- U.S. Meat Animal Research Center, Agricultural Research Service, U.S. Department of Agriculture, Clay Center, NE, USA
The ovary and placenta are dynamic structures requiring constant modification both structurally and through cell–cell communication capabilities. The extracellular matrix and basement membranes are primarily composed of a milieu of glycosaminoglycans, including heparan sulfate and hyaluronan. Heparanase (HPSE) and hyaluronidases (HYAL) are responsible for degrading heparan sulfate and hyaluronan, respectively. Therefore, the objective of this study was to evaluate the relationship of SNPs distinct to HPSE, HYAL1, and HYAL2 with measurements of reproduction and production traits in swine. Single trait associations were performed on a Landrace–Duroc–Yorkshire population using SNPs discovered and identified in HPSE, HYAL1, and HYAL2. Analyses were conducted on an extended pedigree and SNPs were found to be associated with reproductive and production traits. Prior to multiple-testing corrections, SNPs within HPSE were weakly associated (P < 0.03) having additive effects with age at puberty (−2.5 ± 1.08 days), ovulation rate (0.5 ± 0.24 corpora lutea), and number of piglets born alive (0.9 ± 0.44 piglets). A HYAL1 and two HYAL2 SNP were nominally associated (P ≤ 0.0063) with number of piglets born alive after multiple-testing corrections (effects between 1.02 and 1.44 piglets), while one of the same HYAL2 markers maintained a modest association (P = 0.0043) having a dominant effect with number of piglets weaned (1.2 ± 0.41 piglets) after multiple-testing correction. Functionally, HPSE and HYAL1 and 2 have been shown to participate in events related to ovarian and placental activity. SNPs from these studies could potentially assist with understanding genetic components underlying sow lifetime productivity as measured by piglet survivability based on number born alive and number weaned, thereby contributing to a greater number of pigs/sow/year.
Introduction
Glycosaminoglycans contribute to a variety of tissue structures and comprise a large portion of the extracellular matrix. One significant glycosaminoglycan is heparan sulfate, which binds to and assembles extracellular matrix proteins contributing to its infrastructure, as well as other processes including tissue remodeling, vascularization, and metastatic events (Vlodavsky et al., 2002). Heparanase (HPSE) is the predominate endoglycosidase that degrades heparan sulfate. Porcine HPSE is located on SSC8 at approximately 116 cM (Miles et al., 2009). Suggested QTL for total number of piglets born (King et al., 2003), number of piglets born alive (King et al., 2003) and significant QTL for weight of ovary (Rohrer et al., 1999) have been found in this region on SSC8 spanning from ∼90 to 135 cM (Hu et al., 2007).
Breakdown of heparan sulfate by HPSE contributes to structural alterations to not only the extracellular matrix, but also to the basement membrane of tissue structures. Temporal levels of HPSE were evident in granulosa cells and macrophages of human and murine ovaries during the luteal phase and luteal regression, supporting a role for HPSE in extracellular matrix and basement membrane remodeling in the ovary (Haimov-Kochman et al., 2005).
Matrix metalloproteinases in association with HPSE have been implicated in endometrial remodeling during gestation in cattle and baboons ((Hashizume, 2007; D’Souza et al., 2008). In the porcine placenta, HPSE expression was elevated and localized to the cuboidal trophoblast cells (Miles et al., 2009). These findings suggest that HPSE may function to alter and modify the uterine environment and placenta thereby potentially impacting litter size and prenatal survivability.
Another endoglycosidase with implications in placental and ovulatory development is hyaluronidase (HYAL). Interaction between hyaluronan and CD44, its primary receptor, in expanding cumulus cells of the ovary affect oocyte maturation (Yokoo et al., 2010). Furthermore, it has been established that degradation of hyaluronan by HYAL is necessary for normal follicular development and maturation (Tempel et al., 2000). Hyaluronidase 1, 2, and 3 were localized to granulosa cells of murine ovaries (Orimoto et al., 2008) and were involved in induction of apoptosis and follicular atresia independent of their catalytic activity upon hyaluronan.
Increased levels of hyaluronan, along with decreased levels of HYAL3, within the decidua of gestating mice was correlated with spontaneous abortion in comparison to day 7.5 normal pregnancy mice (Cordo-Russo et al., 2009). Vallet et al. (2010) reported a putative role of stromal hyaluronidases in the structural development and angiogenesis of developing pig placentas.
The HYAL1–3 gene family resides on SSC13 (Gatphayak et al., 2004) and map to approximately 27 cM as determined by use of comparative radiation hybrid and linkage maps (Meyers et al., 2005). Within proximity to the HYAL genes, several QTL have been identified with implications to reproductive traits in swine (Hu et al., 2007). Two ovulation rate QTL (Rathje et al., 1997; Bidanel et al., 2008), an age at puberty QTL (Bidanel et al., 2008), and most recently a QTL for testosterone levels (Ren et al., 2009) have been reported in the same region on SSC13. Interestingly, a SNP within HYAL2 was associated with levels of testosterone and androstenone in Norwegian Landrace boars (Moe et al., 2009). Furthermore HYAL2 SNP haplotypes had a negative effect on testosterone levels, which may be indicative of reduced fertility.
Heparanse, HYAL1, and HYAL2 play a role in reproduction and production in swine and are located within close proximity to reported QTL for fertility and production. Therefore, the objective of this study was to determine if SNP within HPSE, HYAL1, or HYAL2 were associated with reproduction and production traits within a Landrace–Duroc–Yorkshire (LDY) resource population.
