- 1INRA UMR1313 Génétique Animale et Biologie Intégrative, Institut National de la Recherche Agronomique, Jouy en Josas, France
- 2Ifremer, Institut Français de Recherche pour l’Exploitation de la Mer, Palavas-les-Flots, France
- 3Sysaaf, Syndicat des Sélectionneurs Avicoles et Aquacoles Français, Rennes, France
Since the middle of the 1990s, parentage assignment using microsatellite markers has been introduced as a tool in aquaculture breeding. It now allows close to 100% assignment success, and offered new ways to develop aquaculture breeding using mixed family designs in commercial conditions. Its main achievements are the knowledge and control of family representation and inbreeding, especially in mass spawning species, above all the capacity to estimate reliable genetic parameters in any species and rearing system with no prior investment in structures, and the development of new breeding programs in many species. Parentage assignment should not be seen as a way to replace physical tagging, but as a new way to conceive breeding programs, which have to be optimized with its specific constraints, one of the most important being to well define the number of individuals to genotype to limit costs, maximize genetic gain while minimizing inbreeding. The recent possible shift to (for the moment) more costly single nucleotide polymorphism markers should benefit from future developments in genomics and marker-assisted selection to combine parentage assignment and indirect prediction of breeding values.
Introduction
Aquaculture is now the fastest growing animal production worldwide, and provides half of the fish for human consumption worldwide (FAO, 2014). Such an important sector would be expected to use the best knowledge-based improvement methods, amongst which selective breeding is of paramount importance. However, Gjedrem et al. (2012) estimated that only 10% of aquaculture production worldwide is based on genetically improved stocks. There may be several reasons for this, but one clear technical weakness of aquaculture regarding the development of optimized selective breeding schemes is the fact that pedigree information is difficult and costly to obtain.
The basic reason is rather straightforward: farmed aquatic animals are all too small at hatching (from a few micrograms in mollusks and crustaceans to ca. 100 mg in salmonids fishes) to be physically tagged.
Then, there were initially two ways for fish genetic studies and breeding programs to deal with the question of pedigrees. The first and simpler solution was not to use a pedigree, using individual selection. In this case, fish are selected solely based on their own individual phenotype (see review in Gjedrem and Thodesen, 2005). Although effective to obtain genetic gain, this method is very limiting for studying genetic variation as: (1) it provides results only after a minimum of two generations, (2) it requires the maintenance of at least two fish lines, selected/control or divergent lines, (3) it limits the evaluation of genetic variation to one trait only, and (4) the precision of realized heritability estimates is low in reasonably-sized two generation experiments (Nicholas, 1980).
The second option to solve the pedigree problem is to use separate rearing of families until a size where tagging is possible, as in the Norwegian salmon breeding program, the first family-based selective breeding program in aquaculture, started in 1972 (Gjedrem, 2010). This was successfully extended to major aquaculture species such as salmonids, tilapias, oyster, or shrimps (Krishna et al., 2011; Thodesen et al., 2011; Gjedrem, 2012; Gjedrem et al., 2012; Zak et al., 2014). Although efficient, this method has three main drawbacks when it comes to estimating genetic parameters of traits. First, as families are reared separately, common environmental effects between tanks may inflate heritability estimates. The second point is that studying genetic variation with separate rearing of progenies requires the pre-existence of the family rearing units–i.e., of the infrastructure of the breeding program. Exploratory studies are then difficult to undertake. The third point is that the number of families is limited to the number of family rearing units used. Then, mating designs are constrained to those where the number of families produced is low for a given number of parents tested, like single pair mating or nested designs, which, unlike factorial designs, do not allow the separation of additive, maternal, common environment, and dominance effects (Becker, 1967).
Therefore, the provision of a method to trace pedigrees in groups of mixed families, with any type of family structure, was expected to be of great interest to study genetic variation of quantitative traits in aquaculture species, and subsequently to set up new types of breeding programs.
The principles of parentage assignment were set up for livestock paternity testing with allozymes (Jamieson, 1965). In fish, the very first trials were done in the 1970s in Israel, also using allozymes, but the number of families that could be discriminated was very low (<10) and the use of the method was limited to carp in one research team (e.g., Brody et al., 1981). The real start of parentage assignment studies in fish was in the 1990s with the availability of microsatellite markers (Herbinger et al., 1995; Estoup et al., 1998).
Technical Aspects of Parentage Assignment
Parentage Assignment Methods
Basically, two computation methods are used for parentage assignment, exclusion-based methods and likelihood-based methods (see Jones et al., 2010 for a review). Exclusion is very simple and makes no hypotheses other than Mendelian segregation of alleles, but is very sensitive to genotyping errors. When error rates are moderate and theoretical assignment power is high, however, genotyping errors can be dealt with by allowing a limited number of allelic mismatches between an offspring and its parents alleles (Vandeputte et al., 2006), and exclusion remains the gold standard of parentage assignment (Yue and Xia, 2014). Exclusion programs used in aquaculture are PROBMAX (Danzmann, 1997), VITASSIGN (Vandeputte et al., 2006), and FAP (Taggart, 2007). Likelihood methods use a different approach, with probabilities. In this case, the most likely couple is chosen as the true one (eventually integrating a genotyping error rate), but the decision rules rely on hypotheses on allelic frequencies. Likelihood methods generally give higher assignment rates than exclusion with low power marker sets, but sometimes give inconsistent results (Herlin et al., 2007; Trong et al., 2013). Using sibship information in calculations can greatly improve the efficiency of the likelihood methods (Wang and Santure, 2009). Likelihood programs used in aquaculture are CERVUS (Kalinowski et al., 2007), PAPA (Duchesne et al., 2002), and PARENTE (Cercueil et al., 2002).
A specific question is also the assignment of polyploids, especially sturgeons (Rodzen et al., 2004) or induced polyploids (Miller et al., 2014), and specific packages have been developed for tetraploids (wHDP; Galli et al., 2011), diploids to octoploids (VITASSIGN-OCTO; Vandeputte, unpublished), as well as a general method to transform polyploid genotypes to pseudo-diploid dominant genotypes (Wang and Scribner, 2014).
