- Department of Biochemistry and Microbiology, University of Victoria, Victoria, BC, Canada
Amphibians are important vertebrates in toxicology often representing both aquatic and terrestrial forms within the life history of the same species. Of the thousands of species, only two have substantial genomics resources: the recently published genome of the Pipid, Xenopus (Silurana) tropicalis, and transcript information (and ongoing genome sequencing project) of Xenopus laevis. However, many more species representative of regional ecological niches and life strategies are used in toxicology worldwide. Since Xenopus species diverged from the most populous frog family, the Ranidae, ~200 million years ago, there are notable differences between them and the even more distant Caudates (salamanders) and Caecilians. These differences include genome size, gene composition, and extent of polyploidization. Application of toxicogenomics to amphibians requires the mobilization of resources and expertise to develop de novo sequence assemblies and analysis strategies for a broader range of amphibian species. The present mini-review will present the advances in toxicogenomics as pertains to amphibians with particular emphasis upon the development and use of genomic techniques (inclusive of transcriptomics, proteomics, and metabolomics) and the challenges inherent therein.
Amphibians have an undeniable, yet understated, role in toxicology. They diverged from other vertebrates 360 million years ago (Frost et al., 2006) and, currently, over 60% of the >6,900 known species are threatened or declining in numbers (AmphibiaWeb, 2012). Recent amphibians are comprised of three orders: Anura (frogs and toads), Caudata (salamanders), and Gymnophiona (caecilians; Dubois, 2004; Frost et al., 2006), of which ~90% of species are Anura. Toxicological studies are primarily on anurans with some studies on caudates. Caecilian representation is lacking. In contrast to the limited ranges of caudate and caecilians, anurans are found on every continent except Antarctica (AmphibiaWeb, 2012).
Amphibians represent the only vertebrate group where a large majority of its members exhibit a life history that includes distinct independent aquatic larval and terrestrial juvenile/adult phases. The transition between the larval and juvenile phases requires substantial or complete remodeling of the organism (metamorphosis) in anticipation of a terrestrial lifestyle. Thus this places amphibians in a unique position for the assessment of toxicological effects in both aquatic and terrestrial environments. Over 10,000 study records on amphibians are currently available on the US Environmental Protection Agency’s ECOTOX database (US EPA, 2012). Ninety percent of records represent aquatic exposures and these are biased toward a single species (Xenopus laevis). The remaining 10% of records comprise terrestrial exposures of frog, toad, and salamander species (US EPA, 2012). Less than 300 records include any mRNA expression data.
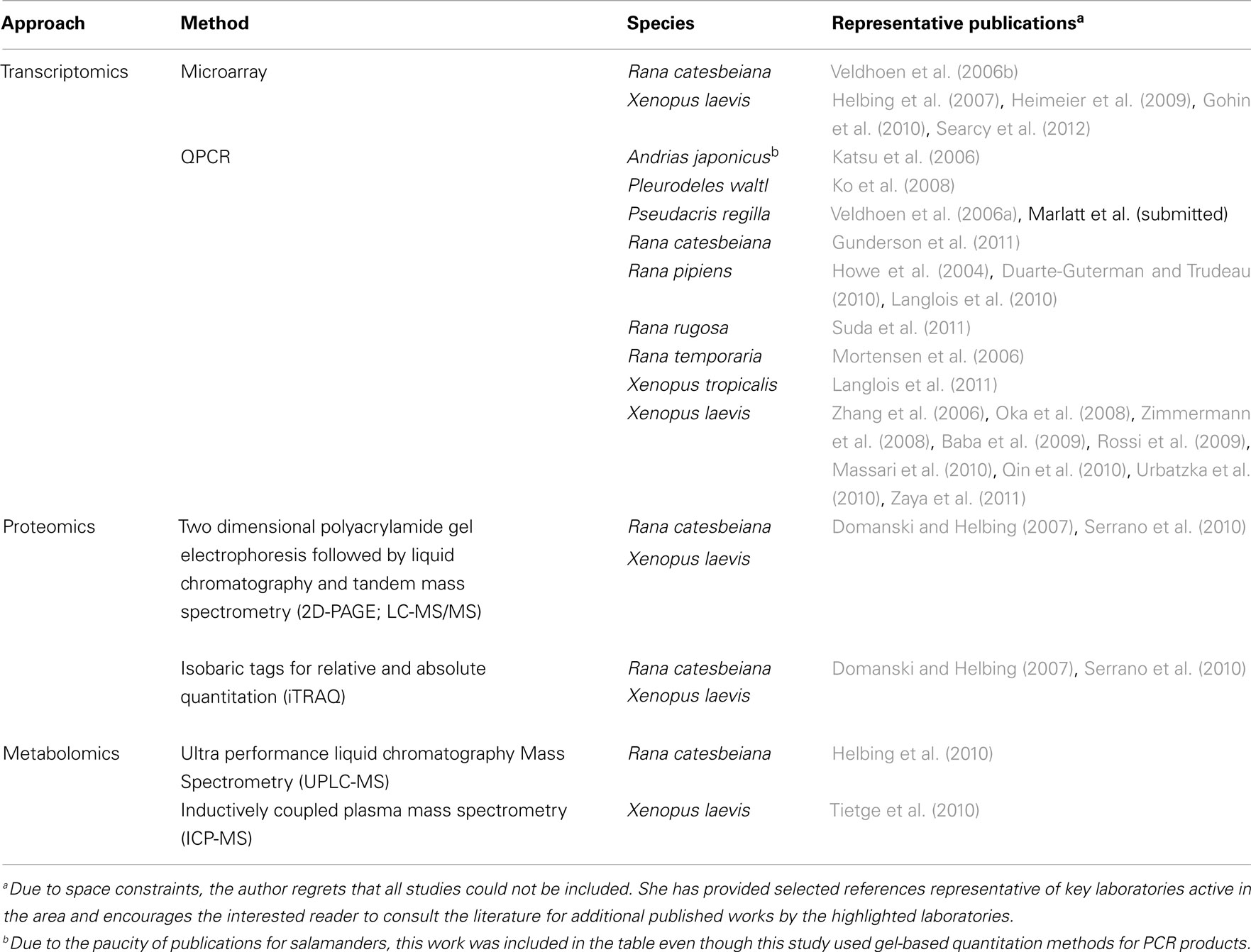
The exquisite sensitivity of frogs to hormones fostered the launching of several initiatives to develop standardized testing methods. Environment Canada is developing a frog tadpole exposure assay that uses native species (Rana catesbeiana and pipiens) and a combination of molecular and morphological criteria (Veldhoen et al., 2006c). The Organization for Economic Cooperation and Development (OECD, 2009) has recently established guidelines for a standardized assay for evaluating thyroid hormone (TH) active chemicals using a X. laevis metamorphosis assay (XEMA) and is in the process of evaluating a multigenerational reproductive assay in X. tropicalis. The XEMA assay has been adapted for X. tropicalis (Mitsui et al., 2006) and served as a template for the development of some native frog metamorphosis assays for R. rugosa (Oka et al., 2009), Bombina orientalis (Park et al., 2010), and Pseudacris regilla (Marlatt et al., submitted). However, the recommended XEMA assay relies upon morphological criteria and the input of toxicogenomic endpoints is not standard practice. Nevertheless, movement toward inclusion of molecular endpoints to reduce assay time and provide greater information regarding test chemical mode of action is evident in the literature (Table 1).
