- 1 Hussman Institute for Human Genomics, University of Miami Miller School of Medicine, Miami, FL, USA
- 2 St. Laurent Institute, Cambridge, MA, USA
- 3 Immunovirology – Biogenisis Group, University of Antioquia, Medellin, Colombia
Prolonged drug use causes long-lasting neuroadaptations in reward-related brain areas that contribute to addiction. Despite significant amount of research dedicated to understanding the underlying mechanisms of addiction, the molecular underpinnings remain unclear. At the same time, much of the pervasive transcription that encompasses the human genome occurs in the nervous system and contributes to its heterogeneity and complexity. Recent evidence suggests that non-coding RNAs (ncRNAs) play an important and dynamic role in transcriptional regulation, epigenetic signaling, stress response, and plasticity in the nervous system. Dysregulation of ncRNAs are thought to contribute to many, and perhaps all, neurological disorders, including addiction. Here, we review recent insights in the functional relevance of ncRNAs, including both microRNAs (miRNAs), and long non-coding RNAs, and then illustrate specific examples of ncRNA regulation in the context of drug addiction. We conclude that ncRNAs are importantly involved in the persistent neuroadaptations associated with addiction-related behaviors, and that therapies that target specific ncRNAs may represent new avenues for the treatment of drug addiction.
Introduction
Evolutionary processes in the human lineage have coupled expanding genome complexity with the acquisition, processing, and distribution of ever increasing amounts of information (Sempere et al., 2006; Mattick, 2007; Berezikov, 2011). Non-coding RNAs (ncRNAs) occupy the center stage of this expanding genome complexity, with the ratio of non-coding genomic sequence to protein coding sequence growing more than 10-fold as primates evolved from simple multicellular organisms (Taft et al., 2007). ncRNAs occupy critical nodes and edges in a majority of physiological networks, frequently participating in feedback loops, stability, and fined-tuned regulatory control. The unique information processing features of ncRNAs permit them to transduce information through heterogeneous molecular machineries with relatively less energy cost than protein networks alone, making them particularly useful in the limited and thermally constrained real estate of the human central nervous system.
In light of these observations, it is not surprising that non-exonic transcripts (those mapping to regions of the genome outside annotated protein coding genes) comprise 2/3 of the total, non-ribosomal, non-mitochondrial RNA in the human brain (Kapranov et al., 2010). Highly articulated nervous system expression of miRNAs (Lagos-Quintana et al., 2002; Schratt et al., 2006; Makeyev et al., 2007) and long non-coding RNAs (lncRNAs; Mercer et al., 2008; Ponjavic et al., 2009) point to the involvement of ncRNAs in key aspects of nervous system homeostasis and plasticity. The ability of addictive stimuli to perturb and disrupt these functions suggests a broad based involvement of ncRNAs in the loss of functional coherence associated with chronic drug use (Chandrasekar and Dreyer, 2009; Hollander et al., 2010). In this review, we consider the participation of ncRNAs in the key nervous system molecular machineries affected by addiction, and the potential mechanisms of their involvement in the dimensions of this complex disorder.
Involvement of ncRNAs in Neuroplasticity and Learning
Emerging evidence suggests that miRNAs and their processing machinery play a critical role in neuroplasticity by regulating protein dynamics in the synapse (Edbauer et al., 2010; Lippi et al., 2011; Siegel et al., 2011; Saba et al., 2012). Given that addiction is considered a maladaptive form of neuroplasticity, identifying miRNAs important in such neuroadaptations may lead to novel insights in addiction research. Demonstrating a role for miRNAs in neuroplasticity, Schratt et al. (2006) showed that miR-134 expression in the dendrites of developing hippocampal neurons is critically involved in dendritic spine formation and plasticity by inhibiting Lim domain-containing protein kinase 1, an important regulator of actin filament dynamics. Following this seminal report, other studies revealed that miR-138, miR-132, and miR-125b play an essential role in dendritic spine formation (Siegel et al., 2009; Edbauer et al., 2010). The role of miRNAs in synapse are not restricted to the those listed above, as recent reports have identified numerous dendritic- and synaptic-enriched miRNAs (Lugli et al., 2008; Eipper-Mains et al., 2011; Saba et al., 2012). Although most of the research thus far has focused on miRNAs, there is some evidence that suggests lncRNAs are also involved in synaptic plasticity. For instance, BC1 and BC200 are lncRNAs that are localized in post-synaptic dendritic compartments where they regulate local gene expression by controlling the activity of specific transcription factors (Lin et al., 2008). Other lncRNAs are also localized in the synapse and neurite extensions (French et al., 2001; Mercer et al., 2008), but their functional role in plasticity remains poorly understood.
In neurons, activity-dependent regulation of miRNAs also contributes extensively to neuroplasticity. Using the marine snail Aplysia, Rajasethupathy et al. (2009) demonstrated that serotonin-induced activation of sensory neurons caused a significant decrease in miR-124, leading to increased cAMP response element-binding (CREB) expression and induction of long-term facilitation. Knockdown of miR-124 also robustly increased serotonin-induced synaptic plasticity and CREB expression, revealing a functional association between miR-124 activity and CREB expression. Additional in vivo studies have demonstrated activity-dependent up regulation of miR-132 and miR-212 following induction of LTP in hippocampal neurons, an increase that was dependent on metabotropic glutamate receptor activation (Wibrand et al., 2010). Consistent with the role of miR-132 in neuronal activation, enhanced expression of miR-132 was observed in a number of behavioral paradigms, such as contextual fear conditioning, odorant exposure, and acute cocaine treatment (Nudelman et al., 2010). In addition, over expressing miR-132 enhances neuronal activity in cortical and hippocampal neurons (Cheng et al., 2007), possibly through miR-132-mediated inhibition of p250GAP, a protein associated with dendritic plasticity (Wayman et al., 2008).