A Crucial Issue: the Assignment Power of Markers Used
However, whatever the method used, the first requirement to be able to use parentage assignment in practice is to obtain high levels of unique assignments, which primarily depends on the assignment power of the marker set used. It depends on the exclusion probabilities of the markers used and on the size of the problem to be solved, the total number of putative parents having an exponential effect on the proportion of unassigned individuals (Vandeputte, 2012). Overestimation of the assignment power of markers is very frequent (Vandeputte et al., 2011), and can be explained by Hardy–Weinberg disequilibrium (Wang, 2007), sampling variance and relatedness of parents (Villanueva et al., 2002; Matson et al., 2008), incomplete genotypes, genotyping errors especially caused by stuttering or size-shift (Sutton et al., 2011; Yue and Xia, 2014), and null alleles (Christie, 2010). In some species groups like mollusks, null alleles may be extremely frequent and problematic (Hedgecock et al., 2004), but the main cause of overestimation of the theoretical assignment power is a widespread inappropriate calculation method (Vandeputte, 2012). Typically, assignment power >0.99 can generally be obtained by 8–15 microsatellite markers in fish crosses involving a few tens or hundreds of parents, and a reasonable option when designing a marker set is to include a few more markers than theoretically needed. This then spares a lot of time by providing easy assignment even if small problems of genotyping errors, inbreeding or null alleles appear. High quality genotyping is also essential, and a recent review by Yue and Xia (2014) gives very useful insights to this question.
Microsatellites and SNPs for Parentage Assignment
Microsatellites, due to their high number and high variability, are the markers that allowed the development of efficient parentage assignment methods. Today, however, SNPs (single nucleotide polymorphisms) use is growing exponentially (Guichoux et al., 2011), but not yet in parentage assignment. It was estimated that ∼6 SNPs give the same assignment power as 1 microsatellite (Glaubitz et al., 2003). Empirical studies tend to suggest that the adequate number of SNP for an efficient panel would be in the 100–450 range (Trong et al., 2013; Lapègue et al., 2014; Nguyen et al., 2014; Sellars et al., 2014). With such numbers, the classical requirement of unlinked markers within a panel cannot be met, thus lowering the real assignment power. SNPs are individually less expensive to genotype than microsatellites, but multiplexing decreased the cost of microsatellites genotyping (Guichoux et al., 2011; Yue and Xia, 2014), and for the moment SNPs remain more expensive due to the number required, but technology is rapidly evolving for SNPs and not for microsatellites. Empirical studies also sometimes reveal quite a high number of genotyping error in SNPs (Trong et al., 2013) and the necessity to test a higher number of SNP markers than expected to select the appropriate ones (Lapègue et al., 2014; Nguyen et al., 2014). However, prospects for development of genomic selection with low-marker density may imply genotyping of a few hundred to several thousand SNPs per fish (Lillehammer et al., 2013), which in this case would be sufficient to provide parentage assignment at no additional cost. The recent shift to SNP markers was, however, efficient to improve assignment at least in some mollusks species which suffered from high numbers of null alleles with microsatellites (Lapègue et al., 2014; Nguyen et al., 2014).
Implementation of Parentage Assignment in Aquaculture
Inbreeding Control
Mass selection is the simplest way to improve traits such as growth or morphology, but bears a high risk of rapid genetic loss, with highly unbalanced families, which was revealed by parentage assignment mostly in mass spawning species (Perez-Enriquez et al., 1999; Waldbieser and Wolters, 1999; Boudry et al., 2002; Brown et al., 2005; Fessehaye et al., 2006; Herlin et al., 2007; Wang et al., 2008), but also in controlled artificial reproduction systems (Saillant et al., 2002; Kaspar et al., 2008).
The impact of different factors (mating design, mating ratio, number of parents per generation, selection pressure, trait heritability, grading practices) were simulated to improve inbreeding control and optimize genetic progress (Gjerde et al., 1996; Dupont-Nivet et al., 2006; Loughnan et al., 2013; Domingos et al., 2014), including optimal contribution selection, which requires pedigree knowledge (Sonesson, 2005; Skaarud et al., 2011).
Estimation of Genetic Parameters
Estimation of heritability and genetic correlations allows to evaluate expected genetic gains and to design breeding programs. This is maybe where the possibility to access pedigree information by genotyping gave the most important and fruitful contribution to date to aquaculture genetics.
Optimization of mixed family designs for genetic parameters estimation was done by Vandeputte et al. (2001) for strain effects, Dupont-Nivet et al. (2002) for heritability, and Sae-Lim et al. (2010) for genotype by environment (G × E) interaction. After several feasibility studies with few families (Herbinger et al., 1995, 1999; Saillant et al., 2002), heritabilities were estimated in a growing number of fishes species for growth (Wilson et al., 2003; Saillant et al., 2006; Dupont-Nivet et al., 2008; Pierce et al., 2008; Wang et al., 2008; Gheyas et al., 2009; Domingos et al., 2013; Whatmore et al., 2013), processing traits (Kocour et al., 2007; Navarro et al., 2009; Saillant et al., 2009; Haffray et al., 2012a), flesh color (Norris and Cunningham, 2004), muscle fiber diameter (Vieira et al., 2007), deformities (Bardon et al., 2009), disease resistance (Guy et al., 2006; Antonello et al., 2009) or sex ratio (Vandeputte et al., 2007), and in shrimps and mollusks for growth (Jerry et al., 2006; Lucas et al., 2006; Kong et al., 2013; Nguyen et al., 2014) or meat yield in mussel (Nguyen et al., 2014). Heritabilities obtained in mixed family rearing are often higher than those recorded in separate rearing, which may be linked to the general absence of between-family environmental variance due to family mixing, although non-genetic maternal effects may persist in mixed family rearing (Haffray et al., 2012b), with possible upward biases on heritability estimates. Explicit comparisons of the same families in mixed or separate rearing design concluded that separate family rearing induced much higher levels of between-families environmental effects (Herbinger et al., 1999; Ninh et al., 2011).
Mixed family rearing also allowed the estimation of G × E interactions between rearing systems (Dupont-Nivet et al., 2008, 2010c; Navarro et al., 2009; Kvingedal et al., 2010; Domingos et al., 2013; Sae-Lim et al., 2013; Vandeputte et al., 2014), density or rearing temperatures (Saillant et al., 2006), plant-based vs marine feeds (Pierce et al., 2008; Le Boucher et al., 2013; Bestin et al., 2014), and even separated vs mixed fish family rearing designs (Ninh et al., 2011).
A limiting factor of such studies is that as fish are generally tagged to maximize individual information collection, individual performances are not available before physical tagging, thus limiting genetic studies on early stages. However, recent advances allow individual tagging at 200–400 mg (Ferrari et al., 2014), which should change this matter of fact.