Amphibians are used in two general ways in the context of toxicology: in laboratory exposure settings where individual chemicals or complex mixtures are tested and in the field setting. Although availability of appropriate life stages is year round for some species (e.g., Xenopus tadpoles can be bred on demand, R. catesbeiana tadpoles can be collected from the wild and housed in aquatics facilities year round), many have limited availability throughout the year. Moreover, field sampling of threatened or endangered species necessitates the development of non-lethal sampling methods (fin biopsies) combined with molecular analyses (Veldhoen and Helbing, 2005). Efforts have also been made to combine transcript analysis with cultured tail fin biopsies for rapid screening of chemicals and effluents (Hinther et al., 2010).
Toxicogenomics are best suited for identifying and evaluating factors categorized as sublethal deleterious effects that influence survival and recruitment; the primary factors contributing to amphibian population declines (Hayes et al., 2010). Such factors include: stress, susceptibility to disease, climate change, and environmental pollutants. Typically, molecular responses precede morphological endpoints giving early indications of response and modes of action. To date, application of toxicogenomics to amphibians has been extremely limited due to restrictions in resources/expertise and the difficulty in obtaining consensus on which toxicologically important species to develop large-scale genomics resources for. Approaches for transcriptomics, proteomics, and metabolomics pertaining to frogs has been previously reviewed elsewhere (Helbing et al., 2010). Research efforts have largely focused on the evaluation of endocrine disruption in frogs, primarily with respect to xenoestrogens and TH-active chemicals, including hormonal cross-talk and their complex interactions with environmental factors (Table 1). Sex reversal and/or intersex conditions in response to chemical exposures have been reported and some laboratories have begun to examine their molecular basis (Table 1). The absolute dependence of frog tadpoles upon TH during metamorphosis into a juvenile frog (Shi, 2000) provides for the most comprehensive and drastic response known to the hormone; although all vertebrates require THs for development, nervous system function, and metabolism (Oppenheimer, 1999). Indeed, the use of frog tadpoles as surrogate species for TH disruption in mammals has been explored at the molecular level (Searcy et al., 2012).
Even where more resources are available (for, e.g., commercially available Xenopus oligo microarrays), restrictions in cost and lack of utility across species platforms (Helbing et al., 2010) have greatly limited application of toxicogenomics tools beyond quantitative real time polymerase chain reaction (QPCR; Table 1). It is notable that very few relevant studies have been performed using salamanders and none with caecilians (Table 1). For labs having the necessary expertise, efforts have concentrated upon the production of transcript-based biomarkers (although some data on proteomics and metabolomics have been published; Table 1), definition of baseline responses to model hormones, identification of appropriate sampling times for molecular biomarker use, and determination of natural variation (Table 1).
