- 1State Key Laboratory of Crop Biology, Shandong Key Laboratory of Crop Biology, Shandong Agricultural University, Tai’an, China
- 2Key Laboratory of Crop Genomics and Genetic Improvement, National Maize Improvement Center of China, China Agricultural University, Beijing, China
- 3School of Biological Science and Technology, University of Jinan, Jinan, China
Violaxanthin de-epoxidase (VDE) has a critical role in the carotenoid biosynthesis pathway, which is involved in protecting the photosynthesis apparatus from damage caused by excessive light. Here, a VDE gene in maize, ZmVDE1, was cloned and shown to have functional domains in common with the gramineous VDE protein. Candidate gene association analysis indicated that no polymorphic sites in ZmVDE1 were significant association with any of the examined carotenoid-related traits at P = 0.05 in an association panel containing 155 maize inbred lines. Nucleotide diversity analysis of VDE1 in maize and teosinte indicated that its exon had less genetic variation, consistent with the conserved function of VDE1 in plants. In addition, dramatically reduced nucleotide diversity, fewer haplotypes and a significantly negative parameter deviation for Tajima’s D test of ZmVDE1 in maize and teosinte suggested that a potential selective force had acted across the ZmVDE1 locus. We further identified a 4.2 Mb selective sweep with low recombination surrounding the ZmVDE1 locus that resulted in severely reduced nucleotide diversity on chromosome 2. Collectively, natural selection and the conserved domains of ZmVDE1 might show an important role in the xanthophyll cycle of the carotenoid biosynthesis pathway.
Introduction
Crop domestication and breeding, which have been ongoing for > 10,000 years, represent evolutionary experiments that have radically altered wild species to meet human needs (Hufford et al., 2012). Archeological (Piperno et al., 2009) and genetic (van Heerwaarden et al., 2011) evidence indicates that maize (Zea mays ssp. mays) was domesticated roughly 10,000 years ago in southern Mexico from Balsas teosinte (Zea mays ssp. parviglumis). Maize domestication involved a radical phenotypic transformation, resulting in an unbranched plant with numerous exposed seed attached to a cob in 20 rows or more. The dramatic morphological changes from teosinte to maize likely involved alterations in only a few significant genes with large effects (Tian et al., 2009).
Recent quantitative trait loci (QTL) mapping identified five regions of the maize genome with large effects on basic morphology (Doebley et al., 1990; Doebley and Stec, 1991), two of which have now been studied in great detail. One locus, teosinte branched1 (tb1), which was a major contributor to the increase in apical dominance during maize domestication, has been successfully cloned (Doebley et al., 1995, 1997; Wang et al., 1999). A transposable element inserted 60 kb upstream of tb1 acts as an enhancer of gene expression and partially explains the increased apical dominance in maize compared with teosinte (Studer et al., 2011). A single genetic locus, teosinte glume architecture1 (tga1), has been identified as a QTL controlling the formation of the hardened protective covering on teosinte kernels that has mostly been lost in maize (Dorweiler et al., 1993). A single amino acid change within tga1, which belongs to the squamosa promoter-binding protein-like transcription factor (SBP-domain) family of transcriptional regulators, is responsible for this radical difference (Wang et al., 2005). Molecular evolutionary analyses have indicated that this region was the target of selection during maize domestication (Wang et al., 2005). Subsequently, a few genes underwent a process of genetic modification to meet the needs of humans, including an increase in harvestable yield and better kernel quality (Tian et al., 2009). Four of the six genes involved in the starch biosynthesis pathway show evidence of selection (Whitt et al., 2002).
Carotenoids are natural plant pigments that are widely found in plants, although among cereals maize is the only major crop that contains appreciable amounts of carotenoids (Wurtzel, 2004). There are two generalized classes of carotenoids: carotenes and xanthophylls. Carotenoids are an important source of vitamin A, antioxidants and photo-protectants in plants (Goff and Klee, 2006). Antheraxanthin and violaxanthin, two xanthophylls are precursors of abscisic acid (ABA), which is essential for seed formation and induction of primary dormancy (Kermode, 2005). The carotenoid biosynthetic pathway is well studied and the enzymes involved in carotenogenesis have been documented in maize and other species (Giuliano et al., 2008; Li et al., 2009). In addition, association analyses of four key candidate genes involved in carotenoid biosynthesis revealed that natural polymorphisms at these loci are significantly associated with either carotenoid concentration or composition (Harjes et al., 2008; Yan et al., 2010; Zhou et al., 2012; Fu Z.Y. et al., 2013). Among these key candidate genes, PSY1 experienced strong selection during maize domestication and improvement from white to yellow kernels, but after this selection bottleneck, there was very little sequence variation in PSY1 within the yellow maize germplasm (Palaisa et al., 2004).
In the carotenoid biosynthetic pathway, violaxanthin de-epoxidase (VDE), a key enzyme of the xanthophyll cycle, has an important role in protecting the photosynthesis apparatus from damage, as excess light catalyzes the conversion of violaxanthin to zeaxanthin through antheraxanthin under high light. Zeaxanthin and antheraxanthin can transfer excess energy from chlorophyll, which is dissipated as heat and through scavenging reactive oxygen species, thus protecting the photosynthetic apparatus from photo damage (Eskling et al., 1997; Muller et al., 2001). In addition, lutein and zeaxanthin are associated with a reduced risk of cataract development and age-related macular degeneration (Abdel-Aal et al., 2013). VDE is a member of the lipocalin family (Bugos and Yamamoto, 1996) and has been isolated and purified from several plant species, such as romaine lettuce (Rockholm and Yamamoto, 1996) and spinach (Arvidsson et al., 1996), but has not yet been isolated from maize.
In this study, we cloned a gene encoding VDE, ZmVDE1, by a comparative genomic approach. Using Functional domain analysis of ZmVDE1, we confirmed that the VDE domain in Z. mays is highly consistent with the graminaceous plants. Furthermore, nucleotide diversity analysis and Tajima’s D test of VDE1 in maize and teosinte indicated that a potential selective force had acted across the ZmVDE1 locus. In addition, we identified a 4.2 Mb selective sweep with low recombination surrounding the ZmVDE1 locus that resulted in severely reduced nucleotide diversity on chromosome 2.
Materials and Methods
Plant Materials and Phenotyping
A maize association panel consisting of 155 inbred lines (Yang et al., 2010) was used to detect associations between the nucleotide polymorphisms of ZmVDE1 and carotenoid content as well as composition in maize kernels. Eighty-nine inbred lines from the 155 lines in the association panel and a set of 44 teosinte accessions from the CIMMYT gene bank (Supplementary Table S1) were used to resequence ZmVDE1 in this study. The maize panel was planted in one-row plots with two replications in a randomized complete block design in Beijing, China during the summer of 2005, 2006, and 2007 and the winter of 2007 in Hainan, China. Each plot was 4 m long. Rows were spaced 0.67 m apart, and plants were grown at a planting density of 45,000 plants/ha; seeds were produced via self-pollination of each plant in the plot (Fu Z.Y. et al., 2013). The maize kernel carotenoids were measured after harvesting. The best linear unbiased predictors (BLUPs) for individual traits in each line were calculated using SAS 8.02 (SAS Institute, 1999) in the association mapping populations. BLUPs for each line across environments were used for the overall analysis (Fu Z.Y. et al., 2013).
