What are Moonlighting Proteins?
Moonlighting proteins comprise a class of multifunctional proteins in which a single protein performs multiple physiologically relevant biochemical or biophysical functions that are not due to gene fusions, multiple RNA splice variants, or pleiotropic effects (Jeffery, 1999). Classic examples include soluble enzymes that also bind to DNA or RNA to regulate translation or transcription (Figure 1A) (reviewed in Commichau and Stulke, 2008) or have a second function as structural proteins in the lens of the eye (crystallins) (Figure 1B) (Wistow and Piatigorsky, 1987; Piatigorsky and Wistow, 1989). Other typical examples are cytosolic enzymes that moonlight as cytokines, chaperones, cytoskeletal components, DNA compactors, adhesins or scaffolds, as well as a chloride transporter that regulates the function of another transmembrane channel, ribosomal proteins that double as translation factors, and a DNA binding protein that becomes a component of the extracellular matrix (for reviews see Nobeli et al., 1996; Jeffery, 2003a,b, 2009, submitted; Piatigorsky, 2007; Gancedo and Flores, 2008; Huberts and van der Klei, 2010; Henderson and Martin, 2011, 2013; Guo and Schimmel, 2013). Moonlighting proteins are found in mammals, yeast, worms, bacteria, plants, viruses, archea and many other organisms. The online MoonProt Database, which includes information about those moonlighting proteins for which biochemical or biophysical evidence supports the presence of at least two biochemical functions in one polypeptide chain, includes over 270 proteins (Mani et al., 2015). It is likely that many other proteins also have additional functions that have not yet been found. In this opinion piece, I argue that there are currently many reasons for studying moonlighting proteins.
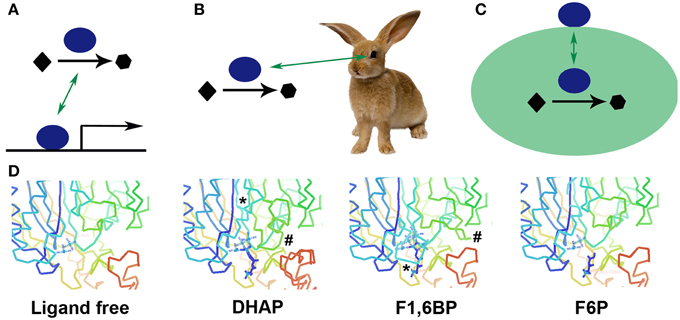
Figure 1. Cartoons illustrating how a protein can moonlight. A protein can catalyze an enzymatic reaction and also (A) bind to DNA and serve as a transcription factor, (B) have a noncatalytic role in the lens of the eye (crystallins), or (C) be placed on the surface of a cell to function as a receptor. Another type of moonlighting is illustrated in (D) where an active site undergoes significant remodeling, or movement of loops (marked by * and #) to bring in different catalytic amino acids to perform different two different catalytic functions. (D) made Using Coot (Emsley et al., 2010) and PDB IDs 3T2B, 3T2C, 3T2D, and 3T2E.
Many Moonlighting Proteins are Involved in Disease
A growing number of moonlighting proteins have been found to play key roles in disease, but there is space to mention only a few examples here. Phosphoglucose isomerase, an enzyme in glycolysis, is also a cytokine (autocrine motility factor) that plays a role in breast cancer metastasis (Watanabe et al., 1991). Alcohol acetaldehyde dehydrogenase/Listeria adhesion protein (LAP) enables Listeria monocytogenes to bind to intestinal epithelial cells and aids in infection (Jagadeesan et al., 2010). Enolase (Knaust et al., 2007; Agarwal et al., 2008; Castaldo et al., 2009), phosphoglycerate kinase (Boone et al., 2011; Fulde et al., 2013), and glyceraldehyde 3-phosphate dehydrogenase (Pancholi and Fischetti, 1992; Seifert et al., 2003; Bergmann et al., 2004; Jobin et al., 2004; Barbosa et al., 2006; Hurmalainen et al., 2007; Matta et al., 2010) are cytosolic enzymes that have a second role in pathogenic bacteria as a cell surface receptor for collagen, fibronectin, or plasminogen (Figure 1C). Adhesion of the pathogen to the host by binding to collagen and fibronectin, components of the host extracellular matrix, aid in colonization of the host. Binding to plasminogen enables its conversion to the active protease plasmin, which is used to degrade host proteins and aid in tissue invasion. In general, these cytoplasmic/cell surface moonlighting proteins can be important in infection, virulence, or immune responses, and some can be potential vaccination targets. Two papers in this Research Topic discuss additional examples of moonlighting proteins involved in disease and how they can be targets for the development of novel therapeutics (Rasch et al., 2015; Henderson and Kaiser, in press).
