- 1 Laboratory of Immunobiology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA
- 2 Department of Medicine, Harvard Medical School, Boston, MA, USA
- 3 Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, USA
- 4 Department of Physiology, The University of Hong Kong, Hong Kong, China
- 5 Department of Pediatrics, Harvard Medical School, Boston, MA, USA
The αβ T cell receptor (TCR) is a multimeric complex whose β chain plays a crucial role in thymocyte development as well as antigen recognition by mature T lymphocytes. We report here crystal structures of individual β subunits, termed N15β (Vβ5.2Dβ2Jβ2.6Cβ2) and N30β (Vβ13Dβ1Jβ1.1Cβ2), derived from two αβ TCRs specific for the immunodominant vesicular stomatitis virus octapeptide (VSV-8) bound to the murine H-2Kb MHC class I molecule. The crystal packing of the N15β structure reveals a homodimer formed through two Vβ domains. The Vβ/Vβ module is topologically very similar to the Vα/Vβ module in the N15αβ heterodimer. By contrast, in the N30β structure, the Vβ domain’s external hydrophobic CFG face is covered by the neighboring molecule’s Cβ domain. In conjunction with systematic investigation of previously published TCR single-subunit structures, we identified several conserved residues forming a concave hydrophobic patch at the center of the CFG outer face of the Vβ and other V-type Ig-like domains. This hydrophobic patch is shielded from solvent exposure in the crystal packing, implying that it is unlikely to be thermodynamically stable if exposed on the thymocyte surface. Accordingly, we propose a dimeric pre-TCR model distinct from those suggested previously by others and discuss its functional and structural implications.
Introduction
The αβ T cell receptor (TCR) is a multimeric complex consisting of the antigen-binding αβ clonotypic heterodimer and six invariant CD3 subunit dimers (Sun et al., 2001 and references therein). Specific interaction between the TCRαβ heterodimer and an antigenic peptide bound to an MHC molecule (pMHC) triggers downstream signaling, resulting in T cell activation (Rudolph et al., 2006; Acuto et al., 2008; Smith-Garvin et al., 2009). TCR signaling is also critical for thymocyte differentiation (Ghendler et al., 1998; Mariathasan et al., 1999; Love et al., 2000; Haks et al., 2002). In early thymic development, the first major checkpoint is referred to as β-selection. During this process TCRβ chain gene rearrangement occurs in the absence of TCRα chain gene rearrangements (Von Boehmer et al., 2003). The expressed TCRβ chain associates with the invariant pre-Tα (pTα), which leads to the formation of pre-TCR on the cell surface. Signaling through the pre-TCR terminates TCRβ locus rearrangement, rescues cells from apoptosis and induces massive proliferation. This process eventually enables CD4−CD8− double-negative (DN) cells to differentiate into CD4+CD8+ double-positive (DP) cells, facilitating TCRα gene rearrangement and thus generating a large αβTCR repertoire (Von Boehmer et al., 2003). DP thymocytes undergo positive selection and negative selection following self–MHC interaction with their αβTCR. In the former case, DP cells mature into single positive (SP) thymocytes and may egress into the periphery (Starr et al., 2003; Juang et al., 2010). In the latter case, cell death occurs and those apoptotic thymocytes are purged from the T lineage repertoire (Gallegos and Bevan, 2006; Ashton-Rickardt, 2007; Griesemer et al., 2010).
The development of αβ lineage thymocytes and the antigen recognition of mature T cells are critically dependent on the TCRβ chain. Certain germline-encoded amino acids within the TCR Vβ domain promote “generic” recognition of MHC molecules during the thymic selection process (Scott-Browne et al., 2009). The germline-encoded TCR Vβ–MHC interactions have evolved to pre-determine, or at least strongly influence, the conserved diagonal TCR–MHC binding mode (Feng et al., 2007). The Vβ domain’s unique contribution has also been shown in providing a structural basis for immunodominance (Ishizuka et al., 2008; Wang et al., 2008). Immunodominant CTL responses to EBV infection incorporate a particular rearranged β chain generated during thymic development, favoring the survival, and expansion of T cells bearing it (Argaet et al., 1994; Gras et al., 2009). Another example involves the conservation of one Vβ domain in the context of highly variable Vα usage, as found in B6 mice responding to the immunodominant Db-restricted NP366–374 influenza A peptide following PR8 viral infection (Zhong and Reinherz, 2004; Zhong et al., 2007). The above observations raise the interesting possibility as to whether the Vβ recognition itself during β-selection can mediate a physiologic binding event in a manner analogous to certain VH domains in antibody molecules (Ward et al., 1989).
The alternative view regarding β-selection is that it is an autonomous signaling process driven by the oligomerization of pre-TCR’s extracellular domain (Yamasaki et al., 2006; Yamasaki and Saito, 2007). Very recently, Pang et al., 2010 determined the crystal structure of a pre-TCR and, following crystal packing analysis, proposed a model for autonomous dimerization of pre-TCR. In this model, a pTα domain’s ABED face contacts the β chain’s Cβ domain to form a pTα/Cβ module, similar to the Cα/Cβ module seen in the αβTCR heterodimer. In addition, the other face of the pTα domain (the CFG face) interacts with a Vβ domain from the neighboring molecule in the crystal lattice, resulting in a dimeric pre-TCR. In this way, the “head-to-tail” pre-TCR dimer referred to by the authors, should it adopt this topology in vivo, would sit almost parallel to the cellular membrane surface such that the Vβ domain becomes virtually unable to access any potential ligand (Pang et al., 2010). This contrasts with the upright model of the mature αβTCR complex, which is arrayed on the T cell surface with six CDR loops pointing upward to facilitate immune recognition (Sun et al., 2004).
We report here crystal structures of two β subunit ectodomains, N15β (Vβ5.2Dβ2Jβ2.6Cβ2), and N30β (Vβ13Dβ1Jβ1.1Cβ2), derived from N15 and N30 αβTCRs (Chang et al., 1997; Goyarts et al., 1998). We have compared these structures with the previously published structure of a TCR β subunit (Vβ8.2Jβ2.1Cβ1; Bentley et al., 1995) and several others in the protein data bank. In the structures surveyed, a few conserved residues were identified on the CFG face of Vβ Ig-like domain. In the biological heterodimeric αβTCR these residues are located at the Vα/Vβ interface. In the Vβ structures studied, the same residues are shielded from exposure by crystal packing in one of several possible ways. Based on these observations, we have proposed an alternative pre-TCR dimer model that is more similar to the heterodimeric αβTCR, and discussed its biological significance.