Candidate Gene Cloning
According to the zeaxanthin, antheraxanthin and violaxanthin inter conversion pathway (EMP, Enzymes and Metabolic Pathways database1), the protein sequence of the VDE homologous gene in Arabidopsis thaliana, AVDE1, was retrieved from TAIR2. The sequence homologous to AVDE1 in maize was obtained via BLAST using AVDE1 protein sequence against the maize genomic sequence in the Maize Genetics Genomics Database (maizeGDB3). The related information for ZmVDE1, including nucleotide sequence, transcript and gene structure, was obtained from the Gramene database4. In addition, DNA motif was predicted using the PLACE5 and PlantCARE6 databases.
Phylogenetic Tree Construction
To further investigate the genetic relationship of the VDE genes among different species, reference sequences were retrieved from the non-redundant protein sequence database using protein tool BLAST at NCBI7. And then the alignment file was constructed using ClustalW in MEGA 5.0. Phylogenetic tree was constructed using neighbor-joining criterion in MEGA 5.0 with 1,000 bootstrap tests for every node (Tamura et al., 2011). The uniform rates was applied for rates among sites. Gaps or missing data was treated as complete deletion.
Quantification of Carotenoids
Multiple self-pollinated ears from each line for each replication were combined for high-performance liquid chromatography (HPLC) analysis. Measured metabolites included lutein, zeaxanthin, β-cryptoxanthin, α-carotene and β-carotene. Carotenoids were extracted as described in Chander et al. (2008) and quantified by standard regression against external standards (Kurilich and Juvik, 1999). External standard curves were constructed with eight serial dilutions and with three repeats for each dilution (R2≥ 0.99). The five carotenoids were separated on a reversed-phase C30 column (YMC Carotenoid, CT99S05-2546WT C30, 4.6 nm × 25 cm, 5 μm; Waters) at 30°C at 1.8 ml/min for the mobile phase, 75:20:5 (v/v/v) acetonitrile/methanol/dichloromethane, by scanning at 450 nm with a reference wave of 360 nm and were identified by the retention time of each standard. The peak times for lutein, zeaxanthin, β-cryptoxanthin, α-carotene and β-carotene were 8.37 min, 10.71, 19.94, 27.61, and 37.61 min, respectively. All phenotypic data were generated with ChemStation software (Agilent Technologies) (Xu et al., 2012).
Genotyping and Sequence Analysis of ZmVDE1
Two primers that spanned most of the gene were used in conjunction with two additional sets of primers to sequence ZmVDE1 in 89 maize lines and 44 teosinte entries (Supplementary Table S2). A 30-μl aliquot of the resulting PCR product from each line was sequenced directly using an ABI3730 sequencer. The sequences were assembled using ContigExpress in Vector NTI Advance 10 (Invitrogen), aligned using MUSCLE (Edgar, 2004) and manually corrected with BioEdit (Hall, 1999). Additionally, to determine the expression level of ZmVDE1, 557,955 polymorphic sites with a minor allele frequency (MAF) ≥0.05 and missing rate <25% were generated by RNA-Sequencing maize kernels collected 15 days after pollination (DAP) from 368 maize inbred lines (Fu J.J. et al., 2013). Using the expression levels of ZmVDE1, we looked for expression QTL and analyzed their correlations with carotenoid content. Subsequently, >120,000 single nucleotide polymorphisms (SNPs) distributed across chromosome 2 in the 368 maize lines were used for testing whether there were selective sweeps around ZmVDE1.
Candidate Gene Association Mapping
The polymorphic sites including SNPs and insertions or deletions (InDels) of ZmVDE1 with MAF ≥0.05 in 89 maize lines from the 155-line association panel were extracted using TASSEL 2.1 (Bradbury et al., 2007). Associations between the polymorphic sites and five carotenoid-related traits were carried out using a mixed linear model (Yu et al., 2006), which incorporated population structure and kinship (Yang et al., 2010) in TASSEL 2.1.
Nucleotide Diversity and Tests for Selection
Nucleotide diversity (π) and Tajima’s D statistic were calculated for ZmVDE1 and chromosome 2 using DNaSP version 5.0 (Librado and Rozas, 2009). The scaled per-nucleotide recombination parameter Rn (Hudson, 1987) is the length-weighted mean of Rn across sweep region estimated from 368 maize lines.
Minimum Spanning Tree
The 89 maize inbred lines and 44 teosinte entries for the two subspecies Z. mays ssp. mexicana and Z. mays ssp. parviglumis (Supplementary Table S1) were used to construct a minimum spanning tree for ZmVDE1. Arlequin version 3.5 (Excoffier and Lischer, 2010) was used to calculate the minimum spanning tree nucleotide polymorphisms. Arlequin’s distance matrix output was used in Hapstar-0.6 (Teacher and Griffiths, 2011) to draw the minimum spanning tree.
Results
Cloning, Phylogenetic Analysis and Expression Pattern of ZmVDE1
A BLAST search with AVDE1 protein sequences from A. thaliana (AT1G08550) against the maize genomic sequences in the maizeGDB database resulted in three hits for homologous genes, GRMZM2G027219 on chromosome 2, GRMZM2G701673 on chromosome 10 and GRMZM2G408706 on chromosome 8. The maize homologous gene GRMZM2G027219 had the highest identity with AVDE1 (E-value = 4.734 × 10-134, 81.95% amino acid identity) and was referred to as ZmVDE1. ZmVDE1 from maize inbred line B73 has five exons, and the full-length cDNA sequence is 1,761 base pairs (bp), encoding 446 amino acids. The length of the 5′untranslated region (UTR) and 3′UTR are 148 and 272 bp, respectively (Supplementary Figure S1). Motif Scan analysis (Sigrist et al., 2010) showed that the VDE domain (amino acid 163–360) is present in the mature protein of ZmVDE1. Using the PLACE and PlantCARE databases, we also found the following putative DNA motifs, which are probably involved in light responsiveness, in the regulatory region and introns of ZmVDE1 (Supplementary Figure S1): I-box, G-box, GT-1 motif, TCT-motif, and MNF1-motif.
Functional domain analysis of this genomic fragment in InterProScan and EBI QuickGO confirmed that the VDE domain in Z. mays is highly consistent with those of the graminaceous plants, such as Triticum aestivum, Sorghum bicolor, and Oryza sativa (Figure 1A). To further investigate the genetic relationship of the VDE proteins among different species, phylogenetic analysis of these homologs was conducted in graminaceous plants, herbaceous plants and oil crops (Figure 1B). ZmVDE1 is grouped with all the graminaceous plant VDEs and is more closely related to the VDE of Sorghum bicolor than to other graminaceous crops. VDEs from other plants included oil crops and herbaceous plants in the another cluster.