Identifying Novel Biochemical Pathways
The growing number of intracellular enzymes or chaperones that are being found to moonlight on the cell surface raises several questions. How do intracellular/cell surface proteins get secreted? How do they become attached to the cell surface? In addition to the well-known Sec pathway, there are several non-canonical secretion pathways, but a secretion method in which a large portion of each protein remains inside the cell while some of it is partitioned to the cell surface has not been identified. This may involve a novel variation of a known secretion pathway or an as yet unknown secretion pathway. In addition, these moonlighting proteins do not contain known signals for attaching to the cell surface. Again, this could involve a new version of a known mechanism or an as yet unknown mechanism for cell surface attachment. With the increasing problem of antibiotic resistance, finding a method to inhibit the targeting of these proteins to the pathogen surface might lead to an alternative method to decrease the ability of bacteria to bind to and degrade host tissues. Understanding more about these cellular processes, whether a new variation of a known process or a completely new process, could provide new target(s) for developing therapeutics to treat infections. Similarly, the study of other moonlighting proteins could lead to the identification of additional previously unknown cellular processes.
Importance for Systems Biology
Some moonlighting proteins serve as a connection between multiple biochemical pathways or a switch between pathways, and help the cell to respond to changes in its environment. Many biosynthetic enzymes moonlight as regulators of transcription or translation and serve as a feedback mechanism to regulate the amount of enzyme synthesized in a biochemical pathway in response to changes in the cellular concentration of a product of the pathway. For example, thymidylate synthase is also an RNA binding protein (Chu et al., 1991), and E. coli putA is also a DNA binding transcriptional repressor (Wood, 1981; Ostrovsky de Spicer et al., 1991; Ostrovsky de Spicer and Maloy, 1993). Other moonlighting proteins provide a mechanism to switch between biochemical pathways in response to changing cellular conditions. Aconitase/IRE binding protein helps the cell respond to changes in cellular iron concentration (Kennedy et al., 1992; Philpott et al., 1994; Chen et al., 2005; Banerjee et al., 2007). Studies of the structures and functions of moonlighting proteins like these can help elucidate how proteins switch activities in response to changes within or surrounding the cell, whether it's due to changes in pH, substrate availability, cellular concentrations of metal ions, or other factors.
Novel Mechanisms of Protein Function
Studies of the structures and functions of some moonlighting proteins have already added to our knowledge of the diverse mechanisms by which a protein can perform multiple functions and how protein structure can change in response to changes in its environment (examples in Jeffery, 2004a,b). The results of two recent studies illustrate the sometimes drastic conformational changes that proteins can undergo and add to our understanding of protein structure and function in general.
Two thermophilic fructose-1,6-bisphosphate aldolase/phosphatase enzymes use a single active site pocket to catalyze two reactions in gluconeogenesis, an aldol condensation of dihydroxyacetone phosphate and glyceraldehyde 3-phosphate and dephosphorylation of fructose 1,6-bisphosphate (Du et al., 2011; Fushinobu et al., 2011). Three loops move into and out of the active site bringing in a different set of catalytic amino acids, and altering the binding of several active site metal ions, in order to bind the second substrate and perform the second catalysis (Figure 1D).
RfaH is a transcription factor that interacts with RNA polymerase (RNAP) to reduce pausing and increase processivity, and it is also a translation factor. In order to perform the second task, its C-terminal domain (CTD) undergoes complete refolding (Schweimer et al., 2012). In an all-alpha conformation, the CTD masks the RNAP binding surface on the N-terminal domain (NTD) until the NTD binds specific operons in DNA. Then the CTD is released from the NTD and surface of the NTD can bind to RNAP. The CTD then refolds into an all-beta conformation and recruits ribosomes by binding to the ribosomal S10 protein, to potentiate translation of RfaH-controlled operons, which contain weak ribosome binding sites.