Materials and Methods
Cloning, Expression and Purification of N15β and N30β
Gene encoding the extracellular domain of N15β and N30β subunits was cloned into the expression vector pET-11d (Novagen). The DNA sequence of N15β and N30β constructs were terminated immediately after the cysteine of the respective TCRβ subunit involved in interchain disulfide formation with the cysteine itself mutated to serine. The recombinant plasmid was then transformed into Escherichia coli strain BL21 (DE3) followed by the induction of N15β and N30β expression using 1 mM isopropyl-β-D-thiogalactopyranoside. Inclusion bodies were isolated and solubilized in 6 M guanidine–HCl. The refolding of protein was achieved by further dilution of the inclusion bodies in a cold buffer containing 100 mM Tris–HCl (pH 8.0), 5.4 M guanidine–HCl, 1 M L-arginine–HCl, 1 mM reduced glutathione, and 0.1 mM oxidized glutathione, followed by dialysis against 20 mM Tris–HCl (pH 8.0), 10 mM NaCl at 4°C for 12 h. Insoluble particles were removed by centrifugation at high speed. The refolding mixture was concentrated prior to sequential purification of refolded proteins on a Sephacryl S-300 column and a Superdex 75 column (GE Healthcare).
Crystallization and Structure Determination
Crystals were grown at room temperature by the hanging-drop vapor diffusion method. Equal volumes of protein solution (15 mg/ml in 20 mM HEPES pH 7.5, 100 mM NaCl) and reservoir solution (100 mM HEPES pH 7.0, 20% PEG3350 for N15β, or 100 mM Tris–HCl pH 8.5, 20% PEG4000 for N30β) were mixed and crystals appeared in 1–2 days. 15% glycerol was added in reservoir buffer as a cryoprotectant solution to soak crystals prior to freezing. Diffraction data were collected at beamline 24-ID-E at the Argonne National Laboratories.
The structures were determined by molecular replacement method. TCRβ structures taken from heterodimeric αβTCR models with PDB accession code 1NFD and 1LP9 were used as the templates for searching N15β and N30β structures, respectively. The solution of N30β structure could only be obtained when Vβ and Cβ domains were used as the templates separately. The models were refined using refmac5 (Vagin et al., 2004) or phenix (Adams et al., 2010), viewed and adjusted by Coot (Emsley and Cowtan, 2004), and validated by MolProbity (Davis et al., 2007). The data collection and refinement statistics are listed in Table 1. The coordinates of the N15β and N30β structures have been deposited in the Protein Data Bank with the code 3Q5Y and 3Q5T, respectively.
Results
Structures of N15β and N30β
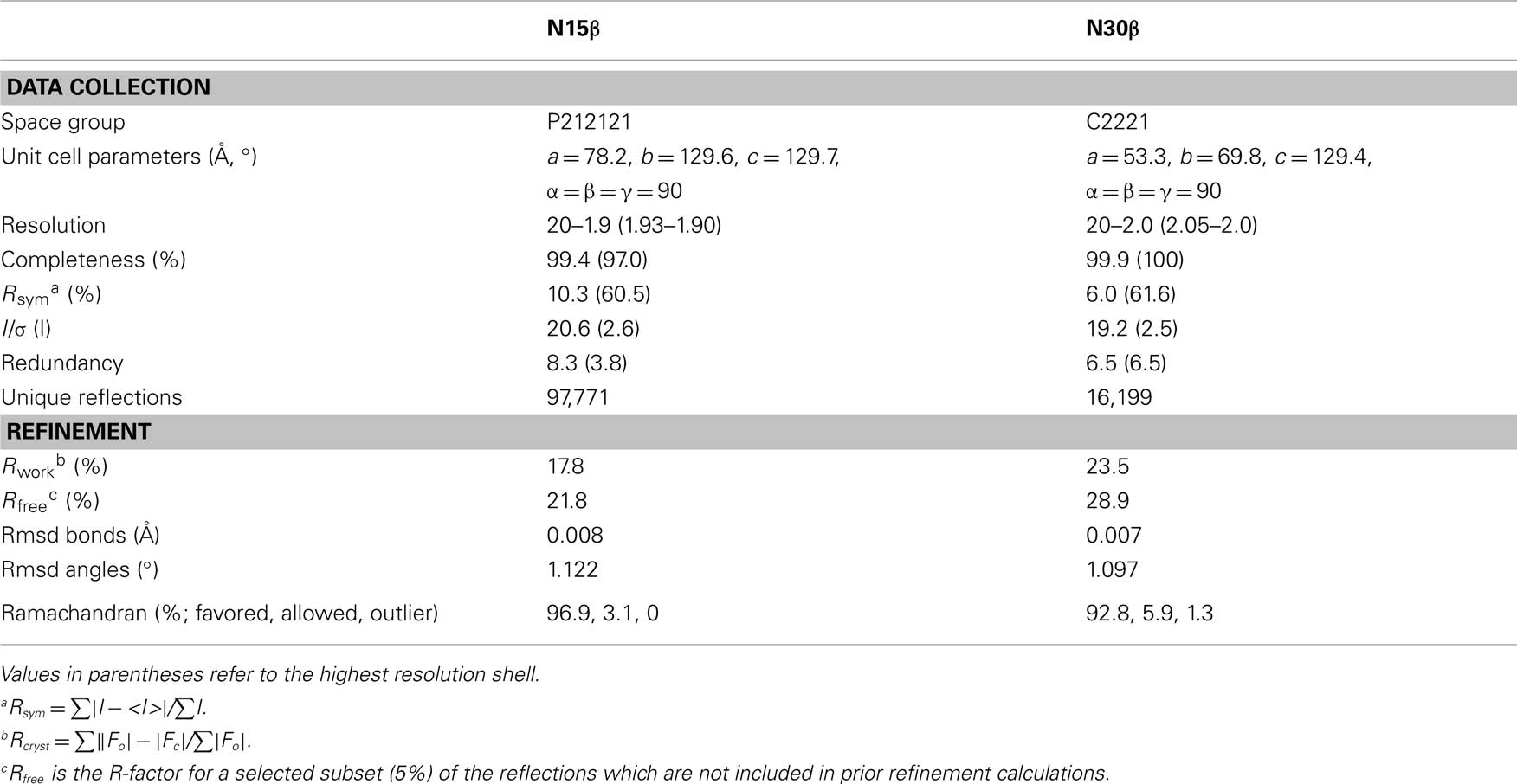
The 240-residue N15 and N30 β subunits were expressed in E. coli and crystallized readily. Their structures were determined using molecular replacement, with R-factor of 0.178 and Rfree of 0.218 for N15β to 1.9 Å, and 0.235 and 0.289 for N30β to 2.0 Å, respectively (see Table 1 for statistics). As shown in Figure 1, each of the models encompasses two Ig-like domains, Vβ and Cβ.