Most of the toxicogenomic studies that have been published so far are laboratory-based using X. laevis strains that are akin to the white mouse of the frog world. Xenopus are Pipidae; one of 38 families within the Anura (Frost et al., 2006). Pipidae are distantly related to most Anura and diverged from the largest family, the Ranidae (the true frogs), ~200 million years ago (Sumida et al., 2004). Marked differences in life histories (for, e.g., Xenopus remain aquatic after metamorphosis, whereas Ranids become terrestrial), species sensitivities (Relyea and Jones, 2009), and genome compositions (discussed below) have prompted a call for developing toxicogenomics tools and approaches applicable to environmentally relevant species (Denslow et al., 2007).
Amphibian toxicogenomics has largely been driven through the adoption of tools made available through disciplines outside of toxicology, namely developmental, cell, and molecular biology. This provided, firstly, gene information from X. laevis, and, subsequently, a genome from X. (Silurana) tropicalis. However, this has not been without difficulty. X. laevis has been the most-used amphibian toxicological model due to the ease of husbandry in laboratory settings. However, genetically, X. laevis is pseudotetraploid, derived from an ancient tetraploid lineage with incomplete diploidization across a large portion of the genome (Mable et al., 2011). In fact, Pipidae have the highest number of polyploid species identified within amphibians (Mable et al., 2011). This created significant problems in initiating a genome sequencing project for this species due to the bioinformatic challenge of assembling a tetraploid genome de novo. Fortunately, a diploid Xenopus species did exist with similar husbandry benefits in this family. Thus the sequencing of the first frog genome was performed on X. tropicalis and completed in 2010 (Hellsten et al., 2010). The problem for toxicology is that the availability of the X. tropicalis genome information is now driving scientists to use this as a test species for use in toxicogenomics. Although an enormous opportunity, it is not clear how suitable this species will be as a representative of native frog species or amphibians in general.
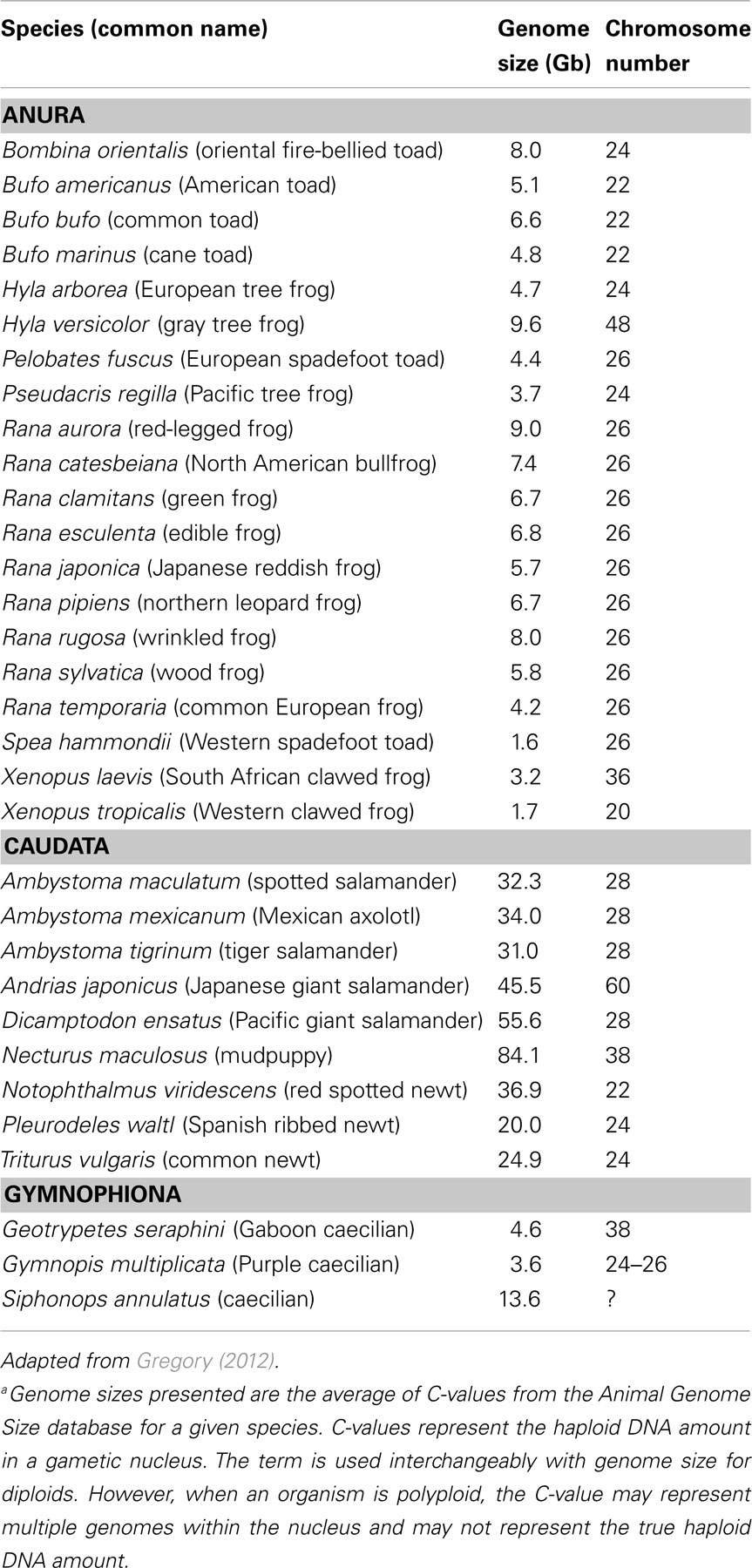
Amphibians present a wide range of species diversity. They contain keystone species within a plethora of ecosystems throughout the world. They are important food sources for humans and wildlife, instrumental in pest control, and serve as sensitive indicators within a variety of ecosystems. With the exception of R. catesbeiana which is distributed worldwide, amphibians tend to have regional representation. Therefore, toxicological evaluations have often tended toward regionalism as well [for example, common frog ecotox species are R. rugosa (Japan), R. temporaria (Europe), and R. pipiens (North America)]. Moreover, amphibians have representatives of different sex determination systems (e.g., XX/XY, ZW/ZZ; Eggert, 2004) that could influence sensitivity to environmental contaminants. Genome organization is similar within amphibian subgroups, but varies substantially between subgroups. In addition to polyploidy in a few amphibian species, the genome sizes of amphibians span four orders