In addition, an increasing number of intrinsically disordered proteins (IDPs) and proteins with intrinsically disordered domains have been found to be moonlighting proteins. The functions of intrinsically disordered proteins often involve interacting with another protein, and the disordered binding region undergoes induced folding upon binding to the protein partner. Some intrinsically ordered domains have different functions by using alternate conformations to interact with multiple protein partners (Tompa et al., 2005). In some cases, these different binding modes and partners result in opposite effects on a pathway, for example by activating one binding partner but inhibiting another.
Predicting Protein Functions
Hundreds of thousands of protein sequences are available due largely to the efforts of the genome projects. During annotation of sequence databases, the functions of most new proteins are predicted through amino acid sequence homology to proteins of known function. The ability of proteins to moonlight complicates this procedure.
First, if a protein is homologous to a moonlighting protein, the protein might have both, one, or none of the functions of the moonlighting protein. For example, two versions of aconitase are found in mammalian cells, in the cytoplasm and the mitochondria. Both aconitases catalyze the transformation of citrate to isocitrate in the citric acid cycle, but only the cytoplasmic protein is also an mRNA binding protein. The mitochondrial protein has a different second function in mitochondrial DNA maintenance (Kennedy et al., 1992; Philpott et al., 1994; Chen et al., 2005; Banerjee et al., 2007). Another moonlighting protein, delta 2 crystallin in the duck eye lens is the same protein as the ubiquitous urea cycle enzyme arginosuccinate lyase. The duck delta 1 crystallin, which has 89% amino acid sequence identity to the delta 2 crystallin, does not have catalytic activity (Piatigorsky et al., 1988; Barbosa et al., 1991; Chiou et al., 1991; Graham et al., 1996; Piatigorsky and Horwitz, 1996). Therefore, sequence homologs of moonlighting proteins might not be moonlighting proteins, so sequence homology alone might not be sufficient for correct prediction of protein function.
The second complication is that it is currently unclear how to identify all the functions of a moonlighting protein from its sequence or structure. Many function prediction algorithms aim to identify a single function for a protein. Three papers in this Research Topic and another recent report have used lists of known moonlighting proteins to test current programs and develop novel ways of predicting multiple functions (Khan et al., 2014; Khan and Kihara, 2014; Chapple et al., 2015; Hernández et al., 2015; Irving et al., in press). Our MoonProt Database can provide a test set for the further development of protein function prediction programs (Mani et al., 2015).
Additional knowledge about which proteins have multiple functions and roles in multiple biochemical pathways, multiprotein complexes or signaling pathways could also help in the prediction of protein function from comparative expression studies, protein-protein interaction analysis, gene knockout experiments, and other proteomics projects (Jeffery, 2014).
Evolution of Protein Function and Design of Proteins with Novel Functions
How a second functional site evolved on a protein structure, as well as how regulation of protein expression, activity, and switching between functions evolved are interesting questions. In some cases, proteins were adopted for a second use without much change in physical features, but in other cases the evolution of new binding sites, new conformational changes, or other features were required. The paper by the DeLuna group in this Research Topic describes gene duplication in the evolution of moonlighting proteins (Espinosa-Cantu et al., in press).
Information about how moonlighting proteins evolved and information about their structures and functions can also be used to aid the design of proteins with new biochemical functions because they serve as examples of how to start with a stable protein fold as a scaffold and add a new functional site. They also provide examples of combining two functional sites in one polypeptide chain, which might be useful in the design of multifunctional protein therapeutics (or proteins used in manufacturing), to deliver one polypeptide drug that can perform multiple functions with those functional sites present at a selected stoichiometry.