Implementation of Breeding Programs
Concepts used and implementation
The first proposal to use parentage assignment in breeding at an acceptable cost was an improvement of within-family selection called “walk-back selection” (Doyle and Herbinger, 1995). A two-step process of assignment was suggested and tested to achieve a minimal number of selected candidates per family (Herbinger et al., 1995).
Since this date, public organizations and breeding companies initiated selection programs using parentage assignment in sturgeons (France, USA), Atlantic salmon (Ireland, Norway, Scotland), tilapia (Philippines), halibut (Norway and Scotland), rainbow trout (France), cod (Norway), gilthead sea bream (France, Greece, Spain), turbot (France), European sea bass (France, Greece), meagre and red drum (France), Asian sea bass (Singapore, Indonesia, Australia), and shrimps (Australia, Thailand, Mexico, Equator, Central and South America). This list may be incomplete and represents the present informal expert knowledge of the authors. Little information is publicly available in these programs but mass selection, family-based selection (often BLUP: best linear unbiased prediction) or a combination of both are used to improve growth, processing yields, quality traits and disease resistance according to different schemes (Figure 1).
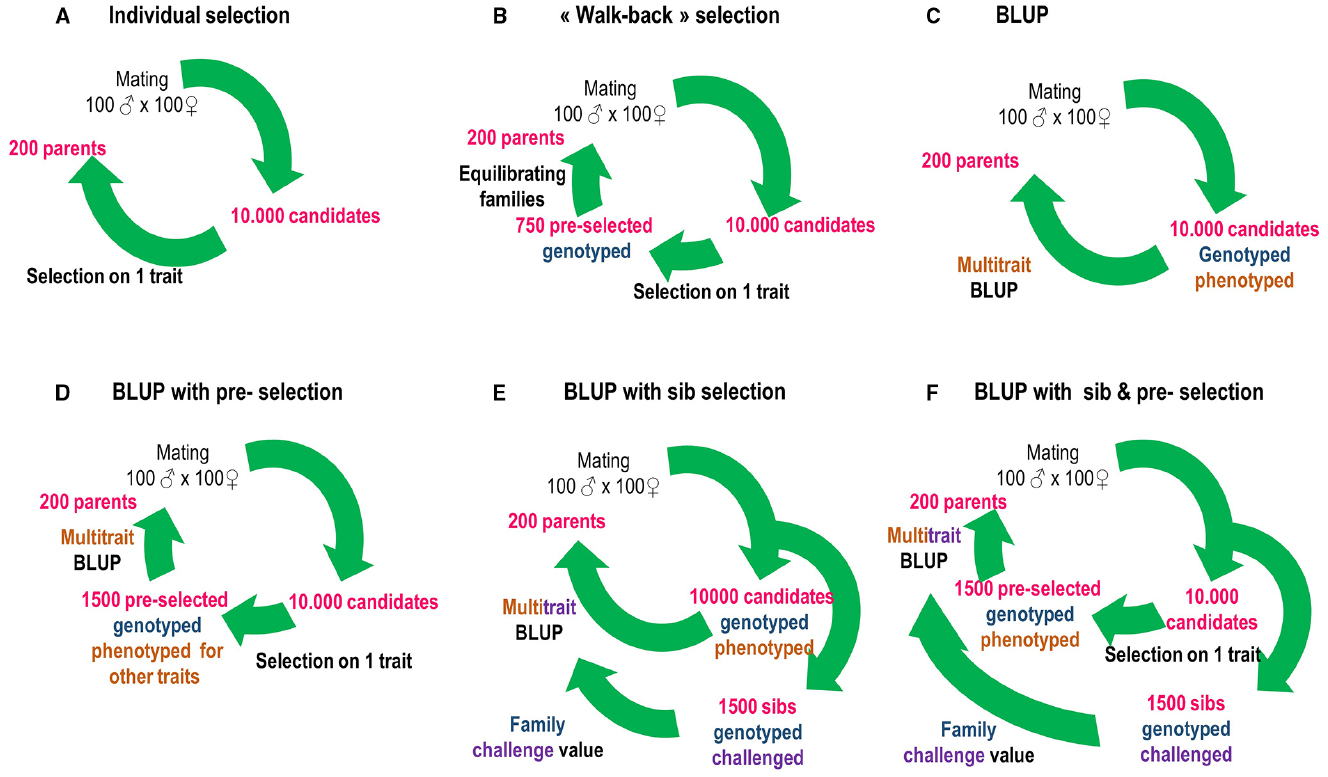
Figure 1. Possible uses of genotyping to evolve from individual selection to BLUP (best linear unbiased prediction) and/or sib selection in aquaculture breeding. Numbers are examples and have to be adapted for different species, traits, and breeding designs. (A) Under individual selection candidates are selected based on their own performance. (B) Under walk-back selection, animals selected on their individual performance are genotyped, and a subset of those with balanced family representation is used as broodstock. (C) Under BLUP, all animals are genotyped and phenotyped (possibly for several traits) and selected based on a BLUP breeding value combining phenotypic and pedigree information. (D) BLUP with pre-selection is similar but multi-trait phenotyping and genotyping is done only one a subset of the population pre-selected for its own performance on one trait (usually growth). (E) Under BLUP with sib selection, unselected sibs of the candidates are submitted to a lethal challenge (e.g., disease, processing yields) and genotyped and family values are incorporated in breeding value evaluation for the lethal traits. (F) BLUP with sib and pre-selection combines panels (D,E).
Key parameters to choose to develop a breeding program using parentage assignment are not only the genotyping cost (12–20 Euros per individual), but also the capacity to produce a large number of families in one batch to avoid tank effects, the true assignment efficiency, as well as the availability of tools such as rapid mass genotyping capacities (specially for species with short generation interval), individual tagging to improve traceability and facilitate data collection, automated database systems to collect, store and link performances to tags, DNA samples and pedigrees, optimized genetic softwares to rank and mate candidates to maximize genetic progress and minimize inbreeding. Use of parentage assignment is not only “genetic tagging,” but requires a complete re-optimization of breeding programs.
