- The Flowering Lab, School of Biological Sciences, University of Auckland, Auckland, New Zealand
The timing of the transition to flowering is carefully controlled by plants in order to optimize sexual reproduction and the ensuing production of seeds, grains, and fruits. The genetic networks that regulate floral induction are best characterized in the temperate eudicot Arabidopsis in which the florigen gene FT plays a major role in promoting the transition to flowering. Legumes are an important plant group, but less is known about the regulation of their flowering time. In the model legume Medicago truncatula (Medicago), a temperate annual plant like Arabidopsis, flowering is induced by prolonged cold (vernalization) followed by long day lengths (LD). Recent molecular-genetic experiments have revealed that a FT-like gene, MtFTa1, is a central regulator of flowering time in Medicago. Here, we characterize the three Medicago FRUITFULL (FUL) MADS transcription factors, MtFULa, MtFULb, and MtFULc using phylogenetic analyses, gene expression profiling through developmental time courses, and functional analyses in transgenic plants. MtFULa and MtFULb have similarity in sequence and expression profiles under inductive environmental conditions during both vegetative and reproductive development while MtFULc is only up regulated in the apex after flowering in LD conditions. Sustained up regulation of MtFULs requires functional MtFTa1 but their transcript levels are not affected during cold treatment. Overexpression of MtFULa and MtFULb promotes flowering in transgenic Arabidopsis plants with an additional terminal flower phenotype on some 35S:MtFULb plants. An increase in transcript levels of the MtFULs was also observed in Medicago plants overexpressing MtFTa1. Our results suggest that the MtFULs are targets of MtFTa1. Overall, this work highlights the conserved functions of FUL-like genes in promoting flowering and other roles in plant development and thus contributes to our understanding of the genetic control of the flowering process in Medicago.
Introduction
Flowering time is an important adaptive trait in crop plants because of its major effect on plant yield and productivity (Jung and Muller, 2009; Putterill et al., 2013). However, the genetic network that regulates the transition to flowering is best understood in the small Brassicaceous annual weed, Arabidopsis (Srikanth and Schmid, 2011; Andres and Coupland, 2012). In Arabidopsis, at least six pathways transduce external and internal cues and regulate flowering time by converging on floral integrator genes such as FT, SOC1, and LFY (Srikanth and Schmid, 2011; Andres and Coupland, 2012). For example, extended winter cold (vernalization) leads to epigenetic silencing of an important floral repressor FLC (Kim et al., 2009). Thus, after winter, FLC inhibition of FT and SOC1 is relieved, allowing the photoperiod pathway via the activator gene, CO to up-regulate them which promotes flowering in the long days (LDs) of spring. The commitment to flower is associated with the development of inflorescence meristem which grows indeterminately producing floral primordia on its flanks. Among the genes implicated to regulate reproductive meristem identity include LFY, SOC1, other MADS-box transcription factors FUL, AGL24, AP1, and CAL as well as the flowering repressor TFL1 (Fornara et al., 2010; Srikanth and Schmid, 2011; Posé et al., 2012; Torti et al., 2012).
Arabidopsis FUL is a member of the euFUL clade, one of the three core eudicot clades, euFUL, euAP1 and AGL79, in the AP1/SQUA/FUL lineage of MADS-box genes (Litt and Irish, 2003; Berbel et al., 2012; Pabón-Mora et al., 2012). It has several functions through Arabidopsis development including flowering time control, inflorescence meristem identity and carpel development (Mandel and Yanofsky, 1995a; Hempel et al., 1997; Gu et al., 1998; Ferrándiz et al., 2000a; Melzer et al., 2008; Torti et al., 2012; Balanzà et al., 2014). The age and ambient temperature flowering-time pathways regulate Arabidopsis FUL expression via the action of SPL proteins and FT and LD photoperiods stimulate FUL expression (Wang et al., 2009; Kim et al., 2012; Balanzà et al., 2014). FUL has partially redundant roles with SOC1 in promoting Arabidopsis flowering as single ful mutants have a mild delay to flowering, but the absence of SOC1 function in the ful soc1 double mutant causes a later flowering phenotype (Melzer et al., 2008; Torti et al., 2012; Balanzà et al., 2014). A recent report suggests that FUL might act by forming heterodimers with MADS proteins SVP and SOC1 to antagonize floral repression by FLC and SVP (Balanzà et al., 2014). The role of FUL-like genes in flowering-time control in other plants is not well understood overall, but progress is being made (Pabón-Mora et al., 2012). For example, the AP1/FUL gene from the basal eudicot opium poppy promotes the transition to flowering (Pabón-Mora et al., 2012) and VRN1, a monocot AP1/FUL-like gene, is an important regulator of flowering in response to vernalization in the temperate grasses wheat and barley (Trevaskis et al., 2007; Distelfeld et al., 2009).
Despite good progress, major gaps remain in our current understanding of flowering networks in many other plants such as the Fabaceae family (Putterill et al., 2013; Wong et al., 2014). Our study focuses on the small temperate forage legume model plant, Medicago truncatula (Medicago) as it offers a number of advantages including a sequenced genome and tagged mutant lines for forward and reverse genetic screens (Tadege et al., 2009; Young et al., 2011; Putterill et al., 2013). Medicago flowers much more rapidly in LD if these conditions are preceded by vernalization (Clarkson and Russell, 1975; Laurie et al., 2011). The results of our work and others indicate that a Medicago FT gene, MtFTa1, one of five MtFT genes, is expressed in response to vernalization and LD photoperiods and functions as a major integrator gene of both the vernalization and LD photoperiod pathways (Laurie et al., 2011; Yeoh et al., 2011, 2013; Jaudal et al., 2013). However, other key components such as the FLC/MAF clade of floral repressors are missing from Medicago and the photoperiod pathway appears to differ from the Arabidopsis model (Hecht et al., 2005; Putterill et al., 2013; Wong et al., 2014).
To identify additional genes in the Medicago flowering time network, we are taking forward and reverse genetic approaches (Jaudal et al., 2013, 2014; Putterill et al., 2013; Yeoh et al., 2013). In our recent reports, we identified three new early flowering mutants in Medicago (Jaudal et al., 2013; Yeoh et al., 2013). These spring mutants flower rapidly in LD conditions in the absence of vernalization. This correlates with the early onset of FTa1 expression. The spring mutants all have retroelement insertions in or near MtFTa1 which cause elevated expression of MtFTa1 in LD conditions in the absence of vernalization.
Global microarray analysis of gene expression in leaves of the early flowering Medicago spring1 mutant, indicated that a second gene with elevated transcript levels was the MADS-box gene MtFULb, one of three MtFUL genes named for their sequence similarity to FUL (Hecht et al., 2005; Yeoh et al., 2013). Increased MtFULb transcript levels were also observed in transgenic plants over expressing MtFTa1 suggesting that MtFTa1 might control the expression of MtFULb (Yeoh et al., 2013). This has resemblance with Arabidopsis where increased levels of FUL transcript were seen in leaves of plants over-expressing FT (Teper-Bamnolker and Samach, 2005). Consistent with being an FT target, FUL is required for very early flowering in Arabidopsis plants over expressing FT (Teper-Bamnolker and Samach, 2005).
While the functional role of Medicago FUL genes have not been reported, the pea VEG1/PsFULc ortholog of MtFULc has been well characterized (Berbel et al., 2012). VEG1 groups in the AGL79 clade of the AP1/SQUA/FUL lineage (Berbel et al., 2012). veg1 mutants have an extreme non-flowering phenotype. However, this is not due to a defect in floral induction or phase delay, but because VEG1 is required for the specification of secondary inflorescence meristems that produce flowers (Berbel et al., 2012).
In this study, to investigate MtFUL function in flowering, we molecularly characterized the three Medicago FUL-like genes, MtFULa, MtFULb and MtFULc, and MtFTa1 by gene expression profiling in wild type, fta1 mutant and transgenic 35S:MtFTa1 Medicago plants and by transformation into wild type Arabidopsis.
Materials and Methods
Bioinformatics
BLAST searches of Arabidopsis and Medicago protein databases were performed with Arabidopsis FUL (AtFUL) and three previously identified Medicago truncatula FUL-like sequences, MtFULa, MtFULb, and MtFULc (Hecht et al., 2005; Berbel et al., 2012). Alignment of the highest scoring sequences was performed using ClustalW in the Geneious software package [version 8.0.4 available from www.geneious.com (Biomatters, Ltd.)]. The phylogenetic tree was generated using full length protein sequences using the neighbor-joining (NJ) method via bootstrap resampling in the Geneious program. The accession numbers for the protein sequences used are as follows: Medicago MtFULa (Medtr2g461760), MtFULb (Medtr4g109830), MtFULc (Medtr7g016630), MtPIM (Medtr8g066260), MtBM5A (MTR_5g046790), ACJ84407; Arabidopsis AtFUL (At5g60910), AtAGL79 (At3g30260), and AtAP1 (At1g69120); Pisum sativum (garden pea) PsFULa (AAX69065), PsFULb (JN974186), and PsFULc/VEG1 (JN974184). These sequences were obtained from previous studies (Hecht et al., 2005; Berbel et al., 2012; Fourquin et al., 2013), JCVI Medicago genome assembly build Mt4.01, TAIR2, and NCBI3.