Materials and Methods
Animals
A founder animal population generated from Yorkshire × maternal Landrace (YL) was mated with commercially available semen from unrelated Duroc or high-lean Landrace boars. Offspring (Duroc × YL and Landrace × YL) were then reciprocally bred to create a 4-line composite LDY animal (Holl et al., 2004). The LDY population has been maintained by inter se matings for at least nine generations.
A basic corn–soybean meal diet was provided to females and was modified depending upon stage of growth or development. From 12 to 16 week of age, gilts received a grower diet containing 16% protein, which was reduced to 12.5% protein during the breeding phase; diets during gestation and lactation consisted of 16.9% protein, and returned to 12.5% protein post-weaning. Diets were formulated to meet or exceed National Research Council (NRC, 1998) recommendations.
All experimental procedures were reviewed and approved by the U.S. Meat Animal Research Center Animal Care and Use Committee.
SNP Discovery
SNPs in candidate genes were identified by extensive, but not complete gene sequencing (Tables 1 and 2). Briefly, sequencing primers were designed based on data from GenBank for Sus scrofa HPSE (FJ713408), HYAL1 (NM214441), and HYAL2 (NM214440) cDNA. Primers were designed in exons to amplify intronic regions. Location within the gene of forward and reverse primers, are presented in Table 1. Generation and sequencing of the coding regions for HYAL1 and HYAL2 were described previously (Vallet et al., 2010). Genomic DNA was isolated from semen of diverse breeds of unrelated pigs using phenol:chloroform extraction followed by ethanol precipitation. Amplification was performed using AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA), 1× of supplied buffer with 1.5 mM MgCl2, 200 μM dNTPs, 0.33 μM each primer, and 25 ng of genomic DNA in 25 μL reactions. Polymerase chain reaction was performed on a PTC-100 Programmable Thermal Controller machine (MJ Research Inc., Watertown, MA, USA) with appropriate annealing temperatures (Table 1). Quality of amplicon products was verified on a 1.6% agarose gel. The remaining PCR product was prepared for direct sequencing using the amplification primers on an ABI Prism 3730 DNA analyzer (Applied Biosystems). Sequence results were assembled and analyzed for polymorphisms as described previously (Rempel et al., 2010).
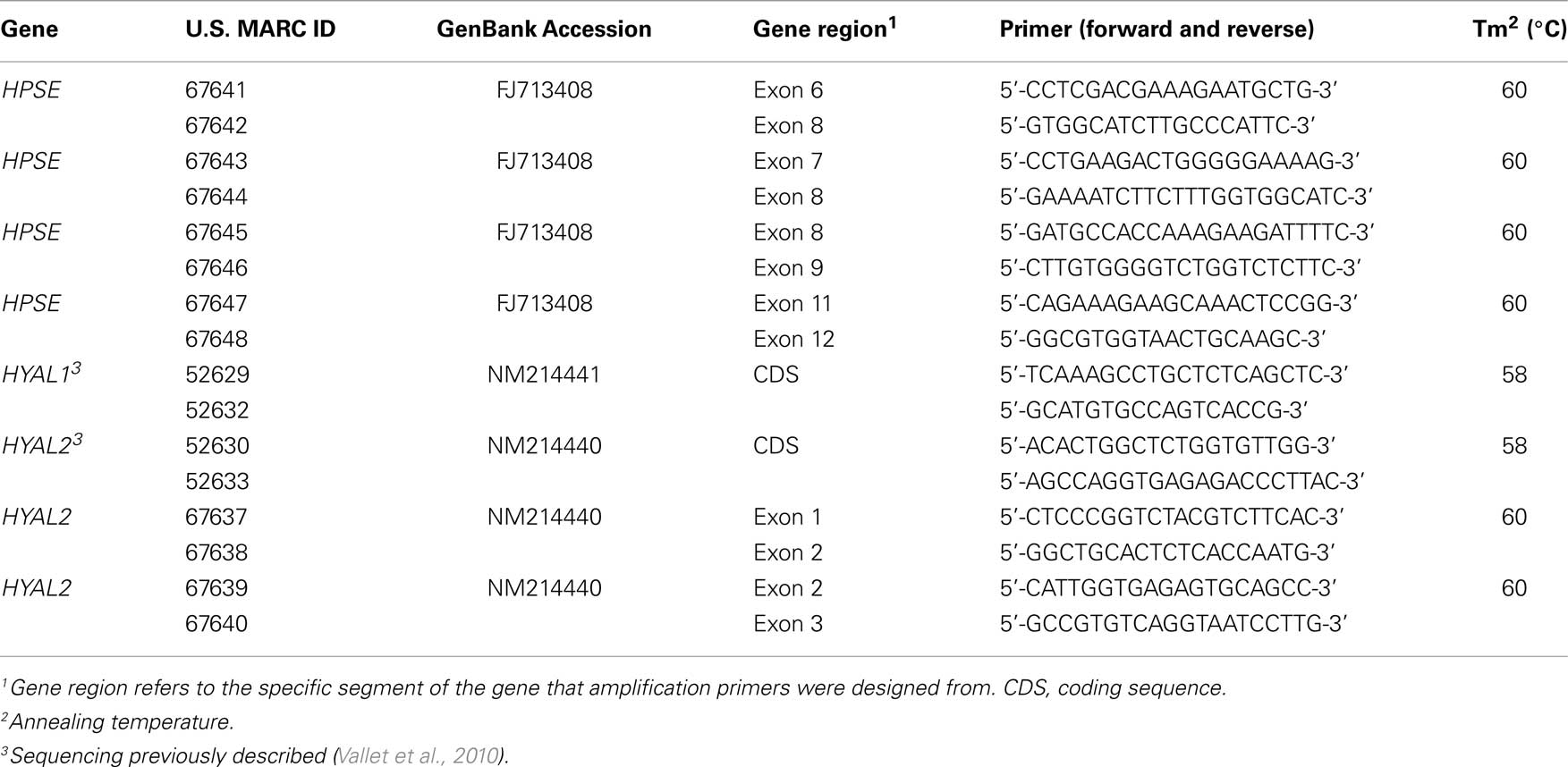
Table 1. Amplification primers for sequencing HPSE, HYAL 1, and HYAL2 for SNP detection and identification.