The superposition of the isolated N15β protein onto the β subunit of our previously published N15 αβTCR heterodimer (PDB code 1NFD) provides one of the first examples demonstrating that there are no significant conformational changes upon TCRβ pairing with TCRα to form an intact αβTCR. If the superposition is based on the Vβ domain, then the two corresponding Cβ domains only have a few degree rigid-body rotation relative to each other (Figure 2 and vide infra). We previously reported that the quaternary arrangement of Vβ–Cβ varies among different TCR molecules (Wang et al., 1998). Our current results suggest that for each individual TCRβ, the quaternary arrangement of Vβ–Cβ domains might be intrinsic, determining at least in part to which α subunit it can pair. A general survey indicates that upon TCR ligation to pMHC, the major conformational changes are limited to the CDR loops of each subunit (Rudolph et al., 2006). The rigidity of the framework module of TCR molecules seems to be essential for signaling following pMHC bindings (Kim et al., 2009).
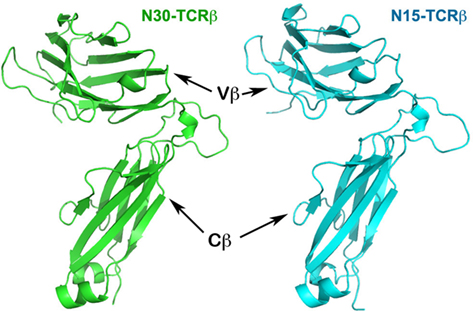
Figure 1. Structures of N30 TCRβ and N15 TCRβ subunits. The two Ig-like domains, Vβ and Cβ, are at the top and bottom, respectively.
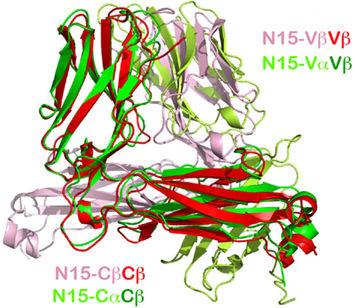
Figure 2. Superposition of the N15β homodimer onto the N15αβ TCR heterodimer. The overlay was based on the Vβ domain in red from the homodimer and in green from the heterodimer (top left in the figure). The pink subunit is the pairing N15β subunit in the homodimer, whereas the lime subunit is the pairing N15α subunit in the heterodimer. Note that in αβTCR heterodimer, Cβ and Cα pair to form a C module, whereas in the N15β homodimer, the two Cβ domains do not contact each other, but instead point in opposite directions.
The Vβ/Vβ Homodimer of N15β is Similar to Vα/Vβ Heterodimer of N15 αβTCR
A particularly intriguing feature of the N15β crystal structure is that two N15β subunits pack to form a Vβ/Vβ homodimer in a fashion very similar to that of the Vα/Vβ module in the αβTCR heterodimer. In each asymmetric unit of the N15β crystal there are four N15β molecules that pair to form two homodimers, using their outer faces of CFG β sheet for contacts. Figure 2 shows the overlay of a crystallographic homodimer of N15β onto the N15 αβTCR heterodimer. The superposition is based on the Vβ domains (in red and green color, respectively, for N15β and N15 αβTCR structures). In this figure, the small movement of the Cβ domain in the TCRβ relative to the αβTCR can be clearly seen. The respective pairing domains, Vβ (in pink color, from the homodimer) and Vα (in lime color, from the heterodimer) are similarly oriented, differing by only 11° from each other (Figure 2). In both the ββ homodimer and αβ heterodimer, the two pairing V-type Ig-like domains combine in an aligned arrangement with their corresponding β strands’ directions at an acute angle to one another.
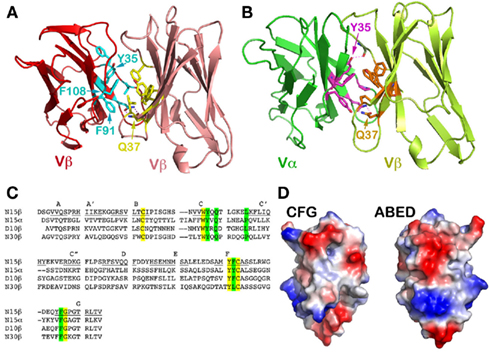
Figure 3. A conserved concave hydrophobic patch in the CFG face of the TCR Vβ domain. (A) In the N15β homodimer, the conserved hydrophobic residues form an interface. The two Gln37 residues from two Vβ protomers within the homodimer form two hydrogen-bonds in the interface, which orient the two domains in a packing pattern with constituent β strands roughly aligned. Note that the numbering of the residues follow the structure of N15αβ TCR (PDB 1NFD); (B) In the N15αβ TCR heterodimer, the conserved hydrophobic residues of the Vβ domain are in contacts with similar residues of Vα domain to form an interface. There are also two hydrogen-bonds between two invariant Gln37 residues, playing the identical role. Furthermore, Vα’s Tyr35 forms a hydrogen bond with the main chain carbonyl group on the Vβ domain, providing an additional force favoring the correct domain/domain orientation register; (C) Selected sequence alignment of several TCR subunits. The yellow shade marks conserved residues that are key for folding, whereas the green shade marks the conserved residues that constitute the hydrophobic interface and the invariant Gln/Gln pairing; (D) Surface representations of the Vβ domain’s CFG and ABED faces. The red color represents negatively charged, blue color positively charged, and gray color hydrophobic residues. The concave hydrophobic patch is obvious at the center of CFG face.
