- 1Program in Human Molecular Genetics, Department of Pediatrics, Children’s Hospital of Chicago Research Center, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA
- 2The Asher Center, Department of Psychiatry and Behavioral Sciences, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA
Genomic imprinting, the preferential expression of maternal or paternal alleles of imprinted genes, is often maintained through expression of imprinted long non-coding (lnc) “antisense” RNAs. These may overlap imprinted transcripts, and are expressed from the opposite allele. Previously we have described brain region-specific imprinted expression of the Dio3 gene in rat, which is preferentially modified by fetal ethanol exposure. The Dio3os (opposite strand) transcript is transcribed in opposite orientation to Dio3 in mouse and human, partially overlaps the Dio3 promoter, and mirrors total Dio3 developmental expression levels. Here, we present that the rat Dio3os transcript(s) exhibits brain region-specific imprinted expression patterns similar to those of Dio3. Rat Dio3os transcript expression is also similarly modified by fetal ethanol exposure. Uniquely, both Dio3 and Dio3os expression occur on the same, rather than opposite, alleles, as determined by strand-specific RT-PCR. Future studies will require direct manipulation of the Dio3os transcript to determine whether the novel paralleling of total and allele-specific expression patterns of this sense/antisense imprinted gene pair reflects an as-yet undefined regulatory mechanism for lncRNA mediated tissue-specific imprinted expression, or rather is a consequence of a more straightforward, but previously undescribed transcriptional coregulation process.
Introduction
Over the past several years, long non-coding RNAs (lncRNAs) have been found to be transcribed across the genome (e.g., Katayama et al., 2005; Huang et al., 2011). Many of these transcripts have a role in the regulation of gene expression, such as via direct overlap with genes or their promoters (reviewed in Amaral and Mattick, 2008) or interaction with and modification of epigenetic chromatin marks (Khalil et al., 2009). Historically, the first lncRNAs were identified within regions of genomic imprinting, characterized by preferential expression of maternal or paternal alleles of imprinted genes. Such imprinted expression is often maintained through expression of “antisense” lncRNAs, which overlap imprinted transcripts and are expressed from the opposite parental allele, potentially “blocking” expression from the “sense” allele on this parental chromosome (reviewed in Peters and Robson, 2008). In addition to direct “antisense” overlap, several imprinted lncRNAs have also been shown to exhibit long-distance cis effects on other genes within the regulatory clusters through interaction with chromatin and DNA modifying proteins (Nagano et al., 2008; Pandey et al., 2008), similar to what is being shown for lncRNAs in non-imprinted regions.
The type 3 deiodinase gene, Dio3, lies at the distal end of a large cluster of imprinted genes. Dio3 is preferentially paternally expressed in most tissues (Charalambous and Hernandez, 2012), as are the three other imprinted coding genes in this cluster (Hagan et al., 2009). In contrast, but consistent with the emerging realization of complex allele-preferential gene expression patterns across the genome, we have recently described brain region-specific imprinted expression of Dio3 in rat (Sittig et al., 2011a). This imprint profile is preferentially modified by fetal ethanol exposure and correlates with behavioral alterations (Sittig et al., 2011b), providing the first evidence of functional consequences of brain region-specific imprinted expression profiles. Although the mechanisms underlying these complex imprint patterns are not yet understood, many possibilities arise through comparison with other imprinted loci, as described above. Immediately adjacent to the Dio3 gene is a lncRNA transcript, Dio3os, that is transcribed in opposite orientation to Dio3 in mouse and human, partially overlaps the Dio3 promoter (Hernandez, 2005), and mirrors total Dio3 developmental and diurnal (Labialle et al., 2008) expression levels. Unlike the large number of large and small non-coding RNA genes within this cluster which are maternally expressed (daRocha et al., 2008), Dio3os has been reported to have biallelic, rather than imprinted, expression in murine embryos (Tierling et al., 2006) and in adult mouse cortex (Labialle et al., 2008). To investigate whether the Dio3os transcript exists in rat and thus might contribute to complex Dio3 expression regulation, we have analyzed the total and allele-specific expression patterns of rDio3os in naive and ethanol-exposed animals. These analyses demonstrate a combination of features and complexities of rDio3 expression that together define a novel category within imprinted, lncRNAs.
Results
Characterization and Brain Region-Specific Imprinting of rDio3os
Transcripts across the upstream rDio3 region were identified by the single poly-A-containing expressed sequence tag (EST) contained in the NCBI database, and by RT-PCR and rapid amplification of cDNA ends (RACE) across the region from placenta and both neonatal and adult rat frontal cortex (FX) and hippocampus (HP). Both unspliced and alternatively spliced transcripts were identified. rDio3os exons overlap those of mouse (Tierling et al., 2006) and extend into the 5′ rDio3 GC rich region, but splicing and exon–intron boundaries are not identical. Mouse and rat show an average of 83% identity across overlapping regions. Alternatively spliced RT-PCR products are indicated in Figure 1A, as are locations of representative primer pairs used for strand-specific RT-PCR (below). We have not been able to identify rDio3os splice variants that extend through and overlap the rDio3 5′UTR and transcription start site, as has been reported in mouse but not humans (Hernandez et al., 2004), although strand-specific RT-PCR has identified the presence of at least an 154 bp opposite strand (OS) transcript 60 bp upstream of the standard rDio3 5′UTR, within the minimal promoter and potentially overlapping an alternative transcription start site identified in keratinocytes (Dentice et al., 2007; Figure 1B, “Z”).
