- 1 VIB Autoimmune Genetics Laboratory, Leuven, Belgium
- 2 Department of Experimental Medicine, University of Leuven, Leuven, Belgium
Mutations in the autoimmune regulator (AIRE) gene lead to autoimmune polyendocrinopathy syndrome type 1 (APS1), characterized by the development of multi-organ autoimmune damage. The mechanism by which defects in AIRE result in autoimmunity has been the subject of intense scrutiny. At the cellular level, the working model explains most of the clinical and immunological characteristics of APS1, with AIRE driving the expression of tissue-restricted antigens (TRAs) in the epithelial cells of the thymic medulla. This TRA expression results in effective negative selection of TRA-reactive thymocytes, preventing autoimmune disease. At the molecular level, the mechanism by which AIRE initiates TRA expression in the thymic medulla remains unclear. Multiple different models for the molecular mechanism have been proposed, ranging from classical transcriptional activity, to random induction of gene expression, to epigenetic tag recognition effect, to altered cell biology. In this review, we evaluate each of these models and discuss their relative strengths and weaknesses.
Introduction
The autoimmune regulator, AIRE (OMIM #607358), has been the focus of intense research since mutations in the gene were identified in 1997 as the cause of autoimmune polyendocrinopathy syndrome type 1 (APS1, OMIM #2400300, also known as autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome; Finnish-German APECED Consortium, 1997; Nagamine et al., 1997). The syndrome is a rare Mendelian disease, found more commonly in isolated populations, with prevalence rates of 1/25000 among Finns (Ahonen, 1985), 1:5600 to 1/9000 among Iranian Jews (Zlotogora and Shapiro, 1992), and 1/14400 among Sardinians (Rosatelli et al., 1998). The disease is clinically classified by the presence of any two out of three primary disorders: hypoparathyroidism, primary adrenocortical failure, and chronic mucocutaneous candidiasis, with additional endocrine failure common (Betterle et al., 1998). Most of these clinical conditions result from progressive autoimmune destruction, with lymphocytic infiltrate and autoantibody production (Betterle and Zanchetta, 2003). The clinical mechanism of enhanced sensitivity toward candidiasis also appears to be autoimmune in nature, with autoantibodies targeted against Th17-associated cytokines neutralizing anti-Candida immunity (Kisand et al., 2010; Puel et al., 2010).
The Functional Role of Aire in Immunological Tolerance
Key insights into the mechanism by which mutations in AIRE affect tolerance have come through the development of Aire-deficient mouse models. Despite relatively mild disease on the C57BL/6 background, each knockout strain develops lymphocytic infiltrate and autoantibodies (Anderson et al., 2002; Kuroda et al., 2005; Su et al., 2008), with more severe clinical presentation on alternative genetic backgrounds (Jiang et al., 2005; Kuroda et al., 2005). A role for Aire in central tolerance was first suggested through experiments demonstrating that expression of Aire is largely restricted to rare cells in the thymic medulla (Bjorses et al., 1999; Heino et al., 1999; Ramsey et al., 2002). Thymic transplant experiments demonstrated that the key non-redundant function of Aire existed within the thymic stroma, and that medullary thymic epithelial cells showed reduced transcription of tissue-restricted antigens (TRAs) in Aire knockout mice (Anderson et al., 2002). While TRA expression in the thymic medulla had been previously documented (Jolicoeur et al., 1994), the finding of a specific molecular mediator to drive this expression suggested an endogenous tolerogenic function. The function of Aire-dependent TRA expression in maintaining immunological tolerance was first demonstrated using a neo-self transgenic system, where TRA expression in the thymus was linked to the efficacy of negative selection of autoreactive thymocytes and consequently to the development of autoimmune disease in the periphery (Liston et al., 2003, 2004). Additional experiments have demonstrated that thymic TRA expression is likewise able to drive regulatory T cell conversion, with the alternative fates (negative selection or regulatory T cell conversion) likely depending on antigen quantity and TCR affinity (Aschenbrenner et al., 2007; Hinterberger et al., 2010; Wirnsberger et al., 2011). Several alternative mechanisms of central tolerance were proposed for Aire (Anderson et al., 2005; Kuroda et al., 2005), however the TRA-transcription function appears to be the most robust, and has been subsequently extended to multiple endogenous self-antigens, including insulin 2, salivary protein 1, desmoglein 3, seminal vesicle secretory protein 2 (SVS2), and interphotoreceptor retinoid-binding protein (IRBP; DeVoss et al., 2006; Yano et al., 2008; Hou et al., 2009; Wada et al., 2011).
A potential role for Aire in peripheral tolerance remains controversial. Several groups have found extra-thymic Aire expression in multiple peripheral locations (Halonen et al., 2001; Kogawa et al., 2002; Gardner et al., 2008), while other studies have found Aire expression to be restricted to thymic epithelial cells (Hubert et al., 2008). Initial experiments suggested that the function of Aire was restricted to the thymic epithelium, as loss of Aire in the thymus was sufficient to induce disease (Anderson et al., 2002) and expression of selected TRAs in the lymph nodes were not Aire-dependent (Kont et al., 2008). However, more recent experiments have suggested that Aire is expressed in the periphery and does have a functional role (Gardner et al., 2008). Aire-expressing stromal cells from the lymph nodes have been demonstrated to express TRAs in a tolerogenic form, and this process has been demonstrated to be Aire-dependent for at least a subset of TRAs (Gardner et al., 2008). Independent experiments have also confirmed that the lymph node stroma mediates TRA immune tolerance (Lee et al., 2007; Nichols et al., 2007; Magnusson et al., 2008; Fletcher et al., 2010), including both Aire-dependent and Aire-independent mechanisms (Cohen et al., 2010). Overall, it is likely that Aire has some function in peripheral tolerance for a subset of TRAs, but that failures in this function are not the primary cause of autoimmunity in APS1.
