- 1 Discipline of Nutrition, Faculty of Medical and Health Sciences, The University of Auckland, Auckland, New Zealand
- 2 Auckland Cancer Society Research Centre, Faculty of Medical and Health Sciences, The University of Auckland, Auckland, New Zealand
- 3 Nutrigenomics New Zealand, Auckland, New Zealand
Selenium (Se) is an important micronutrient that, as a component of selenoproteins, influences oxidative and inflammatory processes. Its’ levels vary considerably, with different ethnic and geographic population groups showing varied conditions, ranging from frank Se deficiencies to toxic effects. An optimum Se level is essential for the maintenance of homeostasis, and this optimum may vary according to life stage, general state of health, and genotype. Nutrigenetic studies of different Se levels, in the presence of genetic variants in selenoproteins, suggest that an effective dietary Se intake for one individual may be very different from that for others. However, we are just starting to learn the significance of various genes in selenoprotein pathways, functional variants in these, and how to combine such data from genes into pathways, alongside dietary intake or serum levels of Se. Advances in systems biology, genetics, and genomics technologies, including genetic/genomic, epigenetic/epigenomic, transcriptomic, proteomic, and metabolomic information, start to make it feasible to assess a comprehensive spectrum of the biological activity of Se. Such nutrigenomic approaches may prove very sensitive biomarkers of optimal Se status at the individual or population level. The premature cessation of a major human Se intervention trial has led to considerable controversy as to the value of Se supplementation at the population level. New websites provide convenient links to current information on methodologies available for nutrigenetics and nutrigenomics. These new technologies will increasingly become an essential tool in optimizing the level of Se and other micronutrients for optimal health, in individuals and in population groups. However, definitive proof of such effects will require very large collaborative studies, international agreement on study design, and innovative approaches to data analysis.
Introduction
Selenium (Se) is an essential human micronutrient, whose status varies significantly across different populations and different ethnic groups (Cook et al., 2005; Kant and Graubard, 2007; Bleys et al., 2008; Johnson et al., 2010). Se enters the food chain through plants, and the amount and bioavailability of Se in the soil typically reflects the plant level. Se is usually provided by the diet in humans, but may also be derived from drinking water, environmental pollution, and supplementation (Robinson, 1989; Darlow et al., 1995; Navarro-Alarcon and Cabrera-Vique, 2008; Schrauzer and Surai, 2009). It is generally considered to be low in New Zealand soils, and is almost certainly at sub-optimal levels in a typical New Zealand diet (Logan, 1988, 1991; Robinson, 1989; Thomson and Robinson, 1990, 1996; Levander, 1991; Dolamore et al., 1992; Darlow et al., 1995; Duffield et al., 1999; De Jong et al., 2001; Karunasinghe et al., 2004; Thomson, 2004a,b; Thomson et al., 2007). In contrast, the diets of the majority of Americans (Bleys et al., 2008), and of many but not all Europeans (Hesketh et al., 2006; Hoeflich et al., 2010; Steinbrecher et al., 2010), are considered to be nutritionally adequate for Se. While parts of China may be deficient and need innovative methods of increasing status (Yang et al., 2007; Gao et al., 2009; Shi et al., 2010), indoor burning of coal may also lead to Se toxicity in other Chinese populations (Guijian et al., 2007; Xue et al., 2010).
As with other micronutrients, sub-optimal intakes of Se may contribute to the development and severity of chronic diseases (Zeisel, 2010). Equally, too high a level of Se is associated with toxicity (Reid et al., 2004; Navarro-Alarcon and Cabrera-Vique, 2008; Gore et al., 2010; Hon et al., 2010; Rusyniak et al., 2010). The definition of Se nutritional adequacy varies across populations, and has changed with time (Levander, 1991; Thomson, 2004a; Bleys et al., 2008; Fairweather-Tait et al., 2010). This is a more general part of a continuous process, of re-assessing dietary requirements and safe upper limits of micronutrients. Early definitions related optimal Se status to the levels necessary to saturate the activity of the glutathione peroxidase 1 (GPx-1) enzyme (Thomson, 2004a; Ashrafi et al., 2007b; Lei et al., 2007; Papp et al., 2007; Steinbrenner and Sies, 2009; Flohe et al., 2010). However, the recognition of a wide range of selenoproteins, many of which may play a more fundamental or equally important role in human health as compared with GPx-1, has significantly changed thinking (Castellano et al., 2008; Allmang et al., 2009; Arbogast and Ferreiro, 2009; Kipp et al., 2009; Lescure et al., 2009; Rayman, 2009; Arner, 2010; Hawkes and Alkan, 2010; Zhang et al., 2010a,b). The importance is being recognized, not only of considering the overt health effects of extreme micronutrient deficiency or excess, but also more subtle effects occurring at the genetic, genomic, or epigenomic levels (Davis and Milner, 2006; Van Ommen et al., 2008; van Ommen et al., 2010a,b; Zeisel, 2010).
There are more than 30 genes that affect Se uptake, metabolism, and excretion (Mithen, 2007; Allmang et al., 2009; Schomburg and Schweizer, 2009). The complexity of selenoprotein pathways and ways in which the genes interact with one another, is now well documented (Castellano et al., 2008; Allmang et al., 2009; Rayman, 2009; Schomburg and Schweizer, 2009; Figure 1). With recent advances in genetics, genomics, and systems biology technologies, it is becoming feasible to assess the biological action of a micronutrient in multiple metabolic pathways, including interactions with other nutrients, in relation to genotype (Bermano et al., 2007; Joost et al., 2007; Méplan et al., 2007; 2009; Cooper et al., 2008; Williams et al., 2008; Méplan, 2011; Penney et al., 2010; van Ommen et al., 2010b). Systems biology approaches to determining optimal levels of micronutrients such as Se may complement public health approaches (Rayman, 2008; Platz, 2010). Two important developments may, in the future, allow us to optimize Se intake to individual human requirements (Van Ommen, 2007). We are now able, at a relatively low cost, to estimate selenoprotein genotype in relation to uptake and metabolism (Joost et al., 2007; Méplan et al., 2007; Williams et al., 2008; Rayman, 2009). Additionally, we are able to understand the consequences of human health status and genetic variation for expression of key Se-related genes (Hesketh and Villette, 2002; Hesketh et al., 2006; Hesketh, 2008). The new molecular technologies pave the way for personalized nutrition and optimal status of micronutrients such as Se. These approaches are embodied in the relatively new fields of nutrigenetics and nutrigenomics. This review will consider the implications of both of these fields for optimizing the status of Se, and other micronutrients.
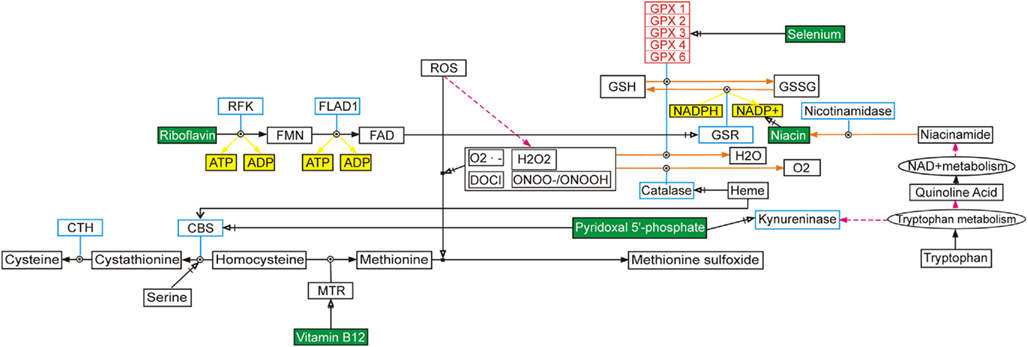
Figure 1. Illustration of the complexity of gene-gene interactions, using selenium and selenoproteins as an example. This selenium pathway can be found at http://www.wikipathways.org/index.php/Pathway:WP15 and the figure has been redrawn from van Ommen et al. (2010a).
Functions of Selenoproteins in Humans
Se is present in several selenoproteins, as the amino acid selenocysteine (Se-Cys; Castellano et al., 2008; Allmang et al., 2009; Arbogast and Ferreiro, 2009; Kipp et al., 2009; Lescure et al., 2009). Functions of a number of the selenoproteins are now known, and imply a role for sub-optimal Se status in a number of human diseases including cancer and epilepsy, and sub-optimal physiological functions, including thyroid and muscle function, and spermatogenesis (Ashrafi et al., 2007a; Flohe, 2007; Agamy et al., 2010; Duntas, 2010).
Approximately half of the characterized selenoproteins have been shown to protect the cell against the action of reactive oxygen species (ROS; Hawkes and Alkan, 2010; Higuchi et al., 2010). GPx enzymes are necessary to regulate intracellular concentration of hydroperoxides, and thought to play a role in antioxidant defense mechanisms (Baliga et al., 2007; Bartel et al., 2007; Bermano et al., 2007; Lei et al., 2007; Savaskan et al., 2007; Méplan et al., 2008). Thioredoxin reductase (TR), together with thioredoxin, forms a redox system with multiple roles, including redox regulation of transcription factors and provision of reducing equivalents for the synthesis of deoxyribonucleotides for DNA synthesis (Ferguson et al., 2006; Papp et al., 2007). TR also carries the function of reducing ubiquinone-10 to regenerate the antioxidant ubiquinol-10 (Nordman et al., 2003; Xia et al., 2003; Madeja et al., 2005). This is an important reaction in preventing peroxidation of lipids, a complex process whereby polyunsaturated fatty acids undergo oxidation to yield lipid hydroperoxides. During the peroxidative pathway, several end-products are formed, including malondialdehyde, pentane, isoprostanes, and cholesterol oxides among others (Nordman et al., 2003). Both the 15-kDa selenoprotein, Se-15 and SePP appear to have an antioxidant function (Méplan et al., 2007, 2009; Higuchi et al., 2010; Penney et al., 2010). SePP contains most of the Se in plasma and appears to play a key role in regulating transport of Se into other tissues (Méplan et al., 2009). Selenophosphate synthetase 2 (SPS2) also plays a key Se donor role (Xu et al., 2007a,b; Schoenmakers et al., 2010).