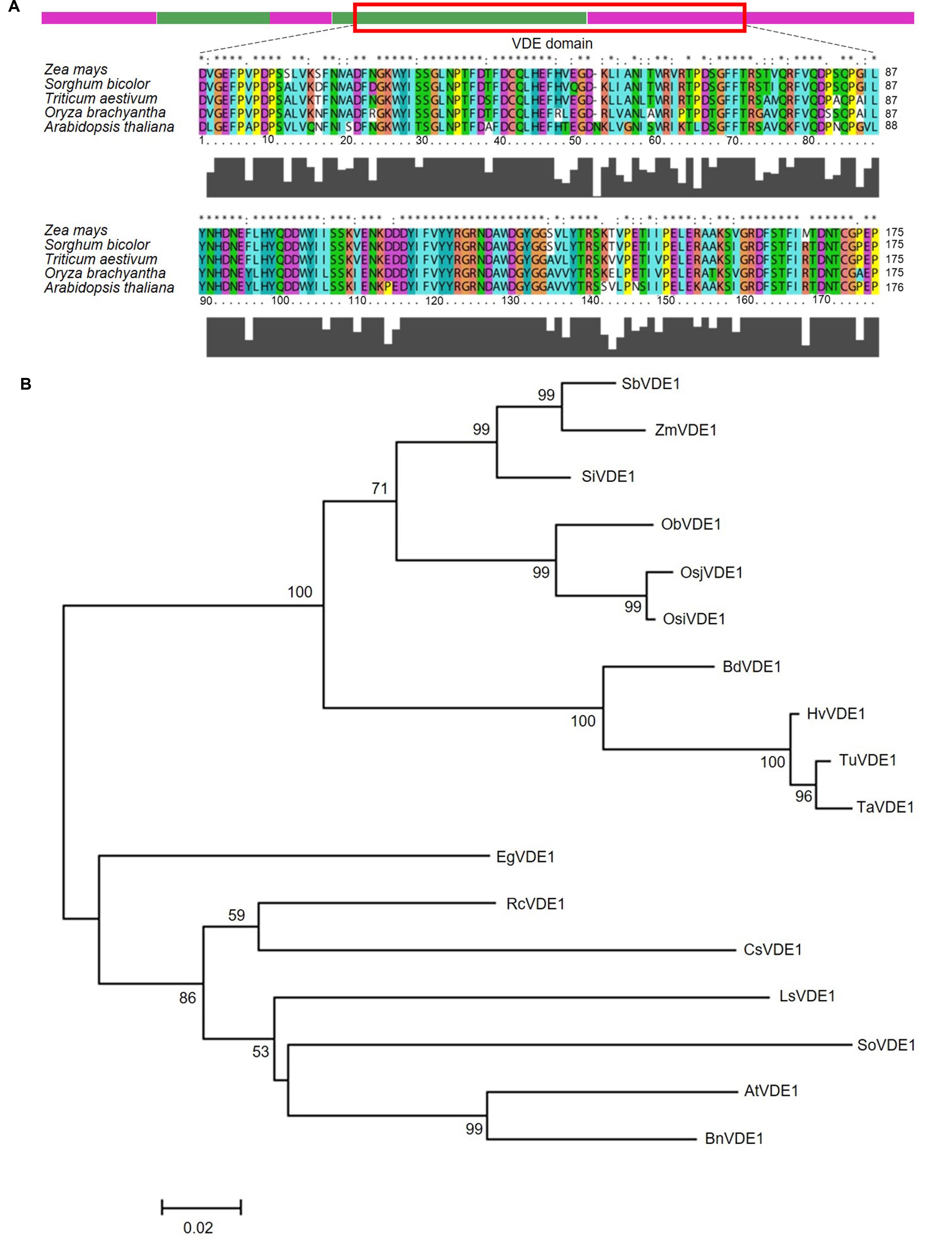
FIGURE 1. Characterization of ZmVDE1. (A) Schematic representation of the ZmVDE1 protein and an amino acid alignment of the VDE domains from different plants. The VDE domain is indicated by a red box. Purple and green bars represent the five exons. Gray squares below the sequence indicate the extent of sequence similarity. (B) Phylogenetic tree of the VDE proteins. The tree was constructed on the basis of genes homologous with ZmVDE1 by a neighbor-joining tree. Numbers above branches indicate the percentage of bootstrap values calculated from 1000 replicates. The scale bar gives a rough indication of sequence divergence. Sb, Sorghum bicolor; Zm, Zea mays; Si, Setaria italic; Ob, Oryza brachyantha; Osj, Oryza sativa L. ssp. Japonica; Osi, Oryza sativa L. ssp. Indica; Bd, Brachypodium distachyon; Hv, Hordeum vulgare; Tu, Triticum urartu; Ta, Triticum aestivum; Eg, Elaeis guineensis; Rc, Ricinus communis; Cs, Cucumis sativus; So, Spinacia oleracea; Ls, Lactuca sativa; At, Arabidopsis thaliana; Bn, Brassica napus.
Based on 53 samples ranging in developmental age from fertilization to maturity and consisting of the embryo, endosperm and whole-seed tissues from a previous study (Chen et al., 2014), we detected the expression pattern of ZmVDE1 during maize kernel development. ZmVDE1 was mainly expressed in the late stages of the embryo, endosperm and whole seed. Moreover, ZmVDE1 at most time points was expressed at a higher level in the endosperm than in the embryo samples (Supplementary Figure S2). Additionally, Stelpflug et al. (2015) had enhanced their previously published microarray-based gene atlas of maize inbred line B73 (Sekhon et al., 2011, 2012) to include 79 distinct replicated samples that had been analyzed using RNA-sequencing. The expression pattern of ZmVDE1 in the endosperm and whole-seed tissues was similar to the findings of Chen et al. (2014). Therefore, we inferred that ZmVDE1 is active in the late stages of seed development, in accordance with its function as an important synthetase involved in nutrient accumulation in maize kernels.
Association between ZmVDE1 and Carotenoid-Related Traits in Maize Kernels
To investigate the nucleotide polymorphisms of ZmVDE1 in maize, the full-length sequence including the promoter, 5′UTR, exons, 3′UTR and most of the introns was obtained from 89 inbred lines. Our re-sequencing results identified five nucleotide polymorphisms with MAF ≥0.05, two InDels and three SNPs, in a 3,724 bp genomic region (Figure 2A). On average, one in every 745 bp was polymorphic. Additionally, the distribution of the nucleotide diversity was not even across ZmVDE1. Among these polymorphic sites, only one SNP (SNP283) was located in the coding region, which affected residue 65 (which encodes a p.Val65Ile alteration) with no change in charge or hydrophobicity. The limited number of nucleotide polymorphisms within ZmVDE1 across 89 maize inbred lines illustrates that ZmVDE1 has an important role in the maize kernel with a conserved function.