Figure 3 depicts the key interface residues in the N15 Vβ/Vβ structure (Figure 3A) and those in the Vα/Vβ interface in the N15 αβTCR structure (Figure 3B). Both interfaces involve Tyr35, Gln37, Phe91, and Phe108, as well as Leu43 in Vβ or Pro43 in Vα. The three aromatic residues along with Leu43 (or Pro43) form a hydrophobic center at the CFG interface. These residues are conserved in the variable domains of the TCR (Chothia et al., 1988; as shown for selected TCR variable domains in Figure 3C) and antibody molecules (Chothia et al., 1998). Figure 3D is an electrostatic surface representation of Vβ domain’s CFG face and the opposite ABED face for comparison. Whereas the exposed ABED face is fully covered by positive and negative charges, by contrast, a concave hydrophobic patch at the center of CFG outer face is readily discernible. Any exposed hydrophobic patch this size may not be stable, necessitating shielding from the solvent environment. On the other hand, the sole hydrophilic residue on the CFG face within this patch, the Gln37, plays a distinct role. In both the N15β and N15 αβTCR structures, this Gln37 on the C strand forms two hydrogen-bonds with its dimeric counterpart near the bottom of the interface between two V-domains (Figures 3A,B). This Gln has been reported to be invariant in variable domains of TCRs and antibodies (Chothia et al., 1988, 1998). Presumably the hydrogen-bonds offered by Gln37 provide a correct register for the two variable domains to dock. In Figure 3B the Vα domain’s Tyr35 forms an additional direct hydrogen bond with the main chain carbonyl oxygen of Gln106 of the Vβ domain near the top of the Vα/Vβ interface. This is also a conserved feature of the αβTCR Vα/Vβ interface in general. In the Vβ/Vβ interface of the N15β structure, the Tyr35’s side chain appears slightly far from Gln106’s carbonyl oxygen (around 4.5 Å) for a hydrogen bond (Figure 3A). It is interesting to note that there is a HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] solvent molecule intercalated into the Vβ/Vβ interface adjacent to Tyr35 for each of two N15β homodimers in the crystal structure, which prevents Tyr35 from forming a hydrogen bond to carbonyl oxygen of Gln106 (Figure A1 in Appendix). Notwithstanding, the N15β Vβ/Vβ homodimer is very similar overall to the N15 αβTCR Vα/Vβ heterodimer.
There are structural differences, however, between the Vβ/Vβ homodimers and Vα/Vβ heterodimers. For example, using the buried area option in the CNS program package (Brunger et al., 1998), we calculate that the surface buried area for the N15 Vβ/Vβ homodimer is 1,291 Å2, significantly smaller than 1,523 Å2 for the N15 Vα/Vβ heterodimer. Moreover, the complementary fitness index, or Sc value (Lawrence and Colman, 1993) of the Vβ/Vβ homodimer and Vα/Vβ heterodimer is 0.469 and 0.545, respectively, indicating a poorer fit at the homodimer interface relative to that of the heterodimer. This differential result reflects the self-association affinity difference between these dimers. Notably, the asymmetric TCR molecule has a much sharper bending angle between Vβ and Cβ domains compared to that between Vα and Cα domains (Wang et al., 1998). Thus, when two N15β molecules’ Vβ domains form a Vβ/Vβ homodimer, the two corresponding Cβ domains do not meet to form a C module like that of Cα/Cβ in the αβTCR molecule, as evident in Figure 2. These data suggest that it is possible to form a homodimer between two TCR β subunits through their Vβ domains, but that the homodimer so formed is substantially less stable than the αβTCR heterodimer.
The observed Vβ/Vβ homodimer in the crystal lattice may well be of biological significance. For instance, using RAG-2−/− deficient mice for functional studies, it was found that the introduction of a TCRβ transgene, but not a TCRα transgene restores T cell development, allowing thymocytes to transit from the DN to the DP stage. These DP cells were observed to express monomeric TCRβ chains in association with CD3Γδε but not ζζ, as well as small amounts of β homodimers (Shinkai et al., 1993). We shall come back to these points later.
The shielding of the Exposed Hydrophobic CFG Face of the Vβ Domain is a Common Feature in Other Crystal Structures that Contain TCRβ
There is only one molecule in the asymmetric unit of the N30β crystals. The symmetry-related molecules pack very differently from those of the N15β crystal, with one molecule’s Cβ domain packing onto the other molecule’s Vβ domain as shown in Figure 4A. Despite the apparent difference, the hydrophobic surface of the N30β Vβ domain (the CFG face) is also shielded from solvent exposure by the ABED face of a neighboring molecule’s Cβ domain. It is not surprising to observe the involvement of the Cβ domain’s ABED face; this face is normally engaged in the contacts with the Cα domain’s ABED face to form the C module in αβTCR molecules. One prominent feature in the N30β structure is that the aromatic ring of Phe128 of this Cβ domain pokes into the concave hydrophobic patch of the Vβ domain from the symmetry-related molecule for interaction (Figure 4A). That may explain the higher Sc value (0.540) for this interface as opposed to 0.469 in the N15β Vβ/Vβ interface, and why this peculiar packing is favored.
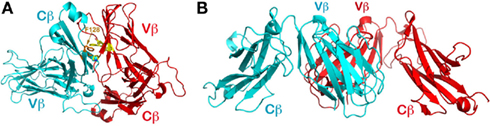
Figure 4. Packing patterns observed in other structures containing TCR β chain. (A) The CFG face of the N30 TCRβ is shielded by the neighboring molecule’s ABED face. Note that F128 of the Cβ subunit pokes into the concave hydrophobic patch of Vβ’s CFG face; (B) The orthogonal packing between two Vβ domains’ CFG faces frequently observed in crystal structures containing TCRβ. In the figure is the first solved TCRβ structure (PDB code 1BEC).
The observations on N15β and N30β structures led us to carry out an extensive survey in the protein data bank to investigate whether, in general, a TCR variable domain’s hydrophobic CFG face shown in Figure 3D is shielded from solvent exposure by crystal packing in various individual (i.e., non-heterodimeric) TCR subunit structures. In retrospect, the first glimpse at a TCR was derived from the structure of a β subunit (Vβ8.2Jβ2.1Cβ1, with PDB code of 1BEC; Bentley et al., 1995). Over the last 15 years, dozens of TCR single-subunit structures have been deposited in the data bank, including TCRβ, also TCRα, and even TCRδ. Many of these TCR β subunit structures are in complex with ligands such as superantigens. In the structure of 1BEC, the hydrophobic surface of the Vβ domain’s CFG face was indeed shielded by crystal packing (Figure 4B). However, in that structure the two contacting Vβ domains’ β strands are not aligned in the same manner as seen in the N15β and N15 αβTCR structures. Rather, in the 1BEC structure, molecules are in an orthogonal packing pattern, similar to the CD2/CD58 adhesion molecule pair (Wang et al., 1999). Interestingly, the majority of those TCRβ structures, be they in isolation or in complex with ligands, have an orthogonal packing in crystals to shield their hydrophobic CFG faces from solvent exposure.