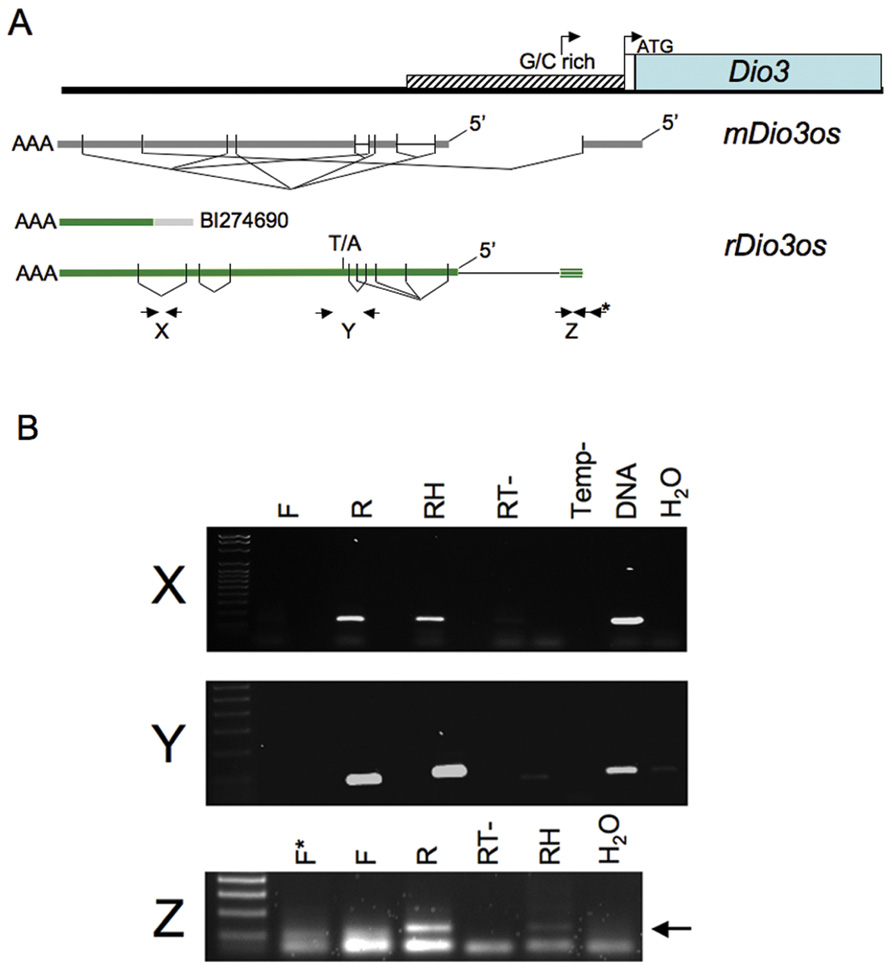
FIGURE 1.(A) Genomic organization of rDio3os transcripts across the upstream rDio3 region. The single rDio3os EST along with alternatively spliced RT-PCR and RACE products are shown in comparison to published mouse Dio3os exons (Hernandez, 2005) and the 5′ Dio3 genomic region (Dentice et al., 2007). rDio3 transcriptional (arrow) and translational (ATG) start sites are indicated. Locations of representative primer pairs used for strand-specific RT-PCR (X, Y, Z) are indicated (bold arrows), as is a T/A SNP used for allele-specific expression analysis. (B) Representative strand-specific RT-PCR of adult male rat hippocampal (X, Y) or placental (Z) total RNA using primers specific for opposite strand expression (reverse primer: R) or sense expression (forward primer: F) across the rDio3 upstream region shows expression is derived exclusively from the opposite strand. Controls: Positive: Random hexamer-primed RT-PCR (RH), which will copy potential transcripts from either strand; genomic DNA (DNA). Negative: Reverse transcriptase deficient (RT-) or template deficient (Temp-) RT-PCR reactions; template-deficient (H2O) PCR reactions.
Upstream Expression is Derived Exclusively from the Opposite Strand to rDio3
The EST BI274690, ~3–4 kb upstream of rDio3 (inclusive), does not encode an open reading frame but contains a poly-A sequence, strongly suggesting that its direction of transcription is opposite to that of rDio3 and that it represents the rDio3os transcript rather than an extended rDio3 5′UTR variant. We identified this poly-A sequence in RACE products as well. However, early reports of rDio3 expression based on Northern blot hybridization from rat brain (Tu et al., 1999) suggested the presence of alternative, longer (>2.1 kb) rDio3 transcripts, with later studies in human tissue demonstrating DIO3 transcription overlapping canonical DIO3 promoter sequences, as well as identifying a longer, 4.8 kb transcript that hybridized exclusively to the 5′ but not 3′ regions of DIO3 and was thus suggested not to encode the DIO3 protein (Hernandez et al., 2004). This 4.8 kb transcript would be predicted to overlap the 5′ end of the mDio3 5′UTR-overlapping mDio3os splice variant (Hernandez et al., 2002) and would be expected to extend at least 2 kb further 5′, depending on splicing profiles, therefore giving it further Dio3os overlap potential. However, in these experiments, (double-stranded) cDNA rather than allele-specific probes were utilized, and thus it is possible that the DIO3 promoter sequence within the extended DIO3 transcript actually hybridized with DIO3os transcripts instead. To confirm that transcription across the 5′ rDio3 region identified herein in fact occurs exclusively in the opposite orientation to rDio3, we performed strand-specific RT-PCR across multiple sites, including X, Y, and Z (Figure 1A), in several different tissue regions/developmental time points (e.g., adult HP, Figure 1B). As indicated in Figure 1B, RT-PCR product was obtained only when primers complementary to the rDio3os transcript were used. OS transcription was confirmed for both unspliced (e.g., across the EST BI274690, “X”) and spliced regions (“Y”).
Imprinted rDio3os Expression Arises from the Same Allele(s) as the Adjacent rDio3 Transcript
The rDio3os transcript had been reported to lack imprinted expression in mouse embryos (Tierling et al., 2006) and in adult mouse cortex (Labialle et al., 2008). However, we have recently discovered that imprinting of the rDio3 transcript exhibits brain regional, strain, and developmental specificity (Sittig et al., 2011a), with the greatest differentials occurring in fetal FX, where imprinted, paternal expression was observed in reciprocal crosses, relaxing to biallelic expression in adult FX, and in adult HP, where rDio3 expression was imprinted and derived from the maternal allele in one cross but biallelic in the reciprocal cross. As is customary for antisense transcripts in imprinted loci, we expected that rDio3os expression would be oppositely imprinted from rDio3, or equally biallelic. To determine whether the rDio3os transcript also exhibited complex imprinted expression patterns, we identified single-nucleotide polymorphisms (SNPs) within the rDio3os transcribed region between Sprague-Dawley (SD) and Brown-Norway (BN) rat strains. Focusing on a T/A SNP distant from G/C-rich or repetitive sequence regions (Figure 1A), we then examined expression across this SNP in animals derived from reciprocal crosses of these strains (S × B or B × S), where the first strain represents the maternal allele. We performed allele-specific expression analysis by strand-specific RT-PCR followed by direct sequencing on dissected frontal cortices and hippocampi from fetal and adult animals. As our prior experiments determined that expression across the region derived exclusively from the OS direction, we also performed direct sequencing on products isolated following random hexamer (RH)-primed RT-PCR reactions from these regions, as well as across several SNPs in placenta.
As shown in Figure 2, rDio3os expression in brain is imprinted in a strain, region, and developmental-specific manner paralleling that of rDio3 (Sittig et al., 2011a). Specifically, both rDio3 and rDio3os show relatively biallelic expression patterns in adult FX from both S × B and B × S crosses, comparable to the biallelic expression reported in adult mouse cortex (Labialle et al., 2008). In the adult HP from the B × S cross, both rDio3 (Sittig et al., 2011a) and rDio3os are also biallelic. In placenta, the predominant, unspliced rDio3os transcripts, which comprise ~80% of total rDio3os, are also biallelically expressed (58:42 s.d. 5.6, n = 3), consistent with what had been observed for Dio3 in rat (Sittig et al., 2011a) and mouse (Yevtodlyenko et al., 2002), although others have reported preferential paternal mDio3 expression (Tsai et al., 2002).
