- 1 Division of Gastroenterology and Hepatology, Department of Internal Medicine, School of Medicine, Keio University, Tokyo, Japan
- 2 Division of Pharmacotherapeutics, Faculty of Pharmacy, Keio University, Tokyo, Japan
MicroRNAs (miRNAs) are small non-coding RNAs that function as endogenous silencers of numerous target genes. Hundreds of miRNAs have been identified in the human genome. miRNAs are expressed in a tissue-specific manner and play important roles in cell proliferation, apoptosis, and differentiation. Aberrant expression of miRNAs may also contribute to the development and progression of human hepatobiliary and pancreatic cancers. Recent studies have shown that some miRNAs play roles as tumor suppressors or oncogenes in hepatobiliary and pancreatic cancers. miR-122, let-7 family, and miR-101 are down-regulated in hepatocellular carcinoma (HCC), suggesting that it is a potential tumor suppressor of HCC. miR-221 and miR-222 are up-regulated in HCC and may act as oncogenic miRNAs in hepatocarcinogenesis. miRNA expression profiling may be a powerful clinical tool for diagnosis and regulation of miRNA expression could be a novel therapeutic strategy for hepatobiliary and pancreatic cancers. In this review, we summarize current knowledge about the roles of important tumor suppressor microRNAs and oncogenic microRNAs in hepatobiliary and pancreatic cancers.
The Biogenesis of microRNAs
MicroRNAs (miRNAs) are ∼22 nucleotide (nt) non-coding RNAs that can post-transcriptionally downregulate the expression of various target genes. Currently, ∼1400 human miRNAs have been identified in the human genome, and each miRNA potentially controls hundreds of target genes. miRNAs are expressed in a tissue-specific manner and play important roles in cell proliferation, apoptosis, and differentiation. Moreover, recent studies have shown a link between aberrant expression of miRNAs and the development of cancer (Saito et al., 2009a; Calin and Croce, 2007; Cho, 2007).
In animals, miRNA genes are generally transcribed by RNA polymerase II (pol II) to form primary transcripts (pri-miRNAs). Pol II transcribed pri-miRNAs are capped with 7-methylguanosine and polyadenylated. The nuclear RNase III enzyme Drosha and its co-factor Pasha (also known as DGCR8) process pri-miRNAs into ∼60 nt precursor miRNAs (pre-miRNAs) which form an imperfect stem–loop structure. Pre-miRNAs are transported into the cytoplasm by the RAN GTP-dependent transporter exportin 5 and are subsequently cleaved by the cytoplasmic RNase III enzyme Dicer into mature miRNAs which are then loaded into the RNA-induced silencing complex (RISC). The miRNA/RISC complex downregulates specific gene products by translational repression via binding to partially complementary sequences in the 3′ untranslated regions (3′-UTRs) of the target mRNAs or by directing mRNA degradation via binding to perfectly complementary sequences.
Identification of miRNA target genes is critical to determining the miRNA functions. There are several databases of human miRNA target predictions using different algorithms such as microRNA.org1 miRBase2 PicTar3 and TargetScan4 It is generally believed that conserved perfect 6–8 base-pair (bp) matches between the 5′ end of the mature miRNA and the 3′ UTR of the predicted target mRNA (called “seed” matches) are the most reliable way of determining miRNA targets. Experimental confirmation using protein expression and luciferase reporter assays for the miRNA targets is necessary to accurately identify the target genes of miRNAs.
Regulatory Mechanisms of miRNA Expression
Since miRNAs can have large-scale effects through regulation of a variety of genes during mammalian development and carcinogenesis, an understanding of the regulatory mechanisms controlling miRNA expression, which is quite limited at present, is important. There are several reports of transcription factors binding to the promoter regions of specific miRNA genes and activating the transcription of pri-miRNAs, resulting in increased expression of mature miRNAs. c-Myc binds to the regulatory region of the miR-17-92 cluster and increased expression of c-Myc leads to the activation of the miRNAs in the cluster (O’Donnell et al., 2005). The location of miRNA genes is also an important factor in the regulation of miRNA expression. Many miRNAs are located in cancer-associated genomic regions that are frequently involved in chromosomal abnormalities, such as loss of heterozygosity (LOH), amplification, and breakpoints (Calin et al., 2004). Chromosomal abnormalities during carcinogenesis could lead to widespread differential expressions of miRNAs in human cancer cells.