Plant Material and Growth Conditions
Medicago truncatula (Medicago) wild type R108_C3 (R108), the fta1 Tnt1 insertion mutant NF3307 and the FTa1 over expression line 35S:FTa1 in the R108 background were used in this study (Laurie et al., 2011). R108 belongs to the Medicago truncatula Gaertn (barrel medic) ssp. tricycla. Scarification, germination, and seed vernalization (V) prior to growth in LD conditions (16 h of light/8 h of dark) and cultivation of Medicago plants were done as described previously (Laurie et al., 2011; Yeoh et al., 2013). For plants that were vernalized as seedlings and then grown in warm LD [depicted as vernalized seedlings in long days (VSLDs) conditions], seedling vernalization was done as described previously (Jaudal et al., 2014) with minor modifications such that the seedlings were grown in LD at 22°C for 11–14 days, then vernalized by exposure to cold at 4°C for 14 days in LD and then transferred to warm (22°C) LD conditions until they flowered. Flowering time of Medicago plants was measured in days after planting of germinated seeds unless otherwise indicated in the text.
Arabidopsis thaliana wild type Columbia (Col) and transgenic plants over expressing MtFUL (35S:MtFULa, 35S:MtFULb, and 35S:MtFULc) in the Col background (this work) were used in this study. These overexpression gene constructs were made by amplifying the cDNAs of the MtFUL genes from R108 RNA and inserting these into the plant transformation vector, pB2GW7 (Karimi et al., 2002) to create expression clones using the GATEWAY TECHNOLOGY (GW) kit (Invitrogen, Corporation, USA) according to the manufacturer’s instructions. The forward and reverse primers, with the ATG translation start codon underlined, used for GW cloning are: GW_MtFULa GGGGACAAGTTTGTACAAAAAAGCAGGCTATATTATGGGGAGAGGAAGGGTG, GGGGACCACTTTGTACAAGAAAGCTGGGTAGGACTAATTAAGCATCCAAGGT; GW_MtFULb GGGGACAAGTTTGTACAAAAAAGCAGGCTAATAATGGGGAGGGGAAGAG, GGGGACCACTTTGTACAAGAAAGCTGGGTCAAATGTACGTAATTATCTTTTTCTC; GW_MtFULc GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATTCATCATCATGGGAAGGG, GGGGACCACTTTGTACAAGAAAGCTGGGTATCTAGTTGGTGAGATGATGGAG. Arabidopsis Col plants were infiltrated with Agrobacterium GV3101 carrying the constructs. T1 and T2 seeds were sterilized for 5 min in 4% bleach, washed, and stratified at 4°C for 3 days prior to planting on rock wool or soil with nutrient media and grown in LD conditions. Transgenic plants were selected by spraying with Basta. Plants were genotyped by PCR to confirm the presence of the 35S:MtFULa, 35S:MtFULb, 35S:MtFULc, and the Basta transgenes. Genotyping primers for 35S:MtFULa transgenic plants were 35S-F CACTGACGTAAGGGATGACG with MtFula_TC182438-R TGGGCGTTGCCATGGGTTTGAC; for 35S:MtFULb the GW_MtFULb primers above were used; for 35S:MtFULc the primers were newqRT-F AGGGCAAGGACATTGCAGGAGCA and newqRT-R TGGTGGTAGCACCTCTGGCTGACAA, and for Basta the primers were Basta F-2 GCGTTCAAAAGTCGCCTAAG and Basta-R GAAGTCCAGCTGCCAGAAAC. Flowering time of Arabidopsis plants was measured in total number of rosette and cauline leaves at flowering.
RNA Extraction and Reverse Transcriptase-qPCR (RT-qPCR) Analysis
RNA extraction, cDNA synthesis, and RT-qPCR were performed on Arabidopsis and Medicago samples as previously described (Laurie et al., 2011; Yeoh et al., 2013). The identity of the PCR amplicons was checked by DNA sequencing. In the Medicago developmental time courses, flower buds were small unopened flowers, while flowers were open flowers. Medicago gene expression is presented as mean ± SE, where n = 3 biological replicates (unless stated otherwise). Each replicate consists of pooled material from 3 individual plants. For MtFTa1 and MtFULb expression in leaf under VSLD, the time points of 45 and 62 days are derived from 1 biological replicate. The Medicago data were normalized to the housekeeping gene, protodermal factor 2 (PDF2). The forward and reverse primers used are PDF2: GTGTTTTGCTTCCGCCGTT, CCAAATCTTGCTCCCTCATCTG; MtFTa1: GTAGCAGTAGGAATCCACTAGC, ACACTCACTCTCGGTTGATTTCC; MtFULa: GGCCCAACTTGAGCAGCAAAATGAGG, TGGGCGTTGCCATGGGTTTGAC; MtFULb: AGAGCACGCAAAACTCAAGGCT, AGCTCTTTGAGACCTAAACCATCCAA; MtFULc: AGGGCAAGGACATTGCAGGAGCA, TGGTGGTAGCACCTCTGGCTGACAA. The Arabidopsis data were normalized to the housekeeping gene At2g32170 amplified using the primers TCCTTTTTCATCGACACTGC and CCATATGTGTCCGCAAAATG. Arabidopsis gene expression is derived from one sample consisting of ∼3 cauline leaves from the same plant and presented as mean ± SE, where n = 3 PCR technical replicates.
Results
Analysis of the Sequence of Three MtFUL-Like Genes in Medicago
Three Medicago FUL-like sequences (MtFULa, MtFULb, and MtFULc) were previously identified in Medicago EST and Genomic databases (Hecht et al., 2005). However, translation of the MtFULb sequence indicated that it encoded a protein of 148 amino acids (aa) that was truncated at the C-terminus compared to Arabidopsis FUL (FUL) and the other MtFULs. To investigate the transcript sequence further, we carried out 3′RACE. This indicated that there was an error in the Genbank sequence and that the corrected sequence encoded a protein of 232 aa which is comparable in size to FUL and MtFULa (242 aa and 236 aa, respectively) and MtFULc which is slightly longer at 256 aa. FUL shares 64.4% identity with MtFULa, 64.6% identity with MtFULb and 47.1% identity with MtFULc. MtFULa and MtFULb are 69.1% identical to each other and 48.2 and 49.4% identical to MtFULc, respectively. MtFULa, MtFULb, and MtFULc are predicted to encode MADS transcription factors as they have a conserved MADS-box domain, intervening (I-box) region, and keratin-like (K-box) motif (Parenicova et al., 2003; Figure 1A). The first few amino acids in the C-terminal region are quite conserved among Arabidopsis FUL and MtFUL proteins but the remainder of the sequences are divergent except for the L/MPPWML motif near the very end of the region, which was shown to be conserved in other eudicot FUL sequences (Litt and Irish, 2003).
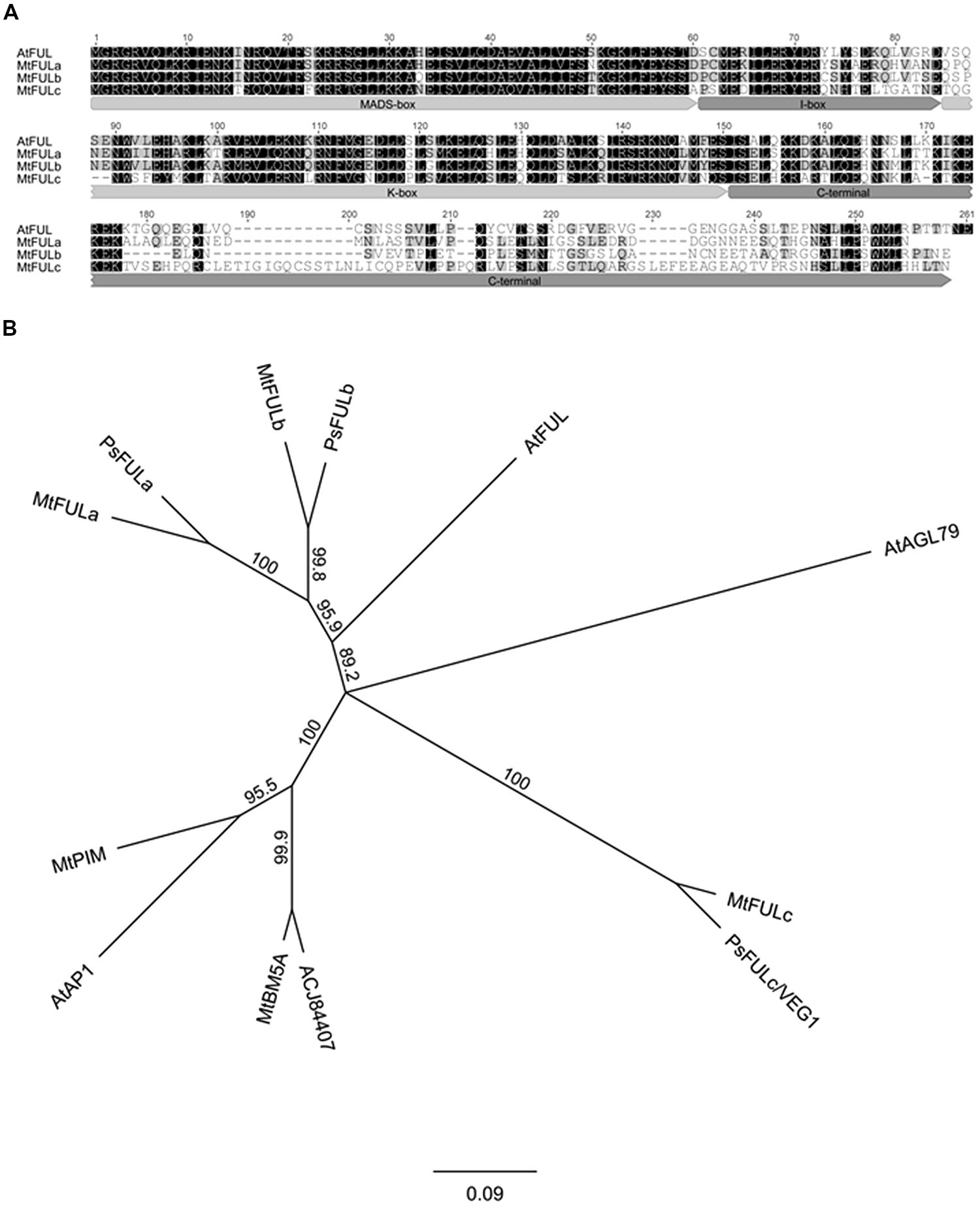
FIGURE 1. Alignment and phylogenetic analysis of FUL - like proteins from Medicago, pea, and Arabidopsis. (A) Alignment of the predicted MtFULa, MtFULb, MtFULc, and AtFUL protein sequences. Typical MIKC domains of MADS box transcription factors are marked. Amino acids with black shading are identical in all sequences, those in gray are similar residues (B) A consensus phylogenetic tree based on the full-length amino acid alignment of MtFULa, MtFULb, MtFULc, FUL - like proteins, and related MADS box transcription factors. The tree was generated using the neighbor-joining (NJ) method via bootstrap resampling with support threshold of 55%. The numbers indicate the bootstrap values based on 1000 replicates. At: Arabidopsis thaliana, Mt: Medicago truncatula, Ps: Pisum sativum.