Summary
The examples above illustrate just some of the ways in which the continuing study of moonlighting proteins is important: developing novel therapeutics, identifying previously unknown cellular processes, elucidating the connections among biochemical pathways, understanding novel protein mechanisms, improving the prediction of protein functions, and providing information about the evolution of protein structure and function as well as examples for the design of new proteins. The references listed above and especially the collection of papers in this Research Topic are recommended for providing more in-depth examples and analysis of these topics.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agarwal, S., Kulshreshtha, P., Bambah Mukku, D., and Bhatnagar, R. (2008). Alpha-enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim. Biophys. Acta 1784, 986–994. doi: 10.1016/j.bbapap.2008.03.017
Banerjee, S., Nandyala, A. K., Raviprasad, P., Ahmed, N., and Hasnain, S. E. (2007). Iron-dependent RNA-binding activity of Mycobacterium tuberculosis aconitase. J. Bacteriol. 189, 4046–4052. doi: 10.1128/JB.00026-07
Barbosa, M. S., Bao, S. N., Andreotti, P. F., de Faria, F. P., Felipe, M. S., dos Santos Feitosa, L., et al. (2006). Glyceraldehyde 3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein, involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect. Immun. 74, 382–389. doi: 10.1128/IAI.74.1.382-389.2006
Barbosa, P., Wistow, G. J., Cialkowski, M., Piatigorsky, J., and O'Brien, W. E. (1991). Expression of duck lens delta-crystallin cDNAs in yeast and bacterial hosts. Delta 2-crystallin is an active argininosuccinate lyase. J. Biol. Chem. 266, 22319–22322.
Bergmann, S., Rohde, M., and Hammerschmidt, S. (2004). Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 72, 2416–2419. doi: 10.1128/IAI.72.4.2416-2419.2004
Boone, T. J., Burnham, C. A., and Tyrrell, G. J. (2011). Binding of group B streptococcal phosphoglycerate kinase to plasminogen and actin. Microb. Pathog. 51, 255–261. doi: 10.1016/j.micpath.2011.06.005
Castaldo, C., Vastano, V., Siciliano, R. A., Candela, M., Vici, M., Muscariello, L., et al. (2009). Surface displaced alfa-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb. Cell Fact. 8, 14–38. doi: 10.1186/1475-2859-8-14
Chapple, C. E., Herrmann, C., and Brun, C. (2015). PrOnto database: GO term functional dissimilarity inferred from biological data. Front. Genet. 6:200. doi: 10.3389/fgene.2015.00200
Chen, X. J., Wang, X., Kaufman, B. A., and Butow, R. A. (2005). Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Nature 307, 714–717. doi: 10.1126/science.1106391
Chiou, S. H., Lo, C. H., Chang, C. Y., Itoh, T., Kaji, H., and Samejima, T. (1991). Ostrich crystallins. Structural characterization of delta-crystallin with enzymic activity. Biochem. J. 273, 295–300.
Chu, E., Koeller, D. M., Casey, J. L., Drake, J. C., Chabner, B. A., Elwood, P. C., et al. (1991). Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 88, 8977–8981. doi: 10.1073/pnas.88.20.8977
Commichau, F. M., and Stulke, J. (2008). Trigger enzymes: bifunctional proteins active in metabolism and in controlling gene expression. Mol. Microbiol. 67, 692–702. doi: 10.1111/j.1365-2958.2007.06071.x
Du, J., Say, R. F., Lü, W., Fuchs, G., and Einsle, O. (2011). Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase. Nature 478, 534–537. doi: 10.1038/nature10458
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. doi: 10.1107/S0907444910007493
Espinosa-Cantu, A., Ascencio, D., Barona-Gomez, F., and DeLuna, A. (in press). Gene duplication and the evolution of moonlighting proteins. Front. Bioeng. Biotech. Res. Top.
Fulde, M., Bernardo-García, N., Rohde, M., Nachtigall, N., Frank, R., Preissner, K. T., et al. (2013). Pneumococcal phosphoglycerate kinase interacts with plasminogen and its tissue activator. Thromb. Haemost. 111, 401–416. doi: 10.1160/TH13-05-0421
Fushinobu, S., Nishimasu, H., Hattori, D., Song, H. J., and Wakagi, T. (2011). Structural basis for the bifunctionality of fructose-1,6-bisphosphate aldolase/phosphatase. Nature 478, 538–541. doi: 10.1038/nature10457
Gancedo, C., and Flores, C. L. (2008). Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 72, 197–210. doi: 10.1128/MMBR.00036-07
Graham, C., Hodin, J., and Wistow, G. (1996). A retinaldehyde dehydrogenase as a structural protein in a mammalian eye lens. Gene recruitment of eta-crystallin. J. Biol. Chem. 271, 15623–15628. doi: 10.1074/jbc.271.26.15623
Guo, M., and Schimmel, P. (2013). Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 9, 145–153. doi: 10.1038/nchembio.1158
Henderson, B., and Kaiser, F. (in press). Moonlighting bacterial virulence factors are novel therapeutic targets for bacterial infections. Front. Bioeng. Biotech. Res. Top.