Epigenetic alterations such as DNA methylation and histone modification play critical roles in chromatin remodeling and regulation of gene expression in mammalian development and in human diseases. Many miRNAs are expressed in a tissue and tumor specific manner, implying that some miRNAs are subject to epigenetic control. We have recently shown that miR-127, which is embedded in a CpG island, is strongly induced by the treatment with the DNA demethylating agent 5-aza-2′-deoxycytidine (5-Aza-CdR) and the histone deacetylase (HDAC) inhibitor 4-phenylbutyric acid (PBA), indicating that some miRNA genes are controlled by epigenetic alterations in their promoter regions and can be activated by chromatin modifying drugs (Saito et al., 2006, 2009b).
MicroRNAs expression profiles can be used to classify the developmental lineages and differentiation stages of tumors. Interestingly, miRNA expression profiles are more accurate for tumor classification than conventional mRNA profiling (Lu et al., 2005). Furthermore, recent studies have demonstrated the association of miRNA expression signatures with prognostic and disease progression factors in human malignancies (Calin and Croce, 2006). These findings indicate that miRNA expression profile might be a powerful clinical tool for the diagnosis and prognosis of malignancies.
Important Tumor Suppressor and Oncogenic miRNAs in HCC
Hepatocellular carcinoma (HCC) is the most common type of liver cancer. Most cases of HCC are secondary to either chronic hepatitis or hepatic cirrhosis caused by a viral infection (hepatitis B or C) or alcoholism. HCC accounts for 85–90% of all primary liver cancers and is one of the most lethal cancers, and affects many of the world’s population (Farazi and DePinho, 2006). The lethality of liver cancer may cause from its resistance to existing anticancer agents, a lack of biomarkers, and underlying liver disease that limits the use of chemotherapeutic drugs. In addition, the molecular pathogenesis of HCC remains poorly understood.
As shown in Table 1, some miRNAs are overexpressed in HCCs, indicating that they may have roles as oncogenes accelerating the development of HCC. Some miRNAs are down-regulated in HCCs, suggesting that they may act as tumor suppressors. miRNAs have critical roles in human hepatocarcinogenesis and that aberrant expression of miRNAs may contribute to the initiation and progression of HCC.
miR-122
miR-122 is specifically expressed and highly abundant in the human liver. Recent studies have reported that miR-122 may modulate and facilitate replication of hepatitis C virus (HCV), suggesting that miR-122 could be a target for antiviral intervention (Jopling et al., 2005; Li et al., 2011).
miR-122 can modulate cyclin G1 expression in HCC-derived cell lines and an inverse correlation between miR-122 and cyclin G1 expression exists in HCCs, indicating that cyclin G1 is a target of miR-122 (Gramantieri et al., 2007). In patients of HCC, lower miR-122 levels were associated with a shorter time to recurrence, whereas higher cyclin G1 expression was related to a lower survival, suggesting that miR-122 might represent an effective molecular target for HCC (Fornari et al., 2009).
miR-122 also modulates Bcl-w expression by directly targeting binding site within the 3′-UTR. The cellular mRNA and protein levels of Bcl-w were repressed by elevated levels of miR-122, which subsequently led to reduction of cell viability and activation of caspase-3, suggesting that Bcl-w is a direct target of miR-122 that functions as an endogenous apoptosis enhancer in HCC cells (Lin et al., 2008). Ma et al. (2010) generated a recombinant adenoviral vector expressing miR-122 (Ad-miR-122). Infection of tumor cells with Ad-miR-122 resulted in inhibition of growth of cancer cells. This antitumor activity was related to the induction of apoptosis and/or cell-cycle arrest in cancer cells. Infection with Ad-miR-122 resulted in decreased expression of Bcl-w and CCNG1 in cancer cells. Induction of miR-122 via adenoviral vector could be a promising strategy for cancer treatment (Ma et al., 2010).
A distintegrin and metalloprotease family 10 (ADAM10), serum response factor (SRF), and insulin-like growth factor 1 receptor (Igf1R) that promote tumorigenesis were validated as targets of miR-122. ADAM10, SRF, and Igf1R were up-regulated in primary human HCCs compared with the matching liver tissue, suggesting that the loss of multifunctional miR-122 contributes to the malignant phenotype of HCC cells (Bai et al., 2009).