Genotyping
Genomic DNA was isolated from tail tissue using the Wizard DNA extraction kit (Promega, Madison, WI, USA) or leukocytes using a saturated salt extraction method (Miller et al., 1988). SNPs were genotyped using primer extension on the Sequenom MassArray system (San Diego, CA, USA). Amplification and probe primers for a subset (n = 12) of all discovered SNP were designed using the MassArray Assay Designer software (Sequenom). Ten microliter PCR reactions contained 10 ng of genomic DNA, 0.5 U HotStar Taq (Qiagen, Valencia, CA, USA), 1× of supplied buffer with 3.5 mM MgCl2, 250 μM dNTPs (Invitrogen, Carlsbad, CA, USA), and 100 nM forward and reverse tailed primers (Integrated DNA Technologies, Coralville, IA, USA). The primer extension reaction used 0.625–1.25 μM probe primer (Integrated DNA Technologies) and was performed according to the manufacturer’s recommendations for iPLEX chemistry (Sequenom).
Phenotypic Traits
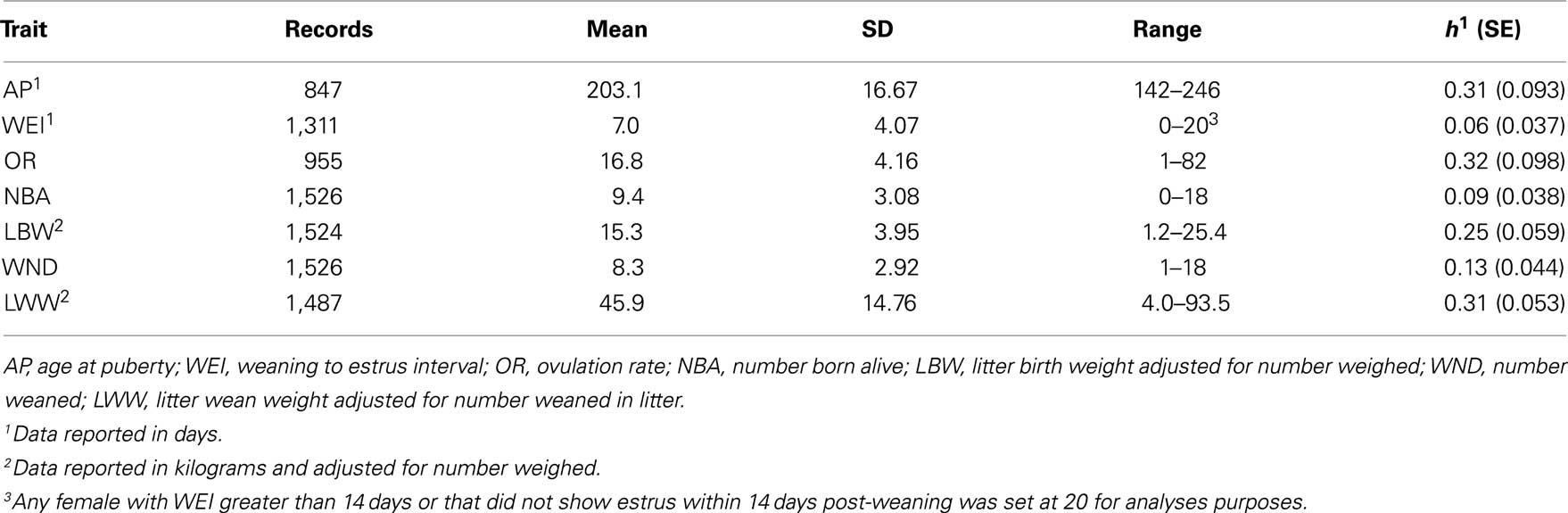
Phenotypic data were collected from the LDY population from 2004 to 2009 for association analyses (Table 3). Age at puberty was defined as the first detected standing estrus beginning at approximately 150 days of age and continued through 260 days of age. Estrus was detected using once daily fenceline exposure, with mature boars, while herdsmen applied back pressure to determine receptivity. Breeding females were bred by artificial insemination on a subsequent estrus during a set breeding season. Females were relocated into individual farrowing crates at approximately 110 days of gestation. Data on the total number of piglets born, number of piglets born alive, and litter birth weight were collected from sows at farrowing. Average lactation length was 18 days, at which time dams were separated from the piglets. Data were recorded for number of piglets weaned and litter weight at weaning. Following weaning sows were relocated and placed in group pen environments (20 sows/pen). Weaning to estrus interval was measured as the number of days following weaning until estrus was detected using back pressure by a herdsman during fenceline boar exposure. If a female failed to show estrus within 14 days post-weaning a value of 20 days was given for statistical purposes. Ovulation rate was determined as the number of corpora lutea at time of slaughter following an estrus event.
Statistical Analyses
Genotypic probabilities of the 12 SNPs for all animals in the extended pedigree (n = 3,056) were calculated using GenoProb software (Thallman et al., 2002), an allelic peeling algorithm program that can be used to predict ordered genotype distributions on pedigreed animals with incomplete genotypic data (Thallman et al., 2001a,b). GenoProb predicts both the missing genotypes and corrects genotyping errors.
In the preliminary analysis each trait was analyzed independently by MTDFREML (Boldman et al., 1995). The following details the models used to generate heritabilities and associations for SNP markers tested within the current dataset.