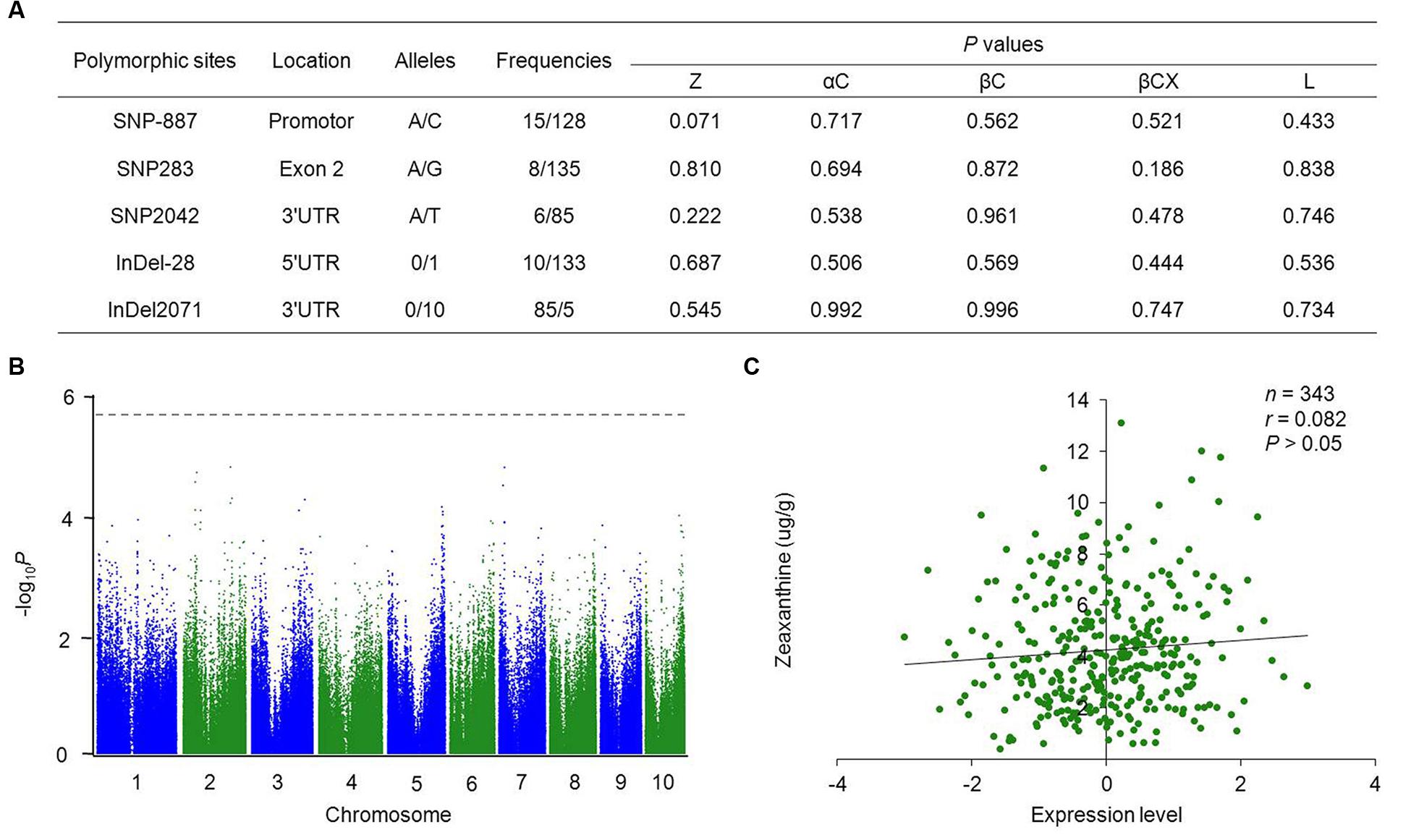
FIGURE 2. Associations between ZmVDE1 and carotenoid-related traits in maize kernels. (A) Summary of significant polymorphisms from candidate gene–based association studies. The first base of the start codon is defined as position 1. All the P values were from a mixed linear model controlling for population structure and individual relatedness in TASSEL 2.1. Z, zeaxanthin; αC, α-carotene; βC, β-carotene; βCX, β-cryptoxanthin; L, lutein. (B) Manhattan plot of eQTL for ZmVDE1. The dashed horizontal line depicts the Bonferroni-adjusted significance threshold (1.8 × 10-6). eQTL, expression quantitative trait loci. (C) Plots of correlation between the level of ZmVDE1 expression and zeaxanthin content in maize kernels. The x axis represents the normalized expression level of ZmVDE1 in kernels collected at 15 DAP (Fu J.J. et al., 2013). The y axis represents the level of zeaxanthin. The r value is a Pearson correlation coefficient.
Association analysis with a mixed linear model that controlled for both population structure and individual relatedness revealed that no polymorphic sites in ZmVDE1 showed a significant association with any of the analyzed carotenoid-related traits at the P = 0.05 level (Figure 2A). Previous studies have shown the importance of transcript abundance in the control of carotenoid profiles (Aluru et al., 2008; Harjes et al., 2008; Bai et al., 2009). Thus, we tested the correlation between the polymorphisms identified in the DNA sequence and the ZmVDE1 expression level to investigate whether ZmVDE1 can regulate any carotenoid-related traits at the level of expression. At P < 1.8 × 10-6, there was no statistical correlation between DNA sequence polymorphisms of ZmVDE1 and its expression level (Figure 2B). In addition, expression of ZmVDE1 was not correlated with phenotypic variation for any measured carotenoid trait (Figure 2C, Supplementary Figure S3). Thus the limited natural variation of ZmVDE1 makes no significant contribution to carotenoid variation.
Nucleotide Diversity of ZmVDE1 during Maize Domestication
To examine the levels of genetic diversity in ZmVDE1, we sequenced the promoter, 5′UTR, exons and introns of ZmVDE1 in 44 teosinte entries consisting of two subspecies, Zea mays ssp. parviglumis and Zea mays ssp. mexicana (Supplementary Table S1). Together with the ZmVDE1 sequences from 89 maize lines, 15 polymorphic sites were identified (Supplementary Table S3). When we used a sliding window of 100 bp with a step size of 25 bp to calculate nucleotide diversity, we noted that nucleotide diversity was not equally distributed in either maize or teosinte, with exons containing less genetic variation (Figure 3A). In most parts of ZmVDE1, the average π over all maize lines was lower than that in teosinte, especially for the promoter and exon 5 regions, which had dramatically reduced diversity in maize (πM/πT= 0.087 and πM/πT= 0.115, respectively) (Figure 3B). This finding suggests that the promoter and exon 5 in ZmVDE1 might have been under selection during the domestication of maize from teosinte.
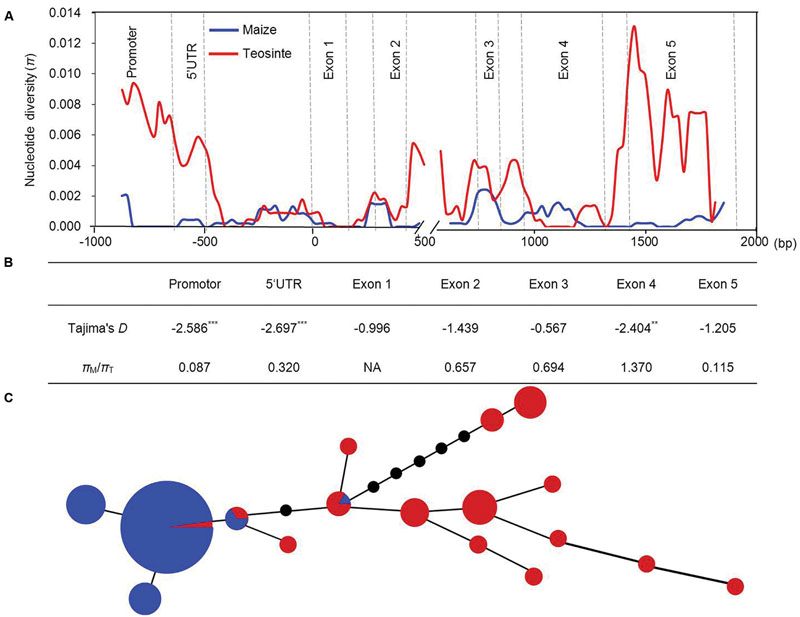
FIGURE 3. Sequence diversity of the ZmVDE1 locus between maize and teosinte. (A) Nucleotide diversity revealed by comparisons between 89 maize lines and 44 teosinte entries across the ZmVDE1 locus. Nucleotide diversity (π) for teosinte and maize was shown. (B) Tajima’s D test for non-neutral evolution and the ratios of π in maize (πM) to π in teosinte (πT) are shown. **P < 0.01; ***P < 0.001. (C) A minimum-spanning tree for ZmVDE1 including 89 diverse maize sequences and 44 diverse teosinte sequences. Each haplotype group is represented by a circle with a color, and circle sizes represent the number of lines within the haplotype. Red and blue represent teosinte and maize, respectively.
To further investigate the evidence of selection during maize domestication, Tajima’s D test was conducted for the promoter, 5′UTR and exon regions of ZmVDE1 in maize and teosinte. Tajima’s D test showed that promoter, 5′UTR and exon 4 regions of ZmVDE1 had significantly negative parameter deviations (P < 0.01) (Figure 3B), which is consistent with directional selection (Jiao et al., 2012). The results suggest that a selective sweep might have occurred in those regions, leading to reduced nucleotide diversity across the ZmVDE1 locus in maize.