Tsai et al. (2009) have shown that miR-122 is significantly down-regulated in HCCs with intrahepatic metastasis and negatively regulates tumorigenesis. Restoration of miR-122 in metastatic cells significantly reduced in vitro migration, invasion, and anchorage-independent growth as well as in vivo tumorigenesis, angiogenesis, and intrahepatic metastasis. They have further shown that one of the miR-122 targets, a disintegrin, and metalloprotease 17 (ADAM17) is involved in metastasis. Silencing of ADAM17 resulted in a dramatic reduction of in vitro migration, invasion, in vivo tumorigenesis, angiogenesis, and local invasion in the livers of nude mice, indicating that miR-122 plays a role in the intrahepatic metastasized HCC by the suppression of angiogenesis via regulation of ADAM17 (Tsai et al., 2009).
These studies demonstrate that miR-122 is significantly down-regulated in HCCs and has multiple functions as a tumor suppressor miRNA during hepatocarcinogenesis. miR-122 is a promising target for HCC treatment as well as a diagnostic and prognostic marker for the progression of HCC. miR-122 alone or in combination with anticancer drugs can be a promising therapeutic strategy against HCC. Further studies are necessary to develop miRNA-based therapy for HCC treatment. Since miR-122 can modulate HCV replication, the status of HCV infection in HCC patients should be detected carefully.
let-7 family
The let-7 family plays critical roles in tumorigenesis by functioning as potential tumor suppressor. The expression of let-7g was markedly decreased in HCC cells. Proliferation of HCC cell line was significantly inhibited after the transfection of let-7g. Concurrently, the mRNA and protein levels of c-Myc were found significantly decreased in HCC cells after transfection of let-7g. let-7g may act as a tumor suppressor gene that inhibits HCC cell proliferation by downregulating c-Myc (Lan et al., 2011). In addition, the level of let-7g was significantly lower in metastatic HCCs compared to metastasis-free HCCs. The low expression level of let-7g in tumor was predictive of poor survival in HCC patients. Type I collagen alpha2 (COL1A2) was validated as a direct target of let-7g, suggesting that let-7g may suppress HCC metastasis through targeting COL1A2 (Ji et al., 2010).
Bcl-xL, an anti-apoptotic member of the Bcl-2 family, is identified as a target of let-7c and let-7g. Human HCC tissues with low expression of let-7c displayed higher expression of Bcl-xL protein than those with high expression of let-7c, suggesting that low let-7 miRNA expression contributes to Bcl-xL overexpression. let-7 miRNAs negatively regulate Bcl-xL expression and induce apoptosis in human HCCs (Shimizu et al., 2010).
miR-101
Expression level of miR-101 was significantly decreased in HCC cell lines and HCC tissues compared with their non-tumor counterparts. Ectopic expression of miR-101 dramatically suppressed the ability of HCC cells to form colonies in vitro and to develop tumors in nude mice. In addition, miR-101 repressed Mcl-1 expression as its target oncogene. These results indicate that miR-101 may exert its proapoptotic function via targeting Mcl-1 (Su et al., 2009). Li et al. (2009) have reported that miR-101 was significantly down-regulated in HCC tissues compared with the matching non-tumor liver tissues. They also showed that miR-101 repressed the expression of v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene, a key component of the activator protein-1 (AP-1) transcription factor. In in vitro invasion and migration assays, enhanced miR-101 expression inhibited the invasion and migration of cultured HCC cells. These findings suggest that miR-101, which is aberrantly expressed in HCC, could repress the expression of the FOS oncogene and may play an important role as a tumor suppressor in HCC (Li et al., 2009).