These antioxidant effects and other actions mean that one of the key activities of many selenoproteins will be to reduce DNA damage, either directly through interacting with free radicals, or more indirectly, through enhanced cellular DNA repair capacity (Baliga et al., 2007). Both these events can be measured in DNA from white blood cells using the single cell gel electrophoresis (COMET) assay. At least in a canine model, Waters et al. (2003, 2005) used this approach to show a “U” shaped curve for Se requirements to protect against DNA damage, and others have recognized the importance of this concept (Chiang et al., 2009). We have shown that an increasing serum Se level up to 100 ng/ml in serum decreases DNA damage, and enhances the activity of DNA repair-associated enzymes such as GPx-1 and TR (Karunasinghe et al., 2004; Ferguson et al., 2006). However, because our Auckland, New Zealand population has generally low Se levels, we could not define an optimal New Zealand level from these data.
Chronic inflammation has been shown to be causal in many of the prevalent disease of the Western world, including cancer, heart disease, and diabetes (Sakr et al., 2007; Zhang et al., 2010a,b). Effects on inflammatory response are among the other key activities identified for selenoproteins. Selenoprotein S (Se-S) appears to be involved in the regulation of the pro-inflammatory response (Curran et al., 2005), which can be monitored by measuring levels of circulating pro-inflammatory cytokines such as interleukin-1β (IL-1β), interleukin-6 (IL-6), and tumor necrosis factor-α (TNF-α). More generally, four selenoproteins, GPx-4, SePP, selenoprotein S (SEL-S), and selenoprotein 15 (Sep-15) have been functionally linked to the inflammatory processes of 5-lipoxygenase (5-LO) metabolism, cytokine regulation, and endoplasmic reticulum (ER) stress. Single nucleotide polymorphisms (SNPs) identified in these selenoprotein genes have shown genotype-dependent modulation of inflammatory status and oxidative stress (Sakr et al., 2007; Chen et al., 2009; Kipp et al., 2009; Schomburg and Schweizer, 2009; Zhang et al., 2010a,b).
In common with other micronutrients, the physiological effect of any given amount and/or source of Se depends on a series of physical, chemical, and physiological processes, including the amount ingested, the meal matrix, digestion, absorption, metabolism, and excretion. It does not act independently (Fairweather-Tait et al., 2010), but acts in conjunction with other micronutrients, including zinc, folate, vitamins D, E, B2, B6, and B12, directly or indirectly in the innate immune response, oxidative stress response, and DNA metabolism (Erickson et al., 2000; Georgieff, 2007; Maggini et al., 2007). Each of these processes involves a complex interaction among genes, gene products, and environmental factors.
Nutrigenetics and Selenium
Until recently, micronutrient research was conducted under the assumption that the underlying mechanisms are the same in all humans. A significant effort is now being made to characterize the effects of variants of key micronutrient genes within the human population (Mathers and Hesketh, 2007; Méplan et al., 2007; van Ommen et al., 2010b). Somewhere around 600 enzymes for which micronutrients are cofactors are known in the human proteome, and the genetic diversity of most of them has not been extensively characterized (Gladyshev et al., 2004; Lobanov et al., 2009). In relation to the specific example of Se, the current nutritional guidelines for this micronutrient may be ideal for only a relatively small number of people. The field of nutrigenetics studies the effect of genetic variations on the interaction between diet and health (Joost et al., 2007). It recognizes the implications of genetic variability, and seeks to optimize human diets for disease prevention, based on identification of key points of human variation and/or effects of dietary components on the expression of key genes. Human genetic variation is projected to include about 25 million SNPs (Johnson, 2009), and an unknown, but possibly even larger, number of small insertion/deletions (indels) plus CNVs (Erickson, 2010; Lee and Scherer, 2010). Those SNPs that are important to gene function include non-synonymous SNPs that may affect enzyme or protein properties, regulatory SNPs that alter gene expression, intron splicing, or RNA stability. All of these changes will profoundly influence the way in which the body metabolizes and utilizes nutrients such as Se (Mathers and Hesketh, 2007; van Ommen et al., 2010b; Zeisel, 2010).
An increasing number of Se studies have used nutrigenetic approaches, in considering the functionality of various SNPs in selenoproteins, and the effects of such functional SNPs on individual Se requirements (Villette et al., 2002; Bermano et al., 2007; Mathers and Hesketh, 2007; Méplan et al., 2007, 2008, 2010; Cooper et al., 2008; Hesketh, 2008; Peters et al., 2008; Lietz and Hesketh, 2009; Sutherland et al., 2010; Xiong et al., 2010). There is also one study suggesting that CNVs may also be important for Se metabolism and function (Amar et al., 2010). Three typical examples, relevant to SNPs, follow:
– Leukotrienes produced from inflammatory cells mediate the inflammatory response and their synthesis has been shown to be 5-LO dependent (Villette et al., 2002). Fatty acid hydroperoxides are potential activators of 5-LO, so the reduction of these intracellular hydroperoxides to hydroxides by GPx-4 and subsequent 5-lipoxygenase (5-LO) inhibition is proposed to be a key reaction capable of influencing the intensity and duration of an immune response. A SNP identified as a C → T variation located at position 718 in the 3′ untranslated region (3′UTR) of the human GPx-4 gene has been described, and shown to contribute to a genotype-dependent functionally significant difference in levels of 5-LO metabolites (Villette et al., 2002). A genotype-dependent role for GPx-4 in lipoxygenase metabolism that is linked to the inflammatory response as a function of 5-LO activation was proposed.
– A role for SEL-S in the translocation of misfolded proteins from the ER to the cytosol for ubiquitin dependent proteasomal degradation and the regulation of the production of pro-inflammatory cytokines has been described (Curran et al., 2005). These authors proposed that the accumulation of misfolded proteins promotes ER stress resulting in the activation of nuclear factor kappa β (NFκβ) and transcription of pro-inflammatory cytokines. The removal of misfolded proteins is thought to prevent ER stress and the pro-inflammatory response. The expression of SEL-S has been shown to be up-regulated in response to pro-inflammatory cytokines, itself down regulating pro-inflammatory cytokine production. Investigation of the functional influence of a SNP described as a G → A variation located at position 105 in the SEL-S promoter region that mediates SEL-S expression in response to pro-inflammatory cytokines demonstrated that a reduction in SEL-S expression and translocation of misfolded proteins resulted in the persistence of the pro-inflammatory response measured as plasma levels of IL-1β, IL-6, and TNF-α (Curran et al., 2005).
– Sep-15 is thought to be involved in the quality control of ER protein folding (Korotkov et al., 2001; Labunsky et al., 2006; Labunskyy et al., 2007, 2009). More specifically, it has been shown to associate with the protein folding sensor UDP–glucose:glycoprotein glucosyltransferase (UGT) that regulates the calnexin cycle in the ER (Labunsky et al., 2006). A role for Sep-15 in the modulation of UGT activity or the assessment of protein structure is proposed, and supported by the finding that Sep-15 expression is increased in response to the accumulation of unfolded proteins in the ER (Labunsky et al., 2006). Two SNPs located in the SECIS portion of the 3′UTR of Sep-15 have been identified that influence Sep-15 translation and responsiveness to selenium intake (Hu et al., 2001; Kumaraswamy et al., 2002; Penney et al., 2010; Sutherland et al., 2010).
The above examples are just a small proportion of those available studies associating variant SNPs in genes for selenoproteins, in conjunction with low Se status, with increased susceptibility to various disease or physiological conditions. Until now, most of these studies have considered a single micronutrient (Se) and one or several variants in a gene or pathway at a time. However, many of these early studies had small study numbers, and not greatly significant p values. For example, Villette et al. (2002), studied only 66 individuals, among which differences significant at p < 0.002 were found. This contrasts unfavorably with the numbers of subjects now being utilized to prove single gene associations with disease. For example, a recent meta-analysis increased the number of confirmed Crohn’s disease susceptibility loci to 71 (Franke et al., 2010). This publication utilized a meta-analysis of six genome-wide association studies (GWAS), comprising 6,333 affected cases and 15,056 controls. Confirmation was provided by studying a further 15,694 cases, 14,026 controls, and 414 parent–offspring trios, with p < 1 × 10−7 providing a cut off point for statistical significance. If these sorts of numbers are now considered essential for single gene associations with disease, how do we even envisage the increased orders of magnitude of numbers that would be necessary to definitively prove gene–gene or gene–diet interactions?
There is increasing recognition of the complexity and interactions among genes, gene pathways, and multiple micronutrients (Pico et al., 2008; Schomburg and Schweizer, 2009; van Ommen et al., 2010b). An open source description of gene pathways for various micronutrients, including Se, is publically available at http://micronutrients.wikipathways.org. (Pico et al., 2008). This offers a search option for pathways, genes and metabolites, a pathway editor, different download options, and web services for programmatic access. A systems biology approach has been suggested as necessary to integrate and data from multiple systems and technologies (Hesketh, 2008; Allmang et al., 2009; Schomburg and Schweizer, 2009). New methods are becoming available for combining the interaction of effects of genotype with dietary factors alongside measurements of factors that influence transcript levels of multiple genes (Parts et al., 2011). While such novel analytical methods have been applied to factors that affect gene expression in a yeast dataset, they await application to human studies at this time.