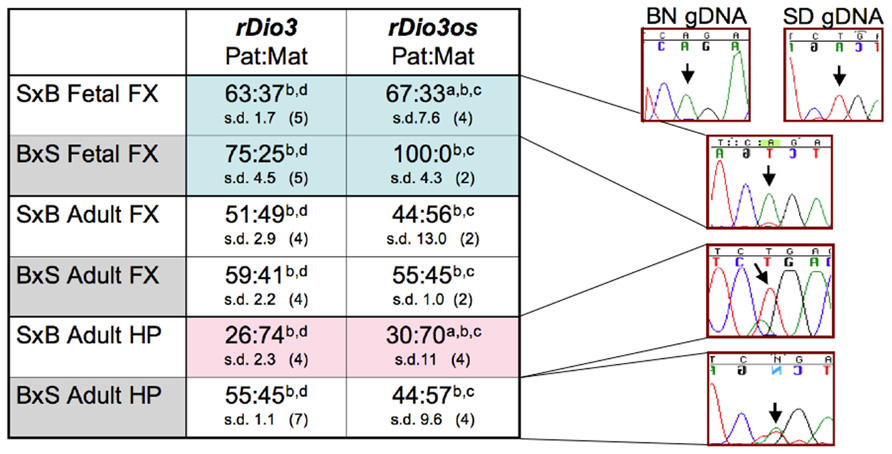
FIGURE 2. Allele-specific expression analysis demonstrates that rDio3os expression is imprinted in a strain, region, and developmental-specific manner paralleling that of rDio3. Here, various imprinted expression patterns are shown for fetal and adult male hippocampus (HP) and frontal cortex (FX), respectively. rDio3 imprinted expression is reproduced from our previous work (Sittig et al., 2011a). Relative rDio3os expression across a SNP between Sprague-Dawley (SD; T allele) and Brown-Norway (BN; A allele) animals was determined following (a) strand-specific or (b) random hexamer-primed RT-PCR, followed by (c) direct sequencing or (d) pyrosequencing analysis (Sittig et al., 2011a). For each cross, the maternal strain is given first (S × B or B × S). For allele-specific expression, paternal:maternal (Pat:Mat) expression ratios are given, along with representative preferential paternal, maternal or biallelic sequence traces. Strand-specific allele-specific analysis of rDio3os samples performed in duplicate-quadruplicate. s.d., standard deviation.
Surprisingly, however, not only is rDio3os imprinted in both S × B and B × S fetal FX, and in S × B adult HP as is rDio3 (Sittig et al., 2011a), but the expression arises from the same allele as that of rDio3 (Figure 2). Analysis of the spliced placental rDio3os transcripts suggest that these, also, exhibit imprinted expression, redolent of the conflicted status of placental murine Dio3 expression. The more common (“Y”) variant demonstrates preferential paternal expression (86:14 s.d. 20, n = 3; p > 0.05 compared to unspliced biallelic transcript), whereas a minor alternative splice variant exhibits exclusively maternal expression in two of three placenti. In placenta, we also observe unspliced biallelic and spliced paternal expression across a downstream SNP, although its location distant from splice junctions and adjacent to a region of repetitive element sequence homology precluded similar examination in brain samples, which have markedly lower rDio3os expression levels than does placenta. Thus, unlike the typical sense/antisense arrangement in imprinted genetic loci, where the antisense transcript arises from the opposite allele from that of the sense transcript, in brain and potentially in placenta, both rDio3 and rDio3os transcripts arise from the same allele.
Total rDio3os Expression Tracks rDio3: Strain and Brain Region-Specific Effects
The standard model for sense and antisense expression in imprinted genomic loci is that, in general, expression of the antisense transcript occludes expression of sense transcripts, through direct overlap (Williamson et al., 2011) or in trans as a negative regulator of expression through interaction with complexes at the gene promoter (Shin et al., 2008). Expression of such sense and antisense genes is thus usually complementary, or inversely proportional. In contrast, most lncRNAs located near coding genes have been found to independently transcribed (Liao et al., 2011). Of the ~2% coregulated with coding genes, most are not found in the “head-to-head” orientation of the Dio3/Dio3os transcripts (Mercer et al., 2008; Liao et al., 2011), as currently characterized (Hernandez et al., 2004). As Dio3os total expression has been reported to be roughly correlated with Dio3 in vivo during development, adulthood (Hernandez et al., 2002; Labialle et al., 2008), and in tissue culture (Kester et al., 2006) for mouse and human, respectively, we wanted to determine whether this coregulation extended to the regional and strain-specific total expression pattern observed for rDio3 in brain. Furthermore, we aimed to determine if rDio3os exhibited transcriptional response to fetal alcohol exposure similar to that of rDio3 (Sittig et al., 2011b), despite its novel imprinted, homo-allelic expression profile and unusual genomic organization.
Initially, we examined total rDio3os expression from micro-dissected rat brain regions (Figure 3). We found that despite the low levels of expression of this transcript we were able to achieve reliable amplification using concentrated cDNA samples in a semi-quantitative RT-PCR assay, whereas we were unable to generate useable standard curves or reproducible relative expression profiles for rDio3os using real-time qRT-PCR amplification cocktails. Previously, we had found that in the adult male S × B rat brain, total rDio3 mRNA expression levels are lower in the HP than in the FX, but these regions exhibit similar total rDio3 expression levels in the B × S cross (Sittig et al., 2011a). We confirmed a similar pattern for rDio3os, wherein rDio3os expression is significantly reduced in the male adult HP compared with that in the FX in the S × B cross, but is statistically indistinct between regions in the B × S cross.