miR-221 and miR-222
miR-221 is overexpressed in human HCC as well as in other malignancies. p27 Was identified as a target of miR-221 and miR-222 (le Sage et al., 2007). Fornari et al. (2008) have reported that the cyclin-dependent kinase inhibitor p57 is also a direct target of miR-221. Indeed, downregulation of both p27 and p57 occurs in response to miR-221 transfection into HCC-derived cells and a significant upregulation of both p27 and p57 occurs in response to anti-miR-221 transfection. They suggest that miR-221 has an oncogenic function in hepatocarcinogenesis by targeting p27 and p57, hence promoting proliferation by controlling cell-cycle inhibitors (Fornari et al., 2008). miR-221 also targets Bmf, a proapoptotic BH3-only protein, and inhibits apoptosis of cells. miR-221 overexpression is associated with a more aggressive phenotype of HCC (Gramantieri et al., 2009). In addition, DNA damage-inducible transcript 4 (DDIT4), a modulator of mTOR pathway, was identified as a target of miR-221, indicating an important contribution for miR-221 in hepatocarcinogenesis and suggest a role for DDIT4 dysregulation in this process (Pineau et al., 2010). Taken together, these findings indicate that miR-221 simultaneously affects multiple pro-oncogenic pathways in hepatocarcinogenesis.
Garofalo et al. (2009) have reported that miR-221 and miR-222 are overexpressed in HCC cells, as compared with normal liver cells. They also show that miR-221 and miR-222 induce TRAIL resistance and enhance cellular migration through the activation of the AKT pathway and metallopeptidases by targeting PTEN and TIMP3 tumor suppressors and that the MET oncogene is involved in miR-221 and miR-222 activation through the c-Jun transcription factor (Garofalo et al., 2009). The protein phosphatase 2A subunit B (PPP2R2A) was identified as a target of miR-222. miR-222 overexpression is common in HCC and could confer metastatic potentials in HCC cells, possibly through activating AKT signaling (Wong et al., 2010).
These studies strongly suggest that miR-221 and miR-222 are oncogenic miRNAs that play critical roles in the initiation and progression of human HCC. The use of synthetic inhibitors of miR-221 and miR-222 may be a promising approach to HCC treatment.
Aberrant Expression of miRNAs in Cholangiocarcinoma
Meng et al. (2006) have reported that miR-21, miR-141, and miR-200b were highly overexpressed in malignant cholangiocytes. They show that miR-21 modulates gemcitabine-induced apoptosis by phosphatase and tensin homolog deleted on chromosome 10 (PTEN)-dependent activation of PI 3-kinase signaling. This study suggests that alterations in miRNA expression contribute to tumor growth and response to chemotherapy (Meng et al., 2006).
They also provide evidence that overexpression of IL-6 cause upregulation of let-7a and reduction of miR-370 in malignant human cholangiocytes. let-7a Contributes to the constitutively increased phosphorylation of STAT-3 by a mechanism involving the neurofibromatosis 2 (NF2) gene. The oncogene mitogen-activated protein kinase kinase kinase 8 (MAP3K8) was identified as a target of miR-370. Thus, let-7a modulates IL-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. In addition, IL-6 may contribute to tumor growth by modulation of expression of selected miRNAs, such as miR-370. Aberrantly expressed miRNA or their targets will provide mechanistic insight and therapeutic targets for cholangiocarcinoma (Meng et al., 2007, 2008).
Aberrant Expression of miRNAs in Pancreatic Cancer
Pancreatic ductal adenocarcinoma (PDAC) is still the fourth leading cause of cancer-related deaths in Western countries with increasing incidence. Neither effective prognostic markers nor therapies exist for PDAC.