The ability of ncRNAs to regulate transcription factors and chromatin remodeling proteins represent additional mechanisms to influence long-term neuroadaptations involved in memory formation. For example, miR-324 and miR-369, two miRNAs implicated in cocaine-induced neuroplasticity (Schaefer et al., 2010), have been shown to modulate transcription factors (MEF2 and FosB) important in reward-related learning and memory (Hiroi et al., 1997; Pulipparacharuvil et al., 2008). Additionally, miR-132 regulation of chromatin remodeling factors methyl CpG binding protein 2 (MeCP2), p300, and Jumonji and ARID domain protein 1 A (JARID1A) in the suprachiasmatic nucleus is important in neuroadaptations associated with circadian rhythm (Alvarez-Saavedra et al., 2011). Although no study to date has examined lncRNAs in learning and memory paradigms, lncRNAs could potentially be involved in long-term neuroadaptations, as they have been shown to regulate transcriptional factors, DNA methylation, and histone modifications (Rinn et al., 2007; Houseley et al., 2008; Yu et al., 2008; Khalil et al., 2009).
The studies reviewed above clearly show the importance of miRNAs (and potentially lncRNAs) in synaptic plasticity and learning and memory. However, many questions remain for future studies to better understand the role of ncRNAs in neuroplasticity. For example, the relationship between ncRNA-mediated plasticity and psychiatric disorders, such as addiction, has raised a number of intriguing questions: Are the same ncRNAs that are involved in plasticity also involved in addiction, or are other addiction-related ncRNAs involved, whose functions have yet to be determined? What are the specific targets of ncRNAs and how do these interactions contribute to the neuroadaptations associated with addiction? These are just a few questions that are currently being addressed to understand the complex relationship between ncRNA-mediated plasticity and addiction.
Emerging Role for miRNAs in Addiction
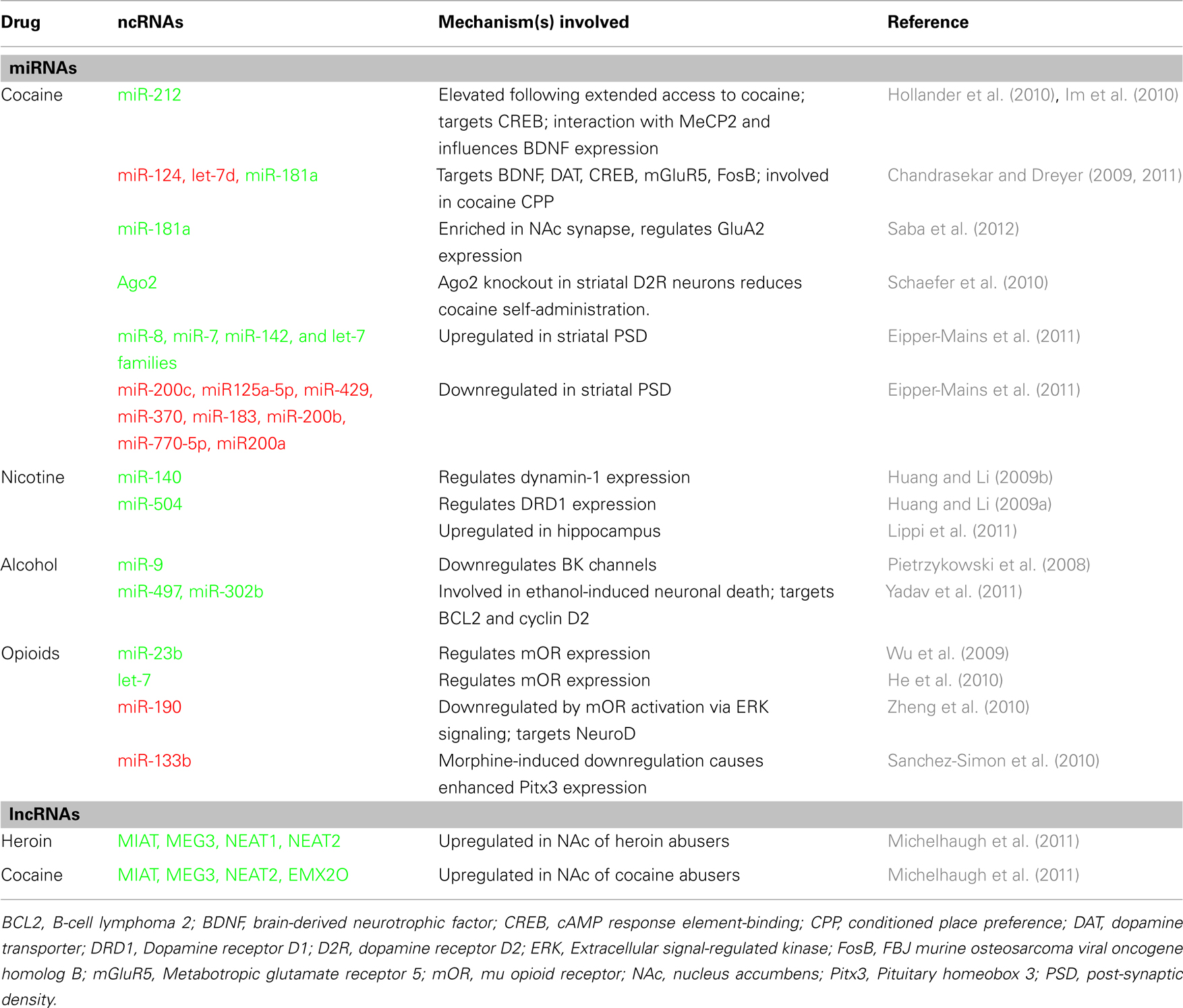
Drugs of abuse induce persistent changes in neuroplasticity by usurping gene regulatory mechanisms, in turn leading to addiction. Given the role of miRNAs in gene regulation and synaptic plasticity, recent studies have begun to examine the involvement of miRNAs in response to drugs of abuse (Table 1). Here, we highlight some of the recent findings that illustrate the importance of miRNAs in drug-induced synaptic plasticity, drug-seeking behaviors, and tolerance to several abused drugs.