Evolution of the ZmVDE1 Locus in Maize and Teosinte
To further investigate the genetic signature of positive selection in the ZmVDE1 region in more detail, we selected 120,204 high-quality SNPs on chromosome 2 with MAF ≥0.05 from a panel of 368 diverse maize inbred lines (Fu J.J. et al., 2013). Using a sliding window of 500 SNPs with a step size of 100 SNPs to calculate the nucleotide diversity and conduct Tajima’s D test, we found that the maize inbred lines had severely reduced nucleotide diversity (π = 0.123) across a 4.2 Mb region surrounding the ZmVDE1 locus relative to other segments on chromosome 2 (Figure 4A). In addition, the statistic for Tajima’s D test was notably negative for a selective sweep in the 368 inbred lines, suggesting an excess of low-frequency polymorphisms relative to the expectation that might have resulted from the selection on the selective sweep in this population (Figure 4A). The large interval (4.2 Mb) associated with this selective sweep might be related to the low SNP or gene density and the recombination rate. As expected, no significant reduction in SNP or gene density was observed in the 4.2 Mb region surrounding the ZmVDE1 locus. There are 478 SNPs in the 4.2 Mb selective sweep with one in every 8,786 bp being polymorphic, and there are 45 genes in the large interval, with one every 93.3 kb (Figure 4B). In contrast, there is one polymorphic nucleotide every 1,951 bp and one gene every 49.5 kb in other regions of chromosome 2 (Figure 4B). The nucleotide estimate for the population recombination rate indicated that the recombination rate across this sweep region (Rn = 0.0757) was 1.59-fold lower than it was at the known selection target tga1 (Rn = 0.1205). These results suggested that few recombination events can be detected in this selective sweep because of the relatively low levels of polymorphism and that the low rate of recombination has contributed to the size of the sweep.
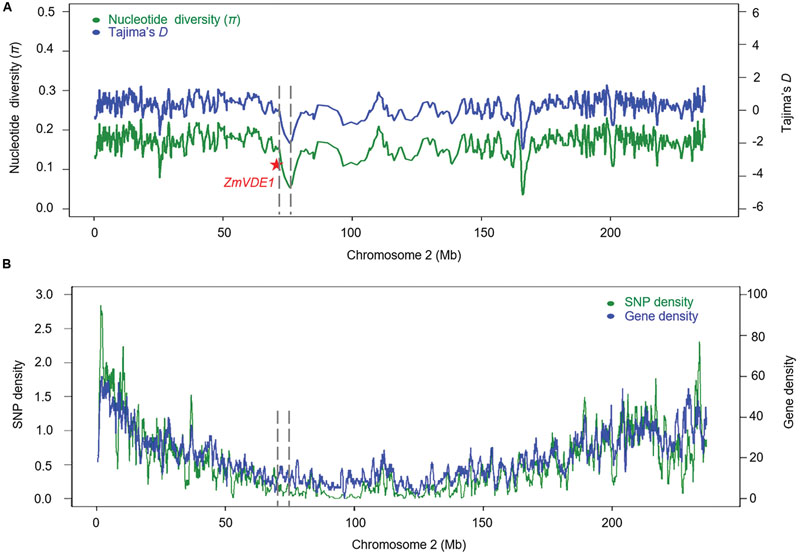
FIGURE 4. Nucleotide variation on chromosome 2 in 368 maize inbred lines. (A) Nucleotide diversity (π) and Tajima’s D for maize chromosome 2. The red star represents ZmVDE1. The dash line represents the selection sweep region of 4.2 Mb. (B) SNP density (green) and gene density (blue) calculated using a 1 Mb sliding window with a 100-kb step.
To gain deeper insights into the evolution of ZmVDE1, its full-length sequence including the promoter, 5′UTR, exons, 3′UTR and most of the introns from 89 Chinese elite inbred lines and 44 additional teosinte entries (Supplementary Table S1) was used to construct a minimum-spanning tree. In total, we identified five haplotypes for the 89 diverse maize lines, of which Hap1 predominated, and 15 haplotypes for 44 teosinte entries (Supplementary Table S4). With these haplotypes, a minimum-spanning tree was built (Figure 3C). The 17 haplotypes formed two distinguishable clusters: a maize haplotype cluster and a teosinte haplotype cluster. The maize haplotype cluster contained almost all the maize lines and two teosinte entries, whereas the teosinte haplotype cluster was composed of teosinte entries with one additional maize line. These results revealed that VDE1 was domesticated with fewer haplotypes in maize comparing with which in teosinte. (Figures 3A,B), the nucleotide diversity was much higher in teosinte than in maize.
Discussion
Violaxanthin de-epoxidase carries out an important role in the photo-protective xanthophyll cycle by catalyzing the formation of non–provitamin A carotenoids such as zeaxanthin that function during the strong light response (Rockholm and Yamamoto, 1996; Li et al., 2009). However, important questions about whether the expression of this protein was selected for during the domestication of maize are still unanswered. In this study, we used association analysis, nucleotide diversity detection and evolutionary analyses to confirm the significance of ZmVDE1 in maize domestication.
VDE Has a Conserved Function in Plants
In maize, ZmVDE1 contains five putative circadian DNA motifs, I-box (GATA motif), G-box, GT-1 motif, TCT-motif and MNF1-motif (Supplementary Figure S1). These motifs are thought to be involved in light regulation and circadian rhythm expression (Borello et al., 1993; Piechulla et al., 1998; Xu et al., 2003; Kidokoro et al., 2009). In addition to the light-regulated motifs, the other most important motifs in introns were cis-acting elements involved in plant responses to environmental stresses, including MYB2CONSENSUSAT(YAACKG), CBFHV (RYCGAC), RAV1AAT (CAACA), CCAATBOX1 (CCAAT), WBOXATNPR1(TTGAC), GT1GMSCAM4(GAAAAA), MYB1 AT (WAACCA), MYBCORE (CNGTTR), MYCCONSENSUSAT (CANNTG) and WRKY71OS (TGAC), which indicated that the expression of ZmVDE1 might also be regulated by transcription factors related to stress tolerance. All of these motifs showed high identities with other gramineous VDEs such as those from T. aestivum and Oryza sativa, especially in the regions corresponding to the mature proteins (Gao et al., 2013).
Violaxanthin de-epoxidase has a conserved function converting violaxanthin to antheraxanthin and then to zeaxanthin in the xanthophyll cycle, which is a photo-protection mechanism conserved among photosynthetic plants and algae to protect the photosynthetic apparatus from excess light (North et al., 2005). Meanwhile, downstream of the xanthophyll cycle, violaxanthin and zeaxanthin are the intermediate products in the biosynthesis pathway of ABA, which regulates the development process, seed maturation, stomatal closure and stress responses (Finkelstein et al., 2002; Zhu, 2002; Wang and Song, 2008). Therefore, VDE and zeaxanthin epoxidase (ZEP) have been the targets of genetic manipulation to better understand the carotenoid synthetic pathway. Römera et al. (2002) found that the amount of violaxanthin was diminished dramatically, and in some cases the amount of the monoepoxy intermediate antheraxanthin was increased, as a consequence of the genetic manipulation of transformation with sense and antisense constructs encoding zeaxanthin epoxidase, even though the total carotenoid content and ABA content were both increased. Thus VDE might regulate the synthesis of ABA (Owen et al., 1992; Marin et al., 1996; Pogson et al., 1996; Audran et al., 1998). The enhanced activity of VDE under excess light could reduce the content of ABA, perhaps by acting on the xanthophyll cycle and regulating the synthesis of the ABA precursor.