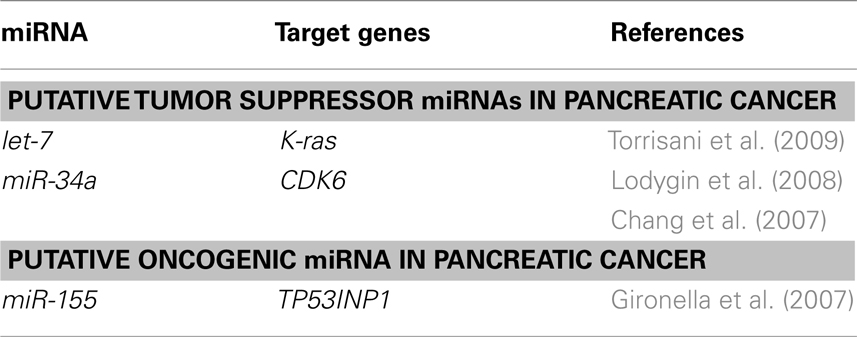
As shown in Table 2, let-7 miRNA expression is remarkably reduced in PDAC samples, as compared with adjacent tissue. Restoring let-7 levels in cancer-derived cell lines strongly inhibits cell proliferation, K-ras expression, and mitogen-activated protein kinase activation (Torrisani et al., 2009). Chang et al. (2007) have demonstrated that miR-34a is commonly deleted in human cancers and frequently absent in pancreatic cancer cells. miR-34a is directly transactivated by the tumor suppressor p53. Expression of miR-34a causes dramatic reprogramming of gene expression and promotes apoptosis (Chang et al., 2007). miR-34a expression is silenced in several types of cancer including pancreatic cancer due to aberrant CpG methylation of its promoter. Re-expression of miR-34a in pancreas carcinoma cell line induced senescence and cell-cycle arrest at least in part by targeting CDK6. These results show that miR-34a represents a tumor suppressor gene which is inactivated by CpG methylation in pancreatic cancer (Lodygin et al., 2008).
Gironella et al. (2007) have shown that oncogenic miRNA miR-155 is overexpressed in PDAC cells and repress tumor protein 53-induced nuclear protein-1 (TP53INP1) as its target. TP53INP1 expression is dramatically reduced in PDAC and this decrease occurs early during pancreatic cancer development (Gironella et al., 2007; Table 2).
Besides miR-155, several miRNAs including miR-21, miR-221, and miR-222 have been reported to be up-regulated in pancreatic cancer, suggesting that they are potential oncogenic miRNAs (Lee et al., 2007).
Perspectives and Conclusion
The distinct connection between aberrant expressions of miRNAs and the development of hepatobiliary and pancreatic cancers indicates that miRNAs can be promising therapeutic targets. A recent study have shown that chemically engineered oligonucleotides, termed locked-nucleic-acid (LNA)-modified miRNAs, were found to be specific inhibitors of endogenous miRNAs in non-human primates (Elmen et al., 2008). They could be used to silence oncogenic miRNAs, such as miR-221 and miR-222 as a strategy for HCC treatment. Regulation of miRNA expression by epigenetic therapy using DNA methylation inhibitors and histone deacetylase (HDAC) inhibitors also has clinical promise for the treatment of human malignancies. In addition, miRNA expression profiling may be a powerful clinical tool for diagnosis of hepatobiliary and pancreatic cancers.
Since the link between miRNAs and hepatobiliary and pancreatic cancers has only just begun to be understood and the number of miRNA genes identified is increasing, there could be a large number of therapeutic targets for HCCs, cholangiocarcinomas, and pancreatic cancers. Further studies are necessary to investigate whether miRNA-mediated therapy can be an effective approach for the chemoprevention of hepatobiliary and pancreatic cancers.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a Grant-in-Aid for Young Scientists A (23680090 to Yoshimasa Saito) and a Grant-in-Aid for Scientific Research B (22300169 to Hidekazu Suzuki) from the Japan Society for the Promotion of Science (JSPS), a grant from the Smoking Research Foundation (to Hidekazu Suzuki), the Keio Gijuku Academic Development Fund (to Yoshimasa Saito and Hidekazu Suzuki), and a Nateglinide Memorial Toyoshima Research and Education Fund (to Hidekazu Suzuki).
Footnotes
References
Bai, S., Nasser, M. W., Wang, B., Hsu, S. H., Datta, J., Kutay, H., Yadav, A., Nuovo, G., Kumar, P., and Ghoshal, K. (2009). MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 284, 32015–32027.
Calin, G. A., and Croce, C. M. (2006). MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866.
Calin, G. A., and Croce, C. M. (2007). Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J. Clin. Invest. 117, 2059–2066.
Calin, G. A., Sevignani, C., Dumitru, C. D., Hyslop, T., Noch, E., Yendamuri, S., Shimizu, M., Rattan, S., Bullrich, F., Negrini, M., and Croce, C. M. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 101, 2999–3004.
Chang, T. C., Wentzel, E. A., Kent, O. A., Ramachandran, K., Mullendore, M., Lee, K. H., Feldmann, G., Yamakuchi, M., Ferlito, M., Lowenstein, C. J., Arking, D. E., Beer, M. A., Maitra, A., and Mendell, J. T. (2007). Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26, 745–752.