Optimization of breeding schemes using parentage assignment
One main target for optimization has been the limitation of numbers genotyped, using two-way nested models for partial pedigrees (Li et al., 2003), or extreme phenotypes with family effect considered as a fixed effect (Morton and Howarth, 2005). BLUP selection normally requires the knowledge of performance and pedigree on all candidates, which is not the case in Figures 1D,F. In these cases, the loss of selection efficiency (compared to BLUP with pedigree known on all candidates) depends on selection intensity and genetic parameters (Chapuis et al., 2010; Dupont-Nivet et al., 2010b; Sonesson et al., 2011). In addition, issues linked to mixing of families were studied, such as methods to limit non-genetic maternal effect in salmonids (Haffray et al., 2012b), effect of grading practices to limit cannibalism on family contributions in barramundi (Loughnan et al., 2013), and the importance to consider male maturation status to estimate heritability of growth more accurately (Dupont-Nivet et al., 2010a). Ninh et al. (2011) and Sae-Lim et al. (2013) compared expected genetic gains with different systems of evaluation (mixed/separate families and impact of G × E interactions), while Haffray et al. (2013) proposed application of ultrasound tomography to predict processing yields on live candidates to limit the use of slaughtered sibs.
Global Appraisal and Perspectives
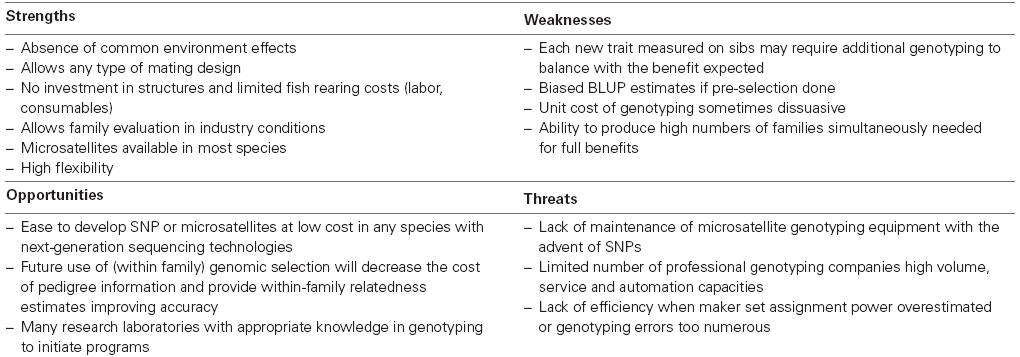
The rapid increase of publications using parentage assignment in the last decade shows how powerful this method is to estimate genetic parameters in any species and rearing system. It avoids the initial investment in separate family rearing units and limits associated biases, even more in species with high larval mortality, small larval size, and initial live feeding. Applications are strongly driven by reproductive constraints linked to the need to simultaneously produce enough families (Table 1). The cost/information ratio has to be maximized with adequate management of variance sources (number of parents, initial representation of families, or groups of spawns), mating design, and number of individuals genotyped.
Optimal investment in parentage assignment is a balance between the reduction of investment and operational costs needed for the separate family rearing and the cost of genotyping, which presently limits the application of parentage assignment to mass selection and family-based selection on a limited number of traits. Moreover, any new trait that cannot be recorded on the live candidate and has to be measured on sibs then requires additional genotyping with a cost/benefit ratio to estimate case by case, and to compare with the possible use of indirect criteria.
A major benefit of parentage assignment is that it allows high selection pressure (<3%) to be applied in commercial conditions, while still controlling inbreeding. The knowledge of pedigree also allows an increase in selection accuracy (and then a higher selection gain) on all traits, as well as selection on lethal traits which cannot be done by individual selection. This technology also allows to easily combine sanitary protection of the breeding nucleus and sib testing in commercial environments. Parentage assignment offers simplicity and flexibility in the life of the breeding program that can be easily adapted to new traits, new mating schemes, different number of candidates. This is critical, especially at the initiation of domestication, for “niche” species or in developing countries, where the need for separate rearing system has often prevented any investment in selective breeding in the past, or has fixed the architecture of the breeding programs.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work received funding from the European Union’s Seventh Framework Programme (FP7 2007-2013) under grant agreement no. 613611 (FISHBOOST).
References
Antonello, J., Massault, C., Franch, R., Haley, C., Pellizzari, C., Bovo, G., et al. (2009). Estimates of heritability and genetic correlation for body length and resistance to fish pasteurellosis in the gilthead sea bream (Sparus aurata L.). Aquaculture 298, 29–35. doi: 10.1016/j.aquaculture.2009.10.022
Bardon, A., Vandeputte, M., Dupont-Nivet, M., Chavanne, H., Haffray, P., Vergnet, A., et al. (2009). What is the heritable component of spinal deformities in the European sea bass (Dicentrarchus labrax)? Aquaculture 294, 194–201. doi: 10.1016/j.aquaculture.2009.06.018
Becker, W. A. (1967). Manual of Procedures in Quantitative Genetics. Pullman, WA: Washington State University Press.
Bestin, A., Dupont-Nivet, M., Haffray, P., Médale, F., Quillet, E., Vandeputte, M., et al. (2014). “Genotype by diet interactions on growth and processing traits in rainbow trout (Oncorhynchus mykiss), European sea bass (Dicentrarchus labrax), gilthead sea bream (Sparus aurata) and meagre (Argyrosomus regius) fed diets with almost complete substitution of both fish meal and fish oil by vegetal ingredients,” in Proceedings of the 10th World Congress on Genetics Applied to Livestock Production, Vancouver, Canada.
Boudry, P., Collet, B., Cornette, F., Hervouet, V., and Bonhomme, F. (2002). High variance in reproductive success of the Pacific oyster (Crassostrea gigas, Thunberg) revealed by microsatellite-based parentage analysis of multifactorial crosses. Aquaculture 204, 283–296. doi: 10.1016/S0044-8486(01)00841-9
Brody, T., Wohlfarth, G., Hulata, G., and Moav, R. (1981). Application of electrophoretic genetic markers to fish breeding. IV. Assessment of breeding value of full-sib families. Aquaculture 24, 175–186. doi: 10.1016/0044-8486(81)90054-5
Brown, C. R., Woolliams, J. A., and McAndrew, B. J. (2005). Factors influencing effective population size in commercial populations of gilthead seabream, Sparus aurata. Aquaculture 247, 219–225. doi: 10.1016/j.aquaculture.2005.02.002
Cercueil, A., Bellemain, E., and Manel, S. (2002). PARENTE: computer program for parentage analysis. J. Hered. 93, 458–459. doi: 10.1093/jhered/93.6.458
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chapuis, H., Vandeputte, M., Dupont-Nivet, M., Haffray, P., and Quillet, E. (2010). “Selection for an improved disease resistance using factorial mating designs and molecular based pedigrees in fish: a simulation study,” in Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany, CDROM paper 210.