Nutrigenomics and Selenium
In the past, direct measurement of plasma micronutrients or biomarkers have been used to quantify micronutrient status. Nutrigenomics is the study of the response of humans to food and food components using genomics, proteomics, and metabolomics approaches (Daniel et al., 2008; Hesketh, 2008; Kaput, 2008). These approaches potentially provide powerful new tools to assess the impact of Se status on the health of an individual.
Gene Expression
Patterns and endpoints of gene expression are analyzed through transcriptomics and proteomics. Such analyses enabling an understanding of the mechanisms of action of Se are increasing (Bartel et al., 2007; Chun et al., 2007; Kibriya et al., 2007; Novoselov et al., 2007; Zeng and Botnen, 2007; Hesketh, 2008; Pagmantidis et al., 2008; Carlson et al., 2009; Yuzbasioglu et al., 2009; Bartel et al., 2010). To integrate these datasets, pathways that graphically show gene expression changes in association with Se levels are being developed (van Ommen et al., 2010a,b). Currently, gene expression is incorporated by including enzymes and transcription levels (Joost et al., 2007; Van Ommen et al., 2008). However, this is changing, with new analytical methods appearing (e.g., Parts et al., 2011).
An example of the potential of the approaches to define the relative roles of genetic variations and Se intake in a human population is provided by Méplan et al. (2009). They studied SePP, the major plasma selenoprotein, with both transport and antioxidant functions, which exists in human plasma as two isoforms, of approximately 50 and 60 kDa. They considered the effect of SNPs in the SEPP-1 gene, on the proportions of SePP plasma isoforms in relation to Se supplementation, and disease status. SePP was isolated from the plasma of healthy volunteers, and from colon cancer patients and controls, before and after a 6-week supplementation with 100 μg sodium selenite. The distribution of SePP isoforms was analyzed by Western blotting. In healthy volunteers, the relative abundance of each isoform primarily depended on two SNPs in the SEPP1 gene, but only in the absence of Se supplementation. These are the two functional SNPs, rs3877899, predicted to cause an Ala-to-Thr amino acid change at position 234, and rs7579, located in the 3′-untranslated region of SEPP-1 mRNA. This difference between genotypes disappeared after Se supplementation. A genotype-dependent reduction in the proportion of the 60-kDa isoform was also seen in patients with colorectal cancer as compared with controls.
A useful review on the regulation of selenoprotein gene expression is provided by Schomburg and Schweizer (2009). They draw attention to the expression of selenoproteins as almost classic textbook examples of different levels of regulatory control on protein biosynthesis. That is, gene expression can be regulated during transcription, RNA processing, translation, and posttranslational events, as well as through control of the stability of metabolic intermediates and final products. Closely regulated expression patterns of selenoproteins are present in different tissues, and show differences between male and female individuals. They are often modified during disease or disease processes, including cancer, inflammation, or neurodegeneration.
Proteomics
Méplan (2011) uses Se as an example of a trace element that can influence aging, and how these effects can be studied through transcriptomics and proteomics technologies. Such approaches in animal models have enabled identification of downstream targets of Se in pathways related to age-related diseases. However, future approaches that combine nutrigenomics with longevity studies in humans will be necessary to identify comparable targets in humans.
Traditional Se indexes, such as Se serum or plasma concentrations, do not explain population differences or chemoprotective effects. Mahn et al. (2009) suggested that knowledge of a group of proteins that respond to Se supplementation could be useful in assessing its’ metabolic status in an individual. They used a rat model to consider the effects of dietary supplementation with sodium-selenate on the plasma proteome. Groups of six rats were fed a basic diet supplemented with sodium-selenate at 1.9 μg of Se per gram of food, in comparison with a group on a low Se diet, for 10 weeks. The authors showed significant changes of the levels of fibrinogen, apolipoproteins, haptoglobin, and transthyretin in the high Se group. They suggest that, more generally, changes in the proteomic profile due to Se supplementation could be used in human studies to assess the metabolic status of Se.
Metabolomics
This field defines the metabolic network(s) linked to gene expression. Powerful new metabolomics approaches, coupled with a number of useful web-based tools, permit a comprehensive analysis of metabolites in the context of an integrated evaluation of biological activity (Xia et al., 2008; Forsythe and Wishart, 2009; Wishart, 2009; Wishart et al., 2009; Xia and Wishart, 2010a,b). In these pathways, intracellular mechanisms related to micronutrient activity are linked to plasma and blood cell membrane concentrations of all relevant metabolites, including proteins (Wishart, 2007; Berjanskii et al., 2009; Xia et al., 2009; Xia and Wishart, 2010a,b).
These approaches were used by Fan et al. (2006) to map the metabolites induced by two different forms of Se, selenite, and selenomethionine, in animal models and human studies.
Epigenetics
This has been defined as effects on gene expression in the absence of heritable DNA changes. It is likely that changes in selenoprotein expression result not only from changes in DNA, but also from epigenetic effects (Davis and Milner, 2006; Baliga et al., 2007; Hesketh, 2008). These changes may relate to effects on transcription factor, microRNAs, and DNA methylation (Ferguson, 2009). However, epigenetic effects of Se have not been extensively described to date, at least in humans.
The Use of Nutrigenetic and/or Nutrigenomics Approaches to Provide Insight into the Potential Value or Hazards of SE Supplementation
A considerable literature has linked deficiency of Se to elevated risk of cancer, especially prostate cancer (Clark et al., 1993; Combs et al., 1997; Clark and Jacobs, 1998; Nelson et al., 1999; Duffield-Lillico et al., 2002, 2003a; Karunasinghe et al., 2004). In the Nutritional Prevention of Cancer (NPC) study, supplementation with Se as Se-yeast was shown to reduce the risk of prostate cancer in subjects with low Se nutritional status (Clark et al., 1996, 1998; Duffield-Lillico et al., 2003a,b).
Recently, considerable concerns about possible adverse effects of Se supplementation have been raised by the premature cessation of the SELECT (the Selenium and Vitamin E Cancer Prevention Trial), the largest-ever prostate cancer prevention trial (Hoque et al., 2001; Klein et al., 2001, 2003; Cook et al., 2005; Lippman et al., 2005; Dunn et al., 2009). The rationale for this study was based partly upon early results from the NPC study. Exhaustive attempts were made to recruit large numbers of American men, including ethnic minorities (Cook et al., 2005; Lippman et al., 2005), involving extensive publicity and considerable expense (Thompson et al., 2005). However, the trial was controversially stopped partway through its’ planned course, because of safety concerns (Lippman et al., 2009; Mueller et al., 2009; Schrauzer, 2009).
Even in the NPC study, not all individuals appeared to benefit from Se supplementation (Duffield-Lillico et al., 2003a,b). For example, there is evidence that a high level of serum Se was associated with a slightly elevated risk of aggressive prostate cancer. However, the increased risk only occurred in individuals carrying a certain variant form of the superoxide dismutase (SOD2) gene (Li et al., 2005). In vitro studies have suggested that protection against DNA damage by various Se analogs occurs through the tumor suppressor p53 gene, and would not occur in the presence of the variant enzymes (Zhao et al., 2006; Sarveswaran et al., 2010). Tsavachidou et al. (2009) completed a randomized, placebo-controlled phase IIA study of prostate cancer patients before prostatectomy, and considered effects of selenomethionine on gene expression profiles. They showed that whether events appeared beneficial or detrimental was related to p53 status.
We have previously suggested that Se requirements should be based on a combination of genotype, and biomarkers that predict cancer risk (Karunasinghe et al., 2004; Ferguson et al., 2006). There is no current agreement as to how to define all relevant genetic variants, and how to combine and integrate data for gene–gene and gene–environment interactions, although it is clear that they are very important (van Ommen et al., 2010b). We have shown the application of metabolomic biomarkers to the modulation of inflammation in animal models (Lin et al., 2010), and these approaches are potentially applicable to optimizing the intake of Se in human populations.
Databases Relevant to Nutrigenetic and Nutrigenomic Studies on SE
The nutritional phenotype database (dbNP) is an infrastructural activity which allows storage, processing, and meaningful queries of information on micronutrient-oriented human and animal model intervention studies with all omics components included (genetics, transcriptomics, proteomics, metabolomics, biomarkers), together with a detailed description of the study design (van Ommen et al., 2010a,b). dbNP also allows additional analyses of existing samples from nutritional intervention studies, and encourages the future collection and analysis of samples for this purpose.
A number of databases, such as the Gene Ontology database1, collect relevant information for genes and gene variants. Traditionally the most relevant resource for the relation between genes and (disease) phenotype is the OMIM database (van Ommen et al., 2010a). The Human Variome Project2 has also begun to develop standards for identifying, characterizing, and databasing gene variations, and their association with clinical or other phenotypes (Kohonen-Corish et al., 2010). The micronutrient genomics database is also important3 (van Ommen et al., 2010b), while a number of metabolomics databases are also available (Xia et al., 2008, 2009; Wishart et al., 2009; Xia and Wishart, 2010a,b).
Conclusion
Nutrigenomics and nutrigenetic studies have the power to generate large data sets, but this may be both a strength and a weakness. A review of published human intervention studies in nutrigenomics highlights a number of flaws and inconsistencies in current data interpretation, sizable knowledge gaps, and a need for improved and agreed study designs that are used consistently on an international basis (Wittwer et al., 2011). The combination of different genomics technologies to the same sample allows cellular and physiological changes to be robustly assessed throughout all molecular layers of change. Current developments mean that we are moving toward getting very solid data at the level of the individual or small group of individuals. But, given the variability between and among individuals, how do we then address the issue of power, so that we can be certain that we can reproduce and interpret gene–gene and gene–diet interactions, and recognize their implications?