For age at puberty: yij = μ + animali + bgj + eij
For weaning to estrus interval: yikl = μ + animali + fgk + lactl + eikl
For ovulation rate: yiklm = μ + animali + fgk + lactl + OR agem + eiklm
For number of piglets born alive, litter birth weight, and number of piglets weaned: yikn = μ + animali + fgk + lit siren + eikn where animali is the random effect because of the ith female; bgj is the fixed effect because of the jth birth group the female was born; fgk is the fixed effect because of the kth farrowing group in which the dam farrowed; lactl is the covariate effect of the lth lactation length in days; OR agem is the covariate effect of the mth age in days of the dam at the time ovulation rate was collected; lit siren is the random effect of the nth sire used for artificial insemination. Both litter birth weight and number of piglets weaned included an adjustment for the number of piglets in each litter; number weighed and number weaned in litter, respectively.
The effects were calculated by using the genotypic probabilities computed by GenoProb described elsewhere (Allan et al., 2007; Kuehn et al., 2007). For each SNP, genotypic regressions for additive and dominance were in the model and run in a single-marker analysis. Additive regressions represented the number of copies of one allele (e.g., 0, 1, or 2). Dominance regressors were 0 for both homozygous genotypes and 1 for heterozygous genotypes. Because GenoProb produces probabilities of genotypes, actual regressors for each SNP were derived as the sum of the products of the probability of each genotype (e.g., AA, Aa, aa) and their regressor value (additive: 0, 1, 2 or dominance: 0, 1, 0). Heritability estimates were obtained from models with no SNP included; all subsequent SNP models used this heritability rather than re-estimating. Nominally significant (P < 0.10) SNP from the initial model are reported (Table 4). A Bonferroni correction for multiple-testing (n; *α = 0.10 or **α = 0.05; n = 12 SNP) was used to provide a preliminary measure of importance of these gene regions for the traits tested.
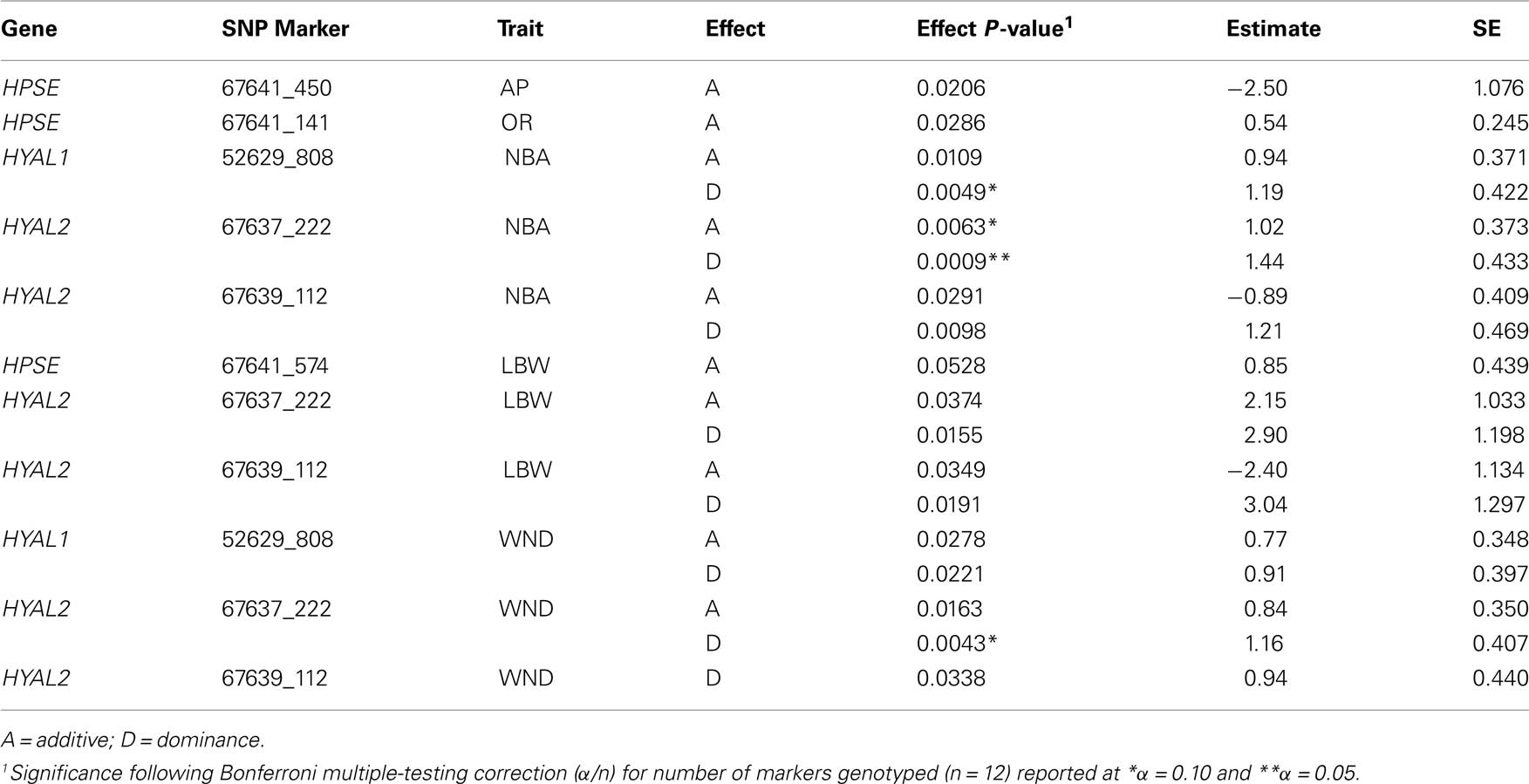
Table 4. Single trait association analysis of SNP with reproduction and production performance traits in a LDY swine population.