Christie, M. R. (2010). Parentage in natural populations: novel methods to detect parent-offspring pairs in large data sets. Mol. Ecol. Res. 10, 115–128. doi: 10.1111/j.1755-0998.2009.02687.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Danzmann, R. G. (1997). PROBMAX: a computer program for assigning unknown parentage in pedigree analysis from known genotypic pools of parents and progeny. J. Hered. 88, 333. doi: 10.1093/oxfordjournals.jhered.a023112
Domingos, J. A., Smith-Keune, C., and Jerry, D. R. (2014). Fate of genetic diversity within and between generations and implications for DNA parentage analysis in selective breeding of mass spawners: a case study of commercially farmed barramundi, Lates calcarifer. Aquaculture 424–425, 174–182. doi: 10.1016/j.aquaculture.2014.01.004
Domingos, J. A., Smith-Keune, C., Robinson, N., Loughnan, S., Harrison, P., and Jerry, D. R. (2013). Heritability of harvest growth traits and genotype × environment interactions in barramundi, Lates calcarifer (Bloch). Aquaculture 402–403, 66–75. doi: 10.1016/j.aquaculture.2014.01.004
Doyle, R. W., and Herbinger, C. M. (1995). “The use of DNA fingerprinting for high-intensity, within family selection in fish breeding,” in Proceedings of the 5th World Congress on Genetics Applied to Livestock Production, Guelph, Ontario, Canada, Vol. 19, 364–371.
Duchesne, P., Godbout, M. H., and Bernatchez, L. (2002). PAPA (package for the analysis of parental allocation): a computer program for simulated and real parental allocation. Mol. Ecol. Notes 2, 191–193. doi: 10.1046/j.1471-8286.2002.00164.x
Dupont-Nivet, M., Chevassus, B., Mauger, S., Haffray, P., and Vandeputte, M. (2010a). Side effects of sexual maturation on heritability estimates in rainbow trout (Oncorhynchus mykiss). Aquac. Res. 41, e878–e880. doi: 10.1111/j.1365-2109.2009.02448.x
Dupont-Nivet, M., Haffray, P., and Vandeputte, M. (2010b). “Effect of culling on bias of BLUP estimates–case of fish species,” in Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany, CDROM paper 429.
Dupont-Nivet, M., Karahan-Nomm, B., Vergnet, A., Merdy, O., Haffray, P., Chavanne, H., et al. (2010c). Genotype by environment interactions for growth in European seabass (Dicentrarchus labrax) are large when growth rate rather than weight is considered. Aquaculture 306, 365–368. doi: 10.1016/j.aquaculture.2010.05.025
Dupont-Nivet, M., Vandeputte, M., and Chevassus, B. (2002). Optimization of factorial mating designs for inference on heritability in fish species. Aquaculture 204, 361–370. doi: 10.1016/S0044-8486(01)00839-0
Dupont-Nivet, M., Vandeputte, M., Haffray, P., and Chevassus, B. (2006). Effect of different mating designs on inbreeding, genetic variance and response to selection when applying individual selection in fish breeding programs. Aquaculture 252, 161–170. doi: 10.1016/j.aquaculture.2005.07.005
Dupont-Nivet, M., Vandeputte, M., Vergnet, A., Merdy, O., Haffray, P., Chavanne, H., et al. (2008). Heritabilities and G × E interactions for growth in the European sea bass (Dicentrarchus labrax L.) using a marker-based pedigree. Aquaculture 275, 81–87. doi: 10.1016/S0044-8486(01)00839-0
Estoup, A., Gharbi, K., SanCristobal, M., Chevalet, C., Haffray, P., and Guyomard, R. (1998). Parentage assignment using microsatellites in turbot (Scophtalmus maximus) and rainbow trout (Oncorhynchus mykiss) hatchery populations. Can. J. Fish. Aquat. Sci. 55, 715–725. doi: 10.1139/f97-268
Ferrari, S., Chatain, B., Cousin, X., Leguay, D., Vergnet, A., Vidal, M. O., et al. (2014). Early individual electronic identification of sea bass using RFID microtags: a first example of early phenotyping of sex-related growth. Aquaculture 426–427, 165–171. doi: 10.1016/j.aquaculture.2014.01.033
Fessehaye, Y., El-bialy, Z., Rezk, M. A., Crooijmans, R., Bovenhuis, H., and Komen, H. (2006). Mating systems and male reproductive success in Nile tilapia (Oreochromis niloticus) in breeding hapas: a microsatellite analysis. Aquaculture 256, 148–158. doi: 10.1016/j.aquaculture.2006.02.024
Galli, A., Greppi, G., Boscari, E., and Bonacina, C. (2011). Parental allocation in Acipenser naccarii: statistical aspects and software development. J. Appl. Ichthyol. 27, 651–654. doi: 10.1111/j.1439-0426.2011.01709.x
Gheyas, A. A., Woolliams, J. A., Taggart, J. B., Sattar, M. A., Das, T. K., McAndrew, B. J., et al. (2009). Heritability estimation of silver carp (Hypophthalmichthys molitrix) harvest traits using microsatellite based parentage assignment. Aquaculture 294, 187–193. doi: 10.1016/j.aquaculture.2009.06.013
Gjedrem, T. (2010). The first family-based breeding program in aquaculture. Rev. Aquac. 2, 2–15. doi: 10.1111/j.1753-5131.2010.01011.x
Gjedrem, T. (2012). Genetic improvement for the development of efficient global aquaculture: a personal opinion review. Aquaculture 344–349, 12–22. doi: 10.1016/j.aquaculture.2012.03.003
Gjedrem, T., Robinson, N., and Rye, M. (2012). The importance of selective breeding in aquaculture to meet future demands for animal protein: a review. Aquaculture 350–353, 117–129. doi: 10.1016/j.aquaculture.2012.04.008
Gjedrem, T., and Thodesen, J. (2005). “Selection,” in Selection and Breeding Programs in Aquaculture, ed. T. Gjedrem (Dordrecht: Springer), 89–111.