While there is great deal of interest and promise in the field, the published human studies of nutrigenetics, nutrigenomics, and selenium to date may be largely considered as “proof-of-concept.” They have demonstrated that selenium at different levels, whether eaten in the normal diet or taken as a supplement, profoundly affects gene expression, proteomics, and metabolic profile. But it is important not to underestimate the challenges remaining in evaluation, interpretation, and integration of results derived from different platforms. There is a strong argument for more concerted actions, larger collaborative study cohorts, and cohesive and collegial research efforts with expert from different disciplines across different countries.
Nutrigenetics and nutrigenomics are comparatively new tools with which to study micronutrients such as Se. A critical evaluation of available data, incorporating omics technologies, strongly suggests that the intake or dietary supplementation with micronutrients such as Se should be optimized at an individual level. However, compared with the study numbers we now require before concluding statistical significance of an individual gene in relation to disease susceptibility, sample sizes reporting gene–diet interactions for Se thus far have been too small to permit definitive conclusions. A previous review in this journal has pointed to the challenges in this field (Zeisel, 2010). Despite the challenges, we suggest that nutrigenetic and nutrigenomic technologies will increasingly become essential tools to study the physiological effects and optimal dietary intake of this important micronutrient in the future. However, international collaborations, agreed study design and analytical methods, alongside new generation genomics technologies will be essential to take the field to the next plateau and begin to release its real potential.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Agamy, O., Ben Zeev, B., Lev, D., Marcus, B., Fine, D., Su, D., Narkis, G., Ofir, R., Hoffmann, C., Leshinsky-Silver, E., Flusser, H., Sivan, S., Soll, D., Lerman-Sagie, T., and Birk, O. S. (2010). Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am. J. Hum. Genet. 87, 538–544.
Allmang, C., Wurth, L., and Krol, A. (2009). The selenium to selenoprotein pathway in eukaryotes: more molecular partners than anticipated. Biochim. Biophys. Acta 1790, 1415–1423.
Amar, S., Ovadia, O., Maier, W., Ebstein, R., Belmaker, R. H., Mishmar, D., and Agam, G. (2010). Copy number variation of the SELENBP1 gene in schizophrenia. Behav. Brain Funct. 6, 40.
Arbogast, S., and Ferreiro, A. (2009). Selenoproteins and protection against oxidative stress: selenoprotein N as a novel player at the crossroads of redox signaling and calcium homeostasis. Antioxid. Redox Signal. 12, 893–904.
Arner, E. S. (2010). Selenoproteins – what unique properties can arise with selenocysteine in place of cysteine? Exp. Cell Res. 316, 1296–1303.
Ashrafi, M. R., Shabanian, R., Abbaskhanian, A., Nasirian, A., Ghofrani, M., Mohammadi, M., Zamani, G. R., Kayhanidoost, Z., Ebrahimi, S., and Pourpak, Z. (2007a). Selenium and intractable epilepsy: is there any correlation? Pediatr. Neurol. 36, 25–29.
Ashrafi, M. R., Shams, S., Nouri, M., Mohseni, M., Shabanian, R., Yekaninejad, M. S., Chegini, N., Khodadad, A., and Safaralizadeh, R. (2007b). A probable causative factor for an old problem: selenium and glutathione peroxidase appear to play important roles in epilepsy pathogenesis. Epilepsia 48, 1750–1755.
Baliga, M. S., Wang, H., Zhuo, P., Schwartz, J. L., and Diamond, A. M. (2007). Selenium and GPx-1 overexpression protect mammalian cells against UV-induced DNA damage. Biol. Trace Elem. Res. 115, 227–242.
Bartel, J., Bartz, T., Wolf, C., Charkiewicz, E., Kuhbacher, M., Pohl, T., and Kyriakopoulos, A. (2007). Activity of the glutathione peroxidase-2. Differences in the selenium-dependent expression between colon and small intestine. Cancer Genomics Proteomics 4, 369–372.
Bartel, J., Charkiewicz, E., Bartz, T., Bartel, J., Schmidt, D., Grbavac, I., and Kyriakopoulos, A. (2010). Metalloproteome of the prostate: carcinoma cell line DU-145 in comparison to healthy rat tissue. Cancer Genomics Proteomics 7, 81–86.
Berjanskii, M., Tang, P., Liang, J., Cruz, J. A., Zhou, J., Zhou, Y., Bassett, E., Macdonell, C., Lu, P., Lin, G., and Wishart, D. S. (2009). GeNMR: a web server for rapid NMR-based protein structure determination. Nucleic Acids Res. 37, W670–W677.
Bermano, G., Pagmantidis, V., Holloway, N., Kadri, S., Mowat, N. a. G., Shiel, R. S., Arthur, J. R., Mathers, J. C., Daly, A. K., Broom, J., and Hesketh, J. E. (2007). Evidence that a polymorphism within the 3′UTR of glutathione peroxidase 4 is functional and is associated with susceptibility to colorectal cancer. Genes Nutr. 2, 225–232.
Bleys, J., Navas-Acien, A., Stranges, S., Menke, A., Miller, E. R. III, and Guallar, E. (2008). Serum selenium and serum lipids in US adults. Am. J. Clin. Nutr. 88, 416–423.
Carlson, B. A., Yoo, M.-H., Sano, Y., Sengupta, A., Kim, J. Y., Irons, R., Gladyshev, V. N., Hatfield, D. L., and Park, J. M. (2009). Selenoproteins regulate macrophage invasiveness and extracellular matrix-related gene expression. BMC Immunol. 10, 57. doi: 10.1186/1471-2172-10-57
Castellano, S., Gladyshev, V. N., Guigo, R., and Berry, M. J. (2008). SelenoDB 1.0: a database of selenoprotein genes, proteins and SECIS elements. Nucleic Acids Res. 36, D332–D338.
Chen, Y.-C., Sosnoski, D. M., Gandhi, U. H., Novinger, L. J., Prabhu, K. S., and Mastro, A. M. (2009). Selenium modifies the osteoblast inflammatory stress response to bone metastatic breast cancer. Carcinogenesis 30, 1941–1948.
Chiang, E. C., Shen, S., Kengeri, S. S., Xu, H., Combs, G. F., Morris, J. S., Bostwick, D. G., and Waters, D. J. (2009). Defining the optimal selenium dose for prostate cancer risk reduction: insights from the u-shaped relationship between selenium status, dna damage, and apoptosis. Dose Response 8, 285–300.
Chun, J. Y., Hu, Y., Pinder, E., Wu, J., Li, F., and Gao, A. C. (2007). Selenium inhibition of survivin expression by preventing Sp1 binding to its promoter. Mol. Cancer Ther. 6, 2572–2580.
Clark, L. C., Combs, G. F. Jr., Turnbull, B. W., Slate, E. H., Chalker, D. K., Chow, J., Davis, L. S., Glover, R. A., Graham, G. F., Gross, E. G., Krongrad, A., Lesher, J. L. Jr., Park, H. K., Sanders, B. B. Jr., Smith, C. L., and Taylor, J. R. (1996). Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. JAMA 276, 1957–1963. [Erratum appears in JAMA 1997 May 21;277, 1520].
Clark, L. C., Dalkin, B., Krongrad, A., Combs, G. F. Jr., Turnbull, B. W., Slate, E. H., Witherington, R., Herlong, J. H., Janosko, E., Carpenter, D., Borosso, C., Falk, S., and Rounder, J. (1998). Decreased incidence of prostate cancer with selenium supplementation: results of a double-blind cancer prevention trial. Br. J. Urol. 81, 730–734.
Clark, L. C., Hixson, L. J., Combs, G. F. Jr., Reid, M. E., Turnbull, B. W., and Sampliner, R. E. (1993). Plasma selenium concentration predicts the prevalence of colorectal adenomatous polyps. Cancer Epidemiol. Biomarkers Prev. 2, 41–46.
Clark, L. C., and Jacobs, E. T. (1998). Environmental selenium and cancer: risk or protection? Cancer Epidemiol. Biomarkers Prev. 7, 847–848; discussion 851–842.
Combs, G. F. Jr., Clark, L. C., and Turnbull, B. W. (1997). Reduction of cancer mortality and incidence by selenium supplementation. Med. Klin. 92(Suppl. 3), 42–45.
Cook, E. D., Moody-Thomas, S., Anderson, K. B., Campbell, R., Hamilton, S. J., Harrington, J. M., Lippman, S. M., Minasian, L. M., Paskett, E. D., Craine, S., Arnold, K. B., and Probstfield, J. L. (2005). Minority recruitment to the selenium and vitamin E cancer prevention yrial (SELECT). Clin. Trials 2, 436–442.
Cooper, M. L., Adami, H.-O., Gronberg, H., Wiklund, F., Green, F. R., and Rayman, M. P. (2008). Interaction between single nucleotide polymorphisms in selenoprotein P and mitochondrial superoxide dismutase determines prostate cancer risk. Cancer Res. 68, 10171–10177.
Curran, J. E., Jowett, J. B. M., Elliot, K. S., Gao, Y., Glaschenko, K., Wang, J., Azim, D. M. A., Cai, G., Mahaney, M. C., Comuzzie, A. G., Dyer, T. D., Walder, K. R., Zimmet, P., Maccluer, W., Collier, G. R., Kissebah, A. H., and Blangero, J. (2005). Genetic variation in selenoprotein S influences inflammatory response. Nat. Genet. 37, 1234–1241.