Next, we performed reciprocal BLAST searches of Arabidopsis and Medicago protein databases with FUL and the three MtFUL-like protein sequences. The top scoring hits with Arabidopsis FUL in the Medicago databases were MtFULa, followed by MtFULb, MtPIM (AP1 ortholog; Benlloch et al., 2006), an additional MADS factor MtBM5A then MtFULc. MtFULa and MtFULb identified each other and AtFUL as top hits in BLAST searches of both databases. MtFULc identified AtFUL as top hit followed by MtBM5A, MtFULb, MtFULa, and MtPIM. This analysis overall confirmed that MtFULa and MtFULb are the most similar proteins to AtFUL in the Medicago databases.
A previous study conducted a phylogenetic analysis of genes that belong to the AP1/SQUA/FUL family, which included the MtFUL-like protein sequences (Berbel et al., 2012). Here, because we identified the full-length MtFULb sequence, in contrast to the truncated version in the GenBank database, and discovered two additional MADS-box genes (MtBM5A and ACJ84407) as a result of BLAST searches with FUL against the recently published Medicago sequence (Mt 4.0 version), we performed a simpler phylogenetic analysis incorporating these changes and including only the top scoring BLAST hits from Arabidopsis and Medicago, and some closely related pea sequences (Figure 1B). Our neighbor joining tree indicates that MtFULa and MtFULb form a sister clade to AtFUL, while MtFULc is more distantly related. All the three Medicago FULs have closely related pea sequence counterparts. MtBM5A and ACJ84407 form a sister clade to AP1 and MtPIM.
Gene Expression Patterns of MtFUL Genes in a Developmental Time Course in Wild Type Medicago in Long Day Conditions
In order to investigate the potential roles of MtFULa, MtFULb, and MtFULc genes in Medicago flowering time regulation, we compared the expression of these genes and the floral integrator gene MtFTa1 through a developmental time course in LD conditions (Figure 2). Gene expression profiles were determined by RT-qPCR on leaves, shoot apices, flower buds, and flowers. Flowering occurred at ∼69 days after planting germinated seeds in these conditions.
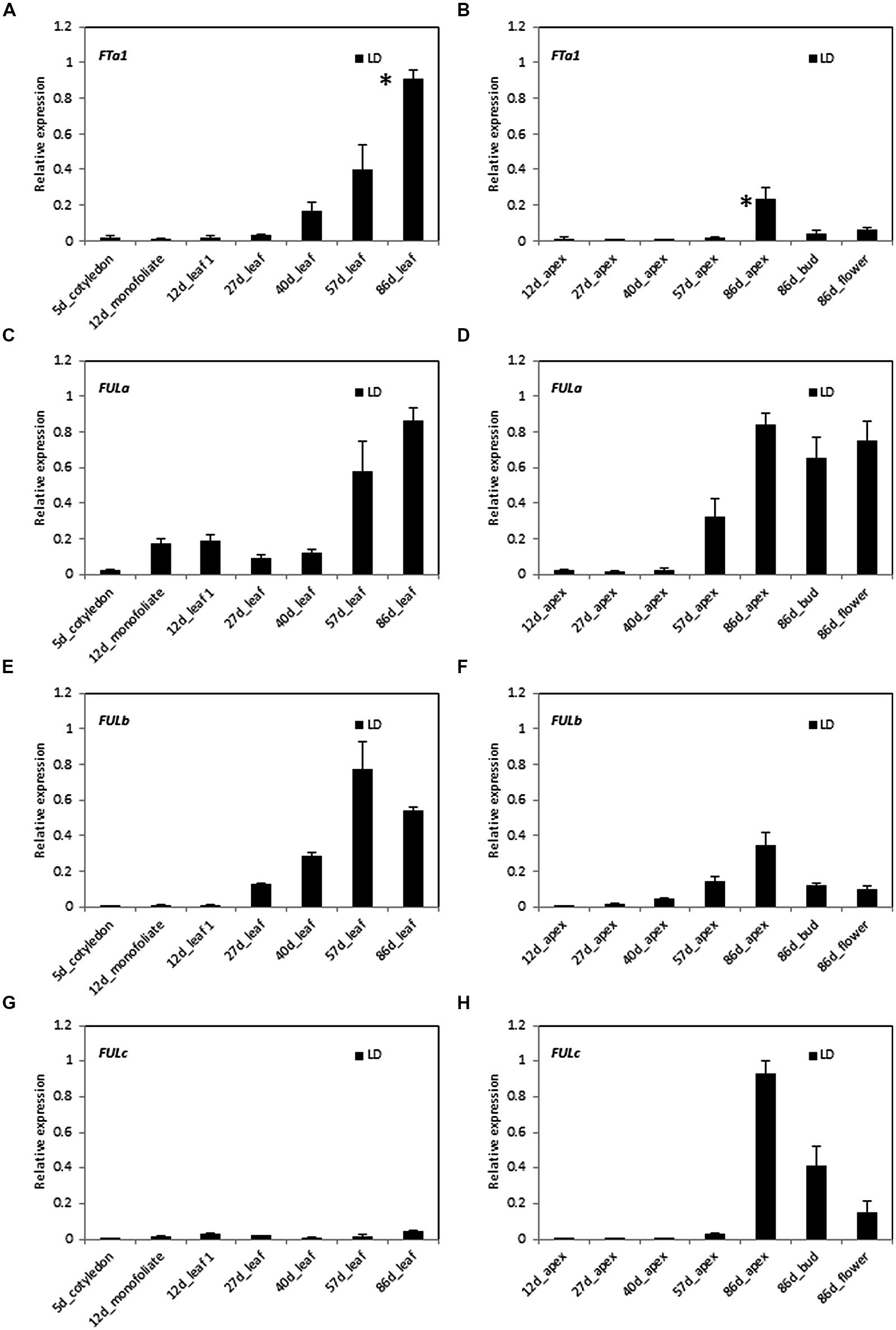
FIGURE 2. Developmental regulation of MtFTa1 and the three MtFUL genes in Medicago wild type R108 plants in long day (LD) conditions. Relative gene expression levels in cotyledons, monofoliate leaves or trifoliate leaves (A,C,E,G) and uppermost apical buds, flower buds, or open flowers (B,D,F,H) at the times shown in days after planting germinated seeds in LD. The * shown in (A,B) indicate that plants had undergone the transition to flowering with floral buds first visible at 69 days after planting of germinated seeds. Tissues were harvested at ZT2. Gene expression was determined using RT-qPCR and the data are shown as the mean ± SE of three biological replicates, which were normalized to PDF2. The data is presented relative to the highest value in all tissues for each gene.
MtFTa1 was slowly up regulated through development prior to flowering in these non-vernalized LD plants as reported previously (Figures 2A,B; Laurie et al., 2011). Its abundance began to noticeably increase in the 27 day-old trifoliate leaf and then rose at each time point thereafter with the highest level detected in the leaf of 86 day-old plants. Expression of MtFTa1 in the shoot apex was much reduced compared to the leaf, with the highest level observed in the 86 day-old apex and reduced amounts in 86 day-old flower buds and flowers.