of magnitude from one-quarter of the human genome (0.9 Gb, Limnodynastes ornatus = ornate burrowing frog) to among the largest known in animals (118 Gb, Necturus lewisi = gulf coast waterdog; Gregory, 2012). The estimated genome sizes and chromosome numbers of commonly used amphibian species in toxicology are presented in Table 2. Thus, coupling toxicological demands with genetics result in logistical and bioinformatic challenges. These have hampered building consensus and concerted effort to further genomics tools.
Despite this, current efforts are focused upon addressing the dearth in (1) application of available toxicogenomics resources to amphibians, and (2) genome sequence information representing amphibian species beyond Xenopus. Access to genome and transcriptome sequence information is critical for the key toxicogenomics approaches today such as microarrays, QPCR, and proteomics techniques. The increasingly recognized role of epigenetic factors in toxicology necessitates the means for analyzing genomes (Bilesimo et al., 2011). De novo high throughput sequencing of transcriptomes (RNA-seq) provides an unprecedented opportunity to obtain sequence and expression information of literally thousands of gene transcripts within a tissue (Martin and Wang, 2011). Access to resources such as Xenbase (www.xenbase.org) and deposition of amphibian expressed sequence tags (ESTs) and individually cloned sequences on publicly accessible databases have made it possible to garner limited information from RNA-seq experiments (Figure 1). However, accurate assembly and bioinformatics evaluation of RNA-seq data requires a genome sequence for the species of interest.
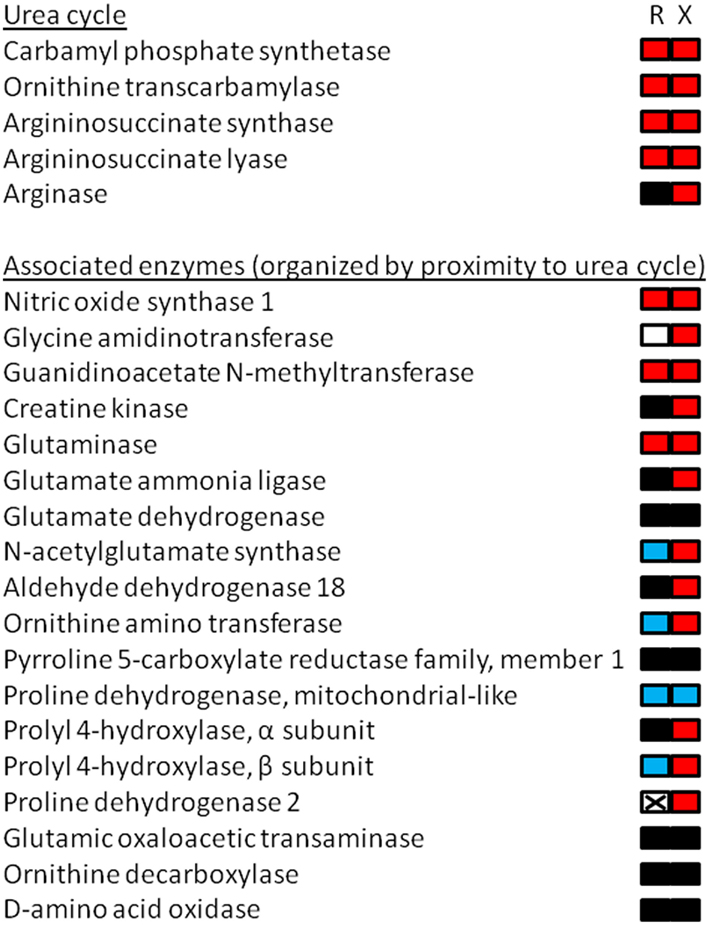
Figure 1. Cartoon depiction of RNA-seq results from the liver of premetamorphic Xenopus laevis and Rana catesbeiana tadpoles focusing upon arginine and proline metabolism including the urea cycle. Tadpoles were exposed to 10 nM 3,5,3′-triiodothyronine (a thyroid hormone) or NaOH vehicle control for 48 h. The animals were treated and maintained in accordance with the guidelines of the Canadian Council on Animal Care. The liver transcriptomes were subjected to RNA-seq using 75 base HiSeq of paired end tagged (PET) libraries. The derived sequence information was assembled using the X. tropicalis genome as a scaffold and the contig identities were determined by a Blastx search against the X. tropicalis genome. The number of read counts (~400 million) was normalized between samples and the relative count frequencies of the indicated pathway components were compared based upon the X. tropicalis arginine and proline metabolism KEGG pathway (xtr00330; www.genome.jp/kegg). The results are depicted as a bipartite rectangle beside the name of the enzymes corresponding with measured transcripts in the RNA-seq experiment that were identified in the KEGG pathway. The left side represents the relative change in transcript abundance levels of Rana (R) and the right side mRNA levels of Xenopus (X) where red is increased, black is no change, and blue is decreased relative to control animals. Non-detected transcript is depicted by a crossed-out white box. Use of the X. tropicalis genome as an assembly scaffold had limited utility since X. laevis and