It is particularly noteworthy that an aligned homodimer was observed in the first TCR Vα domain structure from the αβTCR1934.4 specific for the N-terminal nonapeptide of myelin basic protein bound to the MHC class II molecule I-Au (Fields et al., 1995). The same research group later published a mutant 1934.4Vαmut structure and observed a Vα/Vα homodimer resembling the Vα/Vβ heterodimer of αβTCR 2C and A6 (Li et al., 1997). Moreover, this protein formed homodimers in solution as well (Fields et al., 1994).
Stable, Single-Subunit Molecules Do Not Have an Exposed Hydrophobic Patch
We further sought to carry out comparative investigation of the two co-receptors: CD4 and CD8. CD8 exists either as a CD8αα homodimer or a CD8αβ heterodimer on the T cell surface, whereas CD4 is a monomeric molecule. The N-terminal domains of these two co-receptors are both V-type Ig-like domain. Figures 5A,B are electrostatic surface representations of the N-terminal V-domain CFG face of CD8β and CD4, respectively. The contrast is obvious. Whereas the single-subunit CD4 does not have a hydrophobic patch at the center of the CFG face of its N-terminal domain, CD8β does, analogous to that observed in the TCRβ subunit. This CD8 feature is linked to the fact that CD8β uses its single Ig-like domain to pair with CD8α in order to form a functional heterodimer. This further provides evidence that any membrane distal V-type Ig-like domain in a transmembrane protein is likely to form a dimer to shield its hydrophobic patch, if it exists, at the center of the CFG face from exposure. Moreover, in the case of the CD8αα homodimer the conserved Gln41 plays an identical role as Gln37 for the TCR to form two hydrogen-bonds creating the correct domain–domain interface register (PDB code 1BQH; Kern et al., 1998).
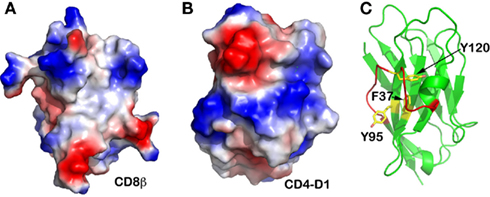
Figure 5. Other immune molecules’ CFG face. (A) CD8β subunit. A hydrophobic “well” is obvious at the center; (B) CD4 N-terminal V-type domain. This single-subunit TCR co-receptor does not have a hydrophobic CFG face at its N-terminal V-type domain; (C) Camelid antibody’s V-type domain. This unique antibody does not have a light chain to pair with the heavy chain. Although there is a hydrophobic “well” at its CFG face, this area is shielded from exposure by an extraordinarily long FG loop (in red color).
It is interesting that camelids possess a functional class of antibodies devoid of light chain (Desmyter et al., 1996). Figure 5C demonstrates that its VH domain does have the conserved aromatic residues Phe37, Tyr95, and Tyr120 (equivalent positions Tyr35, Phe91, and Phe108 in TCRβ, respectively) at the otherwise-exposed hydrophobic patch. However, this camelid molecule has an extraordinarily long FG loop curving into an extra helix that shields off this hydrophobic patch. This structural feature explains why this single heavy-chain antibody is functionally stable in the absence of a V-domain dimeric structure.
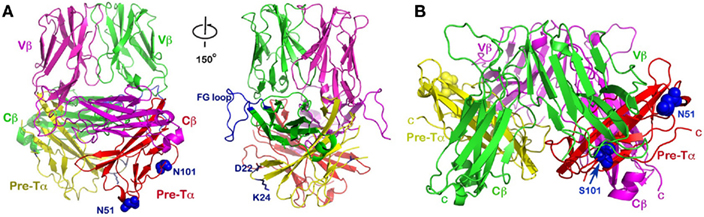
Figure 6. The dimeric pre-TCR models. (A) The upward projecting pre-TCR model was built based on the N15β homodimer as a starting point. This model has a topology similar to that of a mature αβTCR heterodimer with CDR loops pointing up, and thereby available for potential ligand-binding. It also may accommodate CD3 subunits in a somewhat similar manner as αβTCR. Note that the CD3 heterodimer ectodomains can be accommodated adjacent to the Cβ FG loops, one of which is highlighted in blue (left and right panels differ by about 150° rotation around the vertical axis). Pre-TCR residues Asp22 and Lys24 (blue side-chains) were suggested to facilitate pre-TCR oligomerization (Yamasaki et al., 2006). The N-linked glycan attachment sites at N51 and N101 of Pre-Tα are shown in blue space-filling models; (B) The “head-to-tail” flat pre-TCR dimer proposed by Pang et al., 2010. The two Vβ domains point in opposite directions, virtually parallel to the membrane, and hence difficult for any potential ligand interaction. While the human Pre-Tα has only the single N51 glycan addition site, the position of the second glycosylation site of N101 in mouse (S101 in human) is also indicated, as in Figure 6A.
Discussion
The above observation has led to the following key conclusion: for structural stability, a TCR Vβ domain must shield the conserved hydrophobic patch on its CFG face from exposure to solvent. The patterns of shielding can vary, either through Vα/Vβ interaction in the case of the αβTCR heterodimer, Vβ/Vβ aligned homodimeric interaction in the N15β structure, Vβ/Vβ orthogonal pairing in the structure of 1BEC (Bentley et al., 1995), or anti-parallel Vβ/Vβ interaction in the single Vα domain structure 2APB (Cho et al., 2005; not shown here) and in the Vβ/pTα interaction observed in the pre-TCR structure (PDB code: 3OF6; Pang et al., 2010), or Vβ/Cβ interaction in the N30β structure, etc. This set of variations may arise in the absence of a receptor’s membrane anchoring force where the domain orientation is under less constraint. Moreover, receptors’ domain/domain interactions are relatively weak (Wang, 2002; van der Merwe and Davis, 2003), and the non-specific hydrophobic interface is therefore not uniquely defined. In this regard, it is worth re-emphasizing the key role that the invariant Gln37 plays, facilitating the orientation register of Vβ with respect to Vα in the αβTCR heterodimer by providing two specific hydrogen-bonds, as shown in Figure 3B.
Based on the above analysis, we propose an alternative dimeric pre-TCR model (Figure 6A), distinct from the “head-to-tail” “flat” model suggested by Pang et al. (2010; Figure 6B). Our assumption is that the N15β homodimer might represent a physiological dimer existing at an early DN stage of thymocyte development. We superimposed the pre-TCR structure’s Vβ and Cβ domains (PDB code: 3OF6) onto the N15β’s Vβ and Cβ domains, separately, for each of the protomers within the N15β homodimer. This then places the pTα domain along with the Cβ domain as a module without significant steric collision, creating a distinct dimeric pre-TCR as shown in Figure 6A. Given that the Vβ–Cβ bending angle is actually slightly different between the same β subunit in the mature TCR LC13 (PDB code: 1KGC) and in the pre-TCR (PDB code: 3OF6), it is not surprising that this bending angle differs by a few degrees between N15β and LC13β in the pre-TCR.