The MtFULa expression profile had some similarities with MtFTa1 in that its transcript increased prior to flowering, particularly at 57 days with maximum levels detected in the 86 day-old leaf, apex, flower buds, and flowers (Figures 2C,D). However, it was detectable in leaves earlier than MtFTa1, including the monofoliate, which is the first true leaf. It was expressed at lower relative levels in shoot apices early on, but showed a strong increase in level at 57 days and continued to rise in the shoot apex, flower buds, and flowers of 86 day-old plants.
Of the three MtFUL genes, MtFULb had the most similar pattern of expression to MtFTa1 in LD (Figures 2E,F). Its expression strongly increased at 27 days in the leaves and continued to rise to its peak at 57 days then slightly decreased in the 86 day-old plants. In the apical samples, transcript levels increased over time with the highest level detected in the 86 day-old shoot apices, but with lower levels detected in flower buds and flowers. Like MtFTa1, MtFULb transcript levels in apices were generally lower than in leaves at the same time points.
MtFULc was expressed in a strikingly different pattern to MtFTa1, MtFULa, and MtFULb. It was expressed at very low, but detectable levels in leaves throughout development (Figures 2G,H). It was first detectable in shoot apices at 57 days, but showed a very strong elevation in expression in shoot apices of 86 day-old flowering plants and was expressed at high, but slightly reduced relative levels in flower buds and open flowers.
Overall, in summary in LD conditions, transcripts of MtFTa1, MtFULa, and MtFULb were up regulated in leaves and shoot apices prior to the transition to flowering, while MtFULc transcript strongly increased in shoot apices after flowering.
Gene Expression Patterns of MtFUL Genes in a Developmental Time Course in Wild Type Medicago in Vernalized Long Day Conditions
Vernalization accelerates flowering in LD-grown Medicago wild type R108 plants (Laurie et al., 2011; Yeoh et al., 2013). To test if the expression of MtFUL genes were affected by vernalization, germinated seeds were exposed to the cold for 2 weeks followed by planting in soil and growth in warm LD conditions (VLD, Figure 3). This treatment promoted the transition to flowering with plants flowering at ∼32 days after planting. Transcripts of all of the four genes were barely detected in the newly germinated seeds or during the cold treatment, but expression levels began to rise after few days of growth in warm LD conditions prior to flowering.
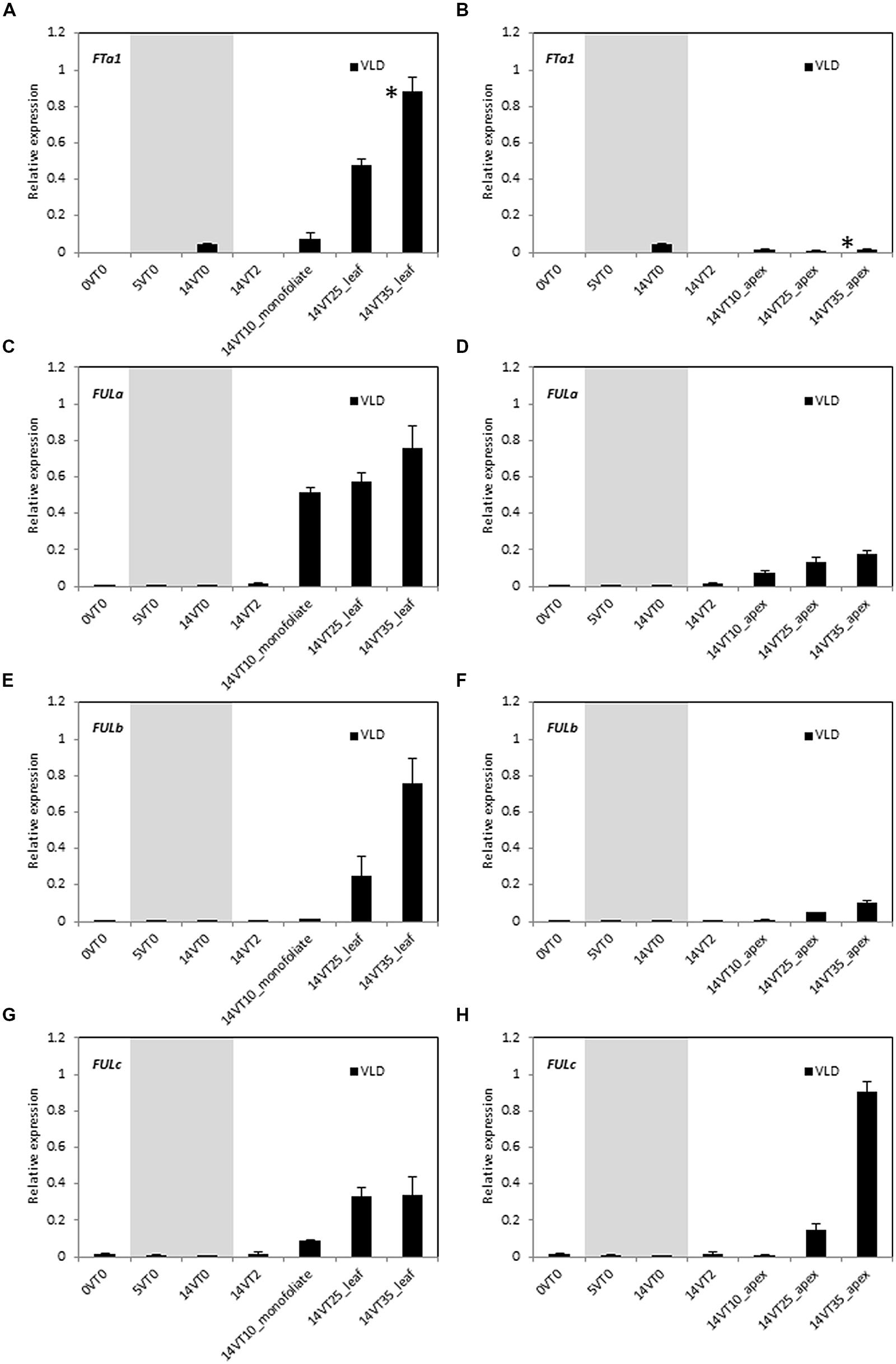
FIGURE 3. Developmental regulation of the three MtFUL genes and the floral integrator gene MtFTa1 in Medicago wild type R108 plants in LD conditions after vernalization of germinated seeds. After germination (0VT0), Medicago seeds were vernalized by exposure to 2 weeks of cold at 4°C in the dark and then planted in pots and transferred to warm (22°C) LD conditions (VLD). Relative gene expression levels in whole seedlings, monofoliate leaves or trifoliate leaves (A,C,E,G) and uppermost apical buds (B,D,F,H) before, during and after vernalization are shown (e.g., 14VT2 signifies 14 days of vernalization and 2 days in warm LD conditions). Tissues were harvested from plants of increasing developmental ages grown in LD. The * shown in (A,B) indicate when plants had undergone the transition to flowering with floral buds first visible at 32 days after planting of the vernalized seeds. Tissues were harvested at ZT2. The gray shading indicates the 14 day vernalization treatment. Gene expression was determined using RT-qPCR and the data are shown as the mean ± SE of three biological replicates, which were normalized to PDF2. The data is presented relative to the highest value in all tissues for each gene.
As expected, MtFTa1 transcript level showed a prominent increase in the 25 and 35 day-old leaves, but was barely detectable in the shoot apex (Figures 3A,B; Laurie et al., 2011). MtFULa transcript level showed a sharp rise, earlier than MtFTa1 and MtFULb, and was greatly increased even in the monofoliate leaf after 10 days in LD conditions. Expression was not as strongly elevated in the apical samples as in the leaves, but a similar profile of expression was seen in the shoot apices (Figures 3C,D). The earlier onset of MtFULa expression in VLD compared with other genes was similar to the results for LD-grown plants (Figure 2). MtFULb transcript again showed a quite similar profile to MtFTa1 showing a sharp rise in the 25 day-old leaf (Figure 3E). It was elevated in the apex at 25 and 35 days but at lower levels than in the leaf samples (Figures 3E,F). MtFULc transcript was most abundant in the 35 day-old apex after flowering, but was detected prior to the transition to flowering, in the 25 day-old apex (Figures 3G,H). The abundance in leaves relative to the apical samples increased in VLD compared to LD.
Gene Expression Patterns of MtFUL Genes in a Developmental Time Course in Wild Type and fta1 Mutant Medicago in Vernalized Seedlings in Long Day Conditions
To examine the dependence of MtFUL gene expression on MtFTa1, we compared the expression of MtFULa, MtFULb, and MtFULc in wild type and fta1 mutant plants over a developmental time course (Figure 4). Seeds of both genotypes were germinated and grown in LD until they were 11–14 days old, then the seedlings were exposed to the cold for 2 weeks in LD and shifted to warm LD conditions (VSLD). Seedling vernalization provided us with the opportunity to test the effect of cold treatment directly on gene expression in young plants, rather than germinated seeds as in the previous experiment. In these conditions, the R108 wild type plants flowered at ∼52 days after the planting of germinated seeds, while the fta1 mutants flowered much later when they were 91 days old. MtFTa1 levels showed a dramatic increase in leaves of wild type plants after the plants were transferred to warm LD conditions and prior to flowering (Figure 4A). Expression levels were very low during the cold treatment of the young seedlings. A very modest increase in expression was detected in apices after plants were grown in warm LD conditions. No MtFTa1 expression was detected in the fta1 mutant. These results are consistent with previous work (Laurie et al., 2011).