Results
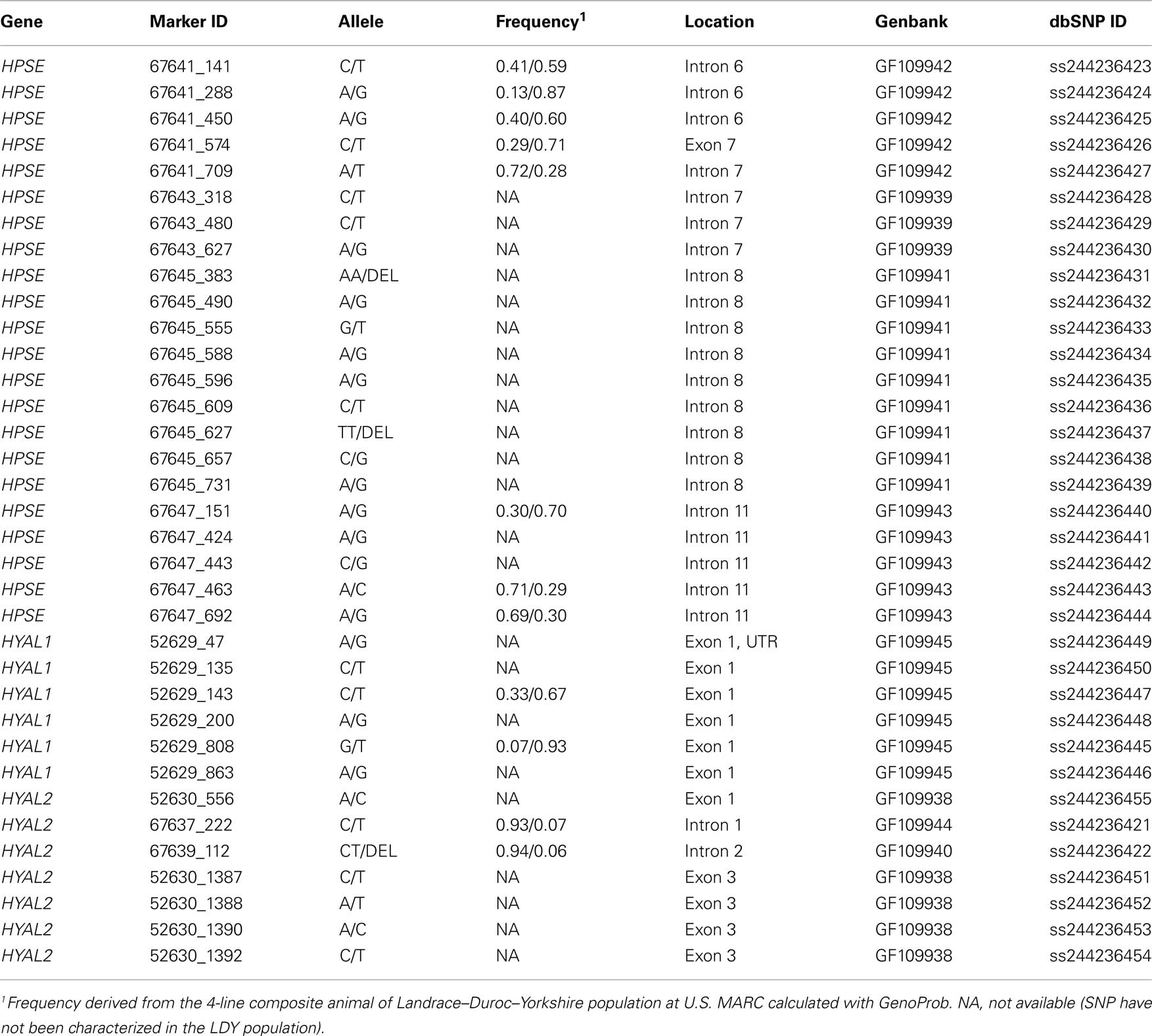
Partial gene sequencing of HPSE, HYAL1, and HYAL2 yielded a total of 35 SNP (Table 2). Three of the identified SNP within HYAL1 (52629_135, 52629_143, and 52629_808) have been previously reported (Gatphayak et al., 2004; Moe et al., 2009) and one SNP within HYAL2 (67637_222) has also been reported (Moe et al., 2009). However, these SNP had not been submitted to the GenBank dbSNP resource therefore all identified SNP were submitted using the nomenclature from the current study. Of the 35 SNPs, 12 were incorporated into an assay using MassArray Assay Design software (Sequenom). The subset of HPSE (8 markers), HYAL1 (2 markers), and HYAL2 (2 markers) SNPs were genotyped in a LDY population and had a minor allele frequency of 0.06 or greater (Table 2).
Summaries of phenotypic data analyzed including mean, SD, ranges, and heritabilities are presented in Table 3. All production and weaning to estrus interval data were collected from females following their first parity. Seven traits were analyzed using single locus analysis. Associations with P-values less than 0.05 prior to Bonferroni corrections are reported in Table 4. Three markers maintained nominal significance (α < 0.10) and one marker maintained significance (α < 0.05) after multiple-test correction (Table 4).
Reproductive traits, age at puberty and ovulation rate, evaluated within the current study had minimal associations (P < 0.03) when initial analyses were performed, but no associations were maintained following correction procedures.