The dimeric model we have proposed here has several advantageous features. Structurally, as discussed above, the two hydrogen-bonds provided by the invariant Gln37 remain intact (Figure 3A). In this sense, the dimeric pre-TCR appears more similar to the mature αβTCR heterodimer. These hydrogen-bonds are absent in the “flat” pre-TCR model proposed by Pang et al., 2010. A more interesting structural feature is that the mouse pTα has two potential glycosylation sites with the NXS sequon, one being Asn51-Gly-Ser at the tip of CC’ loop and the other Asn101-Arg-Ser at the middle of G strand (Figure 6A, left panel). In both cases, the potential glycan adduct would point toward the cellular membrane, perhaps helping the receptor to orient properly on the membrane (Casasnovas et al., 1997). In the human pre-TCR structure (Pang et al., 2010), a clear sugar moiety is seen at Asn51, which is exposed in the Pang’s model as shown in Figure 6B. Since the human pre-TCR has Ser101 in lieu of murine Asn101, there is no glycosylation site at the middle of the human G strand. Were the mouse pre-TCR to assume the same “head-to-tail” dimeric structure as the human pre-TCR, then the glycan attached to Asn101 (in the place of human Ser101) would seriously collide with the Vβ domain from its dimeric partner. This can be seen in Figure 6B where the Ser101 (in blue sphere) in human pre-TCR is buried between pTα (in red) and Vβ (in green).
Functionally, our model would allow the CD3 subunits to be incorporated into the pre-TCR dimer complex in a fashion more analogous to that in the mature αβTCR complex (Sun et al., 2004; Kuhns et al., 2010). By contrast, it is difficult to envision where the CD3 subunits can fit in the “flat” dimeric pre-TCR model. Our model also forces the Vβ domains’ CDR loops to project up and out from the membrane in a mode similar to that seen in the mature αβTCR. Subsequently, with further differentiation, the TCRα subunit might easily replace pTα. Finally, our postulated orientation of the β chain would not exclude the possibility that during β selection Vβ recognition in the form of a dimeric pre-TCR could mediate a physiologic binding event with potential ligands in the thymic development process.
In their pre-TCR crystal structure, Pang et al., 2010 actually observed two potential pre-TCR dimers generated by crystal packing. In addition to the flat “head-to-tail” dimer described above, a side-by-side dimer was also discussed and presented in their Figure A2 in Appendix. This dimer is mediated by Cβ/Cβ interaction and appears to stand on the membrane in a vertical orientation. However, this dimeric model was not regarded by the authors as favorable because it was presumed to sterically block CD3 interaction. By comparison, the dimeric pre-TCR model we have proposed, being more homologous to the mature αβTCR heterodimer, can permit CD3 complex formation as discussed above. This is readily seen from Figure 6A right panel, in which the space beneath and/or adjacent to the protruding FG loop of Cβ could accommodate juxtaposition of the CD3 subunits (Sun et al., 2004; Kuhns et al., 2010). Moreover, we have also noticed that the charged residues Asp22 and Lys24 (or Arg24 in mice) at the tip of AB loop of pTα, which were reportedly involved in pre-TCR oligomerization (Yamasaki et al., 2006), are located below the FG loop where the CD3 subunits may bind (Figure 6A). Mutations of Asp22 and Lys24 (or Arg24 in mice) may alter pre-TCR signaling and/or internalization through modulating effects of CD3-related activation. That being said, whether the CD3 heterodimers are oriented near the Cβ FG loop (Figure 6A) exactly as in the mature TCR complex, or alternatively, that juxtaposition is modified remains to be determined.
In the report by Pang et al., 2010, the authors have provided some experimental evidences, including site-directed mutagenesis, supporting their pre-TCR dimeric model. They demonstrated that three mutations, namely Y35βA, F108βA, and W46αR, affect pre-TCR cell surface expression. It is particularly intriguing that mutation Y35A in the TCRβ completely abrogate cell surface expression of the pre-TCR, but did not affect LC13 αβTCR cell surface expression (Pang et al., 2010). As discussed above, Tyr35 may be involved in hydrogen-bonding to main chain atom of Gln106 to facilitate dimer formation for the ββ homodimer in the pre-TCR as well as the αβTCR heterodimer. Given that in the case of αβTCR, there are many more hydrophilic interactions between Cα and Cβ domains (Wang et al., 1998), the Y35A mutation may be less detrimental than in the pre-TCR where only Tyr35 and Gln37 determine proper homodimeric orientation. On the other hand, Phe108β is within the critical hydrophobic patch we discussed extensively above. It is not surprising to see the lower level expression for the mutant F108βA on the cell surface. The effect of the W46αR mutation is more complicated to interprete. This Trp46 is located at the center of CFG face of pTα domain. In the “head-to-tail” model, this face involves contacts with Vβ domain of the dimeric partner. The W46αR mutation would disrupt the dimer. In our model, the pTα’s CFG face is exposed. However, compared with the pTα’s ABED face, the structure of this CFG face is much less regular. The residue W46αR’s side chain indole ring lies flat onto the pTα domain, and has extensive hydrophobic interaction with nearby Pro42 on the same C strand and His92 from the F strand (Figure A2 in Appendix). Therefore, this Trp46 may serve to stabilize the pTα domain and a mutation of W46R might be detrimental, impacting surface expression.
Whether a pre-TCR monomer–dimer equilibrium might exist on the DN thymocyte surface with one or both structures competent for ligand-binding requires further analysis. Clearly, there is much to be resolved about the structure and function of the pre-TCR through future experimental endeavors.
Conclusion
In this work, we report crystal structures of two TCR β subunits. In conjunction with other similar structures deposited in PDB, we have identified a conserved hydrophobic patch on the CFG face of Vβ domain that is invariably shielded from solvent exposure. In one of our TCR β structures, the N15β, a homodimer is formed between the CFG faces of the two Vβ domains. By incorporating a newly published pre-TCR structure, we propose a pre-TCR dimer model based on this N15β homodimer. We discuss the structural and functional advantages of this model relative to alternative models.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work has been supported by NIH grants to Ellis L. Reinherz and Jia-Huai Wang. We thank our colleagues for helpful discussion. We also thank Rob Meijers for valuable comments.