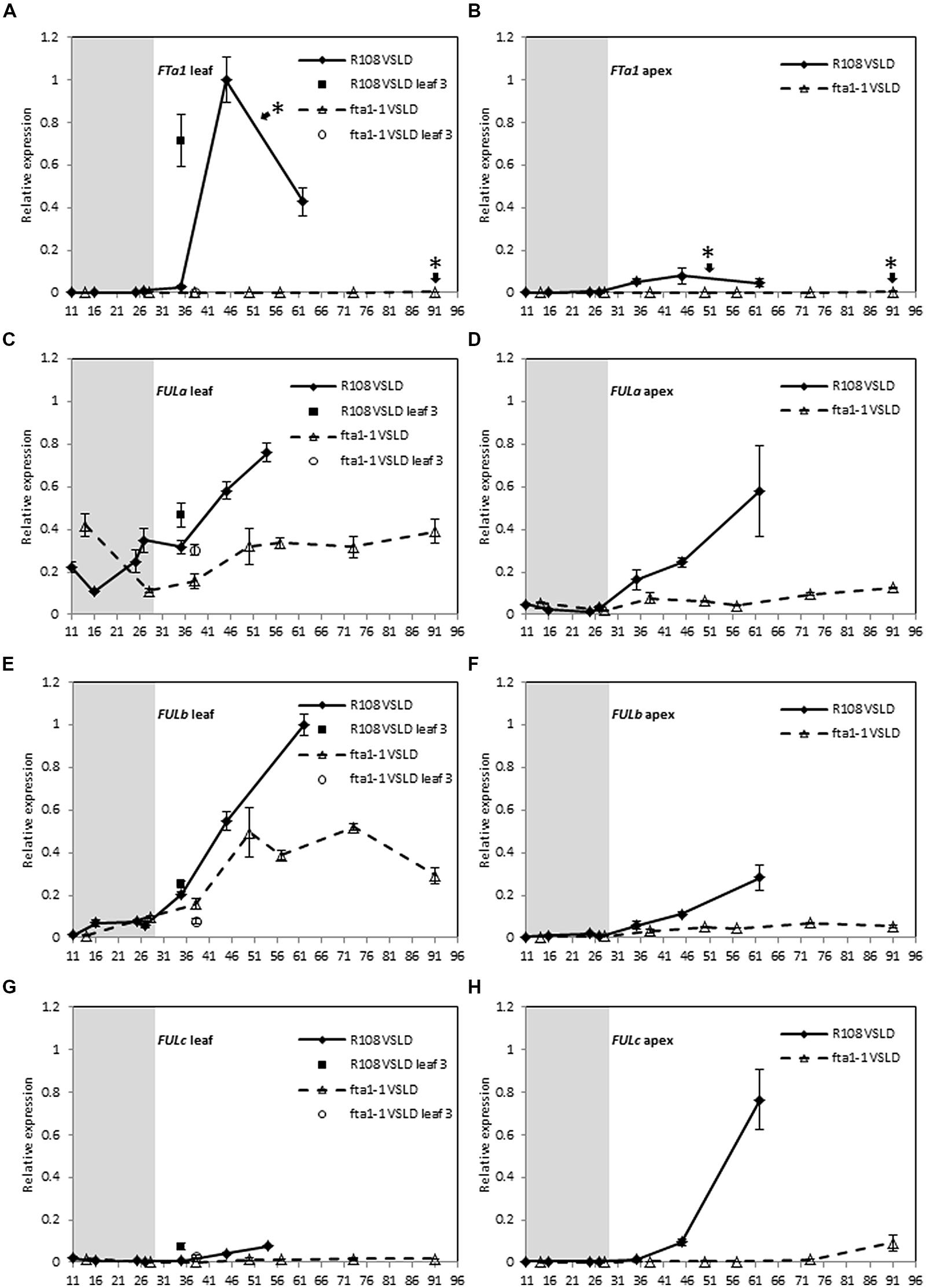
FIGURE 4. Developmental regulation of the three MtFUL genes and the floral integrator gene MtFTa1 in Medicago wild type R108 plants and the fta1 mutant line in LD conditions after vernalization of young seedlings. Seedlings were grown in LD for 11–14 days, then vernalized by exposure to cold for 14 days and then transferred to warm (22°C) LD conditions [vernalized seedlings in long days (VSLDs)]. Relative gene expression levels in trifoliate leaves (A,C,E,G) and uppermost apical buds (B,D,F,H) during the 14 day vernalization treatment (gray-shaded region) and after transfer to warm LD conditions are shown. Tissues were harvested from plants of increasing developmental ages (shown on the x-axis as days after planting of germinated seeds in the soil). The * shown in (A,B) indicate that floral buds were first visible on R108 plants at 52 days after planting of the germinated seeds while 91 days for the fta1 mutant. Tissues were harvested at ZT2. Gene expression was determined using RT-qPCR and normalized to PDF2. The data are shown for R108 as the mean of one to three biological replicates relative to PDF2 with SE of the replicates or technical errors in cases of one biological sample. For fta1 plants, gene expression is presented as the mean of three biological replicates relative to PDF2. Error bars are SE of the replicates. The data is presented relative to the highest value in all tissues for each gene.
Almost similar levels of MtFULa and MtFULb transcripts were observed in the leaves and apices of wild type and the fta1 mutant early in development and during vernalization treatment, with no direct effect of cold on their transcript levels (Figures 4C–F). However, after subsequent growth in warm LD conditions, the expression patterns of MtFULa and MtFULb began to diverge between the wild type and fta1 mutant. MtFULa and MtFULb were continuously up regulated in wild type, which correlated with the increase in MtFTa1 accumulation in the leaves, while expression in the mutant plateaued off. Although the striking difference between MtFTa1 levels in wild type and fta1 mutant was not completely mirrored in the MtFULa and MtFULb profile in leaves, it is evident from the results that MtFTa1 is needed for sustained upregulation of MtFULa and MtFULb. The maximum expression level of both genes was lower in the fta1 mutant compared to wild type in the leaves, and this difference is more pronounced in the shoot apex. In contrast, MtFULc expression levels were very dependent on functional MtFTa1 with almost no expression detected in the fta1 mutant throughout development (Figures 4G,H). A very slight rise in transcript level was seen in the apical samples of fta1 mutant at the time of flowering. There was no direct effect of cold on MtFULc expression.
Gene Expression in Wild Type and 35s:MtFTa1 Transgenic Medicago Plants in Long Day Conditions
The experiments above using the fta1 mutant indicated that all the three MtFUL genes were dependent on MtFTa1 for maximal transcript accumulation. In addition, in previous work we showed that MtFULb levels were elevated in LD plants over expressing MtFTa1 (Yeoh et al., 2013). Here, we examined the effect of 35S:MtFTa1 on MtFULa, MtFULb, and MtFULc levels in young transgenic Medicago plants grown in LD conditions (Figure 5). No floral buds were visible on the plants. MtFTa1 transcript was detected at very high levels in the leaves and apices of the transgenic line compared to wild type R108. The effect of MtFTa1 overexpression on MtFULa levels was quite modest. MtFULa was quite abundant in wild type plants in LD, but its expression level increased particularly in the shoot apex of the transgenic plants. A stronger effect was observed on the MtFULb transcript levels in the leaf of 35S:MtFTa1 plants. The effect on MtFULc transcript levels was quite strong in the leaf, but even more striking was the massive increase of MtFULc levels in the shoot apical samples of the transgenic line compared to wild type.