R. catesbeiana sequences aligned imperfectly to the X. tropicalis genome with R. catesbeiana, not surprisingly, having the least benefit of alignment. Nevertheless, some transcript identities linked to count frequencies were positively confirmed and the data obtained for the urea cycle enzymes, for example, matched well with previous observations (Helbing et al., 1992; Xu et al., 1993; Iwase et al., 1995). This validates the method for transcripts that are identifiable and quantifiable in this way.
We will see more use of X. tropicalis for toxicogenomics; but we must be very careful not to let the genomics drive the toxicology and put resources and effort into developing appropriate tools for a wider range of toxicologically relevant species. Of the greatest urgency is obtaining genome sequence information from the Ranids, since they represent the largest group of amphibians worldwide, and the Caudata due to their unique genome structure. Now more than ever, we need the scaffolds and platforms for more amphibian species to move toxicogenomics forward.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author gratefully acknowledges grant support from the Natural Sciences and Engineering Research Council of Canada (NSERC) and Genome British Columbia. Thanks to N. Veldhoen for assistance in manuscript preparation, A. Carew and G. Taylor (Michael Smith Genome Sciences Centre, Vancouver, BC, Canada) for technical assistance, and N. Veldhoen, S. Maher, A. Carew, and P. Wojnarowicz for helpful discussions.
References
AmphibiaWeb. (2012). AmphibiaWeb: Information on Amphibian Biology and Conservation. Available at: www.amphibiaweb.org [accessed January 12].
Baba, K., Okada, K., Kinoshita, T., and Imaoka, S. (2009). Bisphenol A disrupts Notch signaling by inhibiting gamma-secretase activity and causes eye dysplasia of Xenopus laevis. Toxicol. Sci. 108, 344–355.
Bilesimo, P., Jolivet, P., Alfama, G., Buisine, N., Le Mevel, S., Havis, E., Demeneix, B. A., and Sachs, L. M. (2011). Specific histone lysine 4 methylation patterns define TR-binding capacity and differentiate direct T3 responses. Mol. Endocrinol. 25, 225–237.
Denslow, N. D., Colbourne, J. K., Dix, D., Freedman, J. H., Helbing, C. C., Kennedy, S., and Williams, P. L. (2007). “Selection of surrogate animal species for comparative toxicogenomics,” in Genomic Approaches for Cross-Species Extrapolation in Toxicology, eds W. Benson and R. Di Giulio (Pensacola, FL: SETAC and CRC Press), 33–75.
Domanski, D., and Helbing, C. C. (2007). Analysis of the Rana catesbeiana tadpole tail fin proteome and phosphoproteome during T3-induced apoptosis: identification of a novel type I keratin. BMC Dev. Biol. 7, 94. doi:10.1186/1471-213X-7-94
Duarte-Guterman, P., and Trudeau, V. L. (2010). Regulation of thyroid hormone-, oestrogen- and androgen-related genes by triiodothyronine in the brain of Silurana tropicalis. J. Neuroendocrinol. 22, 1023–1031.
Frost, D. R., Grant, T., Faivovich, J., Bain, R. H., Haas, A., Haddad, C. F. B., De Sa, R. O., Channing, A., Wilkinson, M., Donnellan, S. C., Raxworthy, C. J., Campbell, J. A., Blotto, B. L., Moler, P., Drewes, R. C., Nussbaum, R. A., Lynch, J. D., Green, D. M., and Wheeler, W. C. (2006). The amphibian tree of life. Bull. Am. Mus. Nat. Hist. 297, 8–370.