Gjerde, B., Gjoen, H. M., and Villanueva, B. (1996). Optimum designs for fish breeding programmes with constrained inbreeding mass selection for a normally distributed trait. Livest. Prod. Sci. 47, 59–72. doi: 10.1016/S0301-6226(96)01000-7
Glaubitz, J. C., Rhodes, O. E., and DeWoody, J. A. (2003). Prospects for inferring pairwise relationships with single nucleotide polymorphisms. Mol. Ecol. 12, 1039–1047. doi: 10.1046/j.1365-294X.2003.01790.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guichoux, E., Lagache, L., Wagner, S., Chaumeil, P., Léger, P., Lepais, O., et al. (2011). Current trends in microsatellite genotyping. Mol. Ecol. Res. 11, 591–611. doi: 10.1111/j.1755-0998.2011.03014.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guy, D. R., Bishop, S. C., Brotherstone, S., Hamilton, A., Roberts, R. J., McAndrew, B. J., et al. (2006). Analysis of the incidence of infectious pancreatic necrosis mortality in pedigreed Atlantic salmon, Salmo salar L., populations. J. Fish Dis. 29, 637–647. doi: 10.1111/j.1365-2761.2006.00758.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haffray, P., Bugeon, J., Rivard, Q., Quittet, B., Puyo, S., Allamelou, J. M., et al. (2013). Genetic parameters of in-vivo prediction of carcass, head and fillet yields by internal ultrasound and 2D external imagery in large rainbow trout (Oncorhynchus mykiss). Aquaculture 410–411, 236–244. doi: 10.1016/j.aquaculture.2013.06.016
Haffray, P., Pincent, C., Chapuis, H., Mazeiraud, E., Petit, V., Rault, P., et al. (2012a). Negative genetic trade-offs between production traits and head or bony tissues in large all-female rainbow trout (Oncorhynchus mykiss). Aquaculture 368–369, 145–162 doi: 10.1016/j.aquaculture.2012.09.023
Haffray, P., Vandeputte, M., Petit, V., Pincent, C., Chatain, B., Chapuis, H., et al. (2012b). Minimizing maternal effect in salmonid families mixed since eyed stages and a posteriori DNA-pedigreed. Livest. Sci. 150, 170–178. doi: 10.1016/j.livsci.2012.08.017
Hedgecock, D., Li, G., Hubert, S., Bucklin, K., and Ribes, V. (2004). Widespread null alleles and poor cross-species amplification of microsatellite DNA loci cloned from the Pacific oyster, Crassostrea gigas. J. Shellfish Res. 23, 379–385.
Herbinger, C. M., Doyle, R. W., Pitman, E. R., Paquet, D., Mesa, K. A., Morris, D. B., et al. (1995). DNA fingerprint based analysis of paternal and maternal effects on offspring growth and survival in communally reared rainbow trout. Aquaculture 137, 245–256. doi: 10.1016/0044-8486(95)01109-9
Herbinger, C. M., O’Reilly, P. T., Doyle, R. W., Wright, J. M., and O’Flynn, F. (1999). Early growth performance of Atlantic salmon full-sib families reared in single family tanks versus in mixed family tanks. Aquaculture 173, 105–116. doi: 10.1016/0044-8486(95)01109-9
Herlin, M., Taggart, J. B., McAndrew, B. J., and Penman, D. J. (2007). Parentage allocation in a complex situation: a large commercial Atlantic cod (Gadus morhua) mass spawning tank. Aquaculture 272, S195–S203. doi: 10.1016/j.aquaculture.2007.08.018
Jamieson, A. (1965). The genetics of transferrins in cattle. Heredity 20, 419–441. doi: 10.1038/hdy.1965.54
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jerry, D. R., Preston, N. P., Crocos, P. J., Keys, S., Meadows, J. R. S., and Li, Y. (2006). Application of DNA parentage analyses for determining relative growth rates of Penaeus japonicus families reared in commercial ponds. Aquaculture 254, 171–181. doi: 10.1016/j.aquaculture.2005.10.035
Jones, A. G., Small, C. M., Paczolt, K. A., and Ratterman, N. L. (2010). A practical guide to methods of parentage analysis. Mol. Ecol. Res. 10, 6–30. doi: 10.1111/j.1755-0998.2009.02778.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kalinowski, S. T., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaspar, V., Vandeputte, M., Kohlmann, K., Hulak, M., Rodina, M., Gela, D., et al. (2008). A proposal and case study towards a conceptual approach of validating sperm competition in common carp (Cyprinus carpio L.), with practical implications for hatchery procedures. J. Appl. Ichthyol. 24, 406–409. doi: 10.1111/j.1439-0426.2008.01148.x
Kocour, M., Mauger, S., Rodina, M., Gela, D., Linhart, O., and Vandeputte, M. (2007). Heritability estimates for processing and quality traits in common carp (Cyprinus carpio L.) using a molecular pedigree. Aquaculture 270, 43–50. doi: 10.1016/j.aquaculture.2007.03.001
Kong, N., Li, Q., Yu, H., and Kong, L. F. (2013). Heritability estimates for growth-related traits in the Pacific oyster (Crassostrea gigas) using a molecular pedigree. Aquac. Res. doi: 10.1111/are.12205 [Epub ahead of print].