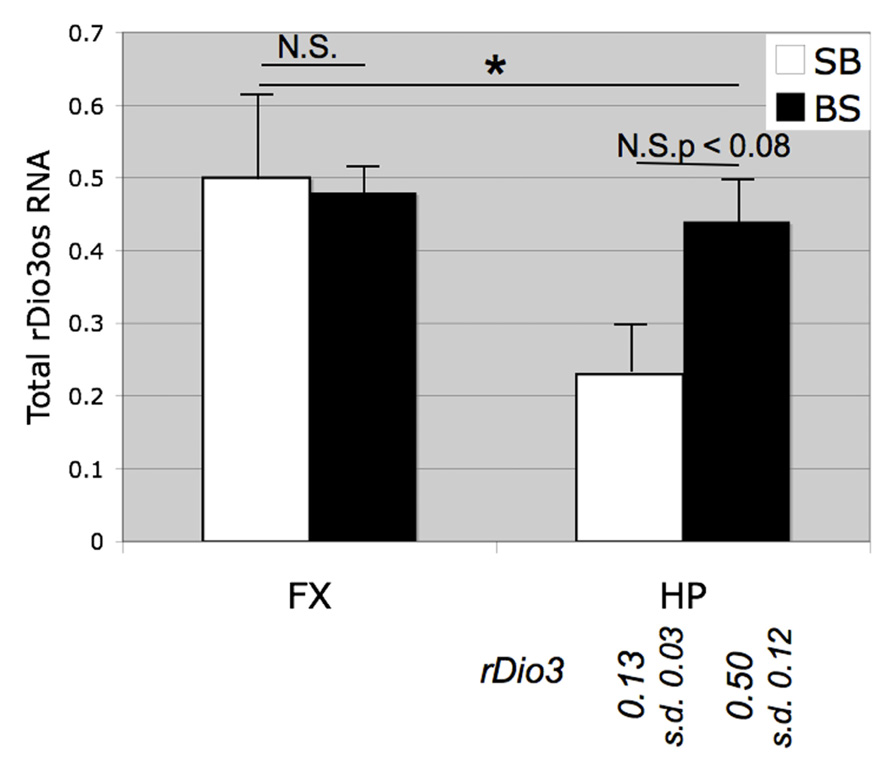
FIGURE 3. Strain and brain region-specific effects on total rDio3os expression. As for rDio3, semi-quantitative RT-PCR from adult male cortex demonstrates no significant difference between animals derived from a B × S cross vs. an S × B cross (p > 0.84), whereas animals from the S × B cross exhibit significantly decreased total rDio3os expression in the hippocampus vs. cortex. S × B adult hippocampi also trend toward decreased rDio3os expression as compared with hippocampi from the B × S cross (p > 0.08). For comparison, total rDio3 adult male hippocampal RNA expression is indicated below bars (from Sittig et al., 2011a; n = 7–8). Samples are normalized to rat beta-actin expression levels. n = 3–4; extreme outliers omitted from analysis. *p > 0.05; N.S., not significant; HP, hippocampus; FX, frontal cortex; s.d., standard deviation.
We then observed that, as with native expression, total rDio3os response to prenatal ethanol exposure mirrors rDio3 expression. Figure 4A illustrates RT-PCR products from representative individual S × B fetal brain regions, demonstrating that expression across the rDio3os transcript roughly correlates with relative rDio3 expression for that individual, as measured by real-time qRT-PCR. Overall, we observed elevated rDio3os expression in ethanol-exposed (E) vs. control (C) FX in three of six female fetal samples, and in three of seven adult samples, by gender (Figure 4B), consistent with the overall increase in rDio3 expression observed in ethanol-exposed FC from these respective groups (Sittig et al., 2011b). The exponential increase in rDio3os expression between E and C groups together and separately by developmental stage is highly significant by Chi-square analysis (p > 0.0001) although the absolute differences in mRNA expression levels between groups are not significant by two-way ANOVA. In contrast, rDio3os expression in adult male HP was decreased following ethanol exposure (Figure 4C), again consistent with the pattern observed for rDio3 (Sittig et al., 2011b). Together, these results demonstrate that total rDio3os expression patterns are regulated in a similar manner to those of rDio3, albeit with somewhat greater variability and to a somewhat lesser degree than the coding transcripts.
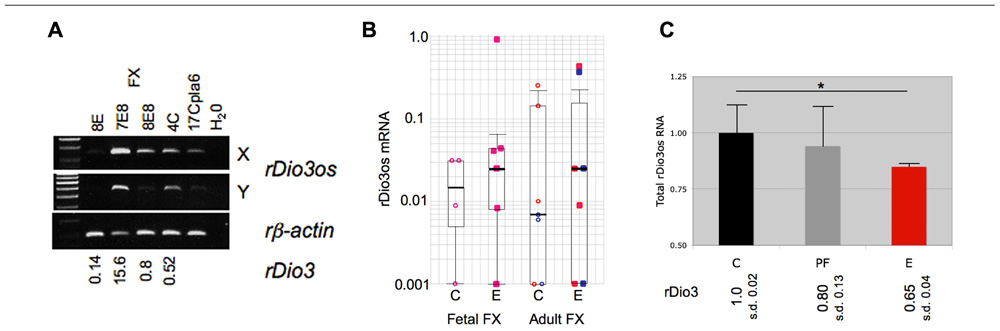
FIGURE 4. Coordinate response of rDio3 and rDio3os to prenatal ethanol exposure. (A) Semi-quantitative RT-PCR from fetal frontal cortex of individual control (C) and ethanol-treated animals (E) demonstrates levels of rDio3os expression tracking total rDio3 expression (relative levels from qRT-PCR in triplicate, normalized to beta-actin; L.S.) for both unspliced (X) and potentially spliced (Y) transcript-specific primer pairs. All animals are from an S × B cross. A placental rDio3os sample shown for comparison. Note that 4C is an upper outlier from controls to allow visualization of expression product. (B) Semi-quantitative RT-PCR from female fetal or adult rat S × B frontal cortex (FC) from control (C) or prenatally ethanol-exposed animals (E). Pink/red: female; blue: male. (C) Semi-quantitative RT-PCR from adult male hippocampus from control (C), pair-fed (PF), and prenatally ethanol-exposed animals (E) demonstrates decreased total rDio3os expression (region Y, shown) in ethanol-exposed animals. Relative rDio3 protein levels (from Sittig et al., 2011b; n = 4–6) are given below each category. As in fetal brain, the direction of change is the same as that for rDio3 although the absolute degree of change is less. Samples run in duplicate, normalized to beta-actin; outliers from each three sample group excluded. *p > 0.05, Student’s t-test. s.d., standard deviation.
Discussion
The present study demonstrates the coordinate expression and imprinting profile of a non-overlapping lncRNA/gene pair. Specifically, we show that the rat Dio3os transcript does not obviously overlap the rDio3 transcript itself, that it is imprinted, and that it is coregulated with rDio3 both at the level of total expression and of imprinted expression. To our knowledge, this represents the first example of paired sense/OS transcripts arising from the same allele. The original paradigm for (long) non-coding RNAs (lncRNAs) was that they were usually found in imprinted gene clusters, overlapped at least one imprinted coding transcript (in antisense orientation), and were themselves reciprocally imprinted to the protein-coding gene (O’Neill, 2005; Peters and Robson, 2008). Subsequent identification of roles for imprinted lncRNAs in the regulation of imprinted expression of non-overlapping genes opened our eyes to the importance of these transcripts as direct functional entities (Nagano et al., 2008; Pandey et al., 2008). In turn, recently published global analyses of lncRNAs has highlighted many non-overlapping lncRNA/gene pairs that frequently show coordinate expression profiles (Mercer et al., 2008; Ponjavic et al., 2009; Liao et al., 2011), with lncRNAs implicated in some cases in regulation of expression of the paired gene (Khalil et al., 2009; Ponjavic et al., 2009). Here, we have shown that, much like Archaeopteryx, the rat Dio3os transcript exemplifies both of these paradigms, carrying features of and serving as a span between the broad and generalized class of lncRNAs, and the more specialized category of imprinted non-coding transcripts.