Elmen, J., Lindow, M., Schutz, S., Lawrence, M., Petri, A., Obad, S., Lindholm, M., Hedtjärn, M., Hansen, H. F., Berger, U., Gullans, S., Kearney, P., Sarnow, P., Straarup, E. M., and Kauppinen, S. (2008). LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899.
Farazi, P. A., and DePinho, R. A. (2006). Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer 6, 674–687.
Fornari, F., Gramantieri, L., Ferracin, M., Veronese, A., Sabbioni, S., Calin, G. A., Grazi, G. L., Giovannini, C., Croce, C. M., Bolondi, L., and Negrini, M. (2008). MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27, 5651–5661.
Fornari, F., Gramantieri, L., Giovannini, C., Veronese, A., Ferracin, M., Sabbioni, S., Calin, G. A., Grazi, G. L., Croce, C. M., Tavolari, S., Chieco, P., Negrini, M., and Bolondi, L. (2009). MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 69, 5761–5767.
Garofalo, M., Di Leva, G., Romano, G., Nuovo, G., Suh, S. S., Ngankeu, A., Taccioli, C., Pichiorri, F., Alder, H., Secchiero, P., Gasparini, P., Gonelli, A., Costinean, S., Acunzo, M., Condorelli, G., and Croce, C. M. (2009). miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16, 498–509.
Gironella, M., Seux, M., Xie, M. J., Cano, C., Tomasini, R., Gommeaux, J., Garcia, S., Nowak, J., Yeung, M. L., Jeang, K. T., Chaix, A., Fazli, L., Motoo, Y., Wang, Q., Rocchi, P., Russo, A., Gleave, M., Dagorn, J. C., Iovanna, J. L., Carrier, A., Pébusque, M. J., and Dusetti, N. J. (2007). Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. U.S.A. 104, 16170–16175.
Gramantieri, L., Ferracin, M., Fornari, F., Veronese, A., Sabbioni, S., Liu, C. G., Calin, G. A., Giovannini, C., Ferrazzi, E., Grazi, G. L., Croce, C. M., Bolondi, L., and Negrini, M. (2007). Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 67, 6092–6099.
Gramantieri, L., Fornari, F., Ferracin, M., Veronese, A., Sabbioni, S., Calin, G. A., Grazi, G. L., Croce, C. M., Bolondi, L., and Negrini, M. (2009). MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin. Cancer Res. 15, 5073–5081.
Ji, J., Zhao, L., Budhu, A., Forgues, M., Jia, H. L., Qin, L. X., Ye, Q. H., Yu, J., Shi, X., Tang, Z. Y., and Wang, X. W. (2010). Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J. Hepatol. 52, 690–697.
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M., and Sarnow, P. (2005). Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309, 1577–1581.
Lan, F. F., Wang, H., Chen, Y. C., Chan, C. Y., Ng, S. S., Li, K., Xie, D., He, M. L., Lin, M. C., and Kung, H. F. (2011). Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A). Int. J. Cancer 128, 319–331.
le Sage, C., Nagel, R., Egan, D. A., Schrier, M., Mesman, E., Mangiola, A., Anile, C., Maira, G., Mercatelli, N., Ciafrè, S. A., Farace, M. G., and Agami, R. (2007). Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 26, 3699–3708.
Lee, E. J., Gusev, Y., Jiang, J., Nuovo, G. J., Lerner, M. R., Frankel, W. L., Morgan, D. L., Postier, R. G., Brackett, D. J., and Schmittgen, T. D. (2007). Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer 120, 1046–1054.
Li, S., Fu, H., Wang, Y., Tie, Y., Xing, R., Zhu, J., Sun, Z., Wei, L., and Zheng, X. (2009). MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology 49, 1194–1202.
Li, Y. P., Gottwein, J. M., Scheel, T. K., Jensen, T. B., and Bukh, J. (2011). MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc. Natl. Acad. Sci. U.S.A. 108, 4991–4996.
Lin, C. J., Gong, H. Y., Tseng, H. C., Wang, W. L., and Wu, J. L. (2008). miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 375, 315–320.