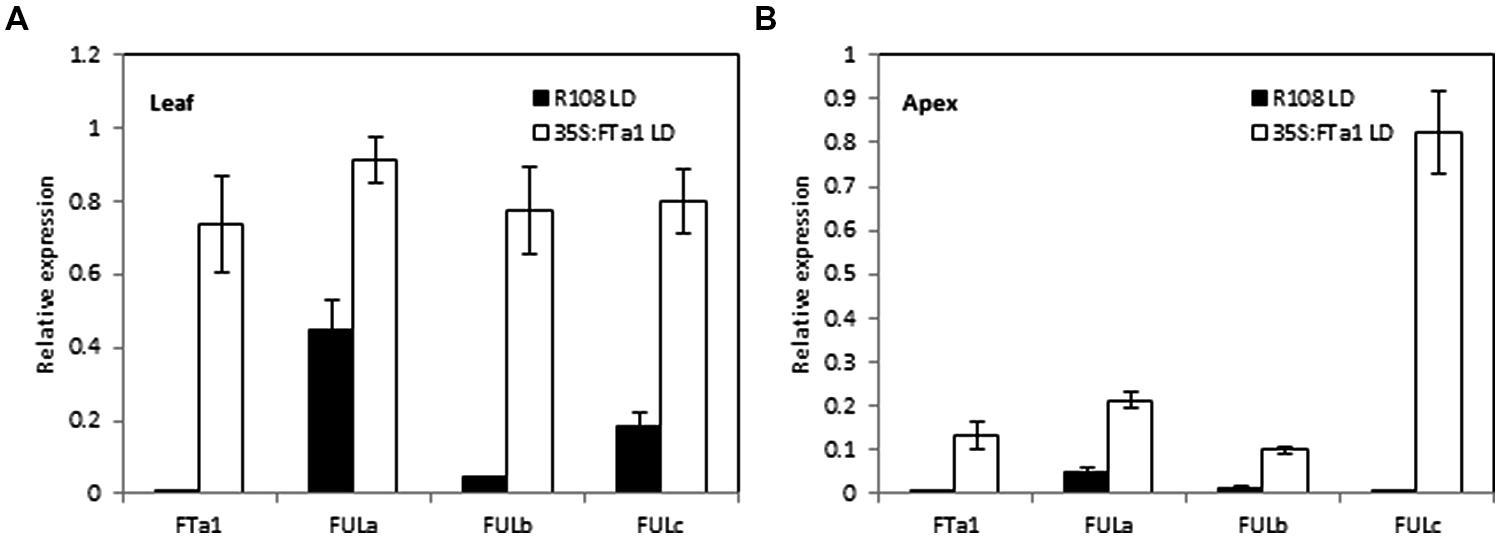
FIGURE 5. Effect of ectopic expression of the MtFTa1 gene on MtFULa, MtFULb, and MtFULc transcript levels in 35S:MtFTa1 transgenic Medicago R108 plants in LD conditions. Relative gene expression levels in trifoliate leaves (A) and uppermost apical buds (B) in 35S:MtFTa1 and wild type R108 (14 day-old) plants grown in LD. Tissues were harvested at ZT2. Gene expression was determined using RT-qPCR and the data are shown as the mean ± SE of three biological replicates, which were normalized to PDF2. The data is presented relative to the highest value in both tissues for each gene.
Effect of 35S:MtFUL Transgenes on Arabidopsis Flowering Time and Inflorescence Development
To investigate the effect of 35S:MtFUL transgenes on Arabidopsis flowering, cDNA clones of MtFULa, MtFULb, and MtFULc genes were obtained by PCR amplification with gene specific primers. The cDNA of each of the three MtFUL genes was then inserted into a GATEWAY binary vector conferring resistance to the herbicide Basta and introduced into Col wild type plants. Transgenic T1 plants (>10 for each construct) were selected by spraying with Basta. T2 seeds were grown up from 6 to 9 independent transgenic lines and Basta-resistant plants were again selected, genotyped and phenotyped (Figures 6 and 7).
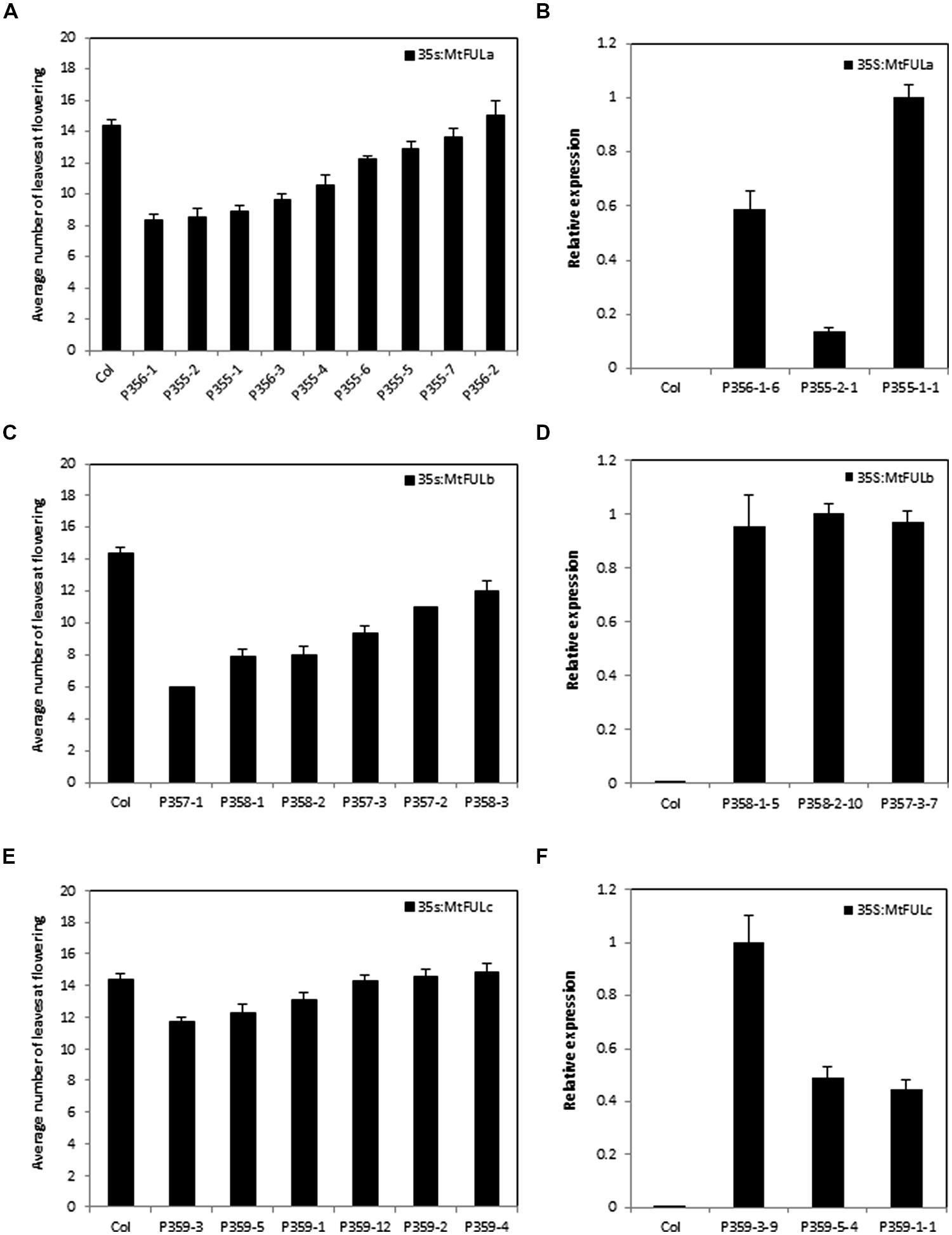
FIGURE 6. Flowering time and gene expression in T2 transgenic Arabidopsis plants with 35S:MtFUL transgenes in LD conditions. Graphs showing the flowering time observed in T2 transgenic families of 35S:MtFULa (A), 35S:MtFULb (C), and 35S:MtFULc (E) compared to wild type Columbia plants. Plants were grown in LD until they flowered. Flowering time was measured as the total number of rosette and cauline leaves at flowering. The T2 flowering time data is presented as the mean ± SE where n = 3–10 plants except for P357-1, 2 where n = 1. Gene expression of MtFULa (B), MtFULb (D), and MtFULc (F) in transgenic 35S:MtFUL Arabidopsis plants compared to wild type Columbia plants. Gene expression was determined in individual T2 plants from representative lines using RT-qPCR and normalized to At2g32170. The data are shown as the mean ± SE of three PCR technical replicates. The data is presented relative to the plant with the highest transgene expression for each over-expression construct.

FIGURE 7. Photographs of plant architecture, inflorescence, and floral phenotypes of T2 Arabidopsis plants with 35S:MtFUL transgenes in LD conditions. (A) 35S:MtFULa, 35S:MtFULb, and 35S:MtFULc plants and wild type Col at the time of the transition to flowering. (B) The terminal flower phenotype observed in some 35S:MtFULb plants compared to Col inflorescence apices. (C) The short stature and low number of siliques produced by some 35S:MtFULb plants due to the premature termination of primary inflorescence development. (D) Small flowers produced by some of the MtFUL transgenics compared to wild type Col. Scale bars are A: 5 mm, B: 1 cm, C,D: 1 mm.
Flowering time was determined for individual plants from each T2 family and for wild type Col by counting the total number of rosette and cauline leaves (Figure 6). A range of flowering times were observed in the MtFULa and MtFULb families with some lines flowering earlier than wild type Col (Figures 6A,C and 7A). The earliest 35S:MtFULa families flowered at ∼8 leaves on average compared to the wild type average of ∼14 leaves, while one 35S:MtFULb plant flowered with six leaves. The 35S:MtFULc lines flowered in a more similar manner to wild type overall, with only two lines slightly earlier than wild type (Figure 6E). RT-qPCR analysis on a selection of the transgenic plants confirmed that the transgenes were overexpressed in the 35S:MtFUL transgenic Arabidopsis plants (Figures 6B,D,F).