Henderson, B., and Martin, A. (2011). Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79, 3476–3491. doi: 10.1128/IAI.00179-11
Henderson, B., and Martin, A. (2013). Bacterial moonlighting proteins and bacterial virulence. Curr. Top. Microbiol. Immunol. 358, 155–213. doi: 10.1007/82_2011_188
Hernández, S., Franco, L., Calvo, A., Ferragut, G., Hermoso, A., Amela, I., et al. (2015). Bioinformatics and moonlighting proteins. Front. Bioeng. Biotechnol. 3:90. doi: 10.3389/fbioe.2015.00090
Huberts, D. H., and van der Klei, I. J. (2010). Moonlighting proteins: an intriguing mode of multitasking. Biochim. Biophys. Acta 1803, 520–525. doi: 10.1016/j.bbamcr.2010.01.022
Hurmalainen, V., Edelman, S., Antikainen, J., Baumann, M., Lähteenmäki, K., and Korhonen, T. K. (2007). Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology 153, 1112–1122. doi: 10.1099/mic.0.2006/000901-0
Irving, H. R., Wong, A., and Gehring, C. (in press). Using search motifs to identify hidden moonlighting centers. Front. Bioeng. Biotech Res. Top.
Jagadeesan, B., Koo, O. K., Kim, K. P., Burkholder, K. M., Mishra, K. K., Aroonnual, A., et al. (2010). LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria, promotes bacterial adhesion to enterocyte-like Caco-2 cells only in pathogenic species. Microbiology 156, 2782–2795. doi: 10.1099/mic.0.036509-0
Jeffery, C. J. (1999). Moonlighting proteins. Trends Biochem. Sci. 24, 8–11. doi: 10.1016/S0968-0004(98)01335-8
Jeffery, C. J. (2003a). Moonlighting proteins: old proteins learning new tricks. Trends. Genet. 19, 415–417. doi: 10.1016/S0168-9525(03)00167-7
Jeffery, C. J. (2003b). Multifunctional proteins: examples of gene sharing. Ann. Med. 35, 28–35. doi: 10.1080/07853890310004101
Jeffery, C. J. (2004a). Molecular mechanisms for multitasking: recent crystal structures of moonlighting proteins. Curr. Opin. Struct. Biol. 14, 663–668. doi: 10.1016/j.sbi.2004.10.001
Jeffery, C. J. (2004b). Moonlighting proteins: complications and implications for proteomics research. Drug Discov. Today Targets 3, 71–78. doi: 10.1016/S1741-8372(04)02405-3
Jeffery, C. J. (2009). Moonlighting proteins—an update. Mol. Biosyst. 5, 345–350. doi: 10.1039/b900658n
Jeffery, C. J. (2014). An introduction to moonlighting proteins. Biochem. Soc. Trans. 42, 1679–1683. doi: 10.1042/BST20140226
Jobin, M. C., Brassard, J., Quessy, S., Gottschalk, M., and Grenier, D. (2004). Acquisition of host plasmin activity by the Swine pathogen Streptococcus suis serotype 2. Infect. Immun. 72, 606–610. doi: 10.1128/IAI.72.1.606-610.2004
Kennedy, M. C., Mende-Mueller, L., Blondin, G. A., and Beinert, H. (1992). Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc. Natl. Acad. Sci. U.S.A. 89, 11730–11734. doi: 10.1073/pnas.89.24.11730
Khan, I., Chen, Y., Dong, T., Hong, X., Takeuchi, R., Mori, H., et al. (2014). Genome-scale identification and characterization of moonlighting proteins. Biol. Direct. 9:30. doi: 10.1186/s13062-014-0030-9
Khan, I. K., and Kihara, D. (2014). Computational characterization of moonlighting proteins. Biochem. Soc Trans. 42, 1780–1785. doi: 10.1042/BST20140214
Knaust, A., Weber, M. V. R., Hammerschmidt, S., Bergmann, S., Frosch, M., and Kurzai, O. (2007). Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis. J. Bacteriol. 189, 3246–3255. doi: 10.1128/JB.01966-06
Mani, M., Chen, C., Amblee, V., Liu, H., Mathur, T., Zwicke, G., et al. (2015). MoonProt: a database of proteins that are known to moonlight. Nucleic Acids Res. 43, D277–D282. doi: 10.1093/nar/gku954
Matta, S. K., Agarwal, S., and Bhatnagar, R. (2010). Surface localized and extracellular Glyceraldehyde-3-phosphate dehydrogenase of Bacillus anthracis is a plasminogen binding protein. Biochim. Biophys. Acta 1804, 2111–2120. doi: 10.1016/j.bbapap.2010.08.004
Nobeli, I., Favia, A. D., and Wool, I. G. (1996). Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 21, 164–165. doi: 10.1016/0968-0004(96)20011-8
Ostrovsky de Spicer, P., and Maloy, S. (1993). PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc. Natl. Acad. Sci. U.S.A. 90, 4295–4298. doi: 10.1073/pnas.90.9.4295
Ostrovsky de Spicer, P., O'Brien, K., and Maloy, S. (1991). Regulation of proline utilization in Salmonella typhimurium: a membrane-associated dehydrogenase binds DNA in vitro. J. Bacteriol. 173, 211–219.