Cocaine and Amphetamine
To investigate the role of miRNAs in cocaine addiction, Chandrasekar and Dreyer (2009) utilized in silico prediction models to identify miRNAs that potentially regulate cocaine-associated genes and discovered a strong prediction for miR-124, let-7d, and miR-181a. Further in vivo studies showed that expression of miR-181a is increased and miR-124 and let-7d are decreased in striatum of rats with a history of chronic cocaine exposure. Subsequent behavioral studies revealed that over expression of miR-124 and let-7d in the nucleus accumbens reduced cocaine conditioned place preference (CPP), whereas over expression of miR-181a enhanced cocaine preference (Chandrasekar and Dreyer, 2011). These effects on cocaine CPP were inversed when expression of the aforementioned miRNAs were inhibited. Interestingly, it was found that altering expression of these miRNAs in the nucleus accumbens modulated the expression of many addiction-related genes (Chandrasekar and Dreyer, 2009). For example, overexpression of miR-124 and let-7d increased dopamine transporter, whereas miR-181a over expression decreased it. Because the dopamine transporter is the primary target of cocaine and is importantly involved in cocaine CPP (Tilley et al., 2009), the observed behavioral changes are likely reflected, in part, by miRNA-regulation of the dopamine transporter. Notably, the expression of several other addiction-related genes, such as Brain-derived neurotrophic (BDNF), CREB, MeCP2, and ΔFosB were also regulated by these miRNAs, thus illustrating the widespread effects on addiction-related gene networks in response to changes in miRNA levels.
Using an extended access model of cocaine self-administration, Hollander et al. (2010) examined the role of miRNAs in regulating compulsive-like cocaine intake. In this study, dorsal striatal miR-212 levels were found to be significantly elevated in rats with a history of extended access to cocaine, but not in rats with short-access to cocaine or in rats receiving non-contingent cocaine exposure (yoked control). Further investigation showed that over expression or knockdown of miR-212 in the dorsal striatum decreased or enhanced cocaine self-administration under extended access conditions, respectively, suggesting that striatal miR-212 is involved in an adaptive response to inhibit escalation of cocaine intake. Reduced motivation to consume cocaine was attributed in part by miR-212-mediated upregulation of CREB, a known antagonistic regulator of cocaine reward (Carlezon et al., 1998). Subsequent studies from the same laboratory revealed that MeCP2 regulates cocaine intake via a homeostatic interaction with miR-212 to influence cocaine-mediated effects on striatal BDNF (Im et al., 2010). By revealing an important miRNA-mediated epigenetic mechanism involved in drug-seeking behaviors, this study raises a number of interesting possibilities because epigenetic factors associated with addiction have recently been the focus of intense investigation (Robison and Nestler, 2011).
Recent studies have examined synaptic expression of miRNAs in response to cocaine. By isolating striatal post-synaptic densities (PSD), Eipper-Mains et al. (2011) identified more than two dozen miRNAs that were significantly altered following chronic cocaine treatment. Interestingly, many of the PSD miRNAs affected by cocaine were found to be members of one of four families (miR-8, miR-7, miR-142, and let-7 families), suggesting that cocaine influences the expression of similar miRNAs with shared synaptic targets. In a similar study, Saba et al. (2012) utilized microarray screening to identify nine enriched and seven depleted miRNAs in the synaptodendritic compartment of the nucleus accumbens. They also revealed that miR-181a, one of the synaptically enriched miRNAs, is increased in reward-related brain regions following cocaine and amphetamine exposure, and miR-181a regulates synaptic plasticity by altering AMPA receptor subunit (GluA2) expression (Saba et al., 2012). Additional reports examining the role of miRNAs in drug-induced synaptic plasticity determined that miR-29a/b was significantly upregulated in addiction-related brain regions in mice with a history of cocaine or amphetamine exposure and plays an pivotal role in synaptic structure and function in vitro (Lippi et al., 2011).
Enzymes that regulate miRNA processing also appear to play a functional role in cocaine addiction. A study by Schaefer et al. (2010) revealed that knockout of argonaute 2 (Ago2, a protein important in miRNA processing) in accumbal dopamine 2 receptor expressing neurons significantly attenuated cocaine self-administration. Further investigation revealed that Ago2 regulates expression of numerous miRNAs in the striatum, and many of the Ago2-dependent miRNAs were predicted to target genes important in cocaine addiction. Consistent with the role of Ago2 in cocaine addiction, Eipper-Mains et al. (2011) showed that chronic cocaine exposure elevates Ago2 expression in the striatum. Dicer, an enzyme responsible for producing mature miRNAs, may also be necessary in cocaine addiction, as previous studies have shown that manipulation of this enzyme affects learning and memory (Konopka et al., 2010) and miRNA expression in striatum (Cuellar et al., 2008).
Nicotine
Utilizing miRNA microarray approach, Huang and Li identified 25 miRNAs that were altered in PC12 cells following nicotine exposure. They also found that miR-140 binds and reduces expression of dynamin-1 (Huang and Li, 2009b), a GTPase that has previously been shown to be important in nicotine dependence (Hwang and Li, 2006). The same research group showed that miR-504 targets a specific dopamine receptor D1 gene containing a single nucleotide polymorphism that has been associated with nicotine dependence (Huang and Li, 2009a). Interestingly, by increasing D1 receptor expression, miR-504 may promote nicotine intake by enhancing dopamine signaling. Finally, chronic injections of nicotine increased several miRNAs in mouse hippocampus, prefrontal cortex, limbic forebrain, and midbrain (Lippi et al., 2011), indicating that nicotine has broad effects on miRNA expression in several addiction-related brain areas, though the implications are not yet clear.
Alcohol
In a seminal set of studies, Pietrzykowski et al. (2008) revealed that alcohol upregulates miR-9 in rat striatum and supraoptic nucleus, two regions important in alcohol tolerance. The increase in miR-9 was found to contribute to alcohol tolerance by preferentially targeting BK channel mRNA isoforms that are sensitive to alcohol. BK channels, large conductance calcium, and voltage-activated potassium channels, are important in neuronal excitability, firing frequency, and neurotransmitter release and have been one of the best described targets for alcohol tolerance (for review see Treistman and Martin, 2009). Thus, miR-9-induced destabilization of alcohol sensitive BK channels likely contributes to alcohol tolerance and addiction by promoting the expression of more tolerant BK channel isoforms. Interestingly, the role of miR-9 in alcohol dependence may not be limited to tolerance, as miR-9 was also found to target other genes that have been implicated in addiction, such as dopamine receptor D2 and histone deacetylase 5 (Pietrzykowski et al., 2008).