Krishna, G., Gopikrishna, G., Gopal, C., Jahageerdar, S., Ravichandran, P., Kannappn, S., et al. (2011). Genetic parameters for growth and survival in Penaeus monodon cultured in India. Aquaculture 318, 74–78. doi: 10.1016/j.aquaculture.2011.04.028
Kvingedal, R., Evans, B. S., Lind, C. E., Taylor, J. J. U., Dupont-Nivet, M., and Jerry, D. R. (2010). Population and family growth response to different rearing location, heritability estimates and genotype × environment interaction in the silver-lip pearl oyster (Pinctada maxima). Aquaculture 304, 1–6. doi: 10.1016/j.aquaculture.2010.02.035
Lapègue, S., Harrang, E., Heurtebise, S., Flahauw, E., Donnadieu, C., Gayral, P., et al. (2014). Development of SNP-genotyping arrays in two shellfish species. Mol. Ecol. Res. 14, 820–830. doi: 10.1111/1755-0998.12230
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Le Boucher, R., Vandeputte, M., Dupont-Nivet, M., Quillet, E., Ruelle, F., Vergnet, A., et al. (2013). Genotype by diet interactions in European sea bass (Dicentrarchus labrax L.): nutritional challenge with totally plant-based diets. J. Anim. Sci. 91, 44–56. doi: 10.2527/jas.2012-5311
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, X., Field, C., and Doyle, R. W. (2003). Estimation of additive genetic variance components in aquaculture populations selectively pedigreed by DNA fingerprinting. Biomet. J. 45, 61–72. doi: 10.1002/bimj.200290016
Lillehammer, M., Meuwissen, T. H., and Sonesson, A. (2013). A low-marker density implementation of genomic selection in aquaculture using within-family genomic breeding values. Genet. Sel. Evol. 45:39. doi: 10.1186/1297-9686-45-39
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Loughnan, S. R., Domingos, J. A., Smith-Keune, C., Forrester, J. P., Jerry, D. R., Beheregaray, L. B., et al. (2013). Broodstock contribution after mass spawning and size grading in barramundi (Lates calcarifer, Bloch). Aquaculture 404–405, 139–149. doi: 10.1016/j.aquaculture.2013.04.014
Lucas, T., Macbeth, M., Degnan, S. M., Knibb, W., and Degnan, B. M. (2006). Heritability estimates for growth in the tropical abalone Haliotis asinina using microsatellites to assign parentage. Aquaculture 259, 146–152. doi: 10.1016/j.aquaculture.2006.05.039
Matson, S. E., Camara, M. D., Eichert, W., and Banks, M. A. (2008). P-LOCI: a computer program for choosing the most efficient set of loci for parentage assignment. Mol. Ecol. Res. 8, 765–768. doi: 10.1111/j.1755-0998.2008.02128.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, P. A., Elliott, N. G., Vaillancourt, R. E., Kube, P. D., and Koutoulis, A. (2014). Genetic diversity and pedigree assignment in tetraploid Pacific oysters (Crassostrea gigas). Aquaculture 433, 318–324. doi: 10.1016/j.aquaculture.2014.06.025
Morton, R., and Howarth, J. M. (2005). Estimation of breeding values using selected pedigree records. Biometrics 61, 422–432. doi: 10.1111/j.1541-0420.2005.040305.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Navarro, A., Zamorano, M. J., Hildebrandt, S., Ginés, R., Aguilera, C., and Afonso, J. M. (2009). Estimates of heritabilities and genetic correlations for growth and carcass traits in gilthead seabream (Sparus auratus L.), under industrial conditions. Aquaculture 289, 225–230. doi: 10.1016/j.aquaculture.2008.12.024
Nguyen, T. T. T., Hayes, B. J., and Ingram, B. A. (2014). Genetic parameters and response to selection in blue mussel (Mytilus galloprovincialis) using a SNP-based pedigree. Aquaculture 420–421, 295–301. doi: 10.1016/j.aquaculture.2013.11.021
Nicholas, F. W. (1980). Size of population required for artificial selection. Genet. Res. 35, 85–105. doi: 10.1017/S0016672300013951
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ninh, N. H., Ponzoni, R. W., Nguyen, N. H., Woolliams, J. A., Taggart, J. B., McAndrew, B. J., et al. (2011). A comparison of communal and separate rearing of families in selective breeding of common carp (Cyprinus carpio). Aquaculture 322–323, 39–46. doi: 10.1016/j.aquaculture.2011.09.031
Norris, A. T., and Cunningham, E. P. (2004). Estimates of phenotypic and genetic parameters for flesh colour traits in farmed Atlantic salmon based on multiple trait animal model. Livest. Prod. Sci. 89, 209–222. doi: 10.1016/j.livprodsci.2004.02.010
Perez-Enriquez, R., Takagi, M., and Taniguchi, N. (1999). Genetic variability and pedigree tracing of a hatchery-reared stock of red sea bream (Pagrus major) used for stock enhancement, based on microsatellite DNA markers. Aquaculture 173, 413–423. doi: 10.1016/S0044-8486(98)00469-4
Pierce, L. R., Palti, Y., Silverstein, J. T., Barrows, F. T., Hallerman, E. M., and Parsons, J. E. (2008). Family growth response to fishmeal and plant-based diets shows genotype × diet interaction in rainbow trout (Oncorhynchus mykiss). Aquaculture 278, 37–42. doi: 10.1016/j.aquaculture.2008.03.017
Rodzen, J. A., Famula, T. R., and May, B. (2004). Estimation of parentage and relatedness in the polyploid white sturgeon (Acipenser transmontanus) using a dominant marker approach for duplicated microsatellite loci. Aquaculture 232, 165–182. doi: 10.1016/S0044-8486(03)00450-2
Sae-Lim, P., Kause, A., Mulder, H. A., Martin, K. E., Barfoot, A. J., Parsons, J. E., et al. (2013). Genotype-by-environment interaction of growth traits in rainbow trout (Oncorhynchus mykiss): a continental scale study. J. Anim. Sci. 91, 5572–5581. doi: 10.2527/jas.2012-5949
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sae-Lim, P., Komen, H., and Kause, A. (2010). Bias and precision of estimates of genotype-by-environment interaction: a simulation study. Aquaculture 310, 66–73. doi: 10.1016/j.aquaculture.2010.10.020
Saillant, E., Dupont-Nivet, M., Haffray, P., and Chatain, B. (2006). Estimates of heritability and genotype-environment interactions for body weight in sea bass (Dicentrarchus labrax L.) raised under communal rearing conditions. Aquaculture 254, 139–147. doi: 10.1016/j.aquaculture.2005.10.018
Saillant, E., Dupont-Nivet, M., Sabourault, M., Haffray, P., Laureau, S., Vidal, M. O., et al. (2009). Genetic variation for carcass quality traits in cultured sea bass (Dicentrarchus labrax). Aquat. Living Resour. 22, 105–112. doi: 10.1051/alr/2009010
Saillant, E., Fostier, A., Haffray, P., Menu, B., Thimonier, J., and Chatain, B. (2002). Temperature effects and genotype-temperature interactions on sex determination in the European sea bass (Dicentrarchus labrax L.). J. Exp. Zool. 292, 494–505. doi: 10.1002/jez.10071
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sellars, M. J., Dierens, L., McWilliam, S., Little, B., Murphy, B., Coman, G. J., et al. (2014). Comparison of microsatellite and SNP DNA markers for pedigree assignment in Black Tiger shrimp, Penaeus monodon. Aquac. Res. 45, 417–426. doi: 10.1111/j.1365-2109.2012.03243.x