Daniel, H., Drevon, C. A., Klein, U. I., Kleemann, R., and Van Ommen, B. (2008). The challenges for molecular nutrition research 3: comparative nutrigenomics research as a basis for entering the systems level. Genes Nutr. 3, 101–106.
Darlow, B. A., Inder, T. E., Sluis, K. B., Nuthall, G., Mogridge, N., and Winterbourn, C. C. (1995). Selenium status of New Zealand infants fed either a selenium supplemented or a standard formula. J. Paediatr. Child Health 31, 339–344.
Davis, C. D., and Milner, J. A. (2006). “Incorporating ‘omics’ approaches to elucidate the role of selenium and selenoproteins in cancer prevention,” in Selenium It’s Molecular Biology and Role in Human Health, 2nd Edn, eds D. L. Hatfield, M. J. Berry, and V. N. Gladyshev (New York: Springer), 367–378
De Jong, N., Gibson, R. S., Thomson, C. D., Ferguson, E. L., Mckenzie, J. E., Green, T. J., and Horwath, C. C. (2001). Selenium and zinc status are suboptimal in a sample of older New Zealand women in a community-based study. J. Nutr. 131, 2677–2684.
Dolamore, B. A., Brown, J., Darlow, B. A., George, P. M., Sluis, K. B., and Winterbourn, C. C. (1992). Selenium status of Christchurch infants and the effect of diet. N. Z. Med. J. 105, 139–142.
Duffield, A. J., Thomson, C. D., Hill, K. E., and Williams, S. (1999). An estimation of selenium requirements for New Zealanders. Am. J. Clin. Nutr. 70, 896–903.
Duffield-Lillico, A. J., Dalkin, B. L., Reid, M. E., Turnbull, B. W., Slate, E. H., Jacobs, E. T., Marshall, J. R., Clark, L. C., and Nutritional Prevention of Cancer Study Group. (2003a). Selenium supplementation, baseline plasma selenium status and incidence of prostate cancer: an analysis of the complete treatment period of the Nutritional Prevention of Cancer Trial. BJU Int. 91, 608–612.
Duffield-Lillico, A. J., Slate, E. H., Reid, M. E., Turnbull, B. W., Wilkins, P. A., Combs, G. F. Jr., Park, H. K., Gross, E. G., Graham, G. F., Stratton, M. S., Marshall, J. R., Clark, L. C., and Nutritional Prevention of Cancer Study Group. (2003b). Selenium supplementation and secondary prevention of nonmelanoma skin cancer in a randomized trial. J. Natl. Cancer Inst. 95, 1477–1481.
Duffield-Lillico, A. J., Reid, M. E., Turnbull, B. W., Combs, G. F. Jr., Slate, E. H., Fischbach, L. A., Marshall, J. R., and Clark, L. C. (2002). Baseline characteristics and the effect of selenium supplementation on cancer incidence in a randomized clinical trial: a summary report of the Nutritional Prevention of Cancer Trial. Cancer Epidemiol. Biomarkers Prev. 11, 630–639.
Dunn, B. K., Ryan, A., and Ford, L. G. (2009). Selenium and vitamin E cancer prevention trial: a nutrient approach to prostate cancer prevention. Recent Results Cancer Res. 181, 183–193.
Duntas, L. H. (2010). Selenium and the thyroid: a close-knit connection. J. Clin. Endocrinol. Metab. 95, 5180–5188.
Erickson, K. L., Medina, E. A., and Hubbard, N. E. (2000). Micronutrients and innate immunity. J. Infect. Dis. 182(Suppl. 1), S5–S10.
Erickson, R. P. (2010). Somatic gene mutation and human disease other than cancer: an update. Mutat. Res. 705, 96–106.
Fairweather-Tait, S. J., Collings, R., and Hurst, R. (2010). Selenium bioavailability: current knowledge and future research requirements. Am. J. Clin. Nutr. 91, 1484S–1491S. [Erratum appears in Am. J. Clin. Nutr. 2010 Oct;92, 1002].
Fan, T. W. M., Higashi, R. M., and Lane, A. N. (2006). Integrating metabolomics and transcriptomics for probing SE anticancer mechanisms. Drug Metab. Rev. 38, 707–732.
Ferguson, L. R. (2009). Epigenetic variation and customising nutritional intervention. Curr. Pharmacogenomics Person Med. 7, 115–124.
Ferguson, L. R., Philpott, M., and Karunasinghe, N. (2006). Oxidative DNA damage and repair: significance and biomarkers. J. Nutr. 136, 2687S–2689S.
Flohe, L. (2010). Changing paradigms in thiology from antioxidant defense toward redox regulation. Meth. Enzymol. 473, 1–39.
Forsythe, I. J., and Wishart, D. S. (2009). Exploring human metabolites using the human metabolome database. Curr. Protoc. Bioinformatics 14, Unit 14.18.
Franke, A., McGovern, D. P., Barrett, J. C., Wang, K., Radford-Smith, G. L., Ahmad, T., Lees, C. W., Balschun, T., Lee, J., Roberts, R., Anderson, C. A., Bis, J. C., Bumpstead, S., Ellinghaus, D., Festen, E. M., Georges, M., Green, T., Haritunians, T., Jostins, L., Latiano, A., Mathew, C. G., Montgomery, G. W., Prescott, N. J., Raychaudhuri, S., Rotter, J. I., Schumm, P., Sharma, Y., Simms, L. A., Taylor, K. D., Whiteman, D., Wijmenga, C., Baldassano, R. N., Barclay, M., Bayless, T. M., Brand, S., Büning, C., Cohen, A., Colombel, J. F., Cottone, M., Stronati, L., Denson, T., De Vos, M., D’Inca, R., Dubinsky, M., Edwards, C., Florin, T., Franchimont, D., Gearry, R., Glas, J., Van Gossum, A., Guthery, S. L., Halfvarson, J., Verspaget, H. W., Hugot, J. P., Karban, A., Laukens, D., Lawrance, I., Lemann, M., Levine, A., Libioulle, C., Louis, E., Mowat, C., Newman, W., Panés, J., Phillips, A., Proctor, D. D., Regueiro, M., Russell, R., Rutgeerts, P., Sanderson, J., Sans, M., Seibold, F., Steinhart, A. H., Stokkers, P. C., Torkvist, L., Kullak-Ublick, G., Wilson, D., Walters, T., Targan, S. R., Brant, S. R., Rioux, J. D., D’Amato, M., Weersma, R. K., Kugathasan, S., Griffiths, A. M., Mansfield, J. C., Vermeire, S., Duerr, R. H., Silverberg, M. S., Satsangi, J., Schreiber, S., Cho, J. H., Annese, V., Hakonarson, H., Daly, M. J., and Parkes, M. (2010). Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42, 1118–1125.
Gao, S., Jin, Y., Hall, K. S., Liang, C., Unverzagt, F. W., Ma, F., Cheng, Y., Shen, J., Cao, J., Matesan, J., Li, P., Bian, J., Hendrie, H. C., and Murrell, J. R. (2009). Selenium level is associated with apoE epsilon4 in rural elderly Chinese. Public Health Nutr. 12, 2371–2376.
Georgieff, M. K. (2007). Nutrition and the developing brain: nutrient priorities and measurement. Am. J. Clin. Nutr. 85, 614S–620S.
Gladyshev, V. N., Kryukov, G. V., Fomenko, D. E., and Hatfield, D. L. (2004). Identification of trace element-containing proteins in genomic databases. Annu. Rev. Nutr. 24, 579–596.
Gore, F., Fawell, J., and Bartram, J. (2010). Too much or too little? A review of the conundrum of selenium. J. Water Health 8, 405–416.
Guijian, L., Liugen, Z., Duzgoren-Aydin, N. S., Lianfen, G., Junhua, L., and Zicheng, P. (2007). Health effects of arsenic, fluorine, and selenium from indoor burning of Chinese coal. Rev. Environ. Contam. Toxicol. 189, 89–106.
Hawkes, W. C., and Alkan, Z. (2010). Regulation of redox signaling by selenoproteins. Biol. Trace Elem. Res. 134, 235–251.
Hesketh, J. (2008). Nutrigenomics and selenium: gene expression patterns, physiological targets, and genetics. Annu. Rev. Nutr. 28, 157–177.
Hesketh, J., Wybranska, I., Dommels, Y., King, M., Elliott, R., Pico, C., and Keijer, J. (2006). Nutrient-gene interactions in benefit-risk analysis. Br. J. Nutr. 95, 1232–1236.
Hesketh, J. E., and Villette, S. (2002). Intracellular trafficking of micronutrients: from gene regulation to nutrient requirements. Proc. Nutr. Soc. 61, 405–414.
Higuchi, A., Takahashi, K., Hirashima, M., Kawakita, T., and Tsubota, K. (2010). Selenoprotein P controls oxidative stress in cornea. PLoS ONE 5, e9911. doi:10.1371/journal.pone.0009911
Hoeflich, J., Hollenbach, B., Behrends, T., Hoeg, A., Stosnach, H., and Schomburg, L. (2010). The choice of biomarkers determines the selenium status in young German vegans and vegetarians. Br. J. Nutr. 104, 1601–1604.
Hon, K.-L. E., Wang, S. S., Hung, E. C. W., Lam, H. S., Lui, H. H. K., Chow, C.-M., Ching, G. K. W., Fok, T.-F., Ng, P.-C., and Leung, T.-F. (2010). Serum levels of heavy metals in childhood eczema and skin diseases: friends or foes. Pediatr. Allergy Immunol. 21, 831–836.
Hoque, A., Albanes, D., Lippman, S. M., Spitz, M. R., Taylor, P. R., Klein, E. A., Thompson, I. M., Goodman, P., Stanford, J. L., Crowley, J. J., Coltman, C. A., and Santella, R. M. (2001). Molecular epidemiologic studies within the selenium and vitamin E cancer prevention trial (SELECT). Cancer Causes Control 12, 627–633.