The great majority of lncRNAs that have been identified within imprinted gene clusters are themselves imprinted, or have tissue-specific imprinted isoforms (Numata et al., 2010), and thus our observation of region-specific imprinting of rDio3os (Figure 2), previously reported to exhibit biallelic expression, might not be considered particularly unusual. Slightly more unusual is the genomic organization of Dio3/Dio3os (Figure 1), which at face value appears to be a bidirectional, head-to-head pairing of sense (coding) and antisense, or OS, transcripts, with no detectable sequence overlap in rat (this manuscript), or human (Hernandez et al., 2004), although a small 5′ overlap has been identified in mouse (Hernandez et al., 2002). This is in contrast to the multiple sense/antisense pairs of coding/non-coding RNAs with significant overlap found within imprinted gene clusters (Katayama et al., 2005; O’Neill, 2005). Whereas there are also many imprinted lncRNAs within these clusters that do not overlap coding transcripts (e.g., Zhang et al., 2011) and which may technically be in antisense orientation to coding genes, including ncRNAs with functional roles on non-overlapping transcripts such as Air (Nagano et al., 2008) and Kcnq1ot1 (Pandey et al., 2008), the distance between these is usually much larger than that between the identified Dio3os and Dio3 transcripts. There does exist a possibility of transcriptional overlap between Dio3os and Dio3, as Hernandez et al. (2004) have identified a putative Dio3os promoter downstream of Dio3, which could facilitate chromatin reorganization across the Dio3 locus. In addition, as neither we (rDio3os) nor others (Hernandez, 2005) have been able to identify the absolute 5′ end of the Dio3os transcript using RACE or other methods, there also remains a likelihood that the full Dio3os transcript flanks the Dio3 gene itself, such that transcription proceeds across Dio3 without overlap in the final processed products. In either case, this organizational structure is highly unusual for imprinted as well as non-imprinted lncRNA/coding RNA gene pairs (Ponjavic et al., 2009), most especially when the latter show evidence of coregulation (Mercer et al., 2008; Liao et al., 2011), as do Dio3/Dio3os (Hernandez et al., 2002; Figures 3 and 4).
Most unusual is our finding that imprinted rDio3os expression originates from the same allele as that of rDio3. Within imprinted clusters, lncRNAs are almost universally transcribed from the allele opposite to the imprinted coding transcripts in the regulatory domains (O’Neill, 2005). To our knowledge, rDio3/rDio3os represent a previously unreported and novel scenario, wherein both an imprinted gene and its immediately adjacent non-coding RNA are transcribed from the same, rather than opposite, parental alleles. The closest comparison to this scenario occurs within the complex Gnas locus, where both Nespas (overlapping Nesp55) and Gnasxl (one of several alternative transcripts of Gnas, along with Nesp55) are paternally expressed, in opposite orientation (Williamson et al., 2006), albeit further apart than Dio3/Dio3os (Williamson et al., 2002). However, unlike Dio3/Dio3os, the expression of Nespas and Gnasxl is not concordant across tissues, although both are expressed in heart and brain (Pasolli et al., 2000; Wroe et al., 2000; Plagge et al., 2004). Furthermore, the Dio3 promoter has not been found to exhibit differential methylation (Tsai et al., 2002), and, as mentioned above, may not itself drive Dio3os expression (Hernandez et al., 2002), whereas expression of Nespas and Gnasxl is regulated by a differentially methylated region (DMR) between them, that controls imprinted expression across the locus (Williamson et al., 2006). Finally, although the imprint of both pairs is maintained by a distant DMR, for Gnasxl this is downstream of Nespas (Frohlich et al., 2010), whereas imprinted mDio3 expression is regulated by a DMR almost 1 Mb distant (Lin et al., 2003).
This novel collection of features suggests that the regulation of expression of Dio3/Dio3os likely differs from the standard paradigm for coding/non-coding genes within imprinted loci, and may be better represented by that observed for coregulated coding/lncRNA pairs. In imprinted loci, expression of the non-coding transcript generally forestalls expression of the coding gene on that allele, either by direct transcriptional (stochastic) interference, often leading to epigenetic changes in the chromatin of overlapped regulatory regions (Williamson et al., 2011), or by recruitment of chromatin modifying factors to (Pandey et al., 2008) or direct interaction with (Nagano et al., 2008) the promoters of neighboring genes. In comparison, about 20% of non-imprinted lncRNAs are coregulated with their neighboring coding gene when in a sense/antisense orientation (Katayama et al., 2005), although coding/lncRNA pairs in all orientations have been described that are coordinately, inversely, or randomly regulated (Mercer et al., 2008). Of course, coregulation of adjacent genes may simply reflect a similar chromatin environment, and the unique expression profile of rDio3/rDio3os may not imply a regulatory role for either in generating that profile. On the other hand, lncRNA members of coregulated gene pairs have, in many instances, been shown to exhibit regulatory function on the coding gene (Katayama et al., 2005; Mondal et al., 2010). They have also, for example, been identified as chromatin-associated RNAs (CARs; Mondal et al., 2010), that positively regulate transcription of neighboring genes via establishing active chromatin structures through interaction with the chromatin. In that study, however, Dio3os was not identified among the CARs, although another ncRNA in the Dlk1-Dio3 locus, Meg3, which exhibits standard opposite imprint expression to Dio3, was. Tellingly, neither has Dio3os been pulled out in several other large-scale studies on lncRNAs, including those identified as associated with chromatin modifying factors (which tend to decrease expression of adjacent genes; Khalil et al., 2009), those identified by evolutionary sequence constraints (Ponjavic et al., 2009), chromatin state (Guttman et al., 2009), expression in brain (Mercer et al., 2008) or across transcriptome analysis (Katayama et al., 2005), and micro-array platforms (Liao et al., 2011). In addition, Dio3os, despite being characterized in the published literature (Hernandez et al., 2004) and listed in lncRNAdb and NONCODE, databases of non-coding RNAs (Amaral et al., 2011; Bu et al., 2012), contains no expression profiling information in these, nor in the Allen Brain Atlas (Lein et al., 2007) and its low level and tissue-specific expression may preclude it from being identified in such screens.