Skaarud, A., Woolliams, J. A., and Gjoen, H. M. (2011). Strategies for controlling inbreeding in fish breeding programs; an applied approach using optimum contribution (OC) procedures. Aquaculture 311, 110–114. doi: 10.1016/j.aquaculture.2010.11.023
Sonesson, A. K. (2005). A combination of walk-back and optimum contribution selection in fish: a simulation study. Genet. Sel. Evol. 37, 587–599. doi: 10.1186/1297-9686-37-7-587
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sonesson, A. K., Gjerde, B., and Robinson, N. (2011). A simple selection scheme to improve disease resistance and growth. Aquaculture 319, 337–341. doi: 10.1016/j.aquaculture.2011.07.009
Sutton, J. T., Robertson, B. C., and Jamieson, I. G. (2011). Dye shift: a neglected source of genotyping error in molecular ecology. Mol. Ecol. Res. 11, 514–520. doi: 10.1111/j.1755-0998.2011.02981.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Taggart, J. B. (2007). FAP: an exclusion-based parental assignment program with enhanced predictive functions. Mol. Ecol. Notes 7, 412–415. doi: 10.1111/j.1471-8286.2006.01616.x
Thodesen, J., Rye, M., Wang, Y., Yang, K. S., Bentsen, H. B., and Gjedrem, T. (2011). Genetic improvement of tilapia in China: genetic parameters and selection responses in growth of Nile tilapia (Oreochromis niloticus) after six generations of multi-trait selection for growth and fillet yield. Aquaculture 322, 51–64. doi: 10.1016/j.aquaculture.2011.10.010
Trong, T. Q., van Bers, N., Crooijmans, R., Dibbits, B., and Komen, H. (2013). A comparison of microsatellites and SNPs in parental assignment in the GIFT strain of Nile tilapia (Oreochromis niloticus): the power of exclusion. Aquaculture 388–391, 14–23. doi: 10.1016/j.aquaculture.2013.01.004
Vandeputte, M. (2012). An accurate formula to calculate exclusion power of marker sets in parentage assignment. Genet. Sel. Evol. 44:36. doi: 10.1186/1297-9686-44-36
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vandeputte, M., Dupont-Nivet, M., Chatain, B., and Chevassus, B. (2001). Setting up a strain testing design for the seabass, Dicentrarchus labrax: a simulation study. Aquaculture 202, 329–342. doi: 10.1016/S0044-8486(01)00782-7
Vandeputte, M., Dupont-Nivet, M., Chavanne, H., and Chatain, B. (2007). A polygenic hypothesis for sex determination in the European sea bass Dicentrarchus labrax. Genetics 176, 1049–1057. doi: 10.1534/genetics.107.072140
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vandeputte, M., Garouste, R., Dupont-Nivet, M., Haffray, P., Vergnet, A., Chavanne, H., et al. (2014). Multi-site evaluation of the rearing performances of 5 wild populations of European sea bass (Dicentrarchus labrax). Aquaculture 424–425, 239–248. doi: 10.1016/j.aquaculture.2014.01.005
Vandeputte, M., Mauger, S., and Dupont-Nivet, M. (2006). An evaluation of allowing for mismatches as a way to manage genotyping errors in parentage assignment by exclusion. Mol. Ecol. Notes 6, 265–267. doi: 10.1111/j.1471-8286.2005.01167.x
Vandeputte, M., Rossignol, M. N., and Pincent, C. (2011). From theory to practice: empirical evaluation of the assignment power of marker sets for pedigree analysis in fish breeding. Aquaculture 314, 80–86. doi: 10.1016/j.aquaculture.2011.01.043
Vieira, V. L. A., Norris, A., and Johnston, I. A. (2007). Heritability of fibre number and size parameters and their genetic relationship to flesh quality traits in Atlantic salmon (Salmo salar L.). Aquaculture 272(Suppl. 1), S100–S109. doi: 10.1016/j.aquaculture.2007.08.028
Villanueva, B., Verspoor, E., and Visscher, P. M. (2002). Parental assignment in fish using microsatellite genetic markers with finite numbers of parents and offspring. Anim. Genet. 33, 33–41. doi: 10.1046/j.1365-2052.2002.00804.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Waldbieser, G. C., and Wolters, W. R. (1999). Application of polymorphic microsatellite loci in a channel catfish Ictalurus punctatus breeding program. J. World Aquac. Soc. 30, 256–262. doi: 10.1111/j.1749-7345.1999.tb00873.x
Wang, C. M., Lo, L. C., Zhu, Z. Y., Lin, G., Feng, F., Li, J., et al. (2008). Estimating reproductive success of brooders and heritability of growth traits in Asian sea bass (Lates calcarifer) using microsatellites. Aquac. Res. 39, 1612–1619. doi: 10.1111/j.1365-2109.2008.02034.x
Wang, J. (2007). Parentage and sibship exclusions: higher statistical power with more family members. Heredity 99, 205–217. doi: 10.1038/sj.hdy.6800984
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J., and Santure, A. W. (2009). Parentage and sibship inference from multilocus genotype data under polygamy. Genetics 181, 1579–1594. doi: 10.1534/genetics.108.100214
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J., and Scribner, K. T. (2014). Parentage and sibship inference from markers in polyploids. Mol. Ecol. Res. 14, 541–553. doi: 10.1111/1755-0998.12210
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Whatmore, P., Nguyen, N. H., Miller, A., Lamont, R., Powell, D., D’Antignana, T., et al. (2013). Genetic parameters for economically important traits in yellowtail kingfish Seriola lalandi. Aquaculture 400–401, 77–84. doi: 10.1016/j.aquaculture.2013.03.002
Wilson, A. J., McDonald, G., Moghadam, H. K., Herbinger, C. M., and Ferguson, M. M. (2003). Marker-assisted estimation of quantitative genetic parameters in rainbow trout, Oncorhynchus mykiss. Genet. Res. Camb. 81, 145–156. doi:10.1017/S0016672302006055
Yue, G. H., and Xia, J. H. (2014). Practical considerations of molecular parentage analysis in fish. J. World Aquac. Soc. 45, 89–103. doi: 10.1111/jwas.12107
Keywords: aquaculture, parentage assignment, selective breeding, microsatellites, SNPs
Citation: Vandeputte M and Haffray P (2014) Parentage assignment with genomic markers: a major advance for understanding and exploiting genetic variation of quantitative traits in farmed aquatic animals. Front. Genet. 5:432. doi: 10.3389/fgene.2014.00432
Received: 12 September 2014; Paper pending published: 20 October 2014;
Accepted: 22 November 2014; Published online: 12 December 2014.
Edited by:
Ross Houston, University of Edinburgh, UKReviewed by:
Jeff Silverstein, United States Department of Agriculture, USAGen H. Yue, National University of Singapore, Singapore
Copyright © 2014 Vandeputte and Haffray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marc Vandeputte, Ifremer/INRA, Chemin de Maguelone, F-34250 Palavas-les-Flots, France e-mail: marc.vandeputte@jouy.inra.fr
 Marc Vandeputte
Marc Vandeputte