Initial marker analyses identified putative associations for production traits including number of piglets born alive, litter birth weight, and number of piglets weaned for HPSE, HYAL1, and HYAL2. However associations maintained after correction for multiple-testing were restricted to number of piglets born alive and number of piglets weaned. A single HYAL1 marker, 52629_808, was nominally (P = 0.0049) associated with number of piglets born alive having a dominant effect of 1.19 ± 0.422 piglets. HYAL2 marker 67637_222 was associated with number of piglets born alive with an additive effect (P = 0.0063) of 1.02 ± 0.373 piglets and a dominant effect (P = 0.0009) of 1.44 ± 0.433 piglets. Similarly, the same HYAL2 marker had a dominant effect (P = 0.0043) of 1.16 ± 0.407 piglets for number of piglets weaned.
Discussion
In the current study, several SNPs were identified from three candidate genes, HPSE, HYAL1, and HYAL2. Biological investigations have shown a link among these genes and reproductive and production traits in swine. Therefore, we investigated the relationship between SNPs of these genes and reproductive and production traits.
Two individual HPSE markers had moderate associations with age at puberty or ovulation rate respectively, prior to correction for multiple-testing. Others (Cassady et al., 2001; Holl et al., 2004) have reported putative QTL for age at puberty on SSC8 within close proximity to the position of HPSE. Likewise a QTL for weight of ovary was also described in a Meishan–White composite population between 116 and 137 cM on SSC8 (Rohrer et al., 1999). An age at puberty study using a White Duroc–Erhualian population detected a peak at 77 cM on SSC8 using a 5% chromosome-wide threshold, but a secondary peak just below the threshold was notable at approximately 125 cM within the region of HPSE (Yang et al., 2008). It is also worth noting that Buske et al. (2006) compiled several fertility-related QTL studies and found a predominant clustering of QTL across SSC8 for reproductive traits in swine. Likely the markers we have genotyped within HPSE were not in strong linkage disequilibrium with the causative mutation. But rather we may infer that our proximity on chromosome 8 was relevant based on our moderate association results and supporting literature from others. Another possible candidate gene is secreted phosphoprotein 1 (SPP1), which has been shown to have an impact on conceptus–embryo interactions during implantation in several mammalian species including swine (Ross et al., 2007; Erikson et al., 2009; Kim et al., 2010) and is located at approximately 112 cM on SSC8. Furthermore polymorphisms within SPP1 have been associated with male fertility traits, sperm motility, and abnormal spermatozoa rate, within Pietrain and Pietrain × Hampshire boars (Lin et al., 2006). Secreted phosphoprotein 1 may also harbor mutations linked to fertility traits, but to date no associations for age at puberty or ovulation rate have been reported for SNP within the SPP1 gene either.
An SNP within HYAL2 had moderate associations with number of piglets born alive and associated with number of piglets weaned following multiple-test corrections within the current study. Moe et al. (2009) identified the same SNP within Duroc and Norwegian Landrace breeds. The minor allele frequency in Duroc was less than 1% and nearly 49% in Norwegian Landrace. The HYAL2 SNP was associated independently and within a haplotype analysis with various androgen levels in the plasma of Norwegian Landrace boars, suggesting a relationship of this SNP with fertility-related traits (Moe et al., 2009). Our genotyping efforts identified a minor allele frequency of 7% for the same SNP within the composite Duroc, Yorkshire, and double Landrace-derived population. Enough genetic discrepancy may exist among the breeds used within these two reports. Nonetheless, the HYAL2 marker, 67637_222, was modestly associated with number of piglets born alive and maintained an association with number of piglets weaned following multiple-testing correction in the current study. The HYAL gene family resides near 27 cM on SSC13. No QTL for number of piglets weaned has been reported on SSC13. However, others (Noguera et al., 2009) have reported a single-dimensional model QTL for number of piglets born alive and total number of piglets born within a Meishan × Iberian population on SSC13 (50 cM) and further described separate epistatic interacting QTLs on SSC13 (66–82 cM) and SSC9 (1–7 cM) for number of piglets born alive using a bi-dimensional modeling system. Both of these reported QTL lie distal to HYAL1–3 on SSC13. In the current study, SNP were analyzed under single trait conditions, but in the future bi-dimensional analyses using the Illumina 60K BeadChip data along with additional independently genotyped SNP data may provide guidance and insight into regions with independent as well as epistatic interactions. These putative studies may confirm involvement of the region where the HYAL gene family resides or provide a more refined region on SSC13 for number of piglets born alive and the highly correlated trait of number weaned within close proximity to the HYAL1–3 genes.
Conclusion
The complexity of the reproductive and production traits evaluated within this study increase the difficulty in ascertaining the impact of single candidate gene markers for HPSE, HYAL1, and HYAL2. However, previous reports identified the importance of these two genes for both ovarian and uterine/placental function, which can have an influence on litter size and piglet survivability. HYAL1 and HYAL2 SNP had putative effects on number of piglets born alive as well as number of piglets weaned based upon limited association levels in the current study. These candidate SNP likely have minimal, if any, linkage disequilibrium with the mutation that affects these traits, however linkage was not assessed in this study due to inherent reduction in statistical power. It is also plausible that these intricate traits are influenced by multiple genes working in a coordinated effort. Future use of genome-wide association analyses for swine production and reproduction traits will further refine regions of interest on SSC8 and SSC13 for greater interrogation that will more than likely lead to discovery of causative nucleotides or responsible genes or gene networks. However the use of SNP within HPSE, HYAL1, and HYAL2 do not appear to be suitable for marker assisted selection for reproduction and production traits within the population tested.
Conflict of Interest Statement
Mention of trade name proprietary product or specified equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable. USDA is an equal opportunity provider and employer.