Mechanisms of Aire-Dependent Tolerance
Despite the crucial function of Aire, the molecular mechanism by which it drives TRA expression in medullary thymic epithelium and lymph node stromal cells remains opaque. The key difficulty is the scale of Aire-dependent gene expression, with dependent TRAs numbering in the order of several hundreds to thousands of genes (Anderson et al., 2002; Derbinski et al., 2005). In this review, we address the various models that have been proposed so far for the elucidation of the mechanism by which Aire regulates its target genes. The models can be envisaged in two broad categories: molecular biology models and cell biology models. The molecular biology models utilize the molecular structure of Aire to explain the mechanism of its target gene activation and include classical transcription activation model, random transcriptional activator model, and epigenetic tag recognition model. In each of these models, Aire is proposed to have direct transcriptional activation properties, driving the expression of target genes. The cell biology models provide an alternative approach to the mechanism of Aire target gene regulation and encompass two contrasting models – “developmental retardation” and “dysregulated death” models – in which both propose Aire to influence the differentiation of medullary thymic epithelial cells to enhance TRA expression. With new studies revealing insights into both the molecular and cellular impacts of Aire expression, this review focuses to evaluate the relative strengths and weaknesses of each of the models.
Molecular Biology Models for Aire Activity
Aire as a classical transcription factor
The first model for Aire transcriptional activity describes Aire as a classical transcription factor, able to bind the promoter sequence of target genes (Figure 1A). Initial protein domain analysis of the human Aire protein identified several functional domains indicative of a transcriptional regulator (Nagamine et al., 1997; Kumar et al., 2001). Most importantly, Aire contains DNA binding domains, which have been mapped to two zinc-finger-binding plant homeodomains (PHD1 and PHD2), and a “human Sp100, Aire1, NucP41/P75, and Drosophila DEAF1 domain” (SAND) domain (Gibson et al., 1998; Bottomley et al., 2001; Kumar et al., 2001). This model is experimentally supported by the ability of Aire to bind DNA via these domains in a sequence-specific manner (Kumar et al., 2001). Gel-shift analysis revealed the SAND domain to bind the motif TTATTA in the presence of guanine residues while the zinc-finger-containing PHD domains were found to bind to the ATTGGTTA sequence (Kumar et al., 2001; Purohit et al., 2005). Furthermore, when tethered to DNA Aire has the capacity to activate transcription (Pitkanen et al., 2000). Also in favor of this model is the finding that Aire is capable of recruiting molecular partners involved in chromatin binding, transcription, mRNA elongation and processing, such as TOP2a, DNA-PK, Ku80, PARP-1, and H2A (Abramson et al., 2010). Importantly, the knockdown of these molecular partners reduces the expression of Aire-dependent genes, indicating a functional role in Aire-mediated transcription (Abramson et al., 2010). The most important support for this model comes from the finding that transgenes using target gene promoters, such as the insulin promoter-driven hen egg lysozyme transgene, also show Aire-dependence (Liston et al., 2004), despite being imbedded in a different chromatin context and using only the core promoter sequence. Likewise the insulin promoter-driven ovalbumin transgene, while being initially characterized as Aire-independent (Anderson et al., 2005), has now been demonstrated to have partial Aire-dependency despite its foreign chromatin context (Su et al., 2008; Hubert et al., 2011).
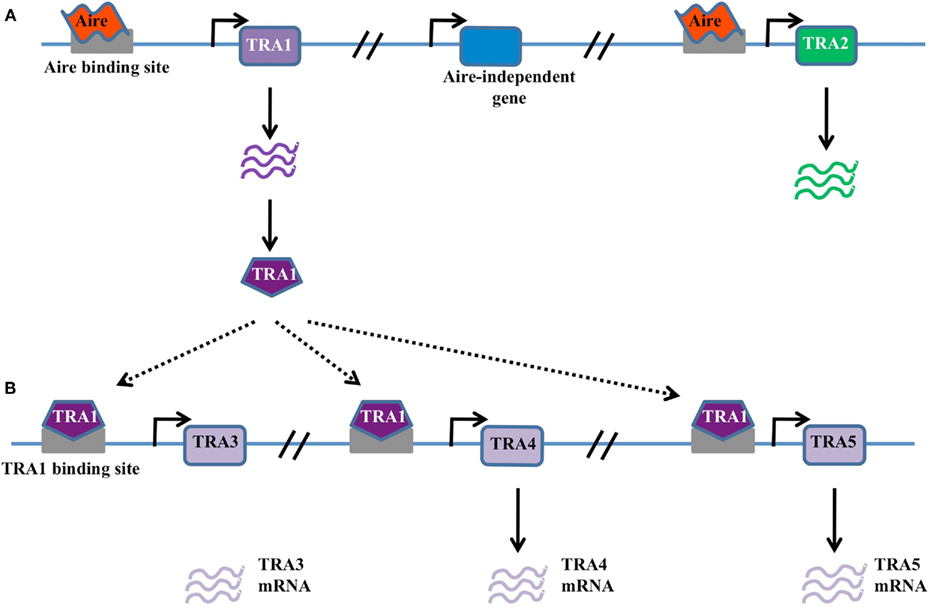
Figure 1. Classical transcription factor model. (A) In this model Aire binds DNA through sequence-specific interaction with promoter elements within TRA genes, such as TRA1 and TRA2. Gene expression is activated in these genes, but not neighboring genes, either through the direct action of Aire or through the action of recruited binding partners. (B) Possible amplification of Aire-dependent targets may occur when Aire-dependent TRAs are themselves transcriptional activators. For example, Aire-dependent TRA1 may in turn drive the expression of TRA3, TRA4, and TRA5. These secondary targets are expressed in an Aire-dependent manner despite the lack of an Aire-binding site in the promoter.