Despite the apparently elusive nature of the Dio3/Dio3os partnership in large-scale screens for lncRNA functionality, the conservation of sequence along with coregulation of total expression between species and maintenance of tissue-specific imprint patterns in wild-type and developmentally substandard conditions described herein, suggest that coregulation is actively maintained and retains the possibility of a role for Dio3os in active regulation of imprinted Dio3 expression. As a coregulated gene pair, Dio3os might enhance or, contrarily, temper expression of Dio3, both scenarios having been observed for coregulated lncRNA/coding gene pairs (Katayama et al., 2005; Mondal et al., 2010; Vance et al., 2011). Multiple potential mechanisms exist for these possibilities, similar to those observed for other lncRNA-based regulation. Whether the Dio3os transcript originates within the Dio3 promoter region or from the flanking, 3′ end of Dio3, active Dio3os transcription across the locus may create an “open” chromatin structure permissive for transcriptional machinery action upon the Dio3 gene. Alternatively, the resultant chromatin structure may limit accessibility, tempering but not occluding Dio3 transcription, as would be expected from an imprinted antisense transcript expressed from the allele opposite to that of gene expression. Direct transcriptional overlap might also regulate the post-transcriptional stability of Dio3 by dsRNA or siRNA formation via pre-processed Dio3os transcripts spanning Dio3, or an as-yet-unidentified Dio3os 5′ exon within the Dio3 3′UTR (Hernandez, 2005). Active Dio3os transcription and presence of the Dio3os transcript itself adjacent to the Dio3 promoter, or overlapping it (Hernandez et al., 2002; Dentice et al., 2007), may prevent binding of an inhibitor protein, thus serving to activate Dio3 expression, or limit binding of a transcription factor or Pol II recruitment, thus moderating expression. Another possibility is that, as for several other lncRNAs, the Dio3os RNA itself may be functional, homologously pairing directly with the Dio3 promoter region and recruiting activating or inhibitory factors, or regionally interacting with chromatin modification factors to epigenetically alter the chromatin landscape, facilitating or tempering Dio3 expression. Any of these models offer the format of an additional layer of transcriptional regulatory control for Dio3, which, while possibly being an evolutionary relic, may in fact be necessary to effect proper control of total Dio3 expression levels, and thus of circulating thyroid hormone levels in the developing and adult animal, in a temporal and regional-specific manner.
These studies demonstrate that the Dio3os transcript is present in rat, and that its structure is generally conserved with that in mouse. As in mouse, total rDio3os levels generally track those of rDio3. Our detailed analyses further demonstrate that total expression similarities for the rDio3 gene and the rDio3os lncRNA are maintained and are similarly influenced by developmental stage, brain region, strain background, and prenatal insult (ethanol exposure).
We further demonstrate that rDio3os expression is also imprinted, as is common for lncRNAs within imprinted clusters. As for total expression, imprinted rDio3os expression patterns vary in line with those of rDio3. Although this could simply be a situation of coregulation of these genes, it is tempting to speculate that rDio3os expression may be positively regulating rDio3 expression, as has been described for other non-imprinted lncRNAs/-adjacent genes (Katayama et al., 2005; Mercer et al., 2008; Mondal et al., 2010).
Future studies including identification of rDio3os transcriptional overlap of rDio3 regulatory regions, as in mouse and humans, and knockdown and truncation of the Dio3os transcript in neuronal tissue will be required to evaluate the requirement of Dio3os expression in Dio3 transcriptional regulation. In comparison with what is known for other lncRNA/coding gene pairs, the direction of Dio3os-mediated regulation of Dio3, if any, will guide further studies addressing the highly unusual mono-allelic expression of these genes and the mechanism of their coregulation or sequential regulation. In turn, understanding these mechanisms may aid in addressing malfunctions in Dio3 expression patterns, among others, and their phenotypic consequences, such as are observed following prenatal ethanol exposure (Sittig et al., 2011b).
Materials and Methods
Animals
Animal procedures and tissue collection and processing were as described (Sittig et al., 2011a,b) Animal procedures were approved by the Northwestern University Animal Care and Use Committee. Adult SD and BN male and female rats (70–85 days of age, Harlan, Indianapolis, IN, USA) were mated (n = 6 SD females, n = 8 BN females) to obtain reciprocal F1 hybrid offspring, B × S and S × B, with first letter of the maternal strain first followed by the first letter of the paternal strain. For the fetal study, dams were sacrificed by decapitation on gestational day 21 (G21). Additional pregnant dams (BN n = 7; SD n = 8) were allowed to give birth and rear litters. One or two males and one or two females from each litter were sacrificed following behavioral testing (Sittig et al., 2011a,b) after 60 days of age. Prenatal ethanol (E) exposure and maternal diet procedures were performed as described previously (Sittig et al., 2011b). Briefly, pregnant females were assigned to a diet group on G8: E dams received an ethanol-containing (5% w/v, 35% ethanol-derived calories) liquid diet between G8 and G21 (Lieber-DeCarli ’82; Bio-Serv, Frenchtown, NJ, USA). Pair-fed (PF) dams received an amount of isocaloric liquid diet (Lieber-DeCarli ’82; Bio-Serv, Frenchtown, NJ, USA) that matched the paired E dam’s diet consumption on the previous day. Liquid diets were replaced with lab chow on G21, while control (C) dams received lab chow and water ad libitum from G1 to G21.
Tissue Collection
Pregnant dams were sacrificed by decapitation on G21 between 1000 and 1200 hours, as previously described (Sittig et al., 2011a,b). Fetal heads were collected directly into RNAlater reagent (Ambion, Austin, TX, USA) and kept at room temperature for 24 h before brain regions were dissected and transferred into fresh RNAlater. Fetal frontal cortices were dissected using a microscope according to the Atlas of Prenatal Rat Brain Development: A/P (1.0–2.2; M/L 0.0–1.5; D/V 0.0–1.0; Altman and Bayer, 1995). Fetal hippocampal dissections included the entire structure. Adult offspring were sacrificed by decapitation 2 weeks after the final behavioral test. FX (A/P 5.2–2.7; M/L 0–3.3; D/V 9.0–5.0) and HP were immediately dissected (Paxinos and Watson, 1997). The left half of each structure was placed on dry ice and the right half was placed into RNAlater. Tissues were stored at -80°C until use.