Acknowledgments
The authors gratefully acknowledge Troy Gramke, Suzy Hassler, Sue Hauver, Mike Judy, Sandra Nejzechleb, and Kris Simmerman for technical assistance; Linda Parnell for manuscript preparation; Jim Wray for sequence submissions; and the U.S. MARC Swine Operations employees for animal management and data collection. Funding for this study was provided by USDA CRIS Project No. 5438-31000-084-00D.
References
Allan, M. F., Thallman, R. M., Cushman, R. A., Echternkamp, S. E., White, S. N., Kuehn, L. A., Casas, E., and Smith, T. P. (2007). Association of a single nucleotide polymorphism in SPP1 with growth traits and twinning in a cattle population selected for twinning rate. J. Anim. Sci. 85, 341–347.
Bidanel, J. P., Rosendo, A., Iannuccelli, N., Riquet, J., Gilbert, H., Caritez, J. C., Billon, Y., Amigues, Y., Prunier, A., and Milan, D. (2008). Detection of quantitative trait loci for teat number and female reproductive traits in Meshan x Large White F2 pigs. Animal 2, 813–820.
Boldman, K. G., Kriese, L. A., Van Vleck, L. D., Van Tassell, C. P., and Kachman, S. D. (1995). A Manual for Use of MTDFREML. A Set of Programs to Obtain Estimates of Variances and Covariances. Lincoln, NE: USDA ARS
Buske, B., Sternstein, I., and Brockmann, G. (2006). QTL and candidate genes for fecundity in sows. Anim. Reprod. Sci. 95, 167–183.
Cassady, J. P., Johnson, R. K., Pomp, D., Rohrer, G. A., Van Vleck, L. D., Spiegel, E. K., and Gilson, K. M. (2001). Identification of quantitative trait loci affecting reproduction in pigs. J. Anim. Sci. 79, 623–633.
Cordo-Russo, R., Garcia, M. G., Barrientos, G., Orsal, A. S., Viola, M., Moschansky, P., Ringel, F., Passi, A., Alaniz, L., Hajos, S., and Blois, S. M. (2009). Murine abortion is associated with enhanced hyaluronan expression and abnormal localization at the fetomaternal interface. Placenta 30, 88–95.
D’Souza, S. S., Fazleabas, A. T., Banerjee, P., Sherwin, J. R., Sharkey, A. M., Farach-Carson, M. C., and Carson, D. D. (2008). Decidual heparanase activity is increased during pregnancy in the baboon (Papio anubis) and in in vitro decidualization of human stromal cells. Biol. Reprod. 78, 316–323.
Erikson, D. W., Burghardt, R. C., Bayless, K. J., and Johnson, G. A. (2009). Secreted phosphoprotein 1 (SPP1, osteopontin) binds to integrin alpha v beta 6 on porcine trophectoderm cells and integrin alpha v beta 3 on uterine luminal epithelial cells, and promotes trophectoderm cell adhesion and migration. Biol. Reprod. 81, 814–825.
Gatphayak, K., Knorr, C., Beck, J., and Brenig, B. (2004). Molecular characterization of porcine hyaluronidase genes 1, 2, and 3 clustered on SSC13q21. Cytogenet. Genome Res. 106, 98–106.
Haimov-Kochman, R., Prus, D., Zcharia, E., Goldman-Wohl, D. S., Natanson-Yaron, S., Greenfield, C., Anteby, E. Y., Reich, R., Orly, J., Tsafriri, A., Hurwitz, A., Vlodavsky, I., and Yagel, S. (2005). Spatiotemporal expression of heparanase during human and rodent ovarian folliculogenesis. Biol. Reprod. 73, 20–28.
Hashizume, K. (2007). Analysis of uteroplacental-specific molecules and their functions during implantation and placentation in the bovine. J. Reprod. Dev. 53, 1–11.
Holl, J. W., Cassady, J. P., Pomp, D., and Johnson, R. K. (2004). A genome scan for quantitative trait loci and imprinted regions affecting reproduction in pigs. J. Anim. Sci. 82, 3421–3429.
Hu, Z. L., Fritz, E. R., and Reecy, J. M. (2007). AnimalQTLdb: a livestock QTL database tool set for positional QTL information mining and beyond. Nucleic Acids Res. 35, D604–D609.
Kim, J., Erikson, D. W., Burghardt, R. C., Spencer, T. E., Wu, G., Bayless, K. J., Johnson, G. A., and Bazer, F. W. (2010). Secreted phosphoprotein 1 binds integrins to initiate multiple cell signaling pathways, including FRAP1/mTOR, to support attachment and force-generated migration of trophectoderm cells. Matrix Biol. 29, 369–382.
King, A. H., Jiang, Z., Gibson, J. P., Haley, C. S., and Archibald, A. L. (2003). Mapping quantitative trait loci affecting female reproductive traits on porcine chromosome 8. Biol. Reprod. 68, 2172–2179.
Kuehn, L. A., Rohrer, G. A., Nonneman, D. J., Thallman, R. M., and Leymaster, K. A. (2007). Detection of single nucleotide polymorphisms associated with ultrasonic backfat depth in a segregating Meishan x White Composite population. J. Anim. Sci. 85, 1111–1119.
Lin, C., Tholen, E., Jennen, D., Ponsuksili, S., Schellander, K., and Wimmers, K. (2006). Evidence for effects of testis and epididymis expressed genes on sperm quality and boar fertility traits. Reprod. Domest. Anim. 41, 538–543.
Meyers, S. N., Rogatcheva, M. B., Larkin, D. M., Yerle, M., Milan, D., Hawken, R. J., Schook, L. B., and Beever, J. E. (2005). Piggy-BACing the human genome II. A high-resolution, physically anchored, comparative map of the porcine autosomes. Genomics 86, 739–752.