Despite the attractive simplicity of the classical transcription factor model, there are several notable disadvantages. Firstly, the utilization of KNKA motif as the binding motif in the SAND domain instead of KDWK or KNW (K/R) in other SAND-containing proteins suggests atypical DNA binding activity (Purohit et al., 2005; Peterson et al., 2008). Secondly, a consensus sequence for DNA binding has not been found among the numerous target genes. Thirdly, the sheer number of target genes (∼200–1000), with highly diverse peripheral expression patterns, make a direct binding function unlikely. Finally, the expression of different Aire-dependent TRA targets in thymic epithelial cells, peripheral lymph node stromal cells (Lee et al., 2007; Gardner et al., 2008), and modified monocytes (Sillanpaa et al., 2004) is not consistent with a fixed promoter recognition site. Nevertheless, the classical transcription factor binding model remains viable, as various modifications could explain these discrepancies, such as the expression of target genes being modulated by the chromatin context in an Aire-independent manner, or the existence of binding partners to Aire that modify target recognition (Abramson et al., 2010). One modification of the model that could explain many of the discrepancies is one where Aire is responsible for initiating the transcription of a small subset of master transcription factors, which then in turn initiate the expression of larger pools of downstream TRAs (Figure 1B). This would enable Aire to drive the expression of large numbers of genes without any apparent conservation within the promoter, and is supported by the finding that diverse tissue-specific master transcription factors, such as Pdx1, are expressed in thymic epithelial cells in an Aire-dependent manner (Gillard et al., 2007).
Aire as a random transcriptional activator
An alternative to the classical transcription factor model is the random transcriptional activator model. Early analysis of the cohort of genes expressed as TRAs suggested that genes were being turned on at low levels in a random manner (Derbinski et al., 2001), similar to the way “leaky” expression of genes is observed in the testes (Derbinski et al., 2001) and stem cells (Miyamoto et al., 2002; Zipori, 2004). In this model, Aire would contribute to the loosening of chromatin structure or general accessibility of genomic DNA in a way which allows non-specific gene expression, or permits otherwise illegitimate expression to be productive (Abramson et al., 2010). The expression of Aire in the testes (Derbinski et al., 2001) and stem cells (Nishikawa et al., 2010), and the observation of infertility in Aire knockout mice (Anderson et al., 2002; Ramsey et al., 2002; Hubert et al., 2009), generated the plausible model that Aire had a DNA accessibility function primarily for proliferative processes, which was co-opted for immunological tolerance in the thymic epithelium. However, the infertility observed in Aire knockout mice reflects autoimmunity against reproductive organs, as it is absent in Aire knockout mice crossed to the Rag knockout background (unpublished observation), and TRA expression in the testes is Aire-independent, unlike thymic epithelial cells (Liston et al., 2004). A primary function for Aire in the cell division process has therefore been discredited. A modified form of the random transcriptional activator hypothesis, however, is still viable, with the observation that Aire-dependent TRAs tend to be found in chromosomal clusters (Derbinski et al., 2005). It is therefore feasible that Aire recognizes sequence-specific DNA regions (as in model 1), or chromatin tags (as in model 3, below) in order to open up transcription of a small genomic region (Figure 2). Without genome-wide knowledge of chromatin boundaries and insulators it is difficult to draw firm conclusions on the existence of these “regional” effects.
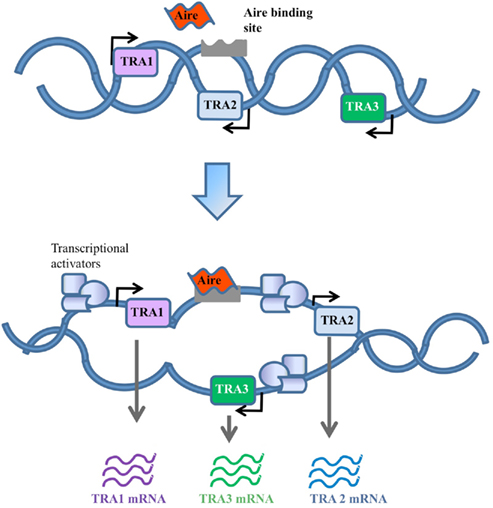
Figure 2. Random gene expression model. In this model Aire contributes to random activation of genes by loosening up the chromatin structure to increase the general accessibility of TRA genes. This would in turn allow the recruitment of transcriptional activators to bind genes that would otherwise be physically restricted.
Aire as an epigenetic tag recognition factor
A third model for the activity of Aire is based on recent biochemical analysis of Aire, indicating a capacity to bind modified histones (Koh et al., 2008; Chignola et al., 2009). Unlike the classical transcription factor model, this epigenetic tag model does not require sequence-specific recognition of target genes, but instead requires a common histone modification. In this scenario Aire would bind the modified histones and activate the transcription of nearby genes (Figure 3), probably through the recruitment of a complex of transcriptional activators and mRNA elongation/processing factors (Abramson et al., 2010). In support of this model, studies have shown that Aire is able to bind to unmethylated K4 on histone 3 (Koh et al., 2008; Org et al., 2008). Furthermore, Aire has been shown to interact with the transcription co-factors CBP (CREB binding protein; Pitkanen et al., 2000), PIAS1 (protein activator of activated STAT1; Ilmarinen et al., 2008), and P-TEFb (positive transcription elongation factor b; Oven et al., 2007), indicating an ability to drive transcription after tethering to the locus (Bjorses et al., 2000; Pitkanen et al., 2000, 2005; Oven et al., 2007; Ilmarinen et al., 2008).
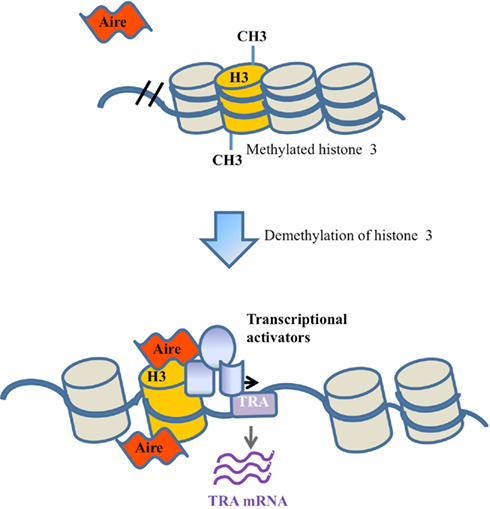
Figure 3. Epigenetic tag recognition model. Under the epigenetic tag model, Aire is recruited to a locus through binding to sequence-independent epigenetic tags, such as unmethylated histone 3. Following demethylation and Aire binding, Aire either directly enhances transcription or operates via the recruitment of other transcriptional activators.
In support of this model is the finding that binding to unmethylated K4 on histone 3 is dependent on the PHD1 domain (Chakravarty et al., 2009; Chignola et al., 2009; Koh et al., 2010), which is evolutionarily conserved among gnathostomes (Saltis et al., 2008) and harbors a high density of the human mutations detected in APS1 patients (Bjorses et al., 1998; Chakravarty et al., 2009). While the importance of the PHD1 domain in the function of Aire has been demonstrated (Koh et al., 2008; Org et al., 2008), it may not reflect Aire’s targeting mechanism as the PHD1 domain has also been implicated in direct DNA binding (Kumar et al., 2001; Purohit et al., 2005). An attractive feature of this model is a putative explanation for the large number of diverse target genes, as many genes throughout the genome would exhibit similar histone modification. Indeed, the range of Aire-dependent genes is actually far smaller than would be predicted from a common histone tag, and the over-expression of a H3K4 demethylase, able to extend the Aire-binding signature, does not extend the range of Aire target genes (Koh et al., 2010). The key weakness with this model is explaining how the specificity of target gene expression is achieved, for example how the core insulin promoter is able to be recognized when placed in a different chromatin context in the form of a transgene (Liston et al., 2004; Hubert et al., 2011). Another weakness is the inability of this model to explain why only small numbers of genes are activated in any given Aire-expressing epithelial cell (Gillard et al., 2007; Villasenor et al., 2008), unless Aire activity itself is limiting. Overall, the biochemical data that Aire binds epigenetic tags are convincing, but it is yet to be determined whether this is the function that primarily drives TRA expression, or whether it is a mechanism of Aire used to increase the stability of interaction with a promoter it binds due to DNA sequence recognition (Ruthenburg et al., 2007).
Cell Biology Models for Aire Activity
“Developmental retardation” model
In contrast to the direct activity models described above, several innovative alternatives have been proposed that postulate an effect of Aire on the cell biology of medullary thymic epithelial cells. The first model is the “developmental retardation” model, where it was proposed that Aire expression keeps thymic epithelial cells in an immature state (Gillard et al., 2007). This model was supported by data reporting Aire-dependent expression of Nanog, Oct4, and Sox2, which are candidates for expression by epithelial progenitor cells in the thymus and are critical for maintaining their multipotentiality (Gillard et al., 2007). This model was also supported by evidence demonstrating that Aire-deficient mice present with a reduced and altered medullary compartment (Gillard et al., 2007) with fewer terminally differentiated epithelial cells in the absence of Aire (Yano et al., 2008). Furthermore, Aire has been found to be actively expressed in early embryogenesis (Nishikawa et al., 2010). Under this model, the expression of Aire in the thymic epithelium would prevent full differentiation into mature epithelial cells, thereby allowing Aire-expressing cells to differentiate into alternative epithelial fates, a process which would initiate the expression of TRAs (Farr et al., 2002).
Ultimately, the “developmental retardation” model for Aire function is incompatible with more recent findings that Aire-expressing thymic epithelial cells are concentrated within the mature CD80hi subpopulation, have a low level of proliferation and a high level of apoptosis (Gabler et al., 2007; Gray et al., 2007; Irla et al., 2008; White et al., 2010). A study utilizing 5-bromo-2′-deoxyuridine (BrdU) incorporation instead reveals that Aire-expressing epithelial cells are terminally differentiated and highly apoptotic (Gray et al., 2007). However, this new evidence does not negate the possibility of a cell biology model for Aire expression of TRAs.
“Dysregulated death” model
An alternative model, that of “dysregulated death,” proposes that Aire expression dysregulates normal cell biology of the thymic epithelial cell, resulting in the partial differentiation toward one or more alternative epithelial fates in a terminal process (Figure 4; Gillard and Farr, 2006; Dooley et al., 2008). The terminal nature of Aire expression may even aid tolerance by facilitating the cross-presentation of TRAs to dendritic cells (Gray et al., 2007). Key support for the “dysregulated death” model comes from numerous studies which demonstrate changes to thymic epithelial cell biology in the absence of Aire. Beyond the well documented decrease in TRA expression, Aire-deficiency can result in morphological changes, altered antigen presentation capacities, and enhanced apoptosis (Anderson et al., 2005; Gray et al., 2007; Dooley et al., 2008; Yano et al., 2008; Hubert et al., 2011). This model has the capacity to explain many of the unique characteristics of Aire-dependent TRA expression, such as the broad repertoire of genes that are activated and the retained ability to activate transgenes utilizing these promoters, as conserved transcriptional pathways would be utilized. The observation that diverse tissue-specific master transcription factors, such as Pdx1, are Aire-dependent supports this hypothesis (Gillard et al., 2007). However this latter property would predict that Aire-dependent TRA expression should cluster TRAs from the same target organ in the same Aire-expressing cells, while this phenomenon is not observed in practice (Gillard and Farr, 2006; Derbinski et al., 2008; Venanzi et al., 2008; Villasenor et al., 2008). The key disadvantage of cell biology-based models is the lack of a known molecular mechanism, as the described molecular properties of Aire are more consistent with a role in transcription than in cell physiology. A putative explanation would involve Aire inducing “dysregulated death” via transcriptional activity, however again a molecular target is not available to explain such behavior.
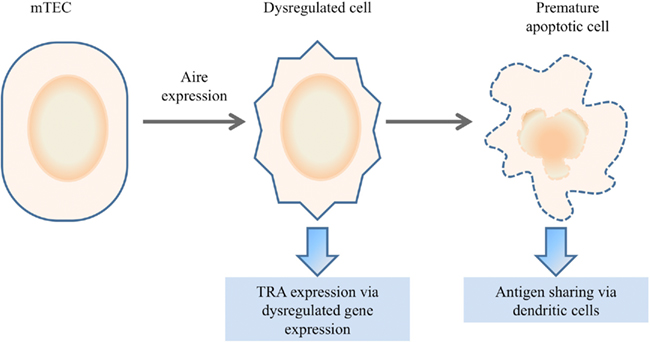
Figure 4. Dysregulated death model. In the dysregulated death model, Aire expression within mTECs results in a terminal degeneration of the normal mTEC biology. Increased plasticity results in partial dedifferentiation or redifferentiation into alternative lineages, resulting in the expression of genes outside the normal mTEC transcriptome. The premature apoptosis of the dysregulated cell may facilitate antigen sharing with dendritic cells for presentation.
Concluding Remarks
The failure of thymic deletion in Aire-deficient mice has revealed the significant role of Aire in promoting clonal deletion of autoreactive organ-specific T cells. The main unresolved issue is the mechanism of transcriptional activation of Aire-dependent TRAs. Despite large advances that have come from the study of Aire-dependent TRA targets, the molecular biology of Aire and the cellular biology of Aire-expressing cells, there is as yet no single coherent model for Aire activity which adequately accounts for all the observed phenomena (refer to Table 1). The provision of new data is required to definitively support a consensus model, whether it be one of the four main models outlined here, a hybrid model or a novel model yet to be proposed.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abramson, J., Giraud, M., Benoist, C., and Mathis, D. (2010). Aire’s partners in the molecular control of immunological tolerance. Cell 140, 123–135.
Ahonen, P. (1985). Autoimmune polyendocrinopathy-candidosis-ectodermal dystrophy (APECED): autosomal recessive inheritance. Clin. Genet. 27, 535–542.
Anderson, M. S., Venanzi, E. S., Chen, Z., Berzins, S. P., Benoist, C., and Mathis, D. (2005). The cellular mechanism of Aire control of T cell tolerance. Immunity 23, 227–239.
Anderson, M. S., Venanzi, E. S., Klein, L., Chen, Z., Berzins, S. P., Turley, S. J., Von Boehmer, H., Bronson, R., Dierich, A., Benoist, C., and Mathis, D. (2002). Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401.
Aschenbrenner, K., D’cruz, L. M., Vollmann, E. H., Hinterberger, M., Emmerich, J., Swee, L. K., Rolink, A., and Klein, L. (2007). Selection of Foxp3(+) regulatory T cells specific for self antigen expressed and presented by Aire(+) medullary thymic epithelial cells. Nat. Immunol. 8, 351–358.
Betterle, C., Greggio, N. A., and Volpato, M. (1998). Clinical review 93: autoimmune polyglandular syndrome type 1. J. Clin. Endocrinol. Metab. 83, 1049–1055.
Betterle, C., and Zanchetta, R. (2003). Update on autoimmune polyendocrine syndromes (APS). Acta Biomed. 74, 9–33.
Bjorses, P., Aaltonen, J., Horelli-Kuitunen, N., Yaspo, M. L., and Peltonen, L. (1998). Gene defect behind APECED: a new clue to autoimmunity. Hum. Mol. Genet. 7, 1547–1553.
Bjorses, P., Halonen, M., Palvimo, J. J., Kolmer, M., Aaltonen, J., Ellonen, P., Perheentupa, J., Ulmanen, I., and Peltonen, L. (2000). Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am. J. Hum. Genet. 66, 378–392.
Bjorses, P., Pelto-Huikko, M., Kaukonen, J., Aaltonen, J., Peltonen, L., and Ulmanen, I. (1999). Localization of the APECED protein in distinct nuclear structures. Hum. Mol. Genet. 8, 259–266.
Bottomley, M. J., Collard, M. W., Huggenvik, J. I., Liu, Z., Gibson, T. J., and Sattler, M. (2001). The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol. 8, 626–633.
Chakravarty, S., Zeng, L., and Zhou, M. M. (2009). Structure and site-specific recognition of histone H3 by the PHD finger of human autoimmune regulator. Structure 17, 670–679.
Chignola, F., Gaetani, M., Rebane, A., Org, T., Mollica, L., Zucchelli, C., Spitaleri, A., Mannella, V., Peterson, P., and Musco, G. (2009). The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res. 37, 2951–2961.
Cohen, J. N., Guidi, C. J., Tewalt, E. F., Qiao, H., Rouhani, S. J., Ruddell, A., Farr, A. G., Tung, K. S., and Engelhard, V. H. (2010). Lymph node-resident lymphatic endothelial cells mediate peripheral tolerance via Aire-independent direct antigen presentation. J. Exp. Med. 207, 681–688.
Finnish-German APECED Consortium. (1997). An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 17, 399–403.
Derbinski, J., Gabler, J., Brors, B., Tierling, S., Jonnakuty, S., Hergenhahn, M., Peltonen, L., Walter, J., and Kyewski, B. (2005). Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 202, 33–45.
Derbinski, J., Pinto, S., Rosch, S., Hexel, K., and Kyewski, B. (2008). Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc. Natl. Acad. Sci. U.S.A. 105, 657–662.
Derbinski, J., Schulte, A., Kyewski, B., and Klein, L. (2001). Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2, 1032–1039.
DeVoss, J., Hou, Y., Johannes, K., Lu, W., Liou, G. I., Rinn, J., Chang, H., Caspi, R. R., Fong, L., and Anderson, M. S. (2006). Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J. Exp. Med. 203, 2727–2735.
Dooley, J., Erickson, M., and Farr, A. G. (2008). Alterations of the medullary epithelial compartment in the Aire-deficient thymus: implications for programs of thymic epithelial differentiation. J. Immunol. 181, 5225–5232.
Farr, A. G., Dooley, J. L., and Erickson, M. (2002). Organization of thymic medullary epithelial heterogeneity: implications for mechanisms of epithelial differentiation. Immunol. Rev. 189, 20–27.
Fletcher, A. L., Lukacs-Kornek, V., Reynoso, E. D., Pinner, S. E., Bellemare-Pelletier, A., Curry, M. S., Collier, A. R., Boyd, R. L., and Turley, S. J. (2010). Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady-state and inflammatory conditions. J. Exp. Med. 207, 689–697.
Gabler, J., Arnold, J., and Kyewski, B. (2007). Promiscuous gene expression and the developmental dynamics of medullary thymic epithelial cells. Eur. J. Immunol. 37, 3363–3372.
Gardner, J. M., Devoss, J. J., Friedman, R. S., Wong, D. J., Tan, Y. X., Zhou, X., Johannes, K. P., Su, M. A., Chang, H. Y., Krummel, M. F., and Anderson, M. S. (2008). Deletional tolerance mediated by extrathymic Aire-expressing cells. Science 321, 843–847.
Gibson, T. J., Ramu, C., Gemund, C., and Aasland, R. (1998). The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem. Sci. 23, 242–244.
Gillard, G. O., Dooley, J., Erickson, M., Peltonen, L., and Farr, A. G. (2007). Aire-dependent alterations in medullary thymic epithelium indicate a role for Aire in thymic epithelial differentiation. J. Immunol. 178, 3007–3015.
Gillard, G. O., and Farr, A. G. (2006). Features of medullary thymic epithelium implicate postnatal development in maintaining epithelial heterogeneity and tissue-restricted antigen expression. J. Immunol. 176, 5815–5824.
Gray, D., Abramson, J., Benoist, C., and Mathis, D. (2007). Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J. Exp. Med. 204, 2521–2528.
Halonen, M., Pelto-Huikko, M., Eskelin, P., Peltonen, L., Ulmanen, I., and Kolmer, M. (2001). Subcellular location and expression pattern of autoimmune regulator (Aire), the mouse orthologue for human gene defective in autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED). J. Histochem. Cytochem. 49, 197–208.
Heino, M., Peterson, P., Kudoh, J., Nagamine, K., Lagerstedt, A., Ovod, V., Ranki, A., Rantala, I., Nieminen, M., Tuukkanen, J., Scott, H. S., Antonarakis, S. E., Shimizu, N., and Krohn, K. (1999). Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem. Biophys. Res. Commun. 257, 821–825.
Hinterberger, M., Aichinger, M., Da Costa, O. P., Voehringer, D., Hoffmann, R., and Klein, L. (2010). Autonomous role of medullary thymic epithelial cells in central CD4(+) T cell tolerance. Nat. Immunol. 11, 512–519.
Hou, Y., Devoss, J., Dao, V., Kwek, S., Simko, J. P., Mcneel, D. G., Anderson, M. S., and Fong, L. (2009). An aberrant prostate antigen-specific immune response causes prostatitis in mice and is associated with chronic prostatitis in humans. J. Clin. Invest. 119, 2031–2041.
Hubert, F.-X., Kinkel, S. A., Crewther, P. E., Cannon, P. Z., Webster, K. E., Link, M., Uibo, R., O’bryan, M. K., Meager, A., Forehan, S. P., Smyth, G. K., Mittaz, L., Antonarakis, S. E., Peterson, P., Heath, W. R., and Scott, H. S. (2009). Aire-deficient C57BL/6 mice mimicking the common human 13-base pair deletion mutation present with only a mild autoimmune phenotype. J. Immunol. 182, 3902–3918.
Hubert, F.-X., Kinkel, S. A., Davey, G. M., Phipson, B., Mueller, S. N., Liston, A., Proietto, A. I., Cannon, P. Z. F., Foreham, S., Smyth, G. K., Wu, L., Goodnow, C. C., Carbone, F. R., Scott, H. S., and Heath, W. R. (2011). Aire regulates transfer of antigen from mTEC to dendritic cells for induction of thymic tolerance. Blood http://bloodjournal.hematologylibrary.org/content/early/2011/04/19/blood-2010-06-286393.long. [Epub ahead of print].
Hubert, F.-X., Kinkel, S. A., Webster, K. E., Cannon, P., Crewther, P. E., Proeitto, A. I., Wu, L., Heath, W. R., and Scott, H. S. (2008). A specific anti-Aire antibody reveals aire expression is restricted to medullary thymic epithelial cells and not expressed in periphery. J. Immunol. 180, 3824–3832.
Ilmarinen, T., Kangas, H., Kytomaa, T., Eskelin, P., Saharinen, J., Seeler, J. S., Tanhuanpaa, K., Chan, F. Y., Slattery, R. M., Alakurtti, K., Palvimo, J. J., and Ulmanen, I. (2008). Functional interaction of AIRE with PIAS1 in transcriptional regulation. Mol. Immunol. 45, 1847–1862.
Irla, M., Hugues, S., Gill, J., Nitta, T., Hikosaka, Y., Williams, I. R., Hubert, F.-X., Scott, H. S., Takahama, Y., Hollander, G. A., and Reith, W. (2008). Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity 29, 451–463.
Jiang, W., Anderson, M. S., Bronson, R., Mathis, D., and Benoist, C. (2005). Modifier loci condition autoimmunity provoked by Aire deficiency. J. Exp. Med. 202, 805–815.
Jolicoeur, C., Hanahan, D., and Smith, K. M. (1994). T-cell tolerance toward a transgenic beta-cell antigen and transcription of endogenous pancreatic genes in thymus. Proc. Natl. Acad. Sci. U.S.A. 91, 6707–6711.
Kisand, K., Boe Wolff, A. S., Podkrajsek, K. T., Tserel, L., Link, M., Kisand, K. V., Ersvaer, E., Perheentupa, J., Erichsen, M. M., Bratanic, N., Meloni, A., Cetani, F., Perniola, R., Ergun-Longmire, B., Maclaren, N., Krohn, K. J., Pura, M., Schalke, B., Strobel, P., Leite, M. I., Battelino, T., Husebye, E. S., Peterson, P., Willcox, N., and Meager, A. (2010). Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J. Exp. Med. 207, 299–308.
Kogawa, K., Nagafuchi, S., Katsuta, H., Kudoh, J., Tamiya, S., Sakai, Y., Shimizu, N., and Harada, M. (2002). Expression of AIRE gene in peripheral monocyte/dendritic cell lineage. Immunol. Lett. 80, 195–198.
Koh, A. S., Kingston, R. E., Benoist, C., and Mathis, D. (2010). Global relevance of Aire binding to hypomethylated lysine-4 of histone-3. Proc. Natl. Acad. Sci. U.S.A. 107, 13016–13021.
Koh, A. S., Kuo, A. J., Park, S. Y., Cheung, P., Abramson, J., Bua, D., Carney, D., Shoelson, S. E., Gozani, O., Kingston, R. E., Benoist, C., and Mathis, D. (2008). Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 105, 15878–15883.
Kont, V., Laan, M., Kisand, K., Merits, A., Scott, H. S., and Peterson, P. (2008). Modulation of Aire regulates the expression of tissue-restricted antigens. Mol. Immunol. 45, 25–33.
Kumar, P. G., Laloraya, M., Wang, C. Y., Ruan, Q. G., Davoodi-Semiromi, A., Kao, K. J., and She, J. X. (2001). The autoimmune regulator (AIRE) is a DNA-binding protein. J. Biol. Chem. 276, 41357–41364.
Kuroda, N., Mitani, T., Takeda, N., Ishimaru, N., Arakaki, R., Hayashi, Y., Bando, Y., Izumi, K., Takahashi, T., Nomura, T., Sakaguchi, S., Ueno, T., Takahama, Y., Uchida, D., Sun, S., Kajiura, F., Mouri, Y., Han, H., Matsushima, A., Yamada, G., and Matsumoto, M. (2005). Development of autoimmunity against transcriptionally unrepressed target antigen in the thymus of Aire-deficient mice. J. Immunol. 174, 1862–1870.
Lee, J. W., Epardaud, M., Sun, J., Becker, J. E., Cheng, A. C., Yonekura, A. R., Heath, J. K., and Turley, S. J. (2007). Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat. Immunol. 8, 181–190.
Liston, A., Gray, D. H., Lesage, S., Fletcher, A. L., Wilson, J., Webster, K. E., Scott, H. S., Boyd, R. L., Peltonen, L., and Goodnow, C. C. (2004). Gene dosage – limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J. Exp. Med. 200, 1015–1026.
Liston, A., Lesage, S., Wilson, J., Peltonen, L., and Goodnow, C. C. (2003). Aire regulates negative selection of organ-specific T cells. Nat. Immunol. 4, 350–354.
Magnusson, F. C., Liblau, R. S., Von Boehmer, H., Pittet, M. J., Lee, J. W., Turley, S. J., and Khazaie, K. (2008). Direct presentation of antigen by lymph node stromal cells protects against CD8 T-cell-mediated intestinal autoimmunity. Gastroenterology 134, 1028–1037.
Miyamoto, T., Iwasaki, H., Reizis, B., Ye, M., Graf, T., Weissman, I. L., and Akashi, K. (2002). Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3, 137–147.
Nagamine, K., Peterson, P., Scott, H. S., Kudoh, J., Minoshima, S., Heino, M., Krohn, K. J., Lalioti, M. D., Mullis, P. E., Antonarakis, S. E., Kawasaki, K., Asakawa, S., Ito, F., and Shimizu, N. (1997). Positional cloning of the APECED gene. Nat. Genet. 17, 393–398.
Nichols, L. A., Chen, Y., Colella, T. A., Bennett, C. L., Clausen, B. E., and Engelhard, V. H. (2007). Deletional self-tolerance to a melanocyte/melanoma antigen derived from tyrosinase is mediated by a radio-resistant cell in peripheral and mesenteric lymph nodes. J. Immunol. 179, 993–1003.
Nishikawa, Y., Hirota, F., Yano, M., Kitajima, H., Miyazaki, J., Kawamoto, H., Mouri, Y., and Matsumoto, M. (2010). Biphasic Aire expression in early embryos and in medullary thymic epithelial cells before end-stage terminal differentiation. J. Exp. Med. 207, 963–971.
Org, T., Chignola, F., Hetenyi, C., Gaetani, M., Rebane, A., Liiv, I., Maran, U., Mollica, L., Bottomley, M. J., Musco, G., and Peterson, P. (2008). The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9, 370–376.
Oven, I., Brdickova, N., Kohoutek, J., Vaupotic, T., Narat, M., and Peterlin, B. M. (2007). AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol. Cell. Biol. 27, 8815–8823.
Peterson, P., Org, T., and Rebane, A. (2008). Transcriptional regulation by AIRE: molecular mechanisms of central tolerance. Nat. Rev. Immunol. 8, 948–957.
Pitkanen, J., Doucas, V., Sternsdorf, T., Nakajima, T., Aratani, S., Jensen, K., Will, H., Vahamurto, P., Ollila, J., Vihinen, M., Scott, H. S., Antonarakis, S. E., Kudoh, J., Shimizu, N., Krohn, K., and Peterson, P. (2000). The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J. Biol. Chem. 275, 16802–16809.
Pitkanen, J., Rebane, A., Rowell, J., Murumagi, A., Strobel, P., Moll, K., Saare, M., Heikkila, J., Doucas, V., Marx, A., and Peterson, P. (2005). Cooperative activation of transcription by autoimmune regulator AIRE and CBP. Biochem. Biophys. Res. Commun. 333, 944–953.
Puel, A., Doffinger, R., Natividad, A., Chrabieh, M., Barcenas-Morales, G., Picard, C., Cobat, A., Ouachee-Chardin, M., Toulon, A., Bustamante, J., Al-Muhsen, S., Al-Owain, M., Arkwright, P. D., Costigan, C., Mcconnell, V., Cant, A. J., Abinun, M., Polak, M., Bougneres, P. F., Kumararatne, D., Marodi, L., Nahum, A., Roifman, C., Blanche, S., Fischer, A., Bodemer, C., Abel, L., Lilic, D., and Casanova, J. L. (2010). Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J. Exp. Med. 207, 291–297.
Purohit, S., Kumar, P. G., Laloraya, M., and She, J. X. (2005). Mapping DNA-binding domains of the autoimmune regulator protein. Biochem. Biophys. Res. Commun. 327, 939–944.
Ramsey, C., Winqvist, O., Puhakka, L., Halonen, M., Moro, A., Kampe, O., Eskelin, P., Pelto-Huikko, M., and Peltonen, L. (2002). Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum. Mol. Genet. 11, 397–409.
Rosatelli, M. C., Meloni, A., Devoto, M., Cao, A., Scott, H. S., Peterson, P., Heino, M., Krohn, K. J., Nagamine, K., Kudoh, J., Shimizu, N., and Antonarakis, S. E. (1998). A common mutation in Sardinian autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients. Hum. Genet. 103, 428–434.
Ruthenburg, A. J., Allis, C. D., and Wysocka, J. (2007). Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30.
Saltis, M., Criscitiello, M. F., Ohta, Y., Keefe, M., Trede, N. S., Goitsuka, R., and Flajnik, M. F. (2008). Evolutionarily conserved and divergent regions of the autoimmune regulator (Aire) gene: a comparative analysis. Immunogenetics 60, 105–114.
Sillanpaa, N., Magureanu, C. G., Murumagi, A., Reinikainen, A., West, A., Manninen, A., Lahti, M., Ranki, A., Saksela, K., Krohn, K., Lahesmaa, R., and Peterson, P. (2004). Autoimmune regulator induced changes in the gene expression profile of human monocyte-dendritic cell-lineage. Mol. Immunol. 41, 1185–1198.
Su, M. A., Giang, K., Zumer, K., Jiang, H., Oven, I., Rinn, J. L., Devoss, J. J., Johannes, K. P., Lu, W., Gardner, J., Chang, A., Bubulya, P., Chang, H. Y., Peterlin, B. M., and Anderson, M. S. (2008). Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J. Clin. Invest. 118, 1712–1726.
Venanzi, E. S., Melamed, R., Mathis, D., and Benoist, C. (2008). The variable immunological self: genetic variation and nongenetic noise in Aire-regulated transcription. Proc. Natl. Acad. Sci. U.S.A. 105, 15860–15865.
Villasenor, J., Besse, W., Benoist, C., and Mathis, D. (2008). Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc. Natl. Acad. Sci. U.S.A. 105, 15854–15859.
Wada, N., Nishifuji, K., Yamada, T., Kudoh, J., Shimizu, N., Matsumoto, M., Peltonen, L., Nagafuchi, S., and Amagai, M. (2011). Aire-dependent thymic expression of desmoglein 3, the autoantigen in pemphigus vulgaris, and its role in T-cell tolerance. J. Invest. Dermatol. 131, 410–417.
White, A. J., Nakamura, K., Jenkinson, W. E., Saini, M., Sinclair, C., Seddon, B., Narendran, P., Pfeffer, K., Nitta, T., Takahama, Y., Caamano, J. H., Lane, P. J., Jenkinson, E. J., and Anderson, G. (2010). Lymphotoxin signals from positively selected thymocytes regulate the terminal differentiation of medullary thymic epithelial cells. J. Immunol. 185, 4769–4776.
Wirnsberger, G., Hinterberger, M., and Klein, L. (2011). Regulatory T-cell differentiation versus clonal deletion of autoreactive thymocytes. Immunol. Cell Biol. 89, 45–53.
Yano, M., Kuroda, N., Han, H., Meguro-Horike, M., Nishikawa, Y., Kiyonari, H., Maemura, K., Yanagawa, Y., Obata, K., Takahashi, S., Ikawa, T., Satoh, R., Kawamoto, H., Mouri, Y., and Matsumoto, M. (2008). Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self-tolerance. J. Exp. Med. 205, 2827–2838.
Keywords: Aire, transcription factor, tissue-restricted antigens, autoimmunity, tolerance
Citation: Danso-Abeam D, Humblet-Baron S, Dooley J and Liston A (2011) Models of Aire-dependent gene regulation for thymic negative selection. Front. Immun. 2:14. doi: 10.3389/fimmu.2011.00014
Received: 27 March 2011; Paper pending published: 15 April 2011;
Accepted: 21 April 2011; Published online: 09 May 2011.
Edited by:
Rachel R. Caspi, National Institutes of Health, USAReviewed by:
Mitsuru Matsumoto, University of Tokushima, JapanLudger Klein, Ludwig-Maximilians-University, Germany
Mark Anderson, University of California at San Francisco, USA
Copyright: © 2011 Danso-Abeam, Humblet-Baron, Dooley and Liston. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: James Dooley and Adrian Liston, VIB Autoimmune Genetics Laboratory, K.U.Leuven, Onderwijs en Navorsing 2, Campus Gasthuisberg, Herestraat 49, Box 1023, 3000 Leuven, Belgium. e-mail: james.dooley@vib-kuleuven.be; adrian.liston@vib.be