Limited Natural Variations within ZmVDE1 Are Not Associated with Carotenoid-Related Traits
Candidate gene association mapping in DNA from kernels grown in multiple environments indicated that none of the five polymorphisms in ZmVDE1 were significantly associated with carotenoid-related traits in maize kernels. To confirm this result, we also extracted 220 SNPs of ZmVDE1 in 368 maize inbred lines (Fu J.J. et al., 2013) to perform a regional association analysis for carotenoid-related traits. No significant SNPs were identified, consistent with the association analysis in the 89 maize inbred lines. In addition, ZmVDE1 expression was not correlated with any of the measured carotenoid-related traits. One possible explanation for the absence of an association may be the small size of our association panel. It is well known that association mapping studies may be adversely affected by two major interconnected factors, the dominance of a SNP with a lower MAF and a small population size (Manolio et al., 2009). Simulation results of the effect on QTL detection power with increasing population size revealed that a population with 500 lines provides an 80% probability of detecting a gene that explains 3% or more of the phenotypic variation (Yan et al., 2011). So some important genetic variations may not have been detected in our association panel with its relatively small population size. Another explanation for this absence of an association is that ZmVDE1 has an important and conserved role in plants. Additionally, ZmVDE1 was expressed throughout the leaf with highest expression level at maturing and mature zones (Supplementary Figure S4). Moreover, expression of ZmVDE1 in seedling leaves was statistically higher than that in maize kernels (Chen et al., 2014; Stelpflug et al., 2015), similar to that of AtVDE in A. thaliana (North et al., 2005), suggesting that ZmVDE1 also has an important protective role in chloroplasts (Audran et al., 2001).
Previous studies have shown that association mapping results of candidate genes cloned by homology-based cloning is quite different among maize. For example, ZmGS3 and ZmGW2 have co-orthologous relationships with the rice region containing GS3 and GW2, respectively. ZmGS3 was slightly associated with maize kernel length and one-hundred kernel weight (HKW), with the P-values ranging between 0.05 and 0.01, whereas ZmGW2 was significantly associated with kernel size (P < 10-3) (Li et al., 2010a,b). These results for ZmVDE1, ZmGS3 and ZmGW2 imply that the power of an association study is dependent on various factors, such as the gene effect, the genetic architecture of the quantitative trait and the size of association population and so on. So, in addition to the association analysis, we still need other genetic methods to validate gene function.
ZmVDE1 Has Been under Strong Selection during Maize Domestication
Because of the role of the VDE in the xanthophyll cycle, an important photo-protection mechanism, it is easy to understand why ZmVDE1 was a target of the domestication or improvement process in maize. In our study, ZmVDE1 was less diverse at the nucleotide level in maize than in teosinte, especially in the promoter and 5′UTR regions, consistent with a significantly negative Tajima’s D parameter, which may indicate that this region has undergone a recent and strong positive selection. While, significant reductions in diversity were detected in the VDE domain region (πM/πT = 0.079) with negative Tajima’s D parameter (Tajima’s D = -1.576), which may be because that VDE domain variants within these regions might have experienced positive selection during maize domestication and improvement. After this selective force on ZmVDE1, the VDE domain became fixed with no polymorphic sites, as indicated by widespread Chinese maize inbred lines (Figure 2A).
Positive directional selection results in reduced variability in particular regions, which are referred to as selective sweep regions (Tian et al., 2009). According to our results, ZmVDE1 is located in a genomic region of 4.2 Mb that was affected by a selective sweep based on the 368-line maize association population. A larger selected region (5.2 Mb) located on chromosome 2 has been reported, and there was a strong signal for ZmVDE1 in this region (Hufford et al., 2012), which supports our findings. Similar evidence for a domestication-related selection sweep has also been reported in maize. A selective sweep extending 60–90 kb upstream of tb1 indicated that tb1 had a role in the domestication of maize from teosinte between 6,000 and 10,000 years ago (Clark et al., 2004). Maize PSY1 on chromosome 6, which controls kernel color, has undergone very strong selection for the yellow kernel color phenotype. Patterns of diversity in this region are consistent with the occurrence of a large selective sweep (>0.5 Mb) at PSY1 in maize breeding lines (Palaisa et al., 2004). Maize also shows severely reduced nucleotide diversity relative to teosinte across a large selective sweep (1.1 Mb) on chromosome 10 that has been the target of strong selection during maize domestication (Tian et al., 2009). In addition, a study on chromosome 3, which contains the largest selected region (2.2 Mb), using 278 temperate maize inbred lines from different stages of breeding history showed that modern breeding has introduced highly dynamic genetic changes into the maize genome (Jiao et al., 2012). At the same time, artificial selection leads to a reduction in nucleotide diversity and an increase in the proportion of rare alleles (Jiao et al., 2012). Collectively, these results imply that ZmVDE1 is a major domestication- and improvement-related gene involved in the carotenoid biosynthesis pathway. The challenge for the future is to figure out how the evolution and selection of this gene have occurred.
Author Contributions
JX analyzed the data and wrote the manuscript, HY and ZL performed the experiments, XY designed the study, CC advised on the study and manuscript, HL analyzed the data and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31361140362, 31401388) and Shandong Province Outstanding Young Scientists Award Fund (BS2014NY002).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2016.00131
FIGURE S1 | ZmVDE1 gene structure based on the genomic sequence from maize inbred line B73. The red, sky blue, and yellow represent the transcription start and termination position, presumptive motif and coding sequence of ZmVDE1.
FIGURE S2 | Expression patterns of ZmVDE1 in various tissues at different developmental stages in maize line B73. FPKM (red; Stelpflug et al., 2015) and relative expression (blue; Chen et al., 2014) represent the expression level of ZmVDE1 based on these two methods. (A–C) Expression of ZmVDE1 in seed (A), endosperm (B) and embryo (C). FPKM, fragments per kilobase of transcript per million mapped reads.
FIGURE S3 | Plots of correlation between ZmVDE1 expression and carotenoid-related traits in maize kernels. (A–D) The x axis represents the normalized expression of ZmVDE1 in kernels collected at 15 DAP. The y axis represents the level of lutein (A), β-cryptoxanthin (B), α-carotene (C) and β-carotene (D). The r value is a Pearson correlation coefficient.
FIGURE S4 | Expression patterns of ZmVDE1 in maize leaf in maize line B73. Histogram represent the expression level of ZmVDE1 based on Chen et al. (2014).