RNA Isolation and Semi-Quantitative RT-PCR
Total RNA extraction was performed using Trizol reagent (Life Technologies, Gaithersburg, MD, USA) according to the manufacturer’s protocol. Genomic DNA was removed using the TURBO DNA-free kit (Applied Biosystems, Foster City, CA, USA). DNased RNA (1 µg) was reverse transcribed using the Promega ImPromII Reverse Transcription kit (Promega, Madison, WI, USA) and RH primers. Final product was resuspended to 100 µl as per manufacturer’s instructions. For strand-specific RT-PCR, DNased RNA (0.3 µg) was reverse transcribed using rDio3 sense-strand (F) or OS-specific (R) primers (0.6 nM). Two microliters of final product (10 µl) was used directly in subsequent PCR amplification procedures, with addition of 0.2 nM of additional RT primer and 0.5 nM of respective Forward or Reverse primer for second strand syn-thesis and amplification. Controls included reactions without reverse transcriptase (RT-) or template. For semi-quantitative analysis, RH-primed total RNA samples were amplified to obtain signal within the exponential phase of the PCR reaction as follows: rDio3os: 60°C annealing temperature, 37 cycles; beta-actin: 1:4 dilution of cDNA, 55°C annealing temperature, 29 cycles, and analyzed by gel electrophoresis followed by digital imaging (Kodak Gel Logic 200, Rochester, NY, USA) and relative densitometry (Adobe Photoshop 12.0.1, San Jose, CA, USA). rDio3os expres-sion signals were normalized to beta-actin. Primer pairs: rDio3os “X”: F, 5′-CTTGGAGGGCCTGGCATTAAC; R, 5′-AAGACACT-GGCACTACTGGC; “Y”: F, 5′-AACTTTCTCGACCAGAAACC-GC; R, 5′-TAGTATAGGAGTCCGATGGC; “Z”: F, 5′-AAGCTGG-TTAAGGGTGGAGC; F*, 5′-TTGCAACTTGAGCCCTGAGGG; R, 5′-TACACCATTGCCACCACCGACTGC; beta-actin: F, 5′- GCTCCTCCTGAGCGCAAGTA; R, 5′-CTCCTGCTTGCTGATCCACAT.
rDio3os Characterization and Sequencing
Putative rDio3os sequence was identified through BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) alignment of the EST BI274690 with rat chromosome 6 genomic contig reference assembly NW_047772.1 and mouse chromosome 12 reference assembly NC_000078.5. PCR primers for amplification of putative rDio3os transcript were designed manually based on mouse Dio3os exons in splice variant ESTs AY077459 and AY238181. To identify 5′ and 3′ transcript ends, RACE was performed using the Invitrogen GeneRacer kit (Life Technologies, Grand Island, NY, USA). PCR amplified products and rDio3os splice variants were extracted from agarose gels, purified and sequenced using the Children’s Hospital of Chicago Research Center (CHCRC) Core Facility (Chicago, IL, USA) or the Northwestern University Feinberg School of Medicine (FSM) Genetics and Genomics Core Facility (Chicago, IL USA). For SNP identification, BN, SD, and S × B genomic DNA was amplified and sequenced across the region (156500–160800 bp). Reported analyses are using T/A (SD/BN) at 158338 bp. For allele-specific expression analysis, primers flanking the SNP(s) between SD and BN rat strains were used for amplification of cDNA derived from RH-primed RT-PCR or strand-specific RT-PCR (above), with a second round of nested PCR as necessary to generate sufficient product for sequencing. Ratio of allele-specific transcripts were determined by direct measurement of sequence traces in both forward and reverse directions, normalized to genomic DNA allelic ratio (50:50). Additional primers used for nested PCR and sequencing are available upon request.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Kelly Varga for excellent technical assistance with initial rDio3os characterization. This work was funded by U.S. National Institutes of Health (NIH) grant R01 AA017978. At the time of this project, Laura J. Sittig was a predoctoral fellow funded by NIH grant F31 AA018251.
References
Altman, J., and Bayer, S. (1995). Atlas of Prenatal Rat Brain Development. Boca Raton, FL: CRC Press.
Amaral, P. P., Clark, M. B., Gascoigne, D. K., Dinger, M. E., and Mattick, J. S. (2011). lncRNAdb: a reference database for long noncoding RNAs. Nucleic. Acids Res. 39, D146–D151.
Bu, D., Yu, K., Sun, S., Xie, C., Skogerbo, G., Miao, R., et al. (2012). NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 40, D210–D215.
Charalambous, M., and Hernandez, A. (2012). Genomic imprinting of the type 3 thyroid hormone deiodinase gene: regulation and developmental implications. Biochim. Biophys. Acta. doi: 10.1016/j.bbagen.2012.03.015 [Epub ahead of print].
daRocha, S. T., Edwards, C. A., Ito, M., Ogata, T., and Ferguson-Smith, A. C. (2008). Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24, 306–316.
Dentice, M., Luongo, C., Huang, S., Ambrosio, R., Elefante, A., Mirebeau-Prunier, D., et al. (2007). Sonic hedgehog-induced type 3 deiodinase blocks thyroid hormone action enhancing proliferation of normal and malignant keratinocytes. Proc. Natl. Acad. Sci. U.S.A. 104, 14466–14471.
Frohlich, L. F., Mrakovcic, M., Steinborn, R., Chung, U.-I., Bastepe, M., and Juppner, H. (2010). Targeted deletion of the Nesp55 DMR defines another Gnas imprinting control region and provides a mouse model of autosomal dominant PHP-1b. Proc. Natl. Acad. Sci. U.S.A. 107, 9275–9280.
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227.
Hagan, J. P., O’Neill, B. L., Stewart, C. L., Kozlov, S. V., and Croce, C. M. (2009). At least ten genes define the imprinted Dlk-Dio3 cluster on mouse chromosome 12qF1. PLoS ONE 4:e4352. doi: 10.1371/journal.pone.0004352
Hernandez, A., Fiering, S., Martinez, E., Galton, V. A., and St. Germain, D. (2002). The gene locus encoding iodothyronine deiodinase type 3 (Dio3) is imprinted in the fetus and expresses antisense transcripts. Endocrinology 143, 4483–4486.
Hernandez, A., Martinez, M. E., Croteau, W., and St. Germain, D. L. (2004). Complex organization and structure of sense and antisense transcripts expressed from the DIO3 imprinted locus. Genomics 83, 413–424.
Huang, R., Jaritz, M., Guenzl, P., Vlatkovic, I., Sommer, A., Tamir, I. M., et al. (2011). An RNA-seq strategy to detect the complete coding and non-coding transcriptome including full-length imprinted macro ncRNAs. PLoS ONE 6:e27288. doi: 10.1371/journal.pone.0027288
Katayama, S., Tomaru, Y., Kasukawa, T., Waki, K., Nakanishi, M., Nakamura, M., et al. (2005). Antisense transcription in the mammalian genome. Science 309, 1564–1566.