Other alcohol-related studies have identified miRNAs involved in ethanol dependence. Guo et al. reported differential miRNA expression patterns following chronic ethanol exposure and ethanol removal in primary cortical neuron cultures. These results may indicate that different stages of alcohol addiction (maintenance, withdrawal, etc.) have distinct miRNA expression profiles (Guo et al., 2011), information that could be important for the development of new therapeutics to treat alcohol addiction. In another study, miR-497 and miR-302b were found to be involved in ethanol-induced neuronal cell death following chronic ethanol exposure, thereby providing a possible link between miRNAs and neuronal loss associated with chronic alcohol abuse (Yadav et al., 2011). In addition, recent studies using human post-mortem tissue, revealed that 35 miRNAs were significantly upregulated in the prefrontal cortex in alcoholics (Lewohl et al., 2011), again suggesting that chronic alcohol has widespread affects on miRNA expression reward-related brain areas. The development of artificial miRNAs may be a novel approach to treating alcoholism and other forms of addiction, as one recent study showed that targeting neurokinin-1 receptor gene with an artificial miRNA significantly reduced alcohol consumption in mice (Baek et al., 2010).
Opioids
Similar to alcohol, specific miRNAs have been implicated in opioid tolerance and addiction. miRNAs, let-7, and miR-23b suppress mu opioid receptor mRNA expression following long-term morphine treatment (Wu et al., 2009; He et al., 2010), demonstrating a new mechanism that might play an important role in morphine tolerance. In addition, the mu opioid receptor agonist, fentanyl, but not morphine, downregulates miR-190 expression via extracellular signal-regulated kinase (ERK) signaling (Zheng et al., 2010), indicating that specific mu opioid receptor agonists have differential influence on miRNA expression.
Summary
The studies highlighted in this section illustrate a wide range of miRNA-mediated mechanisms involved in addiction. With their ability to regulate addiction-related gene networks, drug-induced plasticity, drug-seeking behaviors, and drug tolerance, miRNAs are ideal therapeutic targets for the treatment of addiction. However, much more research in this nascent field is needed to reveal miRNA targets and mechanisms that contribute importantly to the addicted state. Thus, it seems clear that future studies will reveal ever more complex and intriguing properties of these key ncRNAs in addiction.
Potential Role for Long Non-Coding RNAs in Addiction
Recent large-scale genomic studies have revealed that lncRNAs are one of the most abundant classes of ncRNAs (Jia et al., 2010; Kapranov et al., 2010). Additionally, lncRNAs have been implicated in a number of important cellular processes including gene transcription, RNA processing, and chromatin modifications (Wang and Chang, 2011). Although lncRNAs are highly expressed in the brain (Mercer et al., 2008; Belgard et al., 2011), they remain poorly characterized in this context and their role in addiction is unclear. In an attempt to determine whether lncRNAs are differently expressed in response to chronic drug use, a recent study by Michelhaugh et al. (2011) identified lncRNAs that were altered in heroin abusers by mining existing Affymetrix datasets. Of the 23 lncRNAs identified, MIAT, MEG3, NEAT1, and NEAT2 were upregulated in the nucleus accumbens of heroin abusers compared to control subjects. Preliminary reports from the same research group also found that NEAT2, MIAT, MEG3, and EMX2OS are elevated in the nucleus accumbens of cocaine abusers (Michelhaugh et al., 2011), suggesting similar aberrations in lncRNA expression in response to different drugs of abuse. The lncRNAs listed above have been implicated in a range of cellular processes including cAMP signaling (Zhao et al., 2006), GABA neuron neurogenesis (Mercer et al., 2010), and regulation of genes associated with synaptic plasticity (Bernard et al., 2010), but the functional role of these lncRNAs in addiction remains unknown. Although these initial findings are intriguing, only a small number of lncRNAs were examined, suggesting the need for more comprehensive analysis of transcriptome changes during these critical events, with emphasis on specific cell types and locus specific complexity of lncRNA expression during these changes.
Natural antisense transcripts (NATs), a subset of lncRNAs, are transcripts derived from the opposite strand of many protein coding (sense) genes. NATs bind to sense RNA and/or proteins to regulate transcription and translation. Recently, we demonstrated that BDNF, a gene known to be involved in addiction, is controlled by a conserved long non-coding antisense RNA transcript (BDNF-AS; Modarresi et al., 2012). BDNF-AS suppresses BDNF mRNA expression by altering chromatin structure at the BDNF gene locus. Inhibiting BDNF-AS by siRNA or other methods robustly increased BDNF mRNA and protein expression and enhanced neuronal outgrowth. Given the important role of BDNF in cocaine addiction (for review see Ghitza et al., 2010), it would be interesting to determine if BDNF-AS is dysregulated in response to chronic drug use and contributes to drug-seeking behaviors. Other addiction-related NATs have also been identified (Zhang et al., 2007), but their role in addiction-related neuroadaptations and behaviors merits further research.
Although initial studies have identified a potential involvement of lncRNAs in addiction, many additional questions remain – whether specific lncRNAs are necessary for drug-seeking behaviors, whether different drugs of abuse affect different lncRNAs, whether specific lncRNAs are preferentially expressed in reward-related brain areas, and whether differences across species exist (mice vs. rats vs. humans). Thus, a major research goal is to address these important unanswered questions in order to understand the underlying mechanisms of abused drugs and to identify useful targets for the treatment of addiction.
Epigenetic-ncRNA Interactions: Potential Involvement in Drug-Induced Neuroadaptations
The epigenome consists of DNA methylation and several modifications (acetylation, methylation, phosphorylation, etc.) to specific amino acid residues on histone proteins. Chromatin-modifying complexes play an important role in transcriptional regulation by adding or removing covalent modifications to histone proteins. Several key chromatin-modifying proteins have recently been implicated in neuroadaptations associated with addiction. For example, specific enzymes responsible for histone acetylation, methylation, and DNA methylation in reward-related brain areas are critically involved in cocaine addiction (LaPlant et al., 2010; Maze et al., 2010; Wang et al., 2010). However, it has been largely unclear how these proteins target specific regions of the genome, given that the majority of chromatin-modifying proteins lack DNA binding capacity.