Footnotes
- ^ http://www.plantcyc.org
- ^ http://www.arabidopsis.org
- ^ http://www.maizegdb.org
- ^ http://www.gramene.org/
- ^ http://ppdb.agr.gifu-u.ac.jp/ppdb/cgi-bin/index.cgi
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://www.ncbi.nlm.nih.gov
References
Abdel-Aal, E. M., Akhtar, H., Zaheer, K., and Ali, R. (2013). Dietary sources of lutein and zeaxanthin carotenoids and their role in eye health. Nutrients 5, 1169–1185. doi: 10.3390/nu5041169
Aluru, M., Xu, Y., Guo, R., Wang, Z. G., Li, S. S., and White, W. (2008). Generation of transgenic maize with enhanced provitamin A content. J. Exp. Bot. 59, 3551–3562. doi: 10.1093/jxb/ern212
Arvidsson, P. O., Bratt, C. E., Carlsson, M., and Akerlund, H. E. (1996). Purification and identification of the violaxanthinde-epoxidase as a 43 kDa protein. Photosynth. Res. 49, 119–129. doi: 10.1007/BF00117662
Audran, C., Borel, C., Frey, A., Sotta, B., Meyer, C., Simonneau, T., et al. (1998). Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia. Plant Physiol. 118, 1021–1028. doi: 10.1104/pp.118.3.1021
Audran, C., Liotenberg, S., Gonneau, M., North, H., Frey, A., Tap-Waksman, K., et al. (2001). Localisation and expression of zeaxanthine poxidase mRNA in Arabidopsis in response to drought stress and during seed development. Aust. J. Plant Physiol. 28, 1161–1173. doi: 10.1071/PP00134
Bai, L., Kim, E. H., DellaPenna, D., and Brutnell, T. P. (2009). Novel lycopene epsilon cyclase activities in maize revealed through perturbation of carotenoid biosynthesis. Plant J. 59, 588–599. doi: 10.1111/j.1365-313X.2009.03899.x
Borello, U., Ceccarelli, E., and Giuliano, G. (1993). Constitutive, light-responsive and circadian clock-responsive factors compete for the different l box elements in plant light-regulated promoters. Plant J. 4, 611–619. doi: 10.1046/j.1365-313X.1993.04040611.x
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Bugos, R. C., and Yamamoto, H. Y. (1996). Molecular cloning of violaxanthinde-epoxidase from romaine lettuce and expression in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 6320–6325. doi: 10.1073/pnas.93.13.6320
Chander, S., Meng, Y. J., Zhang, Y. R., Yan, J. B., and Li, J. S. (2008). Comparison of nutritional traits variability in selected eighty-seven inbreds from Chinese maize (Zea mays L.) germplasm. J. Agric. Food Chem. 56, 6506–6511. doi: 10.1021/jf7037967
Chen, J., Zeng, B., Zhang, M., Xie, S. J., Wang, G. K., Hauck, A., et al. (2014). Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 166, 252–264. doi: 10.1104/pp.114.240689
Clark, R. M., Linton, E., Messing, J., and Doebley, J. F. (2004). Pattern of diversity in the genomic region near the maize domestication gene tb1. Proc. Natl. Acad. Sci. U.S.A. 101, 700–707. doi: 10.1073/pnas.2237049100
Doebley, J., and Stec, A. (1991). Genetic analysis of the morphological differences between maize and teosinte. Genetics 129, 285–295.
Doebley, J., Stec, A., and Gustus, C. (1995). Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141, 333–346.
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Doebley, J., Stec, A., Wendel, J., and Edwards, M. (1990). Genetic and morphological analysis of a maize-teosinte F2 population: implications for the origin of maize. Proc. Natl. Acad. Sci. U.S.A. 87, 9888–9892. doi: 10.1073/pnas.87.24.9888
Dorweiler, J., Stec, A., Kermicle, J., and Doebley, J. (1993). Teosinte glume architecture 1: a genetic locus controlling a key step in maize evolution. Science 262, 233–235. doi: 10.1126/science.262.5131.233
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Eskling, M., Arvidsson, P. O., and Akerlund, H. E. (1997). The xanthophyll cycle, its regulation and components. Physiol. Plant. 100, 806–816. doi: 10.1111/j.1399-3054.1997.tb00007.x
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Finkelstein, R. R., Gampala, S. S., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14, S15–S45. doi: 10.1105/tpc.010441
Fu, J. J., Cheng, Y. B., Linghu, J. J., Yang, X. H., Kang, L., Zhang, Z. X., et al. (2013). RNA sequencing reveals the complex regulatory network in the maize kernel. Nat. Commun. 4, 2832–2844. doi: 10.1038/ncomms3832
Fu, Z. Y., Chai, Y. C., Zhou, Y., Yang, X. H., Warburton, M. L., Xu, S. T., et al. (2013). Natural variation in the sequence of PSY1 and frequency of favorable polymorphisms among tropical and temperate maize germplasm. Theor. Appl. Genet. 126, 923–935. doi: 10.1007/s00122-012-2026-0
Gao, Z. M., Liu, Q., Zheng, B., and Chen, Y. (2013). Molecular characterization and primary functional analysis of PeVDE, a violaxanthin de-epoxidase gene from bamboo (Phyllostachys edulis). Plant Cell Rep. 32, 1381–1391. doi: 10.1007/s00299-013-1450-1
Giuliano, G., Tavazza, R., Diretto, G., Beyer, P., and Taylor, M. A. (2008). Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol. 26, 139–145. doi: 10.1016/j.tibtech.2007.12.003
Goff, S. A., and Klee, H. J. (2006). Plant volatile compounds: sensory cues for health and nutritional value? Science 311, 815–819. doi: 10.1126/science.1118446
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98.
Harjes, C. E., Rocheford, T. R., Bai, L., Brutnell, T. P., Kandianis, C. B., Sowinski, S. G., et al. (2008). Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319, 330–333. doi: 10.1126/science.1150255
Hudson, R. R. (1987). Estimating the recombination parameter of a finite population model without selection. Genet. Res. 50, 245–250. doi: 10.1017/S0016672300023776
Hufford, M. B., Xu, X., Heerwaarden, J., Pyhäjärvi, T., Chia, J. M., Cartwright, R. A., et al. (2012). Comparative population genomics of maize domestication and improvement. Nat. Genet. 44, 808–811. doi: 10.1038/ng.2309
Jiao, Y. P., Zhao, H. N., Ren, L. H., Song, W. B., Zeng, B., Guo, J. J., et al. (2012). Genome-wide genetic changes during modern breeding of maize. Nat. Genet. 44, 812–815. doi: 10.1038/ng.2312
Kermode, A. R. (2005). Role of abscisic acid in seed dormancy. J. Plant Growth Regul. 24, 319–344. doi: 10.1007/s00344-005-0110-2
Kidokoro, S., Maruyama, K., Nakashima, K., Imura, Y., Narusaka, Y., Shinwari, Z. K., et al. (2009). The phytochrome-Interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151, 2046–2057. doi: 10.1104/pp.109.147033
Kurilich, A. C., and Juvik, J. A. (1999). Simultaneous quantification of carotenoids and tocopherols in corn kernel extracts by HPLC. J. Liq. Chromatogr. Relat. Technol. 22, 2925–2934. doi: 10.1081/JLC-100102068
Li, F., Tsfadia, O., Wurtzel, E. T., Tzfadia, O., and Wurtzel, E. T. (2009). The phytoene synthase gene family in the Grasses: subfunctionalization provides tissue-specific control of carotenogenesis. Plant Signal. Behav. 4, 208–211. doi: 10.4161/psb.4.3.7798
Li, Q., Li, L., Yang, X. H., Warburton, M. L., Bai, G. H., Dai, J. R., et al. (2010a). Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10:143. doi: 10.1186/1471-2229-10-143
Li, Q., Yang, X. H., Bai, G. H., Warburton, M. L., Mahuku, G., Gore, M., et al. (2010b). Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor. Appl. Genet. 120, 753–763. doi: 10.1007/s00122-009-1196-x
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Manolio, T. A., Collins, F. S., Cox, N. J., Goldstein, D. B., Hindorff, L. A., Hunter, D. J., et al. (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753. doi: 10.1038/nature08494
Marin, E., Nussaume, L., Quesada, A., Gonneau, M., Sotta, B., Hugueney, P., et al. (1996). Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15, 2331–2342.