Pancholi, V., and Fischetti, V., A (1992). A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176, 415–426. doi: 10.1084/jem.176.2.415
Philpott, C. C., Klausner, R. D., and Rouault, T. A. (1994). The bifunctional iron-responsive element binding protein/cytosolic aconitase: the role of active-site residues in ligand binding and regulation. Proc. Natl. Acad. Sci. U.S.A. 91, 7321–7325. doi: 10.1073/pnas.91.15.7321
Piatigorsky, J., and Horwitz, J. (1996). Characterization and enzyme activity of argininosuccinate lyase/delta-crystallin of the embryonic duck lens. Biochim. Biophys. Acta 1295, 158–164. doi: 10.1016/0167-4838(96)00030-1
Piatigorsky, J., O'Brien, W. E., Norman, B. L., Kalumuck, K., Wistow, G. J., Borras, T., et al. (1988). Gene sharing by delta-crystallin and argininosuccinate lyase. Proc. Natl. Acad. Sci. U.S.A. 85, 3479–3483. doi: 10.1073/pnas.85.10.3479
Piatigorsky, J., and Wistow, G. J. (1989). Enzyme/crystallins: gene sharing as an evolutionary strategy. Cell 57, 197–199. doi: 10.1016/0092-8674(89)90956-2
Rasch, J., Theuerkorn, M., Unal, C., Heinsohn, N., Tran, S., Fischer, G., et al. (2015). Novel cycloheximide derivatives targeting the moonlighting protein Mip exhibit specific antimicrobial activity against Legionella pneumophila. Front. Bioeng. Biotech. Res. Top. 3:41. doi: 10.3389/fbioe.2015.00041
Schweimer, K., Mooney, R. A., Landick, R., Artsimovitch, I., and Rösch, P. (2012). An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 150, 291–303. doi: 10.1016/j.cell.2012.05.042
Seifert, K. N., McArthur, W. P., Bleiweis, A. S., and Brady, L. J. (2003). Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: surface localization, enzymatic activity, and protein-protein interactions. J. Microbiol. 49, 350–356. doi: 10.1139/w03-042
Tompa, P., Szász, C., and Buday, L. (2005). Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 30, 484–489. doi: 10.1016/j.tibs.2005.07.008
Watanabe, H., Carmi, P., Hogan, V., Raz, T., Silletti, S., Nabi, I. R., et al. (1991). Purification of human tumor cell autocrine motility factor and molecular cloning of its receptor. J. Biol. Chem. 266, 13442–13448.
Wistow, G., and Piatigorsky, J. (1987). Recruitment of enzymes as lens structural proteins. Science 236, 1554–1556. doi: 10.1126/science.3589669
Keywords: moonlighting proteins, multifunctional, protein evolution, protein structure and function, enzyme function
Citation: Jeffery CJ (2015) Why study moonlighting proteins? Front. Genet. 6:211. doi: 10.3389/fgene.2015.00211
Received: 27 February 2015; Accepted: 31 May 2015;
Published: 19 June 2015.
Edited by:
Yasset Perez-Riverol, European Bioinformatics Institute, UKReviewed by:
Felipe Da Veiga Leprevost, University of Michigan, USAFlorencio Pazos, National Center for Biotechnology, Spain
Copyright © 2015 Jeffery. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Constance J. Jeffery, cjeffery@uic.edu
 Constance J. Jeffery
Constance J. Jeffery