GAR+] phenotype in the stuck fermentation (B). Red circles indicate colonies expressing the [GAR+] phenotype.
The spot plate assay detects growth and it was possible that colonies showing growth on this medium in the replica plating assay may derive from original colonies that are mixtures of [GAR+] and [gar−] cells. Therefore, all the original colonies of the 71 isolates showing good growth on GGM were re-assessed using tip-plating directly comparing the numbers of cells growing on GGM to the permissive yeast medium YPD. Upon quantitative replating 20 of the original populations growing as [GAR+] displayed growth on GGM at a population density greater than 1% with the highest density being 52%. These 20 strains were then tested to confirm that the growth on GGM was due to the induction of [GAR+].
Desiccation of yeast cells has been shown to “cure” the [GAR+] prion state due to loss of the prion conformation and recreation of the wildtype conformational form upon regrowth of the desiccated cells (Tapia and Koshland, 2014). To determine if the 20 isolates showing good growth on GGM were indeed expressing the [GAR+] prion vs. a genomic mutation all 20 isolates were subjected to desiccation for 30 days then media added and cell growth assessed on GGM and control non-selective medium YPD. Seventeen of the 20 strains tested showed loss of the ability to grow on GGM consistent with curing of the prion state with curable isolates identified from each of the 13 wines (Table 2). The remaining three strains showed complete loss of viability following the desiccation process and therefore curing percentage could not be evaluated. However, this analysis showed that the [GAR+] prion can be induced in wines during fermentation demonstrating that prion induction can occur in a native yeast ecosystem. The curable isolates obtained represented 7 of the original 40 wineries.
Induction of the [GAR+] Prion Affects Yeast Fermentation Dominance Factors
To test the hypothesis that prion-induced yeast would be less efficient at depletion of nitrogen from the environment, depletion of amino acid from grape juice was compared in [GAR+] vs. [gar−] derivatives of the same yeast strain, UCD932. The majority of the amino acids were depleted in fermentations with the [gar−] cells within the first 24 h (Table 3). At the 48 h time sample low residual levels of ammonia and gamma-amino butyric acid were present. Higher levels of proline persisted but proline utilization requires the presence of molecular oxygen and these fermentations although mixed were oxygen limited in this strain. In contrast, in the [GAR+] strain depletion lagged significantly behind [gar−] strains with the pattern of consumption at 72 h matching that of the non-induced strain at 48 h (Table 3). This is consistent with the slower start of fermentation in the prion-induced strain in this same Chardonnay juice (Supplemental Figure 1) as the initial lag in onset of fermentation is roughly 24 h longer for the [GAR+] strain. Thus, when grown in pure culture the induction of the prion impacts fermentation initiation but does not affect the ability of the cells to completely ferment available sugar. Interestingly tryptophan levels increased in the medium for both strains suggesting tryptophan synthesized within the cells is released during fermentation and persists in the medium.
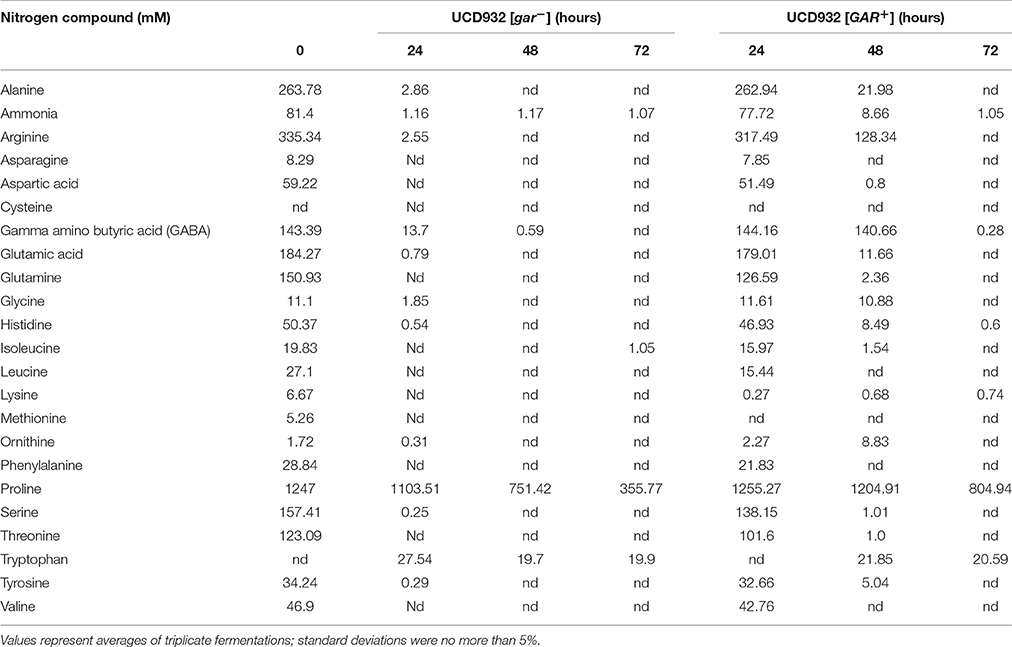
Table 3. Amino acid depletion in [GAR+] and [gar−] cells of UCD932 during Chardonnay juice fermentation.
These studies were conducted using pure cultures of the yeast. However, a decreased rate in nitrogen depletion by the yeast in the presence of bacterial competitors would enhance the ability of the bacteria to proliferate during the early stage of fermentation. We have seen a similar slowdown in early oxygen consumption in [GAR+] yeast strains (Walker, to be reported). Therefore, the benefit to the bacteria from induction of the prion is obvious: they are able to slow yeast metabolism thereby reducing the rate of nutrient and oxygen depletion as well as potentially the production of inhibitory metabolites enabling the bacteria selecting for those bacteria able to proliferate and obtain energy under these oxygen limiting conditions and adapt to the anaerobic conditions of the fermentation. These findings are consistent with analyses of the microbiome of [GAR+] and [gar−] yeast in the presence and absence of the antimicrobial agent sulfur dioxide (Walker et al., 2016).
The Nature of Bacterial Induction
The above studies indicate that bacterial induction is mediated by a diffusible factor produced during the growth of the bacterium in GGM. A clue to the nature of the induction process came from analysis of the cross-feeding hypothesis as the mechanism of growth on GGM. The major acetic acid and lactic acid bacteria metabolites are pyruvate, DL-lactic acid, and acetic acid. To test cross-feeding various concentrations of these acids were spotted onto a lawn of UCD932 yeast on GGM (Figure 7A) at varying concentrations. The spot plates with pyruvate and lactic acid showed no impact on surrounding yeast growth for any of the concentrations evaluated. However, some of the acetic acid spots did show a halo of growth on the plates. This was especially evident for moderate levels of acetic acid. At lower levels no stimulation of growth on GGM occurred. To determine if this was cross feeding or induction, colonies from the growth halo around dots with 200 and 400 μg acetic acid levels were selected, washed, and replated onto GGM (Figure 7B). Yeast around the 200 μg acetic acid dot show evidence of prion induction. Interestingly, the 400 μg sample shows better growth than the no acetic acid treatment control but less than the 200 μg halo.