Gohin, M., Bobe, J., and Chesnel, F. (2010). Comparative transcriptomic analysis of follicle-enclosed oocyte maturational and developmental competence acquisition in two non-mammalian vertebrates. BMC Genomics 11, 18. doi:10.1186/1471-2164-11-18
Gregory, T. (2012). Animal Genome Size Database. Available at: www.genomesize.com [accessed January 12].
Gunderson, M. P., Veldhoen, N., Skirrow, R. C., Macnab, M. K., Ding, W., van Aggelen, G., and Helbing, C. C. (2011). Effect of low dose exposure to the herbicide atrazine and its metabolite on cytochrome P450 aromatase and steroidogenic factor-1 mRNA levels in the brain of premetamorphic bullfrog tadpoles (Rana catesbeiana). Aquat. Toxicol. 102, 31–38.
Hayes, T. B., Falso, P., Gallipeau, S., and Stice, M. (2010). The cause of global amphibian declines: a developmental endocrinologist’s perspective. J. Exp. Biol. 213, 921–933.
Heimeier, R. A., Das, B., Buchholz, D. R., and Shi, Y. B. (2009). The xenoestrogen bisphenol A inhibits postembryonic vertebrate development by antagonizing gene regulation by thyroid hormone. Endocrinology 150, 2964–2973.
Helbing, C., Bailey, C., Ji, L., Gunderson, M. P., Zhang, F., Veldhoen, N., Skirrow, R. C., Mu, R., Lesperance, M., Holcombe, G. W., Kosian, P. A., Tietge, J., Korte, J. J., and Degitz, S. J. (2007). Identification of gene expression indicators for thyroid axis disruption in a Xenopus laevis metamorphosis screening assay Part 1: effects on the brain. Aquat. Toxicol. 82, 227–241.
Helbing, C., Gergely, G., and Atkinson, B. G. (1992). Sequential up-regulation of thyroid hormone beta receptor, ornithine transcarbamylase, and carbamyl phosphate synthetase mRNAs in the liver of Rana catesbeiana tadpoles during spontaneous and thyroid hormone-induced metamorphosis. Dev. Genet. 13, 289–301.
Helbing, C. C., Maher, S. K., Han, J., Gunderson, M. P., and Borchers, C. (2010). Peering into molecular mechanisms of action with frogSCOPE. Gen. Comp. Endocrinol. 168, 190–198.
Hellsten, U., Harland, R. M., Gilchrist, M. J., Hendrix, D., Jurka, J., Kapitonov, V., Ovcharenko, I., Putnam, N. H., Shu, S., Taher, L., Blitz, I. L., Blumberg, B., Dichmann, D. S., Dubchak, I., Amaya, E., Detter, J. C., Fletcher, R., Gerhard, D. S., Goodstein, D., Graves, T., Grigoriev, I. V., Grimwood, J., Kawashima, T., Lindquist, E., Lucas, S. M., Mead, P. E., Mitros, T., Ogino, H., Ohta, Y., Poliakov, A. V., Pollet, N., Robert, J., Salamov, A., Sater, A. K., Schmutz, J., Terry, A., Vize, P. D., Warren, W. C., Wells, D., Wills, A., Wilson, R. K., Zimmerman, L. B., Zorn, A. M., Grainger, R., Grammer, T., Khokha, M. K., Richardson, P. M., and Rokhsar, D. S. (2010). The genome of the Western clawed frog Xenopus tropicalis. Science 328, 633–636.
Hinther, A., Domanski, D., Vawda, S., and Helbing, C. C. (2010). C-fin: a cultured frog tadpole tail fin biopsy approach for detection of thyroid hormone-disrupting chemicals. Environ. Toxicol. Chem. 29, 380–388.
Howe, C. M., Berrill, M., Pauli, B. D., Helbing, C. C., Werry, K., and Veldhoen, N. (2004). Toxicity of glyphosate-based pesticides to four North American frog species. Environ. Toxicol. Chem. 23, 1928–1938.
Iwase, K., Yamauchi, K., and Ishikawa, K. (1995). Cloning of cDNAs encoding argininosuccinate lyase and arginase from Rana catesbeiana liver and regulation of their mRNAs during spontaneous and thyroid hormone-induced metamorphosis. Biochim. Biophys. Acta 25, 139–146.
Katsu, Y., Kohno, S., Oka, T., Mitsui, N., Tooi, O., Santo, N., Urushitani, H., Fukumoto, Y., Kuwabara, K., Ashikaga, K., Minami, S., Kato, S., Ohta, Y., Guillette, L. J. Jr., and Iguchi, T. (2006). Molecular cloning of estrogen receptor alpha (ERalpha; ESR1) of the Japanese giant salamander, Andrias japonicus. Mol. Cell. Endocrinol. 257–258, 84–94.