Hu, Y. J., Korotkov, K. V., Mehta, R., Hatfield, D. L., Rotimi, C. N., Luke, A., Prewitt, T. E., Cooper, R. S., Stock, W., Vokes, E. E., Dolan, M. E., Gladyshev, V. N., and Diamond, A. M. (2001). Distribution and functional consequences of nucleotide polymorphisms in the 3′-untranslated region of the human Sep15 gene. Cancer Res. 61, 2307–2310.
Johnson, A. D. (2009). Single-nucleotide polymorphism bioinformatics: a comprehensive review of resources. Circ. Cardiovasc. Genet. 2, 530–536.
Johnson, C. C., Fordyce, F. M., and Rayman, M. P. (2010). Symposium on “geographical and geological influences on nutrition”: factors controlling the distribution of selenium in the environment and their impact on health and nutrition. Proc. Nutr. Soc. 69, 119–132.
Joost, H.-G., Gibney, M. J., Cashman, K. D., Gorman, U., Hesketh, J. E., Mueller, M., Van Ommen, B., Williams, C. M., and Mathers, J. C. (2007). Personalised nutrition: status and perspectives. Br. J. Nutr. 98, 26–31.
Kant, A. K., and Graubard, B. I. (2007). Ethnicity is an independent correlate of biomarkers of micronutrient intake and status in American adults. J. Nutr. 137, 2456–2463.
Kaput, J. (2008). Nutrigenomics research for personalized nutrition and medicine. Curr. Opin. Biotechnol. 19, 110–120.
Karunasinghe, N., Ryan, J., Tuckey, J., Masters, J., Jamieson, M., Clarke, L. C., Marshall, J. R., and Ferguson, L. R. (2004). DNA stability and serum selenium levels in a high-risk group for prostate cancer. Cancer Epidemiol. Biomarkers Prev. 13, 391–397.
Kibriya, M. G., Jasmine, F., Argos, M., Verret, W. J., Rakibuz-Zaman, M., Ahmed, A., Parvez, F., and Ahsan, H. (2007). Changes in gene expression profiles in response to selenium supplementation among individuals with arsenic-induced pre-malignant skin lesions. Toxicol. Lett. 169, 162–176.
Kipp, A., Banning, A., Van Schothorst, E. M., Meplan, C., Schomburg, L., Evelo, C., Coort, S., Gaj, S., Keijer, J., Hesketh, J., and Brigelius-Flohe, R. (2009). Four selenoproteins, protein biosynthesis, and Wnt signalling are particularly sensitive to limited selenium intake in mouse colon. Mol. Nutr. Food Res. 53, 1561–1572.
Klein, E. A., Thompson, I. M., Lippman, S. M., Goodman, P. J., Albanes, D., Taylor, P. R., and Coltman, C. (2001). SELECT: the next prostate cancer prevention trial. Selenum and vitamin E cancer prevention trial. J. Urol. 166, 1311–1315.
Klein, E. A., Thompson, I. M., Lippman, S. M., Goodman, P. J., Albanes, D., Taylor, P. R., and Coltman, C. (2003). SELECT: the selenium and vitamin E cancer prevention trial. Urol. Oncol. 21, 59–65.
Kohonen-Corish, M. R. J., Al-Aama, J. Y., Auerbach, A. D., Axton, M., Barash, C. I., Bernstein, I., Beroud, C., Burn, J., Cunningham, F., Cutting, G. R., Den Dunnen, J. T., Greenblatt, M. S., Kaput, J., Katz, M., Lindblom, A., Macrae, F., Maglott, D., Moslein, G., Povey, S., Ramesar, R., Richards, S., Seminara, D., Sobrido, M.-J., Tavtigian, S., Taylor, G., Vihinen, M., Winship, I., Cotton, R. G. H., and Human Variome Project Meeting. (2010). How to catch all those mutations – the report of the third Human Variome Project Meeting, UNESCO Paris, May 2010. Hum. Mutat. 31, 1374–1381.
Korotkov, K. V., Kumaraswamy, E., Zhou, Y., Hatfield, D. L., and Gladyshev, V. N. (2001). Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 276, 15330–15336.
Kumaraswamy, E., Korotkov, K. V., Diamond, A. M., Gladyshev, V. N., and Hatfield, D. L. (2002). Genetic and functional analysis of mammalian Sep15 selenoprotein. Meth. Enzymol. 347, 187–197.
Labunsky, V. M., Gladyshev, V. N., and Hatfield, D. L. (2006). “The 15-kDa selenoprotein (Sep15): functional analysis and role in cancer,” in Selenium It’s Molecular Biology and Role in Human Health, 2nd Edn, eds D. L. Hatfield, M. J. Berry, and V. N. Gladyshev (New York: Springer), 141–148.
Labunskyy, V. M., Hatfield, D. L., and Gladyshev, V. N. (2007). The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life 59, 1–5.
Labunskyy, V. M., Yoo, M.-H., Hatfield, D. L., and Gladyshev, V. N. (2009). Sep15, a thioredoxin-like selenoprotein, is involved in the unfolded protein response and differentially regulated by adaptive and acute ER stresses. Biochemistry 48, 8458–8465.
Lee, C., and Scherer, S. W. (2010). The clinical context of copy number variation in the human genome. Expert Rev. Mol. Med. 12, e8.
Lei, X. G., Cheng, W.-H., and Mcclung, J. P. (2007). Metabolic regulation and function of glutathione peroxidase-1. Annu. Rev. Nutr. 27, 41–61.
Lescure, A., Rederstorff, M., Krol, A., Guicheney, P., and Allamand, V. (2009). Selenoprotein function and muscle disease. Biochim. Biophys. Acta 1790, 1569–1574.
Levander, O. A. (1991). Scientific rationale for the 1989 recommended dietary allowance for selenium. J. Am. Diet. Assoc. 91, 1572–1576.
Li, H., Kantoff, P. W., Giovannucci, E., Leitzmann, M. F., Gaziano, J. M., Stampfer, M. J., and Ma, J. (2005). Manganese superoxide dismutase polymorphism, prediagnostic antioxidant status, and risk of clinical significant prostate cancer. Cancer Res. 65, 2498–2504.
Lietz, G., and Hesketh, J. (2009). A network approach to micronutrient genetics: interactions with lipid metabolism. Curr. Opin. Lipidol. 20, 112–120.
Lin, H.-M., Barnett, M. P., Roy, N. C., Joyce, N. I., Zhu, S., Armstrong, K., Helsby, N. A., Ferguson, L. R., and Rowan, D. D. (2010). Metabolomic analysis identifies inflammatory and noninflammatory metabolic effects of genetic modification in a mouse model of Crohn’s disease. J. Proteome Res. 9, 1965–1975.
Lippman, S. M., Goodman, P. J., Klein, E. A., Parnes, H. L., Thompson, I. M. Jr., Kristal, A. R., Santella, R. M., Probstfield, J. L., Moinpour, C. M., Albanes, D., Taylor, P. R., Minasian, L. M., Hoque, A., Thomas, S. M., Crowley, J. J., Gaziano, J. M., Stanford, J. L., Cook, E. D., Fleshner, N. E., Lieber, M. M., Walther, P. J., Khuri, F. R., Karp, D. D., Schwartz, G. G., Ford, L. G., and Coltman, C. A. Jr. (2005). Designing the selenium and vitamin E cancer prevention trial (SELECT). J. Natl. Cancer Inst. 97, 94–102.
Lippman, S. M., Klein, E. A., Goodman, P. J., Lucia, M. S., Thompson, I. M., Ford, L. G., Parnes, H. L., Minasian, L. M., Gaziano, J. M., Hartline, J. A., Parsons, J. K., Bearden, J. D. III, Crawford, E. D., Goodman, G. E., Claudio, J., Winquist, E., Cook, E. D., Karp, D. D., Walther, P., Lieber, M. M., Kristal, A. R., Darke, A. K., Arnold, K. B., Ganz, P. A., Santella, R. M., Albanes, D., Taylor, P. R., Probstfield, J. L., Jagpal, T. J., Crowley, J. J., Meyskens, F. L. Jr., Baker, L. H., and Coltman, C. A. Jr. (2009). Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the selenium and vitamin E cancer prevention trial (SELECT). JAMA 301, 39–51.
Lobanov, A. V., Hatfield, D. L., and Gladyshev, V. N. (2009). Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta 1790, 1424–1428.
Logan, J. W. (1991). Food for New Zealanders: high iodine, low selenium status. N. Z. Med. J. 104, 432.
Madeja, Z., Sroka, J., Nystrom, C., Bjorkhem-Bergman, L., Nordman, T., Damdimopoulos, A., Nalvarte, I., Eriksson, L. C., Spyrou, G., Olsson, J. M., and Bjornstedt, M. (2005). The role of thioredoxin reductase activity in selenium-induced cytotoxicity. Biochem. Pharmacol. 69, 1765–1772.
Maggini, S., Wintergerst, E. S., Beveridge, S., and Hornig, D. H. (2007). Selected vitamins and trace elements support immune function by strengthening epithelial barriers and cellular and humoral immune responses. Br. J. Nutr. 98(Suppl. 1), S29–S35.
Mahn, A. V., Munoz, M. C., and Zamorano, M. J. (2009). Discovery of biomarkers that reflect the intake of sodium selenate by nutritional proteomics. J. Chromatogr. Sci. 47, 840–843.
Mathers, J. C., and Hesketh, J. E. (2007). The biological revolution: understanding the impact of SNPs on diet-cancer interrelationships. J. Nutr. 137, 253S–258S.