Increasing evidence now indicates that chromatin-modifying complexes are directed to their sites of action by lncRNAs. Therefore, it is possible that lncRNAs play an important role in addiction by regulating epigenetic processes. Although no study has examined lncRNA-mediated epigenetic mechanisms in the context of addiction, we speculate that such interactions are important (Figure 1), given that several addiction-related epigenetic factors associate with lncRNAs in tissues outside of the brain (Khalil et al., 2009). For example, in the placenta, lncRNAs Air, and Kcnq1ot1 regulate histone methylation by interacting with the histone methyltransferase G9a (Nagano et al., 2008; Pandey et al., 2008), an epigenetic enzyme that is downregulated in the accumbens following chronic cocaine exposure and is important in cocaine-related behaviors (Maze et al., 2010). Additionally, lncRNA-mediated regulation of histone acetylation, methylation, or DNA methylation, key modifications important in cocaine addiction (for review see, Maze and Nestler, 2011), have also been reported (Rinn et al., 2007; Houseley et al., 2008; Yu et al., 2008; Yap et al., 2010). Interestingly, not only can lncRNAs influence the activity of chromatin-modifying complexes, but evidence now indicates that alterations in epigenetic processes can alter the expression of lncRNAs (Johnson et al., 2009). Whether these specific interactions are important in the brain during drug-seeking behaviors, however, remains to be investigated.
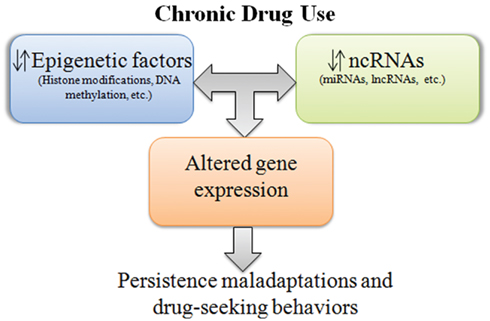
Figure 1. Simplified representation of ncRNA–epigenetic interactions in response to chronic drug use. Long-term use of drugs alters expression of ncRNAs, epigenetic factors, and the interactions between these processes, causing persistent perturbations in gene expression. Chronic aberrations in gene expression are thought to evoke maladaptive neuroadaptations associated with addiction.
MicroRNAs also interact with epigenetic factors important in addiction. As previously described, Im et al. (2010) found that miR-212 influences cocaine seeking by inhibiting MeCP2 in the dorsal striatum. Other epigenetic enzymes that have been implicated in addiction, such as DNA methyltransferase 3A, histone deacetylases 4, and sirtuin 1, are also regulated by specific miRNAs (Chen et al., 2006; Fabbri et al., 2007; Gao et al., 2010), but the significance of these associations in addiction-related behaviors is unknown.
Although the full spectrum of ncRNA-epigenetic associations in the CNS has yet to be seen, these interactions appear to play an essential role in fine-tuning gene expression and proper brain functioning. In drug addiction, it is possible that chronic drug use leads to long-lasting aberrations in ncRNA-mediated epigenetic mechanisms that lead to persistent drug-seeking behaviors. However, additional research is needed to determine the molecular underpinnings involved in ncRNA-epigenetic interactions in the brain and if these interactions contribute importantly to the addicted state.
Future Directions
Although a growing number of reports have implicated miRNAs in addiction-related neuroadaptations, future studies are needed to determine if lncRNAs also play a critical role in drug-seeking behaviors. Given that lncRNAs are the most abundant ncRNA in the brain and critically involved in an array of cellular processes, identifying specific lncRNAs that are regulated by drugs of abuse is likely to be a valuable approach for revealing the underlying mechanisms of addiction. With the recent development of new technologies, such as capture hybridization analysis of RNA targets (CHART) and Chromatin Isolation by RNA Purification (ChIRP; Chu et al., 2011; Simon et al., 2011), it is now possible to identify novel lncRNA-DNA or lncRNA-protein interactions involved in addiction, and future studies using these techniques will determine if these interactions are altered following prolonged drug use. In addition, as recent studies indicate that lncRNAs are differentially expressed in certain brain regions (Mercer et al., 2008; Belgard et al., 2011), identifying preferential expression of specific lncRNAs in reward-related brain areas might lead to new targets for the treatment of addiction. Finally, new therapeutic strategies and delivery approaches that target RNAs are now being explored (Bitko and Barik, 2007; Wood et al., 2007; Hung et al., 2011). Thus, using these new techniques to target ncRNAs holds great potential for treating several psychiatric disorders, including addiction.
Conclusion
Studies over the last several years have established a broad functional context for ncRNAs in the computational matrix of the nervous system. At the cellular level, nervous system signaling networks involve small RNAs at the synapse, where they regulate activity-dependent mRNA translation, and in turn, learning and memory-related plasticity. At the same time, in the nucleus, long RNAs function to provide temporal and spatial information to an array of epigenetic signaling systems. Chronic drug use likely perturbs these networks during the process of addiction in ways that cause a loss of plasticity and in turn establish barriers to the return to homeostasis.
Yet, the complexity of the nervous system suggests that additional layers of ncRNA-mediated events likely occur during the process of addiction. For example, recent evidence suggests that lncRNAs can serve as decoys or storage locations for small RNAs (Tay et al., 2011), in effect competing with targets for the occupancy of effector small RNAs and modulating their downstream effects (Salmena et al., 2011). In neurons this process could function together with anterograde and retrograde transport of RNA–protein vesicles, providing the potential for a link between small RNA mediated translational control at the synapse, and lncRNA-mediated chromatin signaling in the nucleus. Stresses such as repeated increases in intracellular Ca++ levels could reduce the performance of vesicle trafficking and lead to the progressive decoupling of such ncRNA-mediated systems relatively early in the process of addictive maladaptations.