Ko, C. I., Chesnel, A., Mazerbourg, S., Kuntz, S., Flament, S., and Chardard, D. (2008). Female-enriched expression of ERalpha during gonad differentiation of the urodele amphibian Pleurodeles waltl. Gen. Comp. Endocrinol. 156, 234–245.
Langlois, V. S., Carew, A. C., Pauli, B. D., Wade, M. G., Cooke, G. M., and Trudeau, V. L. (2010). Low levels of the herbicide atrazine alter sex ratios and reduce metamorphic success in Rana pipiens tadpoles raised in outdoor mesocosms. Environ. Health Perspect. 118, 552–557.
Langlois, V. S., Duarte-Guterman, P., and Trudeau, V. L. (2011). Expression profiles of reproduction- and thyroid hormone-related transcripts in the brains of chemically-induced intersex frogs. Sex Dev. 5, 26–32.
Mable, B. K., Alexandrou, M. A., and Taylor, M. I. (2011). Genome duplication in amphibians and fish: an extended synthesis. J. Zool. 284, 151–182.
Martin, J. A., and Wang, Z. (2011). Next-generation transcriptome assembly. Nat. Rev. Genet. 12, 671–682.
Massari, A., Urbatzka, R., Cevasco, A., Canesi, L., Lanza, C., Scarabelli, L., Kloas, W., and Mandich, A. (2010). Aromatase mRNA expression in the brain of adult Xenopus laevis exposed to Lambro river water and endocrine disrupting compounds. Gen. Comp. Endocrinol. 168, 262–268.
Mitsui, N., Fujii, T., Miyahara, M., Oka, T., Kashiwagi, A., Kashiwagi, K., Hanada, H., Urushitani, H., Santo, N., Tooi, O., and Iguchi, T. (2006). Development of metamorphosis assay using Silurana tropicalis for the detection of thyroid system-disrupting chemicals. Ecotoxicol. Environ. Saf. 64, 281–287.
Mortensen, A. S., Kortner, T. M., and Arukwe, A. (2006). Thyroid hormone-dependent gene expression as a biomarker of short-term 1,1-dichloro-2,2-bis(p-chlorophenyl)ethylene (DDE) exposure in European common frog (Rana temporaria) tadpoles. Biomarkers 11, 524–537.
Oka, T., Miyahara, M., Yamamoto, J., Mitsui, N., Fujii, T., Tooi, O., Kashiwagi, K., Takase, M., Kashiwagi, A., and Iguchi, T. (2009). Application of metamorphosis assay to a native Japanese amphibian species, Rana rugosa, for assessing effects of thyroid system affecting chemicals. Ecotoxicol. Environ. Saf. 72, 1400–1405.
Oka, T., Tooi, O., Mitsui, N., Miyahara, M., Ohnishi, Y., Takase, M., Kashiwagi, A., Shinkai, T., Santo, N., and Iguchi, T. (2008). Effect of atrazine on metamorphosis and sexual differentiation in Xenopus laevis. Aquat. Toxicol. 87, 215–226.
Organization for Economic Cooperation and Development (OECD). (2009). OECD Guideline for the (Testing) of Chemicals Test No. 231: The Amphibian Metamorphosis Assay (Paris: OECD).
Park, C. J., Kang, H. S., and Gye, M. C. (2010). Effects of nonylphenol on early embryonic development, pigmentation and 3,5,3’-triiodothyronine-induced metamorphosis in Bombina orientalis (Amphibia: Anura). Chemosphere 81, 1292–1300.
Qin, X., Xia, X., Yang, Z., Yan, S., Zhao, Y., Wei, R., Li, Y., Tian, M., Zhao, X., Qin, Z., and Xu, X. (2010). Thyroid disruption by technical decabromodiphenyl ether (DE-83R) at low concentrations in Xenopus laevis. J. Environ. Sci. (China) 22, 744–751.
Relyea, R. A., and Jones, D. K. (2009). The toxicity of roundup original max (R) to 13 species of larval amphibians. Environ. Toxicol. Chem. 28, 2004–2008.
Rossi, F., Bernardini, G., Bonfanti, P., Colombo, A., Prati, M., and Gornati, R. (2009). Effects of TCDD on spermatogenesis related factor-2 (SRF-2): gene expression in Xenopus. Toxicol. Lett. 191, 189–194.
Searcy, B. T., Beckstrom-Sternberg, S. M., Beckstrom-Sternberg, J. S., Stafford, P., Schwendiman, A. L., Soto-Pena, J., Owen, M. C., Ramirez, C., Phillips, J., Veldhoen, N., Helbing, C. C., and Propper, C. R. (2012). Thyroid hormone-dependent development in Xenopus laevis: a sensitive screen of thyroid hormone signaling disruption by municipal wastewater treatment plant effluent. Gen. Comp. Endocrinol. doi:10.1016/j.ygcen.2011.12.036
Serrano, J., Higgins, L., Witthuhn, B. A., Anderson, L. B., Markowski, T., Holcombe, G. W., Kosian, P. A., Korte, J. J., Tietge, J. E., and Degitz, S. J. (2010). In vivo assessment and potential diagnosis of xenobiotics that perturb the thyroid pathway: proteomic analysis of Xenopus laevis brain tissue following exposure to model T4 inhibitors. Comp. Biochem. Physiol. Part D Genomics Proteomics 5, 138–150.