Kester, M. H. A., Kuuiper, G. G. J. M., Versteeg, R., and Visser, T. J. (2006). Regulation of type III iodothyronine deiodinase expression in human cell lines. Endocrinology 147, 5845–5854.
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Faj, A., Morales, D. R., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672.
Labialle, S., Yang, L., Ruan, X., Villemain, A., Schmidt, J. V., Hernandez, A., et al. (2008). Coordinated diurnal regulation of genes from the Dlk1-Dio3 imprinted domain: implications for regulation of clusters of non-paralogous genes. Hum. Mol. Genet. 17, 15–26.
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176.
Liao, Q., Liu, C., Yuan, X., Kang, S., Miao, R., Xiao, H., et al. (2011). Large-scale prediction of long non-coding RNA functions in a coding–non-coding gene co-expression network. Nucleic Acids Res. 39, 3864–3878.
Lin, S. P, Youngson, N., Takada, S., Seitz, H., Reik, W., Paulsen, M., et al. (2003). Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35, 97–102.
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F., and Mattick, J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 105, 716–721.
Mondal, T., Rasmussen, M., Pandey, G. K., Isaksson, A., and Kanduri, C. (2010). Characterization of the RNA content of chromatin. Genome Res. 20, 899–907.
Nagano, T., Mitchell, J. A., Sanz, L. A., Pauler, F. M., Ferguson-Smith, A. C., Reil, R., et al. (2008). The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720.
Numata, K., Kohama, C., Abe, K., and Kiyosawa, H. (2010). Highly parallel SNP genotyping reveals high-resolution landscape of mono-allelic Ube3a expression associated with locus-wide antisense transcription. Nucleic Acids Res. 39, 2649–2657.
O’Neill, M. J. (2005). The influence of non-coding RNAs on allele-specific gene expression in mammals. Hum. Mol. Genet. 14, R113–R120.
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., et al. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246.
Pasolli, H. A., Klemke, M., Kehlenbach, R. H., Wang, Y., and Huttner, W. B. (2000). Characterization of the extra-large G protein a-subunit XLas. J. Biol. Chem. 275, 33622–33632.
Paxinos, G., and Watson, C. (1997). The Rat Brain in Stereotaxic Coordinates, 3rd Edn. San Diego, CA: Academic Press.
Plagge, A., Gordon, E., Dean, W., Bioani, R., Cinti, S., Peters, J., et al. (2004). The imprinted signaling protein XL alpha s is required for postnatal adaptation to feeding. Nat. Genet. 36, 818–826.
Ponjavic, J., Oliver, P. L., Lunter, G., and Ponting, C. P. (2009). Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 5:e1000617. doi: 10.1371/journal.pgen.1000617
Shin, J. Y., Fitzpatrick, G. V., and Higgins, M. J. (2008). Two distinct mechanisms of silencing by the KvDNR1 imprinting control region. EMBO J. 27, 168–178.
Sittig, L. J., Herzing, L. B., Shukla, P. K., and Redei, E. E. (2011a). Parent-of-origin allelic contributions to deiodinase-3 expression elicit localized hyperthyroid milieu in the hippocampus. Mol. Psychiatry 16, 786–787.
Sittig, L. J., Shukla, P. K., Herzing, L. B., and Redei, E. E. (2011b). Strain-specific vulnerability to alcohol exposure in utero via hippocampal parent-of-origin expression of deiodinase-III. FASEB J. 25, 2313–2314.
Tierling, S., Dalbert, S., Schoppenhorst, S., Tsai, C.-E., Oliger, S., Ferguson-Smith, A., et al. (2006). High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics 87, 225–235.
Tsai, C. E., Lin, S. P., Ito, M., Takagi, N., Takada, S., and Ferguson-Smith, A. C. (2002). Genomic imprinting contributes to thyroid hormone metabolism in the mouse embryo. Curr. Biol. 12, 1221–1226.
Tu, H. M., Legradi, G., Bartha, T., Salvatore, D., Lechan, R. M., and Larsen, P. R. (1999). Regional expression of the type 3 iodothyronine deiodinase messenger ribonucleic acid in the rat central nervous system and its regulation by thyroid hormone. Endocrinology 140, 784–790.
Vance, K. W., Bassett, A., Kong, L., Lui, J. L., Marques, A. C. and Oliver, P. L. (2011). “Identification of orthologous long intergenic non-coding RNAs and characterisation of their modes of action,” in Cell Symposia: Regulatory RNAs, Chicago, IL 2011; Abstract P1.11.
Williamson, C. M., Ball, S. T., Dawson, C., Mehta, S., Beechey, C. V., Fray, M., et al. (2011). Uncoupling antisense-mediated silencing and DNA methylation in the imprinted Gnas cluster. PLoS Genet. 7:e1001347. doi: 10.1371/journal.pgen.1001347
Williamson, C. M., Skinner, J. A., Kelsey, G., and Peters, J. (2002). Alternative non-coding splice variants of Nespas, an imprinted gene antisense to Nesp in the Gnas imprinting cluster. Mamm. Genome 13, 74–79.
Williamson, C. M., Turner, M. D., Ball, S. T., Nottingham, W. T., Glenister, P., Fray, M., et al. (2006). Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat. Genet. 38, 350–355.
Wroe, S. F., Kelsey, G., Skinner, J. A., Bodle, D., Ball, S. T., Beechey, C. V., et al. (2000). An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc. Natl. Acad. Sci. U.S.A. 97, 3342–3346.
Yevtodlyenko, A., Carr, M., Patel, N., and Schmidt, J. V. (2002). Analysis of candidate imprinted genes linked to Dlk1-Gtl2 using a congenic mouse line. Mamm. Genome 13, 633–638.
Keywords: imprinting, lncRNA, expression, rat, fetal ethanol exposure, Dio3, Dio3os
Citation: Dietz WH, Masterson K, Sittig LJ, Redei EE and Herzing LBK (2012) Imprinting and expression of Dio3os mirrors Dio3 in rat. Front. Gene. 3:279. doi:10.3389/fgene.2012.00279
Received: 26 July 2012; Accepted: 16 November 2012;
Published online: 06 December 2012.
Edited by:
Peng Jin, Emory University School of Medicine, USAReviewed by:
Peng Jin, Emory University School of Medicine, USAAaron J. Schetter, National Cancer Institute, USA
Copyright: © 2012 Dietz, Masterson, Sittig, Redei and Herzing. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Laura B. K. Herzing, Program in Human Molecular Genetics, Department of Pediatrics, Children’s Hospital of Chicago Research Center, Feinberg School of Medicine, Northwestern University, 255 E. Chicago Ave. Box 211, Chicago, IL 60611, USA. e-mail: l-herzing@northwestern.edu