References
Acuto, O., Di Bartolo, V., and Michel, F. (2008). Tailoring T-cell receptor signals by proximal negative feedback mechanisms. Nat. Rev. Immunol. 8, 699–712.
Adams, P. D., Afonine, P. V., Bunkoczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L. W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C., and Zwart, P. H. (2010). PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221.
Argaet, V. P., Schmidt, C. W., Burrows, S. R., Silins, S. L., Kurilla, M. G., Doolan, D. L., Suhrbier, A., Moss, D. J., Kieff, E., Sculley, T. B., and Misko, I. S. (1994). Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J. Exp. Med. 180, 2335–2340.
Ashton-Rickardt, P. G. (2007). Studying T-cell repertoire selection using fetal thymus organ culture. Methods Mol. Biol. 380, 171–184.
Bentley, G. A., Boulot, G., Karjalainen, K., and Mariuzza, R. A. (1995). Crystal structure of the β chain of a T cell antigen receptor. Science 267, 1984–1987.
Brunger, A., Adams, P., Clore, G., DeLano, W., Gros, P., Grosse-Kunstleve, R., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998). Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921.
Casasnovas, J. M., Springer, T. A., Liu, J. H., Harrison, S. C., and Wang, J.-H. (1997). Crystal structure of ICAM-2 reveals a distinctive integrin recognition surface. Nature 387, 312–315.
Chang, H. C., Smolyar, A., Spoerl, R., Witte, T., Yao, Y., Goyarts, E. C., Nathenson, S. G., and Reinherz, E. L. (1997). Topology of T cell receptor-peptide/class I MHC interaction defined by charge reversal complementation and functional analysis. J. Mol. Biol. 271, 278–293.
Cho, S., Swaminathan, C. P., Yang, J., Kerzic, M. C., Guan, R., Kieke, M. C., Kranz, D. M., Mariuzza, R. A., and Sundberg, E. J. (2005). Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction. Structure 13, 1775–1787.
Chothia, C., Boswell, D. R., and Lesk, A. M. (1988). The outline structure of the T cell αβ receptor. EMBO J. 7, 3745–3755.
Chothia, C., Gelfand, I., and Kister, A. (1998). Structural determinants in the sequences of immunoglobulin variable domain. J. Mol. Biol. 278, 457–479.
Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B. III, Snoeyink, J., Richardson, J. S., and Richardson, D. C. (2007). MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383.
Desmyter, A., Transue, T. R., Ghahroudi, M. A., Thi, M. H., Poortmans, F., Hamers, R., Muyldermans, S., and Wyns, L. (1996). Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat. Struct. Biol. 3, 803–811.
Emsley, P., and Cowtan, K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132.
Feng, D., Bond, C. J., Ely, L. K., Maynard, J., and Garcia, K. C. (2007). Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction “codon”. Nat. Immunol. 8, 975–983.
Fields, B. A., Ober, B., Malchiodi, E. L., Lebedeva, M. I., Braden, B. C., Ysern, X., Kim, J. K., Shao, X., Ward, E. S., and Mariuzza, R. A. (1995). Crystal structure of the Vα domain of a T cell antigen receptor. Science 270, 1821–1824.
Fields, B. A., Ysern, X., Poljak, R. J., Shao, X., Ward, E. S., and Mariuzza, R. A. (1994). Crystallization and preliminary X-ray diffraction study of a bacterially produced T-cell antigen receptor Vα domain. J. Mol. Biol. 239, 339–341.
Gallegos, A. M., and Bevan, M. J. (2006). Central tolerance: good but imperfect. Immunol. Rev. 209, 290–296.
Ghendler, Y., Teng, M. K., Liu, J. H., Witte, T., Liu, J., Kim, K. S., Kern, P., Chang, H. C., Wang, J. H., and Reinherz, E. L. (1998). Differential thymic selection outcomes stimulated by focal structural alteration in peptide/major histocompatibility complex ligands. Proc. Natl. Acad. Sci. U.S.A. 95, 10061–10066.
Goyarts, E. C., Vegh, Z., Kalergis, A. M., Horig, H., Papadopoulos, N. J., Young, A. C., Thomson, C. T., Chang, H. C., Joyce, S., and Nathenson, S. G. (1998). Point mutations in the β chain CDR3 can alter the T cell receptor recognition pattern on an MHC class I/peptide complex over a broad interface area. Mol. Immunol. 35, 593–607.
Gras, S., Burrows, S. R., Kjer-Nielsen, L., Clements, C. S., Liu, Y. C., Sullivan, L. C., Bell, M. J., Brooks, A. G., Purcell, A. W., McCluskey, J., and Rossjohn, J. (2009). The shaping of T cell receptor recognition by self-tolerance. Immunity 30, 193–203.
Griesemer, A. D., Sorenson, E. C., and Hardy, M. A. (2010). The role of the thymus in tolerance. Transplantation 90, 465–474.
Haks, M. C., Pepin, E., van den Brakel, J. H. N., Smeele, S. A. A., Belkowski, S. M., Kessels, H. W. H. G., Krimpenfort, P., and Kruisbeek, A. M. (2002). Contribution of the T cell receptor-associated CD3Γ-ITAM to thymocyte selection. J. Exp. Med. 196, 1–13.
Ishizuka, J., Stewart-Jones, G., van der Merwe, A., Bell, J., McMichael, A., and Jones, E. (2008). The structural dynamics and energetics of an immunodominant T-cell receptor are programmed by its Vβ domain. Immunity 28, 171–182.
Juang, J., Ebert, P. J., Feng, D., Garcia, K. C., Krogsgaard, M., and Davis, M. M. (2010). Peptide-MHC heterodimers show that thymic positive selection requires a more restricted set of self-peptides than negative selection. J. Exp. Med. 207, 1223–1234.
Kern, P. S., Teng, M.-K., Smolyar, A., Liu, J.-H., Liu, J., Hussey, R. E., Spoerl, R., Chang, H.-C., Reinherz, E. L., and Wang, J.-H. (1998). Structural basis of CD8 co-receptor function revealed by crystallographic analysis of a nurine CD8αα ectodomain fragment in complex with H-2Kb. Immunity 9, 519–530.
Kim, S. T., Takeuchi, K., Sun, Z. Y., Touma, M., Castro, C. E., Fahmy, A., Lang, M. J., Wagner, G., and Reinherz, E. L. (2009). The αβ T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 284, 31028–31037.