Muller, K. R., Mika, S., Ratsch, G., Tsuda, K., and Scholkopf, B. (2001). An introduction to kernel-based learning algorithms. IEEE Trans. Neural Netw. 12, 181–201. doi: 10.1109/72.914517
North, H. M., Frey, A., Boutin, J. P., Sotta, B., and Marion-Poll, A. (2005). Analysis of xanthophyll cycle gene expression during the adaptation of Arabidopsis to excess light and drought stress: changes in RNA steady-state levels do not contribute to shortterm responses. Plant Sci. 169, 115–124. doi: 10.1016/j.plantsci.2005.03.002
Owen, M., Gandecha, A., Cockburn, B., and Whitelam, G. (1992). Synthesis of a functional anti-phytochrome single-chain Fv protein in transgenic tobacco. Biotechnology (N. Y.) 10, 790–794. doi: 10.1038/nbt0792-790
Palaisa, K., Morgante, M., Tingey, S., and Rafalski, A. (2004). Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc. Natl. Acad. Sci. U.S.A. 101, 9885–9890. doi: 10.1073/pnas.0307839101
Piechulla, B., Merforth, N., and Rudolph, B. (1998). Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol. Biol. 38, 655–662. doi: 10.1023/A:1006094015513
Piperno, D. R., Ranere, A. J., Holst, I., Iriarte, J., and Dickau, R. (2009). Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. U.S.A. 106, 5019–5024. doi: 10.1073/pnas.0812525106
Pogson, B., McDonald, K. A., Truong, M., Britton, G., and DellaPenna, D. (1996). Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 8, 1627–1639. doi: 10.1105/tpc.8.9.1627
Rockholm, D. C., and Yamamoto, H. Y. (1996). Violaxanthin De-Epoxidase: purification of a 43-Kilodalton lumenal protein from lettuce by lipid-affinity precipitation with Monogalactosyldiacylglyceride. Plant Physiol. 110, 697–703. doi: 10.1104/pp.110.2.697
Römera, S., Lübeckb, J., Kaudera, F., Steigerc, S., Adomata, C., and Sandmannc, G. (2002). Genetic engineering of a zeaxanthin-rich potato by antisense inactivation and co-suppression of carotenoid epoxidation. Metab. Eng. 4, 263–272. doi: 10.1006/mben.2002.0234
Sekhon, R. S., Childs, K. L., Santoro, N., Foster, C., Buell, C. R., Leon, N., et al. (2012). Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol. 159, 1730–1744. doi: 10.1104/pp.112.199224
Sekhon, R. S., Lin, H. N., Childs, K. L., Hansey, C. N., Buell, C. R., Leon, N., et al. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66, 553–563. doi: 10.1111/j.1365-313X.2011.04527.x
Sigrist, C. A., Cerutti, L., Castro, E., Langendijk-Genevaux, P. S., Bulliard, V., Bairoch, A., et al. (2010). PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38, D161–D166. doi: 10.1093/nar/gkp885
Stelpflug, S. C., Sekhon, R. S., Vaillancourt, B., Hirsch, C. N., Buell, C. R., Leon, N., et al. (2015). An expanded maize gene expression atlas based on RNA-sequencing and its use to explore root development. Plant Genome 9. doi: 10.3835/plantgenome2015.04.0025
Studer, A., Zhao, Q., Ibarra, J. R., and Doebley, J. (2011). Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160–1163. doi: 10.1038/ng.942
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Teacher, A. G., and Griffiths, D. J. (2011). HapStar: automated haplotype network layout and visualization. Mol. Ecol. Resour. 11, 151–153. doi: 10.1111/j.1755-0998.2010.02890.x
Tian, F., Stevens, N. M., and Buckler Iv, E. S. (2009). Tracking footprints of maize domestication and evidence for a massive selective sweep on chromosome 10. Proc. Natl. Acad. Sci. U.S.A. 106, 9979–9986. doi: 10.1073/pnas.0901122106
van Heerwaarden, J., Doebley, J., Briggs, W. H., Glaubitz, J. C., Goodman, M. M., Gonzalez, J. J. S., et al. (2011). Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc. Natl. Acad. Sci. U.S.A. 108, 1088–1092. doi: 10.1073/pnas.1013011108
Wang, H., Nussbaum-Wagler, T., Li, B. L., Zhao, Q., Vigouroux, Y., Faller, M., et al. (2005). The origin of the naked grains of maize. Nature 386, 485–488. doi: 10.1038/nature03863
Wang, P. T., and Song, C. P. (2008). Guard-cell signaling for hydrogen peroxide and abscisic acid. New Phytol. 178, 703–718. doi: 10.1111/j.1469-8137.2008.02431.x
Wang, R. L., Stec, A., Hey, J., Lukens, L., and Doebley, J. (1999). The limits of selection during maize domestication. Nature 398, 236–239. doi: 10.1038/18435
Whitt, S. R., Wilson, L. M., Tenaillon, M. L., Gaut, B. S., and Buckler, E. S. (2002). Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 12959–12962. doi: 10.1073/pnas.202476999
Wurtzel, E. T. (2004). Genomics, genetics, and biochemistry of maize carotenoid biosynthesis. Recent Adv. Phytochem. 38, 85–110. doi: 10.1016/S0079-9920(04)80006-6
Xu, S. T., Zhang, D. L., Cai, Y., Zhou, Y., Trushar, S., Farhan, A., et al. (2012). Dissecting tocopherols content in maize (Zea mays L.), using two segregating populations and high-density single nucleotide polymorphism markers. BMC Plant Biol. 12:201. doi: 10.1186/1471-2229-12-201
Xu, Z. F., Chye, M. L., Li, H. Y., Xu, F. X., and Yao, K. M. (2003). G-box binding coincides with increased Solanum melongena cysteine proteinase expression in senescent fruits and circadian-regulated leaves. Plant Mol. Biol. 51, 9–19. doi: 10.1023/A:1020859518877
Yan, J. B., Kandianis, C. B., Harjes, C. E., Bai, L., Kim, E. H., Yang, X. H., et al. (2010). Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 42, 322–327. doi: 10.1038/ng.551
Yan, J. B., Warburton, M., and Crouch, J. (2011). Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci. 51, 433–449. doi: 10.2135/cropsci2010.04.0233
Yang, X. H., Yan, J. B., Shah, T., Warburton, M. L., Li, Q., Li, L., et al. (2010). Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor. Appl. Genet. 121, 417–431. doi: 10.1007/s00122-010-1320-y
Yu, J. M., Pressoir, G., Briggs, W. H., Bi, I. V., Yamasaki, M., Doebley, J. F., et al. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. doi: 10.1038/ng1702
Zhou, Y., Han, Y. J., Li, Z. G., Fu, Y., Fu, Z. Y., Xu, S. T., et al. (2012). ZmcrtRB3 encodes a carotenoid hydroxylase that affects the accumulation of α-carotene in maize kernel. J. Integr. Plant Biol. 54, 260–269. doi: 10.1111/j.1744-7909.2012.01106.x
Keywords: violaxanthin de-epoxidase, nucleotide diversity, selection, domestication, carotenoid, maize
Citation: Xu J, Li Z, Yang H, Yang X, Chen C and Li H (2016) Genetic Diversity and Molecular Evolution of a Violaxanthin De-epoxidase Gene in Maize. Front. Genet. 7:131. doi: 10.3389/fgene.2016.00131
Received: 15 May 2016; Accepted: 07 July 2016;
Published: 26 July 2016.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanCopyright © 2016 Xu, Li, Yang, Yang, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, bio_lih@ujn.edu.cn Cuixia Chen, cxchen@sdau.edu.cn
 Jing Xu
Jing Xu Zhigang Li
Zhigang Li Haorui Yang2
Haorui Yang2 Cuixia Chen
Cuixia Chen Hui Li
Hui Li