Méplan, C. (2011). Trace elements and ageing, a genomic perspective using selenium as an example. J. Trace Elem. Med. Biol. 25(Suppl. 1), S11–S16.
Méplan, C., Crosley, L. K., Nicol, F., Beckett, G. J., Howie, A. F., Hill, K. E., Horgan, G., Mathers, J. C., Arthur, J. R., and Hesketh, J. E. (2007). Genetic polymorphisms in the human selenoprotein P gene determine the response of selenoprotein markers to selenium supplementation in a gender-specific manner (the SELGEN study). FASEB J. 21, 3063–3074.
Méplan, C., Crosley, L. K., Nicol, F., Horgan, G. W., Mathers, J. C., Arthur, J. R., and Hesketh, J. E. (2008). Functional effects of a common single-nucleotide polymorphism (GPX4c718t) in the glutathione peroxidase 4 gene: interaction with sex. Am. J. Clin. Nutr. 87, 1019–1027.
Méplan, C., Hughes, D. J., Pardini, B., Naccarati, A., Soucek, P., Vodickova, L., Hlavata, I., Vrana, D., Vodicka, P., and Hesketh, J. E. (2010). Genetic variants in selenoprotein genes increase risk of colorectal cancer. Carcinogenesis 31, 1074–1079.
Méplan, C., Nicol, F., Burtle, B. T., Crosley, L. K., Arthur, J. R., Mathers, J. C., and Hesketh, J. E. (2009). Relative abundance of selenoprotein P isoforms in human plasma depends on genotype, se intake, and cancer status. Antioxid. Redox Signal. 11, 2631–2640.
Mithen, R. (2007). Effect of genotype on micronutrient absorption and metabolism: a review of iron, copper, iodine and selenium, and folates. Int. J. Vitam. Nutr. Res. 77, 205–216.
Mueller, A. S., Mueller, K., Wolf, N. M., and Pallauf, J. (2009). Selenium and diabetes: an enigma? Free Radic. Res. 43, 1029–1059.
Navarro-Alarcon, M., and Cabrera-Vique, C. (2008). Selenium in food and the human body: a review. Sci. Total Environ. 400, 115–141.
Nelson, M. A., Porterfield, B. W., Jacobs, E. T., and Clark, L. C. (1999). Selenium and prostate cancer prevention. Semin. Urol. Oncol. 17, 91–96.
Nordman, T., Xia, L., Bjorkhem-Bergman, L., Damdimopoulos, A., Nalvarte, I., Arner, E. S. J., Spyrou, G., Eriksson, L. C., Bjornstedt, M., and Olsson, J. M. (2003). Regeneration of the antioxidant ubiquinol by lipoamide dehydrogenase, thioredoxin reductase and glutathione reductase. Biofactors 18, 45–50.
Novoselov, S. V., Kryukov, G. V., Xu, X.-M., Carlson, B. A., Hatfield, D. L., and Gladyshev, V. N. (2007). Selenoprotein H is a nucleolar thioredoxin-like protein with a unique expression pattern. J. Biol. Chem. 282, 11960–11968.
Pagmantidis, V., Meplan, C., Van Schothorst, E. M., Keijer, J., and Hesketh, J. E. (2008). Supplementation of healthy volunteers with nutritionally relevant amounts of selenium increases the expression of lymphocyte protein biosynthesis genes. Am. J. Clin. Nutr. 87, 181–189.
Papp, L. V., Lu, J., Holmgren, A., and Khanna, K. K. (2007). From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid. Redox Signal. 9, 775–806.
Parts, L., Stegle, O., Winn, J., and Durbin, R. (2011). Joint genetic analysis of gene expression data with inferred cellular phenotypes. PLoS Genet. 7, e1001276. doi: 10.1371/journal.pgen.1001276
Penney, K. L., Schumacher, F. R., Li, H., Kraft, P., Morris, J. S., Kurth, T., Mucci, L. A., Hunter, D. J., Kantoff, P. W., Stampfer, M. J., and Ma, J. (2010). A large prospective study of SEP15 genetic variation, interaction with plasma selenium levels, and prostate cancer risk and survival. Cancer Prev. Res. 3, 604–610.
Peters, U., Chatterjee, N., Hayes, R. B., Schoen, R. E., Wang, Y., Chanock, S. J., and Foster, C. B. (2008). Variation in the selenoenzyme genes and risk of advanced distal colorectal adenoma. Cancer Epidemiol. Biomarkers Prev. 17, 1144–1154.
Pico, A. R., Kelder, T., Van Iersel, M. P., Hanspers, K., Conklin, B. R., and Evelo, C. (2008). WikiPathways: pathway editing for the people. PLoS Biol. 6, e184. doi:10.1371/journal.pbio.0060184
Platz, E. A. (2010). Is prostate cancer prevention with selenium all in the genes? Cancer Prev. Res. 3, 576–578.
Rayman, M. P. (2008). Food-chain selenium and human health: emphasis on intake. Br. J. Nutr. 100, 254–268.
Rayman, M. P. (2009). Selenoproteins and human health: insights from epidemiological data. Biochim. Biophys. Acta 1790, 1533–1540.
Reid, M. E., Stratton, M. S., Lillico, A. J., Fakih, M., Natarajan, R., Clark, L. C., and Marshall, J. R. (2004). A report of high-dose selenium supplementation: response and toxicities. J. Trace Elem. Med. Biol. 18, 69–74.
Rusyniak, D. E., Arroyo, A., Acciani, J., Froberg, B., Kao, L., and Furbee, B. (2010). Heavy metal poisoning: management of intoxication and antidotes. EXS 100, 365–396.
Sakr, Y., Reinhart, K., Bloos, F., Marx, G., Russwurm, S., Bauer, M., and Brunkhorst, F. (2007). Time course and relationship between plasma selenium concentrations, systemic inflammatory response, sepsis, and multiorgan failure. Br. J. Anaesth. 98, 775–784.
Sarveswaran, S., Liroff, J., Zhou, Z., Nikitin, A. Y., and Ghosh, J. (2010). Selenite triggers rapid transcriptional activation of p53, and p53-mediated apoptosis in prostate cancer cells: implication for the treatment of early-stage prostate cancer. Int. J. Oncol. 36, 1419–1428.
Savaskan, N. E., Ufer, C., Kuhn, H., and Borchert, A. (2007). Molecular biology of glutathione peroxidase 4: from genomic structure to developmental expression and neural function. Biol. Chem. 388, 1007–1017.
Schoenmakers, E., Agostini, M., Mitchell, C., Schoenmakers, N., Papp, L., Rajanayagam, O., Padidela, R., Ceron-Gutierrez, L., Doffinger, R., Prevosto, C., Luan, J. A., Montano, S., Lu, J., Castanet, M., Clemons, N., Groeneveld, M., Castets, P., Karbaschi, M., Aitken, S., Dixon, A., Williams, J., Campi, I., Blount, M., Burton, H., Muntoni, F., O’donovan, D., Dean, A., Warren, A., Brierley, C., Baguley, D., Guicheney, P., Fitzgerald, R., Coles, A., Gaston, H., Todd, P., Holmgren, A., Khanna, K. K., Cooke, M., Semple, R., Halsall, D., Wareham, N., Schwabe, J., Grasso, L., Beck-Peccoz, P., Ogunko, A., Dattani, M., Gurnell, M., and Chatterjee, K. (2010). Mutations in the selenocysteine insertion sequence-binding protein 2 gene lead to a multisystem selenoprotein deficiency disorder in humans. J. Clin. Investig. 120, 4220–4235.
Schomburg, L., and Schweizer, U. (2009). Hierarchical regulation of selenoprotein expression and sex-specific effects of selenium. Biochim. Biophys. Acta 1790, 1453–1462.
Schrauzer, G. N. (2009). RE: lessons from the selenium and vitamin E cancer prevention trial (SELECT). Crit. Rev. Biotechnol. 29, 81.
Schrauzer, G. N., and Surai, P. F. (2009). Selenium in human and animal nutrition: resolved and unresolved issues. A partly historical treatise in commemoration of the fiftieth anniversary of the discovery of the biological essentiality of selenium, dedicated to the memory of Klaus Schwarz (1914–1978) on the occasion of the thirtieth anniversary of his death. Crit. Rev. Biotechnol. 29, 2–9.
Shi, X. W., Guo, X., Ren, F. L., Li, J., and Wu, X. M. (2010). The effect of short tandem repeat loci and low selenium levels on endemic osteoarthritis in China. J. Bone Joint Surg. Am. 92, 72–80.
Steinbrecher, A., Meplan, C., Hesketh, J., Schomburg, L., Endermann, T., Jansen, E., Akesson, B., Rohrmann, S., and Linseisen, J. (2010). Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol. Biomarkers Prev. 19, 2958–2968.
Steinbrenner, H., and Sies, H. (2009). Protection against reactive oxygen species by selenoproteins. Biochim. Biophys. Acta 1790, 1478–1485.
Sutherland, A., Kim, D.-H., Relton, C., Ahn, Y.-O., and Hesketh, J. (2010). Polymorphisms in the selenoprotein S and 15-kDa selenoprotein genes are associated with altered susceptibility to colorectal cancer. Genes Nutr. 5, 215–223.
Thompson, I. M., Tangen, C. M., Klein, E. A., and Lippman, S. M. (2005). Phase III prostate cancer prevention trials: are the costs justified? J. Clin. Oncol. 23, 8161–8164.
Thomson, C. D. (2004a). Assessment of requirements for selenium and adequacy of selenium status: a review. Eur. J. Clin. Nutr. 58, 391–402.