After bolting, a striking terminal flower phenotype became apparent in some of the 35S:MtFULb lines that was not observed in the 35S:MtFULa, 35S:MtFULc, or wild type Col plants (Figures 7B,C). Plants from independent transgenic 35S:MtFULb families had shorter primary inflorescences than wild type because of the termination of inflorescence development and production of a terminal flower (Figures 7B,C). We also observed that single flowers and floral organs of some transgenic plants were smaller than wild type (Figure 7D). To quantify this, we measured sepal length and petal width on photographs of flowers taken from two-three independent transgenic lines of 35S:MtFULa, 35S:MtFULb, and 35S:MtFULc and wild type Col. A total of 23–30 sepals and 7–10 petals were measured from each group of transgenic lines with 10 and 7 wild type sepals and petals measured, respectively. Sepals from both 35S:MtFULa (1.84 mm ± 0.08 to 0.05 SE) and 35S:MtFULb (1.6 mm ± 0.07 to 0.05 SE) transgenic plants were significantly shorter than wild type Col sepals (2.06 mm ± 0.12 to 0.05 SE). Sepals from 35S:MtFULc flowers (1.97 mm ± 0.06 to 0.05 SE) were not significantly different from Col. Petals from 35S:MtFULa (0.6 mm ± 0.06 to 0.05 SE) and 35S:MtFULb (0.54 mm ± 0.04 to 0.05 SE) plants were also narrower than wild type Col petals (0.72 mm ± 0.08 to 0.05 SE) with 35S:MtFULb sepals significantly different from wild type. Petals from 35S:MtFULc (0.72 mm ± 0.08 to 0.05 SE) were similar to wild type petals. Finally, when seeds were harvested, we observed that all six of the 35S:MtFULb lines had indehiscent (non-shattering) siliques with low numbers of small, dark-colored seeds, one of seven lines of 35S:MtFULa had siliques that were non-shattering, while all six of the 35S:MtFULc lines had siliques that shattered like wild type Col.
Discussion
In this study, the phylogenetic analysis we performed with the three Medicago FUL-like genes using predicted full-length protein sequences is consistent overall with the recent extensive phylogenetic analysis of AP1/SQUA/FUL proteins (Berbel et al., 2012). As shown in their tree, MtFULa and MtFULb form a sister clade to AtFUL, with all three proteins placed in the eudicot FUL clade (Litt and Irish, 2003). Although MtFULc identified Arabidopsis FUL as the top hit in our BLAST searches of the Arabidopsis protein database, we and Berbel et al. (2012) found that it and the orthologous pea VEG1/PsFULc claded with AGL79, a divergent Arabidopsis paralog of FUL (Pabón-Mora et al., 2012). Our BLAST searches of the Medicago protein database identified two new MADS-box proteins that clustered tightly with Arabidopsis AP1/MtPIM (Benlloch et al., 2006). All the three MtFUL genes have closely related pea homologs. The existence of multiple Medicago and pea FUL genes as opposed to one gene in Arabidopsis, appears to be a similar case to our recent identification of duplicated SVP MADS factors in Medicago (Jaudal et al., 2014).
Gene expression profiling indicated that the three MtFUL genes have potential roles in flowering time, inflorescence identity and/or flower development. In LD, transcripts of MtFULa, MtFULb, and the floral integrator MtFTa1 increased in leaves and/or shoot apices of wild type plants prior to the transition to flowering, while MtFULc transcript strongly increased in shoot apices after flowering had occurred. Expression of all of the genes was detected in floral buds and open flowers. Of the three MtFUL genes, MtFULb had the most similar pattern of expression to MtFTa1. However, it is likely that MtFULa and MtFULb genes share partially overlapping functions given their sequence conservation, similar expression profiles and promotive effects on Arabidopsis flowering time. Similar results were observed when the transition to flowering was accelerated in LD by vernalization of germinated seeds. Transcripts levels of MtFTa1 and the three MtFUL genes were very low in the newly germinated seeds and during the cold treatment, but expression levels began to rise prior to flowering and when the plants were grown in warm LD conditions. Thus none of MtFUL genes are directly regulated by cold. This is unlike the temperate grass flowering time regulator, VRN1, from the FUL-like clade in the AP1/SQUA/FUL lineage, whose transcript levels rise during vernalization (Trevaskis et al., 2007; Distelfeld et al., 2009; Berbel et al., 2012).
In Arabidopsis, FUL is most prominently expressed in the inflorescence apex at the transition to flowering and in the valves of the developing carpel (Mandel and Yanofsky, 1995a; Gu et al., 1998). Induction of flowering with LD photoperiods results in a rapid increase in FUL expression in the shoot meristem and in young leaf primordia, from a very low level in rosette leaves (Hempel et al., 1997; Gu et al., 1998; Ferrándiz et al., 2000a). Transcript is not present in early floral primordia, but appears from about stage 3 onward and is also detected in the vasculature of stems and cauline leaves, but not in roots (Gu et al., 1998). In comparison to Arabidopsis FUL, MtFULc has the most similar expression pattern with high expression in the shoot apex, flower buds, and flowers after the transition to flowering. MtFULa and MtFULb have rather a different profile from Arabidopsis FUL in some aspects as they are both expressed abundantly in leaves before the transition to flowering.
Do the MtFUL genes promote the transition to flowering in Arabidopsis? The 35S:MtFULa and 35S:MtFULb gene expression constructs accelerated flowering in wild type Arabidopsis in LD in some independent transgenic lines. The effect of 35S:MtFULa and 35S:MtFULb on Arabidopsis flowering time contrasts with our previous over expression of two MtSVP MADS-box genes that delayed flowering in Arabidopsis (Jaudal et al., 2014). Thus, these findings suggest that MtFULa and MtFULb have functional roles in controlling flowering time as displayed by AtFUL (Gu et al., 1998; Ferrándiz et al., 2000b; Melzer et al., 2008; Balanzà et al., 2014). This is further supported by the fact that sustained upregulation of MtFULa and MtFULb requires MtFTa1, a key regulator of flowering time in Medicago (Laurie et al., 2011). It is highly probable that MtFULa and MtFULb are downstream targets of MtFTa1 because their expression levels were lower in the fta1 mutant compared to wild type and both genes were upregulated in transgenic plants over expressing MtFTa1. As shown in Arabidopsis, FT directly activates the floral identity genes FUL and AP1 (Teper-Bamnolker and Samach, 2005).
Aside from early flowering, a striking additional phenotype was observed in some 35S:MtFULb transgenic Arabidopsis plants, which has resemblance with 35S:AtFUL transgenics (Ferrándiz et al., 2000b). These plants produced a terminal flower, similar to a tfl1 mutant, causing premature termination of inflorescence development. This also bears similarity to over expression of the FUL-related gene Arabidopsis AP1 which results in very early flowering and terminal flowers in transgenic Arabidopsis due to repression of TFL1 (Mandel and Yanofsky, 1995b; Ferrándiz et al., 2000a). Our results indicate that 35S:MtFULb may negatively regulate TFL1 in Arabidopsis causing a tfl1 mutant phenotype. Over expression of a chrysanthemum FUL-like gene CIM8 also accelerated flowering and led to production of terminal flowers in Arabidopsis (Wang et al., 2013). This was accompanied by increased expression of FT, AP1, and LFY, leading the authors to suggest that CIM8 positively regulates FT in Arabidopsis. It is possible that MtFULa and MtFULb regulate MtFTa1 in Medicago because the onset of their expression occurs before MtFTa1 in our time-course experiments. We also observed that siliques of the 35S:MtFULb transgenic plants exhibited an indehiscent (non-shattering) phenotype similar to 35S:AtFUL Arabidopsis siliques due to repression of SHP1/2 genes (Ferrándiz et al., 2000b; Ferrándiz and Fourquin, 2014).
In contrast to MtFULa and MtFULb, overexpression of MtFULc had only very mild effects in promoting flowering time of transgenic Arabidopsis. Thus, it is possible that MtFULc is not involved in regulating floral induction, but in other aspects of plant development, although the Arabidopsis transgenic plants resembled wild type plants in aerial architecture and development. However, this is in fact shown in pea VEG1/PsFULc, which is not involved in flowering time control in pea, but is expressed predominantly during and after the floral transition in the shoot apex where it has an important function in specifying secondary inflorescence meristem identity necessary for compound inflorescence development, rather than the simple raceme of Arabidopsis (Berbel et al., 2012). VEG1 acts downstream of GIGAS (the pea FTa1 gene) because VEG1 was not expressed in gigas mutants (Berbel et al., 2012). This might also be the case for MtFULc because its expression is highly dependent on functional MtFTa1 with almost no expression detected in the fta1 mutant throughout development and overexpression of MtFTa1 also led to upregulation of MtFULc.
The implications of our work overall are that the MtFUL genes are all regulated by MtFTa1 and that MtFULa and MtFULb are likely to play a role in promoting the transition to flowering in Medicago. In addition, we observed previously that MtFULb transcript accumulation in young Medicago plants was increased in leaves by inductive LD photoperiods compared to short days, consistent with a role in flowering time control (Jaudal et al., 2013). On the other hand, MtFULc may have a similar role to pea VEG1/PsMtFULc in compound inflorescence development in Medicago. The overlapping and distinct MtFUL expression patterns in vegetative and reproductive development suggest that they have other roles in Medicago development. The next steps would be to determine the functional roles of the MtFUL genes in Medicago. In particular, it will be interesting to know if the MtFUL genes regulate MtFTa1 and whether they interact with MtSOC1 genes to regulate flowering time and perennial traits.