Miles, J. R., Vallet, J. L., Freking, B. A., and Nonneman, D. J. (2009). Molecular cloning and characterisation of heparanase mRNA in the porcine placenta throughout gestation. Reprod. Fertil. Dev. 21, 757–772.
Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16, 1215.
Moe, M., Lien, S., Aasmundstad, T., Meuwissen, T. H., Hansen, M. H., Bendixen, C., and Grindflek, E. (2009). Association between SNPs within candidate genes and compounds related to boar taint and reproduction. BMC Genet. 10, 32. doi:10.1186/1471-2156-10-32
Noguera, J. L., Rodriguez, C., Varona, L., Tomas, A., Munoz, G., Ramirez, O., Barragan, C., Arque, M., Bidanel, J. P., Amills, M., Ovilo, C., and Sanchez, A. (2009). A bi-dimensional genome scan for prolificacy traits in pigs shows the existence of multiple epistatic QTL. BMC Genomics 10, 636. doi:10.1186/1471-2164-10-636
Orimoto, A. M., Dumaresq-Doiron, K., Jiang, J. Y., Tanphaichitr, N., Tsang, B. K., and Carmona, E. (2008). Mammalian hyaluronidase induces ovarian granulosa cell apoptosis and is involved in follicular atresia. Endocrinology 149, 5835–5847.
Rathje, T. A., Rohrer, G. A., and Johnson, R. K. (1997). Evidence for quantitative trait loci affecting ovulation rate in pigs. J. Anim. Sci. 75, 1486–1494.
Rempel, L. A., Nonneman, D. J., Wise, T. H., Erkens, T., Peelman, L. J., and Rohrer, G. A. (2010). Association analyses of candidate single nucleotide polymorphisms on reproductive traits in swine. J. Anim. Sci. 88, 1–15.
Ren, D. R., Ren, J., Xing, Y. Y., Guo, Y. M., Wu, Y. B., Yang, G. C., Mao, H. R., and Huang, L.-S. (2009). A genome scan for quantitative trait loci affecting male reproductive traits in a White Duroc x Chinese Erhualian resource population. J. Anim. Sci. 87, 17–23.
Rohrer, G. A., Ford, J. J., Wise, T. H., Vallet, J. L., and Christenson, R. K. (1999). Identification of quantitative trait loci affecting female reproductive traits in a multigeneration Meishan-White composite swine population. J. Anim. Sci. 77, 1385–1391.
Ross, J. W., Ashworth, M. D., White, F. J., Johnson, G. A., Ayoubi, P. J., DeSilva, U., Whitworth, K. M., Prather, R. S., and Geisert, R. D. (2007). Premature estrogen exposure alters endometrial gene expression to disrupt pregnancy in the pig. Endocrinology 148, 4761–4773.
Tempel, C., Gilead, A., and Neeman, M. (2000). Hyaluronic acid as an anti-angiogenic shield in the preovulatory rat follicle. Biol. Reprod. 63, 134–140.
Thallman, R. M., Bennett, G. L., Keele, J. W., and Kappes, S. M. (2001a). Efficient computation of genotype probabilities for loci with many alleles: I. Allelic peeling. J. Anim. Sci. 79, 26–33.
Thallman, R. M., Bennett, G. L., Keele, J. W., and Kappes, S. M. (2001b). Efficient computation of genotype probabilities for loci with many alleles: II. Iterative method for large, complex pedigrees. J. Anim. Sci. 79, 34–44.
Thallman, R. M., Cooke, D. B., and Bennett, G. L. (2002). “GenoProb: Computation of genotype and grandparental origin probabilities in complex pedigrees with missing marker data,” in 7th World Congress on Genetics Applied to Livestock Production, pp. Comm. No. 28-09, Montpellier, France.
Vallet, J. L., Miles, J. R., and Freking, B. A. (2010). Effect of fetal size on fetal placental hyaluronan and hyaluronoglucosaminidases throughout gestation in the pig. Anim. Reprod. Sci. 118, 297–309.
Vlodavsky, I., Goldshmidt, O., Zcharia, E., Atzmon, R., Rangini-Guatta, Z., Elkin, M., Peretz, T., and Friedmann, Y. (2002). Mammalian heparanase: involvement in cancer metastasis, angiogenesis and normal development. Semin. Cancer Biol. 12, 121–129.
Yang, G., Ren, J., Li, S., Mao, H., Guo, Y., Zou, Z., Ren, D., Ma, J., and Huang, L. (2008). Genome-wide identification of QTL for age at puberty in gilts using a large intercross F2 population between White Duroc x Erhualian. Genet. Sel. Evol. 40, 529–539.
Keywords: reproductive traits, production traits, heparanase, hyaluronidase, SNP, swine
Citation: Rempel LA, Freking BA, Miles JR, Nonneman DJ, Rohrer GA, Schneider JF and Vallet JL (2011) Association of porcine heparanase and hyaluronidase 1 and 2 with reproductive and production traits in a Landrace–Duroc–Yorkshire population. Front. Gene. 2:20. doi: 10.3389/fgene.2011.00020
Received: 02 March 2011; Accepted: 22 April 2011;
Published online: 04 May 2011.
Edited by:
Jean Michel Claude Elsen, National Institute of Agronomic Research, FranceReviewed by:
Mairead Lesley Bermingham, The University of Edinburgh, UKPierre Mormede, French National Institute for Agricultural Research, France
Copyright: © 2011 Rempel, Freking, Miles, Nonneman, Rohrer, Schneider and Vallet. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Lea A. Rempel, Reproduction Research Unit, U.S. Meat Animal Research Center, Agricultural Research Service, U.S. Department of Agriculture, State Spur 18D, P.O. Box 166, Clay Center, NE 68933-0166, USA. e-mail: lea.rempel@ars.usda.gov