Non-coding RNAs may also play a role in signaling between cells. Circulating exosomes and microvesicles contain many RNA species, and have the ability to traffic these RNAs from one cell type to another (Smalheiser, 2007; Dinger et al., 2008), even supporting the metastatic environment of the soma in some types of cancer. While not yet documented in the context of the nervous system, vesicle based transfer of ncRNAs could offer an additional dimension of intercellular communication in the nervous system. Such vesicles could mediate signaling between neurons and glia, for example, in response to impending cytotoxicity or other stress events. Thus, the versatility of ncRNA-based information processing provides many still unexplored avenues for function in the nervous system and involvement in the stress responses that lead to addictive maladaptations.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Preparation of this review was supported by NIH grants R01 MH083733 and R01 NS063974.
References
Alvarez-Saavedra, M., Antoun, G., Yanagiya, A., Oliva-Hernandez, R., Cornejo-Palma, D., Perez-Iratxeta, C., Sonenberg, N., and Cheng, H. Y. (2011). miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum. Mol. Genet. 20, 731–751.
Baek, M. N., Jung, K. H., Halder, D., Choi, M. R., Lee, B. H., Lee, B. C., Jung, M. H., Choi, I. G., Chung, M. K., Oh, D. Y., and Chai, Y. G. (2010). Artificial microRNA-based neurokinin-1 receptor gene silencing reduces alcohol consumption in mice. Neurosci. Lett. 475, 124–128.
Belgard, T. G., Marques, A. C., Oliver, P. L., Abaan, H. O., Sirey, T. M., Hoerder-Suabedissen, A., Garcia-Moreno, F., Molnar, Z., Margulies, E. H., and Ponting, C. P. (2011). A transcriptomic atlas of mouse neocortical layers. Neuron 71, 605–616.
Berezikov, E. (2011). Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 12, 846–860.
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., Zhang, M. Q., Sedel, F., Jourdren, L., Coulpier, F., Triller, A., Spector, D. L., and Bessis, A. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093.
Bitko, V., and Barik, S. (2007). Intranasal antisense therapy: preclinical models with a clinical future? Curr. Opin. Mol. Ther. 9, 119–125.
Carlezon, W. A. Jr., Thome, J., Olson, V. G., Lane-Ladd, S. B., Brodkin, E. S., Hiroi, N., Duman, R. S., Neve, R. L., and Nestler, E. J. (1998). Regulation of cocaine reward by CREB. Science 282, 2272–2275.
Chandrasekar, V., and Dreyer, J. L. (2009). microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol. Cell. Neurosci. 42, 350–362.
Chandrasekar, V., and Dreyer, J. L. (2011). Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 36, 1149–1164.
Chen, J. F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., Conlon, F. L., and Wang, D. Z. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233.
Cheng, H. Y., Papp, J. W., Varlamova, O., Dziema, H., Russell, B., Curfman, J. P., Nakazawa, T., Shimizu, K., Okamura, H., Impey, S., and Obrietan, K. (2007). microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829.
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E., and Chang, H. Y. (2011). Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678.
Cuellar, T. L., Davis, T. H., Nelson, P. T., Loeb, G. B., Harfe, B. D., Ullian, E., and McManus, M. T. (2008). Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 105, 5614–5619.
Dinger, M. E., Mercer, T. R., and Mattick, J. S. (2008). RNAs as extracellular signaling molecules. J. Mol. Endocrinol. 40, 151–159.
Edbauer, D., Neilson, J. R., Foster, K. A., Wang, C. F., Seeburg, D. P., Batterton, M. N., Tada, T., Dolan, B. M., Sharp, P. A., and Sheng, M. (2010). Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384.
Eipper-Mains, J. E., Kiraly, D. D., Palakodeti, D., Mains, R. E., Eipper, B. A., and Graveley, B. R. (2011). microRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA 17, 1529–1543.
Fabbri, M., Garzon, R., Cimmino, A., Liu, Z., Zanesi, N., Callegari, E., Liu, S., Alder, H., Costinean, S., Fernandez-Cymering, C., Volinia, S., Guler, G., Morrison, C. D., Chan, K. K., Marcucci, G., Calin, G. A., Huebner, K., and Croce, C. M. (2007). MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U.S.A. 104, 15805–15810.
French, P. J., Bliss, T. V., and O’Connor, V. (2001). Ntab, a novel non-coding RNA abundantly expressed in rat brain. Neuroscience 108, 207–215.
Gao, J., Wang, W. Y., Mao, Y. W., Graff, J., Guan, J. S., Pan, L., Mak, G., Kim, D., Su, S. C., and Tsai, L. H. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109.
Ghitza, U. E., Zhai, H., Wu, P., Airavaara, M., Shaham, Y., and Lu, L. (2010). Role of BDNF and GDNF in drug reward and relapse: a review. Neurosci. Biobehav. Rev. 35, 157–171.
Guo, Y., Chen, Y., Carreon, S., and Qiang, M. (2011). Chronic intermittent ethanol exposure and its removal induce a different mirna expression pattern in primary cortical neuronal cultures. Alcohol. Clin. Exp. Res. doi: 10.1111/j.1530-0277.2011.01689.x. [Epub ahead of print].
He, Y., Yang, C., Kirkmire, C. M., and Wang, Z. J. (2010). Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J. Neurosci. 30, 10251–10258.
Hiroi, N., Brown, J. R., Haile, C. N., Ye, H., Greenberg, M. E., and Nestler, E. J. (1997). FosB mutant mice: loss of chronic cocaine induction of Fos-related proteins and heightened sensitivity to cocaine’s psychomotor and rewarding effects. Proc. Natl. Acad. Sci. U.S.A. 94, 10397–10402.
Hollander, J. A., Im, H. I., Amelio, A. L., Kocerha, J., Bali, P., Lu, Q., Willoughby, D., Wahlestedt, C., Conkright, M. D., and Kenny, P. J. (2010). Striatal microRNA controls cocaine intake through CREB signalling. Nature 466, 197–202.
Houseley, J., Rubbi, L., Grunstein, M., Tollervey, D., and Vogelauer, M. (2008). A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 32, 685–695.
Huang, W., and Li, M. D. (2009a). Differential allelic expression of dopamine D1 receptor gene (DRD1) is modulated by microRNA miR-504. Biol. Psychiatry 65, 702–705.
Huang, W., and Li, M. D. (2009b). Nicotine modulates expression of miR-140*, which targets the 3′-untranslated region of dynamin 1 gene (Dnm1). Int. J. Neuropsychopharmacol. 12, 537–546.