Lodygin, D., Tarasov, V., Epanchintsev, A., Berking, C., Knyazeva, T., Körner, H., Knyazev, P., Diebold, J., and Hermeking, H. (2008). Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7, 2591–2600.
Lu, J., Getz, G., Miska, E. A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B. L., Mak, R. H., Ferrando, A. A., Downing, J. R., Jacks, T., Horvitz, H. R., and Golub, T. R. (2005). MicroRNA expression profiles classify human cancers. Nature 435, 834–838.
Ma, L., Liu, J., Shen, J., Liu, L., Wu, J., Li, W., Luo, J., Chen, Q., and Qian, C. (2010). Expression of miR-122 mediated by adenoviral vector induces apoptosis and cell cycle arrest of cancer cells. Cancer Biol. Ther. 9, 554–561.
Meng, F., Henson, R., Lang, M., Wehbe, H., Maheshwari, S., Mendell, J. T., Jiang, J., Schmittgen, T. D., and Patel, T. (2006). Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 130, 2113–2129.
Meng, F., Henson, R., Wehbe-Janek, H., Smith, H., Ueno, Y., and Patel, T. (2007). The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J. Biol. Chem. 282, 8256–8264.
Meng, F., Wehbe-Janek, H., Henson, R., Smith, H., and Patel, T. (2008). Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene 27, 378–386.
O’Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V., and Mendell, J. T. (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843.
Pineau, P., Volinia, S., McJunkin, K., Marchio, A., Battiston, C., Terris, B., Mazzaferro, V., Lowe, S. W., Croce, C. M., and Dejean, A. (2010). miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 264–269.
Saito, Y., Liang, G., Egger, G., Friedman, J. M., Chuang, J. C., Coetzee, G. A., and Jones, P. A. (2006). Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9, 435–443.
Saito, Y., Suzuki, H., and Hibi, T. (2009a). The role of microRNAs in gastrointestinal cancers. J. Gastroenterol. 44(Suppl. 19), 18–22.
Saito, Y., Suzuki, H., Tsugawa, H., Nakagawa, I., Matsuzaki, J., Kanai, Y., and Hibi, T. (2009b). Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene 28, 2738–2744.
Shimizu, S., Takehara, T., Hikita, H., Kodama, T., Miyagi, T., Hosui, A., Tatsumi, T., Ishida, H., Noda, T., Nagano, H., Doki, Y., Mori, M., and Hayashi, N. (2010). The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J. Hepatol. 52, 698–704.
Su, H., Yang, J. R., Xu, T., Huang, J., Xu, L., Yuan, Y., and Zhuang, S. M. (2009). MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 69, 1135–1142.
Torrisani, J., Bournet, B., du Rieu, M. C., Bouisson, M., Souque, A., Escourrou, J., Buscail, L., and Cordelier, P. (2009). let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum. Gene Ther. 20, 831–844.
Tsai, W. C., Hsu, P. W., Lai, T. C., Chau, G. Y., Lin, C. W., Chen, C. M., Lin, C. D., Liao, Y. L., Wang, J. L., Chau, Y. P., Hsu, M. T., Hsiao, M., Huang, H. D., and Tsou, A. P. (2009). MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 49, 1571–1582.
Keywords: microRNA, oncogene, tumor suppressor, epigenetics, hepatocellular carcinoma, cholangiocarcinoma, pancreatic cancer
Citation: Saito Y, Suzuki H, Matsuura M, Sato A, Kasai Y, Yamada K, Saito H and Hibi T (2011) MicroRNAs in hepatobiliary and pancreatic cancers. Front. Gene. 2:66. doi: 10.3389/fgene.2011.00066
Received: 08 July 2011; Accepted: 02 September 2011;
Published online: 16 September 2011.
Edited by:
Alistair R. Forrest, RIKEN Omics Science Center, JapanReviewed by:
Antonio Sorrentino, University of California, San Francisco, USAMarcel E. Dinger, University of Queensland, Australia
Copyright: © 2011 Saito, Suzuki, Matsuura, Sato, Kasai, Yamada, Saito and Hibi. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Hidekazu Suzuki, Division of Gastroenterology and Hepatology, Department of Internal Medicine, School of Medicine, Keio University, 35 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan. e-mail: hsuzuki@a6.keio.jp
 Yoshimasa Saito1,2
Yoshimasa Saito1,2