Kuhns, M. S., Girvin, A. T., Klein, L. O., Chen, R., Jensen, K. D., Newell, E. W., Huppa, J. B., Lillemeier, B. F., Huse, M., Chien, Y. H., Garcia, K. C., and Davis, M. M. (2010). Evidence for a functional sidedness to the αβ TCR. Proc. Natl. Acad. Sci. U.S.A. 107, 5094–5099.
Lawrence, M. C., and Colman, P. M. (1993). Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950.
Li, H., Lebedeva, M. I., Ward, E. S., and Mariuzza, R. A. (1997). Dual conformations of a T cell receptor Vα homodimer: implications for variability in Vα Vβ domain association. J. Mol. Biol. 269, 385–394.
Love, P. E., Lee, J., and Shores, E. W. (2000). Critical relationship between TCR signaling potential and TCR affinity during thymocyte selection. J. Immunol. 165, 3080–3087.
Mariathasan, S., Jones, R. G., and Ohashi, P. S. (1999). Signals involved in thymocyte positive and negative selection. Semin. Immunol. 11, 263–272.
Pang, S. S., Berry, R., Chen, Z., Kjer-Nielsen, L., Perugini, M. A., King, G. F., Wang, C., Chew, S. H., La Gruta, N. L., Williams, N. K., Beddoe, T., Tiganis, T., Cowieson, N. P., Godfrey, D. I., Purcell, A. W., Wilce, M. C., McCluskey, J., and Rossjohn, J. (2010). The structural basis for autonomous dimerization of the pre-T-cell antigen receptor. Nature 467, 844–848.
Rudolph, M. G., Stanfield, R. L., and Wilson, I. A. (2006). How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466.
Scott-Browne, J. P., White, J., Kappler, J. W., Gapin, L., and Marrack, P. (2009). Germline-encoded amino acids in the αβT-cell receptor control thymic selection. Nature 458, 1043–1046.
Shinkai, Y., Koyasu, S., Nakayama, K., Murphy, K. M., Loh, D. Y., Reinherz, E. L., and Alt, F. W. (1993). Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science 259, 822–825.
Smith-Garvin, J. E., Koretzky, G. A., and Jordan, M. S. (2009). T cell activation. Annu. Rev. Immunol. 27, 591–619.
Starr, T. K., Jameson, S. C., and Hogquist, K. A. (2003). Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139–176.
Sun, Z. J., Kim, K. S., Wagner, G., and Reinherz, E. L. (2001). Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 εΓ heterodimer. Cell 105, 913–923.
Sun, Z. Y., Kim, S. T., Kim, I. C., Fahmy, A., Reinherz, E. L., and Wagner, G. (2004). Solution structure of the CD3εδ ectodomain and comparison with CD3εΓ as a basis for modeling T cell receptor topology and signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 16867–16872.
Vagin, A. A., Steiner, R. A., Lebedev, A. A., Potterton, L., McNicholas, S., Long, F., and Murshudov, G. N. (2004). REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195.
van der Merwe, P. A., and Davis, S. J. (2003). Molecular interactions mediating T cell antigen recognition. Annu. Rev. Immunol. 21, 659–684.
Von Boehmer, H., Aifantis, I., Gounari, F., Azogui, O., Haughn, L., Apostolou, I., Jaeckel, E., Grassi, F., and Klein, L. (2003). Thymic selection revisited: how essential is it? Immunol. Rev. 191, 62–78.
Wang, J. (2002). Protein recognition by cell surface receptors: physiological receptors versus virus interactions. Trends Biochem. Sci. 27, 122–126.
Wang, J.-H., Lim, K., Smolyar, A., Teng, M., Liu, J., Tse, A. G., Hussey, R. E., Chishti, Y., Thomson, C. T., Sweet, R. M., Nathenson, S. G., Chang, H. C., Sacchettini, J. C., and Reinherz, E. L. (1998). Atomic structure of an αβ T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody. EMBO J. 17, 10–26.
Wang, J. H., Mallis, R. J., and Reinherz, E. L. (2008). Immunodominant-peptide recognition: beta testing TCRαβ. Immunity 28, 139–141.
Wang, J.-H., Smolyar, A., Tan, K., Liu, J.-H., Kim, M., Sun, Z.-Y. J., Wagner, G., and Reinherz, E. L. (1999). Structure of a heterophilic adhesion complex between human CD2 and CD58 (LFA-3) counter-receptors. Cell 97, 791–803.
Ward, E. S., Gussow, D., Griffiths, A. D., Jones, P. T., and Winter, G. (1989). Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature 341, 544–546.
Yamasaki, S., Ishikawa, E., Sakuma, M., Ogata, K., Sakata-Sogawa, K., Hiroshima, M., Wiest, D. L., Tokunaga, M., and Saito, T. (2006). Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat. Immunol. 7, 67–75.
Yamasaki, S., and Saito, T. (2007). Molecular basis for pre-TCR-mediated autonomous signaling. Trends Immunol. 28, 39–43.
Zhong, W., Dixit, S. B., Mallis, R. J., Arthanari, H., Lugovskoy, A. A., Beveridge, D. L., Wagner, G., and Reinherz, E. L. (2007). CTL recognition of a protective immunodominant influenza A virus nucleoprotein epitope utilizes a highly restricted Vβ but diverse Vα repertoire: functional and structural implications. J. Mol. Biol. 372, 535–548.
Keywords: T cell receptor, beta chain, crystal structure, pre-TCR
Citation: Zhou B, Chen Q, Mallis RJ, Zhang H, Liu J-h, Reinherz EL and Wang J-h (2011) A conserved hydrophobic patch on Vβ domains revealed by TCRβ chain crystal structures: implications for pre-TCR dimerization. Front. Immun. 2:5. doi:10.3389/fimmu.2011.00005
Received: 06 January 2011;
Accepted: 15 February 2011;
Published online: 01 March 2011.
Edited by:
Michael Dustin, New York University, USAReviewed by:
Balbino Alarcon, Superior Council for Scientific Research, SpainRoy Mariuzza, University of Maryland, USA
Christopher Garcia, Stanford University, USA
Copyright: © 2011 Zhou, Chen, Mallis, Zhang, Liu, Reinherz and Wang. This is an open-access article subject to an exclusive license agreement between the authors and Frontiers Media SA, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Dana-Farber Cancer Institute, 44 Binney Street, SM 1030, Boston, MA 02115, USA. e-mail: jwang@red.dfci.harvard.edu