Hung, G., Mazur, C., Wancewicz, E., Norris, D., and Bennet, F. (2011). Second Generation Oligonucleotide CNS Delivery via IT Administration in Mice, Rats and Monkey. Society for Neuroscience, Washington 557.06.
Hwang, Y. Y., and Li, M. D. (2006). Proteins differentially expressed in response to nicotine in five rat brain regions: identification using a 2-DE/MS-based proteomics approach. Proteomics 6, 3138–3153.
Im, H. I., Hollander, J. A., Bali, P., and Kenny, P. J. (2010). MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 13, 1120–1127.
Jia, H., Osak, M., Bogu, G. K., Stanton, L. W., Johnson, R., and Lipovich, L. (2010). Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 16, 1478–1487.
Johnson, R., Teh, C. H., Jia, H., Vanisri, R. R., Pandey, T., Lu, Z. H., Buckley, N. J., Stanton, L. W., and Lipovich, L. (2009). Regulation of neural macroRNAs by the transcriptional repressor REST. RNA 15, 85–96.
Kapranov, P., St Laurent, G., Raz, T., Ozsolak, F., Reynolds, C. P., Sorensen, P. H., Reaman, G., Milos, P., Arceci, R. J., Thompson, J. F., and Triche, T. J. (2010). The majority of total nuclear-encoded non-ribosomal RNA in a human cell is “dark matter” un-annotated RNA. BMC Biol. 8, 149. doi:10.1186/1741-7007-8-149
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., Thomas, K., Presser, A., Bernstein, B. E., van Oudenaarden, A., Regev, A., Lander, E. S., and Rinn, J. L. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672.
Konopka, W., Kiryk, A., Novak, M., Herwerth, M., Parkitna, J. R., Wawrzyniak, M., Kowarsch, A., Michaluk, P., Dzwonek, J., Arnsperger, T., Wilczynski, G., Merkenschlager, M., Theis, F. J., Kohr, G., Kaczmarek, L., and Schutz, G. (2010). MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30, 14835–14842.
Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739.
LaPlant, Q., Vialou, V., Covington, H. E. III, Dumitriu, D., Feng, J., Warren, B. L., Maze, I., Dietz, D. M., Watts, E. L., Iñiguez, S. D., Koo, J. W., Mouzon, E., Renthal, W., Hollis, F., Wang, H., Noonan, M. A., Ren, Y., Eisch, A. J., Bolaños, C. A., Kabbaj, M., Xiao, G., Neve, R. L., Hurd, Y. L., Oosting, R. S., Fan, G., Morrison, J. H., and Nestler, E. J. (2010). Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat. Neurosci. 13, 1137–1143.
Lewohl, J. M., Nunez, Y. O., Dodd, P. R., Tiwari, G. R., Harris, R. A., and Mayfield, R. D. (2011). Up-regulation of MicroRNAs in brain of human alcoholics. Alcohol. Clin. Exp. Res. 35, 1928–1937.
Lin, D., Pestova, T. V., Hellen, C. U., and Tiedge, H. (2008). Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol. Cell. Biol. 28, 3008–3019.
Lippi, G., Steinert, J. R., Marczylo, E. L., D’Oro, S., Fiore, R., Forsythe, I. D., Schratt, G., Zoli, M., Nicotera, P., and Young, K. W. (2011). Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J. Cell Biol. 194, 889–904.
Lugli, G., Torvik, V. I., Larson, J., and Smalheiser, N. R. (2008). Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J. Neurochem. 106, 650–661.
Makeyev, E. V., Zhang, J., Carrasco, M. A., and Maniatis, T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448.
Maze, I., Covington, H. E. III, Dietz, D. M., LaPlant, Q., Renthal, W., Russo, S. J., Mechanic, M., Mouzon, E., Neve, R. L., Haggarty, S. J., Ren, Y., Sampath, S. C., Hurd, Y. L., Greengard, P., Tarakhovsky, A., Schaefer, A., and Nestler, E. J. (2010). Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327, 213–216.
Maze, I., and Nestler, E. J. (2011). The epigenetic landscape of addiction. Ann. N. Y. Acad. Sci. 1216, 99–113.
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F., and Mattick, J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 105, 716–721.
Mercer, T. R., Qureshi, I. A., Gokhan, S., Dinger, M. E., Li, G., Mattick, J. S., and Mehler, M. F. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11, 14. doi:10.1186/1471-2202-11-14
Michelhaugh, S. K., Lipovich, L., Blythe, J., Jia, H., Kapatos, G., and Bannon, M. J. (2011). Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J. Neurochem. 116, 459–466.
Modarresi, F., Faghihi, M. A., Lopez-Toledano, M. A., Fatemi, R. P., Magistri, M., Brothers, S. P., van der Brug, M. P., and Wahlestedt, C. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459.
Nagano, T., Mitchell, J. A., Sanz, L. A., Pauler, F. M., Ferguson-Smith, A. C., Feil, R., and Fraser, P. (2008). The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720.
Nudelman, A. S., DiRocco, D. P., Lambert, T. J., Garelick, M. G., Le, J., Nathanson, N. M., and Storm, D. R. (2010). Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 20, 492–498.
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., Nagano, T., Mancini-Dinardo, D., and Kanduri, C. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246.
Pietrzykowski, A. Z., Friesen, R. M., Martin, G. E., Puig, S. I., Nowak, C. L., Wynne, P. M., Siegelmann, H. T., and Treistman, S. N. (2008). Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron 59, 274–287.
Ponjavic, J., Oliver, P. L., Lunter, G., and Ponting, C. P. (2009). Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 5, e1000617. doi:10.1371/journal.pgen.1000617
Pulipparacharuvil, S., Renthal, W., Hale, C. F., Taniguchi, M., Xiao, G., Kumar, A., Russo, S. J., Sikder, D., Dewey, C. M., Davis, M. M., Greengard, P., Nairn, A. C., Nestler, E. J., and Cowan, C. W. (2008). Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron 59, 621–633.
Rajasethupathy, P., Fiumara, F., Sheridan, R., Betel, D., Puthanveettil, S. V., Russo, J. J., Sander, C., Tuschl, T., and Kandel, E. (2009). Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63, 803–817.