Thomson, C. D. (2004b). Selenium and iodine intakes and status in New Zealand and Australia. Br. J. Nutr. 91, 661–672.
Thomson, C. D., Mclachlan, S. K., Parnell, W. R., Wilson, N., Wohlers, M., Scragg, R., Schaaf, D., and Fitzgerald, E. D. (2007). Serum selenium concentrations and dietary selenium intake of New Zealand children aged 5–14 years. Br. J. Nutr. 97, 357–364.
Thomson, C. D., and Robinson, M. F. (1990). Selenium content of foods consumed in Otago, New Zealand. N. Z. Med. J. 103, 130–135.
Thomson, C. D., and Robinson, M. F. (1996). The changing selenium status of New Zealand residents. Eur. J. Clin. Nutr. 50, 107–114.
Tsavachidou, D., Mcdonnell, T. J., Wen, S., Wang, X., Vakar-Lopez, F., Pisters, L. L., Pettaway, C. A., Wood, C. G., Do, K.-A., Thall, P. F., Stephens, C., Efstathiou, E., Taylor, R., Menter, D. G., Troncoso, P., Lippman, S. M., Logothetis, C. J., and Kim, J. (2009). Selenium and vitamin E: cell type- and intervention-specific tissue effects in prostate cancer. J. Natl. Cancer Inst. 101, 306–320.
Van Ommen, B. (2007). Personalized nutrition from a health perspective: luxury or necessity? Genes Nutr. 2, 3–4.
van Ommen, B., Bouwman, J., Dragsted, L. O., Drevon, C. A., Elliott, R., De Groot, P., Kaput, J., Mathers, J. C., Muller, M., Pepping, F., Saito, J., Scalbert, A., Radonjic, M., Rocca-Serra, P., Travis, A., Wopereis, S., and Evelo, C. T. (2010a). Challenges of molecular nutrition research 6: the nutritional phenotype database to store, share and evaluate nutritional systems biology studies. Genes Nutr. 5, 189–203.
van Ommen, B., El-Sohemy, A., Hesketh, J., Kaput, J., Fenech, M., Evelo, C. T., McArdle, H. J., Bouwman, J., Lietz, G., Mathers, J. C., Fairweather-Tait, S., van Kranen, H., Elliott, R., Wopereis, S., Ferguson, L. R., Méplan, C., Perozzi, G., Allen, L., and Rivero, D. (2010b). The Micronutrient Genomics Project Working Group. The Micronutrient Genomics Project: a community-driven knowledge base for micronutrient research. Genes Nutr. 5, 285–296.
Van Ommen, B., Cavallieri, D., Roche, H. M., Klein, U. I., and Daniel, H. (2008). The challenges for molecular nutrition research 4: the “nutritional systems biology level.” Genes Nutr. 3, 107–113.
Villette, S., Kyle, J. A. M., Brown, K. M., Pickard, K., Milne, J. S., Nicol, F., Arthur, J. R., and Hesketh, J. E. (2002). A novel single nucleotide polymorphism in the 3′ untranslated region of human glutathione peroxidase 4 influences lipoxygenase metabolism. Blood Cells Mol. Dis. 29, 174–178.
Waters, D. J., Shen, S., Cooley, D. M., Bostwick, D. G., Qian, J., Combs, G. F. Jr., Glickman, L. T., Oteham, C., Schlittler, D., and Morris, J. S. (2003). Effects of dietary selenium supplementation on DNA damage and apoptosis in canine prostate. J. Natl. Cancer Inst. 95, 237–241.
Waters, D. J., Shen, S., Glickman, L. T., Cooley, D. M., Bostwick, D. G., Qian, J., Combs, G. F. Jr., and Morris, J. S. (2005). Prostate cancer risk and DNA damage: translational significance of selenium supplementation in a canine model. Carcinogenesis 26, 1256–1262.
Williams, C. M., Ordovas, J. M., Lairon, D., Hesketh, J., Lietz, G., Gibney, M., and Van Ommen, B. (2008). The challenges for molecular nutrition research 1: linking genotype to healthy nutrition. Genes Nutr. 3, 41–49.
Wishart, D. S. (2007). Proteomics and the human metabolome project. Expert Rev. Proteomics 4, 333–335.
Wishart, D. S. (2009). Computational strategies for metabolite identification in metabolomics. Bioanalysis 1, 1579–1596.
Wishart, D. S., Knox, C., Guo, A. C., Eisner, R., Young, N., Gautam, B., Hau, D. D., Psychogios, N., Dong, E., Bouatra, S., Mandal, R., Sinelnikov, I., Xia, J., Jia, L., Cruz, J. A., Lim, E., Sobsey, C. A., Shrivastava, S., Huang, P., Liu, P., Fang, L., Peng, J., Fradette, R., Cheng, D., Tzur, D., Clements, M., Lewis, A., De Souza, A., Zuniga, A., Dawe, M., Xiong, Y., Clive, D., Greiner, R., Nazyrova, A., Shaykhutdinov, R., Li, L., Vogel, H. J., and Forsythe, I. (2009). HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 37, D603–D610.
Wittwer, J., Rubio-Aliaga, I., Hoeft, B., Bendik, I., Weber, P., and Daniel, H. (2011). Nutrigenomics in human intervention studies: current status, lessons learned and future perspectives. Mol. Nutr. Food Res. 55, 1–18.
Xia, J., Bjorndahl, T. C., Tang, P., and Wishart, D. S. (2008). MetaboMiner – semi-automated identification of metabolites from 2D NMR spectra of complex biofluids. BMC Bioinformatics 9, 507. doi: 10.1186/1471-2105-9-507
Xia, J., Psychogios, N., Young, N., and Wishart, D. S. (2009). MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660.
Xia, J., and Wishart, D. S. (2010a). MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 26, 2342–2344.
Xia, J., and Wishart, D. S. (2010b). MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 38(Suppl.), W71–W77.
Xia, L., Nordman, T., Olsson, J. M., Damdimopoulos, A., Bjorkhem-Bergman, L., Nalvarte, I., Eriksson, L. C., Arner, E. S. J., Spyrou, G., and Bjornstedt, M. (2003). The mammalian cytosolic selenoenzyme thioredoxin reductase reduces ubiquinone. A novel mechanism for defense against oxidative stress. J. Biol. Chem. 278, 2141–2146.
Xiong, Y. M., Mo, X. Y., Zou, X. Z., Song, R. X., Sun, W. Y., Lu, W., Chen, Q., Yu, Y. X., and Zang, W. J. (2010). Association study between polymorphisms in selenoprotein genes and susceptibility to Kashin–Beck disease. Osteoarthr. Cartil. 18, 817–824.
Xu, X.-M., Carlson, B. A., Irons, R., Mix, H., Zhong, N., Gladyshev, V. N., and Hatfield, D. L. (2007a). Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem. J. 404, 115–120.
Xu, X.-M., Carlson, B. A., Zhang, Y., Mix, H., Kryukov, G. V., Glass, R. S., Berry, M. J., Gladyshev, V. N., and Hatfield, D. L. (2007b). New developments in selenium biochemistry: selenocysteine biosynthesis in eukaryotes and archaea. Biol. Trace Elem. Res. 119, 234–241.
Xue, W., Wang, Z., Chen, Q., Chen, J., Yang, H., and Xue, S. (2010). High selenium status in individuals exposed to arsenic through coal-burning in Shaanxi (PR of China) modulates antioxidant enzymes, heme oxygenase-1 and DNA damage. Clin. Chim. Acta 411, 1312–1318.
Yang, X.-E., Chen, W.-R., and Feng, Y. (2007). Improving human micronutrient nutrition through biofortification in the soil-plant system: China as a case study. Environ. Geochem. Health 29, 413–428.
Yuzbasioglu, A., Karatas, H., Gursoy-Ozdemir, Y., Saygi, S., Akalan, N., Soylemezoglu, F., Dalkara, T., Kocaefe, Y. C., and Ozguc, M. (2009). Changes in the expression of selenoproteins in mesial temporal lobe epilepsy patients. Cell. Mol. Neurobiol. 29, 1223–1231.
Zeisel, S. H. (2010). A grand challenge for nutrigenomics. Front. Genet. 1:2. doi: 10.3389/fgene.2010.00002
Zeng, H., and Botnen, J. H. (2007). Selenium is critical for cancer-signaling gene expression but not cell proliferation in human colon Caco-2 cells. Biofactors 31, 155–164.
Zhang, J., Dhakal, I. B., Lang, N. P., and Kadlubar, F. F. (2010a). Polymorphisms in inflammatory genes, plasma antioxidants, and prostate cancer risk. Cancer Causes Control 21, 1437–1444.
Zhang, S., Rocourt, C., and Cheng, W.-H. (2010b). Selenoproteins and the aging brain. Mech. Ageing Dev. 131, 253–260.
Keywords: selenium, selenoprotein, nutrigenetics, nutrigenomics
Citation: Ferguson LR and Karunasinghe N (2011) Nutrigenetics, nutrigenomics, and selenium. Front. Gene. 2:15. doi: 10.3389/fgene.2011.00015
Received: 28 January 2011; Paper pending published: 27 February 2011;
Accepted: 21 March 2011; Published online: 25 April 2011.
Edited by:
Sekhar S. Boddupalli, Monsanto Company, USAReviewed by:
Sander Kersten, Wageningen University, NetherlandsTony Merriman, University of Otago, New Zealand
Copyright: © 2011 Ferguson and Karunasinghe. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Lynnette R. Ferguson, Discipline of Nutrition, Faculty of Medical and Health Sciences, The University of Auckland, Private Bag 92019, Auckland 1142, New Zealand. e-mail: l.ferguson@auckland.ac.nz