Author Contributions
JP and MJ: conceived and designed the experiments. MJ, LZ, and CC: performed the experiments. JP, MJ, LZ, and CC: analyzed the data. JP and MJ: wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Wendy Payne, Angel Peng, and Ben Lentzen for technical help. The research was funded by the New Zealand Foundation for Research Science and Technology (www.msi.govt.nz/) contract numbers C10X0816 MeriNET and the New Zealand Marsden Fund (www.royalsociety.org.nz/programmes/funds/marsden/) contract 10-UOA-200.
Footnotes
References
Andres, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Balanzà, V., Martínez-Fernández, I., and Ferrándiz, C. (2014). Sequential action of FRUITFULL as a modulator of the activity of the floral regulators SVP and SOC1. J. Exp. Bot. 65, 1193–1203. doi: 10.1093/jxb/ert482
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Benlloch, R., d’Erfurth, I., Ferrandiz, C., Cosson, V., Beltran, J. P., Canas, L. A.,et al. (2006). Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 142, 972–983. doi: 10.1104/pp.106.083543
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berbel, A., Ferrandiz, C., Hecht, V., Dalmais, M., Lund, O. S., Sussmilch, F. C.,et al. (2012). VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat. Commun. 3:797. doi: 10.1038/ncomms1801
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clarkson, N. M., and Russell, J. S. (1975). Flowering responses to vernalization and photoperiod in annual medics (Medicago spp). Austr. J. Agric. Res. 26, 831–838. doi: 10.1071/AR9750831
Distelfeld, A., Li, C., and Dubcovsky, J. (2009). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12, 178–184. doi: 10.1016/j.pbi.2008.12.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ferrándiz, C., and Fourquin, C. (2014). Role of the FUL-SHP network in the evolution of fruit morphology and function. J. Exp. Bot. 65, 4505–4513. doi: 10.1093/jxb/ert479
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ferrándiz, C., Gu, Q., Martienssen, R., and Yanofsky, M. F. (2000a). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734.
Ferrándiz, C., Liljegren, S. J., and Yanofsky, M. F. (2000b). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289, 436–438. doi: 10.1126/science.289.5478.436
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fornara, F., de Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550.e1–550.e2. doi: 10.1016/j.cell.2010.04.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fourquin, C., del Cerro, C., Victoria, F. C., Vialette-Guiraud, A., de Oliveira, A. C., and Ferrándiz, C. (2013). A change in SHATTERPROOF protein lies at the origin of a fruit morphological novelty and a new strategy for seed dispersal in Medicago genus. Plant Physiol. 162, 907–917. doi: 10.1104/pp.113.217570
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gu, Q., Ferrandiz, C., Yanofsky, M. F., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517.
Hecht, V., Foucher, F., Ferrandiz, C., Macknight, R., Navarro, C., Morin, J.,et al. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137, 1420–1434. doi: 10.1104/pp.104.057018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hempel, F. D., Weigel, D., Mandel, M. A., Ditta, G., Zambryski, P. C., Feldman, L. J.,et al. (1997). Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124, 3845–3853.
Jaudal, M., Monash, J., Zhang, L., Wen, J., Mysore, K. S., Macknight, R.,et al. (2014). Overexpression of Medicago SVP genes causes floral defects and delayed flowering in Arabidopsis but only affects floral development in Medicago. J. Exp. Bot. 65, 429–442. doi: 10.1093/jxb/ert384
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jaudal, M., Yeoh, C. C., Zhang, L., Stockum, C., Mysore, K. S., Ratet, P.,et al. (2013). Retroelement insertions at the Medicago FTa1 locus in spring mutants eliminate vernalization but not long-day requirements for early flowering. Plant J. 76, 580–591. doi: 10.1111/tpj.12315
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jung, C., and Muller, A. E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. doi: 10.1016/j.tplants.2009.07.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Kim, D. H., Doyle, M. R., Sung, S., and Amasino, R. M. (2009). Vernalization: winter and the timing of flowering in plants. Ann. Rev. Cell Dev. Biol. 25, 277–299. doi: 10.1146/annurev.cellbio.042308.113411
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J. J., Lee, J. H., Kim, W., Jung, H. S., Huijser, P., and Ahn, J. H. (2012). The microrNA156-SQUAMOSA promoter binding protein-like3 module regulates ambient temperature-responsive flowering via flowering locus in Arabidopsis. Plant Physiol. 159, 461–478. doi: 10.1104/pp.111.192369
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Laurie, R. E., Diwadkar, P., Jaudal, M., Zhang, L., Hecht, V., Wen, J.,et al. (2011). The Medicago truncatula flowering locus T homologue, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156, 2207–2224. doi: 10.1104/pp.111.180182
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Litt, A., and Irish, V. F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165, 821–833.
Mandel, M. A., and Yanofsky, M. F. (1995a). The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell Online 7, 1763–1771. doi: 10.1105/tpc.7.11.1763
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mandel, M. A., and Yanofsky, M. F. (1995b). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. doi: 10.1038/377522a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Melzer, S., Lens, F., Gennen, J., Vanneste, S., Rohde, A., and Beeckman, T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40, 1489–1492. doi: 10.1038/ng.253
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pabón-Mora, N., Ambrose, B. A., and Litt, A. (2012). Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 158, 1685–1704. doi: 10.1104/pp.111.192104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parenicova, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J.,et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Posé, D., Yant, L., and Schmid, M. (2012). The end of innocence: flowering networks explode in complexity. Curr. Opin. Plant Biol. 15, 45–50. doi: 10.1016/j.pbi.2011.09.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Putterill, J., Zhang, L., Yeoh, C., Balcerowicz, M., Jaudal, M., and Varkonyi Gasic, E. (2013). FT genes and regulation of flowering in the legume Medicago truncatula. Funct. Plant Biol. 40, 1199–1207. doi: 10.1071/FP13087
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tadege, M., Wang, T. L., Wen, J. Q., Ratet, P., and Mysore, K. S. (2009). Mutagenesis and beyond! tools for understanding legume biology. Plant Physiol. 151, 978–984. doi: 10.1104/pp.109.144097
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Teper-Bamnolker, P., and Samach, A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17, 2661–2675. doi: 10.1105/tpc.105.035766
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Torti, S., Fornara, F., Vincent, C., Andrés, F., Nordström, K., Göbel, U.,et al. (2012). Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24, 444–462. doi: 10.1105/tpc.111.092791
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Trevaskis, B., Hemming, M. N., Dennis, E. S., and Peacock, W. J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12, 352–357. doi: 10.1016/j.tplants.2007.06.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J. W., Czech, B., and Weigel, D. (2009). miR156-Regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, Y., Ma, Y., Fu, J., Qi, S., Ma, H., and Dai, S. (2013). Isolation and functional analysis of the ClM8-FRUITFULL-like MADS-box gene from Chrysanthemum lavandulifolium. Sci. Hortic. 161, 125–133. doi: 10.1016/j.scienta.2013.05.045
Wong, A. C. S., Hecht, V. F. G., Picard, K., Diwadkar, P., Laurie, R. E., Wen, J.,et al. (2014). Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 5:486. doi: 10.3389/fpls.2014.00486
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yeoh, C. C., Balcerowicz, M., Laurie, R., Macknight, R., and Putterill, J. (2011). Developing a method for customized induction of flowering. BMC Biotechnol. 11:36. doi: 10.1186/1472-6750-11-36
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yeoh, C. C., Balcerowicz, M., Zhang, L., Jaudal, M., Brocard, L., Ratet, P.,et al. (2013). Fine mapping links the FTa1 flowering time regulator to the dominant spring1 locus in Medicago. PLoS ONE 8:e53467. doi: 10.1371/journal.pone.0053467
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: FRUITFULL, MtFUL, MtFTa1, FT, Medicago, Arabidopsis, flowering time, terminal flower
Citation: Jaudal M, Zhang L, Che C and Putterill J (2015) Three Medicago MtFUL genes have distinct and overlapping expression patterns during vegetative and reproductive development and 35S:MtFULb accelerates flowering and causes a terminal flower phenotype in Arabidopsis. Front. Genet. 6:50. doi: 10.3389/fgene.2015.00050
Received: 08 December 2014; Accepted: 02 February 2015;
Published online: 19 February 2015.
Edited by:
Marta Santalla, Consejo Superior de Investigaciones Cientificas – Mision Biologica de Galicia, SpainReviewed by:
Cristina Ferrandiz, Consejo Superior de Investigaciones Científicas – Instituto de Biologia Molecular y Celular de Plantas, SpainStefan De Folter, CINVESTAV-IPN, Mexico
Copyright © 2015 Jaudal, Zhang, Che and Putterill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanna Putterill, The Flowering Lab, School of Biological Sciences, University of Auckland, Private Bag 92019, Auckland Mail Centre, Auckland 1142, New Zealand e-mail: j.putterill@auckland.ac.nz
 Mauren Jaudal
Mauren Jaudal Lulu Zhang
Lulu Zhang Chong Che
Chong Che Joanna Putterill
Joanna Putterill