Shi, Y.-B. (2000). Amphibian Metamorphosis: From Morphology to Molecular Biology. New York: Wiley-Liss.
Suda, M., Kodama, M., Oshima, Y., Yamamoto, K., Nakamura, Y., Tanaka, S., Kikuyama, S., and Nakamura, M. (2011). Up-regulation of FSHR expression during gonadal sex determination in the frog Rana rugosa. Gen. Comp. Endocrinol. 172, 475–486.
Sumida, M., Kato, Y., and Kurabayashi, A. (2004). Sequencing and analysis of the internal transcribed spacers (ITSs) and coding regions in the EcoR I fragment of the ribosomal DNA of the Japanese pond frog Rana nigromaculata. Genes Genet. Syst. 79, 105–118.
Tietge, J. E., Butterworth, B. C., Haselman, J. T., Holcombe, G. W., Hornung, M. W., Korte, J. J., Kosian, P. A., Wolfe, M., and Degitz, S. J. (2010). Early temporal effects of three thyroid hormone synthesis inhibitors in Xenopus laevis. Aquat. Toxicol. 98, 44–50.
United States Environmental Protection Agency (US EPA). (2012). ECOTOX Database. Available at: http://cfpub.epa.gov/ecotox/quick_query.htm [accessed January 12].
Urbatzka, R., Lorenz, C., Lutz, I., and Kloas, W. (2010). Expression profiles of LHbeta, FSHbeta and their gonadal receptor mRNAs during sexual differentiation of Xenopus laevis tadpoles. Gen. Comp. Endocrinol. 168, 239–244.
Veldhoen, N., Boggs, A., Walzak, K., and Helbing, C. C. (2006a). Exposure to tetrabromobisphenol-A alters TH-associated gene expression and tadpole metamorphosis in the Pacific Tree Frog Pseudacris regilla. Aquat. Toxicol. 78, 292–302.
Veldhoen, N., Skirrow, R., Ji, L., Domanski, D., Bonfield, E. R., Bailey, C. M., and Helbing, C. C. (2006b). Use of heterologous cDNA arrays and organ culture in the detection of thyroid hormone-dependent responses in a sentinel frog, Rana catesbeiana. Comp. Biochem. Physiol. Part D Genomics Proteomics 1, 187–199.
Veldhoen, N., Skirrow, R., Osachoff, H., Wigmore, H., Clapson, D. J., Gunderson, M. P., Van Aggelen, G., and Helbing, C. C. (2006c). The bactericidal agent Triclosan modulates thyroid hormone-associated gene expression and disrupts postembryonic anuran development. Aquat. Toxicol. 80, 217–227.
Veldhoen, N., and Helbing, C. C. (2005). “Monitoring gene expression in Rana catesbeiana tadpoles using a tail fin biopsy technique and its application to the detection of environmental endocrine disruptor effects in wildlife species,” in Techniques in Aquatic Toxicology, Vol. 2, ed. G. K. Ostrander (Boca Raton: CRC Press), 315–327.
Xu, Q., Baker, B. S., and Tata, J. R. (1993). Developmental and hormonal regulation of the Xenopus liver-type arginase gene. Eur. J. Biochem. 211, 891–898.
Zaya, R. M., Amini, Z., Whitaker, A. S., and Ide, C. F. (2011). Exposure to atrazine affects the expression of key genes in metabolic pathways integral to energy homeostasis in Xenopus laevis tadpoles. Aquat. Toxicol. 104, 254–262.
Zhang, F., Degitz, S. J., Holcombe, G. W., Kosian, P. A., Tietge, J., Veldhoen, N., and Helbing, C. C. (2006). Evaluation of gene expression endpoints in the context of a Xenopus laevis metamorphosis-based bioassay to detect thyroid hormone disruptors. Aquat. Toxicol. 76, 24–36.
Keywords: amphibian, frog, molecular techniques, polyploid, endocrine disruptor, estrogen, thyroid hormone, toxicogenomics
Citation: Helbing CC (2012) The metamorphosis of amphibian toxicogenomics. Front. Gene. 3:37. doi: 10.3389/fgene.2012.00037
Received: 31 January 2012; Accepted: 27 February 2012;
Published online: 14 March 2012.
Edited by:
Jonathan Freedman, National Institute of Health, USAReviewed by:
Tara Sabo-Attwood, University of Florida, USARaja S. Settivari, The Dow Chemical Company, USA
Copyright: © 2012 Helbing. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Caren C. Helbing, Department of Biochemistry and Microbiology, University of Victoria, P.O. Box 3055 Stn CSC, Victoria, BC, Canada V8W 3P6. e-mail: chelbing@uvic.ca