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., Goodnough, L. H., Helms, J. A., Farnham, P. J., Segal, E., and Chang, H. Y. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323.
Robison, A. J., and Nestler, E. J. (2011). Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 12, 623–637.
Saba, R., Storchel, P. H., Aksoy-Aksel, A., Kepura, F., Lippi, G., Plant, T. D., and Schratt, G. M. (2012). Dopamine-regulated MicroRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Mol. Cell. Biol. 32, 619–632.
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358.
Sanchez-Simon, F. M., Zhang, X. X., Loh, H. H., Law, P. Y., and Rodriguez, R. E. (2010). Morphine regulates dopaminergic neuron differentiation via miR-133b. Mol. Pharmacol. 78, 935–942.
Schaefer, A., Im, H. I., Veno, M. T., Fowler, C. D., Min, A., Intrator, A., Kjems, J., Kenny, P. J., O’Carroll, D., and Greengard, P. (2010). Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J. Exp. Med. 207, 1843–1851.
Schratt, G. M., Tuebing, F., Nigh, E. A., Kane, C. G., Sabatini, M. E., Kiebler, M., and Greenberg, M. E. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289.
Sempere, L. F., Cole, C. N., McPeek, M. A., and Peterson, K. J. (2006). The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J. Exp. Zool. B Mol. Dev. Evol. 306, 575–588.
Siegel, G., Obernosterer, G., Fiore, R., Oehmen, M., Bicker, S., Christensen, M., Khudayberdiev, S., Leuschner, P. F., Busch, C. J., Kane, C., Hübel, K., Dekker, F., Hedberg, C., Rengarajan, B., Drepper, C., Waldmann, H., Kauppinen, S., Greenberg, M. E., Draguhn, A., Rehmsmeier, M., Martinez, J., and Schratt, G. M. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716.
Siegel, G., Saba, R., and Schratt, G. (2011). microRNAs in neurons: manifold regulatory roles at the synapse. Curr. Opin. Genet. Dev. 21, 491–497.
Simon, M. D., Wang, C. I., Kharchenko, P. V., West, J. A., Chapman, B. A., Alekseyenko, A. A., Borowsky, M. L., Kuroda, M. I., and Kingston, R. E. (2011). The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 108, 20497–20502.
Smalheiser, N. R. (2007). Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol. Direct 2, 35.
Taft, R. J., Pheasant, M., and Mattick, J. S. (2007). The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29, 288–299.
Tay, Y., Kats, L., Salmena, L., Weiss, D., Tan, S. M., Ala, U., Karreth, F., Poliseno, L., Provero, P., Di Cunto, F., Lieberman, J., Rigoutsos, I., and Pandolfi, P. P. (2011). Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147, 344–357.
Tilley, M. R., O’Neill, B., Han, D. D., and Gu, H. H. (2009). Cocaine does not produce reward in absence of dopamine transporter inhibition. Neuroreport 20, 9–12.
Treistman, S. N., and Martin, G. E. (2009). BK Channels: mediators and models for alcohol tolerance. Trends Neurosci. 32, 629–637.
Wang, K. C., and Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914.
Wang, L., Lv, Z., Hu, Z., Sheng, J., Hui, B., Sun, J., and Ma, L. (2010). Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIalpha in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology 35, 913–928.
Wayman, G. A., Davare, M., Ando, H., Fortin, D., Varlamova, O., Cheng, H. Y., Marks, D., Obrietan, K., Soderling, T. R., Goodman, R. H., and Impey, S. (2008). An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. U.S.A. 105, 9093–9098.
Wibrand, K., Panja, D., Tiron, A., Ofte, M. L., Skaftnesmo, K. O., Lee, C. S., Pena, J. T., Tuschl, T., and Bramham, C. R. (2010). Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur. J. Neurosci. 31, 636–645.
Wood, M., Yin, H., and McClorey, G. (2007). Modulating the expression of disease genes with RNA-based therapy. PLoS Genet. 3, e109. doi:10.1371/journal.pgen.0030109
Wu, Q., Zhang, L., Law, P. Y., Wei, L. N., and Loh, H. H. (2009). Long-term morphine treatment decreases the association of mu-opioid receptor (MOR1) mRNA with polysomes through miRNA23b. Mol. Pharmacol. 75, 744–750.
Yadav, S., Pandey, A., Shukla, A., Talwelkar, S. S., Kumar, A., Pant, A. B., and Parmar, D. (2011). miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J. Biol. Chem. 286, 37347–37357.
Yap, K. L., Li, S., Munoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S., Gil, J., Walsh, M. J., and Zhou, M. M. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674.
Yu, W., Gius, D., Onyango, P., Muldoon-Jacobs, K., Karp, J., Feinberg, A. P., and Cui, H. (2008). Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206.
Zhang, Y., Li, J., Kong, L., Gao, G., Liu, Q. R., and Wei, L. (2007). NATsDB: natural antisense transcripts DataBase. Nucleic Acids Res. 35, D156–D161.
Zhao, J., Zhang, X., Zhou, Y., Ansell, P. J., and Klibanski, A. (2006). Cyclic AMP stimulates MEG3 gene expression in cells through a cAMP-response element (CRE) in the MEG3 proximal promoter region. Int. J. Biochem. Cell Biol. 38, 1808–1820.
Keywords: addiction, long non-coding RNA, microRNA, epigenetic, lncRNA, miRNA
Citation: Sartor GC, St. Laurent G III and Wahlestedt C (2012) The emerging role of non-coding RNAs in drug addiction. Front. Gene. 3:106. doi: 10.3389/fgene.2012.00106
Received: 28 February 2012; Accepted: 23 May 2012;
Published online: 22 June 2012.
Edited by:
Leonard Lipovich, Wayne State University, USAReviewed by:
Hongyan Xu, Georgia Health Sciences University, USAMelanie Carless, Texas Biomedical Research Institute, USA
Copyright: © 2012 Sartor, St. Laurent III and Wahlestedt. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Claes Wahlestedt, Hussman Institute for Human Genomics, University of Miami Miller School of Medicine, 1501 Northwest 10th Avenue BRB 407, Miami, FL 33136, USA. e-mail: cwahlestedt@med.miami.edu