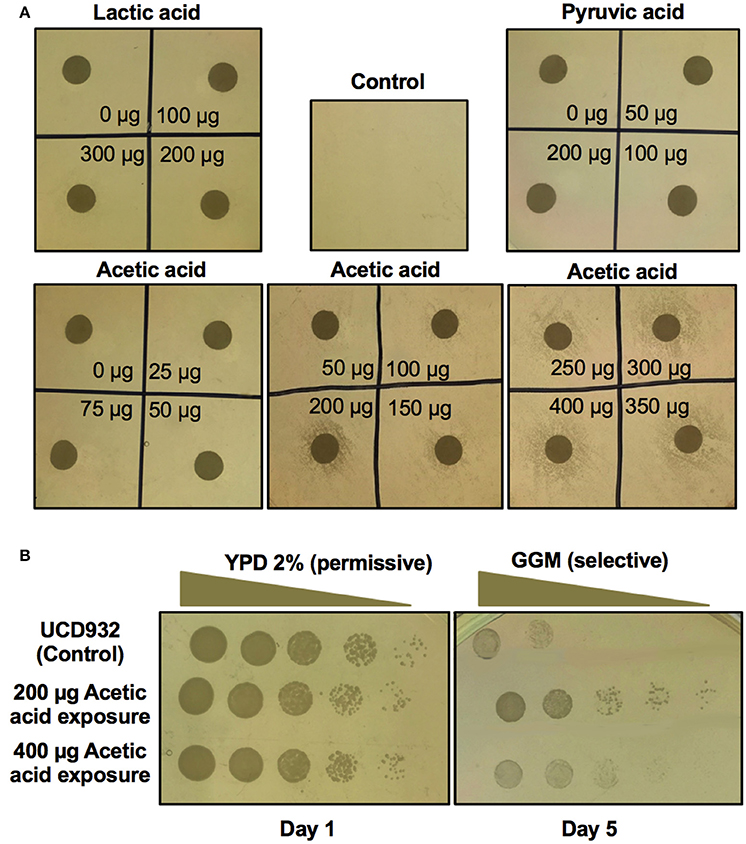
Figure 7. Screen for induction of [GAR+] in Saccharomyces strain UCD932 by bacterial metabolites. Growth of active log phase yeast spread plated on GGM plates with varying levels of lactic acid, pyruvic acid and acetic acid spotted in each quadrant show induction of [GAR+] by acetic acid but no induction by lactic acid or pyruvic acid (A). The colonies from around the inducing concentrations of acetic acid retested on GGM after passage through nonselective YPD media show the most stable induction at 200 μg acetic acid exposure (B). One replicate of quadruplicate experiments is shown.
We next asked if acetic acid alone was sufficient for induction on GGM. To address this question a series of experiments were performed adding acetic acid at varying concentrations to liquid GGM and incubating yeast for 4 h, then washing the cells and plating on GGM (Figure 8). It is evident that even with this treatment at moderate levels of acetic acid growth of the serial dilution culture on GGM extends further than for cells exposed only to GGM during the 4 h incubation period (compare the most dilute sample where roughly 10 colonies are visible for the terminal dilution for the 1 and 2 g/l incubations but not the no acetic acid control). The 3 g/L treatment shows fewer colonies in the final dilution as compared to the 1 and 2 g/L samples. This suggests that induction is reduced at higher acetic acid levels in the environment. There was no difference in growth on YPD, YP glycerol, or YP no carbon source following acetic acid treatment as compared to non-treated cells. A difference in growth was only observable in the presence of the glucose mimetic glucosamine. In order to observe growth on GGM the acetic acid exposure had to occur in the presence of the growth substrate glycerol. Thus, acetic acid appears to be able to induce the prion in the presence of an utilizable alternative growth substrate although less effectively than the induction seen in spent medium.
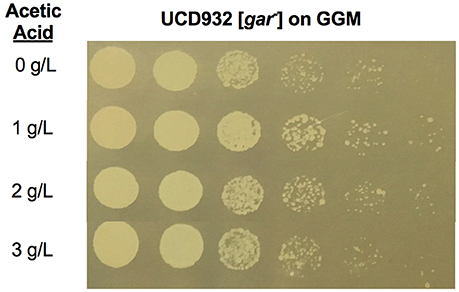
Figure 8. Induction of [GAR+] in Saccharomyces by growth in presence of acetic acid is concentration dependent. Growth of Saccharomyces UCD932 [gar−] strain when exposed to different levels of acetic acid for 4 h in the selective media, GGM then plated on GGM at 1/5 serial dilutions. Prion induction is higher at 1 and 2 g/L as compared to no acetic acid and higher level of acetic acid as evidenced by the presence of colonies in the final dilution.
Two of the strongest inducing bacteria, Lactobacillus kunkeei and A. pasteurianus (strain 175) were grown under a variety of growth conditions and the levels of acetic and lactic acid measured in comparison to a strain of A. pasteurianus that is inhibitory (215) (Table 4). Under all conditions L. kunkeei made higher levels of both acids than either A. pasteurianus strain. L. kunkeei displayed high production of both acids in Chardonnay grape juice. The low acetic acid production in the Acetobacter strains indicates as expected that other energy substrates are being used by these strains under these growth conditions yielding end products other than acetic acid. Oxidation of ethanol is the primary source of acetic acid in the acetic acid bacteria so the observations of low acetic acid production under these inducing conditions is expected. Inoculation with both L. kunkeei and A. pasteurianus 175 yielded similar to slightly higher levels of production for both acids suggesting production is slightly higher than simply additive.
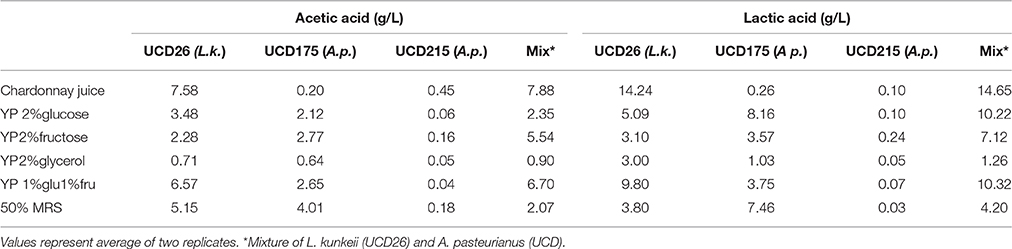
Table 4. Production of acetic and lactic acids by inhibitory Acetobacter pasteurianus (UCD215) and inducing, Lactobacillus kunkeei (UCD26) and Acetobacter pasteurianus (UCD175) and with a mixture of strains 26 and 175 after 48 h of growth in the target medium.
Use of Alternative Substrates by [GAR+] Yeast
The induction of the [GAR+] prion slows yeast metabolism but also enables use of alternative carbon sources in the presence of glucose. If there were no benefit to the yeast of induction of the prion it would seem logical that wine strains would evolve mechanisms to prevent its formation thereby assuring dominance of mixed culture fermentative environments. The retention of the ability to induce the [GAR+] prion in the presence of bacteria suggests the yeast gain some benefit from doing so. Use of an alternative carbon source when continued use of glucose would be toxic is a sound metabolic strategy. Since acetic acid can enter the cell if protonated by simple diffusion its presence could place the cells under cytoplasmic acidification stress and the ability to utilize glucose and fructose fermentatively which also places cells under acidification stress, may be compromised by the presence of acetic acid. In this scenario the reduction in fermentation is to balance the forces leading to cytoplasmic acidification. Spent media and acetic acid induction experiments conducted in control media lacking an alternative substrate did not lead to induction of the prion suggesting that the presence of a usable alternative substrate is important for the establishment of the prion.
Grape juice is an equimolar mixture of glucose and fructose with total sugar levels on the order of ~25% w/v (Boulton et al., 1996). Low concentrations of sucrose, another fermentable sugar, are present but other alternative carbon sources readily utilized by yeast are in short supply. However, both the lactic acid bacteria and acetic acid bacteria can make reduced or oxidized versions of sugars in juice that may represent alternative carbon sources for the yeast. L. kunkeei is a fructophilic lactic acid bacterium able to balance cofactors during metabolism by reducing fructose to mannitol at quite significant levels (Bisson et al., 2016b). Thus, when proliferating in grape juice both acetic acid and mannitol will be present. Similarly the acetic acid bacteria obtain energy from partial oxidation reactions and can oxidize both glucose and fructose again at significant levels providing the yeast with gluconic acid and its further oxidized derivatives as well as oxy-fructose and derivatives (Drysdale and Fleet, 1989; Barbe et al., 2001). An important question was to determine if the induction of the [GAR+] prion enabled utilization of these substrates.
Growth on various substrates of isogenic [GAR+] and [gar−] strains of UCD932 were compared (Figure 9). As expected growth in the presence of glucose (YPD) or in the absence of an added carbon source was identical in the two strains (Figure 9A). Galactose is a substrate showing glucose repression (Carlson, 1987) but use of this substrate is quickly induced on low glucose and the two strain show similar usage of this substrate with the [GAR+] isolate displaying a shorter lag for transit to use of this compound (Figure 9A). The growth of strains was compared in substrates subjected to glucose repression, melibiose, and raffinose (a trisaccharide yielding melibiose) (Neigeborn and Carlson, 1984; Carlson, 1987) and growth on both melibiose and raffinose was better for the [GAR+] isolate. Growth on glycerol, also a non-preferred substrate, was likewise better for the prion-induced strain (Figure 9B). Growth on oxidized (gluconic acid) and reduced (mannitol and sorbitol) substrates of glucose and fructose were also analyzed. In all three cases the [GAR+] strain showed better growth on these substrates (Figure 9C). Induction of the [GAR+] prion therefore enables ready growth on substrates normally subjected to glucose repression including the oxidized and reduced forms of glucose and fructose that can be produced by acetic acid and lactic acid bacteria during growth in grape must.
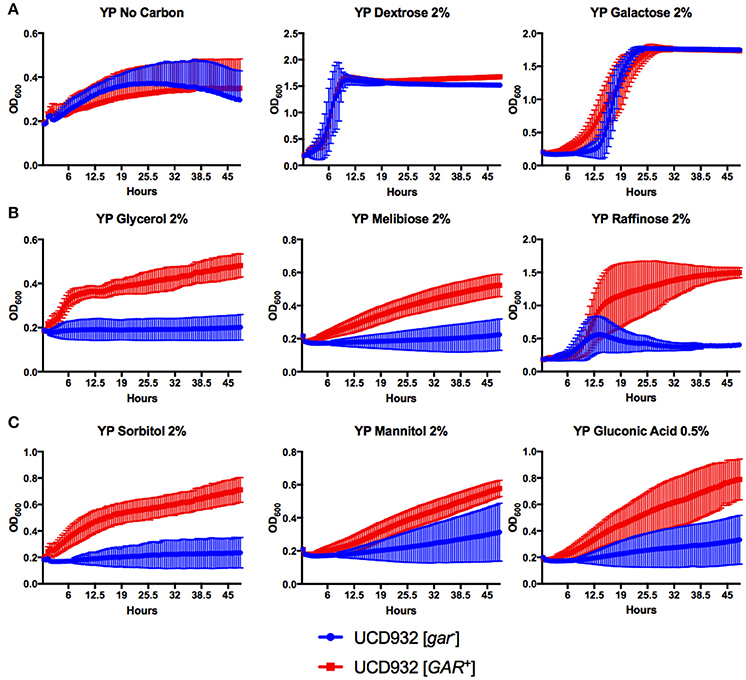
Figure 9. Growth comparisons of UCD932 [GAR+] and UCD932 [gar−] strains on different substrates. No significant differences in the growth of UCD932 [gar−] vs. UCD932 [GAR+] occurred in the absence of added carbon source, or in the presence of glucose or galactose (A). UCD932 [GAR+] displayed a distinct advantage over UCD932 [gar−] when growing on glucose repressible substrates glycerol, melibiose, and raffinose (B). UCD932 [GAR+] displays better growth than UCD932 [gar−] on ecosystem substrates sorbitol, mannitol, and 0.5% gluconic acid (C).
Discussion
The discovery of the bacterial induction of the [GAR+] prion and the accompanying reduction in and modification of metabolic capacity represents a novel form of inter-kingdom communication (Jarosz et al., 2014a). As a consequence of prion induction the bacteria are able to proliferate in the ecosystem and limit the dominance of S. cerevisiae (Walker et al., 2016). Our data presented here confirm the relevance of bacterial induction of the [GAR+] prion in the wine fermentation ecosystem. The ability of yeast to rapidly dominate a fermentative ecosystem is dependent upon the capacity to both deplete the environment of nutrients including molecular oxygen and the production of inhibitory compounds. Cells in which the [GAR+] prion are slower at stripping the environment of amino acids and other nitrogen sources, especially within the first 24–48 h of fermentation when dominance becomes established. Nitrogen uptake is an active proton symport process in yeast requiring ATP and the Pma1 plasma membrane H+ pump to remove accumulating protons from the cytoplasm (Vallejo and Serrano, 1989; Boulton et al., 1996). The ability to deplete amino acids under these conditions is therefore correlated to the ability to rapidly generate the ATP needed for nitrogen uptake. The rapid launch of glycolysis provides the ATP required. In addition uptake of other components is likewise correlated with the strength of the proton gradient across the plasma membrane, a process dependent upon the activity of Pma1 and the generation of energy (Vallejo and Serrano, 1989). The decrease in expression of the Hxt3 glucose transporter which is the main transporter operational during fermentation (Zuchowska et al., 2015) likely slows metabolism sufficiently to alter and the timing of depletion of both nitrogen sources and molecular oxygen leaving these substrates available to other microorganisms present in the ecosystem. Since production of bacterial end products, the acids, also narrows the permissive conditions of the fermentation, the early stage of growth in juice is a race to see which organism can gain the upper hand and create a more favorable environment for their own proliferation and maintenance of a high relative population density.
The induction of the [GAR+] prion allowed significant consumption of sugar substrate with the concomitant production of ethanol and carbon dioxide, when adequate sulfur dioxide was used to inhibit bacterial growth. However, hexoses were not completely depleted (Walker et al., 2016). In contrast in the absence of an antibacterial compound [GAR+] strains were unable to completely ferment available sugar and competing bacteria were evident throughout the fermentation (Walker et al., 2016). It is possible that induction of the [GAR+] prion in and of itself does not necessarily lead to premature arrest of fermentation. Instead we believe that the reduction in competitiveness and ultimate niche dominance stabilizes growth of bacterial communities and it is the end products and metabolites of those communities that are ultimately responsible for cessation of yeast glycolytic activity. The inducing bacteria may or may not be the species ultimately resulting in arrest of yeast fermentation. It is interesting that so many bacterial isolates from problematic wine fermentations can induce the prion state in yeast and it is likely that the reduction in metabolic activity of the yeast enables a broad spectrum of organisms present to adapt to the fermentative conditions. Thus, the benefits to the bacterial community of induction of the yeast [GAR+] prion are obvious.
Less clear are the benefits to the yeast population of induction of the prion state. Under conditions of proton stress reducing the proton output of glycolysis would enable the Pma1 to continue to maintain cytoplasmic pH homeostasis but this would require continued energy production. The metabolic narrowing of the permissive conditions of the fermentation by yeast via the production of ethanol, reduction in redox potential and pH and increase in temperature also narrows the permissive conditions for S. cerevisiae itself. This yeast is more tolerant of these stressors but the narrowing of the niche makes it more sensitive to inhibition by other compounds in the environment, such as organic acids. Acetic acid concentrations that are not toxic in the presence of high sugar become toxic as ethanol accumulates. Induction of the [GAR+] prion by acetic acid suggests that before launching the metabolic powerhouse of glycolysis and the full narrowing of the niche, S. cerevisiae assesses the consequences of doing so and will mitigate the fermentative response if conditions are present that would enhance loss of viability in its narrowed niche.
It is also possible that the yeast benefit from the ability to utilize other energy or carbon sources in the environment under these conditions and the relaxation of glucose repression would aid in maintaining energy levels for Pma1 activity while reducing the proton load from rapid glycolytic metabolism. Conditions are anaerobic which limits metabolism to substrates that can be used fermentatively for energy generation. However, perhaps the yeast may funnel other substrates directly into cell growth dedicating more of the hexoses present for energy production. Substrates normally only catabolized in the presence of molecular oxygen feasibly may be used under these conditions if the reduction or oxidation of other compounds is able to maintain the balance of oxidized and reduced cofactor levels (NAD/NADH; NADP+/NADPH).
That acetic acid can induce the [GAR+] prion poses an attractive model in which the yeast at the onset of growth in fermentative environments assess not only available substrates but also the presence of compounds that may not be immediately toxic but that will place the cells under greater and perhaps lethal stress if typical capacities of glycolysis are launched. It is known that the sensing of external carbon and nitrogen sources is the primary driver of yeast choice of developmental program: growth, quiescence, sporulation, filamentous growth (Zaman et al., 2008; Smets et al., 2010; Wenger et al., 2011), and we now add coexistence to this list of options. This model also explains the differential effect of acetic acid concentrations. At low concentrations coexistence may be feasible and growth and fermentation can proceed in the presence of the inhibitory compound as long as metabolism is tempered by induction of the [GAR+] prion. At higher concentrations the acetic acid may be more toxic to the cells and other adaptations or simple entry into a quiescent phase may be the best course of action for the cells. The presence of acetic acid reduces ethanol tolerance by enhancing cytoplasmic acidification (Pampulha and Loureiro, 1989; Pampulha and Loureiro-Dias, 1989, 1990). However, we propose that the induction of the [GAR+] prion enables Pma1 to reset the pace of glycolysis. Pma1 activity is induced by the presence of glucose (Serrano, 1978). Accumulation of higher than expected proton levels following this induction, as would occur in the presence of diffusion of uncharged organic acids across the plasma membrane, may lead to conformational changes in Pma1 that establish the heritable prion state and decrease glycolytic capacity. That this induction would be heritable makes biological sense as once metabolism is adjusted and pH homeostasis restored a return to activation by external glucose would recreate the potential for cytoplasmic acidification. Further, the reversible nature of prion-based phenotypes also makes this type of regulation of metabolism a sound strategy for the environmental niche. Therefore, the main advantage of the yeast to induction of the prion may be to enable proliferation and survival under conditions made stressful by the metabolic activities of other members of the microbial community. If this model is correct, we predict that compounds other than acetic acid may also induce the prion. Interestingly, in a study comparing wine yeast strain phenotypes to metabolite profiles an association of high acetic acid production in strains of S. cerevisiae (up to 0.9 g/L) was associated with enhanced capacity to survive unfavorable or stressful growth conditions (Franco-Duarte et al., 2016). This level of acetic acid is within the range of induction and, although speculative, this may suggest that some yeast strains evolved to self-induce the prion by production of atypically high levels of acetic acid.
The maximal induction of the [GAR+] prion by 2 g/L acetic acid is also intriguing given what is known about acetic acid inhibition of yeast during grape juice fermentation. In an analysis of impact of added acetic acid to fermentations with subsequent assessment of the ability to restart these sluggish or arrested fermentations, restarts were only successful if the initial dose of acetic acid was 2 g/L or less (Eglinton and Henschke, 1999). Investigation of fermentations inoculated with Gluconobacter oxydans found on average that 1.8 g/L of acetic acid was produced but that the amount of acetic acid varied by the juice (Drysdale and Fleet, 1989). Interestingly this study reported lower concentrations of acetic acid from A. pasteurianus but much stronger inhibition of the fermentation as we have seen with some strains. Phowchinda et al. (1995) obtained a similar result with acetic acid levels of 1–2 g/L impacting yeast growth but only slightly inhibitory compared to higher concentrations. These authors found that a concentration of 10 g/L of acetic acid was needed for complete inhibition of growth. The levels of acetic acid found to be inducing of the prion are similar to the levels made by the inducing bacteria in juice. Although acetic acid seems sufficient to induce the [GAR+] prion on synthetic GGM medium, it is not clear that it is solely responsible for induction in native environments. A broad array of bacteria can induce this state in yeast and not all are acetic acid producers. Also, strain differences within species were noted in ability to induce, which may be a simple consequence of the amount of acetic acid produced, but other factors may also be involved. The impact on proton homeostasis rather than acetic acid per se may be the inducing condition. The establishment of the prion may have as yet unrecognized effects on gene expression. Many interesting questions remain concerning the physiological impacts and benefits to S. cerevisiae of induction of the [GAR+] prion and models other than that proposed here may be equally plausible.
We have shown [GAR+] cells maintain viability longer than their [gar−] counterparts upon depletion of sugar substrates during wine production (Walker et al., 2016). These cells are in a state able to survive the stress of high ethanol and low energy generating reserves, a finding consistent with our model in which the induction of the [GAR+] prion represents an adaptation to the presence and stress of successful competitors in the environment. The enhanced survival of [GAR+] cells may explain the retention of the ability to induce this prion selected for as a means to assure persistence of the genotype within a microbial community is maintained. That the decision to induce a lifestyle of coexistence is mediated by a reversible prion mechanism attests to the evolutionary preference for dominance of permissive environments. Also, during natural induction of the [GAR+] in actual juices the penetrance of the phenotype across the population is at best roughly 50% meaning that at least half or more of the population retains the ability to ferment rapidly under these conditions. Since prion induction is associated with arrest of fermentation these cells likely become metabolically inactivated but may not lose viability perhaps entering a quiescent phase tolerant of ethanol. Indeed during late stationary phases changes in the plasma membrane composition generates a membrane largely impermeable to passive proton and acid species flux but that is also limited in metabolic capacity (Viana et al., 2012). However, should conditions change the fact these cells retain viability may enable this segment of the population to re-launch a selfish fermentation and eliminate competitive microorganisms faster than would be allowed by curing of the prion state.
As attractive as this model is it does not fully explain the ability to utilize alternative carbon sources that accompanies prion establishment. The Hxt3 transporter does not play a vital role in glucose repression other than to assist in glucose uptake and sensing. Deletion of the HXT3 gene has not been reported to lead to glucose repression in any of the mutational studies of this phenomenon. S. cerevisiae utilizable carbon and energy sources that would be alternatives to glucose and fructose are rare in grape juice. This situation changes with respect to the presence and proliferation of the acetic and lactic acid. The acetic acid bacteria are capable of oxidizing glucose and fructose directly in the medium to obtain energy (Švitel and Šturdik, 1995; Barbe et al., 2001), and G. oxydans produced up to 26 g/L of gluconic acid in grape juice and A. pasteurianus up to 3.7 g/L (Drysdale and Fleet, 1989). Gluconic acid is a non-preferred energy source but can be metabolized by S. cerevisiae. [GAR+] strains were able to grow better on this compound than their [gar−] counterparts (Figure 9). The gluconic acid levels produced by G. oxydans and A. pasteurianus in juice decreased 34 and 45% respectively following fermentation of the juice by S. cerevisiae (Drysdale and Fleet, 1989) consistent with potential metabolism of this compound. The lactic acid bacteria use a similar strategy to obtain energy, primarily the reduction of fructose to mannitol in lieu of molecular oxygen (Bisson et al., 2016b). S. cerevisiae can likewise metabolize mannitol but it is a non-preferred carbon and energy source (Maxwell and Spoerl, 1971; Quain and Boulton, 1987) the utilization of which is enhanced with the loss of general transcription factors involved in glucose repression (Chujo et al., 2015). Levels of 10–15 g/L of mannitol may be made by fructophilic lactic acid bacteria such as L. kunkeei (Bisson et al., 2016b). Thus, in the presence of these bacteria alternative, albeit less preferred, carbon and energy sources are available. Use of these compounds under these conditions would be expected to lead to cytoplasmic cofactor imbalance as reduction of oxidized sugar forms and oxidation of reduced forms of these compounds would need to precede metabolism. Although the precise transporters responsible for uptake of these hexose analogs are unknown glucose is a competitive inhibitor of uptake of mannitol (Maxwell and Spoerl, 1971) indicating the HXT transporters are likely involved. It is tempting to speculate that the reduction in Hxt3 levels may in part be done to reduce uptake of these compounds into the cell and thereby minimize cofactor stress. The [GAR+] induced cells were also able to utilize sorbitol more readily than non-induced cells. S. cerevisiae strains can use sorbitol as carbon and energy source but generally only after a long lag period of 2–4 weeks (Sarthy et al., 1994). An alternative explanation for arrest of fermentation may be that as the concentrations of glucose and fructose decrease if mannitol levels remain high mannitol then becomes an effective inhibitor of sugar uptake, perhaps particularly of fructose the transport of which lags behind that of glucose.
Conclusion
The induction of a metabolism-modifying prion by bacterial metabolites represents a new paradigm in regulation of yeast biology. Our data show the relevance of induction of the [GAR+] prion in a native S. cerevisiae ecosystem, grape juice. Induction of the prion hampers early yeast dominance of the fermentation and enables proliferation of bacteria present in the juice. One dominance factor, the rapid depletion of amino acids, is delayed in [GAR+] yeast. The benefits to the bacteria of inducing the prion in yeast competitors is clear. Less clear is the benefit to the yeast but the early presence of high levels of acetic acid may serve as a signal of reduced ethanol tolerance and prion induction therefore assures a segment of the population will persist in the presence of this acid and still be able to proliferate and metabolize available sugar. Many of the details of [GAR+] induction remain to be elucidated. The impact of the prion on plasma membrane functionality and composition likewise merit continued investigation in order to more fully understand the role of [GAR+] induction in yeast physiology.
Author Contributions
VR conducted some of the experiments, prepared figures, participated in discussions of experimental design, and reviewed the final draft. GW conducted some of the experiments, prepared figures, participated in discussions of experimental design, and reviewed the final draft. QF conducted some of the experiments, prepared figures, and participated in discussions of this manuscript. MO, PL, and YL conducted some of the experiments. CJ conducted some of the experiments, participated in discussion of experimental design, prepared tables for the manuscript, supervised MO, PL and YL, and reviewed the final draft. LB lead discussions of the research and design of experiments, wrote the drafts and final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We dedicate this paper to the memory of our colleague Dr. Susan Lindquist, an accomplished and pioneering researcher, valued collaborator, mentor and friend, resilient role model, tireless champion for equity in STEM. This research was supported by a grant from the American Vineyard Foundation. Graham C. Walker and Dan Jarosz are thanked for useful discussions. Nicholas Hung and Haruka Uehara are thanked for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fevo.2016.00137/full#supplementary-material
Supplementary Figure 1. Fermentation performance of Saccharomyces cerevisiae UCD932 [gar −] and [GAR+] strains in filtered Chardonnay juice. Weight loss due to evolved carbon dioxide is represented by blue circles for [gar−] and red squares for [GAR+]. All fermentations were performed at 28°C in triplicate and the fermentation curves represent the average of replicate fermentations. Note error bars are within the markers and not visible in the figure. Pre-inocula of UCD932 without and with the [GAR+] prion were grown to stationary phase (48 h) in 10 mL sterile filtered Chardonnay juice on a roller drum at room temperature (~25°C). Fermentations were carried out in triplicates in filtered Chardonnay juice with starting inoculum of approximately 3 × 105 cells. Flasks were incubated with agitation at 120 rpm at 28°C. Fermentation progress was monitored by measuring weight loss due to carbon dioxide release. The end of fermentation was assessed by Anton Paar instrument and the final residual sugar was measured by Clinitest (Bayer Catalog # AM-2126). This experiment is a replica of that published in Walker et al. (2016) using a Chardonnay juice from a different vintage to show that the impact of the [GAR+] prion occurs across vintages.
References
Albergaria, H., and Arneborg, N. (2016). Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 100, 2035–2046. doi: 10.1007/s00253-015-7255-0
Albergaria, H., Francisco, D., Gori, K., Arneborg, N., and Gírio, F. (2010). Saccharomyces cerevisiae CCMI secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 86, 965–972. doi: 10.1007/s00253-009-2409-6
Almeida, P., Barbosa, R., Zalar, P., Imanishi, Y., Shimiza, K., Turchetti, B., et al. (2015). A population genomics insight into the Mediterranean origins of wine yeast domestication. Mol. Ecol. 24, 5412–5427. doi: 10.1111/mec.13341
Ball, A. J. S., Wong, D. K., and Elliott, J. J. (1976). Glucosamine resistance in yeast. I. A preliminary genetic analysis. Genetics 84, 311–317.
Barbe, J.-C., de Revel, G., Joyeax, A., Bertrand, A., and Lonvaud-Funel, A. (2001). Role of botrytized grape microorganisms in SO2 binding phenomena. J. Appl. Microbiol. 40, 34–42. doi: 10.1046/j.1365-2672.2001.01200.x
Bisson, L. F. (2012). Geographic origin and diversity of wine strains of Saccharomyces. Am. J. Enol. Vitic. 63, 165–176. doi: 10.5344/ajev.2012.11083
Bisson, L. F., Fan, Q., and Walker, G. A. (2016a). Sugar and glycerol transport in Saccharomyces cerevisiae. Adv. Exp. Med. Biol. 892, 125–168. doi: 10.1007/978-3-319-25304-6_6
Bisson, L. F., and Walker, G. A. (2015). “The microbial dynamics of wine fermentation,” in Advances in Fermented Foods and Beverages Improving Quality, Technology and Health Benefits, ed W. Holzapfel (Cambridge, UK: Woodhead Publishing), 434–476.
Bisson, L. F., Walker, G., Ramakrishnan, V., Luo, Y., Fan, Q., Wiemer, E., et al. (2016b). The two faces of Lactobacillus kunkeei: Wine spoilage agent and bee probiotic. Catalyst. doi: 10.5344/catalyst.2016.16002. [Epub ahead of print].
Boles, E., and Hollenberg, C. P. (1997). The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev. 21, 85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x
Boulton, R. B., Singleton, V. L., Bisson, L. F., and Kunkee, R. E. (1996). Principles and Practices of Winemaking. New York, NY: Chapman & Hall.
Branco, P., Francisco, D., Chambon, C., Hébraud, M., Almeida, M. G., Caldeira, J, et al. (2014). Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl. Microbiol. Biotechnol. 98, 843–853. doi: 10.1007/s00253-013-5411-y
Brown, J. C., and Lindquist, S. (2009). A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 23, 2320–2332. doi: 10.1101/gad.1839109
Butzke, C. E. (1998). Survey of yeast assimilable nitrogen status in must from California, Oregon and Washington. Am. J. Enol. Vitic. 49, 220–224.
Carlson, M. (1987). Regulation of sugar utilization in Saccharomyces cerevisiae. J. Bacteriol. 169, 4873–4877. doi: 10.1128/jb.169.11.4873-4877.1987
Chujo, M., Yoshida, S., Ota, A., Murata, K., and Kawai, S. (2015). Acquisition of the ability to assimilate mannitol by Saccharomyces cerevisiae through dysfunction of the general corepressor Tup1-Cyc8. Appl. Environ. Microbiol. 81, 8–16. doi: 10.1128/AEM.02906-14
Comitini, F., Ferretti, R., Clementi, F., Mannuzzu, I., and Ciani, M. (2005). Interactions between Saccharomyces cerevisiae and malolactic bacteria: preliminary characterization of a yeast proteinaceous compound(s) active against Oenococcus oeni. J. Appl. Microbiol. 99, 105–111. doi: 10.1111/j.1365-2672.2005.02579.x
Cray, J. A., Bell, A. N., Bhaganna, P., Mswaka, A. Y., Timson, D. J., and Hallsworth, J. E. (2013). The biology of habitat dominance; can microbes behave as weeds? Microb. Biotechnol. 6, 453–492. doi: 10.1111/1751-7915.12027
Deppenmeier, U., and Ehrenreich, A. (2009). Physiology of acetic acid bacteria in light of the genome sequence of Gluconobacter oxydans. J. Mol. Microbiol. Biotechnol. 16, 69–80. doi: 10.1159/000142895
Drysdale, G. S., and Fleet, G. H. (1989). The effect of acetic acid bacteria upon the growth and metabolism of yeasts during fermentation of grape juice. J. Appl. Bacteriol. 67, 471–481. doi: 10.1111/j.1365-2672.1989.tb02518.x
Eberlein, C., Leducq, J.-B., and Landry, C. R. (2015). The genomics of wild yeast populations sheds light on the domestication of man's best (micro) friend. Mol. Ecol. 24, 5309–5311. doi: 10.1111/mec.13380
Eglinton, J. M., and Henschke, P. A. (1999). Restarting incomplete fermentations: the effect of high concentrations of acetic acid. Aus. J. Grape Wine Res. 5, 71–78. doi: 10.1111/j.1755-0238.1999.tb00155.x
Franco-Duarte, R., Umek, L., Mendes, I., Castro, C. C., Fonseca, N., Martins, R., et al. (2016). New integrative computational approaches unveil the Saccharomyces cerevisiae pheno-metabolomic fermentative profile and allow strain selection for winemaking. Food Chem. 211, 509–520. doi: 10.1016/j.foodchem.2016.05.080
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacteria 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Garcia, D. M., and Jarosz, D. F. (2014). Rebels with a cause: molecular features and physiological consequences of yeast prions. FEMS Yeast Res. 14, 136–147. doi: 10.1111/1567-1364.12116
Gutiérrez, A., Sancho, M., Beltran, G., Guillamon, J. M., and Warringer, J. (2016). Replenishment and mobilization of intracellular nitrogen pools decouples wine yeast nitrogen uptake from growth. Appl. Microbiol. Biotechnol. 100, 3255–3265. doi: 10.1007/s00253-015-7273-y
Hagen, K. M., Keller, M., and Edwards, C. G. (2008). Survey of biotin, pantothenic acid and assimilable nitrogen in winegrapes from the Pacific Northwest. Am. J. Enol. Vitic. 59, 432–436.
Hagman, A., and Piškur, J. (2015). A study on the fundamental mechanism and evolutionary driving forces behind aerobic fermentation in yeast. PLoS ONE 10:e0116942. doi: 10.1371/journal.pone.0116942
Hagman, A., Säll, T., Compagno, C., and Piskur, J. (2013). Yeast “make-accumulate-consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS ONE 8:e68734. doi: 10.1371/journal.pone.0068734
Halfmann, R., Alberti, S., and Lindquist, S. (2010). Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 20, 125–133. doi: 10.1016/j.tcb.2009.12.003
Halfmann, R., and Lindquist, S. (2010). Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science 330, 629–632. doi: 10.1126/science.1191081
Halfmann, R., Jarosz, D. F., Jones, S. K., Chang, A., Lancaster, A. K., and Lindquist, S. (2012). Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482, 363–368. doi: 10.1038/nature10875
Holm Hansen, E., Nissen, P., Sommer, P., Nielsen, J. C., and Arneborg, N. (2001). The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J. Appl. Microbiol. 91, 541–547. doi: 10.1046/j.1365-2672.2001.01426.x
Hinze, H., and Holzer, H. (1985). Accumulation of nitrite and sulfite in yeast cells and synergistic depletion of the intracellular ATP content. Z. Lebensm. Unters. Forch. 180, 117–120. doi: 10.1007/BF01042634
Ingledew, W. M., and Kunkee, R. E. (1985). Factors influencing sluggish fermentations of grape juice. Am. J. Enol. Vitic. 36, 65–76.
Jarosz, D. F., Brown, J. C., Walker, G. A., Datta, M. S., Ung, W. L., Lancaster, A. K., et al. (2014a). Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell 158:1083–1093. doi: 10.1016/j.cell.2014.07.025
Jarosz, D. F., Lancaster, A. K., Brown, J. C., and Lindquist, S. (2014b). An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell 158, 1072–1082. doi: 10.1016/j.cell.2014.07.024
Karpel, J. E., Place, W. R., and Bisson, L. F. (2008). Analysis of the major hexose transporter genes in wine strains of Saccharomyces cerevisiae. Am. J. Enol. Vitic. 59, 265–275.
Kayikci, O., and Nielsen, J. (2015). Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 15:fov068. doi: 10.1093/femsyr/fov068
Kotyk, A., and Knotkova, A. (1989). Uptake of D-glucosamine by Saccharomyces cerevisiae. Folia Microbiol. 34, 1–6. doi: 10.1007/BF02821317
Kruckeberg, A. L. (1996). The hexose transporter family of Saccharomyces cerevisiae. Arch. Microbiol. 166, 283–292. doi: 10.1007/s002030050385
Lafon-Lafourcade, S., Geneix, C., and Ribéreau-Gayon, P. (1984). Inhibition of alcoholic fermentation of grape must by fatty acids produced by yeast and their elimination by yeast ghosts. Appl. Environ. Microbiol. 97, 1246–1249.
Legras, J.-L., Merdinoglu, D., Cornuet, J.-M., and Karst, F. (2007). Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 16, 2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x
Liebman, S. W., and Chernoff, Y. O. (2012). Prions in yeast. Genetics 191, 1041–1072. doi: 10.1534/genetics.111.137760
Liti, G., Carter, D. M., Moses, A. M., Warringer, J., Parts, L., James, S. A., et al. (2009). Population genomics of domestic and wild yeasts. Nature 458, 337–341. doi: 10.1038/nature07743
Luyten, K., Riou, C., and Blondin, B. (2002). The hexose transporters of Saccharomyces cerevisiae play different roles during enological fermentation. Yeast 19, 713–726. doi: 10.1002/yea.869
MacLean, R. C., and Gudelj, I. (2006). Resource competition and social conflict in experimental conflict in experimental populations of yeast. Nature 441, 498–501. doi: 10.1038/nature04624
Mason, A. B., Allen, K. E., and Slaymann, C. W. (2014). C-terminal truncations of the Saccharomyces cerevisiae H+ -ATPase have major impacts on protein concentration, trafficking, quality control and function. Euk. Cell 13, 43–52. doi: 10.1128/EC.00201-13
Matsushita, K., Inoue, T., Adachi, O., and Toyama, H. (2005). Acetobacter aceti possesses a proton-motive-force-dependent efflux system for acetic acid. J. Bacteriol. 187, 4346–4352. doi: 10.1128/JB.187.13.4346-4352.2005
Maxwell, W. A., and Spoerl, E. (1971). Mannitol uptake by Saccharomyces cerevisiae. J. Bacteriol. 105, 753–758.
Monteiro, F. F., and Bisson, L. F. (1991). Amino acid utilization and urea formation during vinification fermentations. Am. J. Enol. Vitic. 42, 199–208.
Mortimer, R. K., Romano, P., Suzzi, G., and Posinelli, M. (1994). Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast 10, 1543–1552. doi: 10.1002/yea.320101203
Nehme, M., Mathieu, F., and Taillandier, P. (2010). Impact of the co-culture of Saccharomyces cerevisiae-Oenococcus oeni on malolactic fermentation and partial characterization of a yeast-derived inhibitory peptidic fraction. Food Microbiol. 27, 150–157. doi: 10.1016/j.fm.2009.09.008
Neigeborn, L., and Carlson, M. (1984). Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108, 845–858.
Newby, G. A., and Lindquist, S. (2013). Blessings in disguise: biological benefits of prion-like mechanisms. Trends Cell Biol. 23, 251–259. doi: 10.1016/j.tcb.2013.01.007
Osbourne, J. P., and Edwards, C. G. (2007). Inhibition of malolactic fermentation by a peptide produced by Saccharomyces cerevisiae during alcoholic fermentation. Int. J. Food Microbiol. 118, 27–34. doi: 10.1016/j.ijfoodmicro.2007.05.007
Pallmann, C. L., Brown, J. A., Olineka, T. L., Cocolin, L., Mills, D. A., and Bisson, L. F. (2001). Use of WL medium to profile native flora fermentations. Am. J. Enol. Vitic. 52, 198–203.
Pampulha, M. E., and Loureiro, V. (1989). Interaction of the effects of acetic acid and ethanol on inhibition of fermentation in Saccharomyces cerevisiae. Biotechnol. Lett. 4, 269–274. doi: 10.1007/BF01031576
Pampulha, M. E., and Loureiro-Dias, M. C. (1989). Combined effect of acetic acid, pH and ethanol on intracellular pH of fermenting yeast. Appl. Microbiol. Biotechnol. 31, 547–550. doi: 10.1007/BF00270792
Pampulha, M. E., and Loureiro-Dias, M. C. (1990). Activity of glycolytic enzymes of Saccharomyces cerevisiae in presence of acetic acid. Appl. Microbiol. Biotechnol. 34, 375–380. doi: 10.1007/BF00170063
Perez, M., Luyten, K., Michel, R., Riou, C., and Blondin, B. (2005). Analysis of Saccharomyces cerevisiae hexose carrier expression during wine fermentation: both low- and high-affinity Hxt transporters are expressed. FEMS Yeast Res. 5, 351–361. doi: 10.1016/j.femsyr.2004.09.005
Pérez-Nevado, F., Albergaria, H., Hogg, T., and Girio, F. (2006). Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 108, 336–345. doi: 10.1016/j.ijfoodmicro.2005.12.012
Pfeiffer, T., Schuster, S., and Bonhoeffer, S. (2001). Cooperation and completion in the evolution of ATP-producing pathways. Science 292, 504–507. doi: 10.1126/science.1058079
Phowchinda, O., and Délia-Dupuy, M. L, Strehaiano, P. (1995). Effects of acetic acid on growth and fermentative activity of Saccharomyces cerevisiae. Biotechnol. Lett. 17, 237–242. doi: 10.1007/BF00127996
Pinu, F. R., Edwards, P. J., Gardner, R. C., and Villas-Boas, S. G. (2014). Nitrogen and carbon assimilation by Saccharomyces cerevisiae during Sauvignon blanc juice fermentation. FEMS Yeast Res. 14, 1206–1222. doi: 10.1111/1567-1364.12222
Quain, D. E., and Boulton, C. A. (1987). Growth and metabolism of mannitol by strains of Saccharomyces cerevisiae. J. Gen. Microbiol. 133, 1675–1684. doi: 10.1099/00221287-133-7-1675
Sarthy, A. V., Schopp, C., and Idler, K. B. (1994). Cloning and sequence determination of the gene encoding sorbitol dehydrogenase from Saccharomyces cerevisiae. Gene 140, 121–126. doi: 10.1016/0378-1119(94)90741-2
Serrano, R. (1978). Characterization of the plasma membrane ATPase of Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 51–63. doi: 10.1007/bf00241470
Shorter, J., and Lindquist, S. (2005). Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6, 435–450. doi: 10.1038/nrg1616
Sicard, D., and Legras, J.-L. (2011). Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. Comptes Rendus Biol. 304, 229–236. doi: 10.1016/j.crvi.2010.12.016
Smets, B., Ghillebert, R., De Snijder, P., Binda, M., Swinnen, E., De Virgilio, C., et al. (2010). Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae cells. Curr. Genet. 56, 1–32. doi: 10.1007/s00294-009-0287-1
Sogawa, K., Watanabe, M., Sato, K., Segawa, S., Ishii, C., Miyabe, A., et al. (2011). Use of MALDI BioTyper system with MALDI-TOF mass spectrometry for rapid identification of microorganisms. Anal. Bioanal. Chem. 400, 1905–1911. doi: 10.1007/s00216-011-4877-7
Spiropoulos, A., Tanaka, J., Flerianos, I., and Bisson, L. F. (2000). Characterization of hydrogen sulfide formation in commercial and natural wine isolates of Saccharomyces. Am. J. Enol. Vitic. 51, 233–248.
Švitel, J., and Šturdik, E. (1995). 2-ketogluconic acid production by Acetobacter pasteurianus. Appl. Biochem. Biotechnol. 53, 53–63. doi: 10.1007/BF02783481
Tapia, H., and Koshland, D. E. (2014). Trehalose is a versatile and long-lived chaperone for desiccation tolerance. Curr. Biol. 24, 1–9. doi: 10.1016/j.cub.2014.10.005
Vallejo, C. G., and Serrano, R. (1989). Physiology of mutants with reduced expression of plasma membrane H+-ATPase. Yeast 5, 307–319. doi: 10.1002/yea.320050411
Viana, T., Loureiro-Dias, M. C., Loureiro, V., and Prista, C. (2012). Peculiar H+ homeostasis of Saccharomyces cerevisiae during the late stages of wine fermentation. Appl. Environ. Microbiol. 78, 6302–6308. doi: 10.1128/AEM.01355-12
Visser, W., Scheffers, W. A., Battenburg-van de Vegte, W. H., and van Dijken, J. P. (1990). Oxygen requirements of yeasts. Appl. Environ. Microbiol. 56, 3785–3792.
Walker, G. A., Hjelmeland, A., Bokulich, N. A., Mills, D. A., Ebeler, S. E., and Bisson, L. F. (2016). Impact of the [GAR+] prion on fermentation and bacterial community composition with Saccharomyces cerevisiae UCD932. Am. J. Enol. Vitic. 67, 296–307. doi: 10.5344/ajev.2016.15092
Wang, C., Mas, A., and Esteve-Zarzoso, B. (2016). The interaction between Saccharomyces cerevisiae and non-Saccharomyces yeast during alcoholic fermentation is species and strain specific. Front. Microbiol. 7:502. doi: 10.3389/fmicb.2016.00502
Wenger, J. W., Piotrowski, J., Nagarian, S., Chiotti, K., Sherlock, G., and Rosenzweig, F. (2011). Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 7:e1002202. doi: 10.1371/journal.pgen.1002202
Wickner, R. B., Shewmaker, F. P., Bateman, D. A., Edskes, H. K., Gorkovskiy, A., Dayani, Y., et al. (2015). Yeast prions: structure, biology, and prion-handling systems. Microbiol. Mol. Biol. Rev. 79, 1–17. doi: 10.1128/MMBR.00041-14
Wieczorke, R., Krampe, S., Weierstall, T., Freidel, K., Hollenberg, C., and Boles, E. (1999). Knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Letts. 464, 123–128. doi: 10.1016/S0014-5793(99)01698-1
Williams, K. M., Liu, P., and Fay, J. C. (2015). Evolution of ecological dominance in yeast species in high-sugar environments. Evolution 69, 2079–2093. doi: 10.1111/evo.12707
Zaman, S., Lippman, S. I., Zhao, X., and Broach, J. R. (2008). How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42, 27–81. doi: 10.1146/annurev.genet.41.110306.130206
Keywords: Saccharomyces, [GAR+], prion, bacterial induction
Citation: Ramakrishnan V, Walker GA, Fan Q, Ogawa M, Luo Y, Luong P, Joseph CML and Bisson LF (2016) Inter-Kingdom Modification of Metabolic Behavior: [GAR+] Prion Induction in Saccharomyces cerevisiae Mediated by Wine Ecosystem Bacteria. Front. Ecol. Evol. 4:137. doi: 10.3389/fevo.2016.00137
Received: 29 August 2016; Accepted: 10 November 2016;
Published: 24 November 2016.
Edited by:
Paul G. Becher, Swedish University of Agricultural Sciences, SwedenReviewed by:
Arne Hagman, VU Brussel and KU Leuven, BelgiumAline Lonvaud, University of Bordeaux, France
Copyright © 2016 Ramakrishnan, Walker, Fan, Ogawa, Luo, Luong, Joseph and Bisson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda F. Bisson, lfbisson@ucdavis.edu
 Vidhya Ramakrishnan
Vidhya Ramakrishnan Gordon A. Walker
Gordon A. Walker  Qingwen Fan
Qingwen Fan Minami Ogawa
Minami Ogawa Peter Luong
Peter Luong C. M. Lucy Joseph
C. M. Lucy Joseph Linda F. Bisson